Two transcription factors positively regulate Fe homeostasis by activating the transcription of the Ib subgroup bHLH genes.
Abstract
The regulation of iron (Fe) homeostasis is critical for plant survival. Although the systems responsible for the reduction, uptake, and translocation of Fe have been described, the molecular mechanism by which plants sense Fe status and coordinate the expression of Fe deficiency-responsive genes is largely unknown. Here, we report that two basic helix-loop-helix-type transcription factors, bHLH34 and bHLH104, positively regulate Fe homeostasis in Arabidopsis (Arabidopsis thaliana). Loss of function of bHLH34 and bHLH104 causes disruption of the Fe deficiency response and the reduction of Fe content, whereas overexpression plants constitutively promote the expression of Fe deficiency-responsive genes and Fe accumulation. Further analysis indicates that bHLH34 and bHLH104 directly activate the transcription of the Ib subgroup bHLH genes, bHLH38/39/100/101. Moreover, overexpression of bHLH101 partially rescues the Fe deficiency phenotypes of bhlh34bhlh104 double mutants. Further investigation suggests that bHLH34, bHLH104, and bHLH105 (IAA-LEUCINE RESISTANT3) function as homodimers or heterodimers to nonredundantly regulate Fe homeostasis. This work reveals that plants have evolved complex molecular mechanisms to regulate Fe deficiency response genes to adapt to Fe deficiency conditions.
Iron (Fe) is of particular importance as a cofactor for a wide variety of proteins in living organisms. In humans, Fe deficiency is one of the major reasons for anemia. A plant diet is a major resource for humans; thus, it is important to clarify the mechanisms of Fe homeostasis in plants. Fe is an indispensable micronutrient for plant growth and development and is involved in many cellular functions, including chlorophyll biosynthesis, photosynthesis, and respiration (Hänsch and Mendel, 2009). Therefore, a decrease in Fe uptake directly affects crop yield and crop quality. Plants take up Fe from the soil, but Fe availability is limited, as its major form is as insoluble ferric hydroxides, especially in calcareous soils, which make up one-third of the world’s cultivated areas.
To cope with low-Fe environments, higher plants have evolved two major strategies for Fe acquisition. Nongraminaceous plants employ a reduction strategy (strategy I) that involves three stages in low-Fe conditions. The acidification of soil by protons extruded by H+-ATPases and phenolic compounds makes Fe(III) more soluble (Santi and Schmidt, 2009; Kobayashi and Nishizawa, 2012). The ferric chelate reductase activity of FERRIC REDUCTION OXIDASE2 (FRO2) is induced by low Fe; this enzyme reduces Fe(III) to Fe(II) (Robinson et al., 1999). The Fe(II) is then transported into roots by the major Fe transporter of plant roots, IRON-REGULATED TRANSPORTER1 (IRT1; Eide et al., 1996; Henriques et al., 2002; Varotto et al., 2002; Vert et al., 2002). In contrast, graminaceous plants use a chelation-based strategy (strategy II; Walker and Connolly, 2008; Morrissey and Guerinot, 2009). These plants release mugineic acid family phytosiderophores, which have high affinity for Fe(III) and efficiently bind Fe(III) in the rhizosphere. The resulting chelate complexes are then transported into plant roots via a specific transport system (Mori, 1999).
The transcriptional regulation of genes involved in Fe uptake under Fe-deficient conditions is intensively controlled by plants. In the past decade, several basic helix-loop-helix (bHLH) proteins have been identified as regulators of the Fe deficiency response. The bHLH protein in tomato (Solanum lycopersicum), FER, was the first characterized to control Fe deficiency responses, and its mutant failed to activate Fe acquisition strategy I (Ling et al., 2002). FE-DEFICIENCY INDUCED TRANSCRIPTION FACTOR (FIT), a homolog of FER in Arabidopsis (Arabidopsis thaliana), is required for the proper regulation of ferric chelate reductase activity and Fe transport into the plant root, and its mutation was lethal when plants were grown in normal soils (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005). However, overexpression of FIT is not sufficient to constitutively induce the expression of its target genes, such as IRT1 and FRO2, in normal growth conditions. In fact, FIT is dually regulated by Fe starvation. At the transcriptional level, FIT is induced by Fe deficiency (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005). At the posttranscriptional level, FIT is actively destabilized and degraded by the 26S proteasome during Fe limitation (Meiser et al., 2011; Sivitz et al., 2011). In addition, FIT can form a heterodimer with members of the Ib subgroup of bHLH proteins (bHLH38/39/100/101) to constitutively activate the transcription of IRT1 and FRO2 (Yuan et al., 2008; Wang et al., 2013). Both FIT and bHLH38/39/100/101 function as positive regulators. In contrast, POPEYE (PYE) was identified as playing a negative role in the Fe deficiency response in Arabidopsis (Long et al., 2010). PYE transcript levels were elevated when plants were subjected to low-Fe conditions. Chip-on-chip experiments revealed that PYE directly targets several genes involved in metal homeostasis, such as NICOTIANAMINE SYNTHASE4 (NAS4), FRO3, and ZINC-INDUCED FACILITATOR1 (ZIF1). In a pye1 mutant, the expression of these target genes is significantly up-regulated in Fe-deficient conditions, suggesting that PYE functions as a negative regulator.
It is unclear how plants perceive Fe status and transmit signals to downstream pathways. In mammals, FBXL5 functions as an Fe sensor and contains a hemerythrin domain (HHE) for Fe binding and an F-box domain for the ubiquitination and degradation of IRP2. The latter governs cellular Fe homeostasis by regulation of the translation and stability of mRNAs involved in Fe homeostasis (Salahudeen et al., 2009; Vashisht et al., 2009). Interestingly, two different groups (Kobayashi et al., 2013; Selote et al., 2015) confirmed that Arabidopsis BRUTUS (BTS) and its rice (Oryza sativa) orthologs, HAEMERYTHRIN MOTIF-CONTAINING REALLY INTERESTING NEW GENE (RING) AND ZINC-FINGER PROTEIN1 (HRZ1) and HRZ2, possess HHE domains for binding Fe. Although they lack an F-box domain similar to that in FBXL5, BTS and HRZ1/2 have RING domains that can mediate the ubiquitination reaction. Functional analysis revealed that BTS and HRZ1/2 negatively regulate the Fe deficiency response in plants. Given that BTS and HRZ1/2 are structurally and functionally similar to FBXL5, they are thought to be potential Fe sensors in plants. Although no substrates for HRZ1/2 have been found, it is established that BTS interacts with three PYE-like proteins (bHLH104, bHLH105/ILR3, and bHLH115) and facilitates the 26S proteasome-mediated degradation of bHLH105/IAA-LEUCINE RESISTANT3 (ILR3) and bHLH115 in the absence of Fe in vitro (Selote et al., 2015). Recently, Zhang et al. (2015) revealed that bHLH104 and bHLH105 positively regulate Fe homeostasis by directly activating the transcription of bHLH38/39/100/101 and PYE. Further investigation is required to explore the functions of the other PYE-like proteins.
Here, through screening regulators upstream of bHLH101, we identified the bHLH transcription factor bHLH34, a homolog of bHLH104 in Arabidopsis. Mutational analysis suggests that bHLH34 and bHLH104 positively regulate the Fe deficiency response. Further analysis reveals that both bHLH34 and bHLH104 directly activate the transcription of bHLH38/39/100/101. In agreement with this, constitutive expression of bHLH101 partially complements the Fe deficiency symptoms of bhlh34bhlh104 double mutants. Protein interaction assays indicate that both heterodimers and homodimers occur between bHLH34, bHLH104, and bHLH105. These data reveal that bHLH34 and bHLH104 play major roles in regulating Fe homeostasis by activating the transcription of bHLH38/39/100/101 under Fe deficiency conditions.
RESULTS
Identification of bHLH34
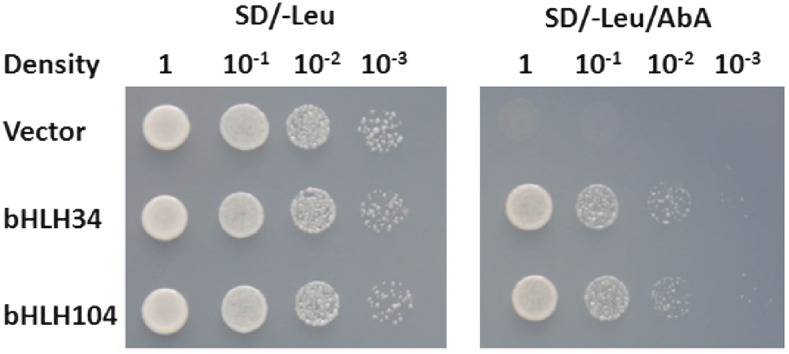
bHLH38/39/100/101 have been characterized as key transcription factors regulating the Fe-deficient response in Arabidopsis, and their transcript levels are also up-regulated by Fe deficiency. To identify transcription factors functioning upstream of bHLH38/39/100/101, yeast one-hybrid screening was performed using the promoter of bHLH101 as bait. From the screening, we obtained nine positive colonies, three of which encoded a bHLH protein, bHLH34. As a close homolog of bHLH34 (Supplemental Fig. S1), bHLH104 has been characterized to bind the promoter of bHLH101 (Zhang et al., 2015). Further experiments confirmed that both bHLH34 and bHLH104, but not the other candidates, can activate the promoter of bHLH101 (Fig. 1), which suggests that both bHLH34 and bHLH104 regulate the bHLH101 gene. Given that other Fe homeostasis-associated transcription factors, such as bHLH38/39/100/101, FIT, PYE, and MYB10/72, are induced by Fe deficiency, we wanted to know whether bHLH34 and bHLH104 are also responsive to Fe deficiency. Transcript abundance analysis indicated that neither bHLH34 nor bHLH104 is affected by Fe deficiency (Supplemental Fig. S2).
Figure 1.
Identification of bHLH34 and bHLH104 by yeast one-hybrid assay. The promoter of bHLH101 was used as bait and bHLH34/104 as prey. The representative growth status of yeast cells is shown on synthetic dextrose medium agar plates without Leu (SD/-Leu) with or without aureobasidin A (AbA) from triplicate independent trails.
bhlh34, bhlh104, and bhlh34bhlh104 Mutant Plants Display Fe Deficiency Symptoms
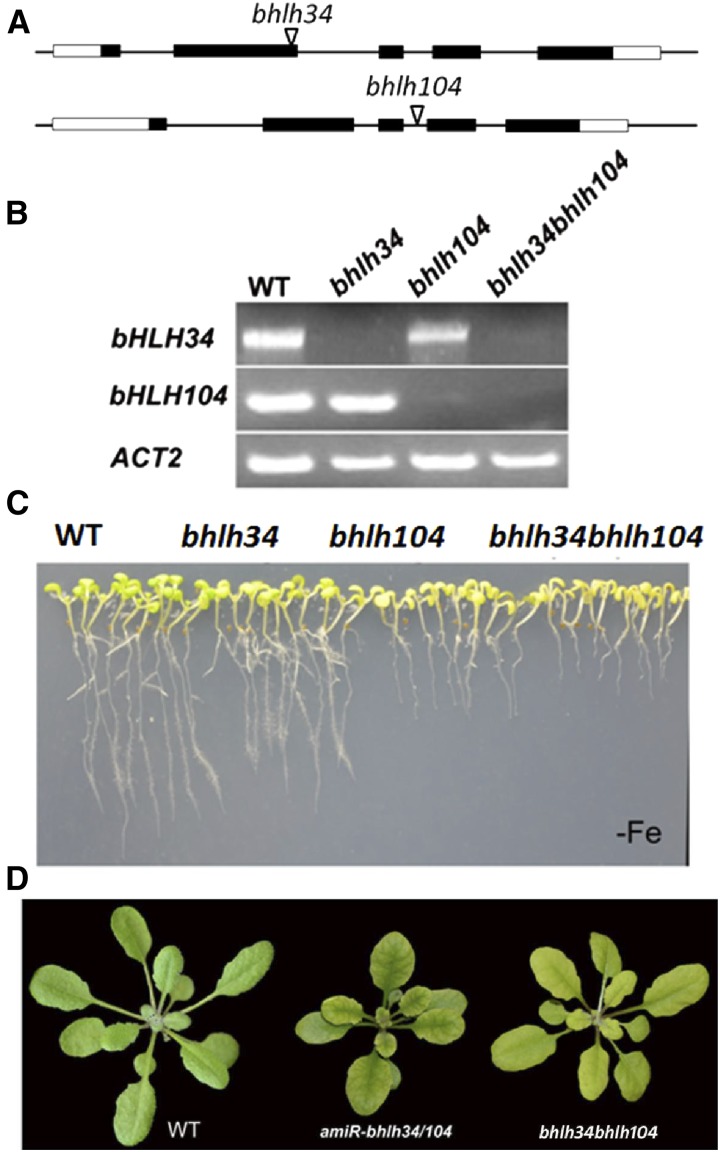
To investigate the functions of bHLH34 and bHLH104, two transfer DNA (T-DNA) insertion alleles for the bHLH34 and bHLH104 genes were obtained from The Arabidopsis Information Resource (TAIR). bhlh34 contains a T-DNA insertion in the second exon and bhlh104 has a T-DNA insertion in the third intron (Fig. 2A). The homozygous lines of bhlh34 and bhlh104 were isolated by PCR, and the positions of the insertions were confirmed. The loss of full-length transcripts in the T-DNA lines were determined by reverse transcription-PCR (Fig. 2B), indicating that both T-DNA lines are knockout mutants. Considering their putative functional redundancy, we constructed bhlh34bhlh104 double mutants by crossing two single knockout mutants. When grown on Fe-sufficient medium, neither single mutants nor double mutants showed any obvious phenotypic differences compared with wild-type plants (Supplemental Fig. S3). However, when they were grown on Fe-free medium, all mutants displayed Fe-deficient symptoms, and the double mutants had the most severe phenotypes (Fig. 2C).
Figure 2.
Characterization of various mutant plants. A, T-DNA insertion position in the corresponding gene. Black boxes indicate the coding sequence. White boxes indicate untranslated regions. Triangles indicate the position of T-DNA. B, The corresponding full-length complementary DNAs (cDNAs) were amplified by reverse transcription-PCR to confirm the mutants. C, Ten-day-old seedlings germinated directly on Fe-deficient (–Fe) medium. D, Phenotypes of amiR-bhlh34/104 and bhlh34bhlh104 plants in soils. Four-week-old plants grown in normal soil are shown. WT, Wild type.
To further confirm the functions of bHLH34 and bHLH104, we designed an artificial microRNA, amiR-bhlh34/104, which is predicted to target both bHLH34 and bHLH104 (Supplemental Fig. S4A), and generated amiR-bhlh34/104 transgenic plants. Quantitative reverse transcription-PCR analysis demonstrated that both bHLH34 and bHLH104 were significantly down-regulated in amiR-bhlh34/104 plants (Supplemental Fig. S4B). As expected, the amiR-bhlh34/104 transgenic plants grown in soils showed interveinal chlorosis in leaves, which is very similar to that of bhlh34bhlh104 double mutants (Fig. 2D). Further phenotype evaluation revealed that the root growth of amiR-bhlh34/104 seedlings was strongly inhibited on Fe-deficient medium, whereas no visible difference was observed between amiR-bhlh34/104 and wild-type plants on normal medium (Supplemental Fig. S4, C and D). These results demonstrate that the amiR-bhlh34/104 plants phenocopy the bhlh34bhlh104 mutants.
Impaired Fe Deficiency Response by Loss of Function of bHLH34 and bHLH104
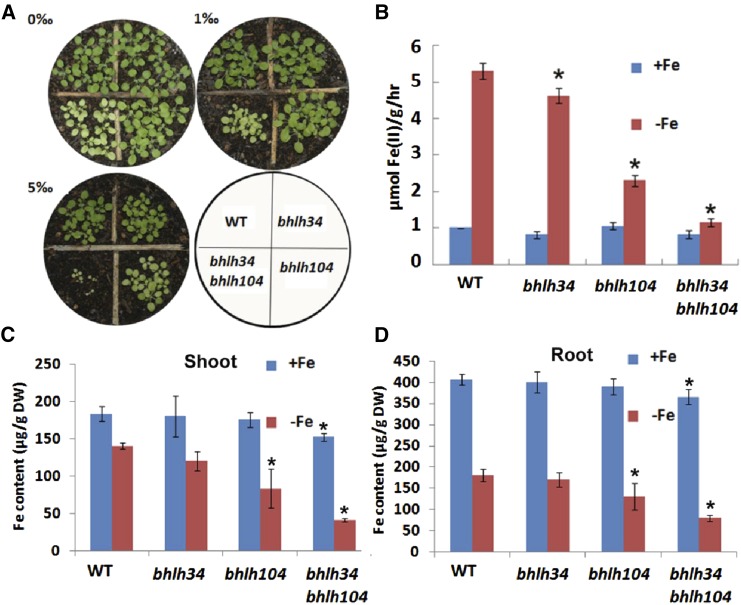
We further determined the Fe-associated phenotypes of plants grown in soils containing various concentrations of calcium oxide (Fig. 3A), which was used to produce alkaline soil in which the Fe availability was limited because of decreased Fe solubility. In normal soil, no visible phenotypic difference was observed between the bhlh34, bhlh104, and wild-type plants, whereas the bhlh34bhlh104 double mutants had small stature and chlorotic leaves. In soil supplemented with 1‰ (w/w) calcium oxide, the bhlh104 mutants showed slight chlorosis. In contrast, 5‰ (w/w) calcium oxide caused leaf chlorosis in all three types of mutants and seedling lethality of the bhlh34bhlh104 double mutants. These data suggest that the loss of function of bHLH34 and bHLH104 inhibits the Fe deficiency response of the bhlh34bhlh104 plants.
Figure 3.
Fe deficiency response of various mutants. A, Three-week-old plants germinated in soil with various concentrations of calcium oxide. B, Iron reductase activity of plants germinated and grown on Fe-sufficient (+Fe) medium for 10 d and then shifted to Fe-deficient (–Fe) medium for 3 d. The ferrozine assay was performed, in triplicate, on 10 pooled plant roots. Significant differences from the wild type (WT) are indicated by asterisks (P < 0.05). C and D, Fe content of shoots and roots. Seedlings were germinated and grown on +Fe medium for 11 d and then shifted to +Fe or –Fe medium for 3 d. Significant differences from the wild type are indicated by asterisks (P < 0.05). DW, Dry weight.
Fe is indispensable for the synthesis of chlorophyll, and Fe deficiency often causes leaf chlorosis with decreased levels of chlorophyll in plants (Terry, 1980). The significant chlorosis of the bhlh34bhlh104 double mutant plants implied a decline in chlorophyll content. We measured chlorophyll levels in plants grown on Fe-sufficient and Fe-deficient media. There were no significant differences under Fe-sufficient conditions. However, under Fe-deficient conditions, the mutants showed significantly reduced chlorophyll levels, and the bhlh34bhlh104 double mutants had the lowest chlorophyll levels (Supplemental Fig. S5).
Fe reductase activity is a typical indicator of Fe deficiency. We analyzed Fe reductase activity using the ferrozine assay (Yi and Guerinot, 1996). In Fe-sufficient conditions, no significant difference in Fe reductase activity was observed between wild-type and mutant plants. In response to Fe deficiency, both wild-type and mutant plants increased their Fe reductase activity. However, the Fe reductase activity of the mutant plants was significantly lower than that of wild-type plants, and the bhlh34bhlh104 double mutants had the lowest Fe reductase activity (Fig. 3B).
To determine whether the loss of function of bHLH34 and bHLH104 causes altered Fe accumulation, the Fe content of wild-type and mutant 2-week-old seedlings was measured. Eleven-day-old seedlings grown under Fe-sufficient conditions were transferred to Fe-sufficient or Fe-deficient medium for another 3 d. Roots and shoots were harvested separately and used for Fe content analysis. The shoots of bhlh34bhlh104 plants had 17% less Fe than the shoots of wild-type plants under Fe-sufficient conditions and 71% less Fe under Fe-deficient conditions (Fig. 3C). Similar results also were observed in roots. The roots of bhlh34bhlh104 plants contained 10% less Fe than the roots of wild-type plants under Fe-sufficient conditions and 56% less Fe under Fe-deficient conditions (Fig. 3D). These data suggest that bHLH34 and bHLH104 are required to maintain Fe homeostasis in both Fe-sufficient and Fe-deficient conditions.
Fe Deficiency-Responsive Genes Are Down-Regulated in the bhlh34, bhlh104, and bhlh34bhlh104 Mutants
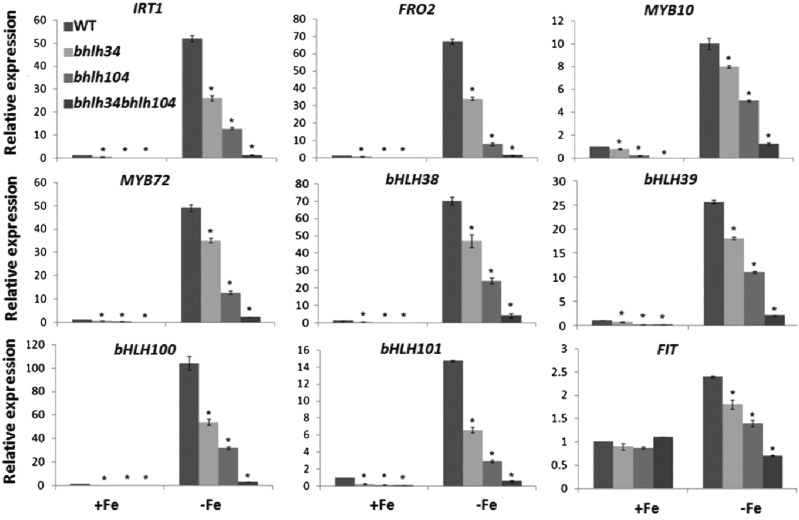
To adapt to Fe-deficient conditions, plants often elevate the expression of Fe uptake-associated genes. Fe deficiency-induced genes FRO2 and IRT1 encode two key root membrane proteins for Fe homeostasis; the former converts Fe(III) to Fe(II), and the latter transports ferrous Fe from soil to root (Robinson et al., 1999; Vert et al., 2002). Given the low Fe levels in the bhlh34bhlh104 mutants, we wanted to know whether the expression of these two Fe uptake-associated genes is altered in the mutants. In response to Fe deficiency, both genes were induced in the roots of wild-type and mutant plants. However, their expression levels were low in the mutants compared with the wild type under both Fe-sufficient and Fe-deficient conditions (Fig. 4). Moreover, their expression levels in bhlh34bhlh104 under Fe-deficient conditions were close to that in the wild type in Fe-sufficient conditions. These alterations suggest that there is decreased Fe uptake in the mutant plants, which is in agreement with the reduction in both Fe reductase activity and Fe accumulation. These data also demonstrate that bHLH34 and bHLH104 directly or indirectly activate the expression of FRO2 and IRT1.
Figure 4.
Expression of Fe deficiency-responsive genes in various mutants. Wild-type (WT) and mutant plants were grown on Fe-sufficient (+Fe) medium for 10 d and then transferred to +Fe or Fe-deficient (–Fe) medium for 3 d. RNA was prepared from root tissues. The data represent means ± sd of three technical repeats from one representative experiment. Significant differences from the corresponding wild type are indicated by asterisks (P < 0.05). DW, Dry weight.
Two MYB transcription factors, MYB10 and MYB72, were identified as Fe deficiency-responsive genes, which are responsible for the activation of expression of the NAS4 gene in Fe-deficient conditions in Arabidopsis (Palmer et al., 2013). Considering the fact that the bhlh34bhlh104 mutants display interveinal chlorosis similar to nas4x-1 mutants (Klatte et al., 2009), we asked whether these two MYB genes are affected in the bhlh34bhlh104 mutants. As expected, compared with the wild type, the mutants show considerably down-regulated transcript abundance of MYB10 and MYB72 regardless of whether Fe is deficient or not (Fig. 4), suggesting that bHLH34 and bHLH104 positively regulate the transcription of MYB10 and MYB72.
The well-known transcription factors for Fe homeostasis in Arabidopsis are four Ib subgroup bHLH transcription factors (bHLH38/39/100/101) and another bHLH transcription factor, FIT, all of which are induced by Fe deficiency (Colangelo and Guerinot, 2004; Wang et al., 2007). Each of bHLH38/39/100/101 can interact with FIT to regulate the expression of Fe uptake-associated genes (Yuan et al., 2008; Wang et al., 2013). To determine the position of bHLH34 and bHLH104 in Fe signaling pathways, it is necessary to investigate their regulatory relationship with these bHLH transcription factors. Gene expression analysis revealed that bHLH38/39/100/101 were strongly down-regulated in the bhlh34bhlh104 mutants under both Fe-sufficient and Fe-deficient conditions, whereas FIT was moderately decreased (Fig. 4). These data imply that bHLH34 and bHLH104 act upstream of bHLH38/39/100/101 and FIT.
In addition, we also determined the expression of genes that are involved in Fe distribution in plants, finding that NAS2, NAS4, ZIF1, FRD3, OPT3, and PYE were down-regulated in the bhlh34bhlh104 mutants in response to Fe deficiency (Supplemental Fig. S6). Taken together, our data suggest that bHLH34 and bHLH104 positively regulate the Fe deficiency response.
Overexpression of bHLH34 and bHLH104 Activates the Fe Deficiency Response
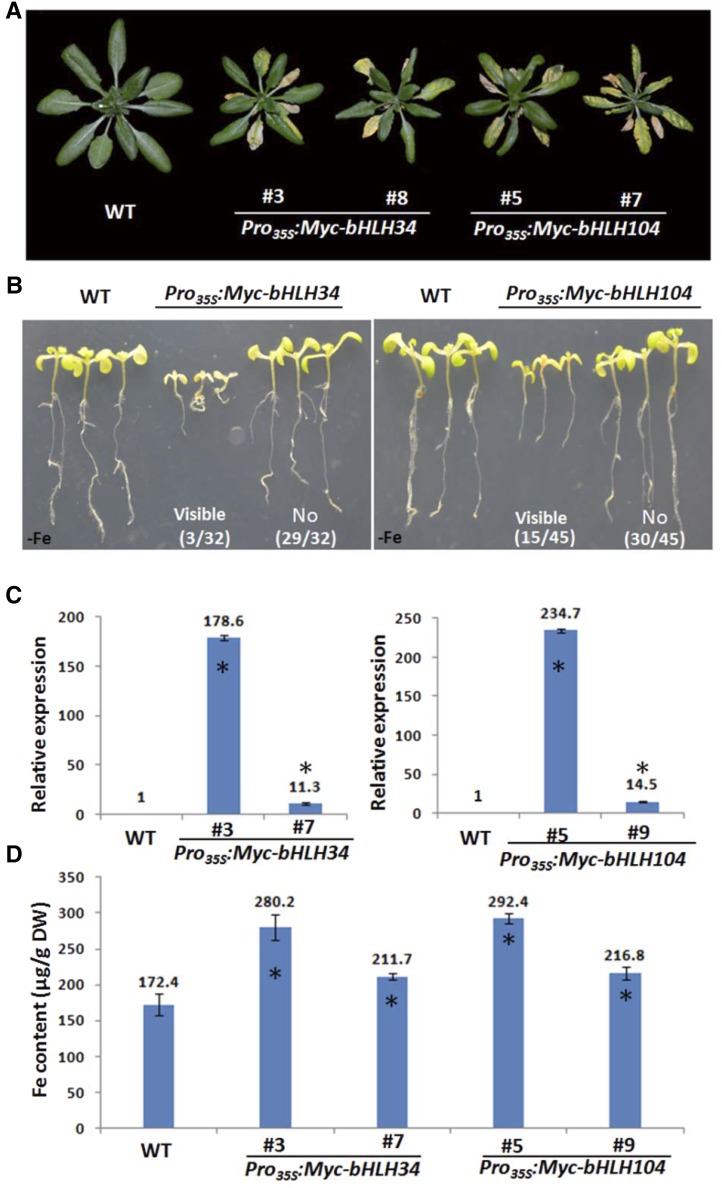
Considering the inhibited Fe deficiency response caused by the loss of function of bHLH34 and bHLH104, we asked whether elevated expression of bHLH34 and bHLH104 would enhance the Fe deficiency response. First, we performed complementation assays. ProbHLH34:Myc-bHLH34 and ProbHLH104:Myc-bHLH104 constructs were introduced into the bhlh34 and bhlh104 mutants, respectively. When grown on Fe deficiency medium, both ProbHLH34:Myc-bHLH34/bhlh34 and ProbHLH104:Myc-bHLH104/bhlh104 plants were comparable with wild-type plants (Supplemental Fig. S7), suggesting that both mutants were rescued. Next, we constructed transgenic plants expressing Pro35S:Myc-bHLH34 and Pro35S:Myc-bHLH104. When grown in normal soils, about 10% of Pro35S:Myc-bHLH34 and 33% of Pro35S:Myc-bHLH104 T0 transgenic plants displayed visible phenotypes (small rosettes and leaf necrosis; Fig. 5A), and no visible phenotype was observed for the other transgenic plants. We further assessed the root phenotypes of T1 plants when grown on Fe deficiency medium, finding that the progeny of T0 plants with visible phenotypes produced short roots whereas the root length of the other progeny was similar to that of wild-type plants (Fig. 5B). In contrast, no visible phenotypes were observed between wild-type and transgenic plants when grown on Fe-sufficient medium for 10 d (Supplemental Fig. S8A). The determination of transgene abundance revealed that the transgenic plants with visible phenotypes have significantly higher transgene levels than those without visible phenotypes (Fig. 5C; Supplemental Fig. S8B), suggesting that the visible phenotypes are closely associated with the transgene levels. For further investigation, Pro35S:Myc-bHLH34#3 and Pro35S:Myc-bHLH104#5 were chosen as representatives of plants with visible phenotypes and Pro35S:Myc-bHLH34#7 and Pro35S:Myc-bHLH104#9 as representatives of plants without visible phenotypes.
Figure 5.
Phenotypes of bHLH34 and bHLH104 overexpression plants. A, Four-week-old plants grown in normal soil. B, Ten-day-old seedlings grown on Fe-deficient (–Fe) medium. Numbers indicate the frequency of transgenic plants with the corresponding phenotypes. C, Relative transcript levels of bHLH34 (left) and bHLH104 (right) in overexpression plants. Significant differences from the wild type (WT) are indicated by asterisks (P < 0.05). D, Fe content of leaves in 4-week-old plants grown in normal soil. Significant differences from the wild type are indicated by asterisks (P < 0.05). DW, Dry weight.
Given that the bhlh34bhlh104 mutants contain less Fe than the wild type, we asked whether overexpression transgenic plants accumulate more Fe than wild-type plants. The leaves of 4-week-old plants grown in normal soil were used for Fe concentration determination. As expected, overexpression plants have higher Fe content than wild-type plants, and the Fe concentration is particularly high in Pro35S:Myc-bHLH34#3 and Pro35S:Myc-bHLH104#5 plants (Fig. 5D). In contrast, the concentration of manganese, copper, and zinc in overexpression plants is comparable to that in the wild type (Supplemental Fig. S9). These data imply that the visible phenotypes may be linked with the high transgene abundance and high Fe concentration.
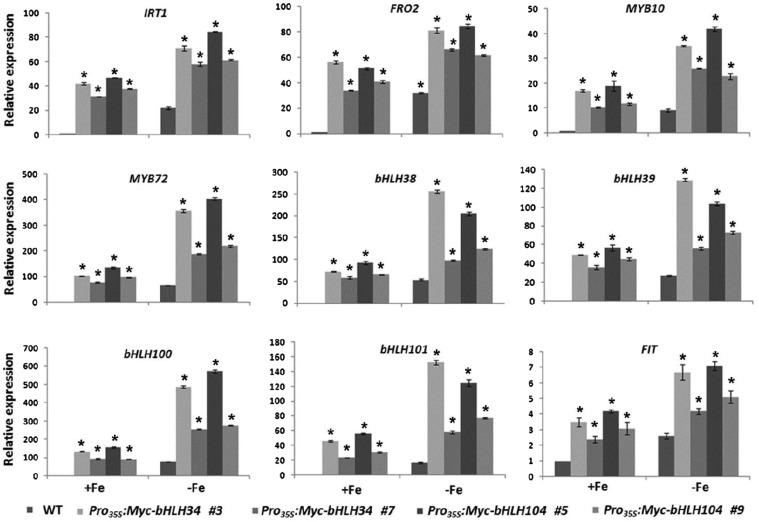
Correspondingly, we examined the transcript abundance of Fe deficiency-responsive genes. In contrast to the bhlh34bhlh104 mutants, overexpression plants constitutively activate FIT, bHLH38/39/100/101, MYB10/72, IRT1, and FRO2 (Fig. 6) as well as NAS2, NAS4, ZIF1, FRD3, OPT3, and PYE (Supplemental Fig. S10). These data suggest that bHLH34 and bHLH104 positively regulate the Fe deficiency response.
Figure 6.
Expression of Fe deficiency-responsive genes in various transgenic plants. Wild-type (WT) and transgenic plants were grown on Fe-sufficient (+Fe) medium for 10 d and then transferred to +Fe or Fe-deficient (–Fe) medium for 3 d. RNA was prepared from root tissues. The data represent means ± sd of three technical repeats from one representative experiment. Significant differences from the corresponding wild type are indicated by asterisks (P < 0.05).
bHLH34 and bHLH104 Directly Regulate the Transcription of bHLH38/39/100/101
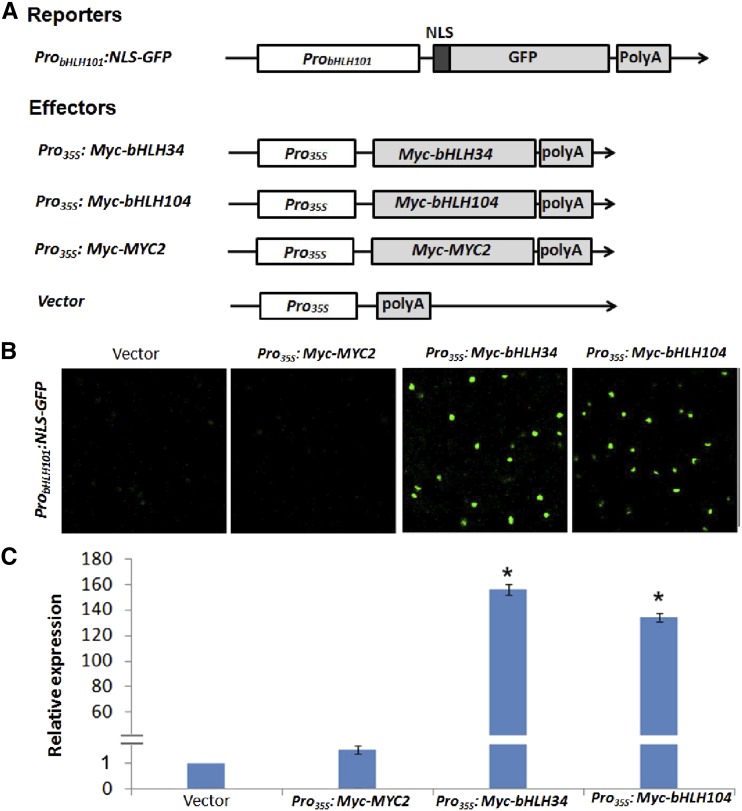
Although bHLH38/39/100/101 and FIT are the major regulators of Fe homeostasis, their transcripts are still up-regulated in response to Fe deficiency. This implies that unidentified transcription factors activate their transcription in Fe-deficient conditions. Our yeast one-hybrid assays indicated that bHLH34 and bHLH104 can bind to the promoter of bHLH101. Considering the fact that the transcript abundance of bHLH38/39/100/101 and FIT is decreased in the bhlh34bhlh104 mutants, we proposed that bHLH38/39/100/101 and FIT are the direct target genes of bHLH34 and bHLH104. To confirm this hypothesis, we designed a reporter-effector transient expression assay system (Fig. 7A; Supplemental Fig. S11A). The promoter of bHLH101 was fused with the GFP reporter gene containing an NLS for construction of the reporter expression cassette, ProbHLH101:NLS-GFP. For the construction of effectors, bHLH34 and bHLH104 were fused with the CaMV 35S promoter to form Pro35S:Myc-bHLH34 and Pro35S:Myc-bHLH104. When the ProbHLH101:NLS-GFP reporter was coexpressed separately with the empty vector (the backbone vector of Pro35S:Myc-bHLH34 and Pro35S:Myc-bHLH104), only a weak GFP signal was observed (Fig. 7B). In contrast, the Pro35S:Myc-bHLH34 or Pro35S:Myc-bHLH104 effector dramatically enhanced the GFP signal of the ProbHLH101:NLS-GFP reporter. To confirm the activation specificity, we also constructed a Pro35S:Myc-MYC2 effector. MYC2 is a bHLH transcription factor with transcription activation activity (Fernández-Calvo et al., 2011). Coexpression assays indicated that use of the Pro35S:Myc-MYC2 effector resulted in as low a GFP signal as was observed for the empty vector (Fig. 7B). We further quantified the GFP transcript levels, which showed that the GFP mRNA abundance was increased considerably when Pro35S:Myc-bHLH34 or Pro35S:Myc-bHLH104 was coexpressed with ProbHLH101:NLS-GFP (Fig. 7C). Correspondingly, we also constructed ProbHLH38:NLS-GFP, ProbHLH39:NLS-GFP, ProbHLH100:NLS-GFP, and ProFIT:NLS-GFP reporters, finding that Pro35S:Myc-bHLH34 and Pro35S:Myc-bHLH104 activated ProbHLH38:NLS-GFP, ProbHLH39:NLS-GFP, and ProbHLH100:NLS-GFP but not ProFIT:NLS-GFP (Supplemental Fig. S11B). These data suggest that both bHLH34 and bHLH104 specifically activate the transcription of bHLH38/39/100/101.
Figure 7.
bHLH34 and bHLH104 activate the promoter of bHLH101. A, Schematic representation of the constructs used for transient expression assays. The reporter construct consists of a bHLH101 promoter, a nuclear localization sequence (NLS) fused with the GFP coding sequence, and a poly(A) terminator. Effector constructs express Myc-bHLH34, Myc-bHLH104, and Myc-MYC2 under the control of the cauliflower mosaic virus (CaMV) 35S promoter. B, bHLH34 and bHLH104 activate the promoter of bHLH101 in transient expression assays. The results are one representative of three biological repeats. C, GFP transcript abundance. In the transient assays, Pro35S:Myc-GUS was expressed as a control. GFP transcript abundance was normalized to GUS transcript. The value with the empty vector as an effector was set to 1. The results are means ± sd of three technical repeats from one of three biological repeats. Significant differences from the empty vector are indicated by asterisks (P < 0.05).
Overexpression of bHLH101 Partially Rescues bhlh34bhlh104 Mutants
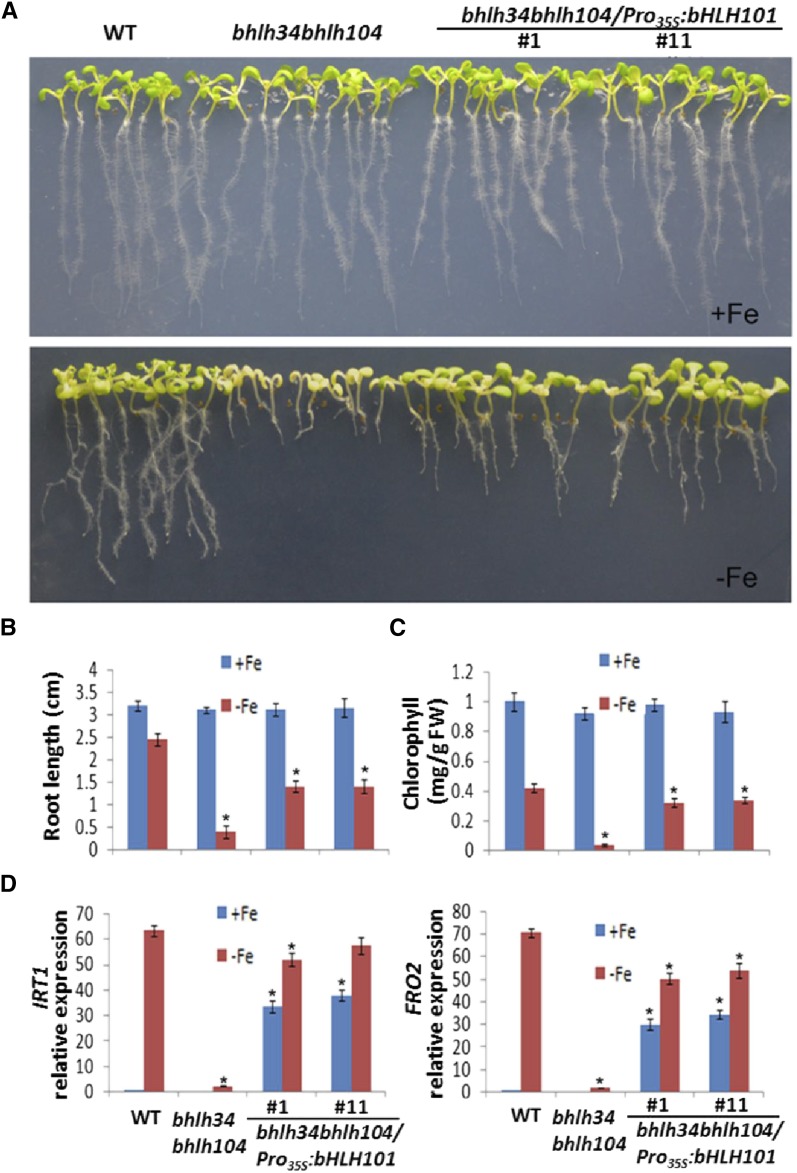
Previous studies have confirmed that the FRO2 and IRT1 genes are regulated directly by bHLH38/39/100/101 and that their expression is decreased drastically in a bHLH39, bHLH100, and bHLH101 triple knockout mutant (Yuan et al., 2008; Wang et al., 2013). Thus, we speculated that the inability of bhlh34bhlh104 to induce bHLH38/39/100/101 is the primary reason for the Fe deficiency symptoms and the reduced expression of FRO2 and IRT1 in the bhlh34bhlh104 mutants. It was expected that elevated expression of bHLH38/39/100/101 would cure or relieve the Fe deficiency symptoms. Because of the functional redundancy of bHLH38/39/100/101 (Wang et al., 2013), we selected bHLH101 as a representative. We employed the CaMV 35S promoter to drive the expression of bHLH101. The Pro35S:bHLH101 cascade was introduced into the bhlh34bhlh104 mutants by Agrobacterium tumefaciens. Of the 78 transgenic plants screened, 42 showed significant relief of the leaf chlorosis symptom. Transcript level analysis indicated that bHLH101 was constitutively overexpressed in the bhlh34bhlh104/Pro35S:bHLH101 plants (Supplemental Fig. S12A). The T2 generation plants from two different transgenic lines, bhlh34bhlh104/Pro35S:bHLH101#1 and bhlh34bhlh104/Pro35S:bHLH101#11, were used for further analyses. When grown for 10 d on Fe-deficient medium, these two lines displayed significant growth advantages compared with the bhlh34bhlh104 mutants (Fig. 8A), including increased root length and chlorophyll content (Fig. 8, B and C). We further assessed the expression of IRT1, FRO2, MYB10, and MYB72, finding that IRT1 and FRO2, but not FIT, MYB10, and MYB72, were elevated in the two transgenic lines exposed to Fe-deficient conditions compared with the bhlh34bhlh104 mutants (Fig. 8D; Supplemental Fig. S12, B–D). These data reveal that the overexpression of bHLH101 partially rescues the bhlh34bhlh104 mutants.
Figure 8.
Overexpression of bHLH101 partially rescues bhlh34bhlh104. A, Ten-day-old seedlings grown on Fe-sufficient (+Fe) or Fe-deficient (–Fe) medium. B, Root length of seedlings on +Fe or –Fe medium. Values are means ± sd of 10 plants for each genotype. C, Chlorophyll content of seedlings on +Fe or –Fe medium. Significant differences from the wild type (WT) are indicated by asterisks (P < 0.05). D, Expression of IRT1 and FRO2. Plants were grown on +Fe medium for 10 d and then transferred to +Fe or –Fe medium for 3 d. RNA was prepared from root tissues. Significant differences from the corresponding wild type are indicated by asterisks (P < 0.05).
bHLH34 Can Interact Physically with bHLH104
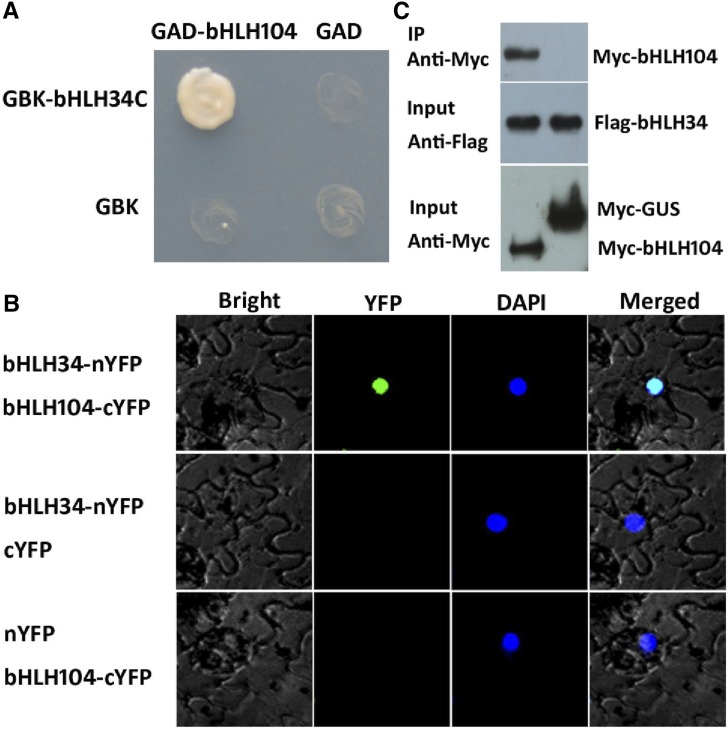
bHLH34 and bHLH104 belong to the IVc bHLH subgroup (Heim et al., 2003). A typical feature of bHLH proteins is the formation of homodimers or heterodimers. To investigate the potential interaction between bHLH34 and bHLH104, we conducted yeast two-hybrid assays (Fig. 9A). When full-length bHLH34 was fused with the binding domain of GAL4 in vector pGBK-T7, it showed strong autoactivation activity. In contrast, the C-terminal truncated versions containing the bHLH domain (bHLH34C) displayed no autoactivation activity; this form was used in the subsequent yeast two-hybrid assays. A protein interaction test in yeast indicated that bHLH34C can interact with bHLH104.
Figure 9.
Interaction between bHLH34 and bHLH104. A, Yeast two-hybrid assays. Representative growth status of yeast cells is shown on synthetic dextrose medium agar plates without Leu/Trp/His/adenine. B, BiFC assays. N. benthamiana leaves were infiltrated with different combinations of the constructs. C, Coimmunoprecipitation (IP) assays. Total protein was immunoprecipitated using Flag antibody, and coimmunoprecipitated protein was then detected using Myc antibody.
Next, we employed bimolecular fluorescence complementation (BiFC) assays to confirm their interaction in plant cells. bHLH34 and bHLH104 were fused with the N-terminal fragment (nYFP) and C-terminal fragment (cYFP) of yellow fluorescent protein, respectively. When bHLH34-nYFP was transiently coexpressed with bHLH104-cYFP, strong YFP fluorescence was visible in the nucleus of epidermal cells in Nicotiana benthamiana leaves, whereas no YFP fluorescence was detected in negative controls (bHLH34-nYFP coexpressed with cYFP or nYFP coexpressed with bHLH104-cYFP; Fig. 9B).
To further confirm whether bHLH34 and bHLH104 form a protein complex in plant cells, we performed coimmunoprecipitation assays (Fig. 9C). bHLH34 and bHLH104 were transiently coexpressed in N. benthamiana leaves. The total proteins were incubated with Flag antibody and A/G-agarose beads and then separated by SDS-PAGE for immunoblotting with Myc antibody. In agreement with the results from BiFC, bHLH34 and bHLH104 were present in the same protein complex. Taken together, these data suggest that bHLH34 could interact physically with bHLH104.
Interaction between bHLH34/bHLH104 and bHLH105
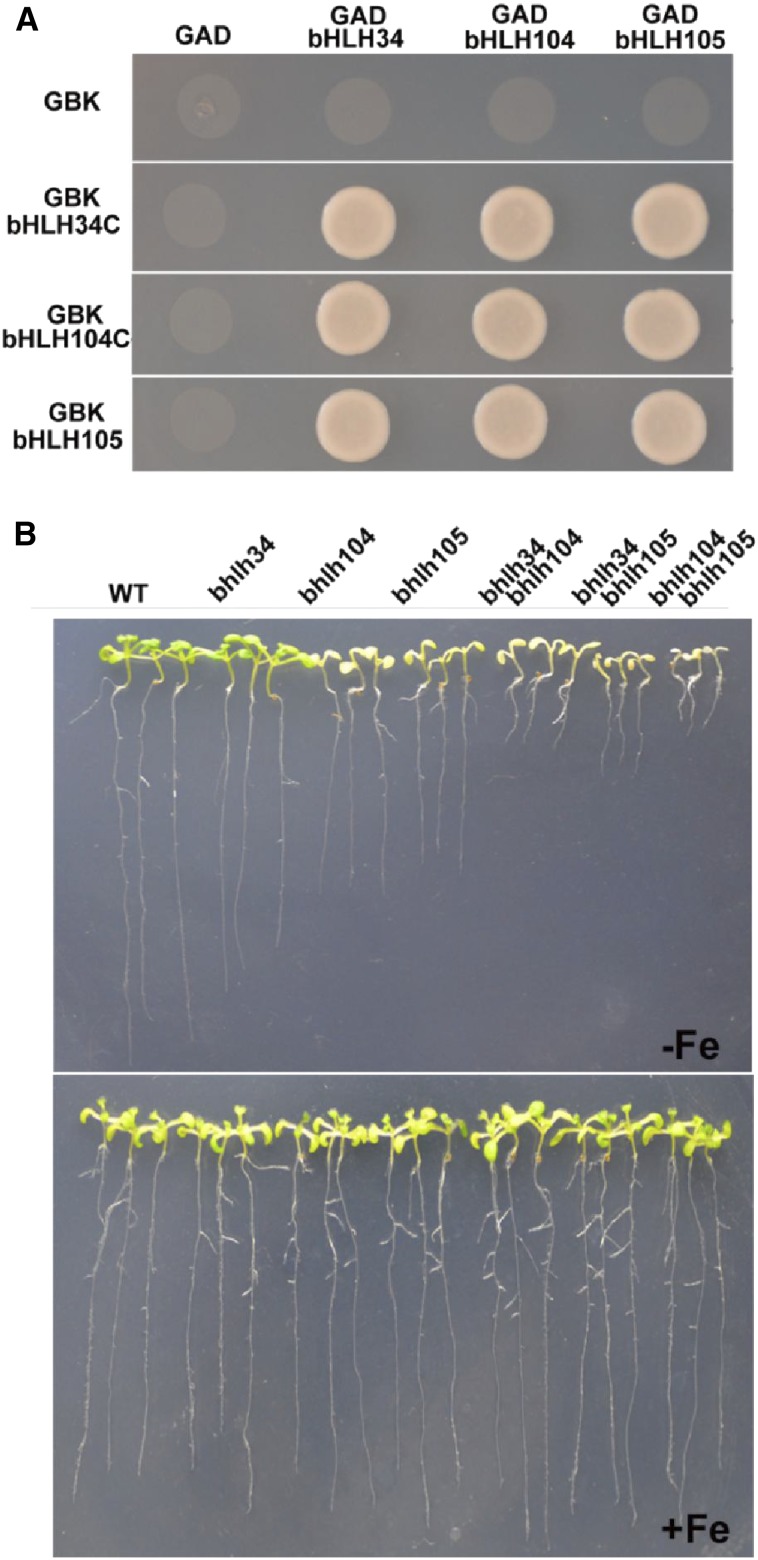
This work confirms the interaction between bHLH34 and bHLH104. A recent study suggested that bHLH104 interacts with bHLH105 and modulates Fe homeostasis in Arabidopsis (Zhang et al., 2015). Considering that these three proteins share the same target genes (bHLH38/39/100/101) and their overexpressors up-regulate the same Fe deficiency-responsive genes, we proposed that they function as heterodimers. To investigate our hypothesis, yeast two-hybrid assays were performed. As shown in Figure 10A, each of these three proteins can interact with itself and the other two proteins, implying that they function as homodimers or heterodimers.
Figure 10.
Interactions between bHLH34, bHLH104, and bHLH105. A, Yeast two-hybrid assays. Interaction was indicated by the ability of cells to grow on synthetic dropout medium lacking Leu/Trp/His/adenine. C-terminally truncated bHLH34 and bHLH104 and full-length bHLH105 were cloned into pGBKT7, and full-length cDNAs of bHLH34, bHLH104, and bHLH105 were cloned into pGADT7. B, Fe deficiency symptoms of various single and double mutants. Ten-day-old seedlings were germinated directly on Fe-deficient (–Fe) or Fe-sufficient (+Fe) medium. WT, Wild type.
To further investigate their genetic interactions, we further produced two double mutants, bhlh34bhlh105 and bhlh104bhlh105. We tried to generate bhlh34bhlh104bhlh105 triple mutants but failed. Then, we evaluated the Fe deficiency tolerance ability of various mutants. We found that, under Fe deficiency conditions, both bhlh34bhlh105 and bhlh104bhlhl105 double mutants displayed the enhanced Fe deficiency symptoms (shorter roots) compared with the three single mutants (Fig. 10B). The expression of Fe deficiency-responsive genes was lower in the bhlh34bhlh105 and bhlh104bhlhl105 double mutants than in the three single mutants (Supplemental Fig. S13). All these data suggest that bHLH34, bHLH104, and bHLH105 play nonredundant but additive roles in modulating Fe homeostasis.
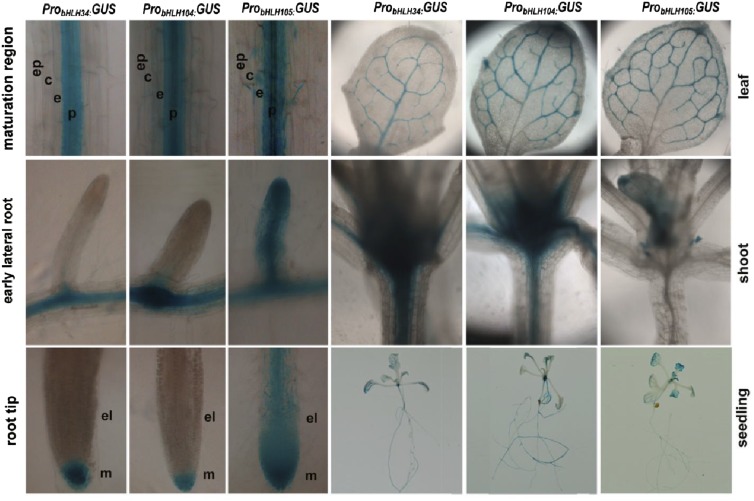
Given their similar molecular functions, we wanted to know whether the expression patterns of these three genes are different. We constructed ProbHLH34:GUS, ProbHLH104:GUS, and ProbHLH105:GUS transgenic plants. Although different expression strengths were observed, bHLH34, bHLH104, and bHLH105 have similar tissue-specific expression patterns in rosette leaves, cauline leaves, stems, and siliques but not in flowers (Supplemental Fig. S14A). When seedlings were further analyzed by microscope, all plants contained GUS staining in the pericycle of the root maturation zone and veins of leaves (Fig. 11). ProbHLH105:GUS was detected in the elongation zone of root tips and early lateral roots, whereas ProbHLH34:GUS and ProbHLH104:GUS were not observed. In contrast, ProbHLH34:GUS and ProbHLH104:GUS were detected in the hypocotyls, whereas ProbHLH105:GUS was not. The difference in tissue-specific expression patterns may explain the functional nonredundancy between bHLH34, bHLH104, and bHLH105. In addition, GUS staining of seedlings exposed to Fe-sufficient and Fe-deficient conditions suggests that their promoter activity is stable irrespective of the Fe status (Supplemental Fig. S14B).
Figure 11.
GUS staining of ProbHLH34:GUS, ProbHLH104:GUS, and ProbHLH105:GUS seedlings. Two-week-old seedlings were used for GUS staining. GUS staining of the root maturation regions, early lateral roots, root tips, leaves, hypocotyls, and whole seedlings is shown. c, Cortex; e, endodermis; el, elongation region of the root tip; ep, epidermis; m, meristem zone of the root tip; p, pericycle.
DISCUSSION
Fe is indispensable for plant growth and development. Fe deficiency often causes delayed growth and reduced photosynthesis and, hence, decreased crop yields. When suffering from a Fe-deficient environment, plants can sense external Fe status and employ transcription factors and intricate mechanisms to regulate the expression of Fe uptake-associated genes, which then facilitate Fe influx from soils to meet the internal demands of the plant. However, excess Fe is toxic to plant cells, owing to the generation of hydroxyl radicals by the Fenton reaction (Thomine and Vert, 2013). Therefore, it is crucial to maintain Fe homeostasis in plants. Plants have evolved sophisticated regulatory mechanisms to maintain Fe homeostasis. It is well known that bHLH38/39/100/101 and FIT have synergistic effects on the regulation of Fe deficiency responses. Their cooverexpression constitutively activates Fe acquisition strategy I in Arabidopsis. It is noteworthy that, like other Fe deficiency-responsive genes, bHLH38/39/100/101 and FIT are induced by Fe deficiency. Thus, to reveal how bHLH38/39/100/101 and FIT are activated in Fe-deficient conditions would provide insights into the mechanism that plants employ to maintain Fe homeostasis. Here, we characterized that both bHLH34 and bHLH104 activate the transcription of bHLH38/39/100/101 and that they nonredundantly regulate Fe deficiency responses in Arabidopsis.
bHLH34 was identified by screening proteins that bound to the promoter of the bHLH101 gene. The bhlh34, bhlh104, and bhlh34bhlh104 mutants display sensitivity to Fe deficiency, including reduced root length, chlorotic leaves, and decreased ferric chelate reductase activity (Figs. 2C and 3B). Correspondingly, amiR-bhlh34/104 plants show phenotypes similar to those of bhlh34bhlh104 mutants (Fig. 2D; Supplemental Fig. S3D). These data indicate that loss of function of bHLH34 and bHLH104 causes sensitivity to Fe deficiency. Fe deficiency sensitivity can be caused by limited Fe uptake or disrupted Fe distribution. For example, irt1, frd1, and fit mutants display severe Fe deficiency sensitivity because of Fe uptake limitation (Robinson et al., 1999; Vert et al., 2002; Colangelo and Guerinot, 2004). In contrast, the frd3 and opt3 mutations enhance Fe deficiency sensitivity by disrupting Fe translocation (Durrett et al., 2007; Zhai et al., 2014). The determination of Fe concentration confirms that the bhlh34bhlh104 mutants contain less Fe than wild-type plants in both Fe-sufficient and Fe-deficient conditions (Fig. 3, C and D), suggesting that limited Fe uptake contributes to the Fe deficiency sensitivity of the bhlh34bhlh104 mutants. We also observed that new leaves of bhlh34bhlh104 mutants are yellower than old leaves (Fig. 2D), whereas the opposite phenomenon occurs in the bHLH34 and bHLH104 overexpression plants (Fig. 5A), implying that bHLH34 and bHLH104 may interrupt Fe distribution between leaves.
Fe deficiency-responsive genes, including IRT1, FRO2, MYB10/72, FIT, and bHLH38/39/100/101, are repressed in the bhlh34bhlh104 mutants (Fig. 4), indicating that the loss of function of bHLH34 and bHLH104 impairs the transduction of Fe deficiency response signaling. Although bhlh34 and bhlh104 mutants display reduced activation of Fe deficiency-responsive genes, the bhlh34bhlh104 mutants have the strongest inhibitory effect on the Fe deficiency-responsive genes. When either ProbHLH34:Myc-bHLH34 or ProbHLH104:Myc-bHLH104 was introduced into bhlh34bhlh104 double mutants, the Fe deficiency symptoms of bhlh34bhlh104 were partially rescued (Supplemental Fig. S15). These data suggest that bHLH34 and bHLH104 have nonredundant functions under Fe-deficient conditions. In contrast to bhlh34 and bhlh104 mutants, bHLH34 and bHLH104 overexpression plants activate the expression of Fe deficiency-responsive genes (Fig. 6). In agreement with this, Fe is overaccumulated in the overexpression plants (Fig. 5D). Taken together, our data suggest that bHLH34 and bHLH104 act as positive regulators in Fe deficiency response signaling.
The bhlh34bhlh104 mutants displayed enhanced Fe deficiency sensitivity. However, unexpectedly, we found that about 10% of Pro35S:Myc-bHLH34 and 30% of Pro35S:Myc-bHLH104 plants showed short roots on Fe-deficient medium, although they accumulated high Fe content and activated Fe deficiency-responsive genes, including bHLH38/39/100/101, MYB10/72, FRO2, and IRT1. In fact, similar observations were made for the pye1 mutant, which up-regulates Fe deficiency-responsive genes and accumulates excessive Fe but displays short roots under Fe deficiency conditions (Long et al., 2010). NAS4 and ZIF1 are significantly up-regulated in the bHLH34 and bHLH104 overexpression plants and particular high when overexpression plants are subjected to Fe deficiency conditions (Supplemental Fig. S10). It is noteworthy that NAS4 and ZIF1 are directly negatively regulated by PYE (Long et al., 2010). As a negative regulator in Fe homeostasis, PYE is also a direct target of bHLH104 and bHLH105 (Zhang et al., 2015). Our results suggest that PYE is positively regulated by bHLH34, bHLH104, and bHLH105. The expression analysis of NAS4 and ZIF1 between bhlh34bhlhl104 and pye1 mutants (Supplemental Fig. S16) indicates that NAS4 and ZIF1 are positively and negatively regulated by bHLH34/bHLH104 and PYE, respectively. Plants have evolved this mechanism to fine-tune Fe homeostasis.
Recently, Zhang et al. (2015) revealed that overexpression of bHLH104 and bHLH105 activates the Fe deficiency-responsive genes and overaccumulation of Fe. However, the Fe deficiency sensitivity of our overexpression plants is completely different from the results of Zhang et al. (2015), who confirmed that bHLH104 overexpression plants produced longer roots than the wild type on Fe-deficient medium. They used Pro35S:bHLH104-GFP as an overexpression construct, whereas we used Pro35S:Myc-bHLH104. To investigate whether the C-terminal fused GFP affects the function of bHLH34 or bHLH104, we constructed Pro35S:Myc-bHLH34-GFP and Pro35S:Myc-bHLH104-GFP transgenic plants. No visible phenotype was found in all Pro35S:Myc-bHLH34-GFP and Pro35S:Myc-bHLH104-GFP transgenic plants compared with the wild type when plants were grown in normal soils. In agreement with the results from Zhang et al. (2015), about 70% of these GFP fusion transgenic plants produced significantly longer roots than the wild type on Fe-deficient medium (Supplemental Fig. S17A). The expression of Fe deficiency-responsive genes in GFP fusion transgenic plants with longer roots is significantly lower than that in Myc fusion transgenic plants with shorter roots, but it is comparable to the Myc fusion transgenic plants with normal roots (Supplemental Fig. S17B). It is noteworthy that the overexpression plants with visible phenotypes have extremely high transgene levels compared with overexpressors with invisible phenotypes (Fig. 5C; Supplemental Fig. S8B). Therefore, it is likely that the extremely high expression of transgene caused abnormal regulation of Fe uptake-associated genes and Fe overload. In any case, it is an efficient approach to generate Fe deficiency-tolerant plants by constitutively expressing bHLH34 or bHLH104 with a C terminus fused with a GFP gene.
Under both Fe-sufficient and Fe-deficient conditions, the transcript levels of bHLH38/39/100/101 are always lower in the bhlh34, bhlh104, and bhlh34bhlh104 mutants than in wild-type plants (Fig. 4). Transient expression assays reveal that both bHLH34 and bHLH104 activate the promoters of bHLH38/39/100/101 (Fig. 7, B and C; Supplemental Fig. S11). These data suggest that bHLH34 and bHLH104 directly and positively regulate the transcription of bHLH38/39/100/101. In agreement with our results, Zhang et al. (2015) showed the direct regulation of bHLH38/39/100/101 by bHLH104. bHLH38/39/100/101 are strongly induced when wild-type plants are exposed to Fe-deficient conditions. However, it is worth noting that, in some mutants where Fe homeostasis is disrupted, such as irt1, frd1, frd3, and fit, bHLH38/39/100/101 transcript abundance is increased even under Fe-sufficient conditions compared with wild-type plants (Wang et al., 2007). The fact that Fe deficiency-responsive genes are regulated by internal Fe demand via shoot-to-root signaling was demonstrated previously (Grusak and Pezeshgi, 1996; Vert et al., 2003). Interestingly, although the bhlh34bhlh104 mutants have lower Fe content than wild-type plants, the induction of bHLH38/39/100/101 by Fe deficiency is strongly inhibited in the bhlh34bhlh104 mutants, suggesting that the transcriptional activation of bHLH38/39/100/101 by internal Fe demand is largely dependent on bHLH34 and bHLH104.
It has been confirmed that bHLH38/39/100/101 have redundant functions in the Fe deficiency response (Yuan et al., 2008; Wang et al., 2013). Our experiments indicate that the constitutive expression of bHLH101 partially rescued the severe Fe deficiency symptoms of bhlh34bhlh104 mutants (Fig. 8), implying that the regulation of bHLH38/39/100/101 by bHLH34 and bHLH104 is required for Fe homeostasis. Because of the imperfect complementation by overexpression of bHLH101, it is possible that the ectopic expression of bHLH101 caused ectopic expression of its target genes, which then led to the abnormal Fe distribution, or that bHLH34 and bHLH104 also regulate other genes that are involved in Fe homeostasis but are not regulated by bHLH101. In fact, we found that the induction of FIT, MYB10, and MYB72 by Fe deficiency is not rescued in bhlh34bhlh104/Pro35S:bHLH101#1 and bhlh34bhlh104/Pro35S:bHLH101#11 plants (Supplemental Fig. S12). It has been reported that MYB10 and MYB72 are required for growth in Fe-limiting conditions by directly regulating the expression of NAS4 (Palmer et al., 2013). NAS genes are responsible for the synthesis of nicotianamine, which plays a role in the intercellular and intracellular distribution of Fe (Takahashi et al., 2003; Klatte et al., 2009). Therefore, the elevated expression of bHLH101 may rescue only the Fe uptake phenotype, but not the Fe distribution phenotype, in the bhlh34bhlh104 mutants. The transcript abundance of bHLH34 and bHLH104 is not induced by Fe deficiency (Supplemental Fig. S2), which implies that they may be regulated at the posttranscriptional level because their target genes are up-regulated by Fe deficiency. To further analyze their protein stability and screen their interaction partners will provide insights into their regulation mechanisms under Fe deficiency conditions. The stronger Fe deficiency symptoms of double mutants than single mutants suggest that bHLH34, bHLH104, and bHLH105 play nonredundant roles in regulating Fe homeostasis. We also observed differential expression patterns among ProbHLH34:GUS, ProbHLH104:GUS, and ProbHLH105:GUS, which might explain the nonredundant roles.
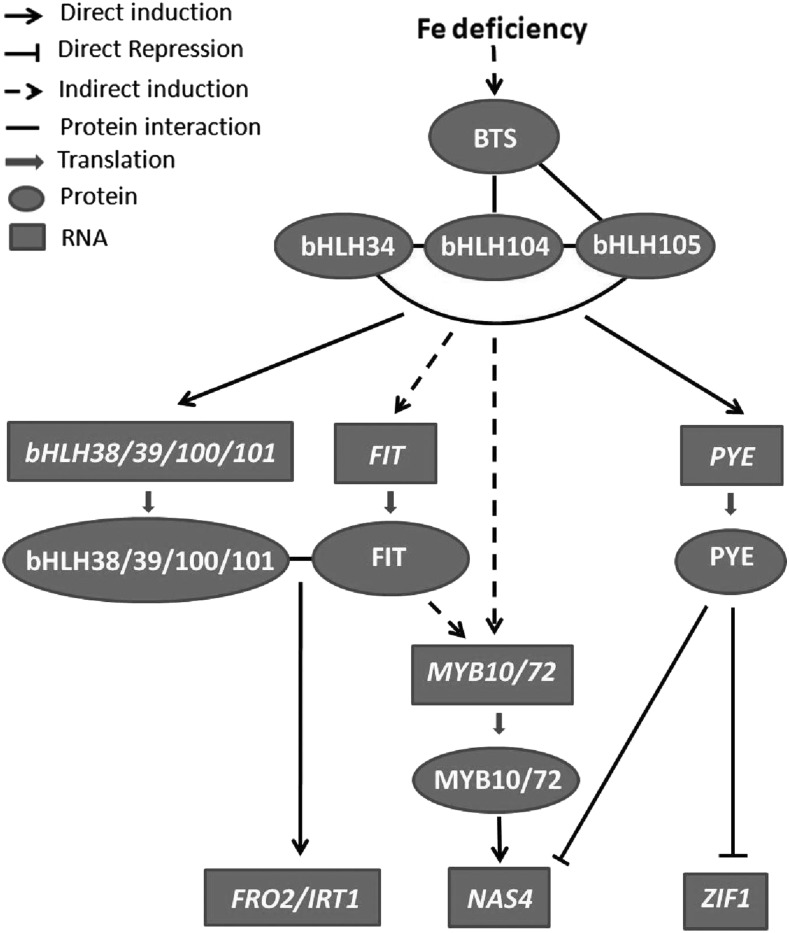
A putative Fe deficiency-responsive signaling pathway is shown in Figure 12. BTS is considered a potential Fe sensor because its structure, its ability to bind Fe, and its capacity to catalyze ubiquitination are similar to the mammalian Fe sensor FBXL5 (Kobayashi et al., 2013; Selote et al., 2015). BTS is induced and its product is stabilized by Fe deficiency; however, it has been confirmed to negatively regulate Fe homeostasis (Long et al., 2010; Selote et al., 2015; Zhang et al., 2015). Long et al. (2010) confirmed that bHLH104 and bHLH105, but not bHLH34, interact with BTS in yeast. Although its interacts with BTS, bHLH104 protein is not affected by BTS (Selote et al., 2015). bHLH34, bHLH104, and bHLH105 function as homodimers or heterodimers. It is likely that the interaction of BTS with bHLH104 competitively interferes with the formation of dimers, which then affects the regulation of downstream genes, such as bHLH38/39/100/101 and PYE. FIT interacts with bHLH38/39/100/101 to activate the expression of IRT1 and FRO2. PYE functions as the negative regulator of ZIF1 and NAS4. MYB10 and MYB72 function redundantly to regulate Fe homeostasis by the direct activation of NAS4. The induction of MYB10/72 by Fe deficiency is partially dependent on FIT, and bHLH34/104/105 are required for the induction of FIT and MYB10/72. It is worth mentioning that the model has no spatial dimension; it reflects interactions globally but is not applicable in any particular cell type, because the expression patterns of the different players do not always overlap. Future research should aim at identifying the distinct modules that are active in the root cortex and epidermis, in the root stele, and in various leaf cell types.
Figure 12.
Fe deficiency-responsive signaling pathway. Fe deficiency stabilizes the BTS protein that interacts with bHLH104 and bHLH105. bHLH34, bHLH104, and bHLH105 can form homodimers or heterodimers to activate the transcription of bHLH38/39/100/101 and PYE. FIT interacts with bHLH38/39/100/101 to activate the transcription of FRO2 and IRT1. PYE regulates the expression of ZIF1 and NAS4 negatively. MYB10 and MYB72 regulate the expression of NAS4 positively. The induction of MYB10/72 by Fe deficiency is partially dependent on FIT; bHLH34/104/105 are required for the induction of FIT and MYB10/72.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 was used as the wild type in this study. The T-DNA insertion lines for bHLH34 (CS411089), bHLH104 (Salk_099496C), and bHLH105 (Salk_043690C) were confirmed using PCR with a T-DNA primer and gene-specific primers (Supplemental Table S1). pye1 was described previously (Long et al., 2010). Plants were grown in long photoperiods (16 h of light/8 h of dark) or in short photoperiods (8 h of light/16 h of dark) at 22°C. Surface-sterilized seeds were stratified at 4°C for 2 to 4 d before being planted on medium. Fe-sufficient medium is one-half-strength Murashige and Skoog (MS) medium with 1% (w/v) Suc, 0.8% (w/v) agar, and 0.1 mm FeEDTA. Fe-deficient medium is the same without FeEDTA.
Plasmid Construction
Standard molecular biology techniques were used for the cloning procedures. Genomic DNA from Arabidopsis was used as the template for amplification of the upstream regulatory promoter sequence for ProbHLH34:GUS, ProbHLH104:GUS, ProbHLH105:GUS, ProFIT:NLS-GFP, ProbHLH38:NLS-GFP, ProbHLH39:NLS-GFP, ProbHLH100:NLS-GFP, and ProbHLH101:NLS-GFP. The NLS was amplified from the pGADT7 plasmid. For Pro35S:Flag-bHLH34, Pro35S:Myc-bHLH34, Pro35S:Myc-bHLH104, Pro35S:bHLH101, Pro35S:Myc-GUS, and Pro35S:Myc-MYC2, the corresponding coding sequences were inserted between the CaMV 35S promoter and poly(A) of the binary vector pOCA30. amiR-bhlh34/104 was integrated into the MIR319a backbone, and the construction strategy was described previously (Liang et al., 2012). Primers used for these constructs are listed in Supplemental Table S1. Arabidopsis transformation was conducted by the floral dip method (Clough and Bent, 1998). Transgenic plants were selected with the use of 50 μg mL−1 kanamycin.
Histochemical GUS Staining
Whole seedlings were immersed immediately in 1.5 mL of staining solution containing 0.5 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (Sigma) in 0.1 m sodium phosphate buffer (pH 7.3) in a microfuge tube. The reaction was performed in the dark at 37°C until a blue indigo color appeared. After the reaction, seedlings were rinsed in 0.1 m sodium phosphate buffer (pH 7.3). The samples were then rinsed twice in 70% (v/v) ethanol to remove chlorophylls.
Fe Concentration Measurement
To determine Fe content, 11-d-old seedlings grown on Fe-sufficient medium were transferred to Fe-sufficient or Fe-deficient medium for 3 d. The shoots and roots were harvested separately and used for Fe measurement. Leaves of 4-week-old plants grown in normal soils were used for the measurement of Fe content. About 100 mg dry weight for roots or shoots was used as one sample, and three samples were used in each independent experiment. Fe content analysis was performed using inductively coupled plasma spectroscopy.
Ferric Chelate Reductase Assays
Ferric chelate reductase assays were performed as described previously (Yi and Guerinot, 1996). Briefly, 10 intact plants for each genotype were pretreated for 30 min in plastic vessels with 4 mL of one-half-strength MS solution without micronutrients (pH 5.5) and then soaked into 4 mL of Fe(III) reduction assay solution [one-half-strength MS solution without micronutrients, 0.1 mm Fe(III)-EDTA, and 0.3 mm ferrozine, pH adjusted to 5 with KOH] for 30 min in darkness. An identical assay solution containing no plants was used as a blank. The purple-colored Fe(II)-ferrozine complex was quantified at 562 nm.
Chlorophyll Measurement
Chlorophyll content was measured in 2-week-old plants grown on Fe-sufficient and Fe-deficient media. All leaves were collected and ground to powder in liquid nitrogen. The powder was resuspended in 80% (v/v) acetone on ice and centrifuged at 10,000g at 4°C for 5 min. Chlorophyll concentrations were calculated from spectroscopy absorbance measurements at 663.2, 646.8, and 470 nm (Lichtenthaler, 1987).
Gene Expression Analysis
One microgram of total RNA extracted using the Trizol reagent (Invitrogen) was used for oligo(dT)18-primed cDNA synthesis according to the reverse transcription protocol (Fermentas). The resulting cDNA was subjected to relative quantitative PCR using the SYBR Premix Ex Taq kit (TaKaRa) on a Roche LightCycler 480 real-time PCR machine, according to the manufacturer’s instructions. ACTIN2 (ACT2) was amplified as an internal control, and gene copy number was normalized to that of ACT2. For the quantification of each gene, at least three biological repeats were used. Each biological repeat contained three technical replicates. One representative result from one biological repeat is shown. The analysis of statistical significance was performed by Student’s t test. The quantitative reverse transcription-PCR primers are listed in Supplemental Table S1.
Yeast Assays
For the yeast one-hybrid assay, the Matchmaker Gold Yeast One-Hybrid Library Screening System (Clontech) was used. The pAbAi vector harboring a 1,000-bp sequence upstream of the bHLH101 gene was integrated into the genome of yeast strain Y1HGold followed by selection on synthetic dextrose medium agar plates without uracil. We screened an Arabidopsis equalized full-length cDNA library on synthetic dextrose medium without Leu supplemented with 100 ng mL−1 aureobasidin A. The aureobasidin A resistance is activated by prey proteins that specifically interact with the bait sequence. Candidate cDNA-harboring vectors were amplified and isolated via Escherichia coli. The sequence of each candidate was analyzed with the help of the TAIR database.
For the yeast two-hybrid assay, C-terminally truncated bHLH34C and bHLH104C containing the bHLH domain and full-length bHLH105 cDNAs were cloned into pGBKT7, and the full-length cDNAs of bHLH34, bHLH104, and bHLH105 were cloned into pGADT7. Growth was determined as described in the Yeast Two-Hybrid System User Manual (Clontech). Primers used for vector construction are listed in Supplemental Table S1. Experiments were repeated three times.
Transient Expression Assays
Plasmids were transformed into Agrobacterium tumefaciens strain EHA105. Agrobacterial cells were infiltrated into leaves of Nicotiana benthamiana by the infiltration buffer (0.2 mm acetosyringone, 10 mm MgCl2, and 10 mm MES, pH 5.6). For the BiFC assay, equal volumes of an A. tumefaciens culture were mixed before infiltration into N. benthamiana leaves. After infiltration, YFP and 4′,6-diamino-phenylindole fluorescence were observed with a confocal laser scanning microscope (Olympus). For transcription activation assay, the final optical density at 600 nm value was 0.1 (an internal control; Pro35S:Myc-GUS), 0.5 (reporter), or 0.5 (effector). After infiltration, plants were placed in the dark at 24°C for 48 h before RNA extraction. The transcript abundance of GFP was normalized to GUS.
Coimmunoprecipitation Assay
Flag-bHLH34 and Myc-bHLH104 (or Myc-GUS) were transiently coexpressed in N. benthamiana leaves. Infected leaves were harvested 48 h after infiltration and used for protein extraction. Flag-fused bHLH34 was immunoprecipitated using Flag antibody, and the coimmunoprecipitated proteins were then detected using Myc antibody.
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: bHLH34 (At3g23210), bHLH104 (At4g14410), bHLH105 (At5g54680), bHLH38 (At3g56970), bHLH39 (At3g56980), bHLH100 (At2g41240), bHLH101 (At5g04150), FIT (At2g28160), MYB10 (At3g12820), MYB72 (At1g56160), FRO2 (At1g01580), IRT1 (At4g19690), NAS2 (At5g56080), NAS4 (At1g56430), ZIF1 (At5g13740), FRD3 (At3g08040), OPT3 (At5g59040), PYE (At3g47640), and ACT2 (At3g18780).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic tree of bHLH proteins involved in Fe homeostasis.
Supplemental Figure S2. Expression of bHLH34 and bHLH104 in response to Fe deficiency.
Supplemental Figure S3. Growth status of wild-type and mutant plants.
Supplemental Figure S4. Analysis of amiR-bhlh34/104 and double mutant plants.
Supplemental Figure S5. Chlorophyll content of mutant seedlings on Fe-sufficient or Fe-deficient medium.
Supplemental Figure S6. NAS2, NAS4, ZIF1, FRD3, OPT3, and PYE transcript levels in various mutant plants.
Supplemental Figure S7. Complementation of bhlh34 and bhlh104 mutants.
Supplemental Figure S8. Analysis of bHLH34 and bHLH104 overexpression plants.
Supplemental Figure S9. Concentration of other metals in overexpression plants.
Supplemental Figure S10. NAS2, NAS4, ZIF1, FRD3, OPT3, and PYE transcript levels in overexpression plants.
Supplemental Figure S11. bHLH34 and bHLH104 activate the promoter of bHLH38/39/100.
Supplemental Figure S12. Analysis of bhlh34bhlh104/Pro35S:bHLH101 plants.
Supplemental Figure S13. Expression of Fe deficiency-responsive genes in various single and double mutants.
Supplemental Figure S14. GUS staining of ProbHLH34:GUS, ProbHLH104:GUS, and ProbHLH105:GUS plants.
Supplemental Figure S15. Partial complementation of bhlh34blhlh104 double mutants by ProbHLH34:Myc-bHLH34 and ProbHLH104:Myc-bHLH104.
Supplemental Figure S16. Expression of NAS4 and ZIF1 in bhlh34blhlh104 and pye1 mutants.
Supplemental Figure S17. Phenotypes of Pro35S:Myc-bHLH34-GFP and Pro35S:Myc-bHLH104-GFP plants.
Supplemental Table S1. Primers used in this article.
Supplementary Material
Acknowledgments
We thank TAIR at Ohio State University for the T-DNA insertion mutants.
Glossary
- Fe
iron
- T-DNA
transfer DNA
- TAIR
The Arabidopsis Information Resource
- NLS
nuclear localization sequence
- CaMV
cauliflower mosaic virus
- BiFC
bimolecular fluorescence complementation
- MS
Murashige and Skoog
- cDNA
complementary DNA
Footnotes
This work was supported by the Youth Innovation Promotion Association of the Chinese Academy of Sciences, the Candidates of the Young and Middle Aged Academic Leaders of Yunnan Province (grant no. 2015HB095), and the Program for the Innovative Research Team of Yunnan Province (grant no. 2014HC017).
References
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrett TP, Gassmann W, Rogers EE (2007) The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol 144: 197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP (2007) Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem Sci 32: 63–70 [DOI] [PubMed] [Google Scholar]
- Grusak MA, Pezeshgi S (1996) Shoot-to-root signal transmission regulates root Fe(III) reductase activity in the dgl mutant of pea. Plant Physiol 110: 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH (2003) Role of duplicate genes in genetic robustness against null mutations. Nature 421: 63–66 [DOI] [PubMed] [Google Scholar]
- Hänsch R, Mendel RR (2009) Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr Opin Plant Biol 12: 259–266 [DOI] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Henriques R, Jásik J, Klein M, Martinoia E, Feller U, Schell J, Pais MS, Koncz C (2002) Knock-out of Arabidopsis metal transporter gene IRT1 results in iron deficiency accompanied by cell differentiation defects. Plant Mol Biol 50: 587–597 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P (2004) FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett 577: 528–534 [DOI] [PubMed] [Google Scholar]
- Klatte M, Schuler M, Wirtz M, Fink-Straube C, Hell R, Bauer P (2009) The analysis of Arabidopsis nicotianamine synthase mutants reveals functions for nicotianamine in seed iron loading and iron deficiency responses. Plant Physiol 150: 257–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Nagasaka S, Senoura T, Itai RN, Nakanishi H, Nishizawa NK (2013) Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat Commun 4: 2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Nishizawa NK (2012) Iron uptake, translocation, and regulation in higher plants. Annu Rev Plant Biol 63: 131–152 [DOI] [PubMed] [Google Scholar]
- Liang G, He H, Li Y, Yu D (2012) A new strategy for construction of artificial miRNA vectors in Arabidopsis. Planta 235: 1421–1429 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Ling H-Q, Bauer P, Bereczky Z, Keller B, Ganal M (2002) The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc Natl Acad Sci USA 99: 13938–13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN (2010) The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22: 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiser J, Lingam S, Bauer P (2011) Posttranslational regulation of the iron deficiency basic helix-loop-helix transcription factor FIT is affected by iron and nitric oxide. Plant Physiol 157: 2154–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S. (1999) Iron acquisition by plants. Curr Opin Plant Biol 2: 250–253 [DOI] [PubMed] [Google Scholar]
- Morrissey J, Guerinot ML (2009) Iron uptake and transport in plants: the good, the bad, and the ionome. Chem Rev 109: 4553–4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CM, Hindt MN, Schmidt H, Clemens S, Guerinot ML (2013) MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoS Genet 9: e1003953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Salahudeen AA, Thompson JW, Ruiz JC, Ma HW, Kinch LN, Li Q, Grishin NV, Bruick RK (2009) An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 326: 722–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi S, Schmidt W (2009) Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol 183: 1072–1084 [DOI] [PubMed] [Google Scholar]
- Selote D, Samira R, Matthiadis A, Gillikin JW, Long TA (2015) Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors. Plant Physiol 167: 273–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz A, Grinvalds C, Barberon M, Curie C, Vert G (2011) Proteasome-mediated turnover of the transcriptional activator FIT is required for plant iron-deficiency responses. Plant J 66: 1044–1052 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Terada Y, Nakai I, Nakanishi H, Yoshimura E, Mori S, Nishizawa NK (2003) Role of nicotianamine in the intracellular delivery of metals and plant reproductive development. Plant Cell 15: 1263–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry N. (1980) Limiting factors in photosynthesis. I. Use of iron stress to control photochemical capacity in vivo. Plant Physiol 65: 114–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomine S, Vert G (2013) Iron transport in plants: better be safe than sorry. Curr Opin Plant Biol 16: 322–327 [DOI] [PubMed] [Google Scholar]
- Varotto C, Maiwald D, Pesaresi P, Jahns P, Salamini F, Leister D (2002) The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J 31: 589–599 [DOI] [PubMed] [Google Scholar]
- Vashisht AA, Zumbrennen KB, Huang X, Powers DN, Durazo A, Sun D, Bhaskaran N, Persson A, Uhlen M, Sangfelt O, et al. (2009) Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326: 718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert GA, Briat JF, Curie C (2003) Dual regulation of the Arabidopsis high-affinity root iron uptake system by local and long-distance signals. Plant Physiol 132: 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EL, Connolly EL (2008) Time to pump iron: iron-deficiency-signaling mechanisms of higher plants. Curr Opin Plant Biol 11: 530–535 [DOI] [PubMed] [Google Scholar]
- Wang HY, Klatte M, Jakoby M, Bäumlein H, Weisshaar B, Bauer P (2007) Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226: 897–908 [DOI] [PubMed] [Google Scholar]
- Wang N, Cui Y, Liu Y, Fan H, Du J, Huang Z, Yuan Y, Wu H, Ling HQ (2013) Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol Plant 6: 503–513 [DOI] [PubMed] [Google Scholar]
- Yi Y, Guerinot ML (1996) Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J 10: 835–844 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397 [DOI] [PubMed] [Google Scholar]
- Yuan YX, Zhang J, Wang DW, Ling HQ (2005) AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res 15: 613–621 [DOI] [PubMed] [Google Scholar]
- Zhai Z, Gayomba SR, Jung HI, Vimalakumari NK, Piñeros M, Craft E, Rutzke MA, Danku J, Lahner B, Punshon T, et al. (2014) OPT3 is a phloem-specific iron transporter that is essential for systemic iron signaling and redistribution of iron and cadmium in Arabidopsis. Plant Cell 26: 2249–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu B, Li M, Feng D, Jin H, Wang P, Liu J, Xiong F, Wang J, Wang HB (2015) The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 27: 787–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.