Abstract
Phosphorylated oligosaccharides (POSs) are produced by the degradation of dolichol-linked oligosaccharides (DLOs) by an unclarified mechanism in mammalian cells. Although POSs are exclusively found in the cytosol, their intracellular fates remain unclear. Our findings indicate that POSs are catabolized via a non-lysosomal glycan degradation pathway that involves a cytosolic endo-β-N-acetylglucosaminidase (ENGase). Quantitative and structural analyses of POSs revealed that ablation of the ENGase results in the significant accumulation of POSs with a hexasaccharide structure composed of Manα1,2Manα1,3(Manα1,6)Manβ1,4GlcNAcβ1,4GlcNAc. In vitro ENGase assays revealed that the presence of an α1,2-linked mannose residue facilitates the hydrolysis of POSs by the ENGase. Liquid chromatography-mass spectrometric analyses and fluorescent labeling experiments show that such POSs contain one phosphate group at the reducing end. These results indicate that ENGase efficiently hydrolyzes POSs that are larger than Man4GlcNAc2-P, generating GlcNAc-1-P and neutral Gn1-type free oligosaccharides. These results provide insight into important aspects of the generation and degradation of POSs.
Keywords: carbohydrate metabolism, endoplasmic reticulum (ER), glycobiology, N-linked glycosylation, oligosaccharide, dolichol-linked oligosaccharides, endo-β-N-acetylglucosaminidase, non-lysosomal glycan degradation, phosphorylated oligosaccharides
Introduction
Dolichol-linked oligosaccharides (DLOs)3 are glycan donor substrates for asparagine (N)-linked glycosylation in mammals (1, 2). The biosynthesis of DLOs starts on the cytosolic side of the endoplasmic reticulum (ER) membrane with the assembly of Man5GlcNAc2-PP-Dol (1). This biosynthetic intermediate is flipped into the ER lumen and is converted to Glc3Man9GlcNAc2-PP-Dol. The fully assembled DLO is then transferred onto nascent polypeptides by the oligosaccharyltransferase (3).
In mammalian cells, the biosynthesis of DLOs is regulated by the supply of glucose. When cells are deprived of glucose, incompletely assembled DLOs are produced (4–9), which are rapidly degraded by a currently unclarified degradation system (2, 10, 11). The enzyme involved in the degradation has long been believed to hydrolyze the pyrophosphate bond of DLOs, releasing phosphorylated oligosaccharides (POSs) into the cytosol (Fig. 1, step i) (10–14). Although previous biochemical studies using paper electrophoresis (11) or anion-exchange chromatography (10, 14) suggest the presence of one phosphate group on POSs, the precise number has not been unequivocally determined, which has hampered the identification of the precise action of the DLO-degrading enzyme.
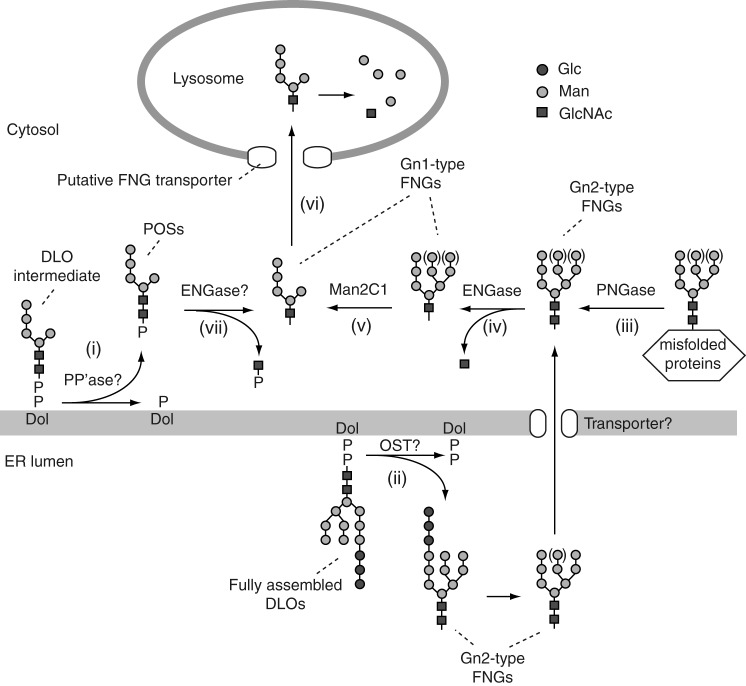
FIGURE 1.
Model of the generation and the non-lysosomal degradation of FNGs and POSs in mammalian cells (2, 35). Biosynthetic intermediates of DLOs, e.g. Man5GlcNAc2-PP-Dol, are presumably degraded by a putative pyrophosphatase (PP'ase), releasing POSs into the cytosol (step i). In the cytosol, Gn2-type FNGs are generated by either the degradation of the fully assembled DLO, presumably by oligosaccharyltransferase (step ii) or the deglycosylation of misfolded glycoproteins by PNGase (step iii). ENGase hydrolyzes the Gn2-type FNGs to produce Gn1-type FNGs and free GlcNAc (step iv). The action of ENGase facilitates mannose trimming of the Gn1-type FNGs by Man2C1 (step v) and lysosomal transport of the FNGs via unidentified FNG transporter (step vi). Although the POSs are not readily transported into lysosomes, the removal of the phosphate group from a POS can initiate the lysosomal transport of the resultant FNGs (23). The hydrolysis of POSs by ENGase may facilitate the catabolism of POSs by removing GlcNAc-P from POSs (step vii).
In addition to POSs, a protein-unbound form of neutral high mannose-type N-glycans (free N-glycans; FNGs), which bare N,N′-diacetylchitobiose at the reducing end (Gn2-type), is produced in the cytosol via two distinct pathways as follows: the degradation of DLOs, presumably by oligosaccharyltransferase in the ER lumen (2, 15, 16), and their transport to the cytosol (Fig. 1, step ii); and the deglycosylation of misfolded glycoproteins by a cytosolic peptide:N-glycanase (PNGase) (Fig. 1, step iii) (2, 17, 18). In the cytosol, the Gn2-type FNGs are processed by an endo-β-N-acetylglucosaminidase (ENGase) (19–21) that hydrolyzes the innermost GlcNAc residue to generate Gn1-type FNGs (Fig. 1, step iv). The action of ENGase is prerequisite for the subsequent trimming of mannose from the FNGs by the α-mannosidase Man2C1 (Fig. 1, step v) (21, 22). This process, which is referred to as non-lysosomal glycan degradation, accelerates the transport of FNGs into lysosomes and their subsequent degradation (Fig. 1, step vi) (23). Although the molecular mechanism responsible for how POSs are catabolized remains unclear, it has been suggested that POSs predominantly accumulate in the cytosol (10, 13, 14).
In this study, we investigated the issue of whether cytosolic ENGase catalyzes the hydrolysis of POSs (Fig. 1, step vii), because an N,N′-diacetylchitobiose core structure is still intact at their reducing ends. Structural analyses of POSs by liquid chromatography-mass spectrometry (LC-MS) showed that the reducing end of the POSs is mono-phosphorylated. Comparisons of POSs produced in wild-type and Engase knock-out (Engase−/−) mouse embryonic fibroblasts (MEFs) revealed that POSs that carry Manα1,2Manα1,3(Manα1,6)Manβ1,4GlcNAcβ1,4GlcNAc accumulate to a significant extent in Engase−/− MEFs. Consistent with this observation, in vitro ENGase assays for POSs with various structures clearly indicated that this enzyme preferentially hydrolyzes POSs that contain an α1,2-linked mannose residue. These results strongly indicate that ENGase efficiently catabolizes POSs that are larger than Man4GlcNAc2-P and generates Gn1-type FNGs and GlcNAc-1-P in the cytosol.
Experimental Procedures
Establishment of MEFs
Wild-type, Engase−/−, and Ngly1−/− Engase−/− MEFs were established and immortalized as described previously (24).
Cell Culture
MEFs (2 × 106 cells/15-cm dish) were seeded and incubated for 24 h at 37 °C in 20 ml of complete Dulbecco's modified Eagle's medium (DMEM), high glucose (Nakalai) supplemented with 10% (v/v) FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin in a 5% CO2 atmosphere. Cells were washed twice with 10 ml of DMEM without glucose (Invitrogen) and then further incubated for 24 h at 37 °C in 20 ml of DMEM without glucose supplemented with either 5 mm (normal glucose condition) or 0.5 mm (low glucose condition) glucose, 10% (v/v) FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin.
Preparation of POSs from MEFs
POSs were prepared essentially as described in a previous report (10). Briefly, MEFs were extracted with 70% (v/v) ethanol, and the soluble fraction was evaporated to dryness. The dried pellet was dissolved in 2.5 ml of water and desalted by passage through a PD-10 column (GE Healthcare) equilibrated with 5% (v/v) ethanol. The desalted sample was adjusted to 10 mm Tris-HCl, pH 7.4, and subjected to anion-exchange column chromatography using a Sep-Pak Accell QMA column (Waters). POSs were eluted from the anion-exchange column with 5 ml of an elution buffer containing 10 mm Tris-HCl, pH 7.4, and 70 mm NaCl.
For the analysis of POSs by LC-MS, the eluates from the anion-exchange column were desalted on PD-10 columns equilibrated with 5% (v/v) ethanol, followed by passing through a 250-μl CM-Sepharose (GE Healthcare) equilibrated with water. The flow-through fraction, which contains POSs, was evaporated to dryness and dissolved in a small volume of water and then subjected to LC-MS analysis.
Pyridylamination
POSs eluted from the anion-exchange chromatography column were dephosphorylated by treatment with recombinant calf intestine alkaline phosphatase (CIP; 1 unit, Roche Applied Science) for 16 h at 37 °C. The dephosphorylated POSs were desalted on an InertSep GC column (150 mg/3 ml; GL Sciences), evaporated to dryness, and labeled with 2-aminopyridine (PA) as described previously (15).
High Performance Liquid Chromatography (HPLC)
Size fractionation HPLC (15) and dual-gradient reversed-phase HPLC (25) were carried out as described previously. PA-glycans were quantitated using PA-Glc6 that is included in PA-glucose oligomer (Takara; 2 pmol/μl) as an external standard.
Preparation of Standard PA-labeled Oligosaccharides
Standard PA-labeled oligosaccharides (PA-Gn2, PA-M1A, PA-M4D, PA-M5B, PA-M6E, and PA-M7E) were prepared as described previously (10). PA-M2A, PA-M3B, PA-M8C, and PA-M9A were purchased from Takara.
LC-MS of POSs
POSs (17 pmol as Man2GlcNAc2-P) obtained from the glucose-deprived wild-type MEFs (10) were analyzed by LC-MS using a quadrupole ion trap time-of-flight mass spectrometer (LCMS-IT-TOF, Shimadzu Corp.) in electrospray ionization (ESI) in the negative ion mode. For analysis, 10-μl aliquots (maintained at 4 °C in the auto sampler) were injected for separation by HPLC, using gradient elution, with a Nexera XR LC system (Shimadzu Corp.) on a Mastro C18 column (100 × 2.1 mm; particle size 3.0 μm) at a flow rate of 0.3 ml min−1, with the column maintained at 40 °C. The chromatographic system used a binary solvent system delivered as a gradient of Solvent A (15 mm ammonium acetate, 10 mm tributylamine solution) and Solvent B (methanol). The initial gradient conditions were 100% A, 0% B for 0.5 min followed by a linear gradient up to 25% B in 8 min, and up to 98% B over the next 4 min. The solvent composition was then held at 98% B for 3 min after which the column was returned to 0% B over the next 5 min, making a total cycle time of 20 min per sample. The following method parameters were used for sample analysis: mass range of m/z 200–1500 in MS; ion source temperature of 200 °C; heated capillary temperature of 200 °C; electrospray voltage of −3.5 kV; electrospray nebulization gas flow 1.5 liters/min; detector voltage 1.7 kV; ion accumulation time 30 ms.
The negative ion mode was used. Mass calibration was carried out using a trifluoroacetic acid sodium solution (2.5 mmol liter−1) from 200 to 1500 Da. Data acquisition and processing used software LCMS Solution 3.80.
To verify component identification, the Formula Predictor software was used (Shimadzu Corp.). Predicting a candidate list based on MS data takes into account a number of variables, including isotopic profile analysis, mass accuracy, and mass resolution of the experimentally derived pseudomolecular ion peak and related fragment ion data.
Large Scale Preparation of the DLOs
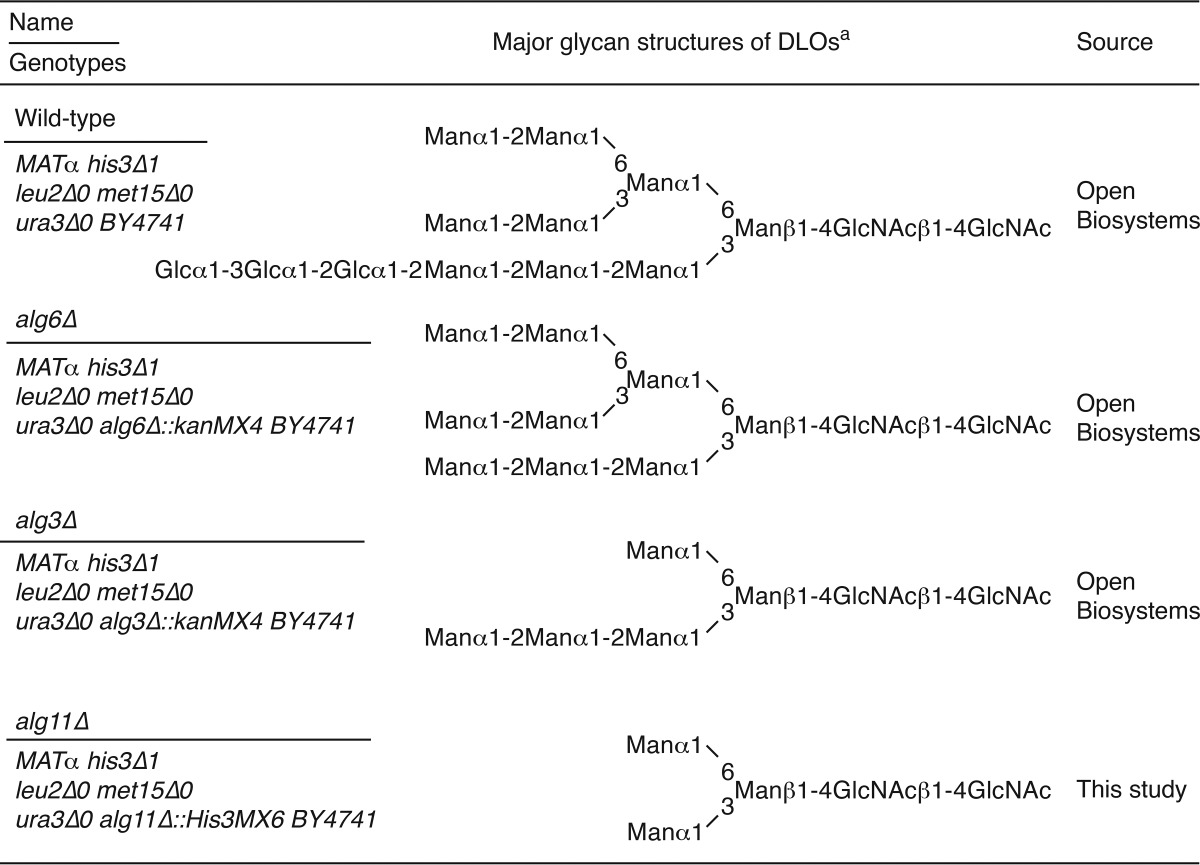
Saccharomyces cerevisiae strains used to prepare DLOs are listed in Table 1. Wild-type, alg6Δ, and alg3Δ cells were transformed with pRS425-GPD-ALG7 to overproduce DLOs (15). The transformants were grown in complete synthetic medium without leucine (26). In contrast, alg11Δ cells were grown in YPD medium (1% yeast extract, 2% polypeptone, 2% glucose). The DLOs were prepared from those strains, and their amounts were quantitated as described previously (15).
TABLE 1.
Yeast strains used in this study and the major glycan structures of their DLOs
Preparation of Detergent-solubilized Mouse Liver Microsomes
Microsomes were prepared from livers of C57BL/6 mice as described previously (27), except that 30% (v/v) glycerol was used instead of 1.3 m sucrose as a cushion to sediment the rough ER by ultracentrifugation. Mouse liver microsomes obtained were homogenized in a homogenization buffer containing 20 mm Hepes-KOH, pH 7.4, 150 mm KCl, 5 mm CoCl2, and 1% (w/v) Triton X-100 and ultracentrifuged at 100,000 × g for 20 min. The supernatant was recovered as detergent-solubilized microsomes. All experimental protocols and procedures were approved by the Ethical Committees on Animal Research of RIKEN.
In Vitro DLO-Pyrophosphatase Assay
DLOs (4 nmol) were dissolved in 25 μl of homogenization buffer and incubated for 16 h at 37 °C with 25 μl of detergent-solubilized microsomes supplemented with glycosidase inhibitors (10 μm swainsonine, 100 μm kifunensine, and 1 mm castanospermine). The reaction was terminated by the addition of 3 volumes of ethanol. After centrifugation at 15,000 × g for 10 min, the supernatant was evaporated to dryness. POSs were purified from the dried pellet and desalted as described above under “Preparation of POSs from MEFs.”
Transfection
Ngly1−/− Engase−/− MEFs (1.0 × 106 cells) were seeded to a 10-cm dish and incubated in 10 ml of complete DMEM without antibiotics for 24 h before transfection. The cells were transfected with 1 μg of pcDNA3.1-mouse ENGase-FLAG (24) using 5 μl of Lipofectamine 2000 (Thermo Fischer Scientific). At 16 h after transfection, the cells were harvested, and the cytosolic fraction was prepared.
Preparation of the Cytosolic Fraction
Ngly1−/− Engase−/− MEFs overexpressing mouse ENGase were washed three times with ice-cold phosphate-buffered saline. The cells obtained from a 10-cm dish were homogenized in 300 μl of cytosol extraction buffer containing 10 mm Hepes-NaOH, pH 7.4, 250 mm mannitol, 5 mm dithiothreitol, 1 mm EDTA, 1× complete protease inhibitor mixture (EDTA-free, Roche Applied Science), and 1 mm Pefabloc (Roche Applied Science). The homogenate was centrifuged at 10,000 × g for 10 min at 4 °C. The supernatant was further ultracentrifuged at 100,000 × g for 60 min at 4 °C to remove the microsomal fraction, and the supernatant was recovered as the cytosol fraction.
To remove endogenous FNGs, the cytosol fraction (400 μl) was concentrated to ∼50 μl using an Amicon Ultra MWCO 50-kDa filter (Millipore) by centrifugation at 10,000 × g for 6 min at 4 °C and diluted with 450 μl of cytosol extraction buffer. After the dilution-concentration step was carried out twice, the final concentrate (∼ 100 μl) was recovered, aliquoted, flash-frozen in liquid nitrogen, and stored at −80 °C until used.
ENGase Activity Assay
The ultrafiltrated cytosol fraction (20 μl) was incubated with 30 μl of ENGase reaction buffer containing 50 pmol of POSs, 50 mm MES-NaOH, pH 6.0, and glycosidase inhibitors (final concentration: 10 μm swainsonine, 100 μm kifunensine, and 1 mm castanospermine) for 1, 2, and 4 h (for Glc3Man9GlcNAc2-P, Man9GlcNAc2-P, and Man5GlcNAc2-P) or 2, 4, and 8 h (for Man3GlcNAc2-P) at 30 °C. For control, Man5GlcNAc2-P was incubated with the ultrafiltrated cytosol fraction without ENGase overexpression for 2 h at 30 °C. The reaction was terminated by the addition of 300 μl of 75% (v/v) ethanol and incubated for 15 min at 0 °C. After centrifugation at 15,000 × g for 10 min at 4 °C, the supernatant was evaporated to dryness. The resulting sample was dissolved in water, desalted on an ion-exchange column containing a stack of AG50-X8 (250 μl, 200–400 mesh, H+ form) and AG1-X2 (250 μl, 200–400 mesh, acetate form) (Bio-Rad), followed by an InertSep GC column (150 mg/3 ml) (GL Sciences). The desalted glycans were fluorescently labeled as described above under “Pyridylamination.”
Fluorescently labeled glycans were analyzed by size-fractionation HPLC. For the detection of PA-labeled Man9GlcNAc1 and Man5GlcNAc1, details for the HPLC condition were described previously (28). For the detection of PA-labeled Glc3Man9GlcNAc1 and Man3GlcNAc1, the following HPLC conditions were used: eluent A, 93% acetonitrile in 0.3% acetate buffer (pH 7.0 adjusted by ammonia); eluent B, 20% acetonitrile in 0.3% acetate buffer (pH 7.0 adjusted by ammonia). The column temperature was 25 °C. The flow rate was 0.8 ml/min. The gradient program was as follows: 0–0.5 min, 1–10% eluent B; 0.5–45 min, 10–55% eluent B; 45–47 min, isocratic 70% eluent B; 47–67 min, isocratic 1% eluent B.
Endoglycosidase Digestion of POSs
To verify the elution positions of Gn1-type FNGs in dual-gradient reversed-phase HPLC (25), POSs (50 pmol) that were obtained as described above under “In Vitro DLO-Pyrophosphatase Assay” were digested with either Endo-H (for Glc3Man9GlcNAc2-P and Man9GlcNAc2-P; Roche Applied Science, 5 milliunits) or Endo-M (for Man5GlcNAc2-P and Man3GlcNAc2-P; Tokyo Chemical Industry, 1 milliunit) in 30 μl of ENGase reaction buffer at 37 °C for 4 h. The reaction was terminated by the addition of 300 μl of 75% (v/v) ethanol, desalted, and fluorescently labeled as described above under “ENGase Activity Assay.”
Results
POSs Are Mono-phosphorylated
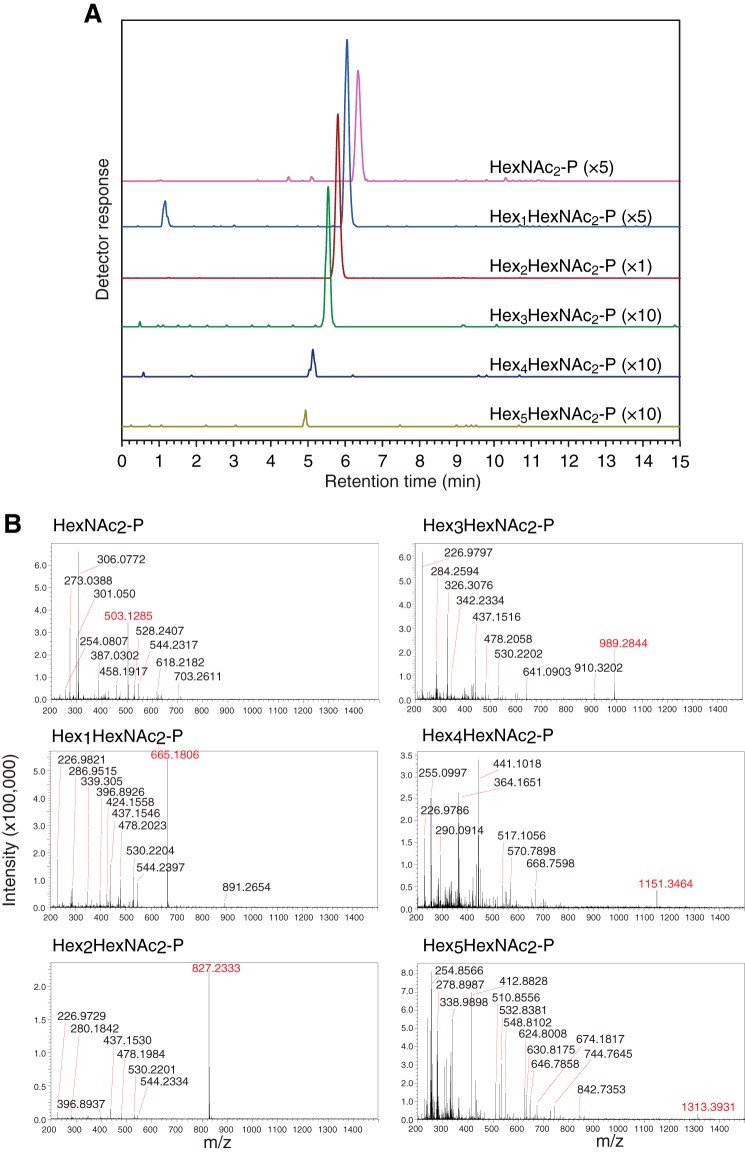
To unequivocally determine the number of phosphate groups of the POSs, POSs that were produced in glucose-deprived wild-type MEFs (10) were analyzed by reversed-phase LC-ESI-MS. In the negative ion mode, singly charged deprotonated molecules [M − H]− corresponding to HexNAc2-P (m/z 503.1285), Hex1HexNAc2-P (m/z 665.1806), Hex2HexNAc2-P (m/z 827.2333), Hex3HexNAc2-P (m/z 989.2844), Hex4HexNAc2-P (m/z 1151.3464), and Hex5HexNAc2-P (m/z 1313.3931) were eluted at around 6.3, 6.0, 5.8, 5.5, 5.1, and 4.9 min in reversed-phase HPLC (Fig. 2). However, no doubly charged deprotonated molecules [M − 2H]2− corresponding to diphosphorylated POSs were detected. These results clearly show that the POSs were mainly mono-phosphorylated, consistent with the predictions made by previous biochemical studies (10, 11, 14).
FIGURE 2.
POSs are mono-phosphorylated. Intact POSs (17 pmol as Man2GlcNAc2-P) prepared from glucose-deprived wild-type MEFs were analyzed by LC-ESI-MS. A, MS chromatograms of Hex0–5HexNAc2-P. Parentheses indicate magnification ratio. B, MS spectra of HexNAc2-P (m/z 503.1285), Hex1HexNAc2-P (m/z 665.1806), Hex2HexNAc2-P (m/z 827.2333), Hex3HexNAc2-P (m/z 989.2844), Hex4HexNAc2-P (m/z 1151.3464), and Hex5HexNAc2-P (m/z 1313.3931). The average mass error for all detected POSs was 0.38 ppm, except that the mass error for Hex5HexNAc2-P was 5.82 ppm. Typical mass accuracy in external calibration was less than 5 ppm.
Ablation of Engase Results in the Accumulation of POSs That Are Larger than Manα1,2Manα1,3(Manα1,6)Manβ1,4GlcNAcβ1,4GlcNAc-P
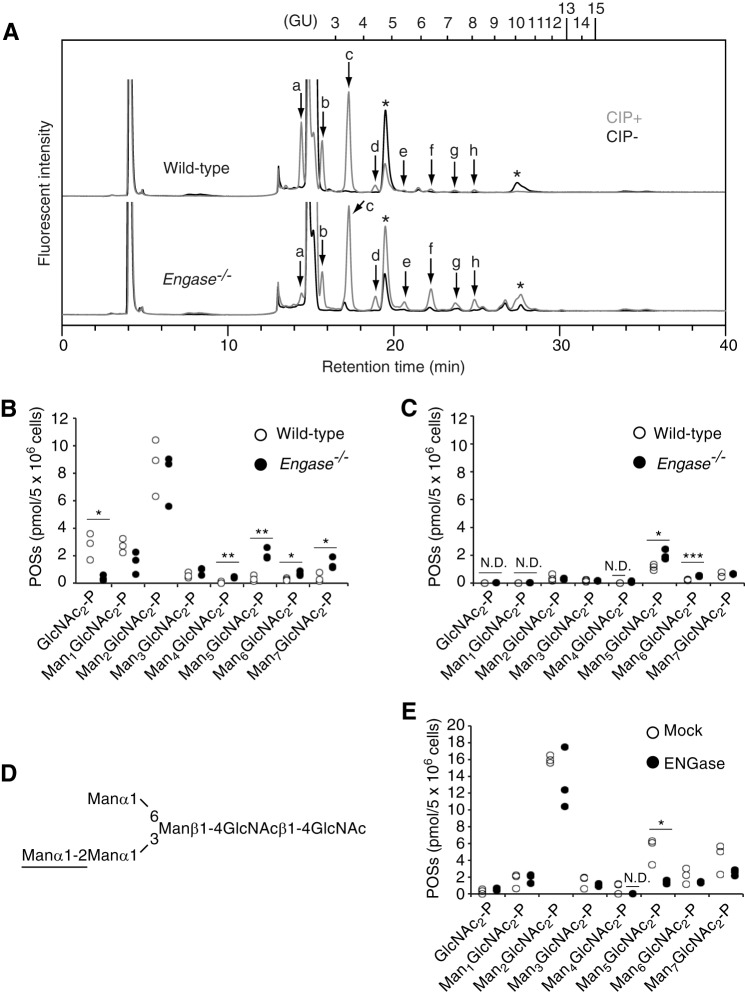
Because ENGase catalyzes the hydrolysis of β1,4-linkage of the N,N′-diacetylchitobiose core of protein-bound (24) and unbound (19, 21) forms of high mannose-type glycans in the cytosol of mammalian cells, we speculated that POSs, which are exclusively localized in the cytosol (10, 13, 14), may also serve as physiological substrates for ENGase. To test this hypothesis, the amounts and glycan structures of the POSs were compared between wild-type and Engase−/− MEFs. The rationale behind this experiment is that if ENGase hydrolyzes POSs, the knock-out of Engase would result in the accumulation of POSs. To detect POSs by a conventional fluorescent labeling method for the reducing end of glycans (29), POSs prepared from glucose-deprived MEFs were treated with or without CIP, fluorescently labeled, and separated by size-fractionation HPLC (Fig. 3A) (10). Fluorescently labeled glycans that appeared in a dephosphorylation-dependent manner (Fig. 3A, peaks a–h) were regarded as POSs. The fluorescently labeled POS-derived glycans thus obtained were fractionated and further analyzed by dual-gradient reversed-phase HPLC (25) to determine their isomeric structures. In both glucose-deprived wild-type and Engase−/− MEFs, Man0–7GlcNAc2-P (Fig. 3B) were detected, and their glycan structures were identical to biosynthetic intermediates of DLOs (Table 2). Quantitative analyses of POSs showed that, in both glucose-deprived wild-type and Engase−/− MEFs, Man2GlcNAc2-P predominantly accumulated, and the knock-out of Engase did not markedly affect the amounts (Fig. 3B). Unexpectedly, however, the amounts of GlcNAc2-P were significantly decreased in Engase−/− MEFs by an unclarified reason (Fig. 3B). By contrast, the amounts of Man4–7GlcNAc2-P were significantly increased in Engase−/− MEFs (Fig. 3B). A similar accumulation of Man4–6GlcNAc2-P was also observed in Engase−/− MEFs that were cultivated under normal glucose conditions (Fig. 3C), indicating that the availability of glucose is not a key factor in regulating the catabolism of Man4–6GlcNAc2-P. The glycan structures of POSs that significantly accumulated in Engase−/− MEFs were found to contain a hexasaccharide structure composed of Manα1,2Manα1,3(Manα1,6)Manβ1,4GlcNAcβ1,4GlcNAc (Fig. 3D). To assess whether the accumulation of these POSs in Engase−/− MEFs is caused by the lack of ENGase, Engase−/− MEFs were transfected with a plasmid-encoding mouse Engase, and POSs that were accumulated in the transfected cells were quantitated. The overexpression of ENGase in Engase−/− MEFs resulted in a significant decrease in Man4,5GlcNAc2-P and showed a propensity for the reduction in Man6,7GlcNAc2-P (Fig. 3E). The partial reduction of these POSs by ENGase overexpression was most likely due to low transfection efficiencies in MEFs (∼50% cells positive in the expression of green fluorescent protein used as a technical control; data not shown). All these results strongly indicate that ENGase is involved in the catabolism of POSs.
FIGURE 3.
POSs carrying a hexasaccharide accumulate in Engase−/− MEFs. A, size-fractionation HPLC of POSs obtained from glucose-deprived wild-type (top panel) and Engase−/− (bottom panel) MEFs. POSs were treated with (+) or without (−) CIP and then fluorescently labeled. Peaks a–h were detected in a CIP-dependent manner. Asterisks indicate peaks derived from labeling reagents. PA-glucose oligomers (degree of polymerization = 3–15) were used as a reference, and the glucose units (GU) were represented on the HPLC chart. B and C, quantification of POSs obtained from MEFs cultured under low glucose (B) and normal glucose (C) conditions. Three independent experiments were carried out. N.D., not detected. *, p < 0.05; **, p < 0.005; and ***, p = 0.001 (Student's t test). D, schematic representation of a hexasaccharide structure found in POSs that were increased in Engase−/− MEFs as compared with wild-type MEFs. An α1,2-linked Man residue important for the action of ENGase (32) is underlined. E, effects of ENGase overexpression in Engase−/− MEFs on POS catabolism. Three independent experiments were carried out. N.D., not detected. *, p = 0.013 (Student's t test).
TABLE 2.
Structures of POSs detected in wild-type and Engase−/− MEFs
ENGase Hydrolyzes POSs in Vitro
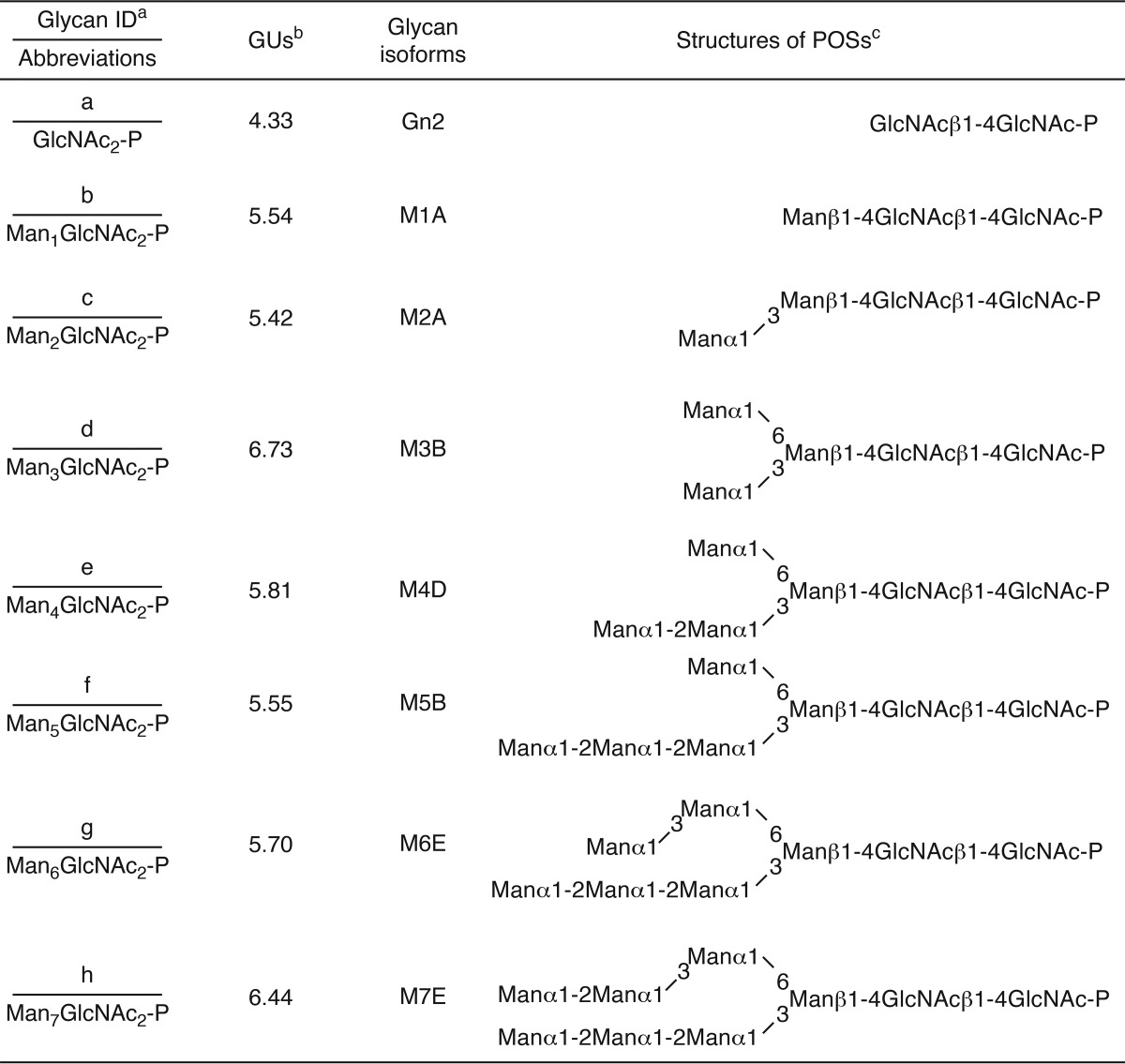
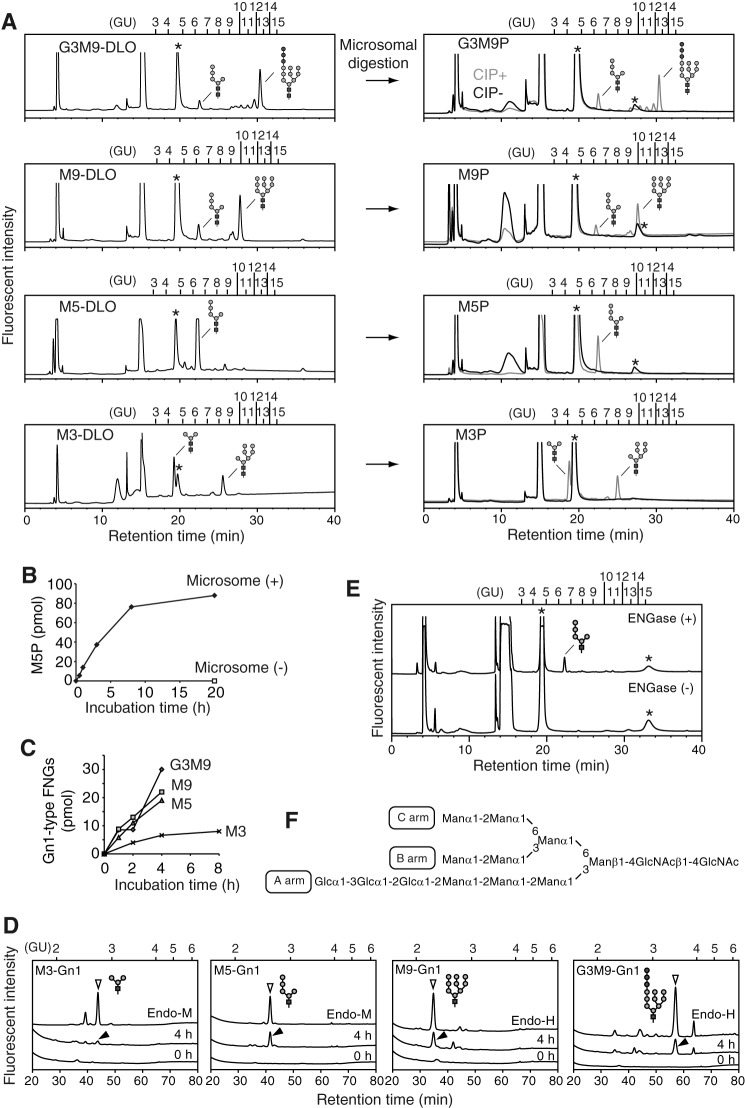
Although our data clearly show that the lack of ENGase causes the accumulation of Man4–7GlcNAc2-P, the issue of whether ENGase catalyzes the hydrolysis of these POSs remained unknown. To address this issue, we carried out in vitro ENGase activity assays. First, we biochemically isolated DLOs with various glycan structures, i.e. Man3GlcNAc2, Man5GlcNAc2, Man9GlcNAc2, and Glc3Man9GlcNAc2 (Fig. 4A, left panels), from various S. cerevisiae strains. POSs with various glycan structures (Glc3Man9GlcNAc2-P, Man9GlcNAc2-P, Man5GlcNAc2-P, and Man3GlcNAc2-P) (Fig. 4A, right panels) were then prepared by incubating the DLO with the detergent-solubilized mouse liver microsomes, which contain DLO-pyrophosphatase activity (Fig. 4B). The POS preparations were fluorescently labeled only after treatment with alkaline phosphatase (Fig. 4A, right panels, CIP+), confirming that the reducing end of the POS preparations is phosphorylated.
FIGURE 4.
ENGase hydrolyzes POSs in vitro. A, size-fractionation HPLC profiles of DLOs extracted from wild-type (Glc3Man9GlcNAc2-PP-dolichol; G3M9-DLO), alg6Δ (Man9GlcNAc2-PP-dolichol; M9-DLO), and alg3Δ (Man5GlcNAc2-PP-dolichol; M5-DLO) cells, which were transformed with a plasmid pRS425-GPD-ALG7 (15), and alg11Δ cells (Man3GlcNAc2-PP-dolichol; M3-DLO) (left panels). The glycan structures shown in the HPLC charts were deduced based on previously reported data (36–41) and further confirmed by the product analyses in the ENGase assay (Table 3). The DLOs were digested with mouse liver microsomes (microsomal digestion) to obtain POSs (G3M9P, M9P, M5P, and M3P, right panels). The resulting POSs were treated with (+) or without (−) CIP before fluorescent labeling. B, time course analysis of the DLO-pyrophosphatase assay in the presence (+) or absence (−) of microsomes using M5P as a substrate. C, time course analyses of the ENGase assay using M3P, M5P, M9P. and G3M9P as substrates. The amounts of Gn1-type FNGs were quantitated by size-fractionation HPLC. D, product analyses for the ENGase assay by dual-gradient reversed-phase HPLC. White arrowheads indicate the elution positions of authentic Gn1-type FNGs generated by Endo-H or Endo-M digestion of POSs. Black arrowheads indicate peaks that are generated by ENGase and co-eluted with the authentic standard glycans. Unassigned peaks generated by endoglycosidase digestions are probably derived from POSs that were co-purified with G3M9P, M9P, M5P, and M3P (see also A, right panels). E, ENGase assay using M5P as a substrate and the cytosolic fraction obtained from either ENGase-transfected (+) or non-transfected (−) Ngly1−/− Engase−/− MEFs as an enzyme source. The reaction mixtures were analyzed by size-fractionation HPLC. F, schematic representation of Glc3Man9GlcNAc2 glycan. The position of the A, B, and C arms of the glycan are indicated. Asterisks in A and E indicate peaks derived from labeling reagents. PA-glucose oligomers (degree of polymerization = 3–15) was used as a reference, and the glucose units (GU) are represented on the HPLC chart.
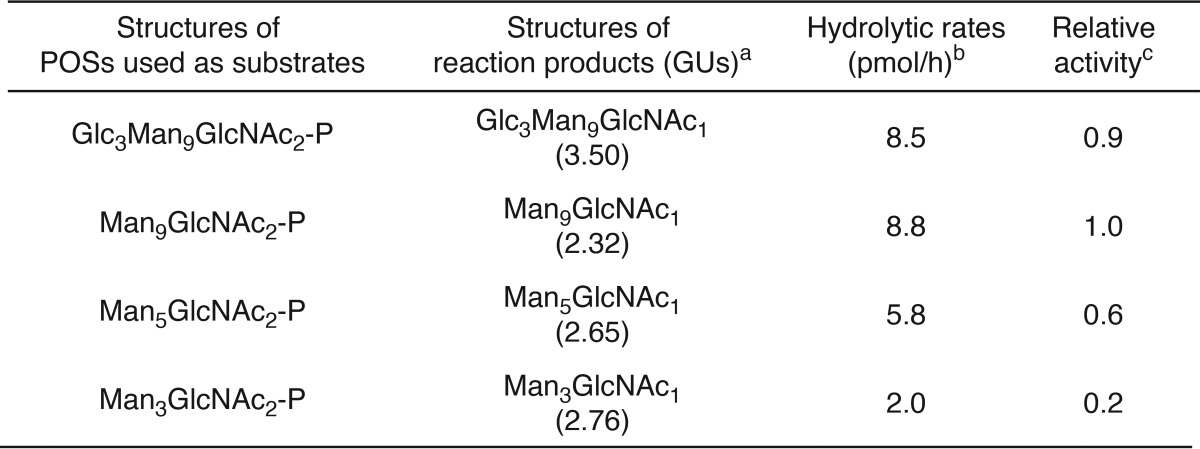
In vitro ENGase assays using the POS preparations and the cytosolic fraction obtained from Ngly1−/− Engase−/− MEFs overexpressing ENGase showed that the order of hydrolytic rates of the POSs was Glc3Man9GlcNAc2-P ≈ Man9GlcNAc2-p > Man5GlcNAc2-P ≫ Man3GlcNAc2-P (Fig. 4C and Table 3). Dual-gradient reversed-phase HPLC analysis (25) of the reaction mixtures confirmed the generation of Gn1-type FNGs, i.e. Glc3Man9GlcNAc1, Man9GlcNAc1, Man5GlcNAc1, and Man3GlcNAc1 (Fig. 4D and Table 3). When the cytosolic fraction obtained from non-transfected Ngly1−/− Engase−/− MEFs was used, no hydrolysis of Man5GlcNAc2-P was detected (Fig. 4E), verifying that the cytosolic fraction of the non-transfected Ngly1−/− Engase−/− MEFs is devoid of degradation activity for POSs. Therefore, our assay system using the cytosol fraction of the ENGase-overexpressing Ngly1−/− Engase−/− MEFs is a reliable one, especially when appropriate glycosidase inhibitors (swainsonine, kifunensine, and castanospermine) are included in the reaction mixture to prevent the glycan trimming of reaction products during the incubation (see “Experimental Procedures” for the detailed conditions). All these results indicate that, although ENGase has a broad substrate specificity for POSs, the presence of α1,2-linked mannose residues at the A arm (Fig. 4F), as well as a branching structure at the B and C arms, facilitates the action of this enzyme.
TABLE 3.
In vitro ENGase activity assay using POSs with various glycan structures
aGlycan structures were determined by the elution positions of both size-fractionation and dual-gradient reversed-phase HPLC and comparison with authentic standard glycans (25). To verify the position of Gn1-type FNGs under the HPLC conditions used, the reaction products were analyzed in parallel with POSs that were digested with Endo-H (for Glc3Man9GlcNAc2-P and Man9GlcNAc2-P) or Endo-M (Man5GlcNAc2-P and Man3GlcNAc2-P).
bThe amounts of reaction products were estimated from peak area of PA-Glc6 in size-fractionation HPLC. The hydrolytic rates were estimated based on the amounts of Gn1-type FNGs at 1 h (Glc3Man9GlcNAc1, Man9GlcNAc1, and Man5GlcNAc1) and 2 h (Man3GlcNAc1) after incubation.
cHydrolytic rates of Man9GlcNAc2-P were set to 1.0.
Discussion
A putative DLO-degrading enzyme has been implicated in the production of POSs (11), whereas the precise cleavage site of DLOs by this enzyme has not been unequivocally determined. Our LC-MS analyses of intact POSs showed that one phosphate group is present (Fig. 2) at the reducing end of POSs (Fig. 3A) (10, 13), providing conclusive evidence that the putative DLO-degrading enzyme is a DLO-pyrophosphatase that splits the pyrophosphate bond of DLOs producing mono-phosphorylated POSs. Although mono-phosphorylated POSs have been identified by MS in bacteria (30) or in milk oligosaccharides (31), to the best of our knowledge this is the first example of the use of MS for the identification of DLO-derived POSs of mammalian origin.
In vitro DLO-pyrophosphatase assays revealed that the enzyme exhibits broad substrate specificities for glycan structures (Fig. 4A, right panels), and even the fully assembled DLO (Glc3Man9GlcNAc2-PP-Dol) can be a substrate in vitro. This result raises the issue of why only limited POS structures, i.e. Man0–7GlcNAc2-P, were detected in MEFs (Fig. 3, B and C) (10, 13). These POSs are produced from both the cytosolic and luminal species of DLOs, which negligibly accumulate in MEFs (10). By contrast, Glc3Man9GlcNAc2-PP-Dol is the predominant DLO at the steady state in MEFs (10), whereas little Glc3Man9GlcNAc2-P is detected even in Engase−/− MEFs (Fig. 3, B and C). Our findings thus imply that the accessibility of the DLO-pyrophosphatase to the fully assembled DLO is somehow limited in vivo via unknown mechanisms. Although it is also possible that the glycan trimming of large POSs by glycosidases may generate small POSs from large POSs, such as Glc3Man9GlcNAc2-P, previous studies have shown that glycan structures of POSs are barely altered by treatment with various glycosidase inhibitors, such as castanospermine (13), kifunensine (13), and swainsonine (10, 13), suggesting that such a possibility is unlikely. Irrespective of mechanisms involved, the identification of the membrane topology of the catalytic site(s) of the DLO-pyrophosphatase will provide deeper insights into important aspects of the regulation of the action of DLO-pyrophosphatase in vivo.
Our data showed that large POSs, i.e. Man4–7GlcNAc2-P, carrying the Manα1,2Manα1,3(Manα1,6)Manβ1,4GlcNAcβ1,4GlcNAc structure tend to accumulate in Engase−/− MEFs (Fig. 3, B and C). The accumulation of these POSs in Engase−/− MEFs is consistent with the finding that, in vitro, ENGase efficiently hydrolyzes POSs bearing α1,2-linked mannose residues located at the A arm (Table 3). In contrast, ENGase hydrolyzes Man3GlcNAc2-P at much slower rates (Table 3). The substrate specificity of ENGase is consistent with the previous report showing that glycans that contain a Manα1,2Manα1,3Manβ1,4GlcNAcβ1,4GlcNAc structure serve as good substrates of ENGase (32). These findings can explain, at least in part, the reason why Man0–3GlcNAc2-P accumulates to a lesser extent when compared with the larger POSs in Engase−/− MEFs. The glycan processing of POSs by ENGase is of biological relevance, because the putative FNG transporter that resides in the lysosomal membrane does not directly transport POSs into lysosomes (33). Therefore, the ENGase-mediated processing of POSs plays a crucial role in initiating their cytosolic clearance.
We noted that the levels of GlcNAc2-P were significantly reduced in Engase−/− MEFs, raising the possibility that the biosynthesis of GlcNAc2-PP-Dol is also compromised in glucose-deprived Engase−/− MEFs. The action of ENGase produces free GlcNAc and GlcNAc-1-P from Gn2-type FNGs and POSs, respectively (Fig. 1). These sugars may be recycled for the biosynthesis of UDP-GlcNAc, which is an essential sugar donor substrate for the assembly of GlcNAc2-PP-Dol. It is also possible that the catabolism of GlcNAc2-P is enhanced in the absence of ENGase. In this regard, it remains to be clarified how small POSs, which are poor substrates of ENGase, are catabolized. A putative POS-phosphatase activity was reported in the microsome fraction from human liver (14), but the molecular nature of this phosphatase is currently unknown. Moreover, we previously found that the induction of bulk autophagy by amino acid starvation reduces the levels of FNGs in the cytosol (34). Further studies will be needed to elucidate the molecular mechanism involved in the catabolism of small POSs.
In summary, the above findings demonstrate that POSs are mono-phosphorylated, thereby implicating the action of a DLO-pyrophosphatase in their production. Moreover, we identified the cytosolic ENGase as a POS-catabolizing enzyme that preferentially hydrolyzes POSs carrying the Manα1,2Manα1,3(Manα1,6)Manβ1,4GlcNAcβ1,4GlcNAc structure. This study provides mechanistic insights into the generation and the catabolism of POSs in mammalian cells.
Author Contributions
Y. H. and T. S. designed the study. Y. H. prepared the samples used for Fig. 2 and also performed the experiments for Figs. 3 and 4 and Tables 2 and 3. C. H. performed the experiments for Figs. 4, C–E, and Table 3. S. Y. and N. D. performed the analyses shown in Fig. 2. Y. H. and T. S. analyzed all the data and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank our laboratory members for fruitful discussions.
This work was supported in part by Grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan 24770134 and 26650040 (to Y. H.) and 25291030 (to T. S.). The authors declare that they have no conflicts of interest with the contents of this article.
- DLO
- dolichol-linked oligosaccharide
- POS
- phosphorylated oligosaccharide
- ENGase
- endo-β-N-acetylglucosaminidase
- MEF
- mouse embryonic fibroblast
- ER
- endoplasmic reticulum
- FNG
- free N-glycan
- PA
- 2-aminopyridine
- CIP
- calf intestine alkaline phosphatase
- Endo
- endoglycosidase.
References
- 1.Kelleher D. J., and Gilmore R. (2006) An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16, 47R–62R [DOI] [PubMed] [Google Scholar]
- 2.Harada Y., Hirayama H., and Suzuki T. (2015) Generation and degradation of free asparagine-linked glycans. Cell. Mol. Life Sci. 72, 2509–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shrimal S., Cherepanova N. A., and Gilmore R. (2015) Cotranslational and posttranslocational N-glycosylation of proteins in the endoplasmic reticulum. Semin. Cell Dev. Biol. 41, 71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doerrler W. T., and Lehrman M. A. (1999) Regulation of the dolichol pathway in human fibroblasts by the endoplasmic reticulum unfolded protein response. Proc. Natl. Acad. Sci. U.S.A. 96, 13050–13055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumann H., and Jahreis G. P. (1983) Glucose starvation leads in rat hepatoma cells to partially N-glycosylated glycoproteins including α1-acid glycoproteins. Identification by endoglycolytic digestions in polyacrylamide gels. J. Biol. Chem. 258, 3942–3949 [PubMed] [Google Scholar]
- 6.Turco S. J., and Pickard J. L. (1982) Altered G-protein glycosylation in vesicular stomatitis virus-infected glucose-deprived baby hamster kidney cells. J. Biol. Chem. 257, 8674–8679 [PubMed] [Google Scholar]
- 7.Rearick J. I., Chapman A., and Kornfeld S. (1981) Glucose starvation alters lipid-linked oligosaccharide biosynthesis in Chinese hamster ovary cells. J. Biol. Chem. 256, 6255–6261 [PubMed] [Google Scholar]
- 8.Gershman H., and Robbins P. W. (1981) Transitory effects of glucose starvation on the synthesis of dolichol-linked oligosaccharides in mammalian cells. J. Biol. Chem. 256, 7774–7780 [PubMed] [Google Scholar]
- 9.Gao N., Shang J., and Lehrman M. A. (2005) Analysis of glycosylation in CDG-Ia fibroblasts by fluorophore-assisted carbohydrate electrophoresis: implications for extracellular glucose and intracellular mannose 6-phosphate. J. Biol. Chem. 280, 17901–17909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harada Y., Nakajima K., Masahara-Negishi Y., Freeze H. H., Angata T., Taniguchi N., and Suzuki T. (2013) Metabolically programmed quality control system for dolichol-linked oligosaccharides. Proc. Natl. Acad. Sci. U.S.A. 110, 19366–19371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cacan R., Hoflack B., and Verbert A. (1980) Fate of oligosaccharide-lipid intermediates synthesized by resting rat-spleen lymphocytes. Eur. J. Biochem. 106, 473–479 [DOI] [PubMed] [Google Scholar]
- 12.Belard M., Cacan R., and Verbert A. (1988) Characterization of an oligosaccharide-pyrophosphodolichol pyrophosphatase activity in yeast. Biochem. J. 255, 235–242 [PMC free article] [PubMed] [Google Scholar]
- 13.Peric D., Durrant-Arico C., Delenda C., Dupré T., De Lonlay P., de Baulny H. O., Pelatan C., Bader-Meunier B., Danos O., Chantret I., and Moore S. E. (2010) The compartmentalisation of phosphorylated free oligosaccharides in cells from a CDG Ig patient reveals a novel ER-to-cytosol translocation process. PLoS ONE 5, e11675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vleugels W., Duvet S., Peanne R., Mir A. M., Cacan R., Michalski J. C., Matthijs G., and Foulquier F. (2011) Identification of phosphorylated oligosaccharides in cells of patients with a congenital disorders of glycosylation (CDG-I). Biochimie 93, 823–833 [DOI] [PubMed] [Google Scholar]
- 15.Harada Y., Buser R., Ngwa E. M., Hirayama H., Aebi M., and Suzuki T. (2013) Eukaryotic oligosaccharyltransferase generates free oligosaccharides during N-glycosylation. J. Biol. Chem. 288, 32673–32684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harada Y., Masahara-Negishi Y., and Suzuki T. (2015) Cytosolic-free oligosaccharides are predominantly generated by the degradation of dolichol-linked oligosaccharides in mammalian cells. Glycobiology 25, 1196–1205 [DOI] [PubMed] [Google Scholar]
- 17.Suzuki T. (2015) The cytoplasmic peptide:N-glycanase (Ngly1)–basic science encounters a human genetic disorder. J. Biochem. 157, 23–34 [DOI] [PubMed] [Google Scholar]
- 18.Hirayama H., Hosomi A., and Suzuki T. (2015) Physiological and molecular functions of the cytosolic peptide:N-glycanase. Semin. Cell Dev. Biol. 41, 110–120 [DOI] [PubMed] [Google Scholar]
- 19.Suzuki T., Yano K., Sugimoto S., Kitajima K., Lennarz W. J., Inoue S., Inoue Y., and Emori Y. (2002) Endo-β-N-acetylglucosaminidase, an enzyme involved in processing of free oligosaccharides in the cytosol. Proc. Natl. Acad. Sci. U.S.A. 99, 9691–9696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato T., Fujita K., Takeuchi M., Kobayashi K., Natsuka S., Ikura K., Kumagai H., and Yamamoto K. (2002) Identification of an endo-β-N-acetylglucosaminidase gene in Caenorhabditis elegans and its expression in Escherichia coli. Glycobiology 12, 581–587 [DOI] [PubMed] [Google Scholar]
- 21.Chantret I., Fasseu M., Zaoui K., Le Bizec C., Sadou Yayé H., Dupré T., and Moore S. E. (2010) Identification of roles for peptide: N-glycanase and endo-β-N-acetylglucosaminidase (Engase1p) during protein N-glycosylation in human HepG2 cells. PLoS ONE 5, e11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki T., Hara I., Nakano M., Shigeta M., Nakagawa T., Kondo A., Funakoshi Y., and Taniguchi N. (2006) Man2C1, an α-mannosidase, is involved in the trimming of free oligosaccharides in the cytosol. Biochem. J. 400, 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saint-Pol A., Bauvy C., Codogno P., and Moore S. E. (1997) Transfer of free polymannose-type oligosaccharides from the cytosol to lysosomes in cultured human hepatocellular carcinoma HepG2 cells. J. Cell Biol. 136, 45–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang C., Harada Y., Hosomi A., Masahara-Negishi Y., Seino J., Fujihira H., Funakoshi Y., Suzuki T., Dohmae N., and Suzuki T. (2015) Endo-β-N-acetylglucosaminidase forms N-GlcNAc protein aggregates during ER-associated degradation in Ngly1-defective cells. Proc. Natl. Acad. Sci. U.S.A. 112, 1398–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki T., Matsuo I., Totani K., Funayama S., Seino J., Taniguchi N., Ito Y., and Hase S. (2008) Dual-gradient high-performance liquid chromatography for identification of cytosolic high-mannose-type free glycans. Anal. Biochem. 381, 224–232 [DOI] [PubMed] [Google Scholar]
- 26.Rose M. D., Winston F., and Hieter P. (1990) Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27.Haga Y., Totani K., Ito Y., and Suzuki T. (2009) Establishment of a real-time analytical method for free oligosaccharide transport from the ER to the cytosol. Glycobiology 19, 987–994 [DOI] [PubMed] [Google Scholar]
- 28.Hirayama H., Seino J., Kitajima T., Jigami Y., and Suzuki T. (2010) Free oligosaccharides to monitor glycoprotein endoplasmic reticulum-associated degradation in Saccharomyces cerevisiae. J. Biol. Chem. 285, 12390–12404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hase S., Ikenaka T., and Matsushima Y. (1979) Analyses of oligosaccharides by tagging the reducing end with a fluorescent compound. I. Application to glycoproteins. J. Biochem. 85, 989–994 [DOI] [PubMed] [Google Scholar]
- 30.Dwivedi R., Nothaft H., Reiz B., Whittal R. M., and Szymanski C. M. (2013) Generation of free oligosaccharides from bacterial protein N-linked glycosylation systems. Biopolymers 99, 772–783 [DOI] [PubMed] [Google Scholar]
- 31.Albrecht S., Lane J. A., Mariño K., Al Busadah K. A., Carrington S. D., Hickey R. M., and Rudd P. M. (2014) A comparative study of free oligosaccharides in the milk of domestic animals. Br. J. Nutr. 111, 1313–1328 [DOI] [PubMed] [Google Scholar]
- 32.Kato T., Hatanaka K., Mega T., and Hase S. (1997) Purification and characterization of endo-β-N-acetylglucosaminidase from hen oviduct. J. Biochem. 122, 1167–1173 [DOI] [PubMed] [Google Scholar]
- 33.Saint-Pol A., Codogno P., and Moore S. E. (1999) Cytosol-to-lysosome transport of free polymannose-type oligosaccharides. Kinetic and specificity studies using rat liver lysosomes. J. Biol. Chem. 274, 13547–13555 [DOI] [PubMed] [Google Scholar]
- 34.Seino J., Wang L., Harada Y., Huang C., Ishii K., Mizushima N., and Suzuki T. (2013) Basal autophagy is required for the efficient catabolism of sialyloligosaccharides. J. Biol. Chem. 288, 26898–26907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki T., and Harada Y. (2014) Non-lysosomal degradation pathway for N-linked glycans and dolichol-linked oligosaccharides. Biochem. Biophys. Res. Commun. 453, 213–219 [DOI] [PubMed] [Google Scholar]
- 36.Reiss G., te Heesen S., Zimmerman J., Robbins P. W., and Aebi M. (1996) Isolation of the ALG6 locus of Saccharomyces cerevisiae required for glucosylation in the N-linked glycosylation pathway. Glycobiology 6, 493–498 [DOI] [PubMed] [Google Scholar]
- 37.Aebi M., Gassenhuber J., Domdey H., and te Heesen S. (1996) Cloning and characterization of the ALG3 gene of Saccharomyces cerevisiae. Glycobiology 6, 439–444 [DOI] [PubMed] [Google Scholar]
- 38.Cipollo J. F., Trimble R. B., Chi J. H., Yan Q., and Dean N. (2001) The yeast ALG11 gene specifies addition of the terminal α,2-Man to the Man5GlcNAc2-PP-dolichol N-glycosylation intermediate formed on the cytosolic side of the endoplasmic reticulum. J. Biol. Chem. 276, 21828–21840 [DOI] [PubMed] [Google Scholar]
- 39.Helenius J., Ng D. T., Marolda C. L., Walter P., Valvano M. A., and Aebi M. (2002) Translocation of lipid-linked oligosaccharides across the ER membrane requires Rft1 protein. Nature 415, 447–450 [DOI] [PubMed] [Google Scholar]
- 40.Absmanner B., Schmeiser V., Kämpf M., and Lehle L. (2010) Biochemical characterization, membrane association and identification of amino acids essential for the function of Alg11 from Saccharomyces cerevisiae, an α1,2-mannosyltransferase catalysing two sequential glycosylation steps in the formation of the lipid-linked core oligosaccharide. Biochem. J. 426, 205–217 [DOI] [PubMed] [Google Scholar]
- 41.Huffaker T. C., and Robbins P. W. (1983) Yeast mutants deficient in protein glycosylation. Proc. Natl. Acad. Sci. U.S.A. 80, 7466–7470 [DOI] [PMC free article] [PubMed] [Google Scholar]