Six Arabidopsis LLM-domain B-GATA transcription factors control greening, hypocotyl elongation, phyllotaxy, floral organ initiation, branching, flowering time and senescence.
Abstract
Leu-Leu-Met (LLM)-domain B-GATAs are a subfamily of the 30-membered GATA transcription factor family from Arabidopsis. Only two of the six Arabidopsis LLM-domain B-GATAs, i.e. GATA, NITRATE-INDUCIBLE, CARBON METABOLISM-INVOLVED (GNC) and its paralog GNC-LIKE/CYTOKININ-RESPONSIVE GATA FACTOR1 (GNL), have already been analyzed with regard to their biological function. Together, GNC and GNL control germination, greening, flowering time, and senescence downstream from auxin, cytokinin (CK), gibberellin (GA), and light signaling. Whereas overexpression and complementation analyses suggest a redundant biochemical function between GNC and GNL, nothing is known about the biological role of the four other LLM-domain B-GATAs, GATA15, GATA16, GATA17, and GATA17L (GATA17-LIKE), based on loss-of-function mutant phenotypes. Here, we examine insertion mutants of the six Arabidopsis B-GATA genes and reveal the role of these genes in the control of greening, hypocotyl elongation, phyllotaxy, floral organ initiation, accessory meristem formation, flowering time, and senescence. Several of these phenotypes had previously not been described for the gnc and gnl mutants or were enhanced in the more complex mutants when compared to gnc gnl mutants. Some of the respective responses may be mediated by CK signaling, which activates the expression of all six GATA genes. CK-induced gene expression is partially compromised in LLM-domain B-GATA mutants, suggesting that B-GATA genes play a role in CK responses. We furthermore provide evidence for a transcriptional cross regulation between these GATAs that may, in at least some cases, be at the basis of their apparent functional redundancy.
Transcription factors of the GATA family contain a type IV zinc-finger DNA-binding domain (C-X2-C-X17-20-C-X2-C), which is predicted to bind the consensus DNA sequence WGATAR (where W is T, or A and R is G or A) (Reyes et al., 2004). The genomes of higher plants encode approximately 30 GATA transcription factors, which are classified as A-, B-, C-, and D-GATAs according to their primary amino-acid sequence, the conservation of their GATA DNA-binding domains, the conservation of protein domains outside of the GATA DNA-binding domain, and their exon-intron structures (Reyes et al., 2004). We recently showed that the Arabidopsis B-GATAs can be further subdivided into two structural subfamilies with either a HANABA TARANU (HAN)- or a Leu-Leu-Met (LLM)-domain located N- or C-terminally of the GATA DNA-binding domain (Behringer et al., 2014; Behringer and Schwechheimer, 2015). The HAN-domain was initially identified as a conserved domain in the floral morphology regulator HAN (Zhao et al., 2004; Nawy et al., 2010; Zhang et al., 2013); the LLM-domain was named after the invariant LLM motif in the conserved C terminus of the respective B-GATAs (Richter et al., 2010; Behringer et al., 2014).
A recent comparative analysis of B-GATAs from Arabidopsis, Solanum lycopersicon (tomato), Hordeum vulgare (barley), and Brachipodium distachyon (Brachipodium) revealed that HAN-domain and LLM-domain B-GATAs have different biological and biochemical activities: the expression of HAN-domain B-GATAs under the control of an LLM-domain B-GATA promoter could not complement LLM-domain B-GATA loss-of-function mutants and the overexpression of HAN- and LLM-domain B-GATAs in Arabidopsis differentially affected plant growth (Behringer et al., 2014; Behringer and Schwechheimer, 2015). The existence of the Brassicaceae-specific GATA23, which has a degenerate LLM-domain, as well as the existence of monocot-specific HAN-domain B-GATAs, reveal that expansions of the B-GATA family have been used during plant evolution to control specific aspects of plant development (De Rybel et al., 2010; Whipple et al., 2010; Behringer and Schwechheimer, 2015).
The Arabidopsis LLM-domain B-GATA family is comprised of GATA15, GATA16, GATA17, and GATA17L (GATA17-LIKE) as well as GNC (GATA, NITRATE-INDUCIBLE, CARBON METABOLISM-INVOLVED; GATA21; hitherto GNC) and GNL (GNC-LIKE/CYTOKININ-RESPONSIVE GATA FACTOR1; hithertobGNL) (Behringer et al., 2014; Behringer and Schwechheimer, 2015). Among these, the paralogous GNC and GNL are already well characterized with regard to their biological function and their upstream regulation: gnc and gnc gnl mutants are defective in greening and chloroplast biogenesis, and they germinate and flower slightly earlier than the wild type (Bi et al., 2005; Richter et al., 2010; 2013a,b; Chiang et al., 2012). Conversely, overexpression of the two GATAs results in a strong increase in chlorophyll accumulation and delayed germination and flowering (Richter et al., 2010; 2013a,b; Behringer et al., 2014). In addition, GNC and GNL overexpression promotes hypocotyl elongation in light-grown seedlings, alterations in leaf shape, and an increased angle between the primary and lateral inflorescences (Behringer et al., 2014). Importantly, deletion or mutation of the LLM-domain of GNC or GNL compromises some but not all phenotypes when these deletion or mutant variants are overexpressed in Arabidopsis and the effects of the overexpression transgenes are compared to the effects of the overexpression of the wild-type proteins (Behringer et al., 2014). The fact that the overexpression of every LLM-domain B-GATA factor tested to date results in highly comparable phenotypes, suggested that LLM-domain B-GATAs have conserved biochemical functions (Behringer et al., 2014).
A range of studies have already addressed the upstream signaling pathways controlling GNC and GNL expression: GNC was initially identified as a nitrate-inducible gene with an apparent role in the control of greening (Bi et al., 2005). GNL was originally identified based on its strong CK- (cytokinin-)regulated gene expression and therefore designated CYTOKININ-RESPONSIVE GATA FACTOR1 (Naito et al., 2007). The expression of GNC and GNL is also controlled by the DELLA regulators of the GA signaling pathway as well as by the light-labile PHYTOCHROME INTERACTING FACTORS (PIFs) of the phytochrome signaling pathway, which are themselves negatively regulated by DELLAs (Richter et al., 2010; Leivar and Monte, 2014). Furthermore, the auxin signaling regulators AUXIN RESPONSE FACTOR2 and AUXIN RESPONSE FACTOR7, the flowering regulator SUPPRESSOR-OF-OVEREXPRESSION-OF-CONSTANS1, and the floral development regulators APETALA3 and PISTILLATA have been implicated in GNC and GNL transcription control (Mara and Irish, 2008; Richter et al., 2010; 2013a,b). Thus, multiple signaling pathways converge on GNC and GNL expression and the two genes control a broad range of developmental responses in plants.
To date, nothing is known about the regulation and biological function of the four Arabidopsis LLM-domain B-GATAs, e.g. GATA15, GATA16, GATA17, and GATA17L. Here, we examine the entire LLM-domain B-GATA gene family through the analysis of single gene insertion mutants and a range of mutant combinations. These analyses reveal mainly overlapping roles for these B-GATAs in the control of previously known B-GATA-regulated processes such as greening, hypocotyl elongation, flowering time, and senescence, but they also reveal previously unknown functions of LLM-domain B-GATAs in phyllotactic patterning, floral organ initiation, and accessory meristem formation. Several of these responses may be mediated by CK signaling and CK treatments induce the expression of all six GATA genes. CK-induced gene expression is partially compromised in LLM-domain B-GATA mutants, suggesting that B-GATA genes play a role in CK responses. Furthermore, we provide evidence for a cross regulation of the different LLM-domain B-GATA genes at the transcriptional level.
RESULTS
Evolutionary Conservation of the LLM-Domain B-GATAs within the Brassicaceae
The Arabidopsis genome encodes six LLM-domain GATA transcription factors, namely GNC (AtGATA21), GNL (AtGATA22), GATA15 (AtGATA15), GATA16 (AtGATA16), GATA17 (AtGATA17), and GATA17L (AtGATA17-LIKE) (Behringer et al., 2014; Behringer and Schwechheimer, 2015). Phylogenetic analyses showed that these proteins can be grouped into three pairs of highly homologous proteins comprised of the, with regard to the length of their N termini, long B-GATAs GNC and GNL and the short B-GATAs GATA15 and GATA16 as well as GATA17 and GATA17L, respectively (Fig. 1, A and B; Supplemental Fig. S1; Behringer et al., 2014; Behringer and Schwechheimer, 2015). It had previously become apparent that the respective subfamilies of long and short B-GATAs were conserved in different plant species but that gene duplications in individual subfamilies may have led to a differential expansion of these subfamilies in different species (Behringer et al., 2014). To gain an insight into the conservation of these GATAs within the family of Brassicaceae, we searched for LLM-domain B-GATAs in the fully sequenced genomes of Arabidopsis lyrata, Capsella rubella, Eutrema salsugineum, and Brassica rapa. We retrieved orthologs for each of the six Arabidopsis B-GATAs within each Brassicaceae species (Supplemental Figs. S1 and S2). Whereas the Arabidopsis lyrata, Capsella rubella, and Eutrema salsugineum genomes contained exactly one apparent ortholog for each of the six Arabidopsis genes, the Brassica rapa genome, known to have undergone hexaploidization followed by genome fractionation, retained at least two orthologs each for GNC, GNL, GATA17, and GATA17L and one ortholog of GATA15 and GATA16 (Supplemental Figs. S1 and S2; Wang et al., 2011; Tang et al., 2012). In summary, we concluded that the six LLM-domain B-GATAs are conserved in the different Brassicaceae examined here, and that four members of the gene family were present in multiple copies in Brassica rapa.
Figure 1.
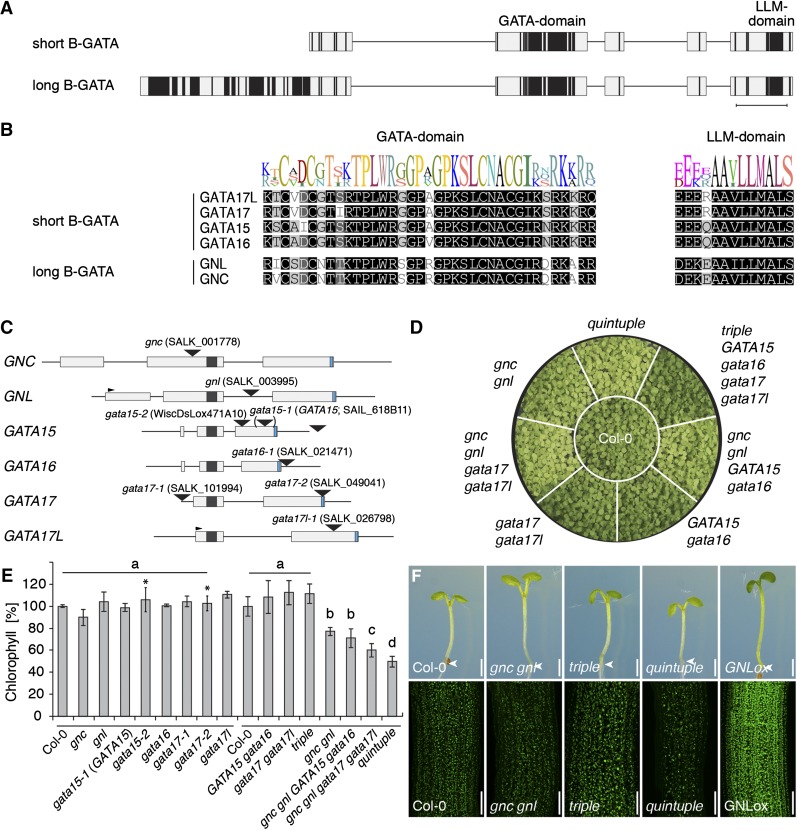
LLM-domain B-GATAs redundantly control greening in Arabidopsis. A, Domain architecture of short and long LLM-domain B-GATAs based on the alignment of family members from Arabidopsis. Protein regions with restricted sequence conservation or gaps in the alignment are depicted as lines; regions with low (≤50%) sequence similarity, gray boxes; regions with high (≥80%) sequence similarity, black boxes. Scale bar = 30 amino acids in a conserved part of the protein. B, MUSCLE alignment with sequence logo of the B-GATA domain and the LLM-domain of the Arabidopsis LLM-domain B-GATAs. C, Schematic representation of the gene models of the six Arabidopsis LLM-domain B-GATA genes with exons (boxes) and introns (lines). The 5′-UTR (untranslated region) of GNL and the 3′-UTR of GATA16 were previously unknown and derived from RNA sequencing data available in the laboratory. The sequences coding for the GATA DNA-binding domain and the LLM-domain are represented as black and blue boxes, respectively. The T-DNA insertion positions are shown. The position in brackets of the GATA15 insertion refers to the original misannotation of this insertion (www.signal.salk.edu). D, Photograph of 14-d-old plants with the genotypes as specified in comparison to the Columbia-0 (Col-0) wild-type control. E, Result of the quantification of chlorophyll A and B from different gata single and complex mutants. The experiment was performed in two rounds, as indicated by the lines above the genotypes, to reduce the complexity of the experiment; the Col-0 wild-type reference was included in both rounds. F, Representative photographs of 7-d-old seedlings (top panel) and confocal images (bottom panel) of these seedlings to visualize differential chloroplast accumulation in these mutants.
Characterization of Insertion Mutants of the Six LLM-Domain B-GATA Genes
All previously published genetic analyses of the LLM-domain B-GATAs genes from Arabidopsis focused on GNC and GNL (Behringer and Schwechheimer, 2015). As yet, no mutants have been described for the remaining four LLM-domain B-GATAs and their biological function remains elusive. We therefore identified homozygous lines of T-DNA insertion mutants for each of the six genes (Fig. 1C; Supplemental Fig. S3) and subsequently analyzed these lines with regard to the effects of the respective mutations on gene expression using quantitative real-time PCR (qRT-PCR) and semi-qRT-PCR (Supplemental Fig. S3B): The analysis of the gnc allele (Salk_001778), which had already been used in several publications, suggested that neither the full-length transcript nor regions downstream from the insertion site were transcribed (Fig. 1C; Supplemental Fig. S3B) (Richter et al., 2010; Bi et al., 2005). Similarly, analyses of the gnl insertion mutant (Salk_003995) using primers spanning the entire transcript or flanking the T-DNA harbored in the intron suggested that the GNL full-length transcript was absent (Fig. 1C; Supplemental Figure S3B) (Bi et al., 2005; Richter et al., 2010). Although one of the selected GATA15 insertion alleles (gata15-1, SAIL_618_B11) was annotated as an in-gene insertion (www.signal.salk.edu), we found no evidence for a change of GATA15 gene expression in the homozygous insertion line (Fig. 1C; Supplemental Fig. S3B). Sequencing of the insertion site then revealed that the insertion was downstream from the gene’s 3′-UTR (untranslated region), indicating that the GATA15 gene was intact in SAIL_618_B11 (Fig. 1C; Supplemental Fig. S3B). The analysis of a second allele (gata15-2, WiscDsLox471A10) with an exon-insertion revealed a strong down-regulation of GATA15 expression in both RT-PCR analyses (Fig. 1C; Supplemental Fig. S3B). In a GATA16 allele with a T-DNA insertion in the gene’s 3′-UTR (gata16-1, Salk_021471), transcript abundance was partially reduced (Fig. 1C; Supplemental Fig. S3B). The abundance of GATA17 was also partially reduced in Salk_101994 (gata17-1), which harbored a T-DNA insertion in the gene’s 5′-UTR and was strongly reduced in SALK_049041 (gata17-2), which carried an insertion in the LLM-domain-encoding gene region (Fig. 1C; Supplemental Fig. S3B). Finally, we detected no transcript in our analyses of a GATA17L exon insertion allele (gata17l-1, Salk_026798) when using primer pairs spanning the insertion or located downstream from the insertion site (Fig. 1C; Supplemental Fig. S3B). When analyzing this allele with a primer combination upstream of the insertion, we were able to detect residual transcript in gata17l-1 but judged that this may not give rise to a fully functional protein since the insertion was upstream of the LLM-domain (Fig. 1C; Supplemental Fig. S3B). We concluded that the expression of the respective GATA genes was absent or at least strongly reduced in alleles for GNC (gnc), GNL (gnl), GATA15 (gata15-2), GATA17 (gata17-2), and GATA17L (gata17l-1) alleles. The gata16-1 and gata17-1 alleles were only partially impaired in the expression of the respective genes whereas the gata15-1 allele was not affected in GATA15 expression. The gata15-2 and gata17-2 alleles became available to us only later in this study and, for strategic reasons, could not be included in the generation of the complex mutant combinations described below. Any of the phenotypes described in the following sections for the more complex mutant combinations were not apparent in the gata15-2 and gata17-2 single mutants. We, therefore, exclude the possibility that these genes are by themselves responsible for any of the phenotypes described here for the higher-order mutants.
LLM-Domain B-GATAs Redundantly Control Greening
We did not observe any obvious defects in the gata single mutants apart from the already well-documented defects in greening of the gnc mutants (Bi et al., 2005; Richter et al., 2010; Chiang et al., 2012). To generate higher-order mutants in the closely related B-GATA genes, we combined the alleles gnc, gnl, gata15-1, gata16-1, gata17-1, and gata17l-1. For simplicity, we will subsequently omit the respective allele numbers. Since the presence of the misannotated gata15-1 mutation does not provide information about the loss of GATA15, we refer to this allele as GATA15.
Through genetic crosses, we obtained gata17 gata17l double mutants, GATA15 gata16 gata17 gata17l, gnc gnl GATA15 gata16, and gnc gnl gata17 gata17l triple and quadruple mutants as well as a gnc gnl GATA15 gata16 gata17 gata17l quintuple mutant. In the following, we will refer to the gata quintuple mutant as the quintuple mutant and to the GATA15 gata16 gata17 gata17l triple mutant that does not include the gnc and gnl alleles as the (complementary) triple mutant. In all other cases, we will specify the allele combinations.
To examine a possible redundant function among the B-GATAs in the control of greening, we analyzed greening and chlorophyll content in 14-d-old light-grown gata mutant plants (Fig. 1, D and E). Although there was no indication for a defect in greening in gata17 gata17l or in the triple mutant, we found that the greening defect of gnc gnl was further enhanced in the gnc gnl gata17 gata17l quadruple mutant and even more in the quintuple mutant (Fig. 1, D and E). At the same time, greening defects were not obvious in any of the gata single mutants (Fig. 1E). In the case of the gata15 and gata17 alleles, this was not due to the fact that weak alleles had been chosen for the complex mutants because also the severe alleles gata15-2 and gata17-2 showed no reduction in chlorophyll levels when compared to the wild type (Fig. 1E).
The further reduction in chlorophyll accumulation in the quintuple mutant was also apparent when we analyzed chlorophyll abundance by fluorescence microscopy; here, we found reduced chloroplast fluorescence when comparing the quintuple mutant to gnc gnl (Fig. 1F). Furthermore, we noted that the greening defect gradually disappeared in gnc gnl mutants after the seedling stage; however, it remained apparent in the quintuple mutants in adult plants (Supplemental Fig. S4). Taken together, this suggested that all mutated GATA genes participate in the control of greening and chlorophyll accumulation and that the defects of mutants defective in only a single gene may be suppressed by the presence of the other family members.
Gene Expression Regulation of the LLM-Domain B-GATAs
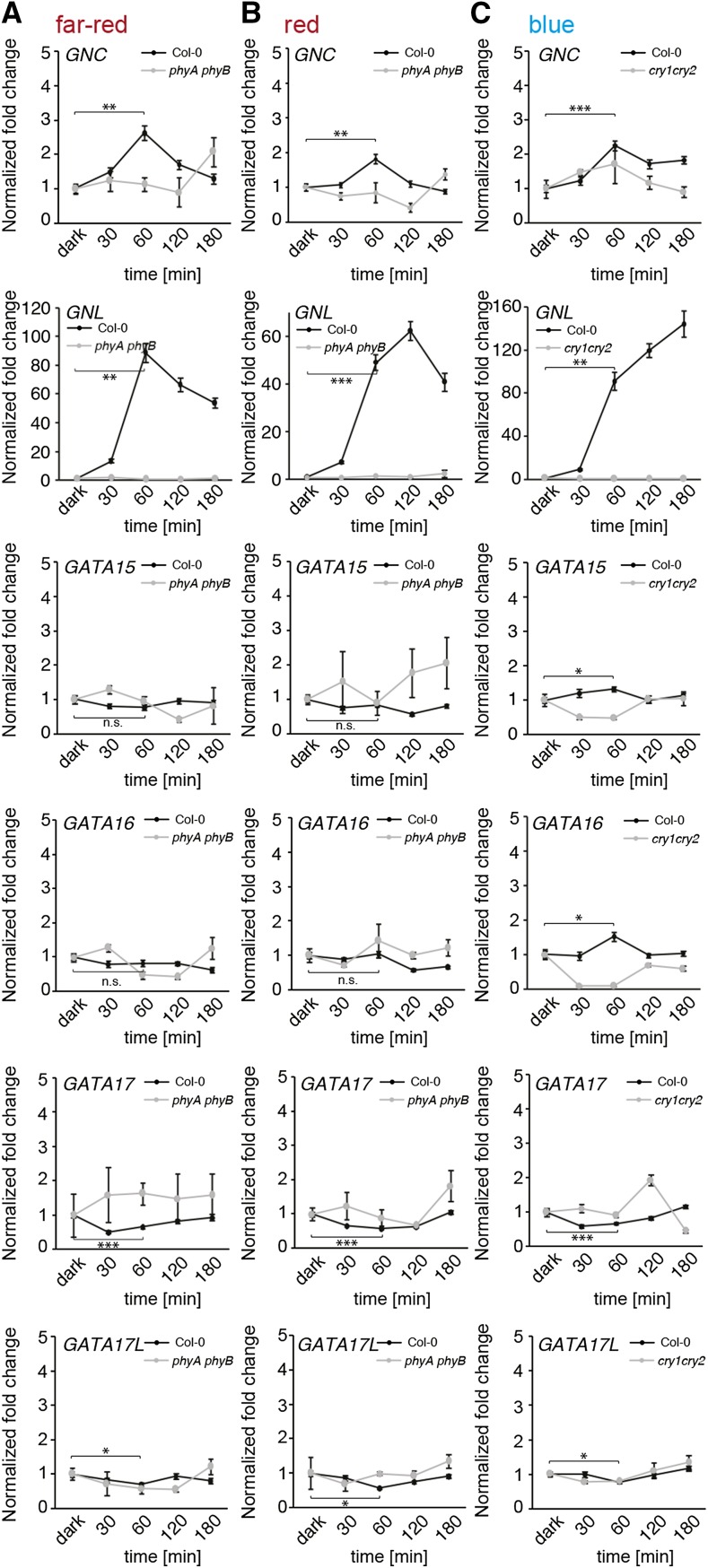
GNC and GNL had been in the focus of several studies based on the strong regulation of their gene expression by light and CK (Bi et al., 2005; Naito et al., 2007; Richter et al., 2010, 2013a,b). We were therefore interested in evaluating to what extent light and CK regulate the expression of the other GATA genes. In the case of light regulation, we found that far-red, red, and blue light strongly induced the expression of GNL, and to some extent the expression of GNC (Fig. 2). In blue light, we noted a subtle but significant induction of GATA15 and GATA16 (Fig. 2C). Importantly, this regulation was not observed in the red and far-red light receptor mutant phyA phyB or in the blue light receptor mutant cry1 cry2 (Fig. 2). Furthermore, light-induced gene expression changes in red light were impaired in the quadruple mutant for the PHYTOCHROME INTERACTING FACTOR (PIF) genes PIF1, PIF3, PIF4, and PIF5 (pifq), indicating that the light-dependent regulation in red light was mediated by the phytochrome and PIF signaling pathways (Supplemental Fig. S5; Leivar et al., 2008).
Figure 2.
GATA gene expression in far-red, red, and blue light conditions. A, B, C, Averages and ses of three replicate qRT-PCR analyses of 4-d-old dark-grown seedlings after transfer to (A) far-red (0.35 μmol s−1 m−2), (B) red (7.2 μmol s−1 m−2), and (C) blue light (4.25 μmol s−1 m−2). The phyA phyB and cry1 cry2 light receptor mutants were used to control the specificity of the respective treatment, and to control for background gene expression. Gene expression data were normalized to the transcript abundance detected in the dark sample of the respective genotype. Student’s t test for selected time points: * P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001; n.s. = not significant.
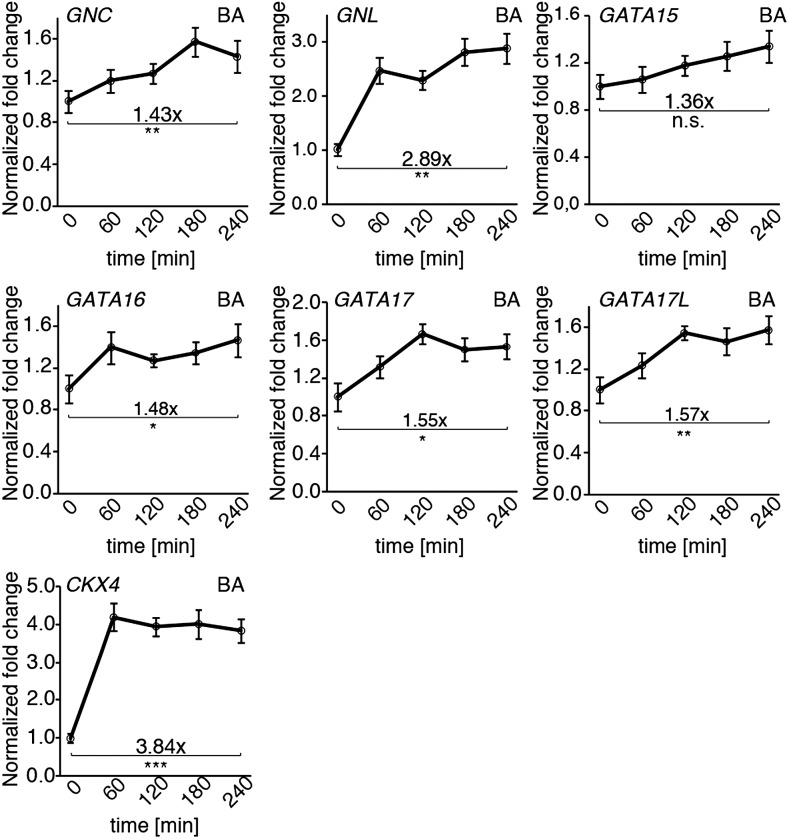
The CK experiments indicated that CK induces the expression of all six GATA genes (Fig. 3). With an approximately 3-fold increase in transcript abundance, this induction was strongest for GNL but could be observed for each of the other GATA genes after a one-to-four h CK treatment (Fig. 3). Thus, CK promotes the expression of all six LLM-domain B-GATAs.
Figure 3.
GATA gene expression is induced by cytokinin. Averages and ses of three replicate qRT-PCR analyses of 10-d-old seedlings that had been treated for the times indicated with 10 μM 6-BA. Expression changes for CKX4 are shown to verify the effectiveness of the hormone treatment. The expression values for each time point were normalized to those of an untreated mock sample. The fold change is shown relative to time point 0, which was set to 1. Student’s t test for selected time points: * P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001; n.s. = not significant.
Mutants of LLM-Domain B-GATAs Are Impaired in Hypocotyl Elongation
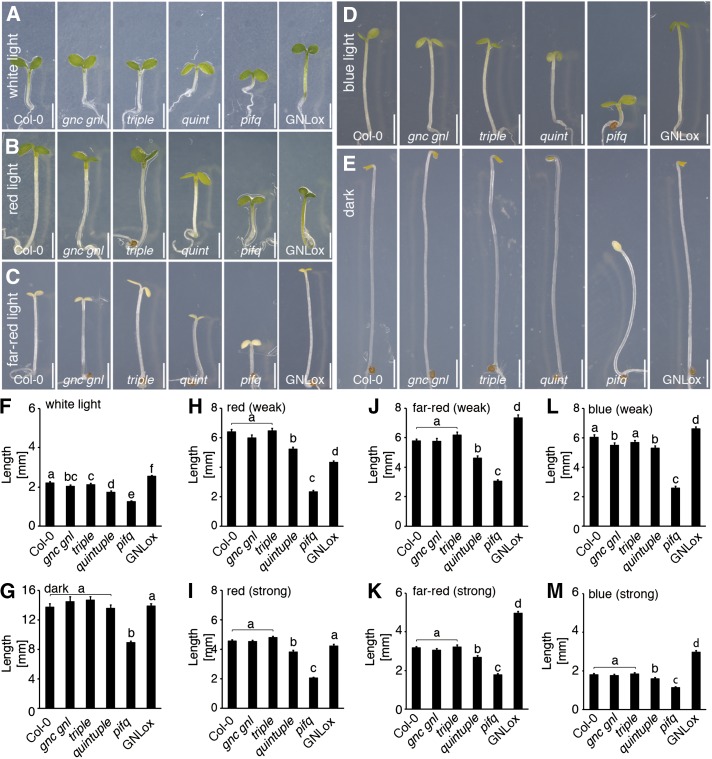
Due to the strong regulation particularly of GNL but also of GNC in far-red, red, and blue light conditions, we examined the contribution of the GATA genes to hypocotyl elongation by analyzing mutant seedlings in the different light conditions in two different light intensities (Fig. 4). Here, we found that quintuple mutant seedlings had a shorter hypocotyl than wild-type seedlings in white light as well as in red, far-red, and blue light (Fig. 4). Notably these phenotypes were not apparent, or at least not significant, in the gnc gnl double mutant or the triple mutant, suggesting that the respective other GATA genes may contribute to the hypocotyl elongation phenotype in these backgrounds (Fig. 4). At the same time, hypocotyl length of dark-grown mutant seedlings was indistinguishable in all genotypes from that observed in the wild type (Fig. 4, E and G). The short hypocotyl phenotype of the GATA gene mutants was also interesting because we previously observed that the LLM-domain B-GATA overexpressors had a longer hypocotyl than the wild type when grown in white light (Behringer et al., 2014). When analyzed in the different light conditions, we found the long hypocotyl phenotype of the GNL overexpression line (GNLox) was also present in far-red and blue light but interestingly not in red light, where hypocotyls were even shorter than in the wild type or the mutants when grown in weak red light conditions (Fig. 4).
Figure 4.
GATA factors contribute to hypocotyl elongation. A, B, C, D, E, Representative photographs; and F, G, H, I, J, K, L, M, averages and ses from hypocotyl measurements of 5-d-old seedlings (n ≥ 50) grown in white light (A and F), weak and strong red light (B, H, I), weak and strong far-red light (C, J, K), weak and strong blue light (D, L, and M), and in the dark (E and G). Scale bar = 2 mm. Student’s t test: datasets with no statistical difference fall in one group and were labeled accordingly.
GATA Gene Mutants Have Differential Gene Expression Defects
The strong induction of GNL gene expression by CK was already repeatedly described and is the reason for the gene’s original designation GNC-LIKE/CYTOKININ-RESPONSIVE GATA FACTOR1 (Naito et al., 2007; Köllmer et al., 2011). In order to examine the role of the GATA genes in CK-responsive gene expression, we performed microarray experiments with 14-d-old wild-type plants, gnc gnl and triple mutants as well as quintuple mutants treated for 60 min with the CK 6-benzylaminopurine (6-BA) or a corresponding mock solvent control (Figs. 5 and 6; Supplemental Tables S1 and S2). We then analyzed the resulting data with regard to differences in basal gene expression (Fig. 5) and with regard to gene expression differences after the CK treatment in comparison to the mock control (Fig. 6). For technical reasons, the experiment was conducted in two rounds. The first experiment included the Col-0 (1) wild-type control, the gnc gnl double mutant, and the quintuple mutant; the second experiment included an independent Col-0 (2) wild-type control and the triple mutant. When we examined the expression of the CK-induced A-TYPE RESPONSE REGULATORS of the CK signaling pathway, we could confirm that the respective CK induction experiments were comparably efficient between the two experiments and in all genotypes tested (Supplemental Fig. S6A). We also examined the microarray dataset with regard to the expression of a previously defined so-called “golden list” of CK-regulated genes (Bhargava et al., 2013). Greater than 70% of the 331 entities derived from this list were significantly CK-regulated in our experiments (Supplemental Table S3). When applying a 2-fold expression change as additional filter criterion, 71 of these 331 entities were identified as up-regulated (Col-0 1 and Col-0 2), and 39 (Col-0 1) and 25 entities (Col-0 2) as down-regulated after CK treatment (Supplemental Fig. S6B; Supplemental Table S3). We thus concluded that the CK treatments in both experiments were similarly efficient and that the data sets could be used for comparative analyses.
Figure 5.
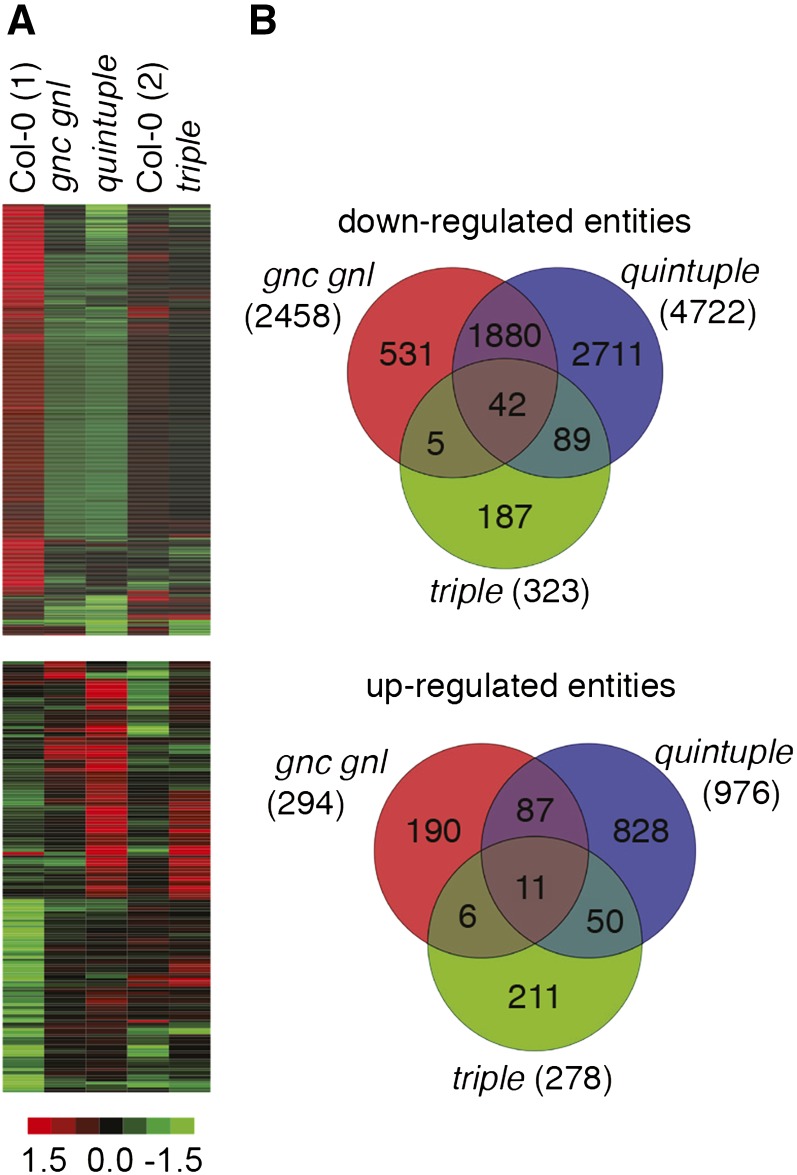
Differential basal gene expression in gata mutants. A, Heat maps of differentially expressed genes from 10-d-old light-grown Col-0 seedlings and the gnc gnl, quintuple, and triple mutants. The experiment was performed in two rounds with two independent wild-type controls referred to as Col-0 (1) and Col-0 (2). Down-regulated and up-regulated genes are shown in the top and bottom panels, respectively. B, Venn diagrams showing the overlap in differential gene expression for down- and up-regulated genes between the three different gata mutant backgrounds using the respective Col-0 wild-type control as a reference. Gene expression identities and data are shown in Supplemental Table S1.
Figure 6.
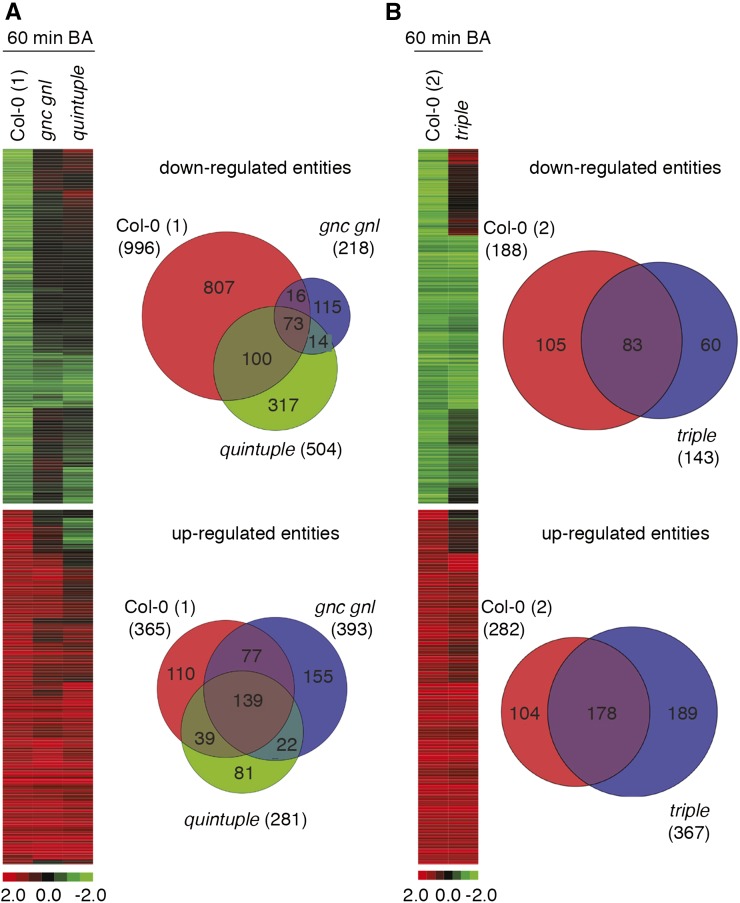
Cytokinin-responsive gene expression is impaired in gata mutants. A, B, Heat maps and Venn diagrams of differentially expressed genes from Col-0 and the gata mutants after 60-min CK treatment. The experiment was performed in two rounds with two independent wild-type controls referred to as Col-0 (1) and Col-0 (2). Down- and up-regulated genes are shown in the top and bottom panels, respectively. Venn diagrams showing the overlap in differential gene expression for genes that were down- and up-regulated following CK treatment in the respective Col-0 wild type and their expression in the respective gata mutants. Gene expression identities and data are shown in Supplemental Table S2.
The comparison of the basal gene expression profiles of the wild-type samples and the mutants indicated that the (untreated) quintuple mutant seedlings had by far the largest number of differentially regulated genes with 4722 down-regulated and 976 up-regulated entities when compared to the Col-0 (1) wild type (Fig. 5; Supplemental Table S1). Furthermore, the gnc gnl double mutant contained 2458 down- and 294 up-regulated entities, respectively (Fig. 5; Supplemental Table S1). In comparison, the number of differentially expressed genes was comparatively minor in the triple mutant, which had only 323 down- and 278 up-regulated entities in this particular experiment and selected physiological growth conditions (Fig. 5).
The comparatively strong gene expression differences in the gnc gnl double and the quintuple mutants, and the fact that both mutants shared the gnc gnl loss-of-function at the genetic level, was reflected by the large overlap between the differentially expressed gene sets of the two genotypes. Specifically, 1922 of the 2458 down-regulated entities from gnc gnl were also down-regulated in the quintuple mutant (Fig. 5B), and 98 of the 294 entities up-regulated in gnc gnl were also up-regulated in the quintuple mutant (Fig. 5B). Conversely, the overlap in gene expression defects between the triple mutant and the quintuple mutant was higher than that between the triple mutant and gnc gnl (Fig. 5B). We therefore concluded that the gene expression differences between the different GATA gene mutants was strongest in the quintuple mutant and that the strong increase in the number of differentially regulated genes exceeded by far the differences seen in the gnc gnl double mutant and the triple mutant. This supported our conclusion from the phenotypic analyses that the GATA genes act in a functionally redundant manner.
Since we had observed increased gene expression of all six GATA genes after CK-treatment, we expected that CK-induced gene expression may be compromised in the gata mutants (Fig. 6). Indeed, of the 996 entities that were down-regulated after CK-treatment in the wild type (Col-0 1), only 173 (18%) and 89 (9%) remained CK-regulated in the quintuple and gnc gnl mutants, respectively (Fig. 6A; Supplemental Table S2). At the same time, of the 188 entities that were down-regulated after CK treatment in the wild-type sample of the second experiment (Col-0 2), 83 (44%) remained CK-regulated in the gata triple mutant (Fig. 6B). The differential effect on gene expression between the different mutants was less pronounced when examining the CK-induced genes. Here, 49% (quintuple mutant), 59% (gnc gnl), and 63% (triple mutant) of the CK-induced entities from the respective wild types remained CK-regulated in the mutants (Fig. 6, A and B). In summary, we concluded that CK-regulated gene expression is impaired in the GATA gene mutants.
GATA Gene Mutants Are Defective in CK-Regulated Developmental Processes
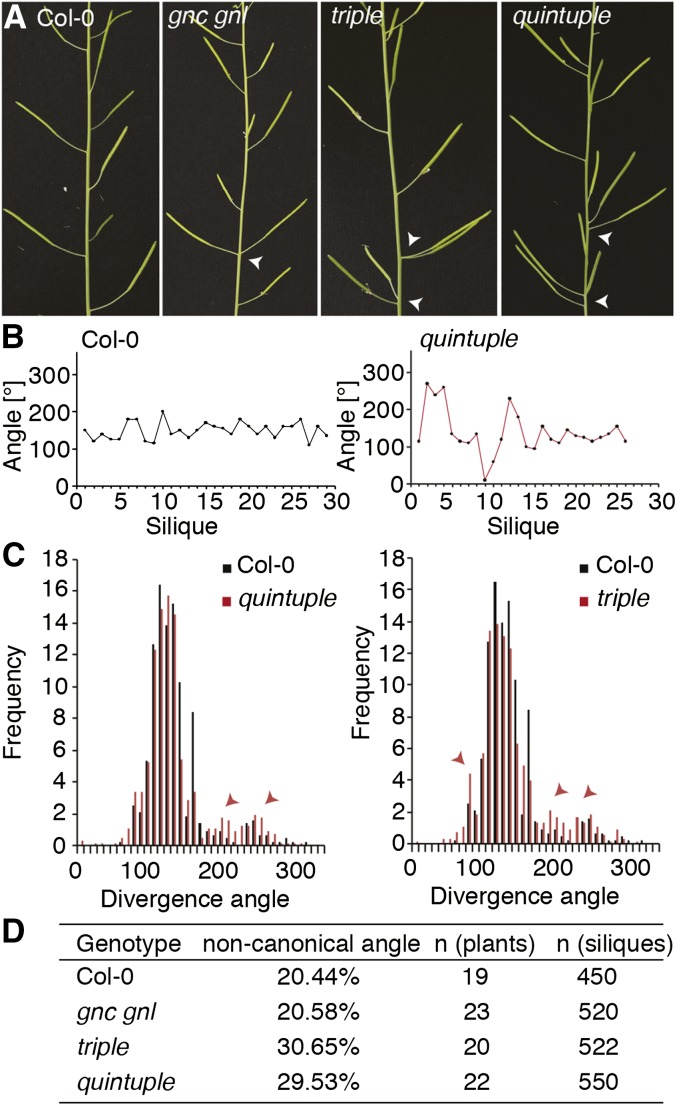
Based on the strong defects of gata mutants in CK-regulated gene expression, we searched for phenotypes that could be explained by defects in CK response. CK signaling was described as a regulator of phyllotactic patterning in the shoot apical meristem of Arabidopsis (Besnard et al., 2014). In line with a role of the LLM-domain B-GATA genes in this process, we observed frequent aberrations from normal phyllotactic patterning in flower positioning in triple and quintuple mutant inflorescences (Fig. 7). Quantitative analyses revealed aberrations from the canonical angle of approximately 137.5° in approximately 30% of the petioles in triple and quintuple inflorescences (Fig. 7D). At the same time, this patterning defect was not apparent in the gnc gnl double mutant. Since we also did not observe a quantitative increase in the severity of this phenotype between the triple and the quintuple mutant, we concluded that GATA16, GATA17, and GATA17L may be responsible for this phenotype (Fig. 7D).
Figure 7.
Phyllotaxis patterns are impaired in GATA gene mutants. A, Representative photographs of mature inflorescences. Arrowheads point at events of aberrant silique positioning. B, Measurement of silique angles along the primary inflorescence of representative wild-type and quintuple mutant plants. Note the particularly strong deviation from the ideal silique angle of 137.5° at the base of the inflorescence. C, Frequency distribution of silique divergence angles in wild-type and gata triple as well as quintuple mutant plants. Arrowheads mark examples for instances where mutant angles strongly deviate from wild-type angles. D, Overview over the number of siliques with a noncanonical angle as observed in the wild type and GATA gene mutants.
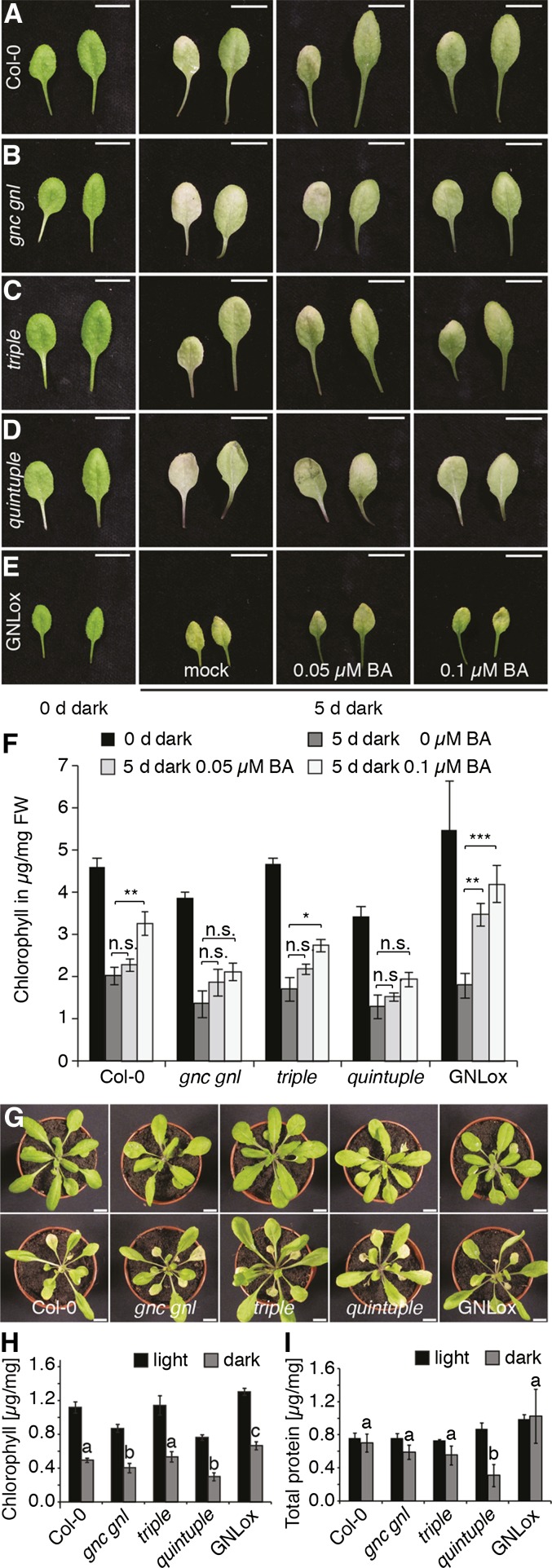
Leaf senescence is another CK-regulated process (Hwang et al., 2012). In the wild type, CK levels are reduced during senescence, and positive or negative interference with the decrease in CK levels can promote and delay senescence, respectively (Hwang et al., 2012). Senescence can be induced by shifting light-grown plants to the dark; we had previously already shown that leaf senescence is delayed in overexpression lines of LLM-domain B-GATAs (Behringer et al., 2014). To examine possible senescence defects in the gata mutants, we floated leaves of wild-type and mutant plants in the dark on a mock solution or on a solution containing 0.05 μM or 0.1 μM 6-BA (Fig. 8, A–F) (Weaver and Amasino, 2001). In line with the expectation that the presence of CK in the medium would suppress the senescence process, we observed decreased senescence in CK-treated wild-type leaves, which we quantitatively assessed by measuring chlorophyll levels in the mock- and CK-treated leaves (Fig. 8F). Here, we found that the effects of the senescence treatment were less efficiently suppressed by CK treatments in the gnc gnl double and in the quintuple mutant than in the wild type or the triple mutant (Fig. 8, A–F). Similarly, we observed a stronger effect on the decreases in chlorophyll and total protein levels in the gnc gnl as well as the quintuple mutant when we transferred light-grown plants for 4 d to the dark (Fig. 8, G–I). As previously reported, GNL overexpressing plants (GNLox) had senescence phenotypes that were antagonistic to the ones observed with the mutants (Fig. 8) (Behringer et al., 2014). In the case of the leaf floating assay, CK-treatment had a stronger effect on greening and chlorophyll abundance in GNLox than in the wild type (Fig. 8, A–F). When we transferred plants from the light to the dark, GNLox seedlings remained visibly green and did not display the strong decrease in total protein abundance that we observed in the wild type or the GATA gene mutants (Fig. 8, G–I). We thus concluded that GATA gene mutants and overexpressors have antagonistic effects with regard to the regulation of senescence, and that GNC and GNL contribute particularly strongly to this phenotype.
Figure 8.
Cytokinin-effects on senescence are impaired in gata mutants. A–E, Representative photographs of leaves from 21-d-old plants. Leave nos. 3 and 5 were detached and kept in the dark for 4 d in liquid medium to monitor senescence in the presence and absence of the cytokinin 6-benzylaminopurin (BA). Scale bar = 1 cm. F, Averages and sd after quantification of chlorophyll abundance of the leaves shown in (A–E) (n = 3). Student’s t test: * P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001; n.s. = not significant. G, Representative photographs of 33-d-old plants that had transferred from the light to the dark for 4 d. Scale bar = 1 cm. H, I, Averages and sds after quantification of chlorophyll measurements (H) and total protein determination (I) of plants shown in (G). n ≥ 3. Student’s t test was performed for dark samples: datasets with no statistical difference fall in one group and were labeled accordingly.
CK induces greening in light-grown seedlings (Kobayashi et al., 2012). To examine to what extent CK can induce chlorophyll accumulation in the gata mutants, we grew plants for 2 weeks in constant light on medium containing CK (5 nm BA) or a corresponding CK-free medium. As expected, chlorophyll content was increased in wild-type plants but this increase was neither significant in the gnc gnl nor the triple or the quintuple mutants (Supplemental Fig. S7). We thus concluded that the GATA factors are required for CK-induced greening in Arabidopsis seedlings.
It was previously reported that hypocotyl elongation could be repressed by incubating dark-grown seedlings in the presence of CK (Su and Howell, 1995). Since the GATA gene mutants had displayed a hypocotyl elongation defect in light-grown seedlings (Fig. 4), we also explored the link between hypocotyl elongation and CK. However, when we tested the effects of CK on hypocotyl elongation in dark-grown seedlings, we did not detect any significant differences in the response between the different genotypes tested (Supplemental Fig. S8). It is thus unlikely that the GATA genes participate in this elongation response.
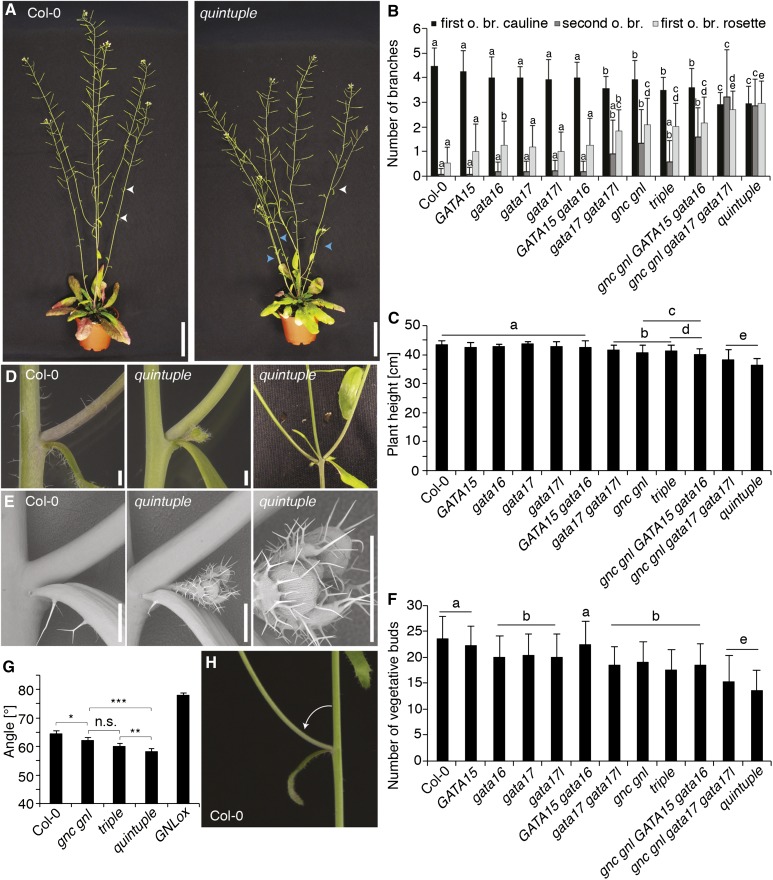
Branching is another phenotype that has been attributed to defects in CK biosynthesis and response (El-Showk et al., 2013). When we analyzed the general habitus of the quintuple mutants, we noted that they had an increased number of secondary branches combined with a reduced plant height (Fig. 9, A–C). Occasionally, we also observed the outgrowth of inflorescences in the axils of lateral inflorescences in the mutants whereas we had not noted this phenotype in the wild type (Fig. 9, D and E). A quantitative analysis of these phenotypes over all gata mutant phenotypes revealed that the number of first- and second-order lateral branches increased with increasing mutant complexity (Fig. 9B). Here, we observed strong effects already in the gata17 gata17l mutant as well as the gnc gnl double mutant but strongest defects in the triple and the quintuple mutant; thus, all five GATA genes may contribute to this phenotype (Fig. 9B). At the same time, plant height was reduced to a significant but still minor level in these mutants, suggesting that decreases in plant height, which may have indirect consequences on the general plant habitus and branching, are not the primary cause for altered branching patterns in the gata mutants (Fig. 9C). It is further important to note that the higher-order GATA gene mutants initiated fewer vegetative buds but had a higher number of growing buds as reflected by the increased branch numbers (Fig. 9F). Thus, axillary bud formation is promoted and bud outgrowth is repressed by the GATAs in the wild type.
Figure 9.
GATA gene mutants have altered branching patterns and branch angles. A, Representative photographs of 7-week-old Col-0 and quintuple mutant plants. Arrowheads indicate positions of second-order lateral branch formation (white) and first-order branches from nodes in the rosette (blue). Scale bar = 5 cm. B, C, and F, Quantitative analysis of branch numbers (first-order branch from cauline leaves, first o. br. cauline; second-order branches, second o. br.; and first-order branches from the rosette, first o. br. rosette). B, Plant height and C, vegetative bud formation along the shoot (F) in the wild-type and gata mutant backgrounds. n = 15. Student’s t test: datasets with no statistical difference fall in one group and were labeled accordingly. D, E, Photographs (D) and scanning electron micrographs (E) of floral meristems and siliques formed in the axils of second-order lateral branches of quintuple mutants, which are typically not found in the wild type. Scale bar = 1 mm. F, Quantitative analysis of branch angles measured on 5-week-old Arabidopsis plants. n = 19. Student’s t test: * P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001; n.s. not significant. G, Representative photographs of lateral inflorescences branching from the primary inflorescence in the wild type. H, The arrows indicate the angles measured for the analysis shown in (G). Scale bar = 1 cm.
Another phenotype that became apparent during our analysis of the gata mutant plant habitus was an increase in the angle formed between the primary inflorescence and the lateral inflorescences (Fig. 9, G and H; Supplemental Fig. S9). The gata mutants had a decreased angle formed between the primary inflorescence and lateral inflorescences and this phenotype was strongest in the quintuple mutant and intermediate in the gnc gnl and triple mutants (Fig. 9, G and H; Supplemental Fig. S9). Interestingly, we had previously observed increased branch angles in GATA overexpression lines such as GNLox (Supplemental Fig. S9) (Behringer et al., 2014). We thus concluded that GATA gene mutants and overexpressors antagonistically regulate branch angles in Arabidopsis.
GATA Genes Regulate Petal and Sepal Numbers as well as Silique Length
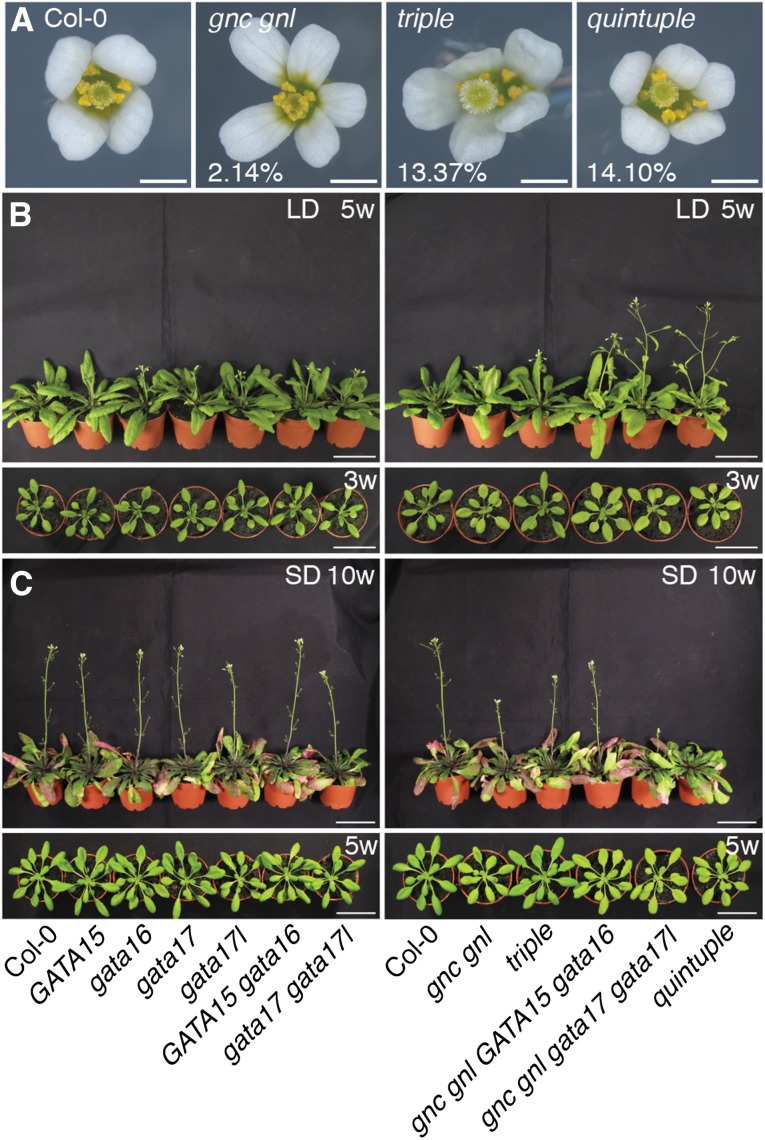
CK signaling has recently been implicated in the control of floral development (Han et al., 2014). When we analyzed gata mutant flowers in more detail, we noted with interest aberrations in floral organ numbers in comparison to the wild type (Fig. 10A). Whereas wild-type flowers have generally four sepals and four petals and only rarely deviate from this pattern (in our study in around 2% of the cases), a strong increase in flowers with more than four (generally five, in rare cases six) petals and sepals became apparent in the higher-order gata mutants (Fig. 10A and Supplemental Fig. S10A). In the gata17 gata17l mutants, we observed flowers with more than four sepals and petals in 12% of all flowers, and this number was slightly enhanced in the triple mutant (13% petals) and the quintuple mutant (14% petals) (Fig. 10A and Supplemental Fig. S10A). Importantly, the single mutants with the clearest apparent effect were the gata17 and gata17l mutants and, inversely, mutations in GNC and GNL contributed only to a minor extent to this phenotype (Supplemental Fig. S10A). In summary, this analysis revealed that GATA17 and GATA17L make a particularly strong contribution to the floral morphology phenotype of the gata mutants.
Figure 10.
gata mutants have impaired floral morphology and flowering time. A, Phenotypic analysis of sepal and petal numbers in the wild-type and the gata gene mutant backgrounds. Percentages indicate the penetrance of the displayed floral phenotype. The wild-type flower phenotype represents about 98% of all flowers analyzed; the frequency of aberrant flowers is similar to that of the gnc gnl mutant. B, C, Representative photographs (side and top views) of wild-type and gata gene mutants grown for 5 weeks in long-day (16 h light/8 h dark) (B) or for 10 weeks in short-day (8 h light/16 h dark) conditions (C). Scale bar = 5 cm.
Since we had previously noted that overexpression lines of LLM-domain B-GATAs have short siliques and a reduced number of seeds per silique, we also analyzed silique length and seed set in the gata mutants (Supplemental Fig. S10, B–D). In line with an antagonistic regulation of these traits in the gata mutants, we measured significant increases in seed set in the siliques of the gnc gnl, the triple as well as the quintuple mutants, and we detected significant increases in silique length in all mutant backgrounds (Supplemental Fig. S10, B–D). We thus concluded that the GATA genes repress silique elongation and seed set in the wild type, at least to a minor extent. A differential contribution of individual GATAs to these phenotypes could, however, not be resolved here.
GATA Gene Mutants Have Antagonistic Effects on Flowering Time in Long and Short Days
Accelerated flowering in plants grown under long-day conditions was another phenotype previously described for gnc gnl mutants (Richter et al., 2010, 2013a,b). Conversely, overexpression of the LLM-domain B-GATAs causes a strong delay in flowering (Richter et al., 2010, 2013a,b; Behringer et al., 2014). To examine the contribution of the other GATA factors to the flowering time phenotype, we grew the gata mutant series in long- and short-day conditions with a 16 h and 8 h light period, respectively (Fig. 10, B and C; Supplemental Fig. S11). Interestingly, we observed a gradual acceleration of flowering in long-day-grown plants that was weakest in the single mutants, more pronounced in the double, triple, and quadruple mutants, and strongest in the quintuple mutants (Fig. 10, B and C; Supplemental Figs. S11 and S12). We thus concluded that all LLM-domain B-GATA genes examined here may contribute to the repression of flowering in long-day-grown plants.
Interestingly, the effects of the reduced GATA gene function on flowering time in short-day conditions were antagonistic to those observed in long day-grown plants in that the complex GATA gene mutants flowered much later than the wild type or the less complex mutant combinations when flowering was counted in days (Fig. 10, B and C; Supplemental Figs. S11 and S12). We thus concluded that the LLM-domain B-GATAs factors regulate flowering time in long- and short-day conditions but, in view of the antagonistic nature of this regulation, their contribution to flowering time control may be complex.
Cross Regulation of GATA Gene Expression
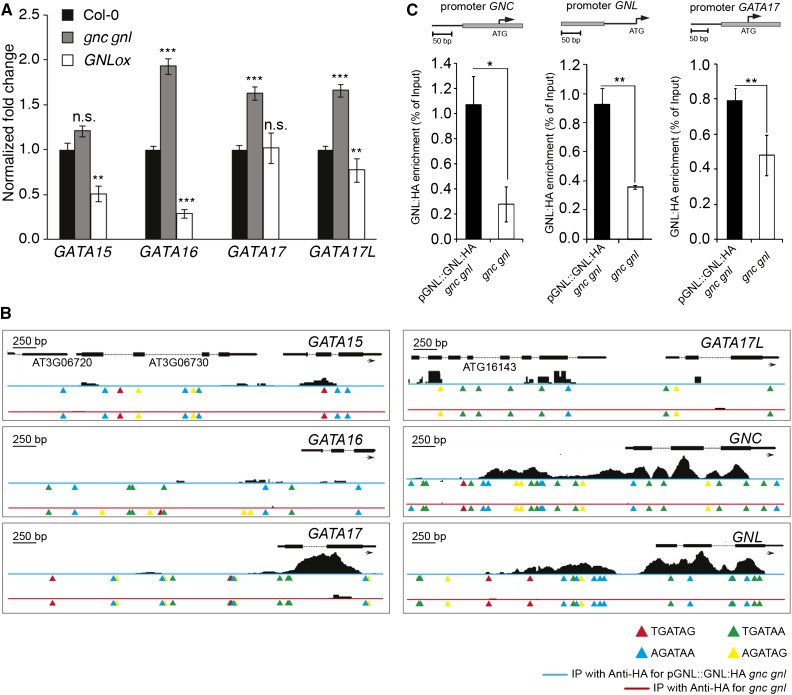
Since several of our genetic analyses had suggested a functional redundancy between the GATA genes, we looked for evidences of a cross regulation of GATA gene expression in the gnc gnl double mutant background. There, we detected indeed an increased transcript abundance of GATA16, GATA17, and GATA17L in the gnc gnl mutant, which could be compensatory for the loss of GNC and GNL (Fig. 11A). At the same time, GATA15, GATA16, and GATA17L gene expression levels were significantly reduced in GNL overexpression lines (GNLox) suggesting that GNL may regulate the expression of these genes.
Figure 11.
GATA gene expression is cross regulated in gnc gnl mutants and GNLox transgenic lines. A, Result from qRT-PCR analyses of the GATA genes GATA15, GATA16, GATA17, and GATA17L in the gnc gnl mutant and GNLox background. Gene expression data were normalized to the transcript abundance detected in the Col-0 wild type. B, Read coverage over the six LLM-domain B-GATA genes and 30-kb upstream region after chromatin immunoprecipitation of GNL:HA from a pGNL::GNL:HA gnc gnl transgenic line (blue line) and the gnc gnl mutant (red line) followed by next-generation sequencing. GATA gene models and, where applicable, the gene models of the neighboring genes are indicated in the figure, different GATA-sequence-containing motifs are indicated below the gene models. C, Result from qRT-PCR analyses for selected amplicons (gray boxes) upstream from the GNC, GNL, and GATA17 gene promoters after chromatin immunoprecipitation of GNL:HA from a pGNL::GNL:HA gnc gnl transgenic line and the gnc gnl mutant. Student’s t tests: * P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001; n.s. = not significant.
Since these observations indicated that molecular mechanisms may exist that compensate for the loss of individual LLM-domain B-GATA genes, e.g. in the gnc gnl mutants, we examined the binding of GNL to the LLM-domain GATA gene promoters after ChIP-seq analysis (chromatin immunoprecipitation followed by DNA sequencing). Here, we found that at least three of the six LLM-domain B-GATA genes were bound by GNL after immunoprecipitation of HA-tagged GNL from transgenic plants expressing a functional GNL:HA transgene from a GNL promoter fragment in the gnc gnl background (pGNL::GNL:HA gnc gnl; Fig. 11B). Through subsequent ChIP-PCR studies targeting selected regions upstream from GNC, GNL, and GATA17, we could confirm this binding through independent analyses (Fig. 11C). Thus, LLM-domain B-GATA factors may directly regulate their own expression as well as that of other family members.
DISCUSSION
In this study, we have characterized the six-membered family of LLM-domain B-GATAs from Arabidopsis. Whereas the family has diverged when compared among Arabidopsis, tomato, barley, and Brachypodium, a clear conservation of this protein family can be observed within the Brassicaceae family (Supplemental Fig. S1; Behringer et al., 2014). Within the Brassicaceae, GNC and GNL, GATA15 and GATA16 as well as GATA17 and GATA17L form closely related LLM-domain B-GATA pairs. In a previous study, we had comparatively analyzed transgenic Arabidopsis lines expressing different members of the LLM-domain B-GATA gene family from Arabidopsis, barley, and tomato, and concluded that these genes are functionally redundant at the biochemical level since their overexpression resulted in highly similar phenotypes (Behringer et al., 2014). Several of the phenotypes analyzed here, namely greening (Fig. 1, D–F), hypocotyl elongation (Fig. 4), plant height, branching patterns (Fig. 9), branch angles (Fig. 9 and Supplemental Fig. S9), silique length, seed set (Supplemental Fig. S10), flowering time (Fig. 10), and senescence (Fig. 8) are strongest in the gata quintuple mutant and weaker, or in some cases not even apparent, in the less complex gata mutant combinations. Thus, in line with this previous study, our present analysis shows that the LLM-domain B-GATA genes from Arabidopsis have a shared biological function in the control of multiple developmental pathways.
Due to the high complexity of the available range of single, double, triple, quadruple, and quintuple mutants, we restricted many of our phenotypic analyses to the gnc gnl double, the complementary triple, and the quintuple mutants. It can nevertheless be suggested, based on the presently available data, that some phenotypes are predominantly controlled by a single LLM-domain B-GATA or rather an LLM-domain B-GATA gene pair. Phyllotactic patterning as assessed here in the inflorescence is impaired in the gata triple but not in the gnc gnl double mutant (Fig. 7). Similarly, the increases in floral organ numbers are already very prominent in the gata17 gata17l double mutants and not much more enhanced in the gata triple or quintuple mutant that includes the gata17 gata17l mutations (Fig. 10A; Supplemental Fig. S10A). Thus, the LLM-domain B-GATA genes have shared overlapping but distinct roles in the control of plant development. The fact that GATA15 gene function is not impaired in the selected GATA15 gene insertion allele does not allow us to draw any direct conclusions on the biological function of this gene based on the data presented here. Furthermore, the fact that the mutant alleles of GATA16 and GATA17 used in the complex mutants are only partially defective in GATA gene expression could suggest that even stronger phenotypes may be observed if loss-of-function alleles were used. The two strong alleles for GATA15 (gata15-2) and GATA17 (gata17-2) isolated here but analyzed only as single mutants may help in the future to understand the biological function of these GATA genes at further depth.
It is interesting that the overexpression lines and the mutants of these GATA genes frequently have antagonistic phenotypes. This study in combination with a previous study identifies such antagonistically regulated phenotypes in the control of greening, hypocotyl elongation, branch angle formation, flowering time, and senescence (Richter et al., 2010; Behringer et al., 2014). Particularly, in the case of transcription factors where the loss-of-function or overexpression may affect the expression of many downstream targets genes and developmental responses triggered by these target genes, one may not expect such a clear antagonistic relationship between mutants with reduced gene functions and the overexpressors. In view of the fact that overexpressors likely control the expression of genes that are normally not targeted by this transcription factor when expressed from their native promoter (off-targets), this observation is even more striking. It may suggest that the GATAs examined here control developmental responses in a dosage-dependent manner together with other critical factors. In this context, the presence and abundance of the GATAs would just modulate the response but would by itself not be sufficient to trigger it. Further analyses will be required to gain a more detailed understanding of the mode of action of the LLM-domain B-GATA factors in the control of gene expression.
We had previously shown that the LLM-domain B-GATAs GNC and GNL repress flowering since their loss-of-function mutants and overexpressors displayed an early and late flowering phenotype, respectively, when plants were grown in long-day conditions (Richter et al., 2010; 2013a,b). We furthermore showed that the control of flowering is partially mediated by the flowering time regulator SUPPRESSOR-OF-OVEREXPRESSION-OF-CONSTANS1, which acts downstream from GNC and GNL and whose promoter can be directly bound by GNC and GNL (Richter et al., 2013a,b). In line with these previous observations, we show here that the complex GATA gene mutants flower even earlier than the gnc gnl mutant, further substantiating our conclusion that LLM-domain B-GATAs are flowering time regulators in Arabidopsis (Fig. 10B; Supplemental Fig. S11). Surprisingly, the complex gata mutants flowered late when grown in short-day conditions and their phenotype was thereby antagonistic to the phenotype observed in long days (Fig. 10C; Supplemental Fig. S9). The reason for this antagonistic flowering time behavior remains to be resolved but a complex scenario may be envisioned. We also do not want to rule out that physiological parameters become limiting in the induction of flowering in the gata mutants. Long-term defects as a result of their reduced greening may play a limiting role during the extended growth in short days with short illumination periods that are not limiting when plants are grown in long days.
Several of our phenotypic analyses suggested that the LLM-domain B-GATAs act in concert to control specific phenotypes. The fact that at least some of the phenotypes were most pronounced in the quintuple mutant and not apparent or only weak in the less complex mutants suggests that the LLM-domain B-GATAs may cross regulate their expression, and that presence and absence of one GATA could interfere with the abundance of other GATA family members. We have analyzed this hypothesis here by examining the expression of the LLM-domain B-GATAs GATA15, GATA16, GATA17, and GATA17L in the gnc gnl mutant and in the GNL overexpression line (Fig. 11A). In both cases, we found the expression of three of the four analyzed genes to be differentially regulated in the two genetic backgrounds suggesting that the GATAs can cross regulate their gene expression and supporting again the hypothesis that the GATAs have a repressive activity on the transcription of the other GATA genes. Further support for such a hypothetical cross regulation came from a ChIP-seq analysis with GNL where we observed a strong binding of GNL, when expressed from its own promoter, to the three GATA gene loci, GNC, GNL, and GATA17 (Fig. 11B). Additional studies are needed to disentangle the interplay of the different LLM-domain B-GATAs in the control of plant growth and the mutual transcriptional control of the different GATA gene family members, and to get a full understanding of their biochemical and biological function during plant growth and development.
MATERIALS AND METHODS
Biological Material
All experiments were performed in Arabidopsis ecotype Columbia. Insertion mutants for GNC (SALK_001778) and GNL (SALK_003995) and their double mutant were previously described (Richter et al., 2010). Insertion mutants for GATA15 (SAIL_618B11 and WiscDsLox471A10), GATA16 (SALK_021471), GATA17 (SALK_101994 and SALK_049041), and GATA17L (SALK_026798) were obtained from the Nottingham Arabidopsis Stock Center. Homozygous single- and higher-order mutant combinations were isolated from segregating populations by PCR-based genotyping. In the context of this analysis, it became apparent that the insertion position for one GATA15 allele (SAIL_618B11) was misannotated and that the correct position of this insertion was downstream of the gene’s 3′-untranslated region. A list of genotyping primers is provided in Supplemental Table S4. In addition, the previously described mutants phyA-211 phyB-9 (Reed et al., 1994), cry1 cry2 (Mockler et al., 1999), and pifq (pif1 pif3 pif4 pif5) were used (Leivar et al., 2008).
Protein Alignment and Phylogeny
LLM-domain B-GATA protein sequences of different Brassicaceae genomes were identified in the ENSEMBL database (http://plants.ensembl.org/index.html). Sequences were filtered for proteins containing the LLM-domain and alignments were performed using the ClustalW2 alignment tool (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Two Brassica rapa genes, Bra037421 and Bra035633, were not included in the analysis because no expressed sequence tags could be detected for these annotations and they may represent pseudogenes (http://www.plantgdb.org/BrGDB/). The phylogenetic tree was generated with MEGA6.06 (http://www.megasoftware.net) using the Neighbor-joining method and the bootstrap method with 1000 bootstrap replications, as well as the Jones-Taylor-Thornton model with gaps/missing data treatment set to pairwise deletion, based on an alignment of trimmed B-GATAs using the entire B-GATA DNA-binding domain and the C termini with the LLM-domain (Supplemental Fig. S2).
Physiological Assays
Unless otherwise stated, all plants were cultivated on sterile 1/2 MS medium without sugar under continuous white light (120 µmol m−2 s−1). For hypocotyl length measurements, seedlings were grown for 5 d in white light (80 µmol m−2 s−1), far-red light (weak: 0.35 μmol s−1 m−2; strong: 0.6 μmol s−1 m−2), red light (weak: 7.2 μmol s−1 m−2; strong: 11 μmol s−1 m−2), and blue light (weak: 4.25 μmol s−1 m−2; strong: 10 μmol s−1 m−2). Hypocotyl length was measured from scanned seedlings using ImageJ software (National Institutes of Health, Bethesda, MD). Chlorophyll measurements were performed with 14-d-old plants and chlorophyll content was determined as previously described and normalized to the chlorophyll content of the wild-type seedlings grown in the same conditions (Inskeep and Bloom, 1985). To visualize chloroplast accumulation and auto-fluorescence, pictures of 7-d-old seedlings were taken with a model no. FV1000 confocal microscope (Olympus, Hamburg, Germany) with an excitation of 405 nm and a detection from 631 to 729 nm. The experiments were repeated three times with comparable outcomes; the result of one representative experiment is shown. For flowering time analyses, plants were grown on soil in 100 μmol m−2 s−1 in 16 h light/8 h dark long-day conditions or in 130 μmol m−2 s−1 8 h light/16 h short-day conditions. For CK treatments, 10-d-old seedlings were preincubated for 4 h in liquid 1/2 MS medium and treatments were started by adding hormone-containing medium to reach a final concentration of 10 μM 6-benzylaminopurin (6-BA). To examine the effects of CK on hypocotyl elongation growth, seedlings were grown in the dark for 5 d on medium containing 0.05 or 0.1 μM 6-BA or a corresponding mock solution. Silique angles were determined as previously described (Besnard et al., 2014). To this end, a round gauge was built in such a way that it could be moved up the main inflorescence to measure the angles of consecutive siliques. The experiment was repeated in two rounds and both datasets were combined. Senescence assays were performed as previously described (Weaver and Amasino, 2001). In brief, detached leaves nos. 3 and 5 of 21-d-old light-grown plants were floated on liquid 1/2 MS in absolute darkness, containing either 0.05 or 0.1 μM BA or a mock solution. After 4 d, chlorophyll was extracted as described above. Alternatively, whole plants were transferred into darkness for 4 d, chlorophyll was determined as described above, and total protein concentration was determined using a Bradford assay. To assess the branching phenotype of gata mutants, the numbers of branches of at least 0.5 cm length were determined from nodes in the rosette and from cauline nodes as well as those of vegetative buds (leaf-bearing nodes) along the shoot. For scanning electron microscopy of floral buds, samples were mounted on prefrozen platforms and pictures were taken with a TM-3000 table top scanning electron microscope (Hitachi, Krefeld, Germany). Numbers of floral organs were counted at the time of anthesis (stage 13). This measurement was repeatedly performed with all plants after new flowers had formed. Siliques were harvested from different plants from a comparable section of the main inflorescence. Length and seed number was determined for each silique separately. To determine flowering, plants were monitored on a daily basis for the presence of a visible inflorescence bud (time to bolting) or petals (flowering time). The numbers of rosette and cauline leaves after bolting were counted.
Microarray Analyses
For microarray analyses, 14-d-old plants grown in continuous white light (120 μmol m−2 s−1) were treated for 60 min with CK (20 μM 6-BA) or a corresponding mock solvent control treatment. Total RNA was extracted with the NucleoSpin RNA Plant Kit (Machery-Nagel, Düren, Germany) and 150 ng total RNA was labeled with Cy3 using the Low Input Quick Amp Labeling Protocol (Agilent Technologies, Böblingen, Germany). Three biological replicate samples were prepared for each genotype and Arabidopsis arrays (V4, design ID 21169; Agilent Technologies) were hybridized at 65°C for 17 h in rotating hybridization chambers (Agilent Technologies). Subsequently, the arrays were washed and scanned using a Microarray Scanner (Agilent Technologies). Total RNA and probe quality were controlled with a Bioanalyzer 2100 (Agilent Technologies). Raw data were extracted using the Feature Extraction software v. 10.7.3.1 (Agilent Technologies). Raw data files were imported into GeneSpring GX (v. 12) and normalized choosing the scale-to-median and baseline-to-median algorithms. Data were then subjected to an ANOVA analysis (P < 0.05) with an S-N-K post hoc test and filtered for genes with fold expression differences to the respective wild-type controls as specified (Supplemental Table S2). Microarray data were deposited as GSE71828 into the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo/).
Quantitative Real Time-PCR
To measure transcript abundance, total RNA was extracted with an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). A quantity of 2 μg total RNA was reverse-transcribed with an oligo(dT) primer and M-MuLV Reverse Transcriptase (Fermentas, St. Leon-Rot, Germany). The transcript levels were detected using a CFX96 Real-Time System Cycler with iQ SYBR Green Supermix (Bio-Rad, Freiburg, Germany). The results were normalized to PROTEIN PHOSPHATASE SUBUNIT2A; AT1G13320 in red and far-red light and to ACTIN2 (AT3G18780) in blue light. Expression in the dark samples was set to one. Normalization for the hormone treatments was performed with ACTIN8 (AT1G49240). Primers for qRT-PCRs are listed in Supplemental Table S4.
ChIP-seq
For ChIP, three biological replicate samples (2 g tissue) of 10-d-old pGNL::GNL:HA gnc gnl or gnc gnl control plants grown on GM medium under constant white light were fixed for 20 min in 1% formaldehyde. The samples were subsequently processed as previously described using anti-HA antibodies (Abcam, Cambridge, UK; Kaufmann et al., 2010) and prepared for DNA sequencing with a MiSeq sequencer (Illumina, San Diego, CA). The data from this analysis are accessible under PRJNA288918 at NCBI-SRA (http://www.ncbi.nlm.nih.gov/sra/). Sequencing reads were then mapped to the Arabidopsis genome (TAIR10) using SOAPv1 (Li et al., 2008) and the following settings: three mismatches, mapping to unique positions only, no gaps allowed, iteratively trimming set to 41–50. Subsequent analysis for peak identification was performed with CSAR, retaining only peaks with an FDR < 0.05 as statistically significant peaks (Muiño et al., 2011). ChIP-seq results for GATA genes were independently verified using ChIP-PCR. The primers for ChIP-PCRs are listed in Supplemental Table S4.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AT5G56860 (GNC, GATA21); AT4G26150 (GNL/CGA1, GATA22); AT3G06740 (GATA15); AT5G49300 (GATA16); AT3G16870 (GATA17); AT4G16141 (GATA17L).
Supplemental Materials
The following supplemental materials are available.
Supplemental Figure S1: Evolutionary conservation of LLM-domain B-GATAs within the Brassicaceae.
Supplemental Figure S2: Sequence alignment of the GATA-domain and the LLM-domain.
Supplemental Figure S3: GATA insertion mutant analysis.
Supplemental Figure S4: Mature plant phenotypes of gata gene mutants.
Supplemental Figure S5: GATA gene expression in red light conditions is PIF-dependent.
Supplemental Figure S6: The effects of CK-induction in the two microarray data sets are comparable.
Supplemental Figure S7: CK cannot efficiently induce greening in GATA gene mutants.
Supplemental Figure S8: The effect of CK on hypocotyl elongation is not impaired in GATA gene mutants.
Supplemental Figure S9: GATA gene dosage affects lateral inflorescence angles.
Supplemental Figure S10: Floral morphology and silique parameters are influenced in GATA gene mutations.
Supplemental Figure S11: Quantitative analysis of flowering time in long- and short-day conditions.
Supplemental Figure S12: GATA gene mutations alter flowering time in long- and short-day conditions.
Supplemental Table S1: Differentially expressed genes in GATA gene mutants.
Supplemental Table S2: Differentially expressed genes in GATA gene mutants after CK-treatment.
Supplemental Table S3: Abundance of CK-regulated genes after CK-treatment in the wild type and GATA gene mutants.
Supplemental Table S4: Primers used in this study.
Supplementary Material
Acknowledgments
C.S. thanks Eilon Shani and Daniel Chamovitz, Department of Molecular Biology and Ecology of Plants, for hosting C.S. at Tel Aviv University during manuscript preparation. Q.L.R., E.B., and C.S. thank Jutta Elgner for her excellent technical support.
Glossary
- GNC
GATA, NITRATE-INDUCIBLE, CARBON METABOLISM-INVOLVED
- GNL
GNC-LIKE/CYTOKININ-RESPONSIVE GATA FACTOR1
- GNLox
GNL overexpressing plants
- HAN
HANABA TARANU
- LLM
Leu-Leu-Met
- PIF
PHYTOCHROME INTERACTING FACTOR
- qRT-PCR
quantitative real-time PCR
Footnotes
Articles can be viewed without a subscription.
References
- Behringer C, Bastakis E, Ranftl QL, Mayer KF, Schwechheimer C (2014) Functional diversification within the family of B-GATA transcription factors through the leucine-leucine-methionine domain. Plant Physiol 166: 293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer C, Schwechheimer C (2015) B-GATA transcription factors—insights into their structure, regulation, and role in plant development. Front Plant Sci 6: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard F, Refahi Y, Morin V, Marteaux B, Brunoud G, Chambrier P, Rozier F, Mirabet V, Legrand J, Lainé S, Thévenon E, Farcot E, et al. (2014) Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature 505: 417–421 [DOI] [PubMed] [Google Scholar]
- Bhargava A, Clabaugh I, To JP, Maxwell BB, Chiang YH, Schaller GE, Loraine A, Kieber JJ (2013) Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-Seq in Arabidopsis. Plant Physiol 162: 272–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi YM, Zhang Y, Signorelli T, Zhao R, Zhu T, Rothstein S (2005) Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J 44: 680–692 [DOI] [PubMed] [Google Scholar]
- Chiang YH, Zubo YO, Tapken W, Kim HJ, Lavanway AM, Howard L, Pilon M, Kieber JJ, Schaller GE (2012) Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol 160: 332–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, van Campenhout J, Overvoorde P, Jansen L, Vanneste S, Möller B, Wilson M, et al. (2010) A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr Biol 20: 1697–1706 [DOI] [PubMed] [Google Scholar]
- El-Showk S, Ruonala R, Helariutta Y (2013) Crossing paths: cytokinin signalling and crosstalk. Development 140: 1373–1383 [DOI] [PubMed] [Google Scholar]
- Han Y, Zhang C, Yang H, Jiao Y (2014) Cytokinin pathway mediates APETALA1 function in the establishment of determinate floral meristems in Arabidopsis. Proc Natl Acad Sci USA 111: 6840–6845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Müller B (2012) Cytokinin signaling networks. Annu Rev Plant Biol 63: 353–380 [DOI] [PubMed] [Google Scholar]
- Inskeep WP, Bloom PR (1985) Extinction coefficients of chlorophyll a and B in n,n-dimethylformamide and 80% acetone. Plant Physiol 77: 483–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Wellmer F, Muiño JM, Ferrier T, Wuest SE, Kumar V, Serrano-Mislata A, Madueño F, Krajewski P, Meyerowitz EM, Angenent GC, Riechmann JL (2010) Orchestration of floral initiation by APETALA1. Science 328: 85–89 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Baba S, Obayashi T, Sato M, Toyooka K, Keränen M, Aro EM, Fukaki H, Ohta H, Sugimoto K, Masuda T (2012) Regulation of root greening by light and auxin/cytokinin signaling in Arabidopsis. Plant Cell 24: 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllmer I, Werner T, Schmülling T (2011) Ectopic expression of different cytokinin-regulated transcription factor genes of Arabidopsis thaliana alters plant growth and development. J Plant Physiol 168: 1320–1327 [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E (2014) PIFs: systems integrators in plant development. Plant Cell 26: 56–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Li Y, Kristiansen K, Wang J (2008) SOAP: short oligonucleotide alignment program. Bioinformatics 24: 713–714 [DOI] [PubMed] [Google Scholar]
- Mara CD, Irish VF (2008) Two GATA transcription factors are downstream effectors of floral homeotic gene action in Arabidopsis. Plant Physiol 147: 707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Guo H, Yang H, Duong H, Lin C (1999) Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126: 2073–2082 [DOI] [PubMed] [Google Scholar]
- Muiño JM, Kaufmann K, van Ham RC, Angenent GC, Krajewski P (2011) ChIP-seq analysis in R (CSAR): an R package for the statistical detection of protein-bound genomic regions. Plant Methods 7: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Kiba T, Koizumi N, Yamashino T, Mizuno T (2007) Characterization of a unique GATA family gene that responds to both light and cytokinin in Arabidopsis thaliana. Biosci Biotechnol Biochem 71: 1557–1560 [DOI] [PubMed] [Google Scholar]
- Nawy T, Bayer M, Mravec J, Friml J, Birnbaum KD, Lukowitz W (2010) The GATA factor HANABA TARANU is required to position the proembryo boundary in the early Arabidopsis embryo. Dev Cell 19: 103–113 [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J (1994) Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol 104: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JC, Muro-Pastor MI, Florencio FJ (2004) The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol 134: 1718–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Bastakis E, Schwechheimer C (2013a) Cross-repressive interactions between SOC1 and the GATAs GNC and GNL/CGA1 in the control of greening, cold tolerance, and flowering time in Arabidopsis. Plant Physiol 162: 1992–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Behringer C, Müller IK, Schwechheimer C (2010) The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev 24: 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Behringer C, Zourelidou M, Schwechheimer C (2013b) Convergence of auxin and gibberellin signaling on the regulation of the GATA transcription factors GNC and GNL in Arabidopsis thaliana. Proc Natl Acad Sci USA 110: 13192–13197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Howell SH (1995) The effects of cytokinin and light on hypocotyl elongation in Arabidopsis seedlings are independent and additive. Plant Physiol 108: 1423–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Woodhouse MR, Cheng F, Schnable JC, Pedersen BS, Conant G, Wang X, Freeling M, Pires JC (2012) Altered patterns of fractionation and exon deletions in Brassica rapa support a two-step model of paleohexaploidy. Genetics 190: 1563–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, Huang S, Li X, et al. (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43: 1035–1039 [DOI] [PubMed] [Google Scholar]
- Weaver LM, Amasino RM (2001) Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiol 127: 876–886 [PMC free article] [PubMed] [Google Scholar]
- Whipple CJ, Hall DH, DeBlasio S, Taguchi-Shiobara F, Schmidt RJ, Jackson DP (2010) A conserved mechanism of bract suppression in the grass family. Plant Cell 22: 565–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhou Y, Ding L, Wu Z, Liu R, Meyerowitz EM (2013) Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell 25: 83–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Medrano L, Ohashi K, Fletcher JC, Yu H, Sakai H, Meyerowitz EM (2004) HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell 16: 2586–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.