Protein kinase CIPK23 and channel subunit AtKC1 are both crucial for the modulation of potassium channel AKT1 as well as for the response to low-potassium stress.
Abstract
In Arabidopsis (Arabidopsis thaliana), the Shaker K+ channel AKT1 conducts K+ uptake in root cells, and its activity is regulated by CBL1/9-CIPK23 complexes as well as by the AtKC1 channel subunit. CIPK23 and AtKC1 are both involved in the AKT1-mediated low-K+ (LK) response; however, the relationship between them remains unclear. In this study, we screened suppressors of low-K+ sensitive [lks1 (cipk23)] and isolated the suppressor of lks1 (sls1) mutant, which suppressed the leaf chlorosis phenotype of lks1 under LK conditions. Map-based cloning revealed a point mutation in AtKC1 of sls1 that led to an amino acid substitution (G322D) in the S6 region of AtKC1. The G322D substitution generated a gain-of-function mutation, AtKC1D, that enhanced K+ uptake capacity and LK tolerance in Arabidopsis. Structural prediction suggested that glycine-322 is highly conserved in K+ channels and may function as the gating hinge of plant Shaker K+ channels. Electrophysiological analyses revealed that, compared with wild-type AtKC1, AtKC1D showed enhanced inhibition of AKT1 activity and strongly reduced K+ leakage through AKT1 under LK conditions. In addition, phenotype analysis revealed distinct phenotypes of lks1 and atkc1 mutants in different LK assays, but the lks1 atkc1 double mutant always showed a LK-sensitive phenotype similar to that of akt1. This study revealed a link between CIPK-mediated activation and AtKC1-mediated modification in AKT1 regulation. CIPK23 and AtKC1 exhibit distinct effects; however, they act synergistically and balance K+ uptake/leakage to modulate AKT1-mediated LK responses in Arabidopsis.
As one of the essential macronutrients, potassium (K) plays crucial roles in plant development and growth by regulating cell electroneutrality, membrane potential, osmolality, and enzyme activity (Clarkson and Hanson, 1980; Leigh and Wyn Jones, 1984). K is present in plant cells in the cation (K+) form, the concentration of which is approximately 100 mm in the cytoplasm and even higher in vacuoles. However, available free K+ for plants is limited in soils, and the concentration is typically within the micromolar range (Schroeder et al., 1994; Maathuis, 2009). Therefore, plants evolved mechanisms to adapt to a suboptimal availability of K, including morphological and physiological alterations (Schachtman and Shin, 2007; Wang and Wu, 2013).
Under limited K+ conditions (below 0.5 mm), plants absorb K+ through high-affinity K+ transporters as well as some K+ channels (Epstein et al., 1963; Maathuis and Sanders, 1997; Véry et al., 2014). In Arabidopsis (Arabidopsis thaliana), the high-affinity K+ transporter HAK5 (Rubio et al., 2000; Nieves-Cordones et al., 2010) and the Shaker inward K+ channel AKT1 (Lagarde et al., 1996; Hirsch et al., 1998; Spalding et al., 1999; Ivashikina et al., 2001) have been considered two major components that conduct K+ uptake in root cells under low-K+ (LK) conditions (Gierth et al., 2005; Pyo et al., 2010; Nieves-Cordones et al., 2014). The transcription of HAK5 is induced by K+ deficiency (Gierth et al., 2005), which is an important mechanism in plant responses to LK stress (Wang and Wu, 2013). Recent reports showed that HAK5 and its ortholog in Dionaea muscipula (DmHAK5) are also regulated by the protein kinase CIPK23 (Ragel et al., 2015; Scherzer et al., 2015).
In contrast, AKT1 is subjected mainly to posttranslational regulation. The regulatory pathway of AKT1 was elucidated based on a forward genetic screening of Arabidopsis low-K+ sensitive (lks) mutants (Xu et al., 2006). LKS1 was found to encode a Ser/Thr protein kinase, CIPK23, that is induced transcriptionally by LK stress (Xu et al., 2006). CIPK23 interacts with the Ca2+-binding proteins CBL1 and CBL9 at the plasma membrane (PM), where CIPK23 phosphorylates AKT1. AKT1 is then activated and mediates K+ uptake in Arabidopsis root cells (Li et al., 2006; Xu et al., 2006). This CBL1/9-CIPK23-AKT1 pathway is an important mechanism in the Arabidopsis response to LK stress. A similar mechanism was also identified in other plant species (for review, see Véry et al., 2014), such as rice (Oryza sativa; Li et al., 2014). Subsequently, a 2C-type protein phosphatase, AIP1, was identified that interacts with and inactivates the AKT1 channel (Lee et al., 2007). Therefore, a phosphorylation/dephosphorylation regulatory mechanism for the AKT1 channel was proposed (Lee et al., 2007). In addition, the Ca2+-binding protein CBL10 interacts directly with AKT1 and negatively modulates AKT1 activity by competing with CIPK23 to bind AKT1 (Ren et al., 2013).
Heterotetramerization is considered another important mechanism for K+ channel regulation, which has been widely characterized in animals and plants (Patel et al., 1997; Ottschytsch et al., 2002; Lebaudy et al., 2008). An intact Shaker-type K+ channel consists of four identical or different α-subunits forming homomeric or heteromeric K+ channels. Heterotetramerization can generate multiform K+ channels with diverse properties in activation kinetics and voltage dependence, which may confer flexibility to cells in response to environmental stimuli. In Arabidopsis, AKT1 is modulated by another Shaker α-subunit, AtKC1, which is electrically silent (Reintanz et al., 2002). By forming the AKT1/AtKC1 heteromeric K+ channel, AtKC1 inhibits AKT1 conductance and negatively shifts AKT1 voltage dependence (Duby et al., 2008; Geiger et al., 2009; Wang et al., 2010). Thus, AtKC1 acts as a general modulator and negatively regulates many inward Shaker K+ channels in Arabidopsis (Jeanguenin et al., 2011). Furthermore, AtKC1 itself is also regulated. Two SNARE proteins, SYP121 and VAMP721, interact with AtKC1 and modulate the activity of the AKT1/AtKC1 heteromeric K+ channel at the PM (Honsbein et al., 2009; Zhang et al., 2015).
In this study, to further explore the mechanism of the CBL1/9-CIPK23-AKT1 pathway in the Arabidopsis response to LK stress, suppressors of lks1 (cipk23) were screened. We isolated the suppressor of lks1 (sls1) mutant, which can suppress the leaf chlorosis phenotype of the lks1 mutant under LK conditions. This suppression resulted from a gain-of-function mutation in AtKC1 (AtKC1D) of the lks1 mutant. AtKC1D exhibited enhanced inhibition of AKT1 channel activity and restricted K+ leakage from the AKT1 channel under LK conditions, which endowed Arabidopsis with tolerance to LK stress. In this study, we explored the link between CIPK23 and AtKC1 in modulating the AKT1-mediated LK response.
RESULTS
Isolation of the sls1 Mutant from Ethyl Methanesulfonate-Mutagenized lks1-2
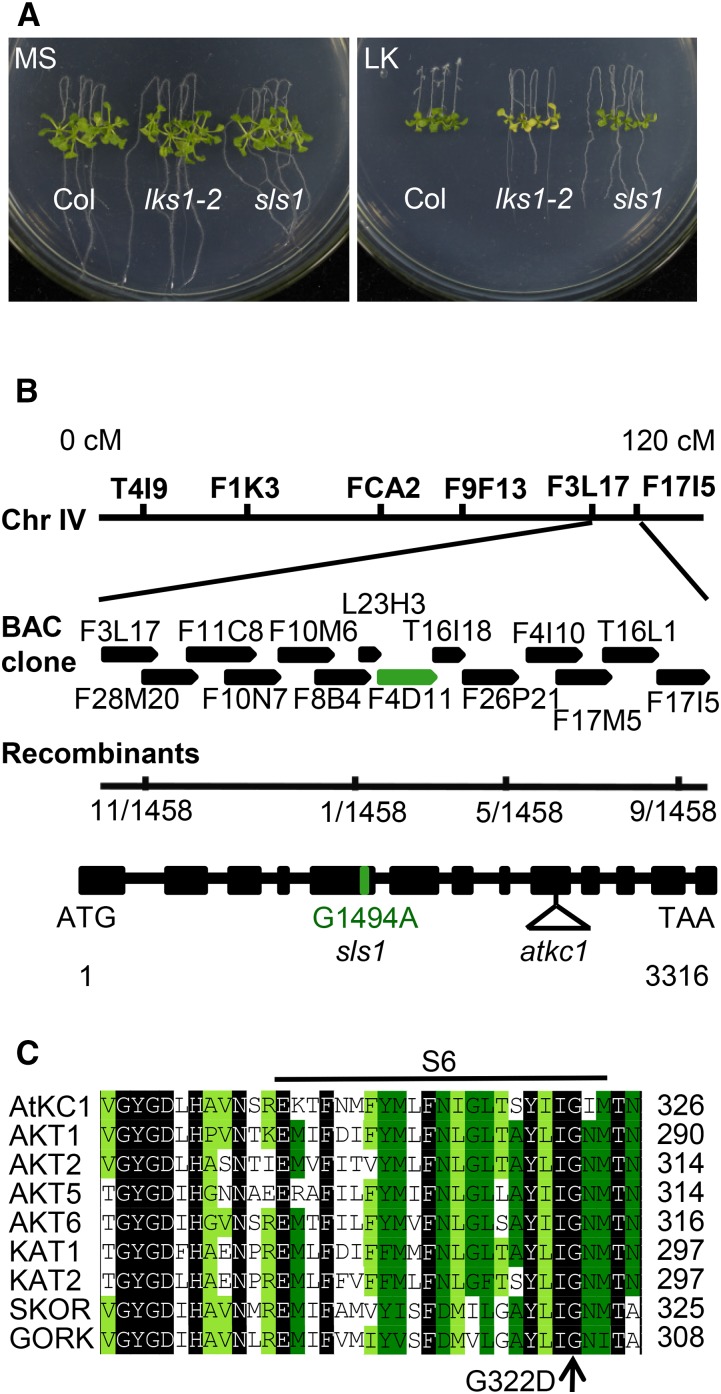
Using a forward genetic approach, our previous study isolated the lks1 mutant, which exhibits a LK-sensitive phenotype on LK (100 μm K+) medium (Xu et al., 2006). LKS1 encodes the protein kinase CIPK23. By interacting with the Ca2+-binding protein CBL1 or CBL9, LKS1/CIPK23 phosphorylates the Shaker K+ channel AKT1 and modulates AKT1-mediated K+ uptake in Arabidopsis root (Xu et al., 2006). To identify new components involved in this LKS1/CIPK23-regulated K+ uptake pathway, a mutant population was screened for sls from ethyl methanesulfonate-mutagenized lks1-2 plants (in the Columbia [Col] background). The lks1-2 mutant showed an obvious shoot chlorosis phenotype compared with the wild type after the plants were transferred to LK medium for 10 d (Xu et al., 2006). In this study, using this LK transfer assay, we screened suppressor mutants for those with a leaf phenotype similar to that of wild-type plants under LK conditions. Finally, the sls1 mutant was isolated. When grown on LK medium for 10 d, the shoots of sls1 remained green, similar to wild-type plants (Col), while the lks1-2 shoots turned yellow (Fig. 1A). To analyze the mutation in sls1, this mutant was backcrossed with lks1-2. F1 progeny showed a shoot chlorosis phenotype similar to that of lks1-2 (Supplemental Fig. S1). The shoot phenotype of F2 progeny was segregated, and the segregation ratio was 3:1 (yellow shoot:green shoot; Supplemental Fig. S1). Genetic analyses indicated that sls1 harbors a monogenic recessive mutation in a nuclear gene other than lks1-2.
Figure 1.
Phenotype assay of the sls1 mutant and map-based cloning of SLS1. A, Phenotype comparison between wild-type Arabidopsis (Col) and lks1-2 and sls1 mutants. The seeds were germinated on Murashige and Skoog (MS) medium for 4 d, after which the seedlings were transferred to MS or LK (100 μm K+) medium for 10 d. B, Map-based cloning of sls1. The SLS1 gene encodes AtKC1. In the AtKC1 gene structure depicted, the black boxes represent exons and the lines represent introns. The sites of the G1494A point mutation in sls1 and the transfer DNA insertion (SALK_140579) in atkc1 are shown. BAC, Bacterial artificial chromosome. C, Sequence alignment of Arabidopsis Shaker K+ channels using DNAMAN software. The point mutation (G322D) of sls1 is located in the S6 region of the Shaker K+ channel.
Map-based cloning was then performed using the F2 population, which included 1,458 plants from the sls1 and lks1-1 cross (in the Landsberg erecta background); the mutated gene was located within the bacterial artificial chromosome clone F4D11 on chromosome IV (Fig. 1B). Combined with next-generation mapping (Austin et al., 2011), we identified a single-nucleotide mutation (G1494A) in the AtKC1 gene (Fig. 1B) within this region (Supplemental Fig. S2). This mutation caused a change from the Gly at position 322 to Asp (G322D) in the S6 transmembrane region of AtKC1; the Gly-322 amino acid residue is highly conserved in the Shaker K+ channel family (Fig. 1C). For convenience, we named this mutant AtKC1D or AtKC1D in this study.
The Shoot Chlorosis Phenotype of lks1 Is Suppressed by AtKC1D
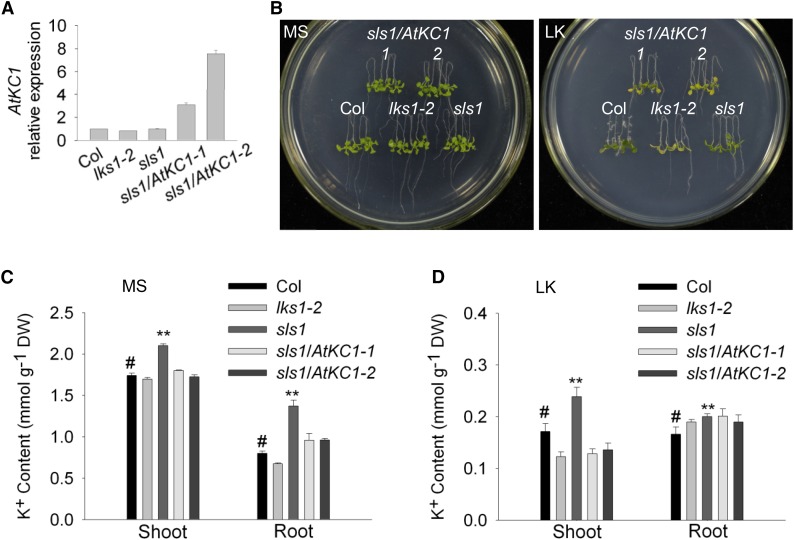
To verify that AtKC1D suppressed the shoot chlorosis phenotype of lks1, the genomic sequence of AtKC1 including the promoter and coding regions was transformed into the sls1 mutant. Two homozygous transgenic lines (sls1/AtKC1-1 and sls1/AtKC1-2) were obtained and used for the LK phenotype test. The expression levels of the AtKC1 gene in different plant materials were determined using real-time PCR (Fig. 2A). As shown in Figure 2B, the two transgenic lines exhibited a similar shoot chlorosis phenotype to that of lks1-2 on LK medium, which indicated that mutation of AtKC1 (AtKC1D) suppressed the LK-sensitive phenotype of lks1-2.
Figure 2.
AtKC1 complements the sls1 phenotype. A, Real-time PCR analysis of AtKC1 relative expression in various plant materials. ACTIN2/8 was used as the internal control. B, Phenotype assay of complementation lines of sls1. Four-day-old seedlings were transferred from MS medium to LK or MS medium and treated for 7 d. C and D, K+ content measurement for various plant materials. The K+ content was measured after the plants were transferred to MS or LK medium for 7 d. Data are shown as means ± se (n = 4). Student’s t test (#, control and **, P < 0.01) was used to analyze statistical significance. DW, Dry weight.
To confirm whether the shoot phenotype is related to K+ content, we measured K+ concentrations in different plant materials. The K+ content in lks1-2 shoot was reduced significantly compared with wild-type plants under LK conditions (Fig. 2D), which resulted in the shoot chlorosis phenotype of lks1-2 on LK medium (Fig. 2B). Surprisingly, the K+ content in the sls1 mutant was much higher than that in both lks1-2 and wild-type plants (Fig. 2, C and D). Under LK conditions, the sls1 mutant not only rescued the LK-sensitive phenotype of lks1-2 (Fig. 2B) but also showed greater accumulation of K+ in shoots compared with wild-type plants (Fig. 2D). The K+ concentrations in sls1/AtKC1-1 and sls1/AtKC1-2 were resumed and very close to the K+ content in lks1-2 (Fig. 2D), which was consistent with their shoot phenotype on LK medium (Fig. 2B).
The Phenotype of sls1 Is Independent of lks1-2
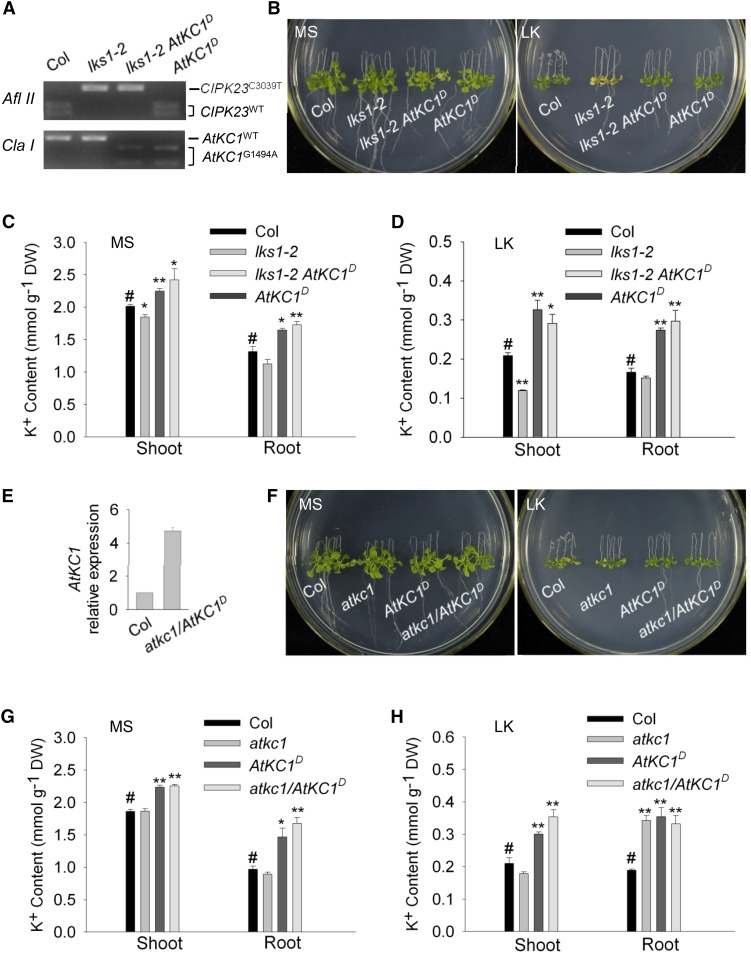
To explore the relationship between lks1-2 and AtKC1D in the sls1 (lks1-2 AtKC1D) mutant, we backcrossed sls1 with wild-type plants and isolated the AtKC1D single mutant. The genotypes of CIPK23 and AtKC1 in different plant materials were confirmed using cleaved-amplified polymorphic sequence (CAPS) markers (Fig. 3A). AtKC1D exhibited a green shoot phenotype on LK medium that was similar to that of sls1 (lks1-2 AtKC1D) (Fig. 3B). In addition, the K+ content in AtKC1D was very close to that in sls1 (Fig. 3, C and D). These results indicated that the LK-insensitive phenotype of sls1 is caused by AtKC1D and is independent of lks1-2.
Figure 3.
The phenotype of sls1 is independent of lks1-2 but dependent on AtKC1D. A, The genotypes of CIPK23 and AtKC1 in different plant materials were confirmed using CAPS markers. The restriction enzymes AflII and ClaI were used to identify the wild-type and mutant forms of CIPK23 and AtKC1, respectively. B, Phenotype comparison among the wild type (Col), lks1-2, sls1 (lks1-2 AtKC1D), and AtKC1D after growth on MS or LK medium for 10 d. C and D, K+ content measurements in the various plant materials described in B. The K+ content was measured after the plants were transferred to MS or LK medium for 10 d. Data are shown as means ± se (n = 4). Student’s t test (#, control, *, P < 0.05 and **, P < 0.01) was used to analyze statistical significance. #, control. E, Real-time PCR analysis of AtKC1 expression in the atkc1/AtKC1D complementation line. F, Phenotype comparison among the wild type (Col), atkc1, AtKC1D, and atkc1/AtKC1D after growth on MS or LK medium for 10 d. G and H, K+ contents in the various plant materials described in F. DW, Dry weight.
According to the phenotype assays (Fig. 3, B–D), AtKC1D may be a gain-of-function mutation of AtKC1. To explore this hypothesis, a loss-of-function mutant of AtKC1 (atkc1) was used as a control. atkc1 (SALK_140579) is a transfer DNA insertion mutant of AtKC1 (Fig. 1B) whose shoot phenotype was similar to that in wild-type plants in LK transfer assays (Wang et al., 2010; Fig. 3F). We generated the lks1-2 atkc1 double mutant and compared its phenotype with that of sls1 (lks1-2 AtKC1D). On LK medium, the lks1-2 atkc1 double mutant exhibited a LK-sensitive phenotype similar to that of lks1-2 (Supplemental Fig. S3), while sls1 (lks1-2 AtKC1D) showed an insensitive phenotype (Supplemental Fig. S3). These results indicated that only AtKC1D, but not atkc1, could suppress the LK-sensitive phenotype of lks1-2.
In addition, we transformed the genomic sequence of AtKC1D, including the promoter and coding regions, into the atkc1 mutant and detected its expression (Fig. 3E). The transgenic line (atkc1/AtKC1D) showed a similar phenotype to that of AtKC1D on LK medium (Fig. 3F), whose K+ content was close to that of AtKC1D (Fig. 3, G and H). In addition, when we prolonged the LK treatment to 12 d, sls1 and AtKC1D both showed a LK-tolerant phenotype (Supplemental Fig. S4). Together, these results suggest that AtKC1D is a gain-of-function mutant of AtKC1 and endows Arabidopsis with tolerance to LK stress.
AtKC1D Enhances the Arabidopsis K+ Uptake Capacity under LK Conditions
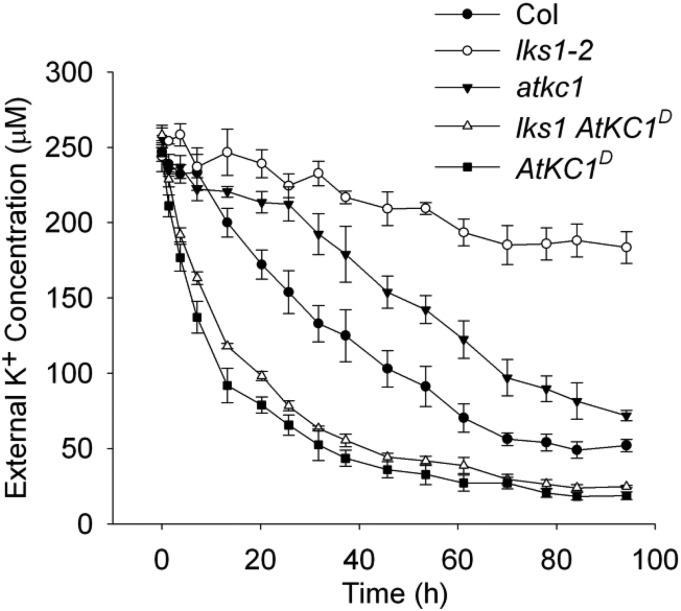
The K+ content measurement indicated that more K+ accumulated in the sls1 (lks1-2 AtKC1D) and AtKC1D mutants compared with wild-type plants under LK conditions (Fig. 3D; Supplemental Fig. S5). We assumed that the AtKC1D mutation may endow Arabidopsis with a higher K+ uptake capacity. To this end, a K+ depletion experiment in hydroponic medium was designed to imitate the LK transfer assay. Five-day-old seedlings were transferred into hydroponic medium at a LK concentration (250 μm), after which the K+ concentration in the medium was measured to reflect the K+ uptake rate in the plants. As shown in Figure 4, lks1-2 showed the lowest K+ uptake rate among all tested materials. In contrast, the K+ uptake rate of sls1 (lks1-2 AtKC1D) was restored to or even higher than wild-type plant levels. The AtKC1D mutant also exhibited a K+ uptake rate similar to that of sls1 (lks1-2 AtKC1D), while the atkc1 mutant showed a lower K+ uptake rate compared with wild-type plants (Fig. 4). These findings demonstrated that, as a gain-of-function mutation, AtKC1D enhances the K+ uptake capacity of Arabidopsis, which results in the LK-tolerant phenotype of AtKC1D and sls1 (lks1-2 AtKC1D) (Fig. 3, B and D).
Figure 4.
AtKC1D enhances Arabidopsis K+ uptake capacity under LK conditions. Comparison is shown for K+ uptake capacity among various plant materials using a K+ depletion experiment. Data are shown as means ± se (n = 3).
The Phenotype of sls1 Is Dependent on AKT1
Considering that both LKS1 and AtKC1 are regulatory components of the K+ channel AKT1, we assumed that the phenotype of sls1 (lks1-2 AtKC1D) may be relevant to AKT1 function. Among the inward-rectifying K+ channels from the Arabidopsis Shaker family, only AKT1 and AtKC1 are abundantly expressed in root tissues (Reintanz et al., 2002). As the regulatory subunit of AKT1, AtKC1 expressed alone remains in the endoplasmic reticulum (ER), but it can be recruited to the PM to regulate AKT1 activity (Duby et al., 2008; Geiger et al., 2009; Honsbein et al., 2009; Wang et al., 2010).
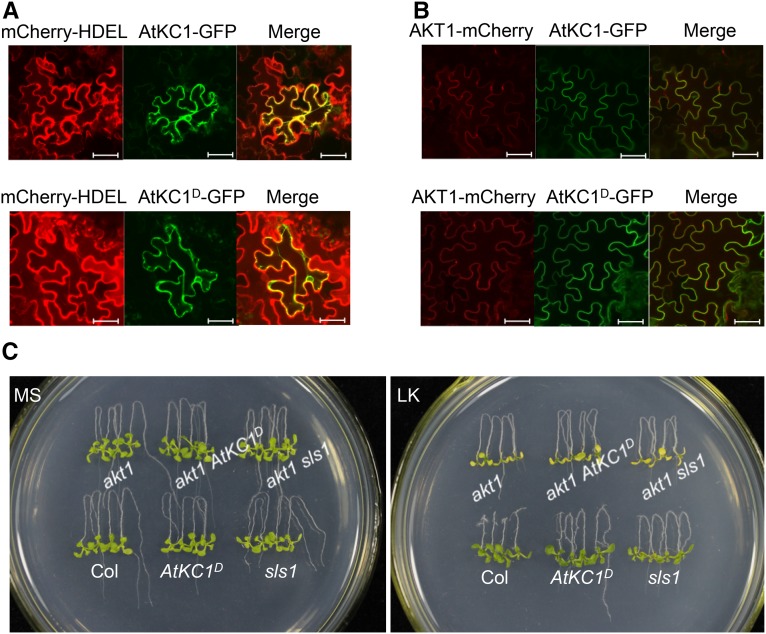
First, we explored whether the G322D point mutation affected AtKC1D localization and the interaction with AKT1. The fusion genes AtKC1-GFP, AtKC1D-GFP, and AKT1-mCherry were cloned into the SUPERR:sXVE vector (Schlücking et al., 2013) driven by an inducible promoter and then transformed into Nicotiana benthamiana leaves. The mCherry-HDEL was used as an ER marker, and empty GFP and mCherry vectors served as controls (Supplemental Fig. S6). As shown in Figure 5A, both AtKC1-GFP and AtKC1D-GFP were localized at the ER. When AtKC1D-GFP was coexpressed with AKT1-mCherry, the green and red fluorescence exhibited obvious overlap at the PM (Fig. 5B). Moreover, the interaction between AKT1 and AtKC1 (or AtKC1D) was confirmed based on bimolecular fluorescence complementation (BiFC) in Xenopus laevis oocytes (Supplemental Fig. S7). Similar to wild-type AtKC1, we found that AtKC1D also interacts with AKT1 and can be recruited to the PM by forming a heteromeric K+ channel with AKT1.
Figure 5.
The phenotypes of sls1 and AtKC1D are dependent on AKT1. A, Subcellular localization assay of AtKC1 and AtKC1D in N. benthamiana leaves. The mCherry-HDEL was used as an ER marker. Bars = 50 μm. B Interaction test between AtKC1 or AtKC1D and AKT1 in N. benthamiana leaves. Bars = 50 μm. C, Phenotype comparison among various plant materials after growth on MS or LK medium for 7 d.
To assess the AKT1 effect in AtKC1D and sls1 mutants, akt1 was crossed into these two mutants. The akt1 atkc1 double mutant also was generated and used as a control (Supplemental Fig. S8). On LK medium, akt1 showed a remarkable shoot chlorosis phenotype similar to the lks1 mutant (Xu et al., 2006). AtKC1D and sls1 were tolerant to LK treatment; however, both akt1 AtKC1D and akt1 sls1 showed LK-sensitive phenotypes similar to that of akt1 under LK conditions (Fig. 5C). These data indicated that AtKC1D could not suppress the LK-sensitive phenotype of akt1, although it rescued the lks1 phenotype. The LK-tolerant phenotype of AtKC1D relies on AKT1 function.
AtKC1D Exhibits Stronger Inhibition on AKT1 Channel Activity
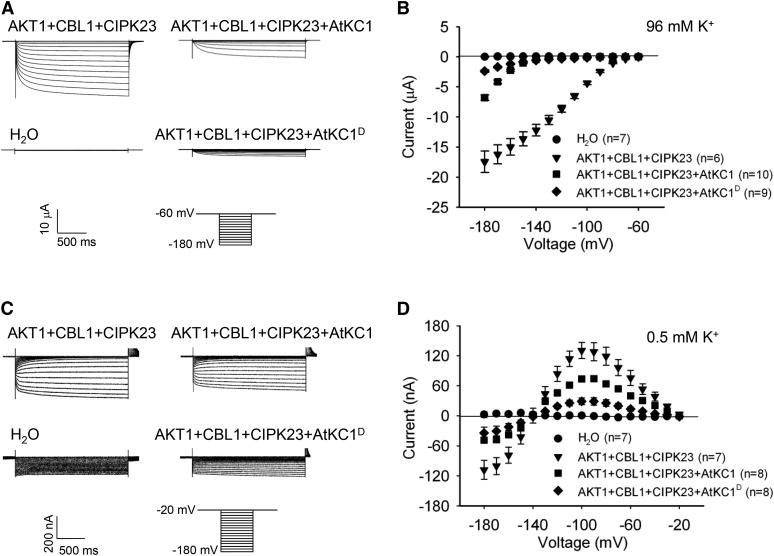
Previous reports showed that AtKC1 inhibits AKT1 activity (Geiger et al., 2009; Wang et al., 2010). In this study, the AtKC1D mutant showed enhanced K+ uptake capacity and accumulated more K+ ions under LK conditions (Fig. 4; Supplemental Fig. S5). We hypothesized that AtKC1D may relieve the inhibition on AKT1 conductance compared with wild-type AtKC1. Thus, AKT1-mediated K+ uptake would be increased in AtKC1D mutants. To explore this hypothesis, we assessed the AtKC1D effect on AKT1 channel activity in X. laevis oocytes. As shown in Figure 6A, when AKT1 and AtKC1 were coexpressed in oocytes, AtKC1 inhibited AKT1-mediated inward K+ currents under hyperpolarization conditions, which was consistent with previous reports (Geiger et al., 2009; Wang et al., 2010). However, compared with wild-type AtKC1, AtKC1D exhibited stronger inhibition on AKT1 conductance (Fig. 6, A and B) and shifted the voltage dependence of AKT1 toward a more negative direction (Table I; Supplemental Fig. S9). The inhibition of AtKC1D on AKT1 currents was dose dependent (Supplemental Fig. S10). These findings indicated that a point mutation in AtKC1D enhances its activity.
Figure 6.
AtKC1D strongly inhibits AKT1 conductance and restricts K+ leakage through AKT1 under LK conditions. A and C, Two-electrode voltage-clamp recordings in X. laevis oocytes. The oocytes were injected with distilled water as the control. The AKT1-expressing oocytes were injected with a RNA mixture of AKT1, CIPK23, and CBL1. The AKT1- and AtKC1 (or AtKC1D)-coexpressing oocytes were injected with a RNA mixture of AKT1, AtKC1 (or AtKC1D), CIPK23, and CBL1. The K+ concentrations in the bath solution were 96 mm (A) and 0.5 mm (C). The voltage protocols, as well as the time and current scale bars for the recordings, are shown. B and D, The current-voltage relationships of time-dependent steady-state K+ currents. The data were derived from the recordings shown in A and C, respectively. The instantaneous currents were subtracted using Clampfit software, and the time-dependent steady-state K+ currents were calculated for current-voltage curve plotting. Data are presented as means ± se.
Table I. Half-activation voltage analysis.
Data are shown as means ± se (n = 4).
| Ratio (1:1)a | Half-Activation Voltage |
|---|---|
| mV | |
| AKT1:water | 121.3 ± 2.2 |
| AKT1:AtKC1 | 177.6 ± 0.7 |
| AKT1:AtKC1D | 222.1 ± 8.8 |
CIPK23 and CBL1 are included in all combinations.
Previous investigations suggested that AtKC1 may inhibit K+ efflux (or leakage) through the AKT1 channel under limited K+ conditions (Geiger et al., 2009; Wang et al., 2010). In this study, we also explored the effect of AtKC1D on AKT1-mediated K+ efflux under low external K+ conditions in X. laevis oocytes. When the K+ concentration in bath solution was reduced to 0.5 mm, we observed significant outward K+ currents (K+ efflux) in AKT1-expressing oocytes (Fig. 6, C and D). This K+ efflux occurred when the test voltages were more positive than the K+ equilibrium potential (about −140 mV under this condition). AtKC1 restrained this AKT1-mediated K+ efflux in AKT1-AtKC1-coexpressing oocytes, and this inhibition was further enhanced by AtKC1D (Fig. 6, C and D). Under LK stress, this increased inhibition of K+ leakage endowed the AtKC1D mutant with enhanced K+ uptake ability compared with wild-type plants (Fig. 4). In addition, we observed that AtKC1D inhibited AKT1-mediated K+ influx when the test voltages were more negative than the K+ equilibrium potential (Fig. 6, C and D).
AtKC1 Cooperates with CIPK23 to Modulate AKT1
LKS1/CIPK23 and AtKC1 are two important regulators of the AKT1 channel. CIPK23 together with CBL1/9 activates AKT1 activity, while AtKC1 inhibits AKT1 activity. Why do plants require these two opposing regulators? How do plants balance these two forms of regulation? In this study, we constructed a double mutant to explore the relationship between CIPK23 and AtKC1 in AKT1 regulation.
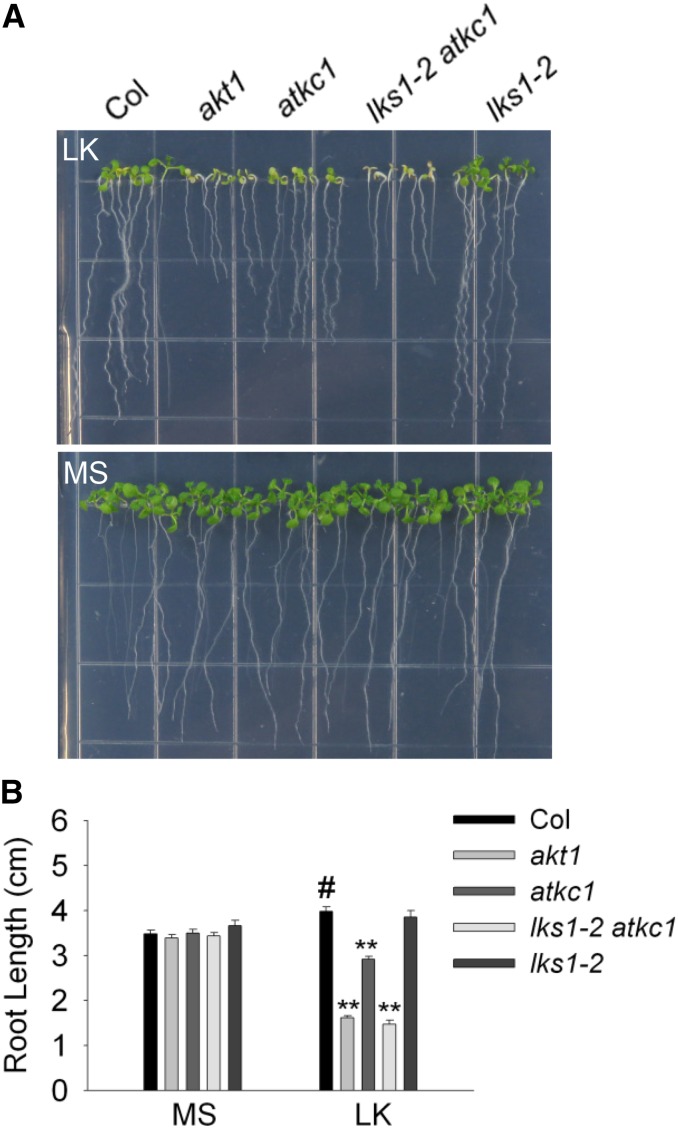
Different plant materials were germinated directly on MS or LK (100 μm) medium, and the phenotype was determined after 10 d. On LK medium, akt1 showed an extremely sensitive phenotype, including shoot chlorosis and root growth inhibition (Fig. 7). Remarkably, lks1 was not as sensitive as akt1 in this LK assay (Fig. 7), although these two mutants showed similar LK-sensitive phenotypes in the LK transfer assays (Xu et al., 2006; Supplemental Fig. S3). The loss of function of AtKC1 led to the LK-sensitive phenotype in the atkc1 mutant, whose phenotype was more sensitive than that of lks1 but less than that of akt1 (Fig. 7). However, the lks1 atkc1 double mutant exhibited a similar LK-sensitive phenotype to akt1 under this condition (Fig. 7). This suggested that CIPK23 and AtKC1 are two major components regulating AKT1 activity and that both are crucial for AKT1 function under LK stress.
Figure 7.
Phenotype assay of various plant materials using a LK germination assay. A, Phenotype assay of various plant materials after the seeds were germinated directly on MS or LK medium for 10 d. B, Root length measurement of various plant materials tested in A. Data are shown as means ± se (n = 20). Student’s t test (#, control and **, P < 0.01) was used to determine statistical significance.
DISCUSSION
Physiological Function of AtKC1 in Plant Cells
AtKC1 has been reported as a modulator of inward Shaker K+ channels (Jeanguenin et al., 2011). It inhibits AKT1 conductance and shifts the voltage dependence of AKT1 toward the negative direction (Geiger et al., 2009; Wang et al., 2010). This negative regulation impairs AKT1-mediated K+ uptake. However, it may suppress K+ leakage through AKT1 in root cells under limited K+ conditions (Geiger et al., 2009; Wang et al., 2010). This explains why disruption of AtKC1 may result in K+ loss from root cells, which leads to a reduction in the K+ uptake capacity in the atkc1 null mutant (Fig. 4). Therefore, the atkc1 mutant exhibits K+ deficiency symptoms, including growth inhibition and shoot chlorosis, when grown on LK medium (Fig. 7; Geiger et al., 2009; Honsbein et al., 2009). In this study, we isolated AtKC1D, a gain-of-function mutant of AtKC1. AtKC1D exhibits stronger inhibition of AKT1 activity compared with the wild-type AtKC1 in oocytes (Fig. 6, A and B). AtKC1D actually enhances the K+ uptake capacity in the AtKC1D mutant, possibly by impairing K+ leakage through AKT1 under LK conditions (Figs. 4 and 6, C and D). Thus, the AtKC1D mutant shows a LK-tolerant phenotype, which requires the presence of AKT1 (Fig. 5C), suggesting that AtKC1D operates via AKT1. These results demonstrate that the modulation of AtKC1 on AKT1 activity is crucial and beneficial to the LK response in Arabidopsis.
However, we also observed that the advantage of the AtKC1D mutant occurs only on LK medium and not on MS medium. The biomass of the AtKC1D mutant is even lower than that of wild-type plants when grown on MS medium (Supplemental Fig. S5). The enhanced inhibition of AKT1 activity may cause a reduction in K+ uptake and growth inhibition in the AtKC1D mutant under high-K+ conditions. Based on these results, AtKC1 activity may be finely tuned in plant cells to balance K+ uptake and K+ leakage through AKT1 channels under different K+ supply levels.
Since AtKC1 is a crucial component in the Arabidopsis response to LK stress, we wondered whether this regulation exists in other plant species. Thus, we searched available data sets for homologs of AtKC1, as well as AKT1, in several main crop species. Phylogenetic analysis revealed that AKT1-like channels exist in all species evaluated. However, AtKC1 homologs are found only in certain species (Supplemental Fig. S11). In some species, such as maize (Zea mays) and barley (Hordeum vulgare), AtKC1 homologs could not be identified (Supplemental Fig. S11). Previous electrophysiological analyses suggested that AtKC1 function in rice is weak (Fuchs et al., 2005; Li et al., 2014), although two putative AtKC1 homologs have been found in the rice genome (Supplemental Fig. S11). It is suggested that the K+ uptake mechanisms mediated by AKT1/AtKC1 homologs are diverse among different crop plants. In recent years, AKT1-like channels as well as their regulatory mechanisms have been analyzed in many crop species (summarized by Véry et al., 2014). However, studies on AtKC1 homologs in crop species are rare, excluding KDC1 in carrot (Daucus carota; Naso et al., 2006, 2009). In future studies, the functional identification of AtKC1 homologs in crop species will be important, as well the K+ uptake mechanism of crops, which will provide information for improvements in crop potassium utilization efficiency (KUE).
The Relationship between AtKC1 and CIPK23 in AKT1 Regulation
The results from LK transfer assays indicated that neither AtKC1D nor atkc1 showed significantly sensitive phenotypes on LK medium (Supplemental Fig. S8). However, when akt1 was crossed with these two mutants, the akt1 AtKC1D and akt1 atkc1 double mutants both exhibited a LK-sensitive phenotype, similar to akt1 (Supplemental Fig. S8), suggesting that AtKC1 function relies on AKT1 activity. In addition, in both the LK transfer and LK germination assays, the lks1 atkc1 double mutant always showed a similar phenotype to that of akt1 under LK conditions (Fig. 7; Supplemental Fig. S3). Therefore, CIPK23 and AtKC1 are two major components that regulate AKT1 activity.
Previous reports showed that K+ deprivation induced the transcription of CIPK23 and AtKC1 (Shin and Schachtman, 2004; Xu et al., 2006). This suggested that the transcriptional regulation of CIPK23 and AtKC1 may be important in Arabidopsis responses to LK stress. However, only the overexpression of CIPK23 enhanced the Arabidopsis K+ uptake capacity and LK tolerance (Xu et al., 2006). Combined with the recent finding that CIPK23 also activates the HAK5 transporter by phosphorylation (Ragel et al., 2015), the LK-tolerant phenotype of CIPK23-overexpressing lines may be due to enhanced K+ uptake from both AKT1 and HAK5. In contrast, the AtKC1-overexpressing lines exhibit a similar shoot phenotype to that of wild-type plants on LK medium (Wang et al., 2010). This may be because the AtKC1 transcription level is already high in roots compared with AKT1 (Reintanz et al., 2002). However, the enhancement of AtKC1 activity (AtKC1D) may result in increased K+ uptake capacity and LK tolerance in the AtKC1D mutant (Figs. 3, B and D, and 4), indicating that regulation at the posttranscriptional level also may be important for AtKC1 function.
It should be noted that various mutant plants exhibit distinct phenotypes in different LK assays. In LK transfer assays, lks1, whose phenotype is similar to that of akt1, was sensitive to LK stress (Xu et al., 2006). However, atkc1 did not show a similar LK-sensitive phenotype to those of lks1 or akt1 on LK medium (Fig. 3F; Supplemental Figs. S3 and S8). On the contrary, in LK germination assays, lks1 did not show a remarkable LK-sensitive phenotype (Fig. 7). While the atkc1 mutant is very sensitive on LK medium, the phenotype is weaker than that of akt1 (Fig. 7). Thus, CIPK23 and AtKC1 are both crucial for AKT1 regulation; however, they exhibit distinct effects and functions in different processes of the LK response. Under LK conditions, CIPK23 promotes AKT1-mediated K+ uptake, while AtKC1 inhibits AKT1-mediated K+ leakage. According to the phenotype assays, CIPK23 is more important in the response to changes in external K+ (from high to low concentrations). AtKC1 is more crucial for Arabidopsis tolerance when the plants suffer LK stress starting from the germination stage. CIPK23 and AtKC1 synergistically modulate AKT1 function and may complement each other to some extent. However, the detailed mechanism between CIPK23 and AtKC1 in planta remains unclear, and further studies are required.
The Conserved Gly in S6 May Act as a Gating Hinge in Plant Shaker Channels
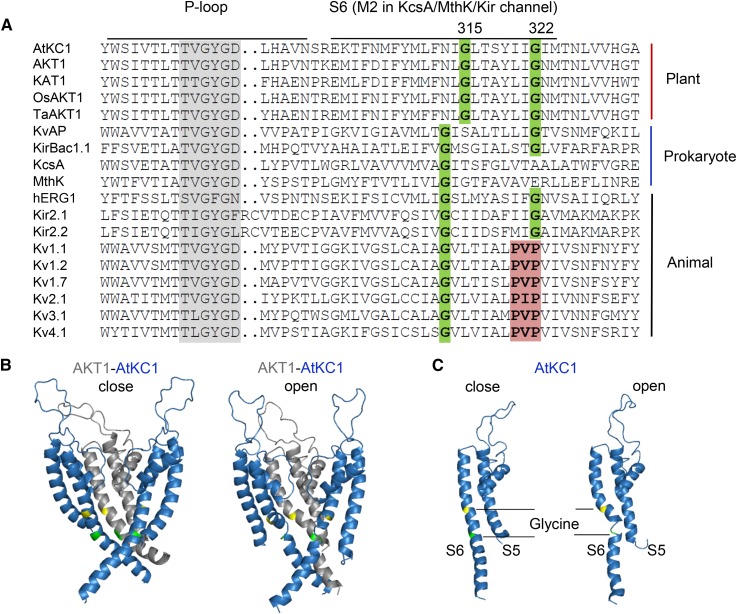
The voltage-dependent K+ selective (Kv) channels are composed of four α-subunits. Each α-subunit contains six transmembrane segments (S1–S6) and one P-loop domain, the so-called 6TM1P. The S5-P-S6 region assembles with three other pore domains into the pore structure forming the K+ permeation pathway. The opening/closing of Kv channels is controlled mainly by S6 bundle crossing. Channel opening involves the bending (tilting or swiveling) of inner S6 helices that allows them to pivot away from the central pore via a hinge-like motion and create a cytosolic opening for K+ diffusion (Labro and Snyders, 2012). The bending of S6 helices is thought to occur at a conserved Gly residue in the middle of the S6 helix (central Gly) and/or at a tandem site occupied by a Gly or a Pro-containing (PxP) motif (Fig. 8A; del Camino et al., 2000; Jiang et al., 2002). Alignment analysis demonstrated that the central Gly is highly conserved in different types of K+ channels from various species (Shealy et al., 2003). Prokaryotic 2TM1P K+ channels, such as KcsA and MthK, contain only the central Gly that acts as a gating hinge for the tilting of the M2 helix (Fig. 8A; Jiang et al., 2002). Besides the central Gly, other Kv channels possess a tandem PxP motif or a Gly more downstream in S6 (Fig. 8A). In animal Shaker-type channels, the PxP motif rather than the central Gly forms the main gating hinge that leads to the swiveling of the bottom portion of S6 during channel opening (del Camino et al., 2000; Long et al., 2005). However, the central Gly is still crucial for the bending of S6 in these channels (Magidovich and Yifrach, 2004; Ding et al., 2005).
Figure 8.
Sequence alignment of K+ selective channels and structural prediction of the AKT1/AtKC1 heteromeric channel. A, Sequence alignment of S6 or M2 regions of K+ selective channels. Two conserved Gly residues (Gly-315 and Gly-322 in AtKC1) are highlighted in green. The PxP motif is labeled in red. Sequences are as follows: KvAP (GI:14601099; Aeropyrum pernix), KirBac1.1 (GI:33357899; Burkholderia pseudomallei), KcsA (GI:21225921; Streptomyces coelicolor), MthK (GI:499179590; Methanothermobacter thermautotrophicus), hERG1/Kv11.1 (GI:4557729; Homo sapiens), Kir2.1 (GI:4504835; H. sapiens), Kir2.2 (GI:23110982; H. sapiens), Kv1.1 (GI:119395748; H. sapiens), Kv1.2 (GI:324021689; H. sapiens), Kv1.7 (GI:25952092; H. sapiens), Kv2.1 (GI:27436972; H. sapiens), Kv3.1 (GI:163792201; H. sapiens), and Kv4.1 (GI:27436981; H. sapiens). B, Structural prediction of the AKT1/AtKC1 heteromeric channel. The three subunits containing only the S5-P-S6 regions are shown for simplification. The three-dimensional structures of the closed (left) and open (right) states are shown. AKT1 and AtKC1 are labeled in gray and blue, respectively. The first conserved Gly residues (Gly-279 in AKT1 and Gly-315 in AtKC1) are highlighted in yellow, and the second conserved Gly residues (Gly-286 in AKT1 and Gly-322 in AtKC1) are highlighted in green. C, Three-dimensional structure of a single AtKC1 subunit.
According to sequence alignment, we found that plant Shaker K+ channels possess two conserved Gly residues (Gly-315 and Gly-322 in AtKC1) at each of the two putative hinge sites in the S6 region (Fig. 8A), although the first Gly is shifted two residues downstream compared with the central Gly in other K+ channels (Fig. 8A). The second Gly in plant Shaker K+ channels is located at the equivalent position of the second Pro of the PxP motif in animal Shaker-type channels (Fig. 8A). The importance of these two Gly residues has been validated in our group by isolating two point mutations in AtKC1. The G315D substitution results in an AtKC1 null mutation that abolishes the function of AtKC1 in AKT1 regulation (Wang et al., 2010). In this study, we also demonstrated that the G322D mutation alters the properties of the AKT1/AtKC1 heteromeric channel (Fig. 6; Table I) and endows Arabidopsis with tolerance to LK stress (Fig. 3; Supplemental Fig. S4).
The structural prediction of the AKT1/AtKC1 heteromeric channel indicated that the second Gly (Gly-286 in AKT1 and Gly-322 in AtKC1) may form a pivoting point for swiveling of the bottom portion of the S6 helix between the open and closed states of the channel (Fig. 8B). This Gly is flexible and breaks the S6 helix in AtKC1 during channel opening (Fig. 8C). A similar phenomenon of the S6 helix break is also observed at the PxP motif in animal Shaker-type channels (del Camino et al., 2000). These findings suggest that the second conserved Gly in the S6 region may function as a hinge point in plant Shaker channel gating.
Variations Endow Channels or Transporters with Plasticity and Diversity
Nutrient elements are absorbed and transported mainly by diversiform ion channels and transporters in plant cells, whose expression levels, as well as transport activities, are highly relevant to plant nutrient use efficiency. Mutagenesis analyses have supported that mutations in crucial regions could significantly alter the transport properties and/or activities of ion channels and transporters. Several studies in Arabidopsis have revealed that different site-specific mutants in the S6 domain cause the different gating properties and K+ dependence of Shaker K+ channels (Johansson et al., 2006; Gajdanowicz et al., 2009). Such mutations or variations endow channels or transporters with plasticity and diversity in ion transport properties and/or activities, which may be utilized in plant nutrient use efficiency improvement (Wang and Wu, 2015).
An important recent finding further supports this assumption. One single-nucleotide polymorphism (SNP) within the nitrate transporter gene NRT1.1B (OsNPF6.5) contributes to the divergence of nitrate use efficiency between two subspecies (indica and japonica) of cultivated rice (Hu et al., 2015). The NRT1.1B-indica variation enhances nitrate uptake and root-to-shoot transport in indica. Introgression of the NRT1.1B-indica allele could significantly improve the nitrogen use efficiency of the japonica variety. Therefore, the NRT1.1B-indica allele could be a crucial candidate in the improvement of nitrogen use efficiency for rice. In this study, we also found that the AtKC1D allele could enhance K+ uptake capacity and endow Arabidopsis with the LK-tolerant phenotype (Figs. 3 and 4), which may be useful for the improvement of KUE in crops.
Previous reports have shown that genotypic differences in KUE exist among different plant varieties (Glass and Perley, 1980; Zhang et al., 1999), which may result from the genotypic variations among K+ channels or transporters. We can assume that searching for natural variations in K+ channels or transporters (such as AKT1 and AtKC1) that control high-KUE traits in plants may be useful for improving crop KUE in the future.
MATERIALS AND METHODS
Mutant Isolation, Plant Materials, and Growth Conditions
To isolate suppressors of lks1, ethyl methanesulfonate-mutagenized M2 seeds of lks1-2 (Col ecotype) Arabidopsis (Arabidopsis thaliana) plants were germinated on MS medium containing 0.9% (w/v) agar and 3% (w/v) Suc at 22°C under constant illumination at 60 μmol m−2 s−1 for 4 d. M2 seedlings were then transferred to LK (100 μm K+) medium to screen for putative suppressors whose shoots remained green on LK medium after 10 d; meanwhile, lks1 showed chlorotic shoots. LK medium was prepared from MS medium with minor modifications (Xu et al., 2006). The putative suppressor mutants were grown under normal conditions in a greenhouse to harvest M3 seeds, the phenotype of which was further confirmed using M3 seedlings on LK medium. The selected M3 mutants were backcrossed with lks1, and the phenotypes of F1 and F2 generations were determined on LK medium for genetic analysis. For seed harvesting and hybridization, Arabidopsis plants were grown in a potting soil mixture (rich soil:vermiculite, 2:1, v/v) and kept in growth chambers at 22°C with illumination at 120 μmol m−2 s−1 for a 16-h daily light period. The relative humidity was approximately 70% ± 5%.
For the LK transfer assays, 4-d-old Arabidopsis seedlings were transferred from MS medium to LK or MS medium and treated for the indicated times described in figure legends. The LK medium used in the transfer assay was the same as described previously (Xu et al., 2006). For the LK germination assay, Arabidopsis seeds were germinated directly on MS or LK medium for 10 d. The LK medium used in the germination assay was the same as described previously (Liu et al., 2013). The K+ concentrations were 100 μm in both types of LK medium.
Map-Based Cloning of SLS1 and Next-Generation Sequencing Analysis
The sls1 (Col background) mutant was crossed with the lks1-1 (Landsberg erecta background) mutant to create mapping populations. A total of 1,458 individual F2 plants showing the sls1 phenotype were selected for chromosomal mapping of SLS1.
DNA samples from 26 individual F3 generation plants of the mapping population were pooled and mixed equally for resequencing. The genomic resequencing data were produced using Illumina HiSeq 2000 and mapped using The Arabidopsis Information Resource 10 (ftp://ftp.arabidopsis.org/home/tair/Sequences/whole_chromosomes/). SNPs were identified using the Monsanto Arabidopsis Polymorphism and Ler Sequence Collections (https://www.arabidopsis.org/browse/Cereon/index.jsp). The analyses of the SNP data and mutation searching were performed according to pervious methods (Austin et al., 2011).
Generation of Arabidopsis Transgenic Plants
ProAtKC1:AtKC1 was ligated into the pCAMBIA1300 vector using the full-length genomic sequence of AtKC1 containing the promoter (2,055 bp), coding region, and untranslated regions. This construct was transformed into the sls1 mutant to obtain independent complementation lines. The ProAtKC1:AtKC1D construct was generated using the QuikChange Lightning Site-Directed Mutagenesis Kit (catalog no. 210518; Agilent Technologies). It was then transformed into the atkc1 mutant to generate transgenic lines. Arabidopsis transformation with Agrobacterium tumefaciens (strain GV3101) was performed using the floral dip method (Clough and Bent, 1998). The T4 homozygous transgenic plants were used for the phenotype test. The genotype and expression level of AtKC1 in transgenic plants were detected using CAPS (Supplemental Fig. S12) and real-time PCR, respectively.
Real-Time PCR Analysis
Total RNA was extracted from Arabidopsis seedlings grown on MS medium using TRIzol reagent (Invitrogen). A total of 4 μg of DNase-treated total RNA was reverse transcribed into complementary DNA (cDNA) using random primers. The cDNA was diluted 70-fold, and 6 μL of diluted cDNA was used as the template in each well for quantitative real-time PCR analysis. The cDNA was amplified using Power SYBR Green PCR Master Mix (Applied Biosystems) on the ABI 7500 thermocycler (Applied Biosystems). The amplification reactions were performed in a total volume of 20 μL, which contained 6 μL of cDNA, 4 μL of forward and reverse primers (1 μm), and 10 μL of SYBR Green premix. The PCR program was as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The ACTIN gene was used as an internal standard for normalization of the test gene expression levels. The primers used in this experiment are listed in Supplemental Table S1. Three replications were performed for each sample. The fluorescence signal was obtained during the PCR annealing step. All experiments were repeated three times.
Subcellular Localization Analysis
The coding sequences of AtKC1, AtKC1D and AKT1 were fused to GFP or mCherry in SUPERR:sXVE:GFPC:Bar or SUPERR:sXVE:mCherryC:Bar, respectively (Schlücking et al., 2013). The constructs and empty vectors were transfected into A. tumefaciens (GV3101) via electroporation using the Bio-Rad Gene Pulser. One day after infiltration by A. tumefaciens, Nicotiana benthamiana leaves were brushed with 100 μm β-estradiol or 0.1% (v/v) ethanol (mock) to induce the expression of target genes (Schlücking et al., 2013). The fluorescence from GFP or mCherry was observed after 3 d using a confocal laser scanning microscope (LSM510; Carl Zeiss).
K+ Content Analysis
The 4-d-old Arabidopsis seedlings were transferred from MS to LK or MS medium and treated for the indicated times described in figure legends. The shoots and roots were harvested separately. The dry weight was measured as an indicator of biomass. Samples were then treated in a muffle furnace at 575°C for 5 h. The ashes were dissolved and diluted in 0.1 m HCl. The K+ concentrations were measured using the 4100-MP AES device (Agilent).
Kinetic Analysis of K+ Uptake
Arabidopsis seeds were germinated on MS medium containing 1% (w/v) agar and 3% (w/v) Suc at 22°C under constant illumination. For K+ depletion experiments, 5-d-old seedlings were collected (the fresh weight of each sample was 0.6 g) and pretreated in one-quarter-strength MS solution (5.15 mm NH4NO3, 0.375 mm MgSO4, 4.7 mm KNO3, 0.31 mm KH2PO4, 0.75 mm CaCl2, and 5.12 mm MES, pH 5.8 adjusted with Tris) at 22°C overnight. The seedlings were then transferred into K starvation solution (7.15 mm NH4NO3, 0.375 mm MgSO4, 0.31 mm NH4H2PO4, 0.75 mm CaCl2, and 5.12 mm MES, pH 5.8) for 4 d. Next, the seedlings were transferred to the K depletion solution (K starvation solution supplemented with 250 μm KCl). The experiments were conducted at 22°C in light, and all samples were shaken on a shaking table during the experiments (Xu et al., 2006; modified). The solution samples were collected at different time points indicated in Figure 4. The K+ concentrations were measured using the 4100-MP AES device (Agilent).
In Vitro Transcription and Expression in Xenopus laevis Oocytes
The coding sequences of AKT1, AtKC1, AtKC1D, CIPK23 (LKS1), and CBL1 were cloned into the pGEMHE vector. The RNAs were transcribed in vitro using the RiboMAX Large Scale RNA Production System-T7 (Promega). Oocytes were isolated from X. laevis and injected with the RNAs. The oocytes were injected with distilled water as a control. The AKT1-expressing oocytes were injected with a RNA mixture of AKT1, CIPK23, and CBL1 (4:2:2 ng in 24 nL). The AKT1- and AtKC1 (or AtKC1D)-coexpressing oocytes were injected with a RNA mixture of AKT1, AtKC1 (or AtKC1D), CIPK23, and CBL1 (96 mm K+, 4:4:2:2 ng in 24 nL; 0.5 mm K+, 4:0.8:2:2 ng in 24 nL). Prior to the voltage-clamp recordings, the injected oocytes were incubated at 17°C in a modified Barth’s solution containing (in mm) 88 NaCl, 1 KCl, 0.91 CaCl2, 0.33 Ca(NO3)2, 0.82 MgSO4, 2.4 NaHCO3, and 10 HEPES-NaOH (pH 7.5), supplemented with gentamycin (0.1 mg mL−1) and streptomycin (0.1 mg mL−1).
BiFC Assays
For the BiFC analyses, the coding sequences of AtKC1 and AtKC1D were fused to the N-terminal fragment of yellow fluorescent protein (YFP), and AKT1 was fused to the C-terminal fragment of YFP in the pGEMKN vector. Oocytes were injected with the RNA mixtures as the indicated combinations (Supplemental Fig. S7) at a ratio of 6:6 ng in 24 nL. BiFC analyses were observed after 50 h using a confocal laser scanning microscope (LSM510; Carl Zeiss).
Two-Electrode Voltage-Clamp Recordings from X. laevis Oocytes
Whole-cell recordings were performed 36 h after RNA injection. A two-electrode voltage-clamp technique was applied using a GeneClamp 500B amplifier (Molecular Devices) at room temperature (approximately 20°C). The microelectrodes were filled with 3 m KCl. The bath solution contained 96 mm KCl, 1.8 mm MgCl2, 1.8 mm CaCl2, 100 μm LaCl3, and 10 mm HEPES-NaOH (pH 7.2). The LK (0.5 mm) bath solution contained d-sorbitol to adjust the osmolality. The whole-cell currents were filtered at 1 kHz and digitized through a Digidata 1322A AC/DC converter using Clampex9.0 software (Molecular Devices).
Structural Analysis
The ion channel templates that ranked best in the open/closed conformations were chosen to model AKT1/AtKC1 structures using the HHpred tool (Söding et al., 2005). The structure of the AKT1/AtKC1 open state was modeled using MlotiK1 protein from Mesorhizobium loti (Protein Data Bank no. 4CHV, chain A) as the template (Kowal et al., 2014). The structure of the AKT1/AtKC1 closed state was modeled using MlotiK1 (Protein Data Bank no. 3BEH, chain A) as the template (Clayton et al., 2008). The tetramer structures were further built in accordance with the template’s subunit topology. The structure models were displayed using PyMOL (http://www.pymol.org/), and the conserved amino acid residues are labeled with different colors.
Sequence data for the genes described in this article can be found in The Arabidopsis Information Resource database (https://www.arabidopsis.org/index.jsp) under the following accession numbers: At4g32650 for AtKC1, At2g26650 for AKT1, At4g22200 for AKT2, At4g32500 for AKT5, At2g25600 for AKT6, At5G46240 for KAT1, At4g18290 for KAT2, At3g02850 for SKOR, At5g37500 for GORK, At1g30270 for CIPK23 (LKS1), and At4g17615 for CBL1. The information for the other genes in the sequence alignment is given in the figure legend.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Genetic analysis of the sls1 mutant.
Supplemental Figure S2. Genome-wide SNP analysis of the sls1 mapping population using the next-generation mapping technique.
Supplemental Figure S3. The atkc1 mutant could not suppress the LK-sensitive phenotype of lks1-2.
Supplemental Figure S4. AtKC1D and sls1 showed a LK-tolerant phenotype on LK medium.
Supplemental Figure S5. Total K+ content and biomass analyses of lks1 AtKC1D and AtKC1D.
Supplemental Figure S6. Subcellular localization of mCherry-HDEL and AKT1-mCherry in N. benthamiana leaves.
Supplemental Figure S7. Protein interaction between AKT1 and AtKC1 or AtKC1D in X. laevis oocytes using the BiFC method.
Supplemental Figure S8. Neither AtKC1D nor atkc1 could suppress the LK-sensitive phenotype of akt1.
Supplemental Figure S9. The open probability and cord conductance analyses of AKT1 in X. laevis oocytes.
Supplemental Figure S10. The dosage effect of AtKC1 or AtKC1D on AKT1 activity.
Supplemental Figure S11. Phylogenetic analysis of AKT1 and AtKC1 homologs in different crops.
Supplemental Figure S12. Molecular verification of various plant materials.
Supplemental Table S1. Primer sequences used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Jörg Kudla (Universität Münster) for providing the SUPERR:sXVE:GFPC:Bar and SUPERR:sXVE:mCherryC:Bar vectors, Dr. Wei-Cai Yang (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for providing the ER marker mCherry-HDEL, and Dr. Emily Limam (University of Southern California) and Dr. Dietmar Geiger (Universität Würzburg) for providing the pGEMHE and pGEMKN vectors, respectively.
Glossary
- K
potassium
- LK
low-K+
- PM
plasma membrane
- Col
Columbia
- CAPS
cleaved-amplified polymorphic sequence
- ER
endoplasmic reticulum
- BiFC
bimolecular fluorescence complementation
- MS
Murashige and Skoog
- KUE
potassium utilization efficiency
- SNP
single-nucleotide polymorphism
- Kv
voltage-dependent K+ selective
- cDNA
complementary DNA
- cRNA
copy RNA
Footnotes
This work was supported by the 973 Project (grant no. 2012CB114203 to W.-H.W.), the National Natural Science Foundation of China (grant no. 31270306 to Y.W., grant no. 30900097 to L.-M.C., and grant no. 31421062 to W.-H.W.), and the 111 Project (grant no. B06003).
Articles can be viewed without a subscription.
References
- Austin RS, Vidaurre D, Stamatiou G, Breit R, Provart NJ, Bonetta D, Zhang J, Fung P, Gong Y, Wang PW, et al. (2011) Next-generation mapping of Arabidopsis genes. Plant J 67: 715–725 [DOI] [PubMed] [Google Scholar]
- Clarkson DT, Hanson JB (1980) The mineral nutrition of higher plants. Annu Rev Plant Physiol 31: 239–298 [Google Scholar]
- Clayton GM, Altieri S, Heginbotham L, Unger VM, Morais-Cabral JH (2008) Structure of the transmembrane regions of a bacterial cyclic nucleotide-regulated channel. Proc Natl Acad Sci USA 105: 1511–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- del Camino D, Holmgren M, Liu Y, Yellen G (2000) Blocker protection in the pore of a voltage-gated K+ channel and its structural implications. Nature 403: 321–325 [DOI] [PubMed] [Google Scholar]
- Ding S, Ingleby L, Ahern CA, Horn R (2005) Investigating the putative glycine hinge in Shaker potassium channel. J Gen Physiol 126: 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duby G, Hosy E, Fizames C, Alcon C, Costa A, Sentenac H, Thibaud JB (2008) AtKC1, a conditionally targeted Shaker-type subunit, regulates the activity of plant K+ channels. Plant J 53: 115–123 [DOI] [PubMed] [Google Scholar]
- Epstein E, Rains DW, Elzam OE (1963) Resolution of dual mechanisms of potassium absorption by barley roots. Proc Natl Acad Sci USA 49: 684–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs I, Stölzle S, Ivashikina N, Hedrich R (2005) Rice K+ uptake channel OsAKT1 is sensitive to salt stress. Planta 221: 212–221 [DOI] [PubMed] [Google Scholar]
- Gajdanowicz P, Garcia-Mata C, Gonzalez W, Morales-Navarro SE, Sharma T, González-Nilo FD, Gutowicz J, Mueller-Roeber B, Blatt MR, Dreyer I (2009) Distinct roles of the last transmembrane domain in controlling Arabidopsis K+ channel activity. New Phytol 182: 380–391 [DOI] [PubMed] [Google Scholar]
- Geiger D, Becker D, Vosloh D, Gambale F, Palme K, Rehers M, Anschuetz U, Dreyer I, Kudla J, Hedrich R (2009) Heteromeric AtKC1·AKT1 channels in Arabidopsis roots facilitate growth under K+-limiting conditions. J Biol Chem 284: 21288–21295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierth M, Mäser P, Schroeder JI (2005) The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol 137: 1105–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass AD, Perley JE (1980) Varietal differences in potassium uptake by barley. Plant Physiol 65: 160–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch RE, Lewis BD, Spalding EP, Sussman MR (1998) A role for the AKT1 potassium channel in plant nutrition. Science 280: 918–921 [DOI] [PubMed] [Google Scholar]
- Honsbein A, Sokolovski S, Grefen C, Campanoni P, Pratelli R, Paneque M, Chen Z, Johansson I, Blatt MR (2009) A tripartite SNARE-K+ channel complex mediates in channel-dependent K+ nutrition in Arabidopsis. Plant Cell 21: 2859–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Wang W, Ou S, Tang J, Li H, Che R, Zhang Z, Chai X, Wang H, Wang Y, et al. (2015) Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat Genet 47: 834–838 [DOI] [PubMed] [Google Scholar]
- Ivashikina N, Becker D, Ache P, Meyerhoff O, Felle HH, Hedrich R (2001) K+ channel profile and electrical properties of Arabidopsis root hairs. FEBS Lett 508: 463–469 [DOI] [PubMed] [Google Scholar]
- Jeanguenin L, Alcon C, Duby G, Boeglin M, Chérel I, Gaillard I, Zimmermann S, Sentenac H, Véry AA (2011) AtKC1 is a general modulator of Arabidopsis inward Shaker channel activity. Plant J 67: 570–582 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R (2002) The open pore conformation of potassium channels. Nature 417: 523–526 [DOI] [PubMed] [Google Scholar]
- Johansson I, Wulfetange K, Porée F, Michard E, Gajdanowicz P, Lacombe B, Sentenac H, Thibaud JB, Mueller-Roeber B, Blatt MR, et al. (2006) External K+ modulates the activity of the Arabidopsis potassium channel SKOR via an unusual mechanism. Plant J 46: 269–281 [DOI] [PubMed] [Google Scholar]
- Kowal J, Chami M, Baumgartner P, Arheit M, Chiu PL, Rangl M, Scheuring S, Schröder GF, Nimigean CM, Stahlberg H (2014) Ligand-induced structural changes in the cyclic nucleotide-modulated potassium channel MloK1. Nat Commun 5: 3106–3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labro AJ, Snyders DJ (2012) Being flexible: the voltage-controllable activation gate of Kv channels. Front Pharmacol 3: 168–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarde D, Basset M, Lepetit M, Conejero G, Gaymard F, Astruc S, Grignon C (1996) Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J 9: 195–203 [DOI] [PubMed] [Google Scholar]
- Lebaudy A, Hosy E, Simonneau T, Sentenac H, Thibaud JB, Dreyer I (2008) Heteromeric K+ channels in plants. Plant J 54: 1076–1082 [DOI] [PubMed] [Google Scholar]
- Lee SC, Lan WZ, Kim BG, Li L, Cheong YH, Pandey GK, Lu G, Buchanan BB, Luan S (2007) A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc Natl Acad Sci USA 104: 15959–15964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh RA, Wyn Jones RG (1984) A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell. New Phytol 97: 1–13 [Google Scholar]
- Li J, Long Y, Qi GN, Li J, Xu ZJ, Wu WH, Wang Y (2014) The Os-AKT1 channel is critical for K+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex. Plant Cell 26: 3387–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Kim BG, Cheong YH, Pandey GK, Luan S (2006) A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc Natl Acad Sci USA 103: 12625–12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LL, Ren HM, Chen LQ, Wang Y, Wu WH (2013) A protein kinase, Calcineurin B-like Protein-Interacting Protein Kinase9, interacts with calcium sensor Calcineurin B-Like Protein3 and regulates potassium homeostasis under low-potassium stress in Arabidopsis. Plant Physiol 161: 266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SB, Campbell EB, Mackinnon R (2005) Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science 309: 897–903 [DOI] [PubMed] [Google Scholar]
- Maathuis FJM. (2009) Physiological functions of mineral macronutrients. Curr Opin Plant Biol 12: 250–258 [DOI] [PubMed] [Google Scholar]
- Maathuis FJM, Sanders D (1997) Regulation of K+ absorption in plant root cells by external K+: interplay of different plasma membrane K+ transporters. J Exp Bot (Spec No) 48: 451–458 [DOI] [PubMed] [Google Scholar]
- Magidovich E, Yifrach O (2004) Conserved gating hinge in ligand- and voltage-dependent K+ channels. Biochemistry 43: 13242–13247 [DOI] [PubMed] [Google Scholar]
- Naso A, Dreyer I, Pedemonte L, Testa I, Gomez-Porras JL, Usai C, Mueller-Rueber B, Diaspro A, Gambale F, Picco C (2009) The role of the C-terminus for functional heteromerization of the plant channel KDC1. Biophys J 96: 4063–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naso A, Montisci R, Gambale F, Picco C (2006) Stoichiometry studies reveal functional properties of KDC1 in plant shaker potassium channels. Biophys J 91: 3673–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieves-Cordones M, Alemán F, Martínez V, Rubio F (2010) The Arabidopsis thaliana HAK5 K+ transporter is required for plant growth and K+ acquisition from low K+ solutions under saline conditions. Mol Plant 3: 326–333 [DOI] [PubMed] [Google Scholar]
- Nieves-Cordones M, Alemán F, Martínez V, Rubio F (2014) K+ uptake in plant roots: the systems involved, their regulation and parallels in other organisms. J Plant Physiol 171: 688–695 [DOI] [PubMed] [Google Scholar]
- Ottschytsch N, Raes A, Van Hoorick D, Snyders DJ (2002) Obligatory heterotetramerization of three previously uncharacterized Kv channel α-subunits identified in the human genome. Proc Natl Acad Sci USA 99: 7986–7991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AJ, Lazdunski M, Honoré E (1997) Kv2.1/Kv9.3, a novel ATP-dependent delayed-rectifier K+ channel in oxygen-sensitive pulmonary artery myocytes. EMBO J 16: 6615–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo YJ, Gierth M, Schroeder JI, Cho MH (2010) High-affinity K+ transport in Arabidopsis: AtHAK5 and AKT1 are vital for seedling establishment and postgermination growth under low-potassium conditions. Plant Physiol 153: 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragel P, Ródenas R, García-Martín E, Andrés Z, Villalta I, Nieves-Cordones M, Rivero RM, Martínez V, Pardo JM, Quintero FJ, et al. (2015) The CBL-interacting protein kinase CIPK23 regulates HAK5-mediated high-affinity K+ uptake in Arabidopsis roots. Plant Physiol 169: 2863–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reintanz B, Szyroki A, Ivashikina N, Ache P, Godde M, Becker D, Palme K, Hedrich R (2002) AtKC1, a silent Arabidopsis potassium channel α-subunit modulates root hair K+ influx. Proc Natl Acad Sci USA 99: 4079–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XL, Qi GN, Feng HQ, Zhao S, Zhao SS, Wang Y, Wu WH (2013) Calcineurin B-like protein CBL10 directly interacts with AKT1 and modulates K+ homeostasis in Arabidopsis. Plant J 74: 258–266 [DOI] [PubMed] [Google Scholar]
- Rubio F, Santa-María GE, Rodríguez-Navarro A (2000) Cloning of Arabidopsis and barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiol Plant 109: 34–43 [Google Scholar]
- Schachtman DP, Shin R (2007) Nutrient sensing and signaling: NPKS. Annu Rev Plant Biol 58: 47–69 [DOI] [PubMed] [Google Scholar]
- Scherzer S, Böhm J, Krol E, Shabala L, Kreuzer I, Larisch C, Bemm F, Al-Rasheid KA, Shabala S, Rennenberg H, et al. (2015) Calcium sensor kinase activates potassium uptake systems in gland cells of Venus flytraps. Proc Natl Acad Sci USA 112: 7309–7314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder JI, Ward JM, Gassmann W (1994) Perspectives on the physiology and structure of inward-rectifying K+ channels in higher plants: biophysical implications for K+ uptake. Annu Rev Biophys Biomol Struct 23: 441–471 [DOI] [PubMed] [Google Scholar]
- Schlücking K, Edel KH, Köster P, Drerup MM, Eckert C, Steinhorst L, Waadt R, Batistič O, Kudla J (2013) A new β-estradiol-inducible vector set that facilitates easy construction and efficient expression of transgenes reveals CBL3-dependent cytoplasm to tonoplast translocation of CIPK5. Mol Plant 6: 1814–1829 [DOI] [PubMed] [Google Scholar]
- Shealy RT, Murphy AD, Ramarathnam R, Jakobsson E, Subramaniam S (2003) Sequence-function analysis of the K+-selective family of ion channels using a comprehensive alignment and the KcsA channel structure. Biophys J 84: 2929–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Schachtman DP (2004) Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc Natl Acad Sci USA 101: 8827–8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33: W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding EP, Hirsch RE, Lewis DR, Qi Z, Sussman MR, Lewis BD (1999) Potassium uptake supporting plant growth in the absence of AKT1 channel activity: inhibition by ammonium and stimulation by sodium. J Gen Physiol 113: 909–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Véry AA, Nieves-Cordones M, Daly M, Khan I, Fizames C, Sentenac H (2014) Molecular biology of K+ transport across the plant cell membrane: what do we learn from comparison between plant species? J Plant Physiol 171: 748–769 [DOI] [PubMed] [Google Scholar]
- Wang Y, He L, Li HD, Xu J, Wu WH (2010) Potassium channel α-subunit AtKC1 negatively regulates AKT1-mediated K+ uptake in Arabidopsis roots under low-K+ stress. Cell Res 20: 826–837 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wu WH (2013) Potassium transport and signaling in higher plants. Annu Rev Plant Biol 64: 451–476 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wu WH (2015) Genetic approaches for improvement of the crop potassium acquisition and utilization efficiency. Curr Opin Plant Biol 25: 46–52 [DOI] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH (2006) A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125: 1347–1360 [DOI] [PubMed] [Google Scholar]
- Zhang B, Karnik R, Wang Y, Wallmeroth N, Blatt MR, Grefen C (2015) The Arabidopsis R-SNARE VAMP721 interacts with KAT1 and KC1 K+ channels to moderate K+ current at the plasma membrane. Plant Cell 27: 1697–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Chen J, Eshetu AT (1999) Genotypic variation for potassium uptake and utilization efficiency in wheat. Nutr Cycl Agroecosyst 54: 41–48 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.