The protease post-translationally fine-tunes a key enzyme in the synthesis of 5-Aminolevulinic acid during photoperiodic growth.
Abstract
5-Aminolevulinic acid (ALA) is the first committed substrate of tetrapyrrole biosynthesis and is formed from glutamyl-tRNA by two enzymatic steps. Glutamyl-tRNA reductase (GluTR) as the first enzyme of ALA synthesis is encoded by HEMA genes and tightly regulated at the transcriptional and posttranslational levels. Here, we show that the caseinolytic protease (Clp) substrate adaptor ClpS1 and the ClpC1 chaperone as well as the GluTR-binding protein (GBP) interact with the N terminus of GluTR. Loss-of function mutants of ClpR2 and ClpC1 proteins show increased GluTR stability, whereas absence of GBP results in decreased GluTR stability. Thus, the Clp protease system and GBP contribute to GluTR accumulation levels, and thereby the rate-limiting ALA synthesis. These findings are supported with Arabidopsis (Arabidopsis thaliana) hema1 mutants expressing a truncated GluTR lacking the 29 N-terminal amino acid residues of the mature protein. Accumulation of this truncated GluTR is higher in dark periods, resulting in increased protochlorophyllide content. It is proposed that the proteolytic activity of Clp protease counteracts GBP binding to assure the appropriate content of GluTR and the adequate ALA synthesis for chlorophyll and heme in higher plants.
In bacteria, archaea, and plants, 5-aminolevulinic acid (ALA), the universal precursor of all tetrapyrroles, is synthesized via the tRNA-dependent C5 pathway (Beale and Castelfranco, 1973; Avissar et al., 1989). Glutamyl-tRNA reductase (GluTR) catalyzes the NADPH-dependent reduction of glutamyl-tRNA(Glu) to produce Glu-1-semialdehyde, which is then converted into ALA by the Glu-1-semialdehyde aminotransferase (GSA-AT). GluTR is the first enzyme in the rate-limiting ALA synthesis and controlled by multiple regulatory mechanisms at the transcriptional and posttranslational levels.
Arabidopsis (Arabidopsis thaliana) GluTR is encoded by three HEMA genes. HEMA1 (AT1G58290) is predominantly expressed in green tissues (Ilag et al., 1994; McCormac et al., 2001; Matsumoto et al., 2004), whereas HEMA2 (AT1G09940) shows a low constitutive expression in all tissues (Ilag et al., 1994; Kumar et al., 1996; Matsumoto et al., 2004). HEMA3 (AT2G31250) is assumed to be a pseudogene (Matsumoto et al., 2004). The transcription of the HEMA genes is controlled by multiple endogenous and environmental factors, e.g. hormones, the endogenous clock, light, and sugars (Ilag et al., 1994; Ujwal et al., 2002; Matsumoto et al., 2004). In addition, ALA synthesis also is controlled by posttranslational feedback cues (Pontoppidan and Kannangara, 1994; Meskauskiene et al., 2001) and plastid-derived retrograde signaling (Kumar et al., 1999; Papenbrock et al., 2000a, 2000b; Cornah et al., 2003; Tanaka and Tanaka, 2007; Stenbaek and Jensen, 2010). The control of tetrapyrrole biosynthesis predominantly optimizes the formation of adequate amounts of chlorophyll (Chl) and heme and prevents accumulation of metabolic intermediates. Due to their photochemical properties, accumulating free tetrapyrroles generate highly reactive singlet oxygen upon illumination and cause severe photooxidative damage (Rebeiz et al., 1988; op den Camp et al., 2003). It is reasonable to predict particularly a multifaceted posttranslational control for rapid and instantaneous changes in the ALA synthesis rate with GluTR acting as major control step for the supply of metabolic precursors (Tanaka and Tanaka, 2007).
Among the posttranslational factors, heme and the tetratricopeptide repeat-containing FLUORESCENT IN BLUE LIGHT (FLU) have been reported to act negatively on ALA synthesis (Pontoppidan and Kannangara, 1994; Meskauskiene et al., 2001). Deficiency of the negative regulator FLU is associated with increasing content of protochlorophyllide (Pchlide), the substrate of NADPH-Pchlide oxidoreductase (POR), in darkness (Meskauskiene et al., 2001). Through interaction with the GluTR1 C terminus, FLU inactivates ALA synthesis in the dark (Goslings et al., 2004). Cofractionation of plastid extracts from photoperiodically grown Arabidopsis plants, which were harvested during the night period, revealed that FLU is assembled with CHL27, a subunit of Mg-protoporphyrin monomethylester cyclase, geranylgeranyl reductase, PORB, PORC, and GluTR1 (Kauss et al., 2012). The inhibitory activity of FLU emphasizes the importance of the Chl-synthesizing branch of tetrapyrrole biosynthesis for the feedback control of ALA synthesis. A crystal structure of a complex between two C-terminal GluTR dimerization domains and a dimer of the tetratricopeptide repeat domains of FLU revealed a shield formed by FLU around GluTR, which possibly is responsible for GluTR inactivation (Zhang et al., 2015).
Reduced heme content by the removal of free Fe2+/Fe3+ leads to an increase in the Pchlide content (Duggan and Gassman, 1974), whereas an accumulation of heme through inactivation of HEME OXYGENASE1 in the hy1 mutant suppresses Pchlide accumulation in dark-grown seedlings (Montgomery et al., 1999). Furthermore, incubation with heme diminished the activity of purified barley (Hordeum vulgare) GluTR (Pontoppidan and Kannangara, 1994; Vothknecht et al., 1996). The N-terminal 30 amino acid residues of mature GluTR were found to be required for heme inhibition and designated heme-binding domain (HBD; Vothknecht et al., 1998; Goslings et al., 2004). Nevertheless, a recently published x-ray structure analysis revealed a dimeric Arabidopsis GluTR1 structure but did not suggest a heme-binding motif of GluTR (Zhao et al., 2014).
Recently, a membrane-bound GluTR-binding protein (GBP; AT3G21200) was identified and suggested to act as an anchor for GluTR at the thylakoid membrane (Czarnecki et al., 2011). It was hypothesized that GBP protects bound GluTR from inactivation by FLU and thereby ensures an ongoing ALA formation for heme production in the dark (Czarnecki et al., 2011). This model supports previous ideas of a spatial separation of heme and Chl synthesis in chloroplasts (Joyard et al., 2009).
Using affinity chromatography, GluTR1 has been identified as one of the substrates for ClpS1 and ClpF that act as substrate selectors for the caseinolytic protease (Clp) in plastids (Nishimura et al., 2013, 2015). Proteases play an important role in embryogenesis, plastid biogenesis, and plant development, but the elucidation of posttranslational control by plastid-localized proteases is in its infancy. The ATP-dependent Clp protease is the most abundant stromal protease system in plastids and homologous to the bacterial Clp protease (Nishimura and van Wijk, 2015). Loss-of-function mutants of the plastid Clp protease complex indicate its essential role during embryogenesis, plant growth, and plastid development (Nishimura and van Wijk, 2015). Knockout mutants of the Clp core subunits can cause developmental arrests. In contrast, clpr2-1, a mutant with 20% of residual ClpR2 content, (Rudella et al., 2006) and the clpc1-1 mutants (Sjögren et al., 2004) used in our experiments are less severely affected, while the clps1 mutant is hardly phenotypically distinguishable from wild-type seedlings with the exception of a low reduction in Chl content (Nishimura et al., 2013). Recently, the thylakoid copper transporter PPA2 was shown to be degraded by the Clp system under copper-replete conditions. However, this degradation did not involve ClpS1, but was dependent on the ClpC1 chaperone and the ClpPR protease core complex (Tapken et al., 2015). Bacterial ClpS homologs function in the selection and delivery of substrates with an N-terminal degradation signal (an N-degron) for degradation by the Clp system. Early observations in yeast led to the formulation of the N-end rule, which states that certain amino acids, when exposed at the N terminus of a protein, act as triggers for degradation (Bachmair et al., 1986). An N-end rule for chloroplasts/plastids in plants is not known but certainly is a possibility (Apel et al., 2010; Rowland et al., 2015).
In this study, we aimed to examine the posttranslational regulation of GluTR1 in further detail in Arabidopsis and focus on the importance of the N-terminal domain in protein-protein interactions. Bimolecular complementation (BiFC) experiments provide evidence for interaction of ClpS1, ClpC1, and GBP with the N terminus of GluTR1. Furthermore, we show results suggesting the control of GluTR turnover by ClpC1 and the Clp protease complex Thus, this study reveals the importance of protein degradation in posttranslational regulation of GluTR.
RESULTS
The N-Terminal HBD of GluTR Interacts with GBP
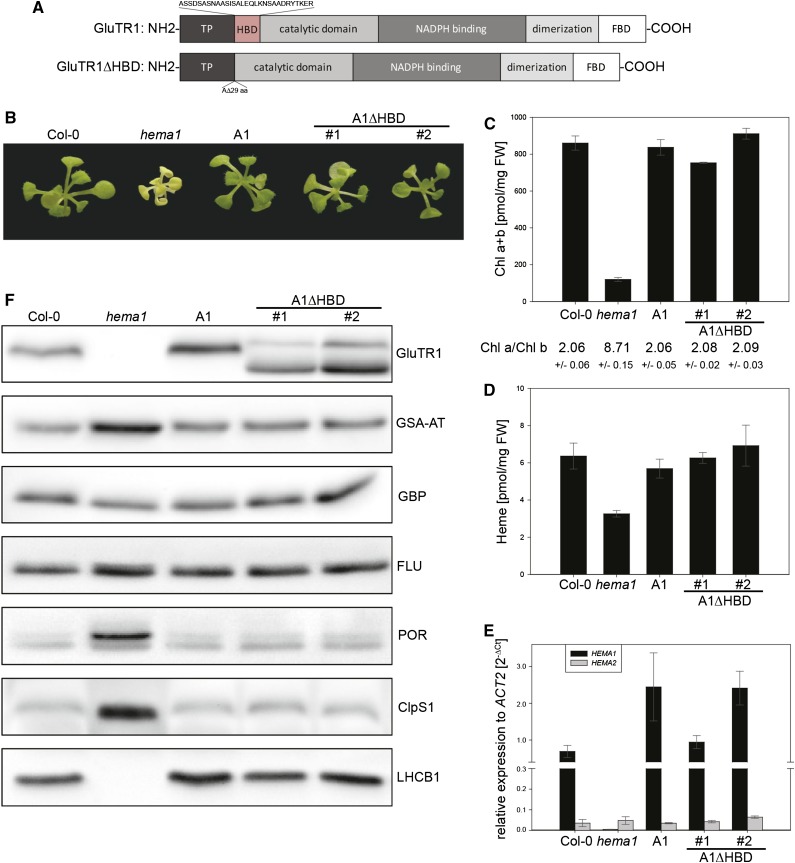
The HEMA1 knockout of Arabidopsis (hema1) shows a pale-green phenotype, does not grow photoautotrophically, and can be maintained on sugar-containing Murashige and Skoog agar, but is not able to produce seeds (Apitz et al., 2014). To elucidate the function of the HBD (Vothknecht et al., 1998) of GluTR, the coding sequence of HEMA1 with a deletion of the corresponding 87 nt was fused with HEMA1 promoter and 5′- as well as 3′-untranslated regions (UTRs), and expressed in the hema1 background to study protein-protein interaction, GluTR stability, and feedback regulation of ALA synthesis (Fig. 1A). Transgenic lines containing pHEMA1:HEMA1Δ87nt are designated A1ΔHBD #1 and A1ΔHBD #2 hereinafter. As complementation control, the entire genomic sequence of HEMA1 was fused with the HEMA1 promoter and UTR sequences and transformed into hema1. The resulting line pHEMA1:HEMA1 is designated A1 from here on.
Figure 1.
Complementation of the hema1 knockout mutant by A1 and A1ΔHBD. A, Schematic overview of the GluTR1 domains (Moser et al., 2001). The transit peptide (predicted by TargetP 1.1; Emanuelsson et al., 2007) is followed by the postulated HBD (Vothknecht et al., 1998; Goslings et al., 2004). Binding of FLU (FBD) is mediated by the C terminus of GluTR1 (Goslings et al., 2004). The truncated GluTR1 expressed in the A1ΔHBD lines misses the HBD. B, The A1ΔHBD lines expressing GluTR1ΔHBD and the control line A1 in comparison with the wild type (Col-0) and hema1. C, Pigment content based on fresh weight (FW) and Chl a/b ratio. D, Heme content related to fresh weight. E, Relative quantification of HEMA1 (black) and HEMA2 (gray) mRNA levels as compared with ACT2. F, Immunoblot analysis of total protein extracts. All analyzed plants were cultivated for 28 d on Suc-containing media under SD conditions (10 h light/14 h dark). Data are given as means ± sd (n = 4).
The homozygous hema1 background and the presence of the HEMA construct were confirmed by PCR genotyping. The control A1 and A1ΔHBD #1 and #2 lines showed full-grown complementation, including wild-type levels of Chl and heme, as well as Chl a/b ratios (Fig. 1, B–D). HEMA1 transcript levels in the control as well as in the A1ΔHBD lines reached at least wild-type expression levels (Fig. 1E) and a slightly increased GluTR1 content (Fig. 1F). The endogenous HEMA2 transcript levels remained largely unaffected (Fig. 1E). The mature, N-terminally truncated GluTR1 has a predicted molecular mass of 49.6 kD and migrated faster in polyacrylamide gels compared with the mature wild-type GluTR1 (52.7 kD; Fig. 1F). In both A1ΔHBD lines, a second band of higher molecular mass was visible and represents the truncated precursor protein including the transit peptide (56.5 kD), indicating that the removal of 29 amino acids at the mature N terminus influences the proteolytic cleavage of the transit peptide. Separation of stroma and thylakoid membrane fractions of isolated plastids from ecotype Columbia (Col-0) and A1ΔHBD seedlings confirmed processed mature variants of GluTR inside plastids, but not the much lower abundant precursor protein (Supplemental Fig. S1). The hema1 mutant showed increased amounts of the tetrapyrrole biosynthetic enzymes GSA-AT and POR, whereas the levels of GBP and FLU were not altered (Fig. 1F). Interestingly, the levels of the ClpS1 adaptor protein increased many fold and Lhcb1 proteins were missing (Fig. 1F). In contrast, A1 as well as A1ΔHBD #1 and #2 had wild-type amount of all analyzed proteins, indicating the successful complementation of hema1.
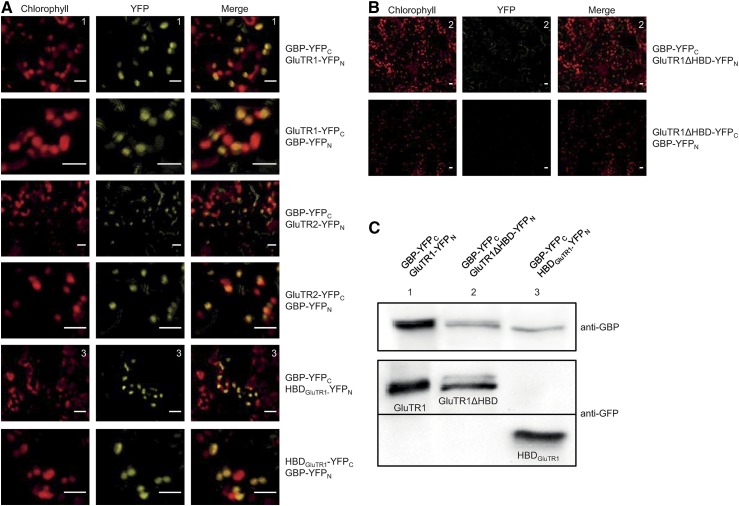
In yeast two-hybrid and BiFC experiments, GBP interacted with GluTR1 and GluTR2 (Czarnecki et al., 2011). To specify the domains of GluTR1 that interact with GBP, YFP halves fused with either truncated mature GluTR1ΔHBD (lacking the first 94 amino acid residues) or HBDGluTR1 were expressed. Coexpression of GBP and HBDGluTR1 resulted in YFP fluorescence, indicating an interaction of GBP with the N terminus of GluTR1 (Fig. 2A). In contrast, simultaneous expression of GBP and mature GluTR1ΔHBD did not exhibit YFP fluorescence, although western-blot analysis confirmed the expression of both fusion proteins with the YFP halves (Fig. 2, B and C).
Figure 2.
BiFC assay of GBP and GluTR exhibits interaction with the N-terminal domain of GluTR1. A, Transient coexpression of GBP with GluTR1, GluTR2, and the HBD of GluTR1 leads to YFP fluorescence in tobacco cells (scale = 10 µm). B, No YFP signal can be observed upon coexpression of GBP and the truncated GluTR1 missing HBD (scale = 10 µm). C, Immunoblot analysis of total protein extracts confirming the transient expression of GBP (with GBP antibody) and GluTR1, GluTR1ΔHBD, and HBDGluTR1 (with GFP antibody).
GBP Delays GluTR Degradation
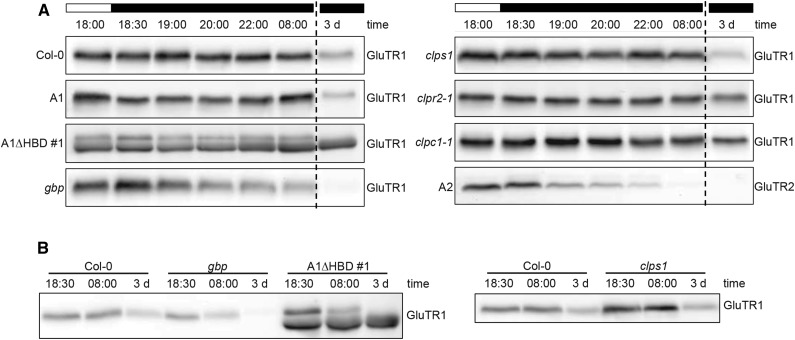
Western-blot analysis showed that the transgenic lines A1ΔHBD #1 and #2 exhibited a higher GluTR1 content compared with the wild type (Fig. 1F) To further investigate GluTR1 protein abundance and stability in adult plants cultivated under short day (SD) conditions, GluTR1 levels were examined in plant extracts harvested at different time points at night as well as after a 3-d dark period (Fig. 3). The GluTR1 content in the wild type and well as control line A1 plants was not altered during the night period, but decreased multifold after a 3-d dark period. The stability of GluTR1 expressed in line A1 resembled that of wild-type GluTR1 during the extended dark period. In contrast, A1ΔHBD displayed protein stability comparable to the wild type during the night phase, but the level of truncated GluTR1 was unaffected after the 3-d dark period (Fig. 3A; results shown only for A1ΔHBD #1 hereinafter). This indicated the HBD is critical for dark-induced degradation. For comparison, expression of HEMA2 under the HEMA1 promotor in hema1 (the line designated A2 hereinafter) leads to rapid GluTR2 degradation during a normal night phase and to undetectable protein levels following 72 h of darkness (Fig. 3).
Figure 3.
GluTR amount during the night and after a prolonged dark period. A, Western-blot analysis of GluTR1 and GluTR2 at the end of the light phase (18:00), during night (18:30–08:00), as well as following a prolonged dark period (3 d). B, Comparison of the GluTR1 protein amounts in A1ΔHBD, gbp, clps1, and Col-0.
The importance of the HBD of GluTR1 for interaction with GBP (Fig. 2, A and B) prompted us to analyze the GluTR1 content in the gbp mutant (SALK_200203) background. Here, GluTR1 levels were rapidly decreased after a few hours of darkness and below detection limit after a 3-d dark period (Fig. 3A). This indicates a reduced protein stability of GluTR1 in gbp due to accelerated degradation in the dark. When the GluTR1 amounts of the different lines were directly compared, the GluTR1 protein content was higher in the complemented line A1ΔHBD than in gbp and the wild type (Fig. 3B). This accumulation of GluTR1ΔHBD was not due to elevated transcript accumulation (see below) but rather due to enhanced protein stability.
Interactions between GluTR and the Adapter Protein ClpS1 and the Chaperones ClpC1 and ClpC2
GluTR1 was recently identified as a substrate of the Clp protease from stromal protein extracts of clps1 clpc1 double null mutants based on ClpS1 affinity purifications (Nishimura et al., 2013). GluTR1 amounts in clps1 (SAIL_326B_G12) were severalfold higher compared with the wild type (Fig. 3B) but decreased to wild-type levels after 3 d of darkness. These findings are consistent with the ClpS1 adaptor aiding in GluTR1 degradation efficiency, without being essential for its degradation. Apart from clps1, the clpc1-1 null mutant (SALK_014058, a knockout for a subunit of the chaperone complex of the Clp protease; Nishimura et al., 2013) and clpr2-1 (SALK_046378, a mutant for a subunit of the core complex; Rudella et al., 2006) were also examined for their GluTR1 content (Fig. 3A). The clpc1-1 and clpr2-1 mutants contained persistently a higher amount of GluTR1 after a 3-d dark period (Fig. 3A), which is not explained by reduced transcript levels of HEMA1 in the clp mutants during extended dark periods (Fig. 3; Supplemental Fig. S2).
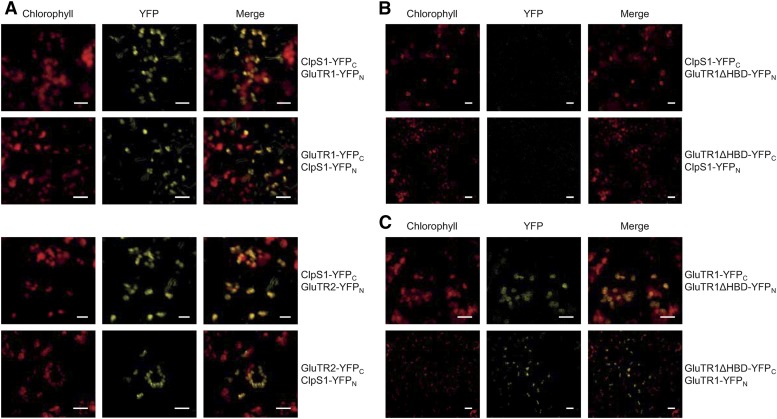
To verify the interaction between ClpS1 and GluTR1 in vivo, BiFC assays were carried out (Fig. 4A). ClpS1 interacts with GluTR1 as well as with GluTR2 (Fig. 4A), suggesting both GluTR proteins as substrates for the Clp protease degradation. To further specify the ClpS1 interaction with GluTR1, split-YFP fusions of the truncated GluTR1 missing 29 amino acids of the HBD and ClpS1 were transiently coexpressed in Nicotiana benthamiana (Fig. 4B). The absence of a YFP fluorescence signal underlined the importance of the N-terminal 29 amino acid residues of mature GluTR1 for the interaction. However, the truncated GluTR1 was still able to form a homodimer, indicating structural integrity of GluTR1ΔHBD (Fig. 4C).
Figure 4.
BiFC assay to specify the interaction of ClpS1 with GluTR1. A, Transient coexpression of ClpS1 with GluTR1 and GluTR2 in tobacco cells leads to the reconstitution of YFP fluorescence. B, No YFP signal can be observed for coexpression of ClpS1 with GluTR1ΔHBD. C, Coexpression of GluTR1 and GluTR1ΔHBD shows fluorescence signals (scale = 10 µm).
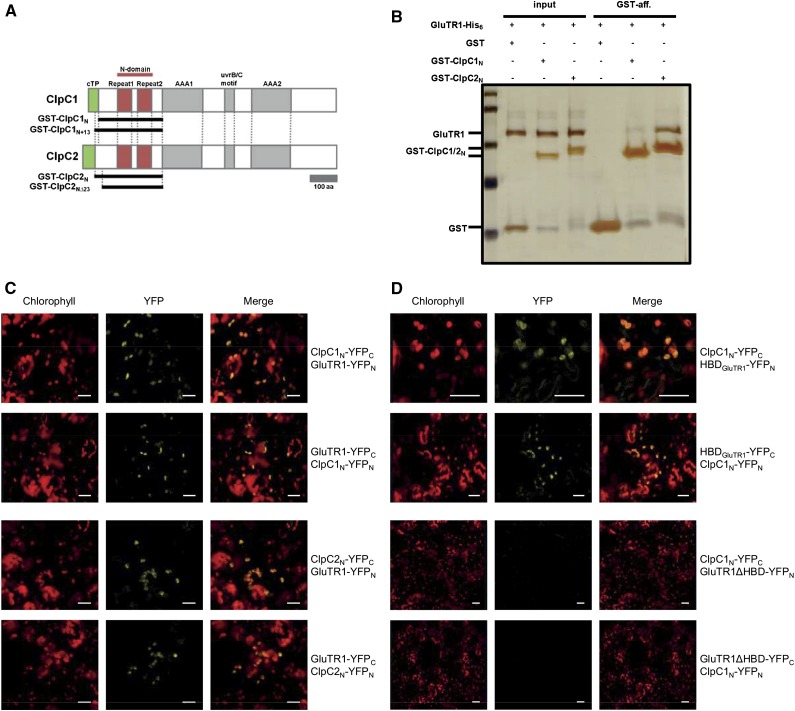
Physical GluTR1-ClpC1/C2 interaction also was demonstrated by pull-down experiments in vitro using recombinant N-domains of ClpC1 and ClpC2 (Fig. 5, A and B), confirming the specific selection of GluTR1 for the Clp-dependent proteolysis. Both ClpC1 and ClpC2 N-terminal domains bound to GluTR1 with ClpC2 displaying higher affinity (Fig. 5B). Longer ClpC1 constructs improved the GluTR1 binding, whereas shorter ClpC2 constructs reduced the interaction (Supplemental Fig. S3A; Fig. 3B), suggesting their N-terminal length as being a determinant for GluTR1 recognition and affinity. This is consistent with information about bacterial chaperone homologs that demonstrated the importance of the N-domain for substrate (and adaptor) interactions. In vivo BiFC assays confirmed the interaction of GluTR1 with ClpC1 and ClpC2 (Fig. 5C), and demonstrated that the HBD of GluTR1 is sufficient for the interaction with ClpC1 (Fig. 5D).
Figure 5.
ClpC chaperones can recognize GluTR1. A, Scheme of ClpC1/2 primary structures. ClpC proteins contain cTP sequences, N-domains with two tandem repeats for ClpS1 and substrate binding, two AAA modules (AAA1/2) for substrate unfolding and translocation, as well as uvrB/C motifs of unknown function. ClpC N-terminal domains fused to GST used in pull-down assays are indicated by black lines. B, GST pull down for GluTR1 interactions with N-terminal domains of ClpC1/2 (ClpC1N and ClpC2N). His-tagged GluTR1 (GluTR1-His6) and GST fusions of ClpC1/2N (GST-ClpC1/2N) were incubated (input lanes) and coeluted (GST lanes) with Laemmli buffer using glutathione sepharose resin. C, In BiFC assays the coexpression of GluTR1 with ClpC1N or ClpC2N leads to a YFP signal (scale = 10 µm). D, HBDGluTR1 is sufficient for the interaction with ClpC1N, while GluTR1ΔHBD does not interact with ClpC1.
Expression of GluTR1 Lacking the N Terminus Leads to Accumulation of Pchlide and Formation of Leaf Necrosis
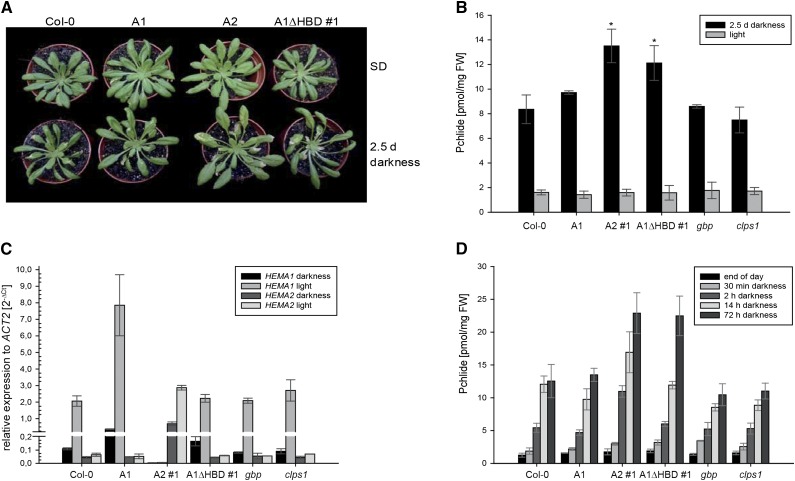
The A1ΔHBD lines, but not the wild type or control lines, showed a necrotic phenotype after a 3-d dark period, followed by 3 d of growth under SD condition (Fig. 6A). The necrotic tissue of A1ΔHBD upon exposure to light is explained by high-level accumulation of the photosensitizer Pchlide (Fig. 6B). The increased Pchlide level in A1ΔHBD after a 3-d dark period cannot not be explained by an inefficient interaction with FLU and therefore lack of down-regulation of GluTR1 activity since the FLU-binding region is not affected in A1ΔHBD (Goslings et al., 2004). Instead, necrosis and increased Pchlide content correlated with the elevated levels of truncated but active GluTR1 during the dark period. Furthermore, increased GluTR1 content is not due to a higher HEMA1 expression in darkness (Fig. 6C) but most likely to diminished proteolysis (Fig. 3).
Figure 6.
The A1ΔHBD line accumulates Pchlide after prolonged dark periods. The wild-type control (Col-0), the three transgenic lines A1, A2, and A1ΔHBD expressing HEMA1, HEMA2, and HEMA1Δ87nt, respectively, under the HEMA1 promoter in the hema1 background, the gbp, and the clps1 null mutant grew under SD conditions and were harvested during light exposure (light) and after 2.5 d of darkness and subsequent illumination under SD (2.5 d darkness). A, Phenotype of Col-0 and the three transgenic lines A1, A2, and A1ΔHBD expressing HEMA1, HEMA2, and HEMA1Δ87nt, respectively. B, Pchlide contents from leaf samples harvested in light under SD (gray) and 2.5 d of darkness (black). C, Relative quantification of HEMA1 (first two columns) and HEMA2 (last two columns) mRNA levels compared with ACT2 levels after 2.5 d of darkness (black) and under SD (gray). D, Pchlide amounts at different time points of the night and after 3 d of darkness. Data are given as means ± sd (n = 4). Asterisks mark significant difference from the wild type (P < 0.05).
A comparable necrotic phenotype has been described for hema1 expressing HEMA2 under the control of the HEMA1 promoter (Apitz et al., 2014). The necroses of this line also correlated with increased accumulation of Pchlide and were hence hypothesized to be a consequence of the photochemical properties of Pchlide upon light exposure (Apitz et al., 2014). Interestingly GluTR2, here exemplified by HEMA2 expression under the HEMA1 promotor in hema1, was degraded faster in darkness (Fig. 3) and caused elevated Pchlide prior to its rapid degradation in the dark. These processes in A2 are in contrast to the slower increase of excessive Pchlide amounts in A1ΔHBD during the dark period (Fig. 6D), which coincides with an attenuated GluTR degradation during the extended dark period in comparison to Col-0 and the GluTR2 and GluTR1 expressing lines (Fig. 3A).
In the A2 line, the missing FLU interaction of GluTR2 entails a more rapid increase of Pchlide accumulation within the first hours of darkness compared with the other analyzed lines (Fig. 6D). Pchlide contents of the A1ΔHBD line significantly increased between 14 and 72 h of darkness compared with the other analyzed lines, reaching similar Pchlide levels as the A2 line during darkness.
The clpr2-1 and clpc1-1 mutants showed an increase of the GluTR1 stability in a 3-d dark exposure (Fig. 3A). These mutants also were characterized by elevated levels of Pchlide, although these two mutants generally were hampered in the accumulation of Chl (Supplemental Fig. S4), which is likely due to a more complex defect in Clp-dependent chloroplast biogenesis (Nishimura et al., 2013).
DISCUSSION
Varying Demands for ALA Require Tight Posttranslational Control of the Two GluTR Isoforms
Continuing environmental changes require modification of photosynthesis rates and, consequently, assembly and disassembly of pigment-containing proteins of the two photosystems. This regulatory adjustment also requires a tight and rapid quantitative and qualitative control of Chl and heme synthesis. The posttranslational control of GluTR activity targets at the rate-determining initiating step into tetrapyrrole metabolism to adjust ALA synthesis to the levels of required end products (Tanaka and Tanaka, 2007). Two functional isoforms of GluTR are expressed in Arabidopsis under developmental and tissue-specific control. GluTR1 is most abundant in green tissue and thus mainly involved in the synthesis of ALA required for Chl synthesis. The corresponding HEMA1 gene is controlled by light and the circadian clock (Ilag et al., 1994; Matsumoto et al., 2004). In contrast, HEMA2 encoding GluTR2 is constitutively expressed at a lower level and is proposed to dedicate GluTR activity mainly in nonphotosynthetic tissue and under heterotrophic conditions (Kumar et al., 1996; Ujwal et al., 2002).
Besides the control of HEMA transcript amounts, the posttranslational regulation of the two GluTR isoforms is important to rapidly adjust ALA synthesis to the varying demands for Chl and heme. In angiosperms, Chl synthesis has to be down-regulated in darkness due to the light dependency of POR (Matsumoto et al., 2004). Thus, the dark repression of GluTR1 by FLU is essential to prevent excessive accumulation of Chl precursor Pchlide in the dark (Meskauskiene et al., 2001). The lack of FLU-mediated GluTR2 inactivation is dispensable and not hazardous due to the low expression of HEMA2 in leaf tissue of Col-0. It is proposed that HEMA2 expression ensures a continuous basal ALA synthesis during darkness in roots and shoots (Kumar et al., 1996; Nagai et al., 2007; Apitz et al., 2014). A decisive function and contribution of GSA-AT to the control of ALA synthesis in higher plants could not be demonstrated so far. However, as soon as GluTR2 is expressed to elevated levels due to HEMA2 expression under the HEMA1 promoter, nonrepressed GluTR2 activity in darkness leads to leaf necroses in plants grown under SD conditions (Apitz et al., 2014).
GluTR1 lacking this N terminus (−29 amino acid residues) entirely complements a hema1 mutant under normal growth conditions. Chl as well as heme content are not compromised (Fig. 1, D and E) and Pchlide accumulates to wild type-like levels during a 14-h night period, indicating a functional FLU repression of GluTR1ΔHBD (Fig. 6D), even though the amount of truncated GluTR1 is slightly elevated (Figs. 1F and 3B).
GluTR Is Substrate of the Clp Protease
In this study, we demonstrate a new posttranslational control mechanism of GluTR activity by protein stability that is executed by the combined action of the Clp protease system and GBP. Both the GBP and the Clp components interact with the N terminus of GluTR1 (Figs. 2 and 4). In our previous study using yeast two-hybrid analysis, we found the GluTR protein lacking the first 91 amino acid residues bound to GBP (Czarnecki et al., 2011), whereas the truncated GluTR expressed in planta for the BiFC assay lacks 94 amino acids. This might explain why we did not see interaction with GBP with the BiFC assay, although the western-blot analysis confirmed the expression of GluTR and GBP each with one of the YFP halves (Fig. 2, B and C). We redefined the length of GluTR-HBD for our BIFC experiments according to the proposed HBD reported by Vothknecht et al., (1998). It is proposed that the three additional residues of the HBD were permissive for the GluTR interaction with GBP.
Previous proteomic data suggested GluTR1 as a target of the Clp proteolytic system (Nishimura et al., 2013). Consistently, GluTR stability in vivo depends on the action of the Clp system. The proteolytic rate of GluTR appears to be controlled by the interdependent interaction of GBP and Clp components with GluTR (Fig. 3). The interaction of ClpS1 and ClpC1 with GluTR1, but not with GluTR1ΔHBD, characterizes a stretch of N-terminal amino acid residues of the mature GluTR1 as an N-degron (Fig. 4, A and B). Consequently, the truncated GluTR1 is more stable during prolonged darkness in comparison to the wild type (Fig. 3A), and enhanced Pchlide accumulation and necroses upon reillumination were observed as result of impaired GluTR1 degradation and elevated ALA synthesis during darkness, respectively (Fig. 6, A and B).
In prokaryotes, ClpS is a key factor in the N-end rule pathway by which the half-life of a protein is controlled according to the identity of the N-terminal amino acid residue (Varshavsky, 1996; Mogk et al., 2007). ClpS binds to the N-degron and delivers the thereby defined substrate to chaperone ClpA (Wang et al., 2008). In Arabidopsis, GluTR1 accumulates at higher levels in clps1 (Fig. 3B), while the dark-dependent degradation is wild type like (Fig. 3A). The data on GluTR accumulation in clps1 and GluTR interaction with Clp adaptor and chaperone are consistent with recent findings of the newly described ClpF subunit of the Clp protease also showing interaction with GluTR (Nishimura et al., 2015). The novel ClpF subunit is an additional substrate selector in plastids for Clp-dependent proteolysis (Nishimura et al., 2015) and acts in concert with ClpS1. It is suggested that both selectors either recognize GluTR in a combined complex or may compensatorily substitute each other. Therefore, GluTR1 content is diminished in dark-incubated clps1 as in the wild type and the A1 line (Fig. 3). In addition, dark-incubated clps1 seedlings contain wild type-like Pchlide levels, which are significantly lower after extended dark periods than those of the clpr2-1 and clpc1-1 mutants (Supplemental Fig. S4). In addition, consistent with the mutual activity of the two selector proteins and the elevated GluTR1 levels, the lack of ClpS1 correlates with enhanced ALA synthesis (Supplemental Fig. S5A) but not with elevated Chl precursor levels (Fig. 6, B and D; Supplemental Fig. S4B). However, when the structure and function of the Clp complex are generally impaired, as observed in clpc1-1 and clpr2-1, GluTR1 is significantly more stable (Fig. 3A) and more Pchlide accumulates in darkness (Supplemental Fig. S4A). It is currently proposed that the action of chaperone ClpC1 is more essential to recognize and direct GluTR1 to protein degradation into the Clp core complex than the other chaperone subunits and the two selector/adaptor proteins (Fig. 5C). It is not excluded that further selective and adaptive mechanisms adjust the rate of GluTR1 proteolysis. Interestingly, proteolysis of GluTR in the gram-negative bacterium Salmonella typhimurium also is controlled by the ClpP system as well as by LON protease (Wang et al., 1999). The degradation depends on the N-terminal region of GluTR. This example shows a striking evolutionary conservation of posttranslational regulation of GluTR for species that produce either heme exclusively or Chl and heme. Moreover, a degron in the N-terminal A domain of the Chl b-synthesizing Chl a oxygenase was identified for the Clp-dependent proteolytic degradation of the enzyme (Nakagawara et al., 2007; Sakuraba et al., 2009), indicating the regulatory importance of the Clp protease for the control of the Chl metabolism and the potential for the identification of additional future substrates among the enzymes of this pathway.
GBP Counteracts the Degradation of GluTR
It was previously shown that lack of GBP results in a minor reduction of ALA synthesis and correlates with reduced heme content rather than with reduced Chl content (Czarnecki et al., 2011). The FLU inhibition of GluTR1 in wild-type plants results in a restricted accumulation of Pchlide in the dark. In gbp, no perturbation of FLU action was observed as no leaf necroses were detected. As seen in A1ΔHBD and A2 (Apitz et al., 2014), leaf necroses were found when GluTR1 activity was not suppressed in the dark and subsequently accumulated Pchlide (Fig. 6, B and D).
Lack of GBP causes enhanced GluTR1 degradation (Fig. 3, A and B) because GluTR is more accessible as a Clp substrate and is apparently less protected against degradation (Figs. 2 and 4). Thus, the lowered GluTR1 content of gbp is most likely caused by its destabilization resulting from an increased accessibility of GluTR to the Clp proteolytic system (Fig. 3). However, future analysis of a gbp/clpc1-1 double mutant would provide the ultimate proof of this interdependent process.
Expression of HEMA2 under the HEMA1 promoter indicates that GluTR2 is faster degraded during the night period than GluTR1 in the wild type (Fig. 3A). Since GluTR2 also interacts with Clp protease subunits (Figs. 4A and 5C), it can be assumed that it is degraded via the Clp system as well. However, the pHEMA1:HEMA2 lines (A2) in the hema background contain the same levels of Chl and heme as wild-type seedlings (Apitz et al., 2014) but accumulate more Pchlide in the daily 14-h dark phase (Fig. 6, B and D). These data confirm that GluTR2 is not controlled by FLU (Apitz et al., 2014). Without FLU-dependent repression of ALA synthesis, the level of Pchlide increases rapidly within 2 h in darkness (Fig. 6D), even though GluTR2 is less stable than GluTR1.
Besides these data on the GluTR interaction with components of the Clp proteolytic system, an important regulatory role of ALA synthesis has been attributed to heme. Heme has been suggested to act as feedback regulator either by direct binding to GluTR or by mediating a posttranslational inhibition of ALA synthesis (Vothknecht et al., 1998). In clps1, slightly reduced Chl and heme levels (Supplemental Fig. S5B; Fig. 5C) correlated with elevated ALA synthesis rates (Supplemental Fig. S5A), so additional feedback regulation by heme cannot be excluded. However, the regulatory mechanism of heme action remains to be clarified. Structural analysis of GluTR1 and GBP (Zhao et al., 2014) did not provide hints for bound heme as a potential mediator of attenuated ALA synthesis (Vothknecht et al., 1998).
CONCLUSION
GluTR stability and enzyme activity are adapted to endogenous and environmental changes by several regulatory proteins. As demonstrated here, GBP and Clp protease counterbalance the activity of GluTR1 for Chl and heme synthesis by binding to its N terminus and thereby influencing the rate of its degradation. It is suggested that binding of GBP protects GluTR1 from degradation, while the Clp system is responsible for GluTR proteolysis. This control is hypothesized to balance GluTR1 activity during dark periods and, likely, also throughout light exposure. With this study, we suggest a more defined function of GBP for a balanced ALA synthesis capacity by preventing excessive proteolysis of GluTR. This regulatory mechanism may be beneficial for adequate heme synthesis (Fig. 7). It will require further work to identify the crucial amino acid residues at the N terminus of GluTR that interact with GBP and the Clp selector and chaperone subunits.
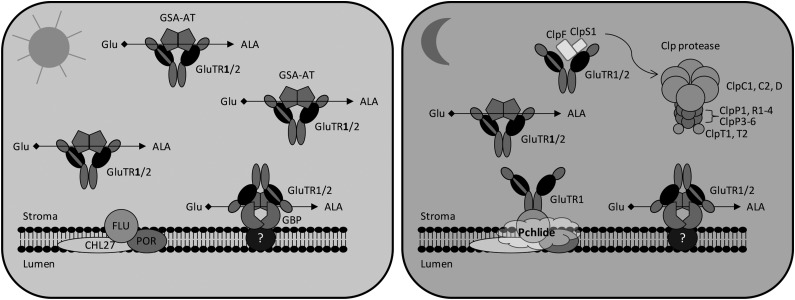
Figure 7.
A model for the role of GBP and the Clp protease activity in regulating GluTR in light- and dark-grown plants. The GSA-AT dimer has been proposed to interact with the V-shaped dimer of GluTR (Moser et al., 2001), allowing an efficient channeling of substrates to form ALA. In leaves, GluTR1 preferentially accumulates compared with the weakly expressed GluTR2 (Matsumoto et al., 2004). A minor amount of GluTR1/2 is bound to the thylakoid membrane via GBP (Czarnecki et al., 2011). During light exposure, the main ALA synthesis is suggested to be located in the stroma, while a minor portion of GluTR is attached to the membrane through interaction with GBP. In darkness, the predominant ALA synthesis is repressed by interaction of GluTR1 with the membrane-localized FLU through an unknown mechanism (Kauss et al., 2012). The binding of GluTR1/2 to GBP allows an ongoing ALA synthesis during darkness for continuous heme formation (Czarnecki et al., 2011). Unwanted GluTR1 and GluTR2 in the stroma are degraded through the Clp protease to prevent the accumulation of Pchlide and necrosis upon illumination (Rebeiz et al., 1988). The N-terminal HBD of GluTR is indicated in black.
MATERIALS AND METHODS
Plant Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown at 22°C, 100 µmol photons m−2 s−1, in 10 h light/14 h dark on soil or Murashige and Skoog medium (4.4 g L−1 Murashige and Skoog, 0.05% [w/v] MES, 1% [w/v] Suc, 0.8% [w/v] agar, pH 5.7). Nicotiana benthamiana plants were grown on soil in 14 h light/10 h dark at 23°C and 200 to 400 µmol photons m−2 s−1.
Generation of Arabidopsis Mutants and Transgenic Lines
The promoter/5′UTR and 3′UTR of HEMA1 were fused through overlap extension PCR using the primers listed in Supplemental Table S1. The fragment was inserted into pCAMBIA3301 using EcoRI and PmlI, and replaced p35S and GUS yielding vector pJA1. The complete HEMA1 coding sequence and a shortened sequence missing 87 bp at the 5′ end were inserted between 5′UTR and 3′UTR using AscI and SbfI. Heterozygous HEMA1 knockout plants (SALK_053036) were transformed and transformants selected on the herbicide BASTA.
DNA and RNA Analysis
DNA extracted with 200 mm Tris, pH 8.0, 150 mm NaCl, 25 mm EDTA, and 0.5% (w/v) SDS and precipitated with isopropyl alcohol was used for PCR genotyping (primers in Supplemental Table S1). One microgram of total RNA extracted using TRIsure (Bioline GmbH) was DNase I treated and reverse transcribed with oligo(dT)18 and RevertAid reverse transcriptase (Thermo Fisher Scientific). cDNA was amplified with SensiMix SYBR No-ROX kit (Bioline GmbH) on a CFX96 Real-Time System (Bio-Rad Laboratories GmbH; primers in Supplemental Table S1). Expression rates were calculated relative to ACTIN2 (ACT2; AT3G18780) according to the 2−ΔΔCT method (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008).
Quantification of Tetrapyrroles
Pchlide and Chl were extracted in alkaline acetone (9:1, 100% acetone:0.2 m NH4OH) and analyzed via HPLC as described (Papenbrock et al., 1999) or spectrophotometrically determined (Lichtenthaler, 1987). Leaf samples for Pchlide determination were fixed with steam for 2 min prior to extraction (Koski and Smith, 1948). Heme was resuspended from the pellet of the alkaline acetone extract two times in 100 µL of acetone/HCl/dimethyl sulfoxide (10:0.5:2, volume). Heme was separated on an Agilent 1290 system with a Poroshell 120 EC-C18 column (2.7 µm; 100 × 3.0 mm; 30°C) at a flow rate of 0.8 mL min−1, eluted with a gradient of solvent A (water, pH 3.2) and solvent B (methanol), and detected by a photodiode array (λ 398 nm, peak width 2.5 Hz; slit width 4 nm).
Determination of ALA Synthesis Rate
After 4-h incubation of 50 mg of seedlings in 5 mL of 50 mm Tris-HCl buffer, pH 7.2, containing 40 mm levulinic acid, 400 µL of homogenate in 1 mL of 20 mm potassium phosphate buffer, pH 6.8, was mixed with 100 µL of ethylacetoacetate and boiled for 10 min. One volume of Ehrlich’s reagent was added, and ALA derivatives were quantified at λ 553 nm (Mauzerall and Granick, 1956).
Immunoblot Analysis
Plant total protein extracts were generated by grinding plant tissue in liquid nitrogen and subsequent heating at 95°C for 5 min in 2% (w/v) SDS, 56 mm Na2CO3, 12% (w/v) Suc, 56 mm DTT, and 2 mm EDTA, pH 8.0. Proteins were separated on 12% or 15% polyacrylamide gels, transferred to Hybond-C membranes (GE Healthcare), and probed with specific antibodies using standard protocols (Sambrook and Russell, 2001). Antibodies for GluTR1, GluTR2, GSA-AT, GBP, and ClpS1 were generated in the lab (Grimm et al., 1989; Hedtke et al., 2007; Czarnecki et al., 2011; Nishimura et al., 2013; Apitz et al., 2014), and those for Lhcb1, POR, and GFP were purchased from Agrisera (Lhcb1 and POR) and Sigma (GFP). The anti-FLU antibody was kindly provided by Prof. K. Apel (Ithaca, NY).
BiFC Assay and BiFC Constructs
After leaf infiltration with Agrobacterium tumefaciens, coding sequences for GluTR1, GluTR2, GBP, HBDGluTR1, GluTR1ΔHBD, ClpS1, ClpC1N, and ClpC2N in the pSPYCE and pSPYNE plasmids (Walter et al., 2004) were expressed in N. benthamiana in darkness for 72 h. The fluorescence of the YFP was detected by a confocal laser scanning microscope (λex 514 nm, λem(YFP) 530–555 nm, λem(Chl) 600–700 nm).
Pull-Down Experiments
The PCR products encoding GluTR1 (in pET21a; Novagen) and various deletion series of ClpC1/2 (in pGEX-5X-1; GE Healthcare) were expressed in the Rosetta (DE3) strain (Novagen). GST fusions were purified through glutathione sepharose 4B resin (GE Healthcare) with 10 mm reduced glutathione (Sigma-Aldrich) and His-tagged proteins with Ni-NTA resin (Qiagen), before they were incubated together in 20 mm Tris-HCl, pH 7.5, 100 mm NaCl and 15% to 20% glycerol for 90 min at 22°C, followed by addition of glutathione sepharose 4B agarose. After 30 min at 4°C, the protein-bound resin was washed five times with 25 mm Tris-HCl, pH 7.5, 2 mm EDTA, 100 mm NaCl, and 0.5% Triton X-100, and the proteins were eluted in Laemmli buffer by heating at 75°C for 5 min, separated on SDS-PAGE gel, and visualized by silver nitrate staining.
Isolation and Fractionation of Arabidopsis Chloroplasts
Fifty grams of Arabidopsis leaves were harvested and disrupted in 500 mL of homogenization buffer (0.45 m sorbitol, 20 mm tricine, pH 8.4, 10 mm EDTA, 10 mm NaHCO3, and 0.1% [w/v] BSA). The homogenate was filtered through Miracloth (Calbiochem) and centrifuged for 2 min at 2,000g. The pellet was resuspended in 1 mL of RB buffer (0.3 m sorbitol, 20 mm tricine, pH 7.6, 5 mm MgCl2, and 2.5 mm EDTA). Intact chloroplasts were purified in a Percoll gradient (50% [v/v] Percoll in RB, centrifuged at 43,400g for 30 min) by overlaying the gradient with the chloroplast suspension. Intact chloroplasts were removed, washed two times in RB buffer, centrifuged (3,300g, 2 min), resuspended in 200 μL of PBS (20 mm sodium phosphate buffer and 150 mm NaCl, pH 7.4), and lysed on ice for 10 min. Stroma and thylakoid fractions were separated by centrifugation at 16,000g for 30 min. Thylakoids were suspended in 2 mL of PBS. Aliquots were washed two times with 1 mL of PBS and finally suspended in 200 μL of PBS.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Truncated GluTR protein can be found in plastids of the transgenic line A1∆HBD
Supplemental Figure S2. HEMA1 mRNA level in clp mutants in comparison to the wild type.
Supplemental Figure S3. The N-terminal region of ClpC chaperones can recognize GluTR1.
Supplemental Figure S4. Pchlide and Chl a/b contents in clp mutants in comparison to the wild type.
Supplemental Figure S5. ALA synthesis rate and heme and pigment content of complemented line A1 and knockout mutants of GBP and ClpS1 in comparison to the wild type.
Supplemental Table S1. Primers used in this work.
Supplementary Material
Acknowledgments
J.A., B.H., and B.G. designed the research; J.A., J.S., A.W., and K.N. performed the experiments; J.A., B.H., K.J.v.W., and B.G. contributed analytic tools and analyzed data; J.A. and B.G. wrote the article.
Glossary
- ALA
5-aminolevulinic acid
- GluTR
glutamyl-tRNA reductase
- GSA-AT
Glu-1-semialdehyde aminotransferase
- Pchlide
protochlorophyllide
- BiFC
bimolecular complementation
- UTR
untranslated region
- SD
short day
Footnotes
This work was supported by the German Research Foundation (Grant GR936 15-1 to B.G.).
Articles can be viewed without a subscription.
References
- Apel W, Schulze WX, Bock R (2010) Identification of protein stability determinants in chloroplasts. Plant J 63: 636–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apitz J, Schmied J, Lehmann MJ, Hedtke B, Grimm B (2014) GluTR2 complements a hema1 mutant lacking glutamyl-tRNA reductase 1, but is differently regulated at the post-translational level. Plant Cell Physiol 55: 645–657 [DOI] [PubMed] [Google Scholar]
- Avissar YJ, Ormerod JG, Beale SI (1989) Distribution of delta-aminolevulinic acid biosynthetic pathways among phototrophic bacterial groups. Arch Microbiol 151: 513–519 [DOI] [PubMed] [Google Scholar]
- Bachmair A, Finley D, Varshavsky A (1986) In vivo half-life of a protein is a function of its amino-terminal residue. Science 234: 179–186 [DOI] [PubMed] [Google Scholar]
- Beale SI, Castelfranco PA (1973) 14 C incorporation from exogenous compounds into -aminolevulinic acid by greening cucumber cotyledons. Biochem Biophys Res Commun 52: 143–149 [DOI] [PubMed] [Google Scholar]
- Cornah JE, Terry MJ, Smith AG (2003) Green or red: what stops the traffic in the tetrapyrrole pathway? Trends Plant Sci 8: 224–230 [DOI] [PubMed] [Google Scholar]
- Czarnecki O, Hedtke B, Melzer M, Rothbart M, Richter A, Schröter Y, Pfannschmidt T, Grimm B (2011) An Arabidopsis GluTR binding protein mediates spatial separation of 5-aminolevulinic acid synthesis in chloroplasts. Plant Cell 23: 4476–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan J, Gassman M (1974) Induction of porphyrin synthesis in etiolated bean leaves by chelators of iron. Plant Physiol 53: 206–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Goslings D, Meskauskiene R, Kim C, Lee KP, Nater M, Apel K (2004) Concurrent interactions of heme and FLU with Glu tRNA reductase (HEMA1), the target of metabolic feedback inhibition of tetrapyrrole biosynthesis, in dark- and light-grown Arabidopsis plants. Plant J 40: 957–967 [DOI] [PubMed] [Google Scholar]
- Grimm B, Bull A, Welinder KG, Gough SP, Kannangara CG (1989) Purification and partial amino acid sequence of the glutamate 1-semialdehyde aminotransferase of barley and synechococcus. Carlsberg Res Commun 54: 67–79 [DOI] [PubMed] [Google Scholar]
- Hedtke B, Alawady A, Chen S, Börnke F, Grimm B (2007) HEMA RNAi silencing reveals a control mechanism of ALA biosynthesis on Mg chelatase and Fe chelatase. Plant Mol Biol 64: 733–742 [DOI] [PubMed] [Google Scholar]
- Ilag LL, Kumar AM, Söll D (1994) Light regulation of chlorophyll biosynthesis at the level of 5-aminolevulinate formation in Arabidopsis. Plant Cell 6: 265–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyard J, Ferro M, Masselon C, Seigneurin-Berny D, Salvi D, Garin J, Rolland N (2009) Chloroplast proteomics and the compartmentation of plastidial isoprenoid biosynthetic pathways. Mol Plant 2: 1154–1180 [DOI] [PubMed] [Google Scholar]
- Kauss D, Bischof S, Steiner S, Apel K, Meskauskiene R (2012) FLU, a negative feedback regulator of tetrapyrrole biosynthesis, is physically linked to the final steps of the Mg(++)-branch of this pathway. FEBS Lett 586: 211–216 [DOI] [PubMed] [Google Scholar]
- Koski VM, Smith JH (1948) The isolation and spectral absorption properties of protochlorophyll from etiolated barley seedlings. J Am Chem Soc 70: 3558–3562 [DOI] [PubMed] [Google Scholar]
- Kumar AM, Csankovszki G, Söll D (1996) A second and differentially expressed glutamyl-tRNA reductase gene from Arabidopsis thaliana. Plant Mol Biol 30: 419–426 [DOI] [PubMed] [Google Scholar]
- Kumar MA, Chaturvedi S, Söll D (1999) Selective inhibition of HEMA gene expression by photooxidation in Arabidopsis thaliana. Phytochemistry 51: 847–851 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic bio-membranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Matsumoto F, Obayashi T, Sasaki-Sekimoto Y, Ohta H, Takamiya K, Masuda T (2004) Gene expression profiling of the tetrapyrrole metabolic pathway in Arabidopsis with a mini-array system. Plant Physiol 135: 2379–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauzerall D, Granick S (1956) The occurrence and determination of delta-amino-levulinic acid and porphobilinogen in urine. J Biol Chem 219: 435–446 [PubMed] [Google Scholar]
- McCormac AC, Fischer A, Kumar AM, Söll D, Terry MJ (2001) Regulation of HEMA1 expression by phytochrome and a plastid signal during de-etiolation in Arabidopsis thaliana. Plant J 25: 549–561 [DOI] [PubMed] [Google Scholar]
- Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K (2001) FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Schmidt R, Bukau B (2007) The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol 17: 165–172 [DOI] [PubMed] [Google Scholar]
- Montgomery BL, Yeh KC, Crepeau MW, Lagarias JC (1999) Modification of distinct aspects of photomorphogenesis via targeted expression of mammalian biliverdin reductase in transgenic Arabidopsis plants. Plant Physiol 121: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser J, Schubert WD, Beier V, Bringemeier I, Jahn D, Heinz DW (2001) V-shaped structure of glutamyl-tRNA reductase, the first enzyme of tRNA-dependent tetrapyrrole biosynthesis. EMBO J 20: 6583–6590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S, Koide M, Takahashi S, Kikuta A, Aono M, Sasaki-Sekimoto Y, Ohta H, Takamiya K, Masuda T (2007) Induction of isoforms of tetrapyrrole biosynthetic enzymes, AtHEMA2 and AtFC1, under stress conditions and their physiological functions in Arabidopsis. Plant Physiol 144: 1039–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawara E, Sakuraba Y, Yamasato A, Tanaka R, Tanaka A (2007) Clp protease controls chlorophyll b synthesis by regulating the level of chlorophyllide a oxygenase. Plant J 49: 800–809 [DOI] [PubMed] [Google Scholar]
- Nishimura K, Apitz J, Friso G, Kim J, Ponnala L, Grimm B, van Wijk KJ (2015) Discovery of a unique Clp component, ClpF, in chloroplasts: a proposed binary ClpF-ClpS1 adaptor complex functions in substrate recognition and delivery. Plant Cell 27: 2677–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Asakura Y, Friso G, Kim J, Oh SH, Rutschow H, Ponnala L, van Wijk KJ (2013) ClpS1 is a conserved substrate selector for the chloroplast Clp protease system in Arabidopsis. Plant Cell 25: 2276–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, van Wijk KJ (2015) Organization, function and substrates of the essential Clp protease system in plastids. Biochim Biophys Acta 1847: 915–930 [DOI] [PubMed] [Google Scholar]
- op den Camp RG, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Göbel C, Feussner I, et al. (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15: 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenbrock J, Mock HP, Kruse E, Grimm B (1999) Expression studies in tetrapyrrole biosynthesis: inverse maxima of magnesium chelatase and ferrochelatase activity during cyclic photoperiods. Planta 208: 264–273 [Google Scholar]
- Papenbrock J, Mock HP, Tanaka R, Kruse E, Grimm B (2000a) Role of magnesium chelatase activity in the early steps of the tetrapyrrole biosynthetic pathway. Plant Physiol 122: 1161–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenbrock J, Pfündel E, Mock HP, Grimm B (2000b) Decreased and increased expression of the subunit CHL I diminishes Mg chelatase activity and reduces chlorophyll synthesis in transgenic tobacco plants. Plant J 22: 155–164 [DOI] [PubMed] [Google Scholar]
- Pontoppidan B, Kannangara CG (1994) Purification and partial characterisation of barley glutamyl-tRNA(Glu) reductase, the enzyme that directs glutamate to chlorophyll biosynthesis. Eur J Biochem 225: 529–537 [DOI] [PubMed] [Google Scholar]
- Rebeiz CA, Montazer‐Zouhoor A, Mayasich JM, Tripathy BC, Wu SM, Rebeiz CC, Friedmann HC (1988) Photodynamic herbicides. Recent developments and molecular basis of selectivity. Crit Rev Plant Sci 6: 385–436 [Google Scholar]
- Rowland E, Kim J, Bhuiyan NH, van Wijk KJ (2015) The Arabidopsis chloroplast stromal N-terminome: complexities of amino-terminal protein maturation and stability. Plant Physiol 169: 1881–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudella A, Friso G, Alonso JM, Ecker JR, van Wijk KJ (2006) Downregulation of ClpR2 leads to reduced accumulation of the ClpPRS protease complex and defects in chloroplast biogenesis in Arabidopsis. Plant Cell 18: 1704–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y, Tanaka R, Yamasato A, Tanaka A (2009) Determination of a chloroplast degron in the regulatory domain of chlorophyllide a oxygenase. J Biol Chem 284: 36689–36699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Sjögren LL, MacDonald TM, Sutinen S, Clarke AK (2004) Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiol 136: 4114–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenbaek A, Jensen PE (2010) Redox regulation of chlorophyll biosynthesis. Phytochemistry 71: 853–859 [DOI] [PubMed] [Google Scholar]
- Tanaka R, Tanaka A (2007) Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol 58: 321–346 [DOI] [PubMed] [Google Scholar]
- Tapken W, Kim J, Nishimura K, van Wijk KJ, Pilon M (2015) The Clp protease system is required for copper ion-dependent turnover of the PAA2/HMA8 copper transporter in chloroplasts. New Phytol 205: 511–517 [DOI] [PubMed] [Google Scholar]
- Ujwal ML, McCormac AC, Goulding A, Kumar AM, Söll D, Terry MJ (2002) Divergent regulation of the HEMA gene family encoding glutamyl-tRNA reductase in Arabidopsis thaliana: expression of HEMA2 is regulated by sugars, but is independent of light and plastid signalling. Plant Mol Biol 50: 83–91 [DOI] [PubMed] [Google Scholar]
- Varshavsky A. (1996) The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci USA 93: 12142–12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vothknecht UC, Kannangara CG, von Wettstein D (1996) Expression of catalytically active barley glutamyl tRNAGlu reductase in Escherichia coli as a fusion protein with glutathione S-transferase. Proc Natl Acad Sci USA 93: 9287–9291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vothknecht UC, Kannangara CG, von Wettstein D (1998) Barley glutamyl tRNAGlu reductase: mutations affecting haem inhibition and enzyme activity. Phytochemistry 47: 513–519 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang KH, Roman-Hernandez G, Grant RA, Sauer RT, Baker TA (2008) The molecular basis of N-end rule recognition. Mol Cell 32: 406–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wilson S, Elliott T (1999) A mutant HemA protein with positive charge close to the N terminus is stabilized against heme-regulated proteolysis in Salmonella typhimurium. J Bacteriol 181: 6033–6041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Zhang F, Fang Y, Chen X, Chen Y, Zhang W, Dai HE, Lin R, Liu L (2015) The non-canonical tetratricopeptide repeat (TPR) domain of Fluorescent (FLU) mediates complex formation with glutamyl-tRNA reductase. J Biol Chem 290: 17559–17565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao A, Fang Y, Chen X, Zhao S, Dong W, Lin Y, Gong W, Liu L (2014) Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its stimulator protein. Proc Natl Acad Sci USA 111: 6630–6635 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.