Abstract
Streptococcus suis is a major endemic pathogen of pigs causing meningitis, arthritis, and other diseases. Zoonotic S. suis infections are emerging in humans causing similar pathologies as well as severe conditions such as toxic shock-like syndrome. Recently, we discovered an IdeS family protease of S. suis that exclusively cleaves porcine IgM and represents the first virulence factor described, linking S. suis to pigs as their natural host. Here we report the identification and characterization of a novel, unrelated protease of S. suis that exclusively targets porcine IgG. This enzyme, designated IgdE for immunoglobulin G-degrading enzyme of S. suis, is a cysteine protease distinct from previous characterized streptococcal immunoglobulin degrading proteases of the IdeS family and mediates efficient cleavage of the hinge region of porcine IgG with a high degree of specificity. The findings that all S. suis strains investigated possess the IgG proteolytic activity and that piglet serum samples contain specific antibodies against IgdE strongly indicate that the protease is expressed in vivo during infection and represents a novel and putative important bacterial virulence/colonization determinant, and a thus potential therapeutic target.
Keywords: antibody, cysteine protease, immunoglobulin G (IgG), proteolytic enzyme, Streptococcus, substrate specificity
Introduction
The Gram-positive bacterium Streptococcus suis commonly colonizes the respiratory and genital tracts of domestic pigs and wild boars, but is also a significant porcine pathogen causing pathologies such as arthritis, abscesses, endocarditis, septicemia, and meningitis (1). Streptococcal meningitis is a common problem in intense pig farming and associated with substantial economic losses. S. suis strains can, based on the structure of capsular polysaccharides, be classified into at least 35 serotypes with S. suis serotype 2 as the significant zoonotic agent causing mainly septicemia and meningitis in humans (2–4). Mortality rates from zoonotic infection range between 3 and 18% (3–6). Transmission of S. suis between humans has not been confirmed, but only few data explaining S. suis functional adaptation to pigs as their dominant host are available (7). The routine use of antibiotics in the swine industry has caused increasing concern with reports of widespread resistance of S. suis strains against tetracyclines, and macrolides, but also cephalosporins, fluoroquinolone, and an emerging resistance against β-lactam antibiotics (2, 8, 9).
Several virulence-associated factors of S. suis have been described, among which the polysaccharide capsule is so far the most prominent one in protecting the pathogen against phagocytosis (10). Numerous surface-bound and secreted proteins of S. suis were identified as homologous to virulence factors from other pathogenic streptococci and shown to exert similar functions (10). Among these are several proteases that appear to be involved in S. suis pathogenicity, but only few have been characterized, e.g. the IL-8 degrading protease SspA, a putative collagenase, and an IgA protease (11). Interestingly, most virulence factors and phagocytosis evasion mechanisms described in pigs address innate immune responses, whereas immune evasion of S. suis from adapted immune responses has so far only been poorly investigated (10).
Recently, a novel protease was added to the list of virulence factors contributing to immune evasion by our identification of an isotype and host-specific IgM protease secreted by S. suis. This IgM endopeptidase, cleaving only porcine IgM, is the first specific prokaryotic IgM protease described. The protease is designated IdeSsuis and is unique in function and size within the IdeS family (C66) of proteases (12). The strict specificity of IdeSsuis for porcine IgM suggests that the enzyme contributes to immune escape during infections of piglets, but probably not in zoonotic infections (7). However, in pigs, IgM is considered the dominating activator of the classical complement cascade (13) and our most recent data show that cleavage of opsonizing IgM by IdeSsuis impairs the activation of the classical complement pathway and leads to improved survival of S. suis in immunized growing piglets (14).
The starting point for the present investigation was the surprising observation of an immunoglobulin (Ig) degrading activity in growth culture supernatants of S. suis strains lacking the IgM cleaving endopeptidase IdeSsuis (10ΔideSsuis). A previously unknown secreted cysteine proteinase was found to account for this activity representing a novel, so far undescribed, type of Ig-cleaving peptidase. Interestingly, the protein has recently been identified in membrane vesicles of S. suis (15). The novel protease provides S. suis with means to deal with IgG-mediated adaptive immune responses and specific opsonizing IgG antibodies and may affect the host-microbe relationship of this emerging pathogen.
Experimental Procedures
Bacterial Strains and Growth Conditions
S. suis strain 10 is a virulent serotype 2 strain that has been used in several studies for mutagenesis and experimental infections of pigs (16). Strain 10M7 was kindly provided by Hilde Smith (AWG, Lelystad, Netherlands) (17). Streptococci were grown on Columbia agar plates with 6% sheep blood or in Bacto Todd-Hewitt broth at 5% CO2 and 37 °C. Escherichia coli strains were cultured in lysogeny broth (LB). When appropriate, antibiotics were added at 50 μg/ml for kanamycin and 20 μg/ml for gentamycin.
Material from Animals
Samples from experimentally infected piglets were drawn within a previous study. The protocol for this animal experiment was approved by the Committee on Animal Experiments of the Lower Saxonian State Office for Consumer Protection and Food Safety in Germany (permit number 33.9-42502-04-07/1243). Collection of blood from conventional piglets for bactericidal assays was registered under N19/14 at the regional office in Saxonia, Germany. The animal studies were performed in strict accordance with the principles and recommendations outlined in the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes (European Treaty Series, no. 123) and the German Animal Protection Law.
Identification of the IgG Degrading Activity
S. suis strain 10M7 cultures were harvested at approximately A600 of ∼0.6 and culture supernatants were sterile-filtrated through a 0.22-μm Express PLUS membrane filter (Millipore) prior to fractionating the culture supernatant with ammonium sulfate to 30% saturation. The resulting precipitate was discarded and ammonium sulfate was added to the remaining supernatant to a final concentration of 50% saturation. The second precipitate was re-suspended in 1/100 of the starting volume with 20 mm BisTris3, pH 6.8, and buffer exchange against the same buffer was performed by HiPrep 26/10 Desalting column (GE Healthcare). The material was further fractionated by FPLC on a HiTrap Q HP column (GE Healthcare). Proteins were eluted by a linear NaCl gradient, and fractions eluted at ∼0.2 m NaCl were found to contain the IgG degrading activity. The active fractions were subjected to size exclusion chromatography (HiPrep 16/60 Sephacryl S-100 HR, GE Healthcare) and the IgG degrading activity eluted at ∼37 ml elution volume. Active fractions were analyzed by SDS-PAGE and protein bands were subjected to mass spectrometry analysis.
Mass Spectroscopy
MALDI-TOF mass spectroscopy was performed by Umeå Protein Analysis Facility (Umeå University). Peptides for MS analysis were prepared by in-gel digestion using trypsin (sequencing grade modified, Promega) and analyzed by ESI LC-MS/MS using an HCT ultra ETD II ion trap instrument (Bruker) linked to an Easy Nano LC system (Proxeon). Processing, deconvolution, and compound detection for the LC-MS/MS datasets was performed using the Data Analysis software (4.0 SP4, Bruker). Database searches using the peaklists files of the processed datasets were performed using the Mascot search engine (Matrixscience) in the bacterial sequences of the NCBInr database. The search parameters permitted a mass error of 0.3 Da for both the MS and the MS/MS mode and variable modifications of methionine by oxidation, of cysteine by propionamide derivation and N-terminal acetylation.
Screening of S. suis Strains for IgG Degrading Capacity
Culture supernatants of 15 different S. suis strains (Table 1) were isolated at A600 of ∼0.6 and concentrated by addition of saturated ammonium sulfate solution to a final saturation of 50%. The precipitate was re-suspended in 1/20 of the starting volume in PBS following buffer exchange against PBS by Zeba Spin Desalting Columns 7K MWCO (Thermo Scientific). The 50% ammonium sulfate precipitation fraction of S. suis culture supernatants were used for IgG degradation analysis.
TABLE 1.
S. suis strains used in this study
| Strain | Capsule type | Source (Reference) |
|---|---|---|
| 10 | cps 2 | 16 |
| A1731/94 | cps 1 | 39 |
| P1/7 | cps 2 | 40 |
| T15 | cps 2 | 16 |
| I9841/1 | cps 2 | 39 |
| 199 | cps 2 (human) | 41 |
| MAC724 | cps 2 (human) | 41 |
| B2795/96 | cps 7 | 39 |
| B2441/96 | cps 2 | 39 |
| A5505/93 | untypeable | 39 |
| V2569/1 | cps 5 | Valentin-Weigand, unpublished data |
| #451 | cps 7 | 42 |
| V3667/1 | cps 7 | Valentin-Weigand, unpublished data |
| A5683/94 | cps 9 | 39 |
| A3286/94 | cps 9 | 39 |
| 5223 | cps 14 | 43 |
| 10M7 | cps 2 | 17 |
| 10 ΔideSsuis | cps 2 | 7 |
| 10 ΔigdE | cps 2 | This publication |
| 10 ΔideSsuis ΔigdE | cps 2 | This publication |
Generation of a ΔigdE Deletion Strain
In-frame deletion of igdE was principally conducted as in Seele et al. (7) with S. suis strain 10. For construction of the thermosensitive vector pSET5_ΔigdE, a 618-bp 5′-igdE amplicon was generated with primers preProIgdEPstI (TCACTGCAGTTTTGGGGAGTAGG) and postSSIgdEBamHI (ATGGATCCCAGTTCAGAACCTC) and a 612-bp 3′-igdE amplicon was generated with primers preEndIgdEBamHI (CGGGATCCAGAGAAAAAAGAGATCC) and postEndIgdEEcoRI (AGGAATTCACCGTTATTGTAGCG). These amplicons were cloned into pSET5 (18) with the restriction enzymes indicated in the names of the primers to generate pSET5_ΔigdE. Deletion clones were confirmed by selective PCR analysis and sequencing of genomic amplicons using primers IgdE-seq_frw (ATTGTATTTGGTGGAGGAG) and IgdE-seq_rev (TTTAGCAGCTAAGTTGATACC).
Sequence Analysis
Sequence analyses were performed using the SIB Bioinformatics Resource Portal Expasy. SignalP 4.0 was run to identify a putative signal peptide and the respective cleavage sites (SignalP) (19). In silico modeling of IgdE to identify the potential active site residues was performed using SWISS-MODEL (20–22). A putative transmembrane region was identified using the consensus prediction web serverTOPCONS (last accession 2015-11-06) (23).
DNA Techniques and Primer Sequences
Primers were designed based on gene SSU_RS08150 in the genome S. suis P1/7. All PCRs were conducted with Phusion Master Mix HF (Thermo Scientific). All obtained plasmids were checked by restriction analyses, PCR, and sequencing.
Cloning of igdE and Generation of IgdE Mutants
The igdE gene lacking the signal peptide coding sequences (encoding amino acid 38–1121) was amplified from chromosomal DNA of S. suis strain 10 as template using primers IgdE-frw_NcoI (GTTTCCATGGATGAAAACTCACATTTACAATCG) and IgdE-rev_NotI (ACGTGCGGCCGCATAAGCTTCGTAC) and cloned into pET_ZZ_1a after digestion with NcoI and NotI (Thermo Scientific). The entire insert was sequenced to verify the cloning and the sequence of igdE. The obtained sequence of igdE was identical to the one of S. suis 05ZYH33 and contained an insertion of 32 amino acids compared with SSU_RS08150 of S. suis P1/7. Directed mutagenesis of the putative active site residues Cys-302 to Ser, His-333 to Ala, and Asp-348 to Ala were performed with QuikChange Lightning Multi-Site-Directed Mutagenesis Kit (Agilent Technologies) and primers IgdE-C302S (AACGTCAGAAAGCGATGAGTGTAGGTTTCAGCACT), IgdE-H333A (CAGAAGGTGTCCCGGCTGCTACAGCGCGTG), and IgdE-D348A (TAAAAAGTGGCACACCATTGCCGGTACAGGTTTTATTACAG) according to the manufacturer's instructions.
IgdEΔC, consisting of the N-terminal 470 amino acids of IgdE was created by digesting full-length igdE in pZZ1a with restriction endonucleases XhoI (Thermo Scientific). The digested plasmids were purified, re-ligated, and transformed into E. coli.
Expression and Purification of Recombinant IgdE
E. coli ArcticExpress (DE3)_RIL (Agilent Technologies) isolates carrying pET_ZZ_1a igdE, igdEC302S, igdEH302A, igdED302A, or igdEΔC were grown to A600 0.6 at 30 °C. Protein expression was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside at 12 °C and incubation was continued for an additional 22 h.
Cells were lysed for crude extracts by BugBuster HT Protein Extraction Reagent (Novagen) or for purification by Stansted high pressure cell disrupter (Stansted Fluid Power) in 50 mm BisTris, pH 7, 0.5 m NaCl, 5% glycerol, 40 μm imidazole. The His-ZZ-tagged protein was purified on HisTrap FF (GE Healthcare) using standard protocols. The tag was removed by enzymatic cleavage by Tev-protease for 20 h at 5 °C followed by a second round of purification on HisTrap FT. The flow-through, containing untagged rIgdE, was collected and buffer exchanged against PBS. Protein concentrations were determined by Nanodrop A280 measurements at appropriate dilutions. Recombinant IgdEΔC was purified as described above with higher yields and purity was compared with full-length rIgdE.
Qualitative IgG Degradation Analyses
To monitor IgG cleavage activity in concentrated and fractionated culture supernatants, samples were incubated with 1% porcine plasma in PBS for 16 h at 37 °C prior to Western blot analyses.
To identify the catalytic type of the IgG protease, the IgG degradation reaction was performed in the presence of 0.1–5 mm AEBSF (Sigma), 0.1–5 mm EDTA, 50–250 μm E-64 (Sigma), 0.1–5 mm Z-LVG-CHN2 (Bachem), 0.1–5 mm iodoacetamide (Sigma), or 1/200 to 1/50 dilution of complete protease inhibitor mixture (Roche).
0.5 mg/ml of porcine IgG (Sigma) in PBS or of other species (human, goat, cow, horse, and mouse; all Sigma) were incubated with either crude extracts of induced E. coli carrying the IgdE constructs mentioned above or 10 nm purified rIgdE for 16 h at 37 °C prior subjection to SDS-PAGE under either reducing or non-reducing conditions. To analyze degradation of endogenous IgG in porcine body fluids the following dilutions were used: 1/10 for heart sac fluid, abdominal cavity fluid, and joint fluid; 1/50 for serum; and undiluted for cerebrospinal fluid. Cerebrospinal and joint fluids were either from piglets with fibrinosuppurative meningitis and synovialitis caused by S. suis infection or from piglets with no lesions. Otherwise IgG degradation analyses were conducted as mentioned above with purified rIgdE and analyzed by both SDS-PAGE and Western blot analyses. Experiments were repeated at least three times and representative analyses are shown.
Quantitative IgG Degradation Analyses
For time course analyses and inhibitor profiles the IgG degradation reaction was performed with 0.25 mg/ml of porcine IgG and 0.2 μm purified IgdEΔC at 37 °C in PBS uninhibited or in the presence of 2.5 or 250 μm AEBSF (Sigma), EDTA, E-64 (Sigma), iodoacetamide (Sigma), or Z-LVG-CHN2 (Bachem). DMSO (Sigma) at 0.275%, corresponding to the DMSO content in 250 μm Z-LVG-CHN2, was used as a solvent control. The reactions were continuously sampled and subjected to SDS-PAGE under reducing conditions. Protein bands were detected by staining with Coomassie FluorTM Orange Protein Gel Stain (Invitrogen). The IgG degradation product was densitometrically quantified by imaging with a LAS4000 imaging system (Fujifilm) and analyzed with Image Studio Version 3.1 (LI-COR Biosciences) software. Uninhibited overnight cleavage was set as 100% relative cleavage for the time course experiment. For the inhibitor profile the initial cleavage rate was calculated from the initial increase of cleavage product (0–25% relative cleavage) and the initial cleavage rate of the uninhibited reaction was set as 100% relative activity. Experiments were at least performed in triplicates. Statistical analyses were performed using GraphPad Prism Version 5.0.
SDS-PAGE and Western Blot Analyses
Samples for SDS-PAGE were prepared with either reducing or non-reducing sample buffer and heated to 95 °C for 5 min. 12% SDS-PAGE was stained with Coomassie Blue (Sigma), Coomassie FluorTM Orange Protein Gel Stain (Invitrogen) or blotted to Hybond-P PVDF membrane (GE Healthcare) for Western blot analyses. Membranes were blocked with 5% dry milk powder in 0.1% PBS-Tween, followed by incubation with horseradish peroxidase-conjugated antibodies. Goat anti-pig IgG-HRP (Thermo Scientific) and goat anti-porcine IgM-HRP (Thermo Scientific) was diluted 1:25,000 and goat anti-porcine IgA-HRP (Thermo Scientific) was diluted 1:12,500. Membranes were thoroughly washed with 0.1% PBS-Tween prior to development with Amersham Biosciences ECL Select Western blotting detection reagent (GE Healthcare) according to manufacturer's instruction and pictured by LAS4000 imaging system (Fujifilm).
N-terminal Edman Sequencing
IgdE processed porcine IgG was separated by SDS-PAGE as previously explained and transferred by semi-dry blotting on Hybond-P PVDF membrane (GE Healthcare) with 50 mm sodium borate, 20% MetOH as blotting buffer. The membrane was stained with Ponceau S (Sigma) and after drying the ∼32 kDa degradation product was tightly cut out. N-terminal Edman sequencing of the degradation product was performed by Proteome Factory (Berlin, Germany) and the sequence W/C PICPACE was obtained. The first position sequence determination was complicated due to a likely contamination yielding a strong tryptophan peak and only a minor peak at the cysteine position. However, in BLAST homology searches the indubitable sequence scored porcine IgG hinge region sequences containing a cysteine residue in the corresponding position. Cysteines were identified by signals of propionamide-modified cysteine; nevertheless, cysteine is not part of the calibration standard.
ELISA
For detection of IgG titers against IgdE Maxisorb® plates (Nunc) were coated with 0.6 μg of rIgdE protein using carbonate buffer. After coating the plates were washed three times with PBS plus 0.1% Tween 20 (PBST) and blocked with 5% milk powder in PBS for 2 h at 37 °C. Every sample and the controls were measured in a duplicate series of four (positive reference serum: six) 2-fold dilutions in PBST starting with a dilution of 1:100. For the detection of IgdE-specific IgG antibodies the plates were incubated with goat anti-pig IgG-HRP (1 mg/ml, Bethyl, A100–105P) at a dilution of 1:10,000 for 1 h at 37 °C. All incubation steps at 37 °C were performed on a shaker and after each incubation step the plates were washed with PBST. The plates were developed with 2,2-azino-di-(3-ethylbenzithiazoline sulfonate) (ABTS) and 0.003% H2O2 as substrate. Absorbance was measured at 405 nm in a microplate reader (Synergy H1, BioTek Instruments GmbH). Optical densities were converted to antibody concentrations through log linear regression analysis after background subtraction. The ELISA units for each sample were defined as the mean of the calculated units for each of the four dilutions of the two series. ELISA values obtained from a serum sample drawn 20 days after experimental infection with S. suis strain 10 was arbitrary set to 100 ELISA units, whereas ELISA values from serum samples derived from colostrum deprived piglets were used as negative control.
Results
S. suis Secretes an IgG-cleaving Enzyme
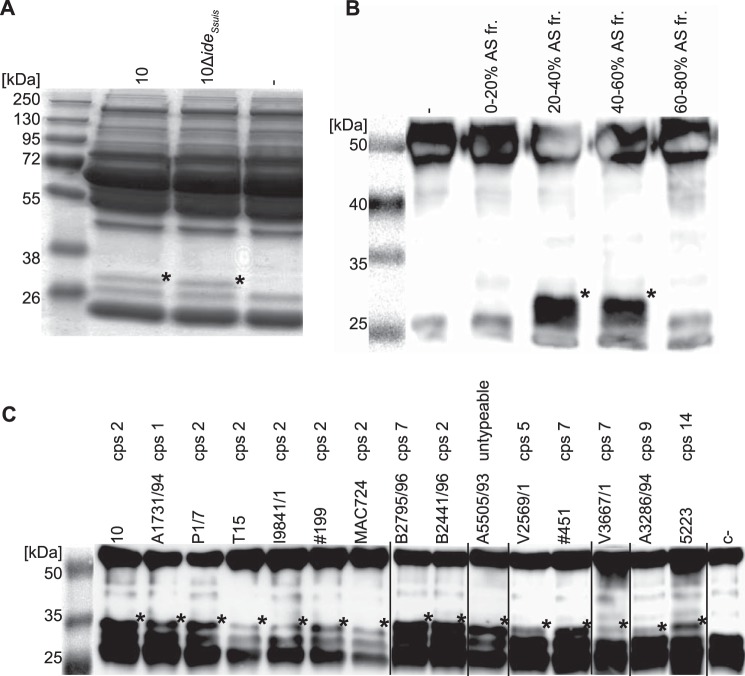
Proteolytic activities of extracellular enzymes of S. suis were analyzed by incubating concentrated supernatants of S. suis strain 10 (16) and the isogenic IgM protease mutant 10ΔideSsuis (7) with 2% porcine plasma as putative substrate. After 3 h of incubation, the reaction was analyzed by reducing SDS-PAGE. Interestingly, the plasma protein band patterns obtained from bacterial culture supernatants of both wild type strain 10 and the isogenic IgM protease mutant 10ΔideSsuis contained an additional protein band of ∼32 kDa, which was absent in the porcine plasma control (Fig. 1A). The 32-kDa band was excised, subjected to MALDI-TOF mass spectroscopy, and identified as an IgG degradation product showing the presence of an IgG proteolytic activity in S. suis. To confirm this finding and to sustain that the proteolytic activity is due to a secreted enzyme of S. suis, growth supernatant of bacterial cultures was fractionated by adding increasing amounts of ammonium sulfate (0 to 80% saturation). Fractions were tested for IgG cleaving activity with porcine plasma and IgG degradation fragments were detected by Western blot using specific polyclonal anti-porcine IgG antibodies. Precipitates of 20 to 60% ammonium sulfate saturation clearly exhibited IgG cleaving activity, demonstrating that the observed IgG proteolytic activity is distinct from the recently described IgM protease IdeSsuis (7) and due to a secreted protein in the culture supernatant of S. suis (Fig. 1B).
FIGURE 1.
IgG degradation activity in culture supernatants of S. suis. A, concentrated (×20) culture supernatants of S. suis strains 10 and 10ΔideSsuis were incubated with 2% porcine plasma for 16 h at 37 °C and analyzed by SDS-PAGE under reducing conditions. A degradation product (*) of ∼32 kDa was observed. B, anti-IgG Western blot analysis of culture supernatant of S. suis strain 10 fractionated by ammonium sulfate precipitation. IgG degradation products (*) were obtained after incubation of 1% porcine plasma with 20–40 and 40–60% ammonium sulfate saturation fractions for 16 h at 37 °C. First lane shows the protein size standard is a photographic image of the membrane before detection of the chemiluminescence signal. C, culture supernatants of S. suis strains of all tested serotypes cleaved IgG. Concentrated (×10) culture supernatants were incubated with 1% porcine plasma for 16 h at 37 °C and analyzed by anti-IgG Western blot. IgG cleavage products (*) at ∼32 kDa of varying intensity were observed in all lanes. Images of different Western blots have been assembled into one figure.
Culture supernatant of 15 different S. suis strains, including several strains of the clinical relevant serotype 2, two human isolates of serotype 2, and a set of strains representing additional serotypes (Table 1) were analyzed by Western blot as described above. For all strains an IgG cleavage product of ∼32 kDa could be detected by Western blot analysis (Fig. 1C), indicating that IgG degradation activity is conserved among different S. suis strains.
Purification and Sequence Characteristics of IgdE, a Novel Protease of S. suis
Prior to a purification trial, protease-enriched ammonium sulfate precipitates were tested for IgG cleaving activity in the presence of class-specific protease inhibitors to preliminary classify the putative IgG degrading protease. Metalloprotease inhibitor EDTA did not affect IgG cleavage, whereas serine protease inhibitor AEBSF interfered moderately with IgG cleavage. Cysteine protease inhibitors E-64, Z-LVG-CHN2, and iodoacetamide, however, all appeared to affect IgG degrading activity (data not shown). Thus, because active site cysteine residues can be inhibited by AEBSF (24, 25), the overall inhibitor profile is most consistent with the assumption that the IgG degrading protease belongs to the class of cysteine proteases. For purification, strain S. suis 10M7 (17), an isogenic mutant of strain 10, was used to avoid masking of low expressed proteins by the in culture supernatants highly abundant muraminidase-released protein. The bacterial culture supernatant was fractionated by ammonium sulfate precipitation and precipitates were subjected to anion-exchange chromatography and size exclusion chromatography. Samples showing IgG cleaving activity were separated by reducing SDS-PAGE. Protein bands were identified by MALDI-TOF mass spectrometry and similarity searches against NCBI databases using a pBLAST algorithm (26). Because the initial characterization of the IgG proteolytic enzyme indicated that the protease is most likely a secreted cysteine protease, sequences of identified proteins were screened for (i) the presence of a cysteine residue within the core of the protein and (ii) the presence of a secretion signal peptide. Beside IdeSsuis that previously has been shown to be IgM specific (7), two (out of 10) identified proteins contained a putative catalytic cysteine residue, but only one contained both a signal peptide sequence predicted by the SignalP algorithm (19) and a core cysteine residue. This protein is annotated as a putative exported protein in the genomic sequences database of S. suis serotype 2 strains (www.sanger.ac.uk) and designated SSU_RS08150 in strain P1/7. SSU_RS08150 encodes a 1121-amino acid protein with a putative transglutaminase core sequence motif located in the N-terminal half of the protein within amino acids 188 to 265. The TOPCONS consensus prediction of membrane protein topology (23) identified a putative transmembrane helix in the C-terminal between approximately amino acids 1059 and 1080. The predicted size of 118 kDa (without signal sequence) is somewhat less than the estimated size of the initially purified protein band after SDS-PAGE (data not shown), which is due to the slightly slower migration of proteins with low pI (pI 4.66) on SDS-PAGE. The full-length protein sequence was used in similarity searches against NCBI databases using a pBLAST algorithm (26). This search revealed no similarities to any known protease of the MEROPS peptidase database (12); no similarities to any eukaryotic protein, but some similarity to hypothetical proteins of Streptococcus porcinus and Streptococcus pseudoporcinus (54% identity in a region of 480–492 amino acid residues) and a hypothetical protein of Streptococcus equi (up to 40% identity in a region of 264 to 406 amino acid residues). The N-terminal part of the protease shows also some similarity to hypothetical proteins of Streptococcus agalactiae and Streptococcus merionis (∼32% identity). The putative protein is in some databases denoted as ribonuclease or ribonucleases G and E, but due to the absence of an experimentally confirmed function and based on the enzymatic activity against porcine IgG, the protein was denoted IgdE for immunoglobulin G degrading enzyme of Streptococcus suis.
IgdE Is a Novel Cysteine Protease
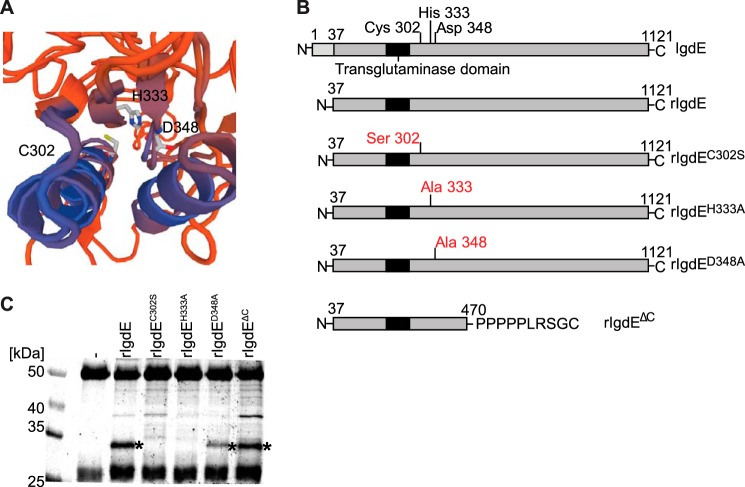
The IgdE sequence reveals the presence of a single cysteine residue in position 302 that, due to the inhibitor profile, is assumed to represent the catalytic site cysteine. Position 302 is located adjacent to the putative transglutaminase domain and in silico three-dimensional modeling (swissmodel.expasy.org) (20–22) of IgdE, using known transglutaminase domain containing proteins as template, allowed for modeling of sequences adjacent to the transglutaminase domain, including a putative active site cleft, showing a potential catalytic triad consisting of cysteine 302, histidine 333, and aspartic acid 348 (Fig. 2A).
FIGURE 2.
Identification of the active site of IgdE. A, in silico modeling of IgdE reveals Cys-302, His-333, and Asp-348 as putative catalytic triad residues. B, schematic illustration of IgdE and the different rIgdE constructs with potential active site residue substitutions and the C-terminal truncation variant. The secretion signal peptide (residues 1–37) is marked in light gray, the transglutaminase domain is boxed, and the potential active site residues and substitutions are indicated. C, 3.3 μm porcine IgG was incubated for 16 h at 37 °C with soluble fractions of E. coli cells expressing different rIgdE constructs. The reactions were analyzed by SDS-PAGE under reducing conditions. IgG cleavage (*) occurred upon incubation with rIgdE, rIgdED348A, and rIgdEΔC but not with rIgdEC302S or rIgdEH333A. The weak protein band of 37 kDa is a contaminant present in lysate preparation and not related to IgdE activity.
For identification of the putative protease domain and catalytic site residues of IgdE several recombinant IgdE constructs were created (Fig. 2B). All three putative catalytic site residues were replaced by site-directed mutagenesis generating mutant proteins, IgdEC302S, IgdEH333A, and IgdED348A, respectively. In addition, a construct lacking the C-terminal part of IgdE was created by an XhoI restriction enzyme cut-back. Crude soluble fractions of E. coli expressing these constructs were incubated with porcine IgG and analyzed by SDS-PAGE. In the presence of recombinant IgdE, the 32-kDa IgG-derived band appeared, demonstrating that the recombinant protein contains IgG proteolytic activity (Fig. 2C, third lane). The IgG cleaving activity could be assigned to the N-terminal part of the protein, as a recombinant protein lacking amino acids 471 to 1121 is sufficient for IgG cleaving activity (Fig. 2C, seventh lane). SDS-PAGE analysis of porcine IgG incubated with the mutant proteins (Fig. 2C, fourth to sixth lanes), revealed that neither rIgdEC302S nor rIgdEH333A exhibited IgG cleaving activity, whereas rIgdED348A showed somewhat reduced IgG cleaving activity. Altogether, these data strongly indicate that these three residues are part of the catalytic site of IgdE.
The initial inhibitor screen was repeated under controlled conditions with purified recombinant IgdEΔC and class-specific protease inhibitors at defined concentrations. Time course experiments of IgG cleavage under uninhibited conditions (Fig. 3) were used to determine initial cleavage rates and as control to quantify significant inhibition by class-specific protease inhibitors. In contrast to classical transglutaminase enzymes, IgdE is not calcium dependent, as the protease is fully active in the presence of EDTA, whereas cysteine class-specific inhibitors iodoacetamide and Z-LVG-CHN2 significantly interfered with IgG proteolytic activity as no cleavage product could be detected at 125-fold molar excess of inhibitor even after 18 h of incubation (Fig. 3C). E-64 did not inhibit purified IgdEΔC in this experimental setting, which might be explained by a narrow active site similar as described for other streptococcal Ig-proteases, like IdeS (27) and IdeSsuis (7). This result differs from the initial inhibitor screen using S. suis culture supernatants and is most likely due to the very high molar excess of E-64 used in these screens with undefined amounts of enzyme concentrations (data not shown). Z-LVG-CHN2 a cysteine protease-specific inhibitor structurally based on the inhibitory reactive site of cystatin C (28–30) completely inhibited IgdEΔC in S. suis culture supernatants and significant inhibition was observed at 125-fold molar excess when using purified enzyme. The inhibition of enzymatic activity with iodoacetamide and Z-LVG-CHN2 and most importantly the lack of enzymatic activity of the IgdEC302S and IgdEH333A mutant variants strongly suggest that IgdE is a novel and so far unique member of the cysteine protease family.
FIGURE 3.
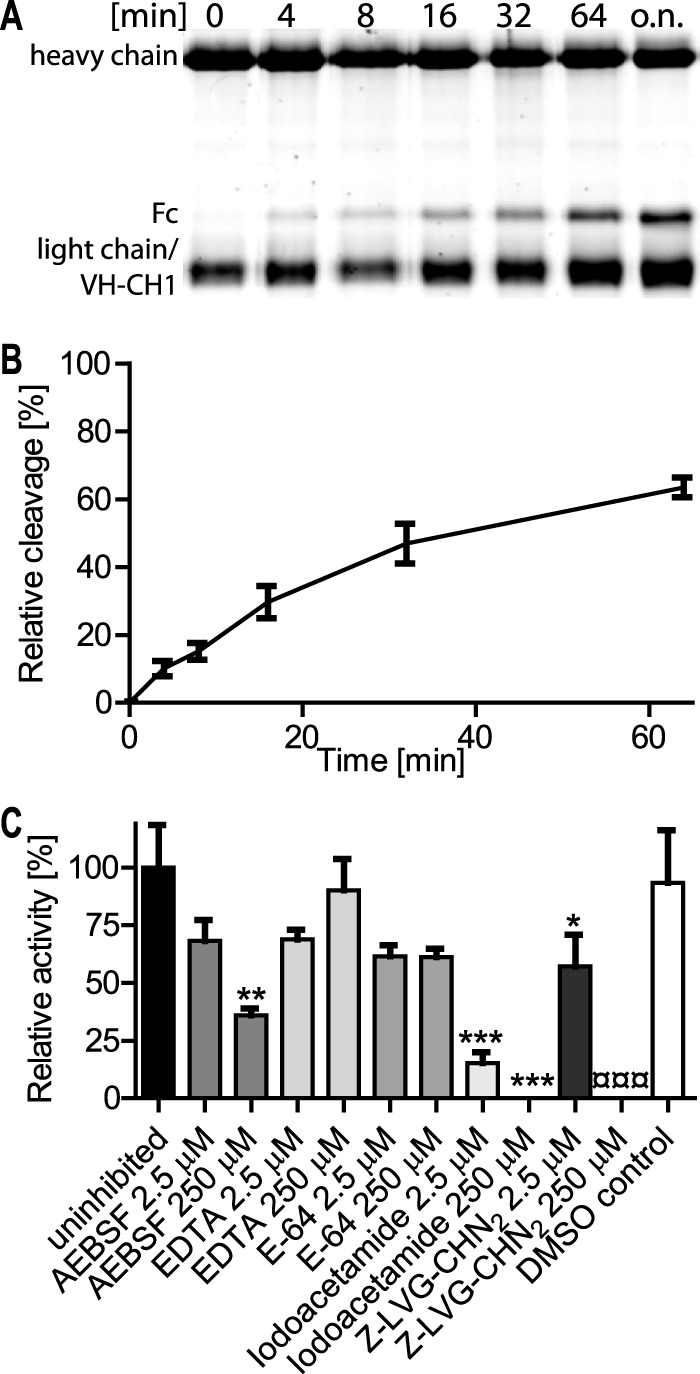
Time course and inhibitor profile of recombinant IgdE. Porcine IgG (1.67 μm) were incubated with 2 μm purified IgdEΔC at 37 °C. The time course of cleavage was monitored by continuous sampling prior to Coomassie Fluor Orange stained SDS-PAGE under reducing conditions. A, a representative gel of a cleavage assay is shown. B, the amount of cleavage product was densitometrically quantified and cleavage obtained after overnight incubation (16 h) was set as 100% relative cleavage (n = 4). C, for the inhibitor profile initial cleavage in the presence of 250 or 2.5 μm of each inhibitor was monitored. Initial cleavage activity of the uninhibited control was set as 100% relative activity. The DMSO control correlates to 250 μm Z-LVG-CHN2. Data are presented as mean ± S.E. of three experiments. Differences to the uninhibited control (*) or DMSO control (¤) were analyzed by Dunnett's multiple comparison test with significance set at p values of <0.05 (*), <0.01 (**) and <0.001 (*** or ¤¤¤) (n = 3).
IgdE Cleaves Porcine IgG with High Specificity in the Hinge Region
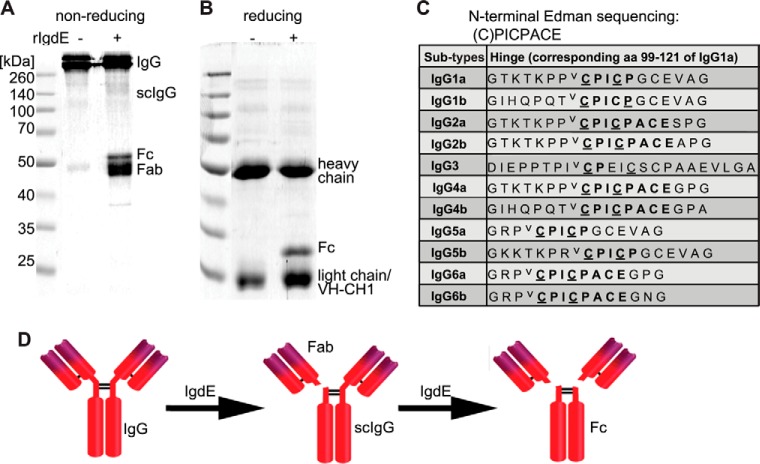
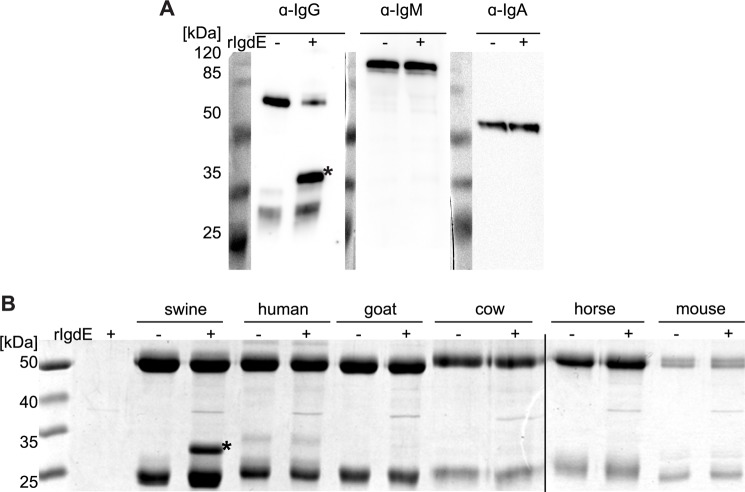
Porcine IgG degradation products were analyzed by reducing and non-reducing SDS-PAGE. The observed band pattern under non-reducing conditions is consistent with a cleavage site located in the hinge region of the heavy chain just N-terminal of the interconnecting disulfide bonds (Fig. 4A). IgG degradation products obtained under reducing conditions (Fig. 4B) were subjected to N-terminal Edman sequencing and the obtained sequence C*PICPACE was found in the hinge region sequences of porcine IgG2ab, IgG4ab, and IgG6ab subtypes. Similar sequences with CPICPGCE as a common motif are present in the hinge region of IgG1ab and IgG5ab subtypes, whereas IgG3 exhibited a CPXXXXC sequence in the hinge region (Fig. 4C). The observed cleavage patterns on reducing and non-reducing SDS-PAGE, in addition to the identified cleavage site, are consistent with a cleavage reaction in which one IgG heavy chain is hydrolyzed just N-terminal of the homo-dimer disulfide bonds, before a second step hydrolysis cleaves the second heavy chain (Fig. 4D). Sequences similar to the IgG hinge region could not be found in the heavy chain sequences of porcine IgA and IgM. Therefore IgdE specificity was further investigated employing specific antibodies against porcine IgG, IgM, and IgA. Porcine plasma incubated with recombinant IgdE was analyzed by Western blot demonstrating that the protease targets IgG in porcine plasma, but does not cleave IgA or IgM (Fig. 5A).
FIGURE 4.
IgdE cleaves the heavy chain of porcine IgG in the hinge region. The reaction was analyzed by non-reducing (A) and reducing (B) Coomassie Blue-stained SDS-PAGE. 3.3 μm IgG were incubated with (+) or without (−) 10 nm purified rIgdE for 16 h at 37 °C. C, hinge region sequences of all porcine IgG subtypes. Cysteine residues believed to be involved in S-S covalent bonds (underlined) and the potential cleavage site (⋁) are marked in the table. D, the observed cleavage pattern and cleavage site proposes a model where one IgG heavy chain is first hydrolyzed by IgdE resulting in one free Fab fragment (Fab) and single cleaved IgG (scIgG) and in a second step the other heavy chain is hydrolyzed resulting in one Fc fragment (Fc) and two Fab fragments.
FIGURE 5.
IgdE is highly specific for porcine IgG. A, 2% porcine plasma was incubated with (+) or without (−) purified 10 nm rIgdE for 16 h at 37 °C. The reaction was analyzed by anti-porcine IgG, IgM, and IgA Western blots under reducing conditions. Only cleavage of IgG was observed (*). B, 0.5 mg/ml of IgG of different species was incubated with (+) or without (−) 10 nm purified rIgdE for 16 h at 37 °C and analyzed SDS-PAGE under reducing conditions. No cleavage of IgG derived from any other species than pig (*) was observed. The weak protein band of 37 kDa is a contaminant present in lysate preparation (see second lane) and not related to IgdE activity. Images of different gels have been assembled into one figure.
The host specificity of the protease was investigated by incubation of purified IgdE with IgG preparations of humans, goat, bovine, horse, and mouse (Fig. 5B). Only pig IgG was found to be a substrate for this novel IgG degrading protease. Thus, IgdE shows a pronounced species specificity and targets only porcine IgG.
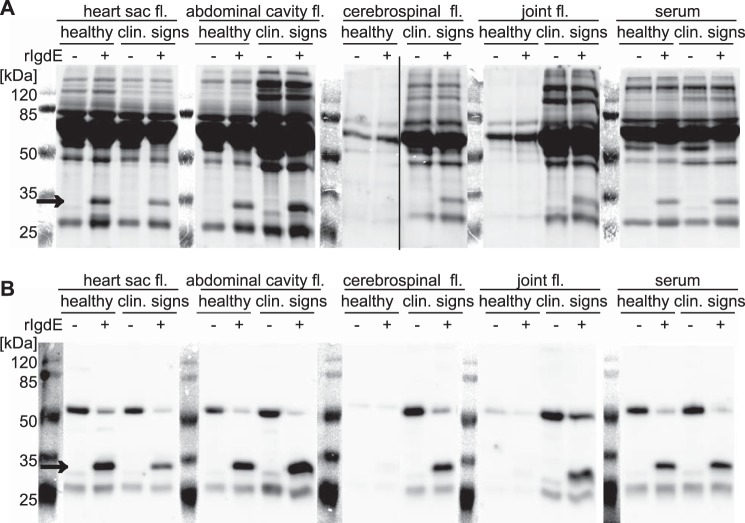
To further evaluate the specificity of IgdE ex vivo, body fluids of healthy or diseased piglets were used. Fluids from heart sac, abdominal cavity, and joint, as well as cerebrospinal fluid and serum were treated with recombinant IgdE and analyzed by SDS-PAGE (Fig. 6A) and Western blot using porcine IgG-specific antibodies (Fig. 6B). No apparent change in the protein band pattern, except for the appearance of the diagnostic IgG cleavage product could be observed in the presence of rIgdE in heart sac, abdominal cavity, and serum samples. Also no IgG, i.e. no cleavage products, were present in joint fluid and cerebrospinal fluids of healthy pigs. In contrast, all body fluids obtained from piglets with clinical signs, e.g. suffering from meningitis and synovialitis, contained IgG as consequence of an inflammatory response and again a single diagnostic 32-kDa band could be observed in all samples treated with rIgdE (Fig. 6). The specificity of IgdE is emphasized by the observation that no additional degradation products could be identified on SDS-PAGE following incubation of porcine body fluids with the protease. Thus, although it cannot be excluded that additional substrates exist, these results indicate that IgdE is a highly specific IgG protease, complementing the previously described IgA and IgM degrading proteases of S. suis (7, 10).
FIGURE 6.
IgdE degrades endogenous IgG in all tested body fluids of healthy pigs and pigs with respective lesions (size indicated with arrows). No other degradation products could be observed. Diluted body fluid samples were incubated with (+) or without (−) 10 nm purified rIgdE for 16 h at 37 °C. The reactions were analyzed by SDS-PAGE (A) and anti-IgG Western blots (B) under reducing conditions. Lanes showing the protein size standard are a photographic image of the membrane before detection of the chemiluminescence signal. Images of different gels have been assembled into one figure.
IgdE Is the Sole IgG Cleaving Enzyme Expressed by S. suis
Strain 10 and strain 10ΔideSsuis, the latter lacking IgM cleaving activity, were used to generate igdE in-frame deletion mutants. IgG and IgM cleaving activities in growth supernatant were determined by Western blot using specific polyclonal anti-porcine IgG or IgM antibodies (Fig. 7). As expected, wild type strain 10 exhibited IgG and IgM cleaving activity (Fig. 7, A and B, third lane), whereas supernatant of the isogenic mutant strain 10ΔigdE only contained IgM cleaving activity and neither IgG nor IgM degradation was detectable in supernatant of double mutant strain 10ΔideSsuis ΔigdE (Fig. 7, A and B, sixth lane). Thus, it appears that IgdE is required and sufficient to cleave porcine IgG and that no other IgG cleaving activity is released under these experimental conditions.
FIGURE 7.

IgdE is necessary for IgG cleavage by S. suis and IgG cleavage is independent of IdeSsuis. Concentrated (×10) supernatants of S. suis strains 10, 10ΔigdE, 10ΔideSsuis, and 10ΔideSsuis ΔigdE were incubated with 1% porcine plasma for 16 h at 37 °C. The reaction was analyzed by anti-IgG Western blot (A) or anti-IgM Western blot (B) under reducing conditions. IgG degradation (*) was only observed when incubated with supernatants of strains 10 and 10ΔideSsuis, meanwhile IgM degradation (*) only was observed upon incubation with supernatants of strains 10 and 10ΔigdE.
In Vivo Expression of IgdE
Weaning piglets are initially protected from bacterial infection through colostrum-derived maternal antibodies (31). Levels of maternal antibodies decrease over time and active IgG-mediated immunity develops at earliest after 2 weeks (31). Accordingly, levels of IgG antibodies in serum of piglets deprived of colostrum do not exceed background values in ELISA measurements. An ELISA was conducted with rIgdE as antigen to investigate different serum samples from piglets for the presence of antibodies directed against this IgG protease. A control serum sample, drawn 20 days after experimental infection with S. suis strain 10, contained high titers against rIgdE and was defined as 100 ELISA units (see “Experimental Procedures”). In contrast to serum of piglets deprived of colostrum that was used as negative control, specific antibodies against IgdE were detectable in seven of nine 5–6-week-old conventional weaning piglets with ELISA units ranging from 36 and 92. These results demonstrate that IgdE is an immunogenic antigen expressed by S. suis in vivo.
Discussion
Pathogenic bacteria have evolved various strategies to colonize and invade their host and a wide variety of virulence factors are employed to promote growth and mediate evasion from host immune responses. To avoid the obvious risk of extinction, pathogenic bacteria have to deal with both innate immune responses, but most importantly also with specific immunoglobulins. Specific Ig is central to the adaptive immune system by initiating the complement-based and/or phagocyte-based immune response. Ig consists of variable antigen-recognizing Fab regions that are linked through a flexible hinge region with the Fc effector part. The Fc region mediates contact with specific receptors on phagocytic cells or triggers the classical pathway of complement by binding C1q. Thus, the hinge region is target for several microbial proteases and examples include IdeS from Streptococcus pyogenes (32), Gingipain K from Porphymonas ginivalis (33), and SspA from Staphylococcus aureus (34). Piglets obtain initial immunity through colostrum and maternal antibodies. Levels of IgG in sow milk have been determined to exceed 95 mg/ml during the first 12 h (35) and mediate protection against bacterial infections after birth in suckling pigs. The importance of maternal IgG is further emphasized by the fact that colostrum-deprived animals in experimental settings suffer of bacterial sepsis with mortality rates up to 90% (31).
The current study describes the identification and characterization of a novel IgG endopeptidase of S. suis that is highly specific for porcine IgG. Similar to the IgM cleaving protease IdeSsuis, the 1121-amino acid large protein carries the proteolytic active domain in the N-terminal part of the protein. A full size protein is not essential for IgG cleavage in vitro, as a truncated IgdE protein consisting of the N-terminal 470 amino acids retains IgG cleavage activity. The function of the C-terminal part of the protein is currently unknown, but based on the prediction of a putative transmembrane helix in the very C-terminal part of IgdE and based on weak structural similarities to transmembrane domains of the Cps2 protein of S. pneumoniae, the C-terminal might mediate association to the bacterial surface. This hypothesis is currently under investigation.
Cleavage of porcine IgG by IgdE occurred just N-terminal of the hinge region cysteine residues that are likely to form covalent disulfide bonds between the two IgG heavy chains. Thus, IgG cleavage results in the formation of a 64-kDa Fc fragment and two Fab fragments. This cleavage pattern is distinct from the IgG endopeptidase IdeS of S. pyogenes that hydrolyzes IgG in the lower hinge region, thereby generating one F(ab′)2 fragment and two identical ½Fc fragments (32) and from IdeSsuis that cleaves IgM C-terminal from the intra-chain disulfide bonds generating free ½Fc fragments (14).
An initial inhibitor screen, performed to facilitate the purification of native IgdE, indicated inhibition of the IgG protease by common cysteine protease inhibitors. The enzyme concentrations in these assays were unknown and the amounts of inhibitors employed are likely to represent a vast molar excess compared with the concentration present in culture supernatant. The inhibitor screen of IgdE was repeated with purified recombinant IgdEΔC at a defined concentration (2 μm) and revealed that the irreversible modification of the catalytic sulfhydryl group by iodoacetamide (36) efficiently interfered with IgdE activity. Likewise strong inhibition was observed using Z-LVG-CHN2, a tripeptide structurally based on the inhibitory site of cystatin C (28) linked to a diazomethyl ketone group to inactivate the sulfhydryl group of the catalytic cysteine (30). Z-LVG-CHN2 has previously been shown to inhibit cysteine proteases, including papain, SpeB, and IdeS (28, 32), but notably failed to inhibit IgM cleavage by IdeSsuis (7). Although this finding strongly suggests that the enzyme belongs to the clan of cysteine proteases, in this assay the enzymatic activity of IgdE was not inhibited by the common cysteine proteinase inhibitor E-64 at 125-fold molar excess (Fig. 3). The lack of inhibitory activity of E-64 has previously been explained by the presence of an unusual narrow active site that sterically hinders the inhibitor to access the active site of IdeS (27) and it is likely that this also might be the case for IdeSsuis and IgdE. The serine protease inhibitor AEBSF showed a moderate inhibition at 125-fold molar excess. This result must, however, not be conflicting with IgdE being a cysteine protease, as inhibition of cysteine proteases with high concentrations of AEBSF and the structurally similar PMSF has been reported (24, 25). Based on these findings the class specificity of IgdE as a cysteine protease was further investigated by site-directed mutagenesis of the proposed catalytic site residues cysteine 302, histidine 333, and aspartic acid 348. Clearly, a replacement of the cysteine with the structurally similar serine residue abolished enzymatic activity of IgdE. Also the replacement of the histidine and aspartic acid residues in the proposed active site affected IgdE activity suggesting that the protease, albeit an unique inhibitor profile, is a novel cysteine protease, unrelated to the IdeS family of immunoglobulin cleaving proteases.
We earlier reported the identification of a host-specific IgM protease, IdeSsuis, with high sequence similarity to group A streptococcal IdeS (7). It was speculated that the variety and functional differences among IgG of humans and pigs would account for the different specificities of these related proteases and furthermore, that the diversity of porcine IgG could have been an obstacle for the evolution of an IgG endopeptidase (7). The identification of a highly specific, but apparently unrelated porcine IgG endopeptidase denounces the latter assumption. However, six diverse IgG subclasses, with at least two allotypes for five of the six subclasses are expressed in pigs (37). Incubation of recombinant IgdE with body fluids of healthy or diseased piglets leads to almost complete degradation of endogenous IgG in the samples indicating that all porcine subclasses might be a substrate for IgdE, albeit the fact that IgG3 exhibits a weakest cleavage site consensus motif of all porcine IgGs. IgG3 has been predicted to have the highest affinity for FcγR (38) and to be the only porcine IgG subclass to bind C1q (13). Thus, IgG3 and IgM are important for complement activation during the early adaptive immune response and the IgM protease IdeSsuis has recently be shown to be a novel complement evasion factor (14). Unfortunately, porcine IgG subclasses are considered to be biochemically inseparable (38) and a reliable purification method has not been described. As a consequence, subclass-specific antibodies are not available (9), which makes a detailed analysis of the hydrolysis of single porcine IgG subclasses currently impossible. It is not unlikely that the protease differs in affinity for single IgG subtypes, which would also explain an apparent uncomplete cleavage after overnight incubation in experimental settings using whole IgG preparations (Fig. 3A). However, given the suggested important role of IgG3 in complement activation it would make sense that S. suis, as endemic pig pathogen, has evolved an IgG-specific endopeptidase to counteract complement activation. The igdE mutant strain, as well as an igdE, ideSsuis double mutant strain will be evaluated in complement activation assays to elucidate the role of IgdE in complement evasion. Respective experiments are currently in progress to investigate this hypothesis.
As already mentioned, S. suis is considered an endemic pathogen of pigs but the underlying molecular basis for the observed species restriction has only poorly been investigated. However, the pronounced specificity of IdeSsuis for porcine IgM (7, 14) shed light on this matter and the current finding of a porcine IgG-specific protease builds further on a model of elaborated mechanisms that have evolved specifically to meet porcine immune responses. IgdE-mediated cleavage of porcine, but not human IgG, indicates that the protease is most likely dedicated to contribute to immune escape during infections of pigs, but probably not during zoonotic infections. Serum samples of healthy pigs contain specific antibodies against IgdE, indicating that the protein is expressed during colonization/infection in vivo and likely to play an important role in the pathogen-host interplay. Further experiments are ongoing to analyze the impact of immunoglobulin cleavage by IgdE on the pathogenesis of S. suis infections and elucidate a putative role of IgdE during colonization of piglets.
In summary, this work describes the identification of a novel, specific and efficient IgG cleaving cysteine protease from S. suis. Thus, S. suis is the first known pathogen cleaving specifically three different immunoglobulin isotypes, IgM (7), IgA (11), and IgG. All S. suis strains investigated possessed the IgG protease activity and future analyses on the function of IgdE in infection models will increase our knowledge of host-specific bacterial immunoglobulin endopeptidases and their role in both colonization and infection of the primary host of the bacteria.
Author Contributions
U. v. P. R., C. S., and C. G. B. conceived the study; U. v. P. R. and C. S. coordinated the study and wrote the paper. C. S. and J. S. designed, performed, and analyzed the experiments; U. v. P. R., C. S., C. G. B., and P. V. W. interpreted the data and revised the article. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
Daisuke Takamatsu (National Institute of Animal Health, Japan) is acknowledged for kindly providing pSET5s. We thank Hilde Smith (DLO-Lelystad, Netherlands) for S. suis strains 10, 10M7, and 5223. Parts of this work were planned and performed by the Umeå Protein Expertise Platform. Mass spectrometry analyses were performed at the proteomics facility at Umeå University. Bioinformatic support was received from BILS/CliC.
This work was supported in part by Carl Tryggers foundation, Insamlingstiftelsen at Umeå University, and the Stiftelsen J.C. Kempes Stipendiefond at Umeå University. The materials from piglets investigated in this work were obtained within a study financed by the German Research Foundation under Grants SFB587 and BA 4730/1-1. A patent application for IgdE has been filed. C. S. and U. v. P. R. are listed as inventors in this application.
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- AEBSF
- 4-(2-aminoethyl)benzenesulfonyl fluoride
- DMSO
- dimethyl sulfoxide
- Z
- benzyloxycarbonyl.
References
- 1.Higgins R., and Gottschalk M. (2005) Streptococcal diseases in Diseases of Swine (Straw B., D'Allaire S., Mengeling W., and Taylor D., eds) 2nd Ed., Iowa State University, Ames, IA [Google Scholar]
- 2.Palmieri C., Varaldo P. E., and Facinelli B. (2011) Streptococcus suis, an emerging drug-resistant animal and human pathogen. Front. Microbiol. 2, 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arends J. P., and Zanen H. C. (1988) Meningitis caused by Streptococcus suis in humans. Rev. Infect. Dis. 10, 131–137 [DOI] [PubMed] [Google Scholar]
- 4.Mai N. T., Hoa N. T., Nga T. V., Linh le D., Chau T. T., Sinh D. X., Phu N. H., Chuong L. V., Diep T. S., Campbell J., Nghia H. D., Minh T. N., Chau N. V., de Jong M. D., Chinh N. T., et al. (2008) Streptococcus suis meningitis in adults in Vietnam. Clin. Infect. Dis. 46, 659–667 [DOI] [PubMed] [Google Scholar]
- 5.Tang J., Wang C., Feng Y., Yang W., Song H., Chen Z., Yu H., Pan X., Zhou X., Wang H., Wu B., Wang H., Zhao H., Lin Y., Yue J., et al. (2006) Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS Med. 3, e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wertheim H. F., Nghia H. D., Taylor W., and Schultsz C. (2009) Streptococcus suis: an emerging human pathogen. Clin. Infect. Dis. 48, 617–625 [DOI] [PubMed] [Google Scholar]
- 7.Seele J., Singpiel A., Spoerry C., von Pawel-Rammingen U., Valentin-Weigand P., and Baums C. G. (2013) Identification of a novel host-specific IgM protease in Streptococcus suis. J. Bacteriol. 195, 930–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holden M. T., Hauser H., Sanders M., Ngo T. H., Cherevach I., Cronin A., Goodhead I., Mungall K., Quail M. A., Price C., Rabbinowitsch E., Sharp S., Croucher N. J., Chieu T. B., Mai N. T., et al. (2009) Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS ONE 4, e6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aarestrup F. (2012) Sustainable farming: Get pigs off antibiotics. Nature 486, 465–466 [DOI] [PubMed] [Google Scholar]
- 10.Fittipaldi N., Segura M., Grenier D., and Gottschalk M. (2012) Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol. 7, 259–279 [DOI] [PubMed] [Google Scholar]
- 11.Zhang A., Mu X., Chen B., Liu C., Han L., Chen H., and Jin M. (2010) Identification and characterization of IgA1 protease from Streptococcus suis. Vet. Microbiol. 140, 171–175 [DOI] [PubMed] [Google Scholar]
- 12.Rawlings N. D., Waller M., Barrett A. J., and Bateman A. (2014) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 42, D503–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler J. E., Zhao Y., Sinkora M., Wertz N., and Kacskovics I. (2009) Immunoglobulins, antibody repertoire and B cell development. Dev. Comp. Immunol. 33, 321–333 [DOI] [PubMed] [Google Scholar]
- 14.Seele J., Beineke A., Hillermann L. M., Jaschok-Kentner B., von Pawel-Rammingen U., and Valentin-Weigand P., and Baums C. G. (2015) The immunoglobulin M-degrading enzyme of Streptococcus suis, IdeSsuis, is involved in a novel complement evasion mechanism. Vet. Res. 46, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haas B., and Grenier D. (2015) Isolation, characterization and biological properties of membrane vesicles produced by the swine pathogen Streptococcus suis. PLoS ONE 10, e0130528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith H. E., Damman M., van der Velde J., Wagenaar F., Wisselink H. J., Stockhofe-Zurwieden N., and Smits M. A. (1999) Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect. Immun. 67, 1750–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith H. E., Vecht U., Wisselink H. J., Stockhofe-Zurwieden N., Biermann Y., and Smits M. A. (1996) Mutants of Streptococcus suis types in expression of muramidase-released protein and extracellular protein induce disease in newborn 1 and 2 impaired germfree pigs. Infect. Immun. 64, 4409–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takamatsu D., Osaki M., and Sekizaki T. (2001) Thermosensitive suicide vectors for gene replacement in Streptococcus suis. Plasmid 46, 140–148 [DOI] [PubMed] [Google Scholar]
- 19.Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) Signal P 4.0: discriminating signal peptides from transmembrane regions. Nature Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 20.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., and Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnold K., Bordoli L., Kopp J., and Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 22.Kiefer F., Arnold K., Künzli M., Bordoli L., and Schwede T. (2009) The SWISS-MODEL repository and associated resources. Nucleic Acids Res. 37, D387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsirigos K. D., Peters C., Shu N., Käll L., and Elofsson A. (2015) The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 43, W401–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vincents B., von Pawel-Rammingen U., Björck L., and Abrahamson M. (2004) Enzymatic characterization of the streptococcal endopeptidase, IdeS, reveals that it is a cysteine protease with strict specificity for IgG cleavage due to exosite binding. Biochemistry 43, 15540–15549 [DOI] [PubMed] [Google Scholar]
- 25.Dunn B. M. (1989) in Proteolytic enzymes: a practical approach (Beynon R. J., and Bond J. S., eds), pp. 57–81, IRL Press at Oxford University Press, Oxford, UK [Google Scholar]
- 26.Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 27.Wenig K., Chatwell L., von Pawel-Rammingen U., Björck L., Huber R., and Sondermann P. (2004) Structure of the streptococcal endopeptidase IdeS, a novel cysteine proteinase with strict specificity for IgG. Proc. Natl. Acad. Sci. U.S.A. 101, 17371–17376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abrahamson M., Ritonja A., Brown M. A., Grubb A., Machleidt W., and Barrett A. J. (1987) Identification of the probable inhibitory reactive sites of the cysteine proteinase inhibitors human cystatin C and chicken cystatin. J. Biol. Chem. 262, 9688–9694 [PubMed] [Google Scholar]
- 29.Björck L., Akesson P., Bohus M., Trojnar J., Abrahamson M., Olafsson I., and Grubb A. (1989) Bacterial growth blocked by a synthetic peptide based on the structure of a human proteinase inhibitor. Nature 337, 385–386 [DOI] [PubMed] [Google Scholar]
- 30.Green G. D., and Shaw E. (1981) Peptidyl diazomethyl ketones are specific inhibitors of thiol proteinases. J. Biol. Chem. 256, 1923–1928 [PubMed] [Google Scholar]
- 31.Drew M. D., and Owen B. D. (1988) The provision of passive immunity to colostrum-deprived piglets by bovine or porcine serum immunoglobulins. Can. J. Anim. Sci. 68, 1277–1284 [Google Scholar]
- 32.von Pawel-Rammingen U., Johansson B. P., and Björck L. (2002) IdeS, a novel streptococcal cysteine proteinase with unique specificity for immunoglobulin G. EMBO J. 21, 1607–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vincents B., Guetsch A., Kostolowska D., von Pawel-Rammingen U., Eick U., Potempa J., and Abrahamson M. (2011) Cleavage of IgG1 and IgG3 by Gingipain K from Porphymonas gingivalis compromises host defense in progressive periodontitis. FASEB J. 25, 3741–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prokesová L., Potuzníková B., Potempa J., Zikán J., Radl J., Hachová L., Baran K., Porwit-Bobr Z., and John C. (1992) Cleavage of human immunoglobulins by serine proteinase from Staphylococcus aureus. Immunol. Lett. 31, 259–265 [DOI] [PubMed] [Google Scholar]
- 35.Klobasa F., Werhahn E., and Butler J. E. (1981) Regulation of humoral immunity in the piglet by immunoglobulin of maternal origin. Res. Vet. Sci. 31, 195–206 [PubMed] [Google Scholar]
- 36.Wallenfels K., and Eisele B. (1968) Stereospecific alkylation with asymmetric reagents. Eur. J. Biochem. 3, 267–275 [DOI] [PubMed] [Google Scholar]
- 37.Butler J. E., and Wertz N. (2006) Antibody repertoire development in fetal and neonatal piglets: XVII. IgG subclass transcription revisited with emphasis on new IgG3. J. Immunol. 177, 5480–5489 [DOI] [PubMed] [Google Scholar]
- 38.Butler J. E., Wertz N., Deschacht N., and Kacskovics I. (2009) Porcine IgG: structure, genetics, and evolution. Immunogen 61, 209–230 [DOI] [PubMed] [Google Scholar]
- 39.Allgaier A., Goethe R., Wisselink H. J., Smith H. E., and Valentin-Weigand P. (2001) Relatedness of Streptococcus suis isolates of various serotypes and clinical backgrounds as evaluated by macrorestriction analysis and expression of potential virulence traits. J. Clin. Microbiol. 39, 445–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacobs A. A., Loeffen P. L., van den Berg A. J., and Storm P. K. (1994) Identification, purification, and characterization of a thiol-activated hemolysin (suilysin) of Streptococcus suis. Infect. Immun. 62, 1742–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baums C. G., Verkühlen G. J., Rehm T., Silva L. M., Beyerbach M., Pohlmeyer K., and Valentin-Weigand P. (2007) Prevalence of Streptococcus suis genotypes in wild boars of Northwestern Germany. Appl. Environ. Microbiol. 73, 711–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Unterweger C., Baums C. G., Höcher M., Fischer L., Weiss A., and Hennig-Pauka I. (2014) Clinics, diagnosis and prophylaxis of a Streptococcus suis serotype 7 farm problem. Berl. Münch Tierarztl. Wochenschr. 127, 194–201 [PubMed] [Google Scholar]
- 43.Wisselink H. J., Joosten J. J., and Smith H. E. (2002) Multiplex PCR assays for simultaneous detection of six major serotypes and two virulence-associated phenotypes of Streptococcus suis in tonsillar specimens from pigs. J. Clin. Microbiol. 40, 2922–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]