A software tool incorporating a range of experimental and data analysis choices in a sequencing-based forward genetics study aids in the design of an optimal procedure for gene identification.
Abstract
The Gene Identification via Phenotype Sequencing (GIPS) software considers a range of experimental and analysis choices in sequencing-based forward genetics studies within an integrated probabilistic framework, which enables direct gene cloning from the sequencing of several unrelated mutants of the same phenotype without the need to create segregation populations. GIPS estimates four measurements to help optimize an analysis procedure as follows: (1) the chance of reporting the true phenotype-associated gene; (2) the expected number of random genes that may be reported; (3) the significance of each candidate gene’s association with the phenotype; and (4) the significance of violating the Mendelian assumption if no gene is reported or if all candidate genes have failed validation. The usage of GIPS is illustrated with the identification of a rice (Oryza sativa) gene that epistatically suppresses the phenotype of the phosphate2 mutant from sequencing three unrelated ethyl methanesulfonate mutants. GIPS is available at https://github.com/synergy-zju/gips/wiki with the user manual and an analysis example.
One of the major challenges in plant research is to achieve phenotypic control in breeding practices. This objective requires an understanding of genes and their phenotypic effects. To address this objective, the reverse genetics approach is popular because it is relatively convenient to perform. However, the conventional forward genetics approach remains effective, especially in cases where the aim is to identify genes that are responsible for a predefined target phenotype, such as economical traits.
The advent of next-generation sequencing enabled fast and cost-effective genotyping, which has significantly accelerated the process of gene identification in sequencing-based forward genetics studies. In general, methods for mapping the locations of phenotype-associated mutations using genetic crosses and genome sequencing were well summarized by Schneeberger (2014). Previous sequencing-based forward genetics studies in plants (Schneeberger et al., 2009; Hong et al., 2010; Austin et al., 2011; Zhang et al., 2014) were typically based on bulked segregant analysis, whereby a mutant is crossed to a wild type to create an F2 (or a BC1F2) population. The F2 population is subsequently screened for the mutant phenotype. The mutants obtained are bulked and analyzed with whole-genome sequencing. Although this approach is much faster than the traditional map-based cloning approach for gene identification, it still requires the creation of a population, which is tedious and time consuming.
Identifying phenotype-associated genes via sequencing unrelated mutants with the same phenotype was first reported by Nordström et al. (2013). In past screenings of random mutagenesis libraries, we also obtained multiple unrelated mutants exhibiting the same phenotype, and further investigations indicated that these same-phenotype mutants harbored allelic mutations in the same gene (Chen et al., 2011). Although it is possible that multiple genes can produce similar phenotypes for a qualitative trait, it is unlikely that two genes produce exactly the same phenotype. This is because most, if not all, genes are pleiotropic. Disruptions of different genes might produce the same major effect, but each will also produce dissimilar side effects. Therefore, by sequencing several mutants of exactly the same phenotype, it is possible to perform direct gene cloning without the need to create segregation populations. This strategy will be more time efficient than bulked segregant analysis and is free from the limitations associated with the need to create a population.
The success of this sequencing-based direct cloning approach is dependent on a range of experimental and analysis choices, including the number of samples being sequenced, the genomic region being sequenced, the sequencing quality and depth, the approach to mapping the sequencing reads onto the genome, variant-calling methods, the approach to filtering unlikely functional variants, and the criterion to report candidate genes. Previous studies have characterized the impact of some of these choices (Ratan et al., 2013; Chilamakuri et al., 2014; Lelieveld et al., 2015). However, it is difficult for an investigator to design an optimal analysis procedure that integrates all the factors that may affect the chance of success by using the sequencing-based direct cloning approach. Furthermore, after the sequencing results are obtained, designing an effective analysis procedure that fits the quality of this particular set of sequencing data lacks well-defined guidance.
To meet this analytical need, we developed the Gene Identification via Phenotype Sequencing (GIPS) software. GIPS estimates four measurements to help optimize an analysis procedure that directly identifies candidate genes by sequencing several unrelated mutants of the same phenotype. These measurements are as follows: (1) the chance of the analysis procedure reporting the true phenotype-associated gene; (2) the expected number of random genes that may be reported; (3) the significance of each candidate gene’s association with the phenotype; and (4) the significance of violating the Mendelian assumption if no gene is reported or if all candidate genes have failed validation.
The use of GIPS is demonstrated by the identification of a rice (Oryza sativa) gene that epistatically suppresses the phenotype of the phosphate2 (pho2) mutant that shows symptoms of phosphate toxicity under a normal phosphate supply. In this example, three unrelated ethyl methanesulfonate (EMS) mutants exhibiting the same phenotype were sufficient for the identification of this gene.
RESULTS
GIPS version 1.0 considers single-nucleotide polymorphisms (SNPs) and small insertions/deletions in unrelated samples. In plant research, forward genetics studies are usually performed using random chemical mutagenesis, which produces unrelated lines of mutants with SNPs (Greene et al., 2003).
The impact of diverse experimental and analysis choices on the chance of an analysis procedure successfully identifying the phenotype-associated gene generally can be summarized into their impact on two sample-wise analysis effectiveness indicators: the sensitivity and specificity with which the analysis procedure under evaluation is able to detect the phenotype-causing variants for each sample. This framework allows the application of different analysis procedures on different samples, which permits fine-tuning of sample-specific analysis procedures based on sample-specific data qualities. With the sample-wise variant detection sensitivities and specificities, the combined study-wise analysis effectiveness indicators (the four measurements outlined previously) can be computed in a recursive form. The algorithm is detailed in the software manual.
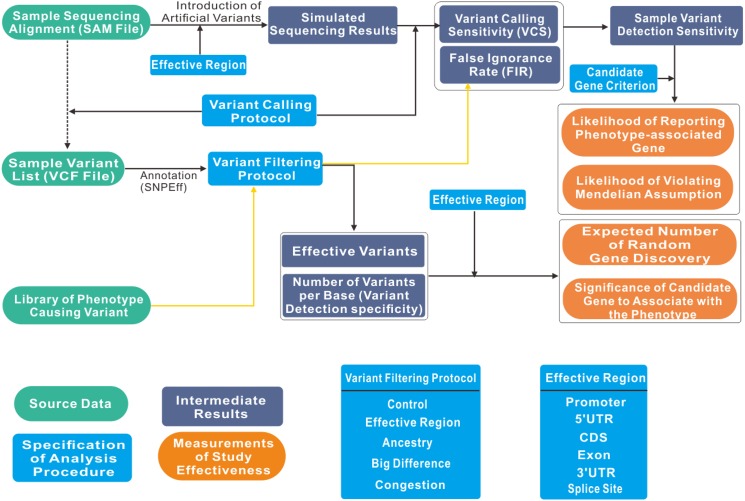
Figure 1 illustrates the general workflow of GIPS, which formally considers seven aspects of an analysis procedure that change the procedure’s chance of success in gene identification. These aspects are as follows: (1) the number of phenotype-exhibiting samples being sequenced; (2) the genomic region being sequenced; (3) the quality and depth distribution of the sequencing data; (4) the choice of software and parameters used to align the sequencing reads; (5) the choice of software and parameters to call variants; (6) the choice of strategies to filter variants that are unlikely to associate with the phenotype; and (7) the criterion to report candidate genes. The impacts of these choices can be estimated from real data or custom specified in a simulation experiment according to the belief of an investigator.
Figure 1.
GIPS workflow. The impacts of diverse experimental and analysis choices on the chance of an analysis procedure to successfully identify the phenotype-associated gene can generally be summarized into their impacts on two sample-wise analysis effectiveness indicators: the sensitivity and specificity for each sample with which the analysis procedure is able to detect the phenotype-causing variants. Sensitivity and specificity are affected by the genomic region being sequenced, the quality and depth distribution of sequencing reads, the choice of software and parameters to align sequencing reads, the choice of software and parameters to call variants, and the choice of strategies to filter variants that are unlikely to associate with the phenotype.
In general, the impact of the genomic region being sequenced and the approach to filtering unlikely causal variants on the variant detection sensitivity of each sample can be estimated by using the same approach as that used to filter a library of known phenotype-causing variants and then computing what proportion of the phenotype-causing variants will be discarded. For humans, the ClinVar database collected 19,334 Mendelian phenotype-associated variants (Landrum et al., 2014). If we assume that disruption of the same functional genomic region (i.e. promoter, exon, splice site, etc.) has the same probability of producing a phenotypic change, ClinVar also may be used as a reference library for higher plants if no appropriate library exists.
The impact of the quality and depth distribution of the sequencing data, the approach to mapping sequencing reads onto the genome, and the variant-calling methods on the variant detection sensitivity of each sample can be estimated by simulating a set of sequencing data with the same quality and depth distribution. The simulated sequencing reads are from a genome containing random artificial (simulated) variants. Therefore, the combined impact of these factors on variant detection sensitivity can be estimated by computing what proportion of the artificial SNPs is detected.
GIPS measures the sample-wise specificities of an analysis procedure by computing the frequency of detected variants per base in the effective genomic region (after all variant filtering steps) for each sample based on its actual sequencing data. Assuming that all detected variants are unrelated to the phenotype, these frequencies are used to compute how many genes are expected, by chance, to accumulate random mutations in multiple samples and pass the candidate gene criterion.
In sequencing-based direct gene cloning, the criterion to report candidate genes is typically a minimal frequency with which a candidate gene is expected to harbor variants in phenotype-exhibiting samples (i.e. M from n samples). The technique of recursive computing allows an efficient summation of probabilities over all possible combinations of M from n samples. Therefore, the four study-wise analysis effectiveness measurements outlined before can be recursively factored into terms that are computable from the sample-wise variant detection sensitivities and specificities. The full GIPS algorithm is detailed in the user manual.
The use of GIPS is demonstrated with a forward genetics study that aims to identify a rice gene that can epistatically suppress the phenotype of Ospho2, which shows symptoms of phosphate toxicity under a normal phosphate supply. Phosphate (Pi) is an essential nutrient for plant growth and development. Pi limitation is generally a constraint on crop yield in cultivated soils (Raghothama, 1999). To identify breeding practices that improve crop nutrient efficiency, it is important to understand the molecular mechanisms of Pi uptake and utilization. The mutation of PHO2 was first described in Arabidopsis. Its phenotype is an overaccumulation of Pi in shoot tissues (Delhaize and Randall, 1995). Arabidopsis PHO2 (AtPHO2) was later characterized as a ubiquitin-conjugating E2 enzyme (Liu et al., 2012). OsPHO2, the AtPHO2 homolog in rice (LOC_Os05g48390), also was identified as an important regulator in rice phosphate translocation and homeostasis, which functions similarly to AtPHO2. The Ospho2 mutant shows leaf tip necrosis and Pi accumulation in mature leaves (Wang et al., 2009; Hu et al., 2011).
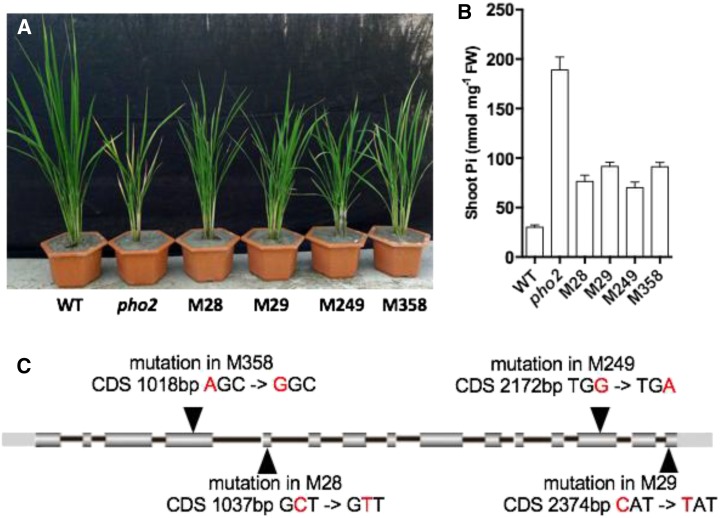
An Ospho2 Tos17 insertion mutant was obtained from the Rice Genome Resource Center, Japan (accession no. NE8536). We derived a homozygous mutant line (HNE8536) from this Ospho2 Tos17 insertion line (Wang et al., 2009). An EMS-induced mutant library was generated from the HNE8536 homozygous Ospho2 mutant line. From the M2 population of approximately 13,000 lines grown in soil, three mutants exhibiting an identical phenotype of Pi tolerance were obtained (M28, M29, and M249; Fig. 2A). Their phenotypes were further validated by measuring their shoot Pi concentrations (Fig. 2B) and by confirming their OsPHO2 Tos17 insertions (data not shown). M28, M29, and M249 seedlings exhibited similar levels of shoot Pi accumulation, which is significantly lower when compared with HNE8536 (Fig. 2B).
Figure 2.
Plant phenotypes, Pi contents, and SNPs detected in the OsPHO1;2 region. A, Phenotypes of 60-d-old plants grown in soil. WT, Wild-type cv Nipponbare; pho2, homozygous pho2 mutant (HNE8536). B, Shoot Pi contents of 40-d-old plants grown in plus-Pi (200 µm) hydroponic medium. Error bars represent se (n = 3). FW, Fresh weight, C, Variations detected in the OsPHO1;2 region. CDS, Coding sequence.
To identify the mutated gene, we sequenced the genomes of M28, M29, and M249 and used GIPS to analyze the results (Table I). As detailed in the user manual, the default analysis procedure identified 28 candidate genes, too many for experimental validation. The study effectiveness indicators computed by GIPS were used to guide the optimization of the analysis procedure. Using the optimized analysis procedure, only one candidate gene was reported (LOC_Os02g56510, or OsPHO1;2) (Table I). This candidate is a known regulator of Pi homeostasis (Secco et al., 2010) and is likely downstream of OsPHO2, as evidenced by the homologous gene function in Arabidopsis (Liu et al., 2012). The SNPs detected in OsPHO1;2 in M28, M29, and M249 were further confirmed with Sanger sequencing (data not shown).
Table I. The candidate gene (LOC_Os02g56510, OsPHO1;2) and the variants it harbors in sequenced samples.
| Sample | Position | Reference | Alternative | Effect |
|---|---|---|---|---|
| M249 | 34,614,218 | G | A | Stop gained |
| M28 | 34,611,907 | C | T | Missense (A:V) |
| M29 | 34,614,585 | C | T | Missense (H:Y) |
In Arabidopsis, the AtPHO1 gene is known to function in root-to-shoot Pi transfer. The Atpho1 mutant shows symptoms of low leaf Pi content and severe Pi deficiency because of defective Pi loading into the xylem (Poirier et al., 1991). Therefore, AtPHO1 has been considered a Pi transporter (Hamburger et al., 2002). A recent study demonstrated that AtPHO1 is a crucial component downstream of AtPHO2. AtPHO2 modulates the degradation of AtPHO1 in endomembranes (Liu et al., 2012). In the rice genome, there are three AtPHO1 homologs. Previous studies identified OsPHO1;2 as a key player in Pi homeostasis (Secco et al., 2010). Data obtained from this study further illustrate that OsPHO1;2 is epistatically downstream of OsPHO2, which suggests that the PHO2-PHO1 regulatory pathway may be conserved between the monocot and dicot plants.
Because the size of the rice genome is approximately 3 times that of the Arabidopsis genome, one of the advantages of performing a forward genetics study is the ability to determine whether there are any additional major components downstream of OsPHO2 in rice. Our results showed that all random rice mutants exhibiting the same rescuing phenotype harbored mutations in the same gene. This observation indicates the lack of other major components downstream of OsPHO2, which provides additional evidence for the conservation of the PHO2-PHO1 pathway. To further validate this conservation, we screened another 5,000 lines from the EMS-induced mutant library based on the HNE8536 homozygous Ospho2 mutant line. One additional mutant (M358), which exhibited the same Pi tolerance phenotype, was obtained. We validated the phenotype of M358 similarly by measuring its shoot Pi concentration and confirming its OsPHO2 Tos17 insertion (Fig. 2). As expected, we found a high-effect mutation, Ser-340→Gly, in OsPHO1;2 in M358 (Fig. 2).
DISCUSSION
Henry et al. (2014) showed an analysis of a broad range of unrelated mutations from a genetic screen in rice. Although this approach is not for identifying causal genes, this study demonstrated an elaborate pipeline for mutation calling. The method of identifying phenotype-associated genes via sequencing unrelated mutants exhibiting the same phenotype was first reported by Nordström et al. (2013), which used F2 populations or M3 (self-pollination) lines. This study demonstrated the possibility of direct cloning of the causal genes without creating segregation populations. In this direction, we provide a software tool that helps optimize the procedure of direct gene cloning via sequencing several unrelated same-phenotype M2 mutant lines. For plants with a long life cycle, such as rice, this approach provides a notable advantage in time.
To identify a phenotype-associated gene, a stricter or more accurate analysis procedure will only consider high-confidence variations that are supported by a significant number of high-quality sequencing reads. Additionally, the subsequent filtering process keeps only variations that are very likely to produce the target phenotype. Such an accurate procedure will identify a smaller number of possible variations that might produce the phenotype in each sample. Consequently, a candidate gene harboring variations in multiple samples will have a higher significance for association with the phenotype. However, such an accurate procedure will also risk ignoring the true phenotype-causing mutations that are not supported by unequivocal evidence. This undesired ignorance can result in the true phenotype-associated gene harboring variations in an insufficient number of samples, which will fail the candidate gene criterion.
On the other hand, an analysis procedure that is more permissive or comprehensive will identify more false-positive variations and/or more less-likely phenotype-causing variations in each sample, which results in a higher chance that random genes may harbor variations in multiple samples. When the candidate gene criterion is met, the significance of a candidate’s association with the phenotype is also lower. Although a comprehensive analysis procedure is less likely to render the true phenotype-associated gene subject to failing the candidate gene criterion, such a procedure is more likely to report phenotype-unrelated candidates, which demands significant extra effort in their validations.
In general, using an analysis procedure that is more accurate whenever possible is recommended for most investigators. Accurate procedures produce highly likely candidates, which minimizes the chance of failure in subsequent experimental validations. GIPS calculates a statistical significance (P value) for each candidate gene for association with the phenotype. If there is at least one unconfirmed candidate gene and it is not counterindicated by other evidence (e.g. evidence that some of its variations might be false-positive calls), it is advisable to validate this candidate first. In this scenario, the study effectiveness measurement chance of reporting the true phenotype-associated gene is not informative. If it is low, it just means that this analysis procedure happens to fit the need of identifying this candidate gene very well. However, if an analysis procedure produces many candidates, this procedure is probably not sufficiently accurate. Investigators are advised to try more aggressive approaches to further increase the confidence of the reported candidates.
It is advisable to use an accurate analysis procedure as long as it can identify biologically sound candidate genes for validation. However, in cases where the protocol is too strict to identify any candidate or if all identified candidates have failed validation, two study effectiveness measurements, the chance of reporting the true phenotype-associated gene and the significance of violating the Mendelian assumption, may provide guidance on the next steps. If the chance of reporting the true phenotype-associated gene is low, the analysis procedure is likely too strict and needs to be relaxed. Investigators may consider validating more candidates reported by a more permissive analysis procedure or sequencing more phenotype-exhibiting samples to increase the support for the phenotype-associated gene.
Conversely, if an investigator has sequenced a large number of phenotype-exhibiting samples and/or has validated many candidates and the phenotype-associated gene is still at large, the study effectiveness measurement, significance of violating the Mendelian assumption, may provide advice on the next steps. If this significance is low (e.g. P > 0.05), there is no compelling evidence that the phenotype is controlled by multiple genes and the investigator is still advised to add samples, validate more candidates, or relax the analysis procedure to identify more candidates. If this significance is high (e.g. P < 0.05), however, the investigator is advised to reexamine the phenotype-exhibiting samples included in the study.
As outlined previously, for a qualitative trait, although it is possible that multiple genes can produce similar phenotypes, it is unlikely that two genes will produce exactly the same phenotype. Therefore, the key to the success of a sequencing-based direct gene-cloning study is arguably the definition of a proper set of phenotype criteria that can identify mutants of the same gene. The stricter the phenotype criteria are, the more likely that the included samples are mutants of the same gene. When GIPS reports a high significance of violating the Mendelian assumption, investigators are advised to reconfirm the phenotypes of the samples included in the study. If there is no doubt, investigators are advised to consider using a stricter set of phenotype criteria for this study that examine more minor phenotypic traits and can distinguish mutants of different but functionally related genes.
In general, the approach of sequencing-based direct cloning in a forward genetics study is expected to gain popularity. The reasons are 2-fold. First, this approach does not require the generation of cross or backcross populations, which significantly accelerates the gene identification process. Although this time advantage may require more effort spent in screening mutant libraries to obtain multiple unrelated mutants of the same phenotype, library screenings in typical forward genetics studies require only bare-eye observation. The cost of screening a larger library is usually acceptable. Furthermore, the rapid development of automated phenotyping technologies facilitates screening large libraries for minor phenotypes that cannot be observed easily (Humplík et al., 2015). Second, this approach does not require the creation of a population and, therefore, is free from related limitations. This approach is readily applicable in the identification of genes that are important in organ development and reproductive development.
In this context, GIPS provides guidance on the effective design and execution of a sequencing-based direct cloning study. It is different from other gene prioritization software, such as ANNOVAR (Wang et al., 2010), which scores genes and variants to provide a rank. These priority scores do not advise, when no phenotype-associated gene can be identified, whether an investigator should change the analysis procedure, validate more candidate genes, add more samples, or reexamine the phenotype criteria used in the study. GIPS implements a probabilistic framework that models the entire process of the sequencing-based direct cloning study. Within this framework, other gene prioritization software focusing on removing genes/variants that are unlikely to associate with the phenotype can be integrated with the GIPS workflow as additional gene/variant filters.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The original pho2 Tos17 insertion mutant and wild-type rice (Oryza sativa ‘Nipponbare’) were obtained from the Rice Genome Resource Center, Japan (http://tos.nias.affrc.go.jp/). The homozygous pho2 mutant (HNE8536) was prepared as described by Wang et al. (2009). Hydroponic and soil pot experiments were performed as described by Zhou et al. (2008).
Forward Genetics Screening
An EMS-induced mutant library was generated based on HNE8536. The M2 population of approximately 13,000 lines was grown from May to October in the Agricultural Experiment Station of Zhejiang University in Changxin, Zhejiang. Twenty-five plants were planted in a 5 × 5 plot for each line. Mutants showing the identical phenotype of Pi tolerance were selected and subjected to further tests in hydroponics, as described by Wang et al. (2014). Photographs were taken for plants grown in soil in the greenhouse of Zhejiang University 60 days after planting.
Whole-Genome Sequencing
Genomic DNA was extracted from leaf tissue using the Qiagen Maxi Kit. For M29, a library of approximately 500-bp fragment size was constructed, and next-generation sequencing of 2x91-bp paired-end reads was performed by the Beijing Genomics Institute Tech Solutions. For M28, M249, and M358, libraries of approximately 500-bp fragment size were constructed, and next-generation sequencing of 2x100-bp paired-end reads was performed by the Hangzhou Guhe Info-Technology. All samples were sequenced using the Illumina HiSeq 2000 platform.
Pi Content Measurement
Shoots of wild-type, HNE8536, M29, M28, M249, and M358 plants grown in plus-Pi (200 µm) hydroponic conditions for 40 d were sampled for Pi content measurement. An inorganic Pi content measurement was performed following the method described by Zhou et al. (2008).
Raw sequencing results can be accessed at the Sequence Read Archive database with accession numbers SRS949736, SRS949738, and SRS949741.
Glossary
- GIPS
Gene Identification via Phenotype Sequencing
- EMS
ethyl methanesulfonate
- SNPs
single-nucleotide polymorphisms
- Pi
phosphate
Footnotes
This work was supported by the National Basic Research Program of China (grant nos. 2012CB944900 and 2011CB100303), the Zhejiang Provincial Natural Science Foundation of China (grant no. LR13C020001), the National Science Foundation of China (grant nos. 31571356 and 30971742), the Ministry of Agriculture of China (grant nos. 2014ZX08009–003 and 2014ZX08001–005), and the Ministry of Education Foreign Experts Bureau of China (grant no. B14027).
References
- Austin RS, Vidaurre D, Stamatiou G, Breit R, Provart NJ, Bonetta D, Zhang J, Fung P, Gong Y, Wang PW, et al. (2011) Next-generation mapping of Arabidopsis genes. Plant J 67: 715–725 [DOI] [PubMed] [Google Scholar]
- Chen J, Liu Y, Ni J, Wang Y, Bai Y, Shi J, Gan J, Wu Z, Wu P (2011) OsPHF1 regulates the plasma membrane localization of low- and high-affinity inorganic phosphate transporters and determines inorganic phosphate uptake and translocation in rice. Plant Physiol 157: 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilamakuri CSR, Lorenz S, Madoui MA, Vodák D, Sun J, Hovig E, Myklebost O, Meza-Zepeda LA (2014) Performance comparison of four exome capture systems for deep sequencing. BMC Genomics 15: 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ (1995) Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol 107: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene EA, Codomo CA, Taylor NE, Henikoff JG, Till BJ, Reynolds SH, Enns LC, Burtner C, Johnson JE, Odden AR, et al. (2003) Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 164: 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger D, Rezzonico E, MacDonald-Comber Petétot J, Somerville C, Poirier Y (2002) Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry IM, Nagalakshmi U, Lieberman MC, Ngo KJ, Krasileva KV, Vasquez-Gross H, Akhunova A, Akhunov E, Dubcovsky J, Tai TH, et al. (2014) Efficient genome-wide detection and cataloging of EMS-induced mutations using exome capture and next-generation sequencing. Plant Cell 26: 1382–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Song HR, Lutz K, Kerstetter RA, Michael TP, McClung CR (2010) Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 21211–21216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Zhu C, Li F, Tang J, Wang Y, Lin A, Liu L, Che R, Chu C (2011) LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiol 156: 1101–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humplík JF, Lazár D, Husičková A, Spíchal L (2015) Automated phenotyping of plant shoots using imaging methods for analysis of plant stress responses: a review. Plant Methods 11: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR (2014) ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42: D980–D985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelieveld SH, Spielmann M, Mundlos S, Veltman JA, Gilissen C (2015) Comparison of exome and genome sequencing technologies for the complete capture of protein-coding regions. Hum Mutat 36: 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TY, Huang TK, Tseng CY, Lai YS, Lin SI, Lin WY, Chen JW, Chiou TJ (2012) PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24: 2168–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström KJ, Albani MC, James GV, Gutjahr C, Hartwig B, Turck F, Paszkowski U, Coupland G, Schneeberger K (2013) Mutation identification by direct comparison of whole-genome sequencing data from mutant and wild-type individuals using k-mers. Nat Biotechnol 31: 325–330 [DOI] [PubMed] [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J (1991) Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol 97: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG. (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Ratan A, Miller W, Guillory J, Stinson J, Seshagiri S, Schuster SC (2013) Comparison of sequencing platforms for single nucleotide variant calls in a human sample. PLoS ONE 8: e55089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K. (2014) Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nat Rev Genet 15: 662–676 [DOI] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jørgensen JE, Weigel D, Andersen SU (2009) SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods 6: 550–551 [DOI] [PubMed] [Google Scholar]
- Secco D, Baumann A, Poirier Y (2010) Characterization of the rice PHO1 gene family reveals a key role for OsPHO1;2 in phosphate homeostasis and the evolution of a distinct clade in dicotyledons. Plant Physiol 152: 1693–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Ying S, Huang H, Li K, Wu P, Shou H (2009) Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J 57: 895–904 [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Ruan W, Shi J, Zhang L, Xiang D, Yang C, Li C, Wu Z, Liu Y, Yu Y, et al. (2014) Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc Natl Acad Sci USA 111: 14953–14958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XC, Millet Y, Ausubel FM, Borowsky M (2014) Next-gen sequencing-based mapping and identification of ethyl methanesulfonate-induced mutations in Arabidopsis thaliana. In Current Protocols in Molecular Biology. John Wiley & Sons, Hoboken, New Jersey: [DOI] [PubMed] [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]