Abstract
The discovery of a handful of conserved signaling pathways that dictate most aspects of embryonic and post-embryonic development of multicellular organisms has generated a universal view of animal development (Perrimon, N., Pitsouli, C., and Shilo, B. Z. (2012) Cold Spring Harb. Perspect. Biol. 4, a005975). Although we have at hand most of the “hardware” elements that mediate cell communication events that dictate cell fate choices, we are still far from a comprehensive mechanistic understanding of these processes. One of the next challenges entails an analysis of developmental signaling pathways from the cell biology perspective. Where in the cell does signaling take place, and how do general cellular machineries and structures contribute to the regulation of developmental signaling? Another challenge is to examine these signaling pathways from a quantitative perspective, rather than as crude on/off switches. This requires more precise measurements, and incorporation of the time element to generate a dynamic sequence instead of frozen snapshots of the process. The quantitative outlook also brings up the issue of precision, and the unknown mechanisms that buffer variability in signaling between embryos, to produce a robust and reproducible output. Although these issues are universal to all multicellular organisms, they can be effectively tackled in the Drosophila model, by a combination of genetic manipulations, biochemical analyses, and a variety of imaging techniques. This review will present some of the recent advances that were accomplished by utilizing the versatility of the Drosophila system.
Keywords: bone morphogenetic protein (BMP), cell signaling, development, Drosophila, epidermal growth factor receptor (EGFR), fibroblast growth factor (FGF), transcription, Wnt signaling, cytonemes, morphogen
A New View on Wnt Signaling: Notum
Wnt proteins control the fine balance between differentiation and proliferation, most notably in stem cell niches (2). Tight regulation of Wnt activity is thus essential. A recent study from the lab of J. P. Vincent identified a mechanism to attenuate Wnt activity through its associated acyl group (3). Covalent attachment of an acyl group to Wnt is mediated by the multi-pass transmembrane protein Porcupine. Once attached, the acyl group provides a molecular handle to associate with the Wntless protein that mediates Wnt trafficking for secretion. The same acyl group then serves as an essential module in recognition of Wnt by the Frizzled receptor.
Notum was originally identified in flies as a secreted protein that functions to attenuate Wnt signaling, but its mechanism of action and the basis for its specificity toward Wnt were debated. Structural analysis of the human and fly Notum protein demonstrated that it has a specific pocket that can fit the acyl group attached to Wnt, but not other acyl groups. Furthermore, it has an overall structure of a hydrolase enzyme. In fact, Notum specifically removes the acyl group bound to Wnt, thus rendering it biologically inactive (3). This work provides the mechanistic basis for the specific attenuation of Wnt signaling by Notum. Because modification of Wnt by acylation is the basis for its normal trafficking and association with the receptor, elimination of this modification by Notum provides an effective way to modulate its activity (Fig. 1).
FIGURE 1.
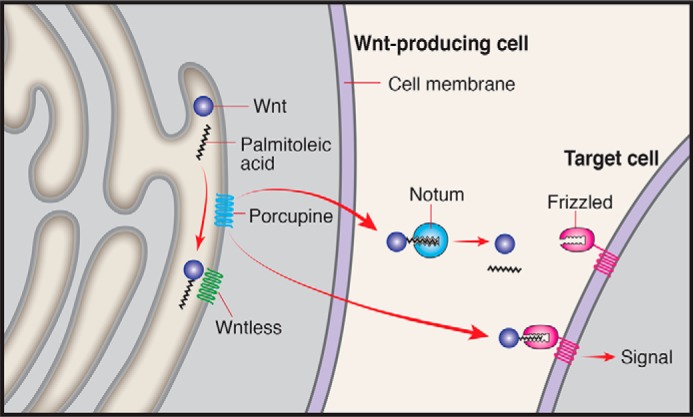
Scheme of Wnt modification by Porcupine, trafficking by Wntless, and inactivation by Notum. Only Wnt that is associated with palmitoleic acid can be trafficked by Wntless, bind, and activate the Frizzled receptor.
Cytonemes and Nanotubes
Because developmental cell communication is triggered by secreted proteins, the way in which these proteins are presented to their respective receptors, for both short-range and long-range signaling, has far-reaching implications on the subsequent signaling steps. Live imaging studies from a variety of systems point to the surprising observation of long and slender actin-based cellular extensions (termed cytonemes) that emanate from epithelial cells (4). The work of Tom Kornberg's lab over the years has argued that these structures provide direct contacts between communicating cells, even ones that are positioned several cell diameters away. Furthermore, Kornberg argued that the contact points can be regarded as signaling synapses, where a ligand from one cell is loaded on receptors in the adjacent cell (5). The Kornberg lab presented evidence indicating that different cytonemes emanating from the same cell make contact with distinct signaling sources, and accordingly, carry discrete receptors. One tissue where these features were particularly prominent was in the tracheal air sac of the Drosophila larva, where separate contacts are made with underlying cells producing bone morphogenetic protein (BMP) or FGF (6).
The physical cytoneme-mediated contact between adjacent cells was clearly demonstrated by complementary expression of split GFP (6). In this experiment, cells at the source of the signal express one part of GFP on the cell surface, and cells at the receiving end express the complementary segment. At the sites where the two cell types make a synapse-like physical contact, the two parts of GFP have the capacity to associate and give rise to fluorescence.
However, the key question was whether these cytonemes actually have a role in transmitting the activating ligands at the “synapse.” Toward this end, the Kornberg lab identified a class of cell adhesion proteins that are not required for cytoneme growth, but are essential for their tight contact with the target at the “synapse.” When the contact was abolished by knockdown of the cell adhesion protein Capricious, activation of the signaling cascade in the receiving cells was compromised, highlighting the important role of cytonemes in signal transduction (6). Although the function of cytonemes in mediating short-range interactions over several cell diameters is more intuitive, it is puzzling to consider how they may mediate long-range activation in a scenario of a morphogen gradient. A novel mechanism for gradual reduction of signaling levels in cells that are farther away from the morphogen source would be required to shape the graded response of the cells in the field.
In other biological contexts, cell extensions may fulfill an opposite role, to restrict the range of ligand distribution. The stem cell microenvironment of the Drosophila ovary and testes (termed niche) provides an extreme case, where the germline stem cells that are in direct contact with the niche receive high levels of the bone morphogenetic protein ligand Dpp, whereas the next cell row makes the switch to the transit-amplifying population and initiates differentiation. A variety of mechanisms have been demonstrated to reduce the effective diffusion of Dpp to facilitate this switch. They include trapping of Dpp by the extracellular matrix and a bistable translational switch for the translation of the Dpp cascade components (7). Still, it is difficult to perceive how these mechanisms would work to restrict the diffusion of Dpp to a single cell row. Again, cell extensions come to the rescue.
In the male testis, the extensions emanate from the germline stem cells toward the niche and are microtubule-based (termed nanotubes). The capacity of the stem cells to take up Dpp and maintain their non-differentiated state depends on the presence and thickness of the nanotubes (8). The inability of adjacent cells that do not produce nanotubes to receive the Dpp signal strongly suggests that the nanotubes provide the conduit for efficient and highly targeted delivery of this cardinal ligand.
Dynamic FGF Ligand Presentation in the Trachea
In many signaling scenarios, the dynamic expression and presentation of the ligand to a ubiquitously expressed receptor provide the spatial and temporal cues for developmental signaling cascades (1). One of the champion ligands in this regard is the Drosophila FGF Branchless (9), which activates the FGF receptor Breathless (10). While Breathless is broadly expressed in all tracheal cells, it is the dynamic expression of Branchless that shapes the structure of the trachea throughout the life cycle of the fly. During embryogenesis, the dynamic and stereotyped expression of Branchless in cells and tissues adjacent to the trachea, which is driven by spatio-temporal developmental cues, attracts the tips of the growing tracheal branches (9). In contrast, during the larval phase, the expression of Branchless is triggered by metabolic signals, attracting the migration of extending secondary branches toward hypoxic tissues (11).
The Krasnow lab has recently demonstrated how dynamic Branchless expression directs two additional phases of tracheal remodeling. During metamorphosis, the larval tracheal branches decay, and the tracheal system is remodeled by newly formed branches. However, the scaffold of the decaying branches shapes the pattern of the new branches. It turns out that in this scenario and in contrast to other cases, Branchless is dynamically expressed within the decaying larval tracheal system itself, ahead of the migrating new progenitor tracheal cells (12). Thus, the stereotyped pattern of the larval tracheal system is preserved and passed on to the newly formed adult system.
The subcellular distribution of Branchless protein turns out to be instructive for tracheal extensions into the muscle tissue during metamorphosis. Mature flight muscles contain T-tubules, which are extended invaginations of the muscle membrane that connect the cell surface with mitochondria that are positioned adjacent to the contracting sarcomeres. The trachea that extend into the muscles invade the tissue through the T-tubules, to reach the vicinity of the mitochondria and facilitate efficient oxygen delivery. However, how are the tracheal branches targeted through this intricate route into the muscle? The Krasnow lab has demonstrated that prior to the tracheal invasion of the muscle, the subcellular localization of Branchless within the muscle loses its broad surface distribution and becomes restricted to the T-tubules, which are rich in basolateral proteins (13). Thus, a switch in intracellular protein targeting of Branchless at the critical time is responsible for the altered migration route of the trachea.
Quantitative Dynamic Signaling Analysis
Development is a dynamic process, where the time element plays a key role. However, many of the analyses rely on preparations of fixed embryos, where different staining protocols provide “snapshots” of a dynamic process. Although the compilation of these snapshots simulates the dynamic processes, the precise timing and the ability to monitor the effect of subtle modifications on timing remain ambiguous. To address this challenge, the Shvartsman group has generated a platform that combines information from live embryos where the time axis is precise, with fixed images. Alterations in the structure of the nuclei during embryonic development, as well as other morphological features, are extracted from a movie of live embryos. These features are then used to place fixed snapshots reliably into the dynamic process (14). This allows the incorporation of multiple parameters into the same platform. When the assays are quantitative, they also permit the investigation of a dynamic process from a quantitative standpoint.
The approach was applied to study the early activation phase of MAP kinase in the Drosophila embryo. Dual phosphorylation of MAPK is a universal consequence of activation by different receptor tyrosine kinases (RTKs), and displays a highly dynamic pattern throughout development. In the Drosophila embryo, each aspect of MAPK activation can be attributed to a single receptor tyrosine kinase (15). In the early embryo, MAPK is induced in two lateral stripes by the EGF receptor, which leads to the induction of target genes such as ind, which is required for specification of neuroblast cell fates. In this case, transient production of a short-range ligand leads to a simple signal interpretation system, where a pulse of a given threshold is sufficient to induce the target gene. This hypothesis could be tested in a mutant background where the rate of activation is reduced, and induction of the target gene is indeed delayed until the same critical active MAPK threshold is reached (16).
Coordination of Patterning and Growth
In many tissues, patterning takes place in parallel to growth. In one of the best studied models, Dpp expressed in a stripe at the center of the wing disc serves as the morphogen source and spreads to neighboring cells up to 30 cell diameters away. The role of graded Dpp distribution in patterning different regions of the wing disc has been well characterized (17, 18). On the other hand, it was noted that at the same time Dpp is also necessary for cell proliferation, but the proliferation rates were similar throughout the wing pouch. Extensive efforts and different models have been put forward to explain how graded Dpp signaling can lead to uniform proliferation (19).
Two recent studies utilized new techniques to critically assess the role of the central Dpp stripe in cell proliferation. The Gibson lab has generated a CRISPR-based2 conditional null background of Dpp at the stripe in the third instar larva. Although patterning is abolished, surprisingly proliferation ensues, suggesting another source of Dpp for proliferation (20). The Affolter lab has generated membrane-tethered “nanobodies” that trap diffusible Dpp on the surface of the disc and eliminate its graded distribution. Again, although patterning is lost, proliferation is retained (21).
Coordination between patterning and growth also takes place at a higher level, where local signals provide graded patterning cues, whereas nutrient-dependent systemic signals facilitate scaling and coordination of organ growth. The final element in the Hippo/Warts pathway is represented by the transcription factor Yorkie, whose entry into the nucleus is highly regulated and induces organ growth. The Struhl lab has shown that the TOR pathway feeds into growth of the wing disc at the level of Yorkie, but utilizes a novel mechanism. After Yorkie enters the nucleus, TOR signaling facilitates the capacity of Yorkie and its cofactor Sd to access their target genes. Upon nutrient deprivation, when the TOR pathway is inactive, although Yorkie accumulates in the nucleus, it is sequestered from its target genes (22). This mechanism contributes to scaling of wing size with nutrient availability.
Synthetic Screens Identify Drug Targets
Cultured Drosophila cells provide an effective platform for cell-based screening of signaling pathway components and their regulation. These cells are especially suited for RNAi screens because they readily take up large double-stranded RNA fragments. Over the past decade, these screens have yielded a wealth of information and provided a new glimpse into the pathways that complemented the data obtained from whole animal studies. However, RNAi treatment results in a variable phenotype, such that when synthetic screens are considered to look for the effects of loss of genes in the background of an absence for another gene, the readouts may be ambiguous.
To overcome this problem, the Perrimon lab has combined CRISPR-based gene knockout with RNAi screens. By creating a uniform null background following genetic inactivation of the primary gene, these cells can now serve as a platform to query the effect of different RNAi molecules. Such a screen was carried out for the tuberous sclerosis complex, representing a family of tumor suppressors. Following elimination of the TSC1 or TSC2 genes, the cells were queried by RNAi screens directed at all kinases and phosphatases. Synthetic interactions reducing cell growth were identified with three candidate genes (23). Identification of interacting genes significantly broadens the range of potential drug targets that could be used and raises the possibility that there may already be approved drugs against some of the interacting genes that were identified by the screen.
Dynamic Transcription Live!
Analysis of transcription serves as one of the cornerstones in our understanding of development. Until recently, this analysis was based on RNA in situ hybridization carried out on fixed tissues. The limitations of this technique include the threshold sensitivity of detection only when sufficient levels of mRNA accumulate, and the ability to monitor mRNA accumulation in snapshots but not the actual transcription. These limitations have now been overcome by the application of the technology of MS2 RNA stem-loop structures and binding of MS-2 coat protein (MS2-CP-GFP) protein. When the 5′-UTR of a gene is tagged with multiple MS2 loops, every nascent transcript can bind the MS2-CP-GFP protein in the course of transcription. The presence of multiple nascent transcripts along a gene gives rise to a fluorescent dot in the nucleus, which can be detected in fixed or live tissue. The most remarkable aspect is the ability to detect transcription live. In addition, the actual quantitative features such as synchrony between cells and the intensity of the dot can be used to determine the density of active RNA polymerase II complexes, and hence the magnitude of transcription (24, 25).
This technology has recently been used by the Levine and Gregor groups to ask how the organization of enhancers, most notably “shadow” enhancers that provide a backup system, impinges on the rate of transcription in vivo. Following the actual transcription rather than the accumulation of mRNA demonstrates that in syncytial embryos, transcription of key genes such as snail actually starts several nuclear division cycles before the mRNA accumulates. In the case of genes that carry two strong enhancer elements, removal of one of the enhancers elevated the level of transcription because less time was “wasted” on a switch between the two enhancers. On the other hand, for genes that carry enhancers of moderate strength, two enhancers were more effective than a single one, because the combined occupancy time was higher (26).
In conclusion, the wealth of information on developmental signaling pathways and their components now brings us to an era where deep understanding of their mechanism of action can be obtained. It is especially exciting to see how the fundamental and conserved signaling cascades can be tweaked to accommodate the specific requirements of the different organs. This modulation is carried out in conjunction with cellular machineries, which allows the organism to tailor the canonical pathways to the unique biology of each and every tissue.
This is the third article in the Thematic Minireview series “Cell Signaling in Simple Organisms.” The author declares that he has no conflicts of interest with the contents of this article.
- CRISPR
- clustered regularly interspaced short palindromic repeats
- TOR
- target of rapamycin.
References
- 1.Perrimon N., Pitsouli C., and Shilo B. Z. (2012) Signaling mechanisms controlling cell fate and embryonic patterning. Cold Spring Harb. Perspect. Biol. 4, a005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clevers H., Loh K. M., and Nusse R. (2014) An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346, 1248012. [DOI] [PubMed] [Google Scholar]
- 3.Kakugawa S., Langton P. F., Zebisch M., Howell S. A., Chang T. H., Liu Y., Feizi T., Bineva G., O'Reilly N., Snijders A. P., Jones E. Y., and Vincent J. P. (2015) Notum deacylates Wnt proteins to suppress signalling activity. Nature 519, 187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramírez-Weber F. A., and Kornberg T. B. (1999) Cytonemes: cellular processes that project to the principal signaling center in Drosophila imaginal discs. Cell 97, 599–607 [DOI] [PubMed] [Google Scholar]
- 5.Kornberg T. B., and Roy S. (2014) Communicating by touch: neurons are not alone. Trends Cell Biol. 24, 370–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy S., Huang H., Liu S., and Kornberg T. B. (2014) Cytoneme-mediated contact-dependent transport of the Drosophila Decapentaplegic signaling protein. Science 343, 1244624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newton F. G., Harris R. E., Sutcliffe C., and Ashe H. L. (2015) Coordinate post-transcriptional repression of Dpp-dependent transcription factors attenuates signal range during development. Development 142, 3362–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inaba M., Buszczak M., and Yamashita Y. M. (2015) Nanotubes mediate niche-stem-cell signalling in the Drosophila testis. Nature 523, 329–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutherland D., Samakovlis C., and Krasnow M. A. (1996) branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell 87, 1091–1101 [DOI] [PubMed] [Google Scholar]
- 10.Klämbt C., Glazer L., and Shilo B. Z. (1992) breathless, a Drosophila FGF receptor homolog, is essential for migration of tracheal and specific midline glial cells. Genes Dev. 6, 1668–1678 [DOI] [PubMed] [Google Scholar]
- 11.Jarecki J., Johnson E., and Krasnow M. A. (1999) Oxygen regulation of airway branching in Drosophila is mediated by branchless FGF. Cell 99, 211–220 [DOI] [PubMed] [Google Scholar]
- 12.Chen F., and Krasnow M. A. (2014) Progenitor outgrowth from the niche in Drosophila trachea is guided by FGF from decaying branches. Science 343, 186–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peterson S. J., and Krasnow M. A. (2015) Subcellular trafficking of FGF controls tracheal invasion of Drosophila flight muscle. Cell 160, 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dsilva C. J., Lim B., Lu H., Singer A., Kevrekidis I. G., and Shvartsman S. Y. (2015) Temporal ordering and registration of images in studies of developmental dynamics. Development 142, 1717–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabay L., Seger R., and Shilo B. Z. (1997) MAP kinase in situ activation atlas during Drosophila embryogenesis. Development 124, 3535–3541 [DOI] [PubMed] [Google Scholar]
- 16.Lim B., Dsilva C. J., Levario T. J., Lu H., Schüpbach T., Kevrekidis I. G., and Shvartsman S. Y. (2015) Dynamics of inductive ERK signaling in the Drosophila embryo. Curr. Biol. 25, 1784–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lecuit T., Brook W. J., Ng M., Calleja M., Sun H., and Cohen S. M. (1996) Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing. Nature 381, 387–393 [DOI] [PubMed] [Google Scholar]
- 18.Nellen D., Burke R., Struhl G., and Basler K. (1996) Direct and long-range action of a DPP morphogen gradient. Cell 85, 357–368 [DOI] [PubMed] [Google Scholar]
- 19.Hariharan I. K. (2015) Organ size control: lessons from Drosophila. Dev. Cell 34, 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akiyama T., and Gibson M. C. (2015) Decapentaplegic and growth control in the developing Drosophila wing. Nature 527, 375–378 [DOI] [PubMed] [Google Scholar]
- 21.Harmansa S., Hamaratoglu F., Affolter M., and Caussinus E. (2015) Dpp spreading is required for medial but not for lateral wing disc growth. Nature 527, 317–322 [DOI] [PubMed] [Google Scholar]
- 22.Parker J., and Struhl G. (2015) Scaling the Drosophila wing: TOR-dependent target gene access by the Hippo pathway transducer Yorkie. PLoS Biol. 13, e1002274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Housden B. E., Valvezan A. J., Kelley C., Sopko R., Hu Y., Roesel C., Lin S., Buckner M., Tao R., Yilmazel B., Mohr S. E., Manning B. D., and Perrimon N. (2015) Identification of potential drug targets for tuberous sclerosis complex by synthetic screens combining CRISPR-based knockouts with RNAi. Sci. Signal. 8, rs9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bothma J. P., Garcia H. G., Esposito E., Schlissel G., Gregor T., and Levine M. (2014) Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. Proc. Natl. Acad. Sci. U.S.A. 111, 10598–10603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lucas T., Ferraro T., Roelens B., De Las Heras Chanes J., Walczak A. M., Coppey M., and Dostatni N. (2013) Live imaging of bicoid-dependent transcription in Drosophila embryos. Curr. Biol. 23, 2135–2139 [DOI] [PubMed] [Google Scholar]
- 26.Bothma J. P., Garcia H. G., Ng S., Perry M. W., Gregor T., and Levine M. (2015) Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo. Elife 4, e07956. [DOI] [PMC free article] [PubMed] [Google Scholar]


