Soil flooding reactivates dormant adventitious root primordia via ethylene-induced reduction in ABA level.
Abstract
Soil flooding is a common stress factor affecting plants. To sustain root function in the hypoxic environment, flooding-tolerant plants may form new, aerenchymatous adventitious roots (ARs), originating from preformed, dormant primordia on the stem. We investigated the signaling pathway behind AR primordium reactivation in the dicot species Solanum dulcamara. Transcriptome analysis indicated that flooding imposes a state of quiescence on the stem tissue, while increasing cellular activity in the AR primordia. Flooding led to ethylene accumulation in the lower stem region and subsequently to a drop in abscisic acid (ABA) level in both stem and AR primordia tissue. Whereas ABA treatment prevented activation of AR primordia by flooding, inhibition of ABA synthesis was sufficient to activate them in absence of flooding. Together, this reveals that there is a highly tissue-specific response to reduced ABA levels. The central role for ABA in the response differentiates the pathway identified here from the AR emergence pathway known from rice (Oryza sativa). Flooding and ethylene treatment also induced expression of the polar auxin transporter PIN2, and silencing of this gene or chemical inhibition of auxin transport inhibited primordium activation, even though ABA levels were reduced. Auxin treatment, however, was not sufficient for AR emergence, indicating that the auxin pathway acts in parallel with the requirement for ABA reduction. In conclusion, adaptation of S. dulcamara to wet habitats involved co-option of a hormonal signaling cascade well known to regulate shoot growth responses, to direct a root developmental program upon soil flooding.
Flooding is among the most commonly occurring abiotic stress factors that affect plant growth and performance (Bailey-Serres et al., 2012). The primary effect of this stressor is a slower gas diffusion (about 10,000-fold less in water than in air; Armstrong et al., 1991), which leads to reduced gas exchange between the plant and its environment, thereby disturbing internal concentrations of oxygen, carbon dioxide, and ethylene (Bailey-Serres and Voesenek, 2008). This results in detrimental effects on cellular metabolic homeostasis, but at the same time these changes in gas concentrations are used as cues to alter gene expression leading to acclimation. In many plant species, genes involved in (hormonal) signaling, photosynthesis, anaerobic metabolism, and secondary metabolism are rapidly modulated in response to flooding (Zhang et al., 2006; Lee et al., 2007; Christianson et al., 2010; Lee et al., 2011; Nanjo et al., 2011; Qi et al., 2012). To prevent or shorten the imminent energy crisis, plant species from wetland habitats show morphological adaptations such as elongation of the shoot, formation of aerenchyma in existing tissues, or development of new, aerenchymatous adventitious roots (ARs), all of which are aimed at improving gas exchange between plant tissues and the atmosphere (Bailey-Serres and Voesenek, 2008).
In flood-tolerant species like marsh dock (Rumex palustris), rice (Oryza sativa), and bittersweet (Solanum dulcamara), new ARs emerge very rapidly upon flooding, as they derive from preformed, dormant primordia (Visser et al., 1996a; Lorbiecke and Sauter, 1999; Dawood et al., 2014). Although AR primordia do not necessarily originate from the pericycle as lateral root (LR) primordia do, on the molecular and physiological level, initiation of AR and LR primordia show considerable similarity, with auxin taking a central role in both cases (Bellini et al., 2014). Mutants defective in the initiation of ARs have been identified in rice and maize (Zea mays; Inukai et al., 2001; Taramino et al., 2007; Kitomi et al., 2011), and the underlying genes of crown rootless1/adventitious rootless1 (CRL1/ARL1; in rice) and rootless concerning crown and seminal roots (RTCS; in maize) were shown to encode orthologous members of the LATERAL ORGAN BOUNDARIES DOMAIN/ASYMMETRIC LEAVES2-LIKE (LBD/ASL) protein family involved in LR initiation in Arabidopsis (Arabidopsis thaliana; Kitomi et al., 2011; Liu et al., 2005). As in the case of the LBD/ASL genes, the expression of RTCS and CRL1 is rapidly induced by application of auxin, and both genes are thought to be direct targets of AUXIN RESPONSE FACTOR (ARF) transcription factors in the auxin signaling pathway.
The process of activation of dormant AR primordia has been studied to a much lesser extent in rice and R. palustris. In both species, ethylene is a key regulator of the process (Visser et al., 1996b, 1996c; Mergemann and Sauter, 2000). In rice, the ethylene that accumulates upon submergence induces formation of the reactive oxygen species (ROS) hydrogen peroxide, which in turn leads to cell death in the epidermal cell layer that covers the AR primordia at the nodes, thereby facilitating their emergence (Mergemann and Sauter, 2000). Treatment with the plant hormone GA alone had little effect on root emergence, while combined application with ethylene, in the form of the ethylene-releasing compound ethephon, increased the number of emerging ARs rapidly. By contrast, abscisic acid (ABA) inhibited root emergence, even when it was combined with ethephon and GA (Steffens et al., 2006). The authors therefore suggested a hormonal model in which the balance between the positive, synergistic action of ethylene and GA and the inhibitory action of ABA regulates the process of epidermal cell death and AR emergence (Steffens et al., 2006). Ethylene and hydrogen peroxide promote epidermal cell death by coregulating a small set of genes (Steffens and Sauter, 2009). Among these, metallothionein MT2b was down-regulated, and this was shown to be sufficient to induce cell death. As this gene encodes a ROS scavenger, its down-regulation seems to be part of a positive feedback loop that auto-amplifies ROS levels. In R. palustris, ethylene accumulation was observed in roots following 24 h of submergence, and inhibition of ethylene production decreased the number of ARs induced by flooding (Visser et al., 1996b). While no change in the endogenous free auxin concentration was detected upon flooding, blocking polar auxin transport with N-1-naphtylphtalamic acid (NPA) decreased the number of ARs after flooding (Visser et al., 1996c). Reconciling these findings, it was found that accumulation of ethylene leads to AR emergence by increasing the sensitivity to auxin in the rooting zone (Visser et al., 1996c).
Although we have first clues about hormones involved in activation of preformed AR primordia in mono- and dicot plants, the way in which they interact to form a signaling pathway is still unclear. Moreover, to advance our understanding of the AR activation process, there is a need to determine the molecular and physiological targets of the hormone signaling pathways involved. We have recently shown that flooding rapidly activates the dormant AR primordia on the stem of the dicot species S. dulcamara (Dawood et al., 2014), and we generated the genomic tools necessary for using this species as a model system (Golas et al., 2013; D’Agostino et al., 2013). To further our understanding about the molecular and physiological mechanisms that enable dormant AR primordia to resume growth and develop into roots upon an environmental trigger, we analyzed the transcriptomic response to flooding as well as the hormonal regulation of the process in S. dulcamara.
RESULTS
Submergence Causes Extensive Transcriptomic Reprogramming
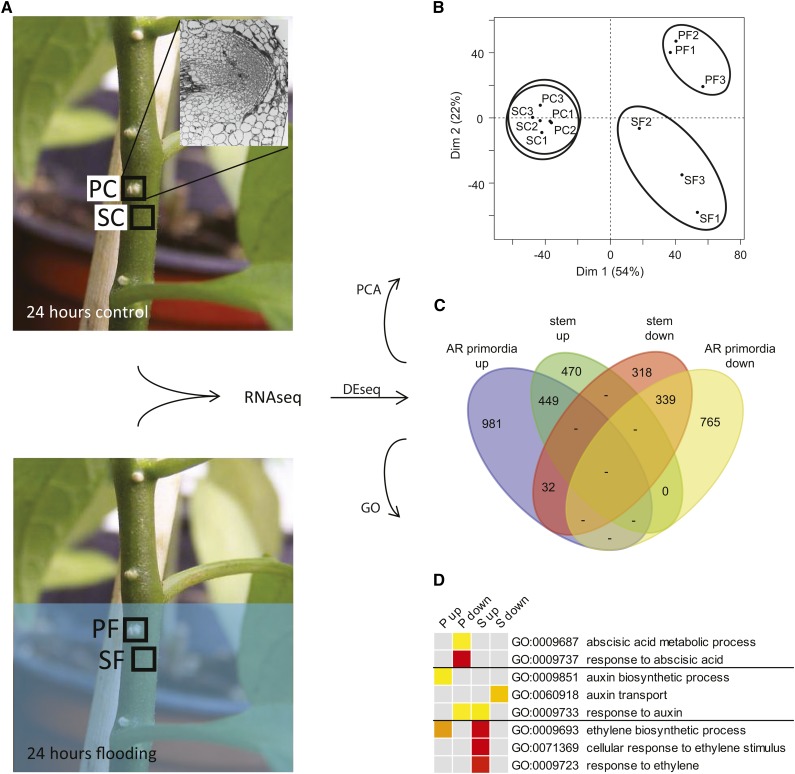
The well-described mechanism for flooding-induced breaking of AR primordia dormancy in rice involves hydrogen peroxide-dependent epidermal cell death, resulting in removal of the physical restriction to primordium growth (Steffens and Sauter, 2009). In S. dulcamara, the mechanism is fundamentally different, as AR primordia are located much deeper, covered by several layers of cortex cells (Fig. 1A, inset; Dawood et al., 2014), and, importantly, the epidermis at the site of the AR primordia already contains numerous cracks surrounded by dead cells before flooding (Supplemental Fig. S1). To characterize the molecular changes occurring during activation of dormant AR primordia in S. dulcamara, plants were flooded to 15 cm above soil level, and submerged AR primordia and bordering stem tissues were dissected separately from plants partially flooded for 24 h and control plants (Fig. 1A). The reads obtained from RNA sequencing (RNAseq) of the samples were subsequently mapped to the S. dulcamara transcriptome assembly (D’Agostino et al., 2013). In this way, we detected expression of 28,035 transcripts, corresponding to 87% of all transcripts in the assembly. After correcting for multiple testing, a total of 3354 genes showed differential expression (fold change ≥ 2, false discovery rate ≤ 0.05) in ARs, stem, or both tissues upon flooding (Supplemental Data Set S1). Principal component (PC) analysis indicated that the main difference between the samples, i.e. PC1, closely correlated to flooding treatment (Fig. 1B). The second largest source of variation, PC2, separated the tissue types, primordium and stem, but only after the flooding treatment, indicating a differential response of the two tissues to flooding. Slightly more genes were up-regulated than down-regulated (1932 versus 1454 genes; χ2, P < 0.001; Fig. 1C), and the overall transcriptomic response was stronger in primordia than in stem tissue (2566 versus 1608 genes; χ2, P < 0.001). Only a small number of genes (32) showed opposite regulation in the two tissues. To validate these RNAseq results, we looked at six genes whose behavior upon flooding was characterized before (Dawood et al., 2014), and found that five showed the expected direction of change in expression while one was below detection level (Supplemental Data Set S2). Furthermore, we selected eight genes from the current data set that appeared up-regulated specifically in the primordia (Supplemental Data Set S2) and tested their expression in a set of samples obtained from a new submergence experiment. All eight genes were significantly up-regulated by flooding in primordia, and only one of them also was slightly but significantly induced in the stem after 24 h of submergence (Supplemental Fig. S2). Together, these results confirm the validity of the RNAseq data.
Figure 1.
Flooding leads to extensive transcriptomic reprogramming in stem and AR primordia of S. dulcamara. A, Analysis pipeline showing the origin and processing of the four sample types (n = 3). Inset in the top left photo represents the composition of a primordium sample. B, PC analysis score plot showing the differentiation between the four sample types. C, Venn diagram of all genes differentially expressed in AR primordia and/or stem tissue upon flooding (fold change ≥ 2, false discovery rate ≤ 0.05). D, GO analysis of hormone-related categories. Color scale indicates significance of enrichment, from false discovery rate = 1 × 10−2 (yellow) to false discovery rate = 1 × 10−6 (red). PC, Primordia control; PF, primordia flooded; SC, stem control; SF, stem flooded; P, primordia; S, stem; up, up-regulated genes; down, down-regulated genes.
Submergence Results in Tissue-Specific Changes to Basic Cytological Processes
To understand what kind of processes take place in the AR primordia and surrounding stem upon flooding, we first applied enrichment analysis of gene ontology (GO) categories. For obtaining a broad overview, these were subsequently merged to reduced GOSlim categories (Supplemental Fig. S3). The analysis showed that stress response-related genes were both up- and down-regulated, but with a bias toward down-regulation, especially in the AR primordia. An example of the latter are genes related to the response to water deprivation (GO:0009414). Commonly up-regulated stress response genes included, as expected, those related to reduced oxygen levels (GO:0070482, 0036293, 0001666, 0034059), such as alcohol dehydrogenase (ADH) and pyruvate decarboxylase (PDC; Supplemental Data Set S3). Analysis of earlier time points indicated that the hypoxic response, i.e. ADH1 and PDC induction, could be detected already 3 h after the onset of submergence (Supplemental Fig. S4). Similar to above, the broad category of metabolism-related genes was both up- and down-regulated, but with a bias toward down-regulation in the AR primordia. This included genes functioning in photosynthesis, secondary metabolism, and lipid metabolism. Clear tissue differentiation was seen concerning genes involved in cellular activity (e.g. translation, protein metabolism, and cellular component organization), which were highly significantly up-regulated in the AR primordia specifically, and genes involved in cell division, growth, and differentiation, which were strongly overrepresented among down-regulated genes in the stem specifically. For example, cyclin (Cyc) genes showed a clear trend toward up-regulation in the primordia and down-regulation in the stem. Seven of the nine identified mitosis-associated CycB orthologs were significantly down-regulated in the latter tissue and none in the primordia. And whereas two of the five CycA orthologs were down-regulated in the stem, one was up-regulated in the primordia, with a similar trend for another CycA and a CycD4 gene (Supplemental Data Set S3). Notably, 18 of 32 oppositely regulated genes (i.e. primordia up and stem down) have a function in DNA replication. Time-series analysis showed that the cell cycle and DNA replication processes started to be affected after 12 to 18 h of submergence, mostly (Supplemental Fig. S4).
Together, these results show that the reaction of primordia and stem tissue to flooding is differentiated at the level of gene functions, especially regarding cellular activity and cell cycle progression. While the stem is reducing its cellular activity, the AR primordia have clearly started reactivating 24 h after the onset of flooding.
Hormone Synthesis and Signaling Pathways Are Affected by Submergence
To identify what kind of signaling processes underpin AR primordia activation upon flooding, we looked in detail at hormone-related genes. GO categories concerning signaling by various hormones were significantly affected (Supplemental Fig. S3). This included a slight up-regulation of the brassinosteroid biosynthesis-related category in the primordia and cytokinin biosynthesis and GA response categories in the stem. A more significant overrepresentation was seen of salicylic acid and jasmonic acid-related genes among the up-regulated genes in the stem, while, in contrast, the latter type was mostly down-regulated in the primordia. Ethylene synthesis and response-related genes were strongly overrepresented among stem- and, to a lesser extent, primordia-up-regulated genes. This included one 1-aminocyclopropane-1-carboxylate (ACC) synthase (ACS), two ACC oxidases (ACO), and orthologs of eight of the 13 ETHYLENE RESPONSE FACTOR-type ERF/AP2 genes from tomato (Solanum lycopersicum) that were shown to be ethylene responsive (Pirrello et al., 2012; Supplemental Data Set S3). Analysis of earlier time points after onset of flooding indicated that induction of three ethylene signaling-associated genes was fast, within 3 h, and peaked earlier in the primordia (after 3 to 6 h) than in the stem (after 12 to 18 h; Supplemental Fig. S4). The ABA response was down-regulated significantly in the primordia, as exemplified by the key ABA-responsive genes identified by Fujita et al. (2009), such as the orthologs of the ABA-regulated PP2CA, ABF, RD26, and RD29B genes (Supplemental Data Set S3) and six of the 13 identified ABA-responsive LATE EMBRYOGENESIS ABUNDANT genes (Cao and Li, 2015; Supplemental Data Set S3). Down-regulation was detected already after 3 h of flooding (Supplemental Fig. S4). Furthermore, auxin biosynthesis genes were up-regulated in the primordia, while the category of auxin response genes was overrepresented among stem-up- and primordia-down-regulated genes. Closer inspection of the differentially expressed gene list, however, revealed that a number of auxin-responsive AUX/IAA genes and orthologs of the auxin-dependent LBD genes that function during LR development in Arabidopsis (i.e. LBD16, LBD29, and the same trend for LBD18; Lavenus et al., 2013; Supplemental Data Set S3) were specifically up-regulated in the primordia. We noticed up-regulation of this auxin-responsive gene expression in the primordia from 6 h of flooding onward (Supplemental Fig. S4). Auxin transport was enriched among stem-down-regulated genes.
Taken together, these results indicate that multiple hormone signaling pathways are likely to be affected by flooding and may thus mediate the response to this environmental cue.
Ethylene Is Necessary and Sufficient for Activation of AR Primordia
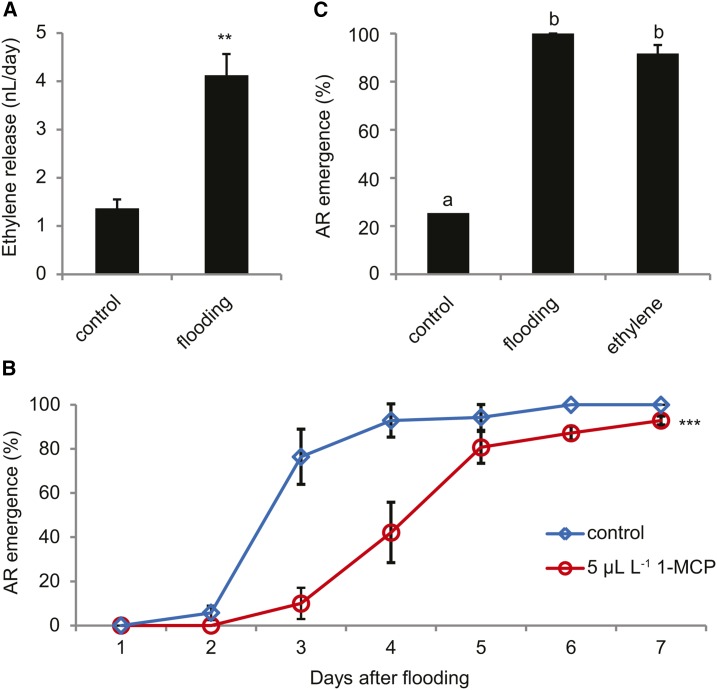
According to the transcript profiling, ethylene, ABA, and auxin signaling were enhanced upon submergence (Fig. 1D). As these hormones have been identified as major regulators of flooding-induced shoot growth responses, we set out to test their role in the signaling pathway regulating root primordium activation. Ethylene emission from the stem and AR primordia was measured by mounting a small cuvette around the lower section of the stem. After partial submergence (including the part of the stem carrying the cuvette), the rate of ethylene release into the cuvette increased significantly (Fig. 2A). To examine the function of ethylene in the activation of AR primordia, the process was studied after treating plants with an ethylene perception inhibitor, 1-methylcyclopropene (1-MCP), for 20 h directly prior to submergence. AR outgrowth was significantly delayed in the ethylene-desensitized plants as compared with control plants (Fig. 2B). To determine to what extent ethylene mediates the submergence cue, we studied the effect of ethylene application on AR emergence and expression of flooding-induced genes in AR primordia. Using high humidity conditions, which facilitate AR emergence (Dawood et al., 2014), root emergence was analyzed 7 d after the start of the hormone treatment. On average, less than 30% of the ARs emerged in control plants, and treatment with ethylene increased AR outgrowth to ∼90%, thus being nearly as effective as the partial flooding treatment (Fig. 2C). The accumulation of seven transcripts that were primordia-specifically induced by flooding (Supplemental Fig. S2) was analyzed 24 h after ethylene treatment. Four of the seven flooding-induced genes were responsive to the hormone in this time frame, while after 48 h two more genes responded to the treatment (Supplemental Fig. S5). Only one gene responded to flooding but was not responsive to ethylene.
Figure 2.
Ethylene is a major regulator of flooding-induced AR primordium activation. A, Release of ethylene from the stem of S. dulcamara after 24 of submergence. Data are means ± se for control (n = 4) and partially flooded plants (n = 6). **, Significantly different from control (Student’s t test, P < 0.01). B, Effect of inhibition of ethylene perception by 1-MCP. The visible growth of ARs was scored daily from 20 primordia per plant. Plants were treated with 0 (control) or 5 µL L−1 1-MCP for 16 h. Data are means ± se (n = 7 plants). ***, Average time to AR emergence 3.3 ± 0.23 [se] and 4.9 ± 0.23 d for control and 1-MCP treatment, respectively (Student’s t test, P < 0.001). C, Effect of treatment with ethylene. Per plant, 15 primordia located on the stem segment up to 15 cm above the soil were analyzed. Plants were kept under high humidity conditions and received no additional treatment (control), partial submergence (flooding), or 4 µL L−1 ethylene (ethylene). Data are means ± se (n = 8). Different letters above the bars indicate significant difference between the treatments (ANOVA with Tukey posthoc, P < 0.05).
In conclusion, the ethylene concentration in the submerged stem parts increases during flooding, and ethylene signaling is necessary and sufficient for emergence of ARs and for a considerable part of the flooding-dependent gene expression changes.
Reduction of ABA Levels Acts Downstream of Ethylene in the AR Activation Pathway
Contrary to the case of ethylene, ABA responses seemed to be down-regulated upon submergence according to the transcriptome analysis. Indeed, ABA levels in the AR primordia and surrounding stem were significantly reduced 24 h after the onset of flooding (Fig. 3A). Consistent with a primary role for ethylene accumulation in the flooding response, a reduction in ABA level in the primordia was also seen upon treatment with this hormone (Fig. 3A). Treatment with ABA prevented AR primordium emergence under flooded conditions (Fig. 3B), while treatment with the carotenoid biosynthesis inhibitor fluridone, which also blocks ABA synthesis (Gamble and Mullet, 1986), showed to be sufficient for AR primordium activation (Fig. 3C). To determine at what stage AR development is blocked by ABA, we made sections 7 d after the start of the flooding plus ABA treatment. AR primordia were developed slightly further than in the nonflooded control situation, but additional pretreatment of the primordia with ABA 1 d before flooding resulted in a near-complete growth inhibition (Supplemental Fig. S6). To determine the cause of the flooding-induced drop in ABA concentration, expression of ABA metabolism genes was studied. ABA accumulation is commonly controlled at the level of synthesis, at the step catalyzed by 9-cis-epoxycarotenoid dioxygenase (NCED)-type enzymes (Schwartz et al., 1997). Both of the identified ABA synthesis-related NCED orthologs were strongly down-regulated 24 h after the start of flooding or ethylene treatment (Fig. 3D; Supplemental Fig. S7). ABA levels may also be regulated at the level of catabolism, at the step catalyzed by ABA 8′-hydroxylases (Kushiro et al., 2004). One of three identified ABA 8′-hydroxylase orthologs was up-regulated in the AR primordia 24 h after the start of flooding. Additional quantitative PCR (qPCR) analysis, however, showed that this effect could not be mimicked by a 24-h ethylene treatment (Fig. 3E). Also, shorter ethylene treatments did not lead to induction of this gene in the primordia (Supplemental Fig. S7).
Figure 3.
ABA maintains AR dormancy and is reduced by flooding and ethylene treatment. A, ABA concentration in primordia and stem of 13-week-old plants after 24 h of various treatments. Around 20 primordia were collected per plant. Data are means ± se (n = 5, each a pool of two plants). Different letters above the bars indicate significant difference between the treatments (ANOVA with Tukey posthoc, P < 0.05). n.a., Not available. B, Effect of ABA treatment on AR emergence and length on 13-week-old plants partially submerged for 7 d. Per plant at least 10 primordia were analyzed. Data are means ± se (n = 5). *, Significantly different from the control, ANOVA, P < 0.05; ***, P < 0.001. C, Effect of ABA synthesis inhibitor fluridone on AR emergence under nonflooded conditions. Per plant 15 primordia were analyzed. Data are means ± se (n = 6). *, Significantly different from the control, ANOVA, P < 0.05. D, Expression of NCED genes upon various treatments. Values for AR primordia and stem are relative to the value of each sample type under control conditions. Data are means ± se (n = 3). *, Significantly different from the control, ANOVA, P < 0.05; ***, P < 0.001. n.a., Not available. E, Expression of an ABA 8′-hydroxylase gene upon various treatments. Values for AR primordia and stem are relative to the value of each sample type under control conditions. Data are means ± se (n = 3). *, Significantly different from the control, ANOVA, P < 0.05. n.a., Not available.
Polar Auxin Transport Is Required for AR Emergence
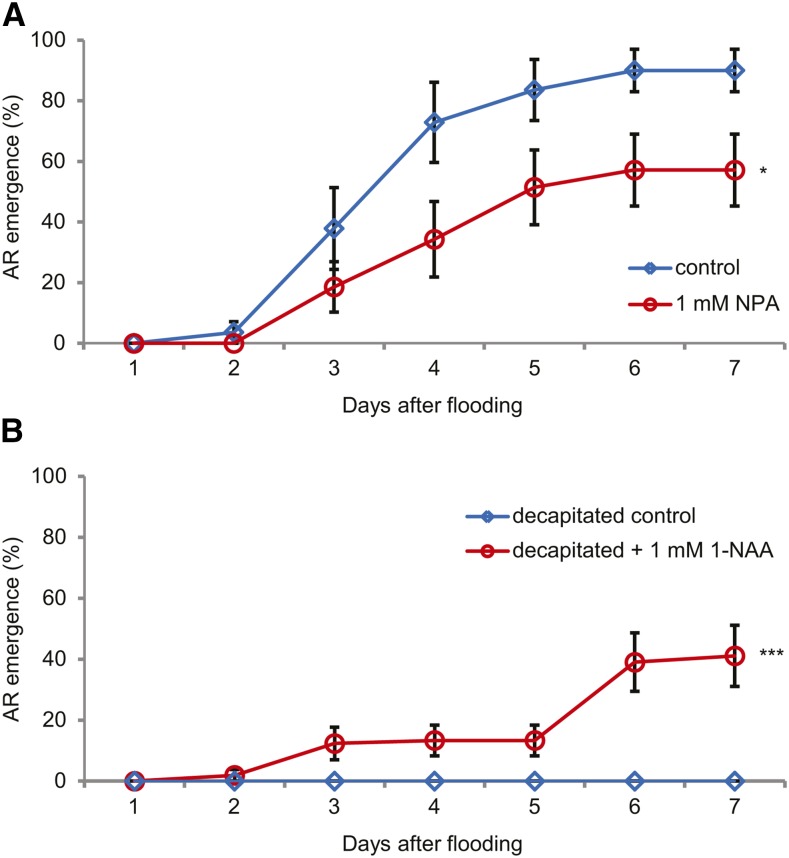
To examine the role of auxin in AR activation, basipetal polar auxin transport was blocked in the stem above the submerged plant parts by local application of NPA. After 7 d only 60% of the ARs had grown out and their emergence was significantly delayed (Fig. 4A). Basipetal auxin transport can also be inhibited by removing the main auxin source from the plant shoot by decapitation. On decapitated plants, ARs did not emerge upon flooding. Complementing the decapitated shoots with the synthetic auxin 1-naphthylacetic acid (1-NAA; applied to the shoot stump) partially rescued the outgrowth of ARs (Fig. 4B). Together, these results support a requirement for shoot-derived auxin to activate the primordia. Direct application of auxin to primordia, however, was not enough to activate AR primordia (AR emergence 25% ± 8% [se] in control, and 23% ± 11% with 25 μm 1-NAA).
Figure 4.
Basipetal auxin transport is required for AR primordium activation. The visible growth of ARs was scored daily from 20 primordia per plant. A, Effect of inhibition of polar auxin transport by NPA. Plants were treated with 0 (control) or 1 mm NPA in water before being submerged. Data are means ± se (n = 7 plants). *, Average time to AR emergence 4.2 ± 0.48 [se] and 5.8 ± 0.56 d for control and NPA treatment, respectively (Student’s t test, P < 0.05). B, Effect of decapitation. Treatments applied: decapitated and complemented with 0 (decapitated control) or decapitated and complemented with 1 mm 1-NAA. Data are means ± se (n = 7 for both decapitated treatments). ***, Emergence of 0% and 41% of ARs after 7 d for control and 1-NAA treatment, respectively (Fisher’s exact test, P < 0.001).
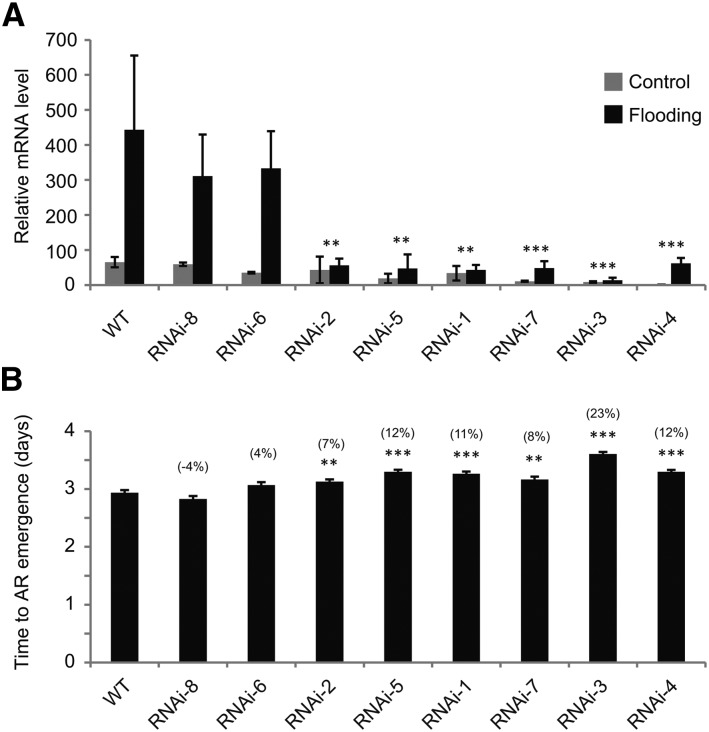
Interestingly, one of the flooding-induced genes, also shown to be responsive to ethylene, was annotated as auxin efflux carrier PIN2: the predicted protein clusters with tomato and Arabidopsis PIN2 in phylogenetic analysis and shows 97% and 89% similarity to these proteins, respectively (Supplemental Data Set S3; Supplemental Fig. S5). Having found an important role for polar auxin transport in AR activation, we analyzed the function of this gene in submergence-induced activation of the AR primordia. RNA interference (RNAi) S. dulcamara lines were generated by Agrobacterium tumefaciens-mediated transformation using a 281-bp 3′-untranslated region fragment of SdPIN2 in a hairpin construct. We selected eight independent RNAi lines and designated them as PIN2-RNAi-1 to -8. The transcript level of SdPIN2 was reduced in six of the lines (Fig. 5A). Emergence of ARs was significantly delayed in all these six RNAi lines, although the delay was limited in absolute terms (up to 0.7 d; Fig. 5B). No delay was found for the two lines that did not show reduced SdPIN2 expression. Thus, flooding- and ethylene-dependent induction of SdPIN2 expression supports the activation of AR primordia upon flooding.
Figure 5.
PIN2 silencing delays activation of AR primordia by flooding. A, Relative expression of SdPIN2 in the AR primordia in control and 24-h submerged plants in the wild type (WT) and PIN2-RNAi lines. Order of lines is according to significance of difference with the wild type. Data are means ± se (n = 2–4). **, Line significantly different from the wild type, two-way ANOVA, P < 0.01; ***, P < 0.001. B, Time to emergence of ARs after start of flooding. Data are means ± se (n = 30 individuals for the wild type and 8–15 for other types; in all cases 15 AR primordia per individual). Difference from the wild type is indicated in parentheses. **, Significantly different from the wild type, one-way ANOVA, P < 0.01; ***, P < 0.001.
DISCUSSION
Plasticity, or environmental responsiveness, is a universal property of life needed to optimize fitness under local circumstances. Here, we uncovered the molecular physiological processes that underlie plastic development of the root system in response to flooding in S. dulcamara. Partial flooding has a severe impact on plant physiology, which explains why the abundance of around 15% of all detected transcripts was modulated by this stress after 24 h in two examined tissues. One of the responses of this plant species to flooding is the reactivation of dormant AR primordia that are constitutively present on the stem (Terras, 1897; Dawood et al., 2014). By comparing the transcriptomic adjustments of AR primordia and regular stem tissue upon flooding, followed up by targeted physiological experiments, insight was gained into the regulatory mechanism behind this morphological acclimation.
Increased Cellular Activity of AR Primordia and Quiescence of Stem Tissue upon Flooding
About a quarter of all observed flooding-induced transcriptomic changes occurred in both the AR primordia and stem (Fig. 1). An example of this is up-regulation of genes involved in hypoxia acclimation, which suggests that the metabolic readjustments to cope with decreased oxygen availability are highly similar in the different types of cells. By contrast, genes involved in cell division and cell growth are regulated more differentially. For example, a significant number of cell wall-modifying enzymes are up-regulated only in the primordia. These genes could have a function in the growth of the primordia that is about to start after 24 h of flooding and requires weakening of existing cell walls to allow for cell enlargement as well as synthesis of new wall material after cell division (Cosgrove, 2005). Also, like in the process of LR emergence, the cortex cells that cover the root primordia will need to separate from each other, and in Arabidopsis this has been shown to require wall modification (Péret et al., 2009). Although we did not detect up-regulation of CycB genes in the primordia at the 24-h time point, there is already a specific induction of many DNA replication-related genes, such as CDC6, a key regulator of DNA replication in eukaryotes (Castellano et al., 2001).
In contrast to the situation in the primordia, a large group of genes encoding cell wall-modifying and cell cycle-related genes is specifically down-regulated in the stem. These findings suggest that cell division and growth are suppressed in the stem upon flooding, such that this tissue enters a state of quiescence. Adhering to a quiescent strategy regarding shoot growth has been shown to be beneficial for plant survival under long-term flooding conditions, potentially because more energy may be available for basal cellular functions and physiological acclimation (Bailey-Serres et al., 2012). The increased cell growth- and division-related activity of AR primordia after 24 h of flooding precedes visual growth of the primordia, but is in accordance with the finding that AR activation is triggered within 24 to 48 h, after which outgrowth occurs even if the water level is lowered (Dawood et al., 2014). The finding that two CycA3 orthologs and a CycD4 are induced suggests that the dormancy involved cell cycle arrest at the G1 to S phase, as has been described for other dormant organs (Horvath et al., 2003).
Ethylene as a Conserved Trigger for Adaptive Responses to Flooding
Upon flooding, diffusion of the gaseous hormone ethylene out of the plant will be reduced due to physical entrapment by the surrounding water layer (Armstrong et al., 1991), and together with sustained ethylene production, as indicated by increased expression of the ethylene biosynthesis genes ACS and ACO, this then leads to increased accumulation of ethylene, shown by the about 3-fold stronger ethylene release from the flooded S. dulcamara stem after 24 h of partial submergence (Fig. 2A). Ethylene and ethylene perception inhibitor treatments showed that this ethylene signal is necessary and sufficient for timely AR primordium activation (Fig. 2, B and C), as it is in rice and R. palustris (Visser et al., 1996b; Lorbiecke and Sauter, 1999). Ethylene functions as the cue for activation of flooding responses in various species and tissues, including positive and negative shoot growth responses and aerenchyma formation (Bailey-Serres and Voesenek, 2008). While ARs may develop from preformed primordia upon flooding, in some species they are formed de novo during the flood period (Negi et al., 2010; Vidoz et al., 2010). Interestingly, this stress-induced AR initiation, too, is stimulated by ethylene, while initiation of LRs, which is highly similar to that of ARs from a developmental point of view, is usually inhibited by ethylene (Negi et al., 2008; Ivanchenko et al., 2008). This highlights the repeated linking of the ethylene pathway to adaptive growth responses during evolution.
Removal of the ABA Dormancy Signal Activates AR Primordia
Ethylene then acts on AR primordium activation through restriction of ABA accumulation (Fig. 3A). This is accomplished via the down-regulation of NCED-type ABA biosynthesis genes (Fig. 3D). In our experiments, flooding, but not ethylene treatment, also led to higher expression of an ABA 8′-hydroxylase gene in the primordia (Fig. 3E). This thus represents a nonessential, ethylene-independent response, which might for example depend on hypoxia as a signal. Fluridone treatment showed that reduction of carotene-derived compounds is sufficient for AR primordium activation (Fig. 3B). Although this may include compounds other than ABA, such as strigolactones, the fact that ABA treatments then prevented AR emergence confirms that reduction of the ABA signal is involved in AR primordium activation (Fig. 3C). A similar negative relation between ethylene and ABA accumulation has been described earlier in shoot growth responses to flooding in R. palustris and rice (Saika et al., 2007; van Veen et al., 2013) and in germinating seeds and seedlings in Arabidopsis (Ghassemian et al., 2000; Cheng et al., 2009; Dong et al., 2011). The negative effect of ethylene on ABA accumulation in Arabidopsis seedlings was accompanied by lower NCED expression and higher ABA 8′-hydroxylase gene expression, as in the signaling cascade described here (Cheng et al., 2009). In rice, ABA treatment hampers flooding-induced AR emergence by preventing epidermal cell death (Steffens and Sauter, 2005), but it has not been reported whether ABA levels actually go down at the site of the preformed AR primordia and whether this is sufficient to mimic the flooding and ethylene responses. ABA also has a prominent role in dormancy initiation and maintenance in seeds and vegetative organ buds (Arend et al., 2009; Nambara et al., 2010; Reddy et al., 2013). Interestingly, a (transient) dormancy phenotype has been reported for LR primordia in Arabidopsis and is thought to occur in situations where LR emergence is not required (e.g. high plant nitrate status; Zhang and Forde, 1998) or unfavorable (e.g. dry or salty environments; Ginzburg, 1966; Duan et al., 2013). Nitrate- and salt-induced LR dormancy has been shown to be mediated by high ABA levels and to become established around the stage of root apical meristem activation, similar to the stage at which S. dulcamara ARs are kept dormant (Signora et al., 2001; De Smet et al., 2003; Duan et al., 2013; Dawood et al., 2014). It should be noted that upon flooding, ABA levels also drop in stem tissue of S. dulcamara, although this tissue becomes less active according to cell cycle-related gene expression. Thus, the response differentiates between the primordia and stem tissues at this point, with additional factors imposing a quiescent state on the stem.
An Essential Role for Auxin in Controlling AR Activation
During submergence, polar auxin transport is required for AR growth in various species (Visser et al., 1996c; Wample and Reid, 1979; McDonald and Visser, 2003; Xu et al., 2005; Vidoz et al., 2010), including S. dulcamara. According to the gene set enrichment analysis, signaling by auxin was stronger in flooded stem tissue but not significantly induced in primordia. Auxin, however, is a hormone with many cell type-specific effects, making it difficult to define a set of generally auxin-responsive genes (Wang and Estelle, 2014). Looking at the set of transcripts responding positively to flooding in the primordia, a number of genes that are regarded as auxin responsive could be identified, such as the LBD genes. Various members of this transcription factor family function in de novo formation of LRs and ARs, downstream of auxin (Inukai et al., 2005; Péret et al., 2009; Benková and Bielach, 2010). During LR development in Arabidopsis, LBD16 and LBD29 activate the cell cycle and induce primordium development (Péret et al., 2009; Lee et al., 2013), and, similarly, the rice LBD29 ortholog CRL1/ARL1 is necessary for AR initiation (Liu et al., 2005; Inukai et al., 2005). Furthermore, the Arabidopsis LBD18 protein activates the expression of the EXPANSIN14 gene to facilitate emergence of the LR (Péret et al., 2009; Lee et al., 2013). We found that expression of LBD16, -18, and -29 orthologs was induced by flooding, specifically in the primordia. Another auxin-responsive gene, PUCHI, is thought to act in parallel with the LBD genes during LR formation in Arabidopsis (Kang et al., 2013), and this gene also is induced in AR primordia of S. dulcamara upon flooding. Taken together, these results reveal considerable similarity between transcriptional programs underlying de novo LR/AR formation in Arabidopsis and rice and reactivation of dormant AR primordia in S. dulcamara. The reason for this is not clear, as root identity is already evident in the dormant S. dulcamara AR primordia (Dawood et al., 2014) and suggests either that cellular patterning of the AR primordia is not yet fully completed and is reinforced upon reactivation or that the auxin/LBD module has functions beyond the initial developmental steps of lateral/adventitious roots as identified in Arabidopsis and rice. It would be interesting to investigate if the same happens in reactivated AR primordia of rice.
In agreement with a role for auxin, treatment with NPA, a polar auxin transport inhibitor, significantly reduced the number of ARs induced by flooding and delayed their emergence (Fig. 4A). In Arabidopsis seedlings, emergence of the LR primordia was dependent on the auxin derived from the first two leaves, and the process was inhibited when they were removed (Bhalerao et al., 2002). Likewise, in S. dulcamara, removing the main source of auxin by decapitating the shoot abolished AR growth, while application of the synthetic auxin 1-NAA on top of the cut stem partially restored it (Fig. 4B). Although the process of auxin transport as a whole was slightly enriched among the flooding-down-regulated genes in the stem, resembling results from tomato (Negi et al., 2010), we observed a specific up-regulation of PIN2 in the AR primordia by flooding and ethylene treatment. This effect was of physiological relevance, as PIN2-silenced lines showed delayed AR emergence (Fig. 5). This may be due to the fact that PIN2 acts, redundantly with PIN1, as the major contributor to basipetal auxin transport to the root meristem and thereby promotes root growth (Blilou et al., 2005). Furthermore, PIN2 is involved in redistribution of auxin from the root tip to the elongation zone and in this role acts downstream of ethylene. Although primary root growth in Arabidopsis is suppressed by ethylene as a result of elevated, supra-optimal levels of auxin in the elongation zone, due to enhanced PIN2 activity (Luschnig et al., 1998), it is possible that in S. dulcamara, enhanced PIN2 activity is necessary for auxin concentrations proximal to the AR tip becoming optimal for growth. The limited phenotypic effect of PIN2 silencing and NPA treatment, compared with decapitation, may be due to partial silencing/inhibition or, in case of the silenced lines, to functional redundancy between the members of the PIN family, as has been demonstrated in primary root and LR development in Arabidopsis (Blilou et al., 2005). Indeed, there is considerable expression of several related PINs in the primordia (Supplemental Data Sets S1 and S3). As basipetal auxin transport is negatively affected by low oxygen levels and low light conditions (Wample and Reid, 1979; Keuskamp et al., 2010), this pathway may mediate integration of additional environmental signals into the flooding response, such as the observed suppression of AR emergence upon complete submergence and shading.
A Model for Hormonal Regulation of AR Activation
Based on our observations, we present a model in which the flooding-dependent ethylene response pathway that activates AR primordia controls two downstream branches, one suppressing signaling by the hormone ABA and one enhancing signaling by auxin. This model fits with the timing of reactivation-related gene expression, as changes in ethylene-, ABA-, and auxin-associated gene expression were observed after 3 to 6 h of flooding, while effects on cell cycle and DNA replication genes were first detected after 12 to 18 h. The finding that ABA reduction is not sufficient to activate primordia in the absence of basipetal auxin transport, as in the decapitated plants (Fig. 4B), and high auxin concentrations by themselves are also not sufficient to activate the primordia indicates that these two pathways act at least partially in parallel. This situation is similar to rice, in which polar auxin transport is also necessary for AR emergence but not sufficient (Lorbiecke and Sauter, 1999; Xu et al., 2005). Future transcript profiling studies into the effect of modifying ABA and auxin signal strength, alone and combined, should shed light on the nonadditive interaction between these two hormones. Additional factors, such as hypoxia, might feed into the model presented here but are not essential for reactivation of the primordia. An important distinction between the S. dulcamara and rice models for AR reactivation is the target tissue: while in rice the epidermal cells are the main target of the ethylene-dependent activation pathway (Steffens and Sauter, 2009), AR dormancy does not seem to depend on mechanical restriction by the epidermis in S. dulcamara. (Supplemental Fig. S1). Cell type-specific modifications will be necessary to determine target tissues in the latter system. In addition, it is currently unclear whether in rice, too, ABA reduction has a central role in mediating the ethylene signal.
The aquatic habit of plants evolved more than 200 times independently, suggesting that flood-adaptive traits can evolve relatively easily (Jackson et al., 2009). The initiation and then reactivation of dormant AR primordia in S. dulcamara is an adaptive novelty within the Solanum genus. One path for such an evolutionary process is the redeployment of preexisting developmental and signaling pathways (True and Carroll, 2002; Bento et al., 2010). This study supports this hypothesis by showing that a hormone cascade known to operate in seeds and seedlings and direct shoot growth responses to flooding in various plant species has been co-opted to control root growth upon flooding in S. dulcamara. Future studies employing detailed comparative genomic analyses may shed light on the molecular basis of this evolutionary process. Also, it will be interesting to study the putative involvement of other hormones in the dormancy breaking process, such as brassinosteroids and jasmonic acid, as these compounds have not been implicated in dormancy breaking processes before. The same applies to strigolactones, which are known to influence LR and AR initiation (Ruyter-Spira et al., 2011; Rasmussen et al., 2012) but might well have an extended function as suggested for the LBD genes.
There is an urgent need to increase crop production. A generally adopted strategy in this regard is to manipulate traits in crop plants, such that they better suit their environment, which will increasingly often include regularly soil-flooded areas. A deep and broad understanding of mechanisms that can potentially contribute to flooding tolerance, including the adaptive mechanisms that have evolved under natural selection, is imperative to the necessary advancements in crop breeding.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Solanum dulcamara (accession A54750008 from Wijchense Ven) was obtained from the Experimental Garden and Genebank, Radboud University, Nijmegen, The Netherlands. Seeds were sown in vermiculite in small round plastic pots of 10 cm height and 13 cm diameter. These were kept in dark at 4°C for 3 d and then grown under standardized greenhouse conditions, with a daily temperature regime of 20 to 23°C (day) and 15 to 18°C (night) and additional light supplied by high-pressure sodium lamps (SON-T; 600W; Philips). Three-week-old seedlings were individually transplanted into 12 × 11 × 11 cm (h × w × d) plastic pots filled with potting soil and kept under the same conditions as above. The plants were watered daily and fertilized once per 2 weeks up to 10 to 12 weeks. For flooding experiments, individual plants were submerged up to 15 cm above the soil in glass containers of 60 × 21 × 21 cm (h × w × d) filled with tap water 1 d before start of the experiment. Emergence of the ARs was scored by eye. All nonflooded hormone and inhibitor treatments were carried out under high humidity conditions. For the high humidity treatment, the inner walls of the glass containers were lined with wet filter paper. Then, plants were placed individually into the containers and the open top was wrapped with plastic foil to obtain high humid conditions inside the container. During this treatment, fresh air, humidified by passing it through a water column using bubble stones, was flushed at 1.75 L min−1 through each glass container to prevent atmospheric gas changes. Humidity was measured with a humidity detector (Testo AG 605-H1), showing levels of 90% to 93% relative humidity. Light and temperature were kept at standard conditions. The control plants were treated with high humidity only. To isolate the primordia samples, external excisions were made around a primordium, which was distinguishable by its white dome-shaped structure. This was followed by peeling off of the primordium (separation occurred at the stem vascular cambium) while minimizing contamination with surrounding stem tissue. To isolate stem samples, a similar section was taken from green stem tissue in which no primordium was present.
RNAseq and Data Processing
For RNAseq analysis, 12 plants were used, of which six were flooded while the rest were kept drained. From these plants, the primordia and stem explants were dissected 24 h after the onset of partial submergence. Samples were collected in three biological replicas, and each sample was a pool from two plants to minimize plant-level biological variation. Control samples were taken at the same time of day to avoid effects of the circadian rhythm. The samples were frozen directly in liquid nitrogen. Total RNA was isolated using the RNeasy plant mini kit (Qiagen). Genomic DNA was removed from the RNA samples using the Ambion TURBO DNA-free kit (Life Technologies). mRNA isolation, cDNA synthesis with random primers, and sequencing on the Illumina HiSeq2000 platform (100 bp, single end, four samples/lane) were performed by Fasteris SA according to standard protocols. The RNAseq read data quality was assessed using fastqc (version 0.10.0) (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). All reads were aligned to the contigs of a transcript-based assembly of the S. dulcamara genome (D’Agostino et al., 2013) using bowtie (version 0.12.7; Langmead et al., 2009). Only uniquely mapping reads were considered further (bowtie parameters: tryhard, best, strata, k = 1, m = 1). Because contigs represented estimated transcription units, we quantified the expression by the number of reads aligning to each contig. These counts were further normalized by the contig length in kilobases to obtain expression values that are comparable between contigs and by a factor that represents the effective library size to make the values comparable between samples. The library size factors were determined by first calculating the samples’ trimmed-means (Robinson and Oshlack, 2010) and rescaling them to an average of 1. The rescaling made the library size factors directly usable for calling differentially expressed genes with DESeq (method = per-condition, sharingMode = maximum, fitType = parametric). Adjusted P values were calculated according to Benjamini and Hochberg (1995). Transcripts were considered to be expressed if the average normalized reads per kilobase per million reads were equal to or larger than 1 for at least one sample type. GO enrichment analysis was done using Blast2Go (Götz et al., 2008). S. dulcamara orthologs of selected Arabidopsis (Arabidopsis thaliana) genes were identified via analysis of Arabidopsis-tomato (Solanum lycopersicum) phylogenetic relationships on the Ensemble Plants platform (http://plants.ensembl.org/index.html) and subsequent determination of the tomato-S. dulcamara orthologous relationship as the bidirectional best hit (D’Agostino et al., 2013).
qPCR Analysis
Total DNA-free RNA was reverse transcribed using a cDNA synthesis kit (iScript cDNA synthesis kit; Bio-Rad). The real-time quantitative PCR was carried using cDNA corresponding to 8 ng (RNAseq conformation experiment) or 15 ng (hormone treatment experiment) of total RNA in a 25-µL PCR reaction containing 400 nm each primer and 12.5 µL of iQ SYBER Green Supermix (Bio-Rad). The PCR reactions were performed in a 96-well thermocycler (Bio-Rad iCycler) by starting with 3 min at 95°C, followed by 45 cycles consisting of 15 s at 95°C and 60 s at 60°C and a melt curve from 65°C to 95°C in 0.5°C increments per 10 s to verify the presence of a specific product. TIP41, SAND, CAC, and Expressed (Expósito-Rodríguez et al., 2008) were used as reference genes. Reference gene stability was confirmed with geNorm software. The quantification of the transcripts level was done as described by Rieu and Powers (2009). Sequences of primers used for qPCR analyses are provided in Supplemental Table S1.
Ethylene Analysis
The side branches and leaves located on the 15-cm stem segment above the soil were removed to be able to fix a cuvette around the stem at 3 d prior to the start of an experiment to eliminate potential effects of wound/touch-induced ethylene production. Cuvettes consisted of an upright glass cylinder that was vertically cut into two, containing inlets and outlets for gas flow, and that could be fixed around a 10-cm segment stem above the soil using Terostat Butyl-IX putty (Imbema Rhiwa B.V.). One day before the onset of flooding, the cuvettes were fixed around the stem segment. The first internode, reaching to about 2 cm above the soil, was not included in the cuvette. Plants were kept in a growth chamber (16/8 h photoperiod at 22°C/20°C, 60% relative humidity) and flooded individually, with the water level reaching to 8 cm above the soil. The inlet and outlet of the cuvette were connected to a laser-based ethylene detector system (ETD-300; Sensor Sense B.V.). Ethylene measurement was essentially done as described before (Nitsch et al., 2012). The ethylene evolution was corrected for flow, weight, and background ethylene in the air. Similar results were obtained with and without water inside the cuvette (half filled), and data therefore were combined. In total, six biological replicas (separate plants) were used per treatment and line.
ABA Analysis
The sample preparation for ABA quantitation analysis was performed according to Priest et al. (2006), with some modifications. Powder of lyophilized tissue (∼5 mg) was extracted with 2 mL of acetone:water (80:20, v/v) in the presence of antioxidant—2,6-di-tert-butyl-4-methylphenol (0.1 mg mL−1) and 100 pmol isotope-labeled internal standard [2H6]-ABA. After centrifugation (15 min, 3000 rpm, 4°C), the supernatant was collected and the pellet was re-extracted with 2 mL of extraction solvent. Pooled supernatants were evaporated to dryness under a vacuum. The extracts were suspended in 1 mL of isopropanol:formic acid (99:1, v/v, pH 3.3) and dried under a vacuum. Reconstitutes in 1 mL of 10% (v/v) methanol containing 0.1% (v/v) formic acid, pH 2.6, were partitioned twice with 1 mL of n-hexane each time. Dried samples were dissolved in 1 mL of 10% (v/v) methanol, containing 0.1% (v/v) formic acid, pH 2.6, and purified using an Oasis-HLB cartridge (150 mg/6 cc; Waters). The sorbent was preconditioned with 3 mL of methanol and equilibrated with 3 mL of methanol containing 0.1% (v/v) formic acid. After loading the sample, the column was washed, and ABA was eluted with two series of eluents, 1 mL of acetonitrile:water:formic acid (50:49.9:0.1, v/v/v) and 2.5 mL of acetonitrile:water:formic acid (90:9.9:0.1, v/v/v), respectively. Both eluates were combined and dried under vacuum. Quantification of ABA metabolites was achieved by liquid chromatography coupled to tandem mass spectometry using an Acquity UPLC system (Waters) coupled with a Xevo TQ-S triple quadrupole mass spectrometer (Waters) as described by Floková et al. (2014), with modifications. Purified samples were reconstructed in 200 µL of mobile phase, filtered with 0.45-µm PTFE membrane filter (Phenomenex), and injected onto Acquity UPLC CSH C18 column (100 × 2.1 mm, 1.7 µm; Waters). Analytes were eluted using a binary gradient, consisting of 15 mm formic acid in water (A) and acetonitrile (B), for 7 min at a flow rate 0.7 mL min−1 and constant column temperature at 45°C. The linear gradient elution was performed as follows: 0 to 0.5 min, 15% (v/v) eluent B; 0.5 to 3.5 min, 15% to 60% eluent B; 3.5 to 4.5 min, 60% to 80% eluent B; and 4.5 to 5.75 min, 80% to 100% eluent B. At the end of gradient, the column was equilibrated to initial conditions for 1.25 min. The effluent was introduced in electrospray ion source of mass spectrometer with optimized operating parameters: capillary voltage 3 kV, cone voltage 25 V, source/desolvation temperature 150°C/600°C, cone/desolvation gas flow 150/600 L h−1, and collision energy 10 V. Compound was quantified in negative mode as [M-H]−, and two diagnostic transition reactions were used to perform multiple reaction monitoring detection (263.15 > 153.1; 263.15 > 219.1). Data were processed by MassLynx software version 4.1 (Waters).
Hormone and Inhibitor Treatments
For ethylene treatment, air was enriched with 4.4 to 5 µL L−1 ethylene, by means of a gas mixer (Bronckhorst High Tech BV). The concentrations in the glass containers were checked before starting the treatments by measuring gas samples with gas chromatography. Treatment was continued for up to 1 d (for gene expression analysis) or for 7 d (for phenotypic analysis).
For 1-MCP treatment, plants were either pretreated with 5 µL L−1 1-MCP or with air in an inflatable glove chamber with a size of ∼600 L (type 108D-X-37-37; Instruments for Research and Industry) overnight before submergence. This concentration of 1-MCP was obtained by dissolving 4.82 g of Ethylbloc (Floralife) in 100 mL of water, which was placed in an open petri dish in a glove bag. Subsequently, plants were flooded individually in glass containers, and the emergence of ARs was scored daily up to 1 week after flooding.
For ABA treatment, (+)-cis, trans-ABA (Duchefa Biochemie) was dissolved in absolute ethanol to 1 m, diluted with distilled water to 10−3 m, and further diluted with 0.1% (v/v) ethanol to maintain equal ethanol concentrations. Thirteen-week-old plants were treated locally with ABA or a control ethanol solution using a 5-mL cuvette fixed onto the stem (5–15 cm high above the soil) with terostat and then flooded to 15 cm above soil level. For pretreatment, stems with primordia were sprayed with 10−3 m ABA or control solution the day before flooding. ARs were analyzed 7 d after the onset of flooding.
For fluridone treatment, fluridone (Duchefa Biochemie) was dissolved in absolute ethanol and diluted to working solutions in 10% (v/v) ethanol. Stems of 12-week-old plants were wetted with fluridone or control solution for seven consecutive days before analysis of AR emergence.
Auxin treatment was performed for three consecutive days using 1-NAA (Duchefa Biochemie) dissolved in a small volume of 1 n NaOH, diluted with water to a final concentration of 25 µm and then sprayed on the lower 15 cm of stem using a spray bottle. Emergence of ARs was scored daily for 1 week after start of the treatment.
For NPA treatment, the main stem, branches, and apical meristems were sprayed with 0 or 1 mm NPA (Duchefa Biochemie), with the lowest 15-cm stem portion of the main stem remaining untreated. NPA solutions were prepared by dissolving NPA in a small volume of 1 n NaOH and then further diluting with water. Control plants were sprayed with a similar concentration of NaOH. Spraying was done daily from 1 d before until 1 d after the onset of flooding. AR emergence was scored daily up to 1 week after flooding.
For the decapitation experiment, plants were decapitated by removing the shoot from 20 cm above soil level. 1-NAA (1 mm) or a control solution in 1% (w/v) microagar (Duchefa Biochemie) was applied to decapitated shoots 1 d before flooding. AR growth was scored daily for 7 d.
Generation and Functional Analysis of SdPIN2-RNAi Lines
To generate transgenic RNAi SdPIN2 lines, a fragment of a 281-bp 3′-untranslated region of the comp28330_c0_seq1 of S. dulcamara was amplified using forward primer 5′-CATGGGGAACAGAGACAGAT-3′ and reverse primer 5′-GACTGAAACAATATGAAGGC-3′ and cloned into the destination vector pk7GWIWG2(I) under control of the cauliflower mosaic virus 35S promoter (Karimi et al., 2002). The construct was transformed into Agrobacterium tumefaciens strain GV3101 using freeze-thaw transformation (Chen et al., 1994). Transgenic plants were made by the leaf disc method. In short, leaves were harvested from 3- to 4-week-old S. dulcamara plants, sterilized for 10 min in a solution of 1.5% bleach and 0.01% Tween, and washed four times for 5 min with sterilized demineralized water. The leaf explants were cut without the veins and incubated in a 1:100 diluted bacterial culture (OD600 0.4–0.6) with liquid cocultivation medium of MS20 (20 g L−1 Suc, 4.4 g L−1 Murashige and Skoog with Gamborg B5, and 0.5 g L−1 MES monohydrate, pH 5.8) with growth regulators (2 mg L−1 6-benzylaminopurine, 0.1 mg L−1 1-NAA, and 10 mg L−1 acetosyringone) and kept in dark for 3 d under climate chamber growth conditions. Thereafter, leaf explants were transferred to selective medium of MS20 supplemented with growth regulators (2 mg L−1 6-benzylaminopurine, 0.1 mg L−1 1-NAA, 300 mg L−1 cefotaxime, 300 mg L−1 vancomycin, and 25 mg L−1 kanamycin). The plates were covered with three layers of filter paper and kept for a week in the climate chamber. The filter papers were removed gradually (one per week). Every 2.5 weeks, the explants were transferred on to new fresh selective medium. After ∼7 weeks, the emerged shoots were excised and transferred to MS20 medium supplemented with 300 mg L−1 cefotaxime, 300 mg L−1 vancomycin, 10 mg L−1 kanamycin, and 0.25 mg L−1 indole-3-butyric acid. When roots had formed, the transgenic plants were transferred to the greenhouse and kept under standardized greenhouse conditions. For qPCR analysis of SdPIN2 expression, six propagated plants for each independent transgenic line were used. Six wild-type plants were used as control plants. The primordia were dissected from all the plants at 0 and 24 h after the onset of partial submergence. The total RNA was isolated, DNase treated, and converted to cDNA as described in the previous paragraphs. Relative gene expression was normalized with two reference genes, namely, CAC and SAND.
Statistical Analysis
Fisher’s exact and χ2 tests were used to analyze categorical data as indicated. A two-tailed Student’s t test or univariate ANOVA with uncorrected lsd was used to assess significance of differences between phenotypes under submergence and the control condition and between transgenic lines and the wild type as indicated. PASW Statistics for Windows (version 18; SPSS Inc.) was used for all analyses. qPCR data were log transformed before analysis to correct for heterogeneity of variance (Rieu and Powers, 2009).
Accession Numbers
Raw sequence reads obtained from Illumina sequencing were submitted to the NCBI Short Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under accession SRP020226. The S. dulcamara contigs are available from the Sol Genomics Network Web site (ftp://ftp.solgenomics.net/unigene_builds/single_species_assemblies/Solanum_dulcamara/).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Surface morphology and cell death assay of AR primordia in control conditions.
Supplemental Figure S2. Validation of RNAseq gene expression analysis through reanalysis of genes up-regulated specifically in primordia by flooding.
Supplemental Figure S3. GO analyses.
Supplemental Figure S4. Expression of selected genes at early time points after flooding.
Supplemental Figure S5. Expression of primordia-specific genes upon ethylene treatment.
Supplemental Figure S6. Effect of ABA treatment on AR primordium activation by flooding.
Supplemental Figure S7. Effect of short-term ethylene treatment on ABA metabolism genes.
Supplemental Table S1. Sequences of primers used for qPCR analyses.
Supplemental Data Set S1. RNAseq gene expression analysis results.
Supplemental Data Set S2. Validation of RNAseq gene expression analysis through analysis of known flooding-responsive genes.
Supplemental Data Set S3. Gene expression data for selected genes.
Supplementary Material
Acknowledgments
We would like to thank Gerard van der Weerden (Experimental Garden and Genebank, Radboud University, Nijmegen, The Netherlands) for seed material and the greenhouse staff for taking excellent care of the plants. We appreciate receiving technical assistance from Peter de Groot and Suzanne Hoogstrate, and acknowledge contributions from Marjolein Hunck, Tamara Fitters, Judith Rotink, Emiel Derksen, and Federico Muffato. The use of the Xevo TQ-S equipment was made possible by CAT-AgroFood Shared Research Facilities, Wageningen UR, The Netherlands.
Glossary
- AR
adventitious root
- LR
lateral root
- ABA
abscisic acid
- ROS
reactive oxygen species
- NPA
N-1-naphtylphtalamic acid
- PC
principal component
- RNAseq
RNA sequencing
- GO
gene ontology
- 1-MCP
1-methylcyclopropene
- NCED
9-cis-epoxycarotenoid dioxygenase
- qPCR
quantitative PCR
- 1-NAA
1-naphthylacetic acid
- RNAi
RNA interference
Footnotes
Articles can be viewed without a subscription.
References
- Arend M, Schnitzler JP, Ehlting B, Hänsch R, Lange T, Rennenberg H, Himmelbach A, Grill E, Fromm J (2009) Expression of the Arabidopsis mutant ABI1 gene alters abscisic acid sensitivity, stomatal development, and growth morphology in gray poplars. Plant Physiol 151: 2110–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong W, Justin SHFW, Beckett PM, Lythe S (1991) Root adaptation to soil waterlogging. Aquat Bot 39: 57–73 [Google Scholar]
- Bailey-Serres J, Lee SC, Brinton E (2012) Waterproofing crops: effective flooding survival strategies. Plant Physiol 160: 1698–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J, Voesenek LACJ (2008) Flooding stress: acclimations and genetic diversity. Annu Rev Plant Biol 59: 313–339 [DOI] [PubMed] [Google Scholar]
- Bellini C, Pacurar DI, Perrone I (2014) Adventitious roots and lateral roots: similarities and differences. Annu Rev Plant Biol 65: 639–666 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57: 289–300 [Google Scholar]
- Benková E, Bielach A (2010) Lateral root organogenesis - from cell to organ. Curr Opin Plant Biol 13: 677–683 [DOI] [PubMed] [Google Scholar]
- Bento G, Ogawa A, Sommer RJ (2010) Co-option of the hormone-signalling module dafachronic acid-DAF-12 in nematode evolution. Nature 466: 494–497 [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Eklöf J, Ljung K, Marchant A, Bennett M, Sandberg G (2002) Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J 29: 325–332 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Cao J, Li X (2015) Identification and phylogenetic analysis of late embryogenesis abundant proteins family in tomato (Solanum lycopersicum). Planta 241: 757–772 [DOI] [PubMed] [Google Scholar]
- Castellano MM, del Pozo JC, Ramirez-Parra E, Brown S, Gutierrez C (2001) Expression and stability of Arabidopsis CDC6 are associated with endoreplication. Plant Cell 13: 2671–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Nelson RS, Sherwood JL (1994) Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16: 664–668, 670 [PubMed] [Google Scholar]
- Cheng WH, Chiang MH, Hwang SG, Lin PC (2009) Antagonism between abscisic acid and ethylene in Arabidopsis acts in parallel with the reciprocal regulation of their metabolism and signaling pathways. Plant Mol Biol 71: 61–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JA, Llewellyn DJ, Dennis ES, Wilson IW (2010) Global gene expression responses to waterlogging in roots and leaves of cotton (Gossypium hirsutum L.). Plant Cell Physiol 51: 21–37 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- D’Agostino N, Golas T, van de Geest H, Bombarely A, Dawood T, Zethof J, Driedonks N, Wijnker E, Bargsten J, Nap JP, et al. (2013) Genomic analysis of the native European Solanum species, S. dulcamara. BMC Genomics 14: 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawood T, Rieu I, Wolters-Arts M, Derksen EB, Mariani C, Visser EJW (2014) Rapid flooding-induced adventitious root development from preformed primordia in Solanum dulcamara. AoB Plants 6: plt058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Signora L, Beeckman T, Inzé D, Foyer CH, Zhang H (2003) An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J 33: 543–555 [DOI] [PubMed] [Google Scholar]
- Dong H, Zhen Z, Peng J, Chang L, Gong Q, Wang NN (2011) Loss of ACS7 confers abiotic stress tolerance by modulating ABA sensitivity and accumulation in Arabidopsis. J Exp Bot 62: 4875–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L, Dietrich D, Ng CH, Chan PMY, Bhalerao R, Bennett MJ, Dinneny JR (2013) Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25: 324–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floková K, Tarkowská D, Miersch O, Strnad M, Wasternack C, Novák O (2014) UHPLC-MS/MS based target profiling of stress-induced phytohormones. Phytochemistry 105: 147–157 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, et al. (2009) Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol 50: 2123–2132 [DOI] [PubMed] [Google Scholar]
- Gamble PE, Mullet JE (1986) Inhibition of carotenoid accumulation and abscisic acid biosynthesis in fluridone-treated dark-grown barley. Eur J Biochem 160: 117–121 [DOI] [PubMed] [Google Scholar]
- Ghassemian M, Nambara E, Cutler S, Kawaide H, Kamiya Y, McCourt P (2000) Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12: 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginzburg C. (1966) Formation des racines laterales dormantes chez Zygophyllum dumosum. C R Acad Sci Paris 263: 909–912 [Google Scholar]
- Golas TM, van de Geest H, Gros J, Sikkema A, D’Agostino N, Nap JP, Mariani C, Allefs JJHM, Rieu I (2013) Comparative next-generation mapping of the Phytophthora infestans resistance gene Rpi-dlc2 in a European accession of Solanum dulcamara. Theor Appl Genet 126: 59–68 [DOI] [PubMed] [Google Scholar]
- Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36: 3420–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath DP, Anderson JV, Chao WS, Foley ME (2003) Knowing when to grow: signals regulating bud dormancy. Trends Plant Sci 8: 534–540 [DOI] [PubMed] [Google Scholar]
- Inukai Y, Miwa M, Nagato Y, Kitano H, Yamauchi A (2001) Characterization of rice mutants deficient in the formation of crown roots. Breed Sci 51: 123–129 [Google Scholar]
- Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, Umemura I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M (2005) Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17: 1387–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanchenko MG, Muday GK, Dubrovsky JG (2008) Ethylene-auxin interactions regulate lateral root initiation and emergence in Arabidopsis thaliana. Plant J 55: 335–347 [DOI] [PubMed] [Google Scholar]
- Jackson MB, Ishizawa K, Ito O (2009) Evolution and mechanisms of plant tolerance to flooding stress. Ann Bot (Lond) 103: 137–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang NY, Lee HW, Kim J (2013) The AP2/EREBP gene PUCHI co-acts with LBD16/ASL18 and LBD18/ASL20 downstream of ARF7 and ARF19 to regulate lateral root development in Arabidopsis. Plant Cell Physiol 54: 1326–1334 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Keuskamp DH, Pollmann S, Voesenek LACJ, Peeters AJM, Pierik R (2010) Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc Natl Acad Sci USA 107: 22740–22744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitomi Y, Ito H, Hobo T, Aya K, Kitano H, Inukai Y (2011) The auxin responsive AP2/ERF transcription factor CROWN ROOTLESS5 is involved in crown root initiation in rice through the induction of OsRR1, a type-A response regulator of cytokinin signaling. Plant J 67: 472–484 [DOI] [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J 23: 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Roberts I, Guyomarc’h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L (2013) Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci 18: 450–458 [DOI] [PubMed] [Google Scholar]
- Lee HW, Kim MJ, Kim NY, Lee SH, Kim J (2013) LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J 73: 212–224 [DOI] [PubMed] [Google Scholar]
- Lee MO, Hwang JH, Lee DH, Hong CB (2007) Gene expression profile for Nicotiana tabacum in the early phase of flooding stress. J Plant Biol 50: 496–503 [Google Scholar]
- Lee SC, Mustroph A, Sasidharan R, Vashisht D, Pedersen O, Oosumi T, Voesenek LACJ, Bailey-Serres J (2011) Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol 190: 457–471 [DOI] [PubMed] [Google Scholar]
- Liu H, Wang S, Yu X, Yu J, He X, Zhang S, Shou H, Wu P (2005) ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J 43: 47–56 [DOI] [PubMed] [Google Scholar]
- Lorbiecke R, Sauter M (1999) Adventitious root growth and cell-cycle induction in deepwater rice. Plant Physiol 119: 21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR (1998) EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev 12: 2175–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald MP, Visser EJW (2003) A study of the interaction between auxin and ethylene in wild type and transgenic ethylene-insensitive tobacco during adventitious root formation induced by stagnant root zone conditions. Plant Biol 5: 550–556 [Google Scholar]
- Mergemann H, Sauter M (2000) Ethylene induces epidermal cell death at the site of adventitious root emergence in rice. Plant Physiol 124: 609–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Okamoto M, Tatematsu K, Yano R, Seo M, Kamiya Y (2010) Abscisic acid and the control of seed dormancy and germination. Seed Sci Res 20: 55–67 [Google Scholar]
- Nanjo Y, Maruyama K, Yasue H, Yamaguchi-Shinozaki K, Shinozaki K, Komatsu S (2011) Transcriptional responses to flooding stress in roots including hypocotyl of soybean seedlings. Plant Mol Biol 77: 129–144 [DOI] [PubMed] [Google Scholar]
- Negi S, Ivanchenko MG, Muday GK (2008) Ethylene regulates lateral root formation and auxin transport in Arabidopsis thaliana. Plant J 55: 175–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi S, Sukumar P, Liu X, Cohen JD, Muday GK (2010) Genetic dissection of the role of ethylene in regulating auxin-dependent lateral and adventitious root formation in tomato. Plant J 61: 3–15 [DOI] [PubMed] [Google Scholar]
- Nitsch L, Kohlen W, Oplaat C, Charnikhova T, Cristescu S, Michieli P, Wolters-Arts M, Bouwmeester H, Mariani C, Vriezen WH, et al. (2012) ABA-deficiency results in reduced plant and fruit size in tomato. J Plant Physiol 169: 878–883 [DOI] [PubMed] [Google Scholar]
- Péret B, De Rybel B, Casimiro I, Benková E, Swarup R, Laplaze L, Beeckman T, Bennett MJ (2009) Arabidopsis lateral root development: an emerging story. Trends Plant Sci 14: 399–408 [DOI] [PubMed] [Google Scholar]
- Pirrello J, Prasad BCN, Zhang W, Chen K, Mila I, Zouine M, Latché A, Pech JC, Ohme-Takagi M, Regad F, et al. (2012) Functional analysis and binding affinity of tomato ethylene response factors provide insight on the molecular bases of plant differential responses to ethylene. BMC Plant Biol 12: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest DM, Ambrose SJ, Vaistij FE, Elias L, Higgins GS, Ross ARS, Abrams SR, Bowles DJ (2006) Use of the glucosyltransferase UGT71B6 to disturb abscisic acid homeostasis in Arabidopsis thaliana. Plant J 46: 492–502 [DOI] [PubMed] [Google Scholar]
- Qi XH, Xu XW, Lin XJ, Zhang WJ, Chen XH (2012) Identification of differentially expressed genes in cucumber (Cucumis sativus L.) root under waterlogging stress by digital gene expression profile. Genomics 99: 160–168 [DOI] [PubMed] [Google Scholar]
- Rasmussen A, Mason MG, De Cuyper C, Brewer PB, Herold S, Agusti J, Geelen D, Greb T, Goormachtig S, Beeckman T, et al. (2012) Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiol 158: 1976–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy SK, Holalu SV, Casal JJ, Finlayson SA (2013) Abscisic acid regulates axillary bud outgrowth responses to the ratio of red to far-red light. Plant Physiol 163: 1047–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu I, Powers SJ (2009) Real-time quantitative RT-PCR: design, calculations, and statistics. Plant Cell 21: 1031–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter-Spira C, Kohlen W, Charnikhova T, van Zeijl A, van Bezouwen L, de Ruijter N, Cardoso C, Lopez-Raez JA, Matusova R, Bours R, et al. (2011) Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol 155: 721–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saika H, Okamoto M, Miyoshi K, Kushiro T, Shinoda S, Jikumaru Y, Fujimoto M, Arikawa T, Takahashi H, Ando M, et al. (2007) Ethylene promotes submergence-induced expression of OsABA8ox1, a gene that encodes ABA 8′-hydroxylase in rice. Plant Cell Physiol 48: 287–298 [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JAD, McCarty DR (1997) Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276: 1872–1874 [DOI] [PubMed] [Google Scholar]
- Signora L, De Smet I, Foyer CH, Zhang H (2001) ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J 28: 655–662 [DOI] [PubMed] [Google Scholar]
- Steffens B, Sauter M (2005) Epidermal cell death in rice is regulated by ethylene, gibberellin, and abscisic acid. Plant Physiol 139: 713–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens B, Sauter M (2009) Epidermal cell death in rice is confined to cells with a distinct molecular identity and is mediated by ethylene and H2O2 through an autoamplified signal pathway. Plant Cell 21: 184–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens B, Wang J, Sauter M (2006) Interactions between ethylene, gibberellin and abscisic acid regulate emergence and growth rate of adventitious roots in deepwater rice. Planta 223: 604–612 [DOI] [PubMed] [Google Scholar]
- Taramino G, Sauer M, Stauffer JL Jr, Multani D, Niu X, Sakai H, Hochholdinger F (2007) The maize (Zea mays L.) RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and post-embryonic shoot-borne root initiation. Plant J 50: 649–659 [DOI] [PubMed] [Google Scholar]
- Terras JA. (1897) The relation between the lenticels and adventitious roots of Solanum dulcamara. Transactions of the Botanical Society of Edinburgh 21: 341–353 [Google Scholar]
- True JR, Carroll SB (2002) Gene co-option in physiological and morphological evolution. Annu Rev Cell Dev Biol 18: 53–80 [DOI] [PubMed] [Google Scholar]
- van Veen H, Mustroph A, Barding GA, Vergeer-van Eijk M, Welschen-Evertman RAM, Pedersen O, Visser EJW, Larive CK, Pierik R, Bailey-Serres J, et al. (2013) Two Rumex species from contrasting hydrological niches regulate flooding tolerance through distinct mechanisms. Plant Cell 25: 4691–4707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidoz ML, Loreti E, Mensuali A, Alpi A, Perata P (2010) Hormonal interplay during adventitious root formation in flooded tomato plants. Plant J 63: 551–562 [DOI] [PubMed] [Google Scholar]
- Visser EJW, Blom CWPM, Voesenek LACJ (1996a) Flooding-induced adventitious rooting in Rumex: morphology and development in an ecological perspective. Acta Bot Neerl 45: 17–28 [Google Scholar]
- Visser EJW, Bögemann GM, Blom CWPM, Voesenek LACJ (1996b) Ethylene accumulation in waterlogged Rumex plants promotes formation of adventitious roots. J Exp Bot 47: 403–410 [Google Scholar]
- Visser E, Cohen JD, Barendse G, Blom C, Voesenek L (1996c) An ethylene-mediated increase in sensitivity to auxin induces adventitious root formation in flooded Rumex palustris Sm. Plant Physiol 112: 1687–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wample RL, Reid DM (1979) The role of endogenous auxins and ethylene in the formation of adventitious roots and hypocotyl hypertrophy in flooded sunflower plants (Helianthus annuus). Physiol Plant 45: 219–226 [Google Scholar]
- Wang R, Estelle M (2014) Diversity and specificity: auxin perception and signaling through the TIR1/AFB pathway. Curr Opin Plant Biol 21: 51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Zhu L, Shou H, Wu P (2005) A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol 46: 1674–1681 [DOI] [PubMed] [Google Scholar]
- Zhang H, Forde BG (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]
- Zhang ZX, Zou XL, Tang WH, Zheng YL (2006) Revelation on early response and molecular mechanism of submergence tolerance in maize roots by microarray and suppression subtractive hybridization. Environ Exp Bot 58: 53–63 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.