A seed-to-seedling gene coexpression network reveals homeodomain leucine zipper I transcription factor AtHB13 as a novel key regulator affecting primary root length during late seedling establishment.
Abstract
The transition from a quiescent dry seed to an actively growing photoautotrophic seedling is a complex and crucial trait for plant propagation. This study provides a detailed description of global gene expression in seven successive developmental stages of seedling establishment in Arabidopsis (Arabidopsis thaliana). Using the transcriptome signature from these developmental stages, we obtained a coexpression gene network that highlights interactions between known regulators of the seed-to-seedling transition and predicts the functions of uncharacterized genes in seedling establishment. The coexpressed gene data sets together with the transcriptional module indicate biological functions related to seedling establishment. Characterization of the homeodomain leucine zipper I transcription factor AtHB13, which is expressed during the seed-to-seedling transition, demonstrated that this gene regulates some of the network nodes and affects late seedling establishment. Knockout mutants for athb13 showed increased primary root length as compared with wild-type (Columbia-0) seedlings, suggesting that this transcription factor is a negative regulator of early root growth, possibly repressing cell division and/or cell elongation or the length of time that cells elongate. The signal transduction pathways present during the early phases of the seed-to-seedling transition anticipate the control of important events for a vigorous seedling, such as root growth. This study demonstrates that a gene coexpression network together with transcriptional modules can provide insights that are not derived from comparative transcript profiling alone.
Plants undergo a number of developmental phase transitions during their life cycle. These transitions are controlled by distinct genetic circuits that integrate endogenous and environmental cues (Rougvie, 2005; Amasino, 2010; Huijser and Schmid, 2011). The correct timing of events occurring in the postembryonic developmental phase transitions (i.e. germination, the heterotrophic-to-autotrophic transition, juvenile vegetative to adult vegetative, and vegetative to reproductive) is critical for plant survival and reproduction. The transition from seed to seedling is mediated by germination, which is a complex process that starts with imbibition and is completed with radicle emergence. Seed germination is a crucial process in seedling establishment, as it marks a functional point of no return. Once germination has commenced, the consumption of reserves accumulated during seed maturation is necessary for energy production to ensure heterotrophic growth (Fait et al., 2006; Carrera et al., 2007; Bassel et al., 2008). This reserve mobilization phase occurs prior to the greening of the cotyledons and results in depletion of the storage reserves, making the shift from heterotrophic to autotrophic metabolism necessary for successful seedling establishment (Mansfield and Briarty, 1996; Allen et al., 2010). Despite the profound impact of seedling performance on crop establishment and yield, relatively little is known about the molecular processes underlying the transition from seed to seedling, or from heterotrophic to autotrophic growth. This transition is decisive for plants to enter a natural or agricultural ecosystem and is an important basis for crop production.
Once germination has started, the mobilization of stored reserves is essential to provide the growing seedling with energy and building blocks before it becomes (photo)autotrophic. The importance of energy metabolism to support germination and seedling growth is evident from primary metabolite profiling of early germination (Fait et al., 2006) and from studies that show inhibited seedling growth in mutants defective in seed lipid mobilization (Fulda et al., 2004). Moreover, evidence from gene expression profiling studies in Arabidopsis (Arabidopsis thaliana) suggests that the transcriptome required for seed germination and seedling growth is already present in the mature dry seed that has just completed development and maturation (Cadman et al., 2006; Finch-Savage et al., 2007).
Application of α-amanitin, an inhibitor of transcription that targets RNA polymerase II, appears to allow completion of the germination process until radicle protrusion but inhibits subsequent seedling growth, while inhibitors of translation prevent the progress of germination from the start (Rajjou et al., 2004). This suggests that transcriptional changes during the germination process are required to accommodate postgermination growth. Thus, it appears that in the successive developmental stages between seed maturation and seedling growth, the transcriptome is one developmental step ahead of the proteome and the physiology. This conclusion was corroborated by the observation that light, perceived by phytochrome B in the seed, generated a downstream transdevelopmental phase signal (mediated by the ABSCISIC ACID INSENSITIVE3 [ABI3] gene), which, apparently, preconditions seedlings to their most likely environment (Mazzella et al., 2005). Seedling emergence, therefore, depends, at least partly, on inherent seed characteristics.
Although germination has been studied for many years, a significant advancement of knowledge regarding the complex germination process was not attained until sequence information and omics technologies became widely available. In Arabidopsis, a number of studies utilizing sometimes high-resolution, transcriptomic approaches to investigate the time course of seed germination have made major contributions (Holdsworth et al., 2008a, 2008b; Narsai et al., 2011; Dekkers et al., 2013). However, there is a general lack of similar studies following the completion of seed germination (viz. the beginning of radicle protrusion and subsequent seedling establishment). Similar studies of the transcriptome during this phase of growth, therefore, may provide not only a global view of gene expression patterns, including biological function enrichment, but also a predictive dimension once coexpressed gene sets have been identified. For example, a time-resolved transcriptional study of the time course of seed development reported predictions of gene regulatory networks, identifying regulators of seed development (Belmonte et al., 2013). In this context, seedling growth stages until the appearance of the first root hairs, or beyond, have not been studied in meaningful detail. This implies that potentially regulatory changes in the transcriptome have not yet been associated with seedling establishment.
The main objective of this study was to identify regulatory factors to reveal signal transduction routes that are involved in the seed-to-seedling transition. Studies using large transcriptome data sets have demonstrated the correlation of gene expression (Usadel et al., 2009; Bassel et al., 2011; Dekkers et al., 2013; Verdier et al., 2013). Coexpressed genes have a greater likelihood of being involved in a common biological condition or developmental process (Aoki et al., 2007; Usadel et al., 2009; Bassel et al., 2011; Verdier et al., 2013). In addition, transcriptional modules, such as those identified for seed development (Belmonte et al., 2013), are likely to identify regulatory circuits of a process by the association of overrepresented DNA sequence motifs with coexpressed transcription factors (TFs; Orlando et al., 2009b). Here, we show that the assessment of global transcript changes across developmental stages from the mature dry seed to the seedling stage of fully opened cotyledons provides a comprehensive view of the biological processes involved in seedling development and establishment. Analysis of these developmental stages enabled us to identify informative gene sets, such as stage peak transcripts and dominant expression patterns (DPs). This was achieved by a comprehensive analysis of 21 transcriptomes of the seed-to-seedling phase transition. Mutant analysis revealed a regulatory profile controlling the seed-to-seedling transition, which was functionally validated. Also, several biological functions could be attributed to seedling establishment, including photosynthesis and root growth. The identification of these robust expression patterns will provide an essential resource to better understand the seed-to-seedling transition.
RESULTS
Transcriptomic Changes during the Seed-to-Seedling Transition
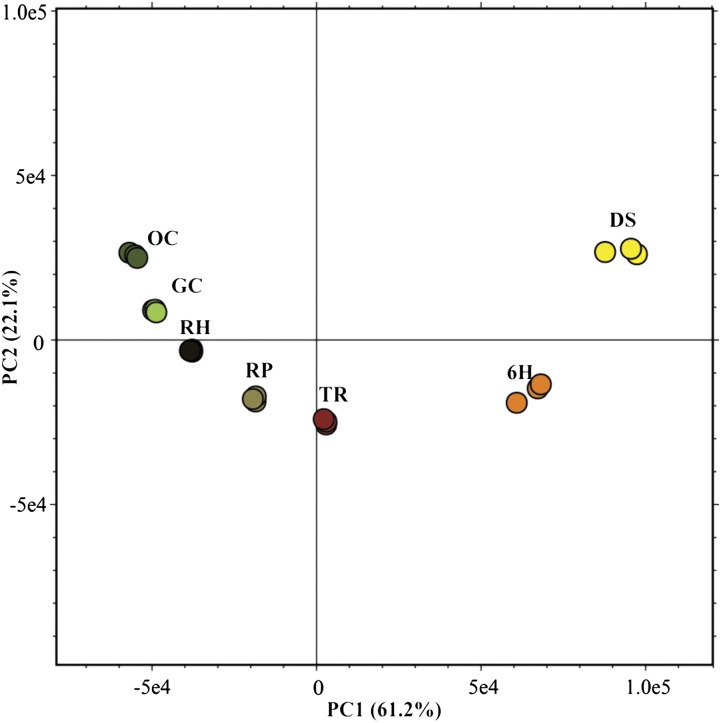
The transcript abundance of Arabidopsis Columbia-0 (Col-0) of seven developmental stages during the seed-to-seedling transition (Fig. 1) was analyzed using a high-density Affymetrix array (Aragene.st1.1). This array encompasses the complete Arabidopsis transcriptome. Principal component analysis (PCA) was used to compare the overall variation in gene expression levels among the seven developmental stages, using the entire transcriptome with three biological replicates for each developmental stage (Fig. 2). Each developmental stage was clearly distinct from the other stages. The proximity of the replicates in the PCA plot highlights the robustness of the experimental setup and data-processing steps and shows that this is a powerful data set in which to study the seed-to-seedling transition.
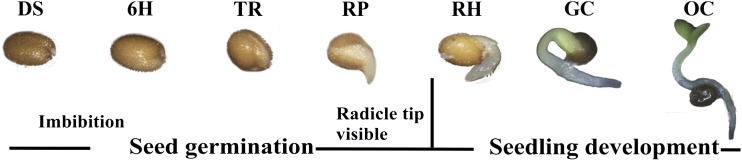
Figure 1.
Subdivision of the seed-to-seedling developmental stages: DS, dry seeds; 6H, 6-h imbibed; TR, testa rupture; RP, radicle protrusion; RH, root hair; GC, greening cotyledons; and OC, cotyledons fully opened.
Figure 2.
PCA plot of transcript abundance of the seven seed-to-seedling developmental stages. PCA-derived score plots PC1 (61.2%) and PC2 (22.1%) are shown for the microarray data representing all of the seed-to-seedling developmental stages.
Of the complete transcriptome, 19,130 (69%) transcripts were differentially expressed in at least two developmental stages during the seed-to-seedling transition. Pearson correlation was applied to these transcripts, and this indicated distinct temporal profiles (Supplemental Fig. S1; Supplemental Data Set S1). When successive developmental stages were compared, considerable changes could be observed. The greatest change in the number of transcripts was observed in the comparison between DS and 6H (over 7,000 genes up-regulated and 4,000 genes down-regulated). Other sets of transcripts showed more moderate changes: from 6H to TR (5,600 genes up and 4,768 genes down), TR to RP (840 genes up and 1,107 genes down), RP to RH (1,596 genes up and 1,824 genes down), RH to GC (1,543 genes up and 1,407 genes down), and GC to OC (2,515 genes up and 3,272 genes down; Supplemental Fig. S2). Highly expressed genes in DS were enriched for Gene Ontology (GO) terms related to heat response and lipid storage, whereas high expression at 6H was related to nucleotide binding and structural constituent of ribosome, suggesting the activation of translational activity. Gene sets of the other comparisons were associated with such processes as cell cycle, protein synthesis, DNA processing, and transcription (Supplemental Table S1). For example, GO terms such as RNA processing and nucleic acid binding were enriched for genes highly expressed in the TR, RP, and RH stages, whereas chloroplast envelope, endomembrane system, and ribosome biogenesis were enriched for genes highly expressed in GC and OC stages.
Transcriptomic Analysis Identifies Sets of Developmentally Regulated Processes during the Seed-to-Seedling Transition
The gene clusters suggested the temporal expression of developmentally regulated transcripts. The results show transcripts that specifically peaked at each developmental stage and 10 DPs across all seed-to-seedling developmental stages.
Stage Peaking Gene Sets
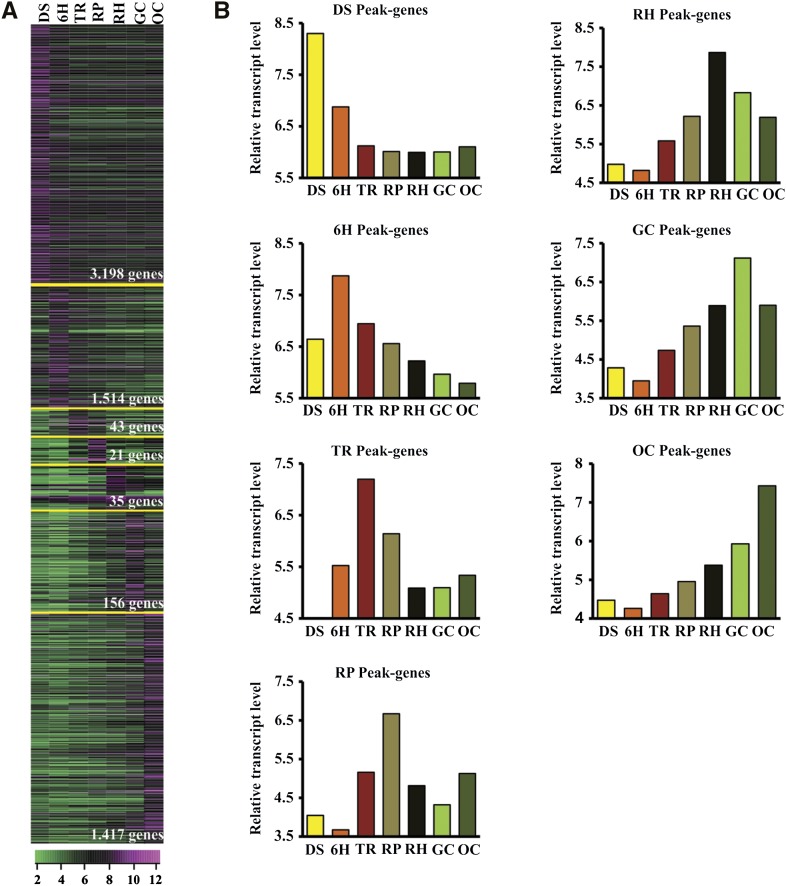
The complexity of the data sets suggests a coordinated shift in gene expression at the developmental stages of the transition. Because of this complexity, we identified genes that peaked (P < 0.01, Bonferroni adjusted) at a particular stage, derived from the subset of 19,130 transcripts. This analysis illustrates that the different sets of genes display peaks of expression at different developmental stages, which is suggestive of their relevance for stage-specific developmental functions (Supplemental Table S2). Interestingly, the clusters of developmentally regulated transcripts grouped into specific stages and formed a wave of transcript abundance, moving from a quiescent dry seed to a growing seedling (Fig. 3). These clusters may thus govern the progression of the genetic program toward seedling establishment. Analysis of the peaking genes resulted in 6,384 transcripts that showed significant levels of differential expression with a single peak across the seed-to-seedling development stages. Of 6,384 transcripts, 50% showed a maximum transcript expression in DS and 24% in 6H, whereas in TR, RP, and RH, less than 2% displayed maximum expression (0.6% at TR, 0.3% at RP, and 0.5% at RH). GC and OC displayed maximum expression of around 22% and 2%, respectively (Fig. 3; Supplemental Table S2). The number of peaking transcripts for each developmental stage indicated that transcript abundance can be grouped in three distinct clusters: (1) DS and 6H; (2) TR, RP, and RH; and (3) GC and OC, implying two major transitions. This complex pattern of gene activity observed during the seed-to-seedling transition can help to determine the fundamental molecular processes involved in seedling establishment and, hence, to predict seed and seedling quality by monitoring gene expression during seed germination and seedling establishment.
Figure 3.
Overview of the expression patterns of transcripts peaking at selected seed-to-seedling transition stages. A, Differentially expressed gene clustering based on the peak of expression at the selected seed-to-seedling stage. B, General tendency and average abundance of transcripts at each stage. This analysis shows the maximum expression of transcripts at different stages, indicating the stage-specific maximum activity of the transitionally regulated transcripts.
DPs
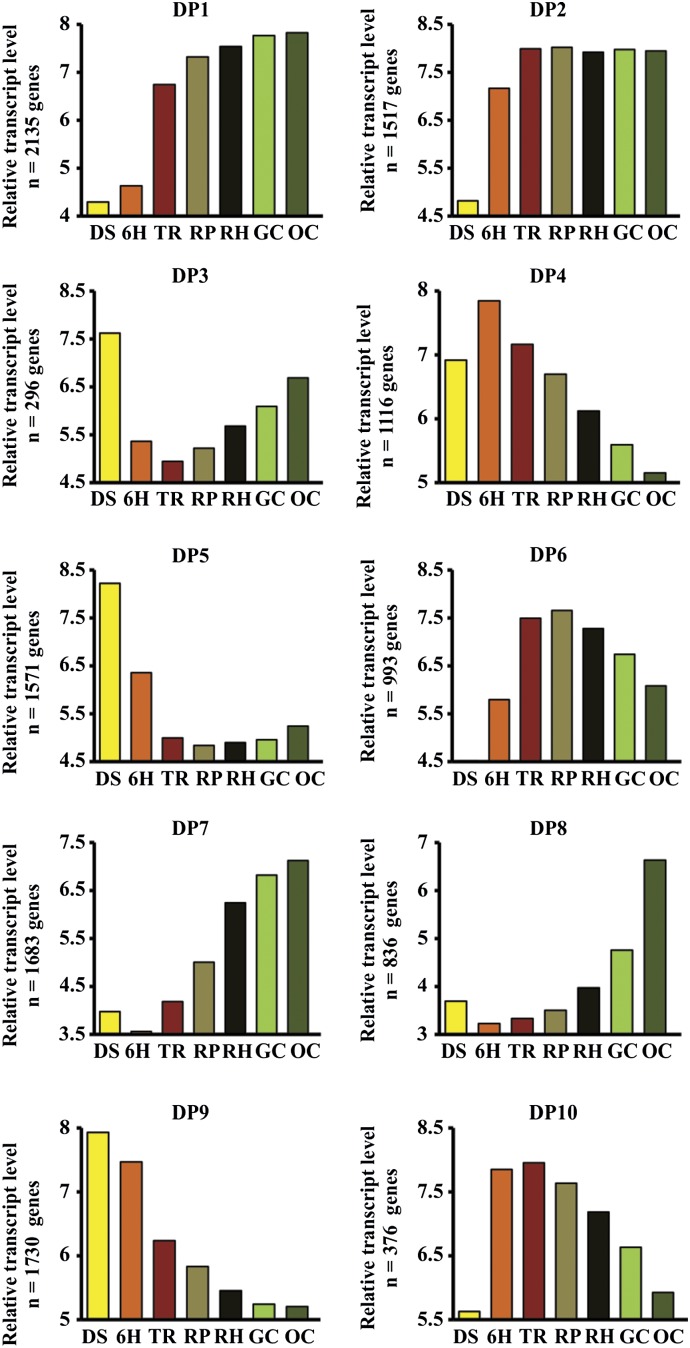
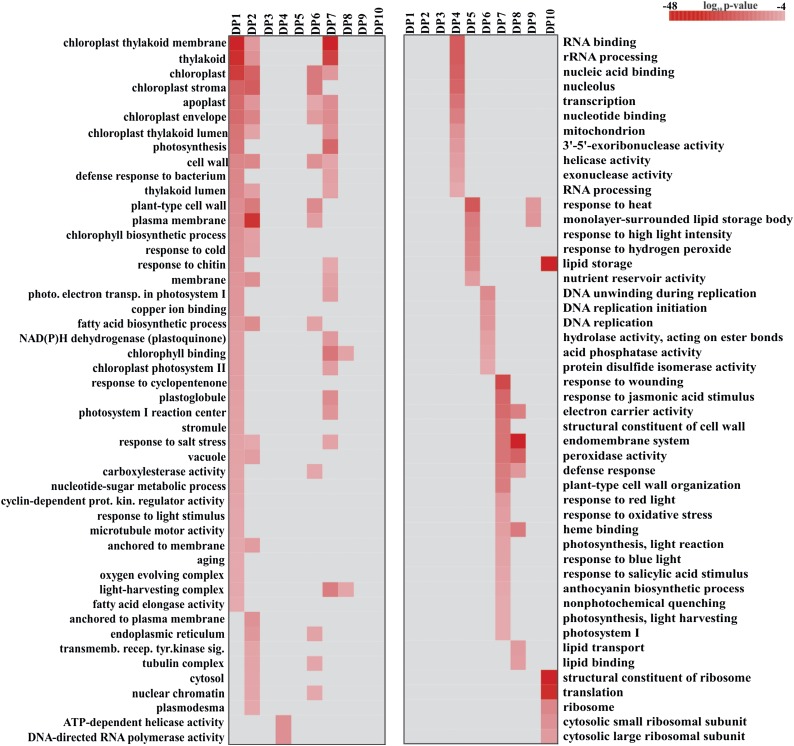
To determine how transcript abundance changes during the seed-to-seedling transition, we also clustered transcripts from the subset of 19,130 transcripts into 10 DPs (Fig. 4; Supplemental Table S2) using the Fuzzy K-Means clustering method (Orlando et al., 2009b). Five of the coexpressed gene sets consisted of transcripts with high expression at only one stage (DP3, DP4, DP5, DP8, and DP9), whereas the other five coexpressed gene sets were expressed across several developmental stages. These expression patterns suggest the occurrence of processes related to specific stages of the seed-to-seedling transition. Predictive functions of these processes were determined for each of the DPs by an analysis of enriched GO terms (P < 0.0001) and metabolic processes (P < 0.001; Fig. 5; Supplemental Table S2). For example, the DP1 gene set was overrepresented for genes with GO terms such as fatty acid activity, chlorophyll biosynthesis, and photosynthesis (Fig. 5). Another gene set, DP7, was significantly enriched for GO terms and metabolic processes associated with photosynthesis and carbon metabolism, including chloroplast and thylakoid structure, photorespiration, Calvin cycle, and gluconeogenesis (Fig. 5; Supplemental Table S2). DP6 and DP10 consisted of transcripts highly expressed in the TR, RP, and RH stages. DP6 and DP10 were significantly enriched for DNA unwinding and ribosome-related transcripts (Fig. 5). The DPs displayed a transient variation of transcript abundance across the seed-to-seedling developmental stages, which suggests a complex network of transcripts that control the major transitions in seedling development.
Figure 4.
DPs of gene expression during the seed-to-seedling transition. Ten DPs were found using Fuzzy K-Means clustering of the 50% most variant transcripts from the data set of 19,130 transcripts that showed a significant expression difference in at least one developmental stage. Bar graphs represent averages of mRNA expression levels in each stage (left to right, mature seeds to cotyledons fully opened).
Figure 5.
Heat map showing the P value significance of GO enrichment terms for DPs (Supplemental Table S2). GO terms listed are for biological process and/or cellular components that were overrepresented for each DP gene set. P values were calculated by their hypergeometric distribution.
In summary, the predicted pathways associated with the peaking genes and DP data sets can be described by GO terms and metabolic processes (Supplemental Table S2). DS peak genes were significantly enriched for GO terms known to be associated with events that occur during seed development, maturation, and abscission, including ubiquitin-protein ligase activity and response to heat. 6H peak genes were overrepresented for processes associated with high levels of transcriptional activity, such as RNA binding and nucleic acid binding. Gene sets of TR, RP, and RH were not significantly enriched for GO terms and metabolic processes, because only a few transcripts peaked at these stages. GC-specific transcripts, however, were highly enriched for microtubule motor activity genes (mitotic kinesins) that are associated with the cell cycle. Transcripts peaking at OC were enriched for processes supporting tissue growth, such as transport and lipid binding (Supplemental Fig. S3). Moreover, DP gene sets associated with transition regions (DP1, DP2, DP6, DP7, and DP10) were significantly enriched for cell differentiation, proliferation, and photosynthesis. DP2, DP6, and DP10 probably undergo cell divisions with subsequent cellularization but no photosynthesis. Photosynthesis and chlorophyll binding, however, were enriched in DP1 and DP7. Gene expression related to photosynthesis might start already at the TR stage (DP1) with a second wave of transcript abundance starting at RP (DP7; Fig. 4).
A Coexpression Gene Regulatory Network of the Seed-to-Seedling Transition
To identify regulatory processes that control the seed-to-seedling transition, an unweighted gene coexpression network analysis was carried out. The Pearson correlation threshold was set at 0.98 according to Freeman et al. (2007). This threshold was used to determine connections between edges and nodes in the network, resulting in a network consisting of 6,896 nodes with 99,762 edges. Network visualization was carried out in Cytoscape using the organic layout (Fig. 6; Supplemental Data Set S2). Temporal expression profiles corresponding to the different coexpression gene sets were clearly separated. These distinct regions are associated with the three main stages that dominate the transition: DS, 6H, and OC. As we were interested in the mechanisms that govern the transition from a quiescent to a photoautotrophic state, we investigated which DPs were most representative for this transition. For this purpose, each DP identified previously was mapped on the seed-to-seedling network (Fig. 7).
Figure 6.
Unweighted gene coexpression network for the seed-to-seedling transition. Circles (nodes) represent transcripts, and lines (edges) represent significant transcriptional interactions between the transcripts. Temporal analysis of network clusters was performed by coloring each gene by its maximum expression across seed-to-seedling transition developmental stages (peaking gene sets).
Figure 7.
DPs mapped on the seed-to-seedling network. Nodes (transcripts) are colored in red according to the DP to which they belong, showing expression regions on the network for the different DPs (Fig. 4).
Regions of transcriptional interaction associated with phase transitions were observed for the DP1, DP2, DP6, DP7, and DP10 clusters. DP2 and DP10 showed gene expression patterns increasing from DS to 6H, whereas DP1 and DP6 showed an increase from 6H to TR (Fig. 4). This suggests that these four DPs are associated with phase transitions in seed germination rather than early seedling growth. Therefore, of these five DPs (DP1, DP2, DP6, DP7, and DP10), DP7 is predicted to bridge the gap between germination and the seedling stage (Fig. 7). Therefore, DP7 is the most representative gene set associated with the transition from germination (6H) to early seedling establishment (OC). The DP7 set is unique in that the transcript abundance increased after RP and kept increasing to higher levels thereafter (Fig. 4). The expression pattern of DP7 suggests that a common set of transcripts is uniformly up-regulated across the seedling developmental stages. It also may indicate that a distinct regulatory process during RP affects the abundance of over 1,600 transcripts that subsequently control seedling establishment.
Predicted Regulatory Circuitry Controlling Transcripts Expressed during the Transition from Germinated Seed to Established Seedling
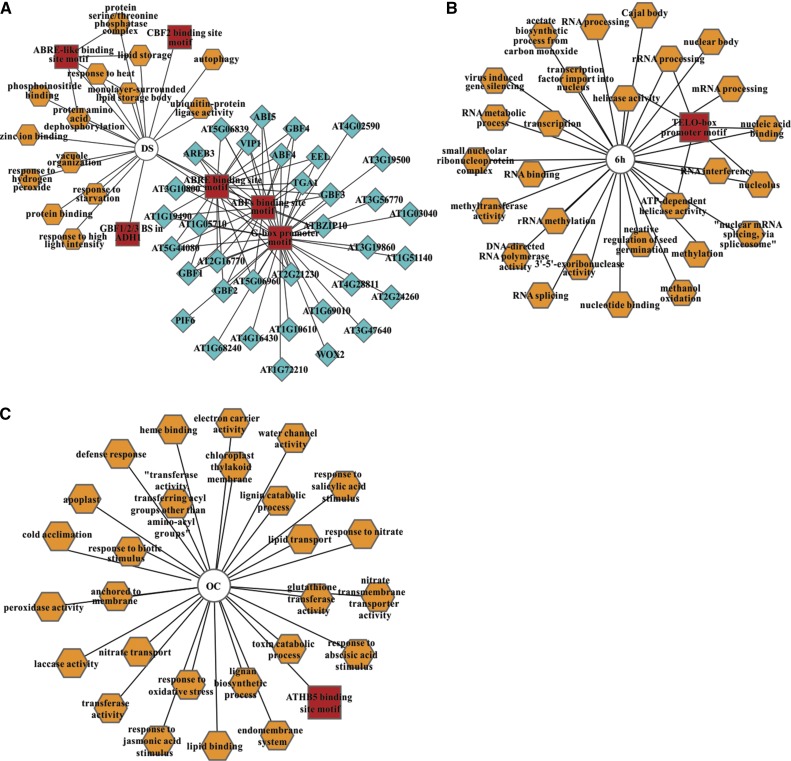
To define a gene regulatory network that might control the transition from a quiescent to an autotrophic photosynthesizing state, we inferred a predicted transcriptional module that links TFs with their potential coexpressed target transcripts, using ChipEnrich and Cytoscape. Transcriptional modules of stage-specific transcripts (DS and OC) link enriched ABRE, G-box, ABFs, and AtHB5 DNA sequence motifs (homeodomain leucine zipper I [HD-Zip I]) with known or predicted TFs that are present in mature seeds and seedlings (Fig. 8; Supplemental Table S3). The transcriptional module for DS is enriched for G-box, ABRE, and ABF DNA motifs, which are associated with bZIP TFs known to function in seed maturation (Gutierrez et al., 2007). The transcriptional module for OC showed enrichment for the AtHB5 DNA motif known to be associated with seedling development (Johannesson et al., 2003). Transcriptional modules built from the other seed-to-seedling transition gene sets may give additional input to identify potential regulatory circuits that govern phase transitions within the network.
Figure 8.
Transcriptional modules predicted to regulate the seed-to-seedling transition. DNA motifs (red rectangles) and GO terms (orange octagons) that are significantly overrepresented (P < 0.001) within the peak transcript sets (circles) are shown together with coexpressed TFs (light blue diamonds). Transcriptional modules were predicted for DS (A), 6H (B), and OC (C) peak transcripts (Supplemental Table S2). P values were calculated by their hypergeometric distribution.
We also built transcriptional modules for each DP (Supplemental Figs. S4 and S5). These showed an evident overlap in enriched DNA motifs and TFs identified in the stage-specific gene sets. This observation suggests that seedling establishment processes are regulated in a dynamic manner, as shown by the DPs. The transcriptional module for the coexpressed gene sets identified as being in the transitory region from germinated seed to the photoautotrophic seedling (DP7) was predicted to be enriched for the AtHB5 DNA motif associated with the three homeodomain-Leu zipper TFs AtHB13, AtHB20, and AtHB23. These TFs are members of subclass α (AtHB3, AtHB13, AtHB20, and AtHB23) within the class I homeodomain-Leu zipper TFs, with a putative role in leaf development (Henriksson et al., 2005). However, none of these have been related previously to a seedling-related developmental phase transition.
AtHB13 Regulates Primary Root Growth
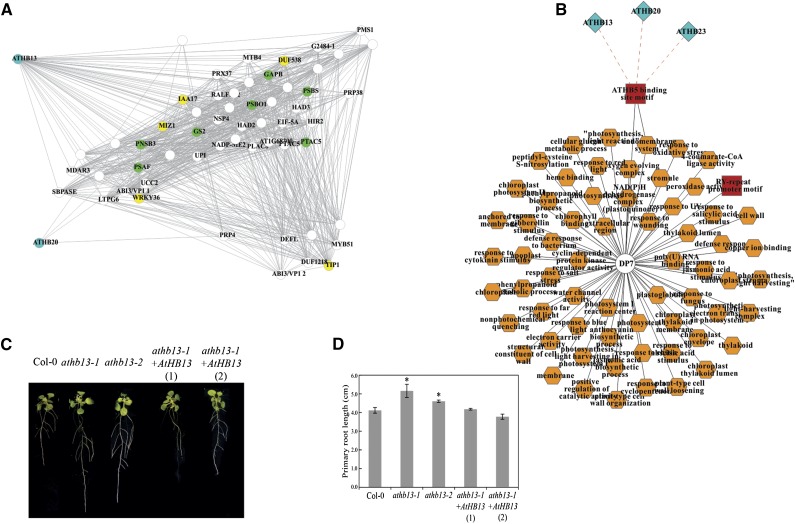
To investigate whether AtHB TFs are present in the transition region, we mapped these TFs on the seed-to-seedling network (Fig. 9). AtHB13 and AtHB20 were in the transition region, whereas AtHB23 was not. Since the inference of coexpression networks and DPs represent two independent methods (see “Materials and Methods”), this result is plausible. To generate a seed-to-seedling coexpression network, a high cutoff for Pearson correlation (0.98) was used, whereas for the DPs, a lower cutoff was used (0.8). Therefore, we conclude that AtHB23 has a weaker interaction with transcripts of the transition region of the seed-to-seedling network than AtHB13 and AtHB20, which showed strong interactions with many transcripts of this region (Fig. 9). In our data set, 58 transcripts were found to be coexpressed with AtHB13 and AtHB20. Within the transition region, many key regulators of photosynthesis and root development interact in the regulation of seeding establishment. Seven genes characterized previously with a role in photosynthesis and five with a role in root development were identified from this coexpression analysis (Fig. 9). Photosynthesis-related genes included PHOTOSYSTEM I SUBUNIT F (PSAF; At1g31330), GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE B SUBUNIT (GAPB; At1g42970), CHLOROPHYLL A-B BINDING FAMILY PROTEIN (CP22; At1g44575), NDH-DEPENDENT CYCLIC ELECTRON FLOW1 (NDF4; At3g16250), PLASTID TRANSCRIPTIONALLY ACTIVE5 (PTAC5; At4g13670), GLUTAMINE SYNTHETASE2 (GS2; At5g35630), and PHOTOSYSTEM II OXYGEN-EVOLVING COMPLEX1 (PSBO1; At5g66570). Root-related genes were AUXIN RESISTANT3 (IAA17; At1g04250), DUF538 (At1g09310), WRKY DNA-BINDING PROTEIN36 (WRKY36; At1g69810), TONOPLAST INTRINSIC PROTEIN1 (TIP1; At2g36830), and MIZU-KUSSEI1 (MIZ1; At2g41660). We phenotyped transfer DNA (T-DNA) insertion mutants for AtHB13 and AtHB20 (athb13-1, athb13-2, and athb20-1) with no previously described function in the seed-to-seedling phase transition. A major effect of both the athb13-1 and athb13-2 knockouts was a significantly increased primary root length relative to the wild type (Fig. 9; Supplemental Fig. S5), whereas athb20-1 did not show a similar root phenotype (Supplemental Fig. S6). Complementation of the athb13-1 mutant restored the wild-type phenotype. This shows that AtHB13 negatively regulates root growth during seedling establishment. AtHB20 also could be a good candidate to interact with AtHB13 in the control of primary root length, as expression of these genes is highly correlated. Furthermore, the seed-to-seedling network showed that AtHB20 is not correlated with so many genes related to root development as AtHB13. This could be a possible explanation for why the knockout of AtHB20 did not exhibit a phenotype. This example demonstrates how a transcriptional module together with a coexpression network can generate insights that are not immediately apparent from simple transcript profile comparisons.
Figure 9.
Validation of the seed-to-seedling gene coexpression network showing the homeodomain-Leu zipper AtHB13 playing a negative role in primary root elongation. A, Gene network interactions of AtHB13 and AtHB20 as derived from the seed-to-seedling network. Green nodes represent genes known to be associated with photosynthesis, whereas yellow nodes represent genes associated with root development. B, Transcriptional modules predicting regulators of the transition region (DP7) of the seed-to-seedling transition. DNA motifs (red rectangle) and GO terms (orange octagons) that are significantly overrepresented (P < 0.001) within the transition gene set (circle) are shown together with coexpressed TFs (light blue diamonds). The transition region gene set possesses a transcriptional module, of which ATHB5 is known to be associated with seedling development. All DPs with DNA motif, TF, and GO term enrichments are listed in Supplemental Table S2. C, Root growth phenotypes of the wild type (Col-0), two AtHB13 knockout lines (athb13-1 and athb13-2), and two complemented lines (athb13-1 + 35S::AtHB13 lines 1 and 2). Roots were scanned at 15 d after transfer to Murashige and Skoog (MS) medium. D, Primary root length comparison at 15 d after transfer to MS medium. Error bars represent the sd of three independent biological replicates for each genotype. Statistical analyses of the mean values were performed using R statistical language (R Development Core Team, 2011), and asterisks denote significant differences with Student’s t test at P < 0.01.
DISCUSSION
To reach an autotrophic state, the seed-to-seedling transition is assumed to temporally and spatially employ various regulatory factors. These regulatory factors modulate the controlling genes and proteins, which will eventually govern seedling establishment. A detailed transcriptome analysis with the inference of coexpression networks and transcriptional modules of the seed-to-seedling transition resulted in the identification of several coregulating modules of gene expression, coinciding with specific developmental stages. Expression profiles for each gene of single stages of the transition matched with known expression profiles (Ding et al., 2006; Tepperman et al., 2006; Hammani et al., 2011; Narsai et al., 2011; Feng et al., 2014), validating the data set. Expression data across the seven developmental stages displayed high reproducibility. Each developmental stage was clearly distinguishable in a PCA plot, which suggests that changes in gene expression are already occurring within 6 h of imbibition. The changes in the number of transcripts, which were highly abundant during the first 6 h of imbibition, demonstrate that transcripts that had accumulated during seed maturation are degraded and that the induction of translation-associated transcripts, involved in germination, may commence directly upon imbibition. This observation confirms that of Dekkers et al. (2013), who showed that the majority of seed maturation-repressed genes are reactivated in the first phase of Arabidopsis seed germination (dry to imbibed seed). Moreover, the enriched GO terms for DS peak transcripts corroborate a previous study of gene coexpression networks of seed maturation in Medicago truncatula (Verdier et al., 2013), in which genes expressed during seed maturation also were associated with GO terms such as lipid binding and heat response. The overrepresented GO terms for 6H peak genes are associated with RNA binding and nucleic acid binding, and these are in agreement with a previous study of mitochondrial biogenesis during seed germination (Law et al., 2012). That study showed that transcripts that increased in abundance in early germination were overrepresented for RNA processing and encode mitochondrial biogenesis functions, which precede crucial bioenergetic and metabolic functions. This observation is in agreement with our 6H peak gene results and postulates that an early transcript expression (6H) may act as a signal to prompt the expression of genes involved in various metabolic functions required for germination and seedling establishment. Clusters of TR, RP, and RH showed only a few peaking genes and a minor change in transcript expression. The GC peak gene set, however, is overrepresented for microtubule motor activity genes (mitotic kinesins), which are associated with the cell cycle (Vanstraelen et al., 2006). Thus, this cluster has genes playing roles in cellular organization and dynamics, chromosome movement, and cytokinesis (Vanstraelen et al., 2006), which prompts us to conclude that, at the GC stage, high cell division activity occurs. Genes peaking at OC are enriched for processes supporting tissue growth, such as transport and lipid binding. Taken together, overrepresentation results for peak gene sets reflect two major transitions in gene activity. These results, furthermore, imply that a transcript preparation for germination and early seedling development is initiated already at early imbibition (6H peak genes) and that an efficient seedling establishment is ensured by additional distinct transcript expression of GC and OC peak genes.
Besides overrepresentation of GO terms for the peak gene sets, changes in transcriptional modules (Belmonte et al., 2013) of these gene sets link to TFs that control functions of the regulatory networks. The transcriptional module for transcripts that accumulated in DS include DNA motifs (G-box, ABRE, and ABFs) linking to bZIP TFs such as ABI5. ABI5 is known to play a role in maturing seeds (Finkelstein et al., 2005) and to be a major regulator of late embryogenesis abundant proteins (Nakashima et al., 2006). Additionally, ABI5 has been shown to play an important role in the expression of an abscisic acid (ABA)-responsive gene, RESPONSIVE TO DESSICATION29B, which contains the ABRE motif (Nakashima et al., 2006). Thus, the occurrence of DS peak genes is in agreement with the known regulatory network in maturing seeds. Transcripts of key regulators of germination are associated with 6H peak genes and are rapidly down-regulated during early seedling establishment. The 6H peak gene data set is enriched for the TELO box motif, which is associated with genes involved in DNA replication (Wang et al., 2011), whereas the transcriptional module of OC peak genes is enriched for the AtHB5 DNA-binding site motif known to be associated with the regulation of ABA responsiveness and seedling development (Johannesson et al., 2003).
The transcriptional modules identify potential regulatory circuits that control processes associated with the GO terms of seed-to-seedling phase transitions. The transcriptional interactions in each of the stage peak genes are distinct, with the DS peak genes, 6H peak genes, and OC peak genes showing greater numbers of highly expressed genes and a transcriptional coordination according to the modules. Interestingly, TR, RP, and RH peak genes, with only a few highly expressed transcripts, suggest that a common transcriptional mechanism may be responsible for seedling establishment, which continues by the activation of additional transcriptional mechanisms (GC peak genes and OC peak genes).
To better understand the seed-to-seedling phase transition, we examined the distribution of these peak genes over the seed-to-seedling coexpression network. Coexpression analysis suggests that stage peak genes capture transcriptional interactions associated with developmental stages from dry seed to fully open cotyledons. The seed-to-seedling network shows three regions of interaction (dry, germination, and seedling states), with a transition region between the germination and seedling stages. It suggests that functional differentiation within the transitional developmental stages occurs mainly through two distinct processes, namely the germination (6H peak genes) and seedling establishment (OC peak genes) states.
Our analysis highlights this network as a powerful tool with which to understand the regulation of the transition from a quiescent dry state to a photoautotrophic one. Moreover, the DP analysis reduces the complexity of coexpression data (Brady et al., 2007; Orlando et al., 2009b) and helps in understanding how transcript abundance changes over successive developmental stages (Brady et al., 2007; Belmonte et al., 2013). For example, the DP1 gene set is overrepresented for GO terms associated with chlorophyll biosynthesis and photosynthesis. Chlorophyll biosynthesis is controlled during the critical initial emergence of seedlings from darkness into light (Huq et al., 2004; Moon et al., 2008). Thus, the DP1 coexpression pattern may serve as a blueprint for chlorophyll biosynthesis and photosynthesis during the seed-to-seedling transition. Two other gene sets (DP6 and DP10) show clusters of transcripts highly expressed in the TR, RP, and RH stages. DP6 and DP10 are overrepresented for DNA unwinding, ribosome, and mitochondrion. The expression of these DPs points to a rapid reestablishment of various biochemical activities to support seedling establishment. The biogenesis of mitochondria is known to provide energy for cellular processes, which is critical for seed germination and seedling establishment (Law et al., 2012; Jiang et al., 2013). Consistent with these observations, transcripts associated with mitochondrion display high expression already after 6 h of imbibition (related to DP10), which suggests that mitochondrial biogenesis is required already at 6H to ensure successful seedling establishment. The predicted regulatory circuitry controlling transcripts expressed during the seed-to-seedling transition appears to be essential for successful early seedling establishment.
The transition region between germination and the seedling stage in the seed-to-seedling network is determined by essentially one DP, namely DP7. The three TFs identified in DP7 are members of the HD-Zip gene family, which are classified into four groups (Henriksson et al., 2005). Of the three identified family members, only two (AtHB13 and AtHB20) displayed strong interactions within the seed-to-seedling network. These TFs are paralogs and have similarities and differences in their response. AtHB13 is involved in cold tolerance (Cabello et al., 2012), whereas AtHB20 has been assigned roles in ABA, salt, and cold responses (Henriksson et al., 2005) as well as seed dormancy (Barrero et al., 2010). AtHB23 and AtHB20 play roles in light signaling (Barrero et al., 2010; Choi et al., 2014), but none of these have been related previously to the seed-to-seedling phase transition.
These TFs represent high-confidence candidates of previously described regulatory factors present in DP7 and probably have a role in coordinating gene partners within this DP (Belmonte et al., 2013). The transcriptional module for DP7, which shows a significant increase from the last stage of germination (RP) to the first stage of seedling establishment (RH) and keeps increasing thereafter, suggests a regulatory process involving over 1,600 transcripts. The coexpression regulatory circuit of AtHB13 suggests that this TF plays a role in the regulation of transcripts related to photosynthesis and root growth. However, there is no evidence for a connection among AtHB13, root growth, and photosynthesis, but a correlation has been reported between photosynthesis and AtHB2, another AtHB family member (Carabelli et al., 1996; Hu et al., 2013), and between root growth and AtHB8 (Baima et al., 1995). In addition, AtHB13 has been described recently to play a crucial role in Arabidopsis development (Ribone et al., 2015). This TF appeared to be vital for pollen germination, the mutant showed defected siliques, and it may also play a negative role in stem elongation. Thus, this suggests that AtHB13 is part of the same regulatory pathway leading to root growth or photosynthesis for successful seedling establishment. This is corroborated by the root phenotypes of athb13-1 and athb13-2 (Fig. 9). To our knowledge, the role of AtHB13 in the regulatory circuit for root development is unknown. The increased root growth of athb13-1 and athb13-2, as compared with Col-0, suggests a negative feedback regulation in root development regulation, as shown previously for AtHB8 in auxin signaling (Baima et al., 2014) and AtHB13 in stem growth (Ribone et al., 2015). Interestingly, the enhanced primary root growth of the mutant represents postseedling establishment growth, whereas the transition studied here largely represents cell expansion-driven processes. This confirms previous conclusions that the transcriptome may be expressed ahead of the prevailing developmental stage, in preparation for what follows (Cadman et al., 2006; Finch-Savage et al., 2007). The repressing activity in root development by AtHB13 may indicate the antagonistic activities of modulating signals toward the completion of seedling establishment.
CONCLUSION
Our seed-to-seedling gene expression network describes global transcriptional interactions for two distinctive developmental states, namely germination and seedling development. Evidently, the seed-to-seedling transition is related to the agronomic trait of seedling establishment and vigor, and understanding the associated transcriptional regulatory network will facilitate studies to ultimately enhance seedling establishment in agriculture. The seed-to-seedling gene expression network provides a template to postulate new hypotheses about transcriptional regulators and their interactions. Many previously described TFs interact in the regulation of the seed-to-seedling transition. How seedling establishment may be improved by this transition depends on the interaction activity of factors promoting signaling across the germination and seedling stages. Our data here suggest that previously unknown regulators identified in the transition region act through known regulatory components to promote information controlling development later in seedlings. Therefore, further investigation of coexpressed genes in the transition region might lead to an answer for the connection between coexpressed genes and seedling establishment.
MATERIALS AND METHODS
Plant Material Collection
Seeds of Arabidopsis (Arabidopsis thaliana accession Col-0 [N60000]) were cold stratified at 4°C in the dark for 72 h in petri dishes using two layers of moistened blue filter paper (Anchor Paper) to break residual dormancy. Germination tests were performed in a growth chamber at 22°C under constant white light. To elucidate the changes in the transcriptome related to the transition from a seed to a photoautotrophic seedling, seven developmental stages were identified: DS, 6H, TR, RP, RH, GC, and OC (Fig. 1).
RNA Extraction
Total RNA was extracted using the hot borate method according to Wan and Wilkins (1994) with some modifications as described previously (Maia et al., 2011). RNA quality and concentration were measured by agarose gel electrophoresis (0.1 g mL−1) and NanoDrop.
Microarray Hybridization
Quality control, RNA labeling, hybridization, and data extraction were performed at ServiceXS. Labeled single-stranded complementary DNA was synthesized using the Affymetrix NuGEN Ovation PicoSL WTA Version 2 Kit and Biotin Module using 50 ng of total RNA as template. The fragmented single-stranded complementary DNA was utilized for hybridization on an Affymetrix ARAGene 1.FIRST array plate. The Affymetrix HWS Kit was used for the hybridization, washing, and staining of the plate. Scanning of the array plates was performed using the Affymetrix GeneTitan scanner. All procedures were performed according to the instructions of the manufacturers (nugen.com and affymetrix.com). The resulting data were analyzed using the R statistical programming environment and the Bioconductor packages (Gentleman et al., 2004). The data were normalized using the RMA algorithm (Irizarry et al., 2003) with the TAIRG v17 cdf file (http://brainarray.mbni.med.umich.edu). Expression data are hosted in the National Center for Biotechnology Information Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=onyxsyycjtaxxux&acc=GSE65394). Validation of the seed-to-seedling transcriptome data set was performed by comparison with previously published expression patterns of genes known to be differentially expressed across seed-to-seedling developmental stages (Supplemental Table S1; Ding et al., 2006; Tepperman et al., 2006; Hammani et al., 2011; Narsai et al., 2011; Feng et al., 2014).
Identification of Coexpression Gene Sets and Transcriptional Modules
Stage-Specific Gene Sets
For the selection of differentially expressed transcripts specific for a given developmental stage, a two-step approach was used. First, Bayesian estimation of temporal regulation (P < 0.001) was used to identify differentially expressed transcripts in at least two different developmental stages (Aryee et al., 2009). At the next step, differentially expressed transcripts were further filtered to determine whether they were specific for a particular stage. The Limma package (Gentleman et al., 2005) was used to check whether the average expression values in a specific stage were significantly (P < 0.01) larger than the expression at other stages. The extracted data sets were hierarchically clustered and visualized in GeneMaths XT.
DP and Transcriptional Module Prediction
DPs were identified as described previously by Orlando et al. (2009b). DPs were identified for the 50% most variant transcripts, corresponding to 9,565 mRNAs. The R function FANNY (http://cran.r-project.org/web/packages/cluster/cluster.pdf) with a minimum Pearson correlation of 0.85 was used to evaluate the number of clusters (K) choices from one to 50 with a cutoff for cluster membership of 0.4. The K choice that yielded the greatest number of transcriptional modules was 10. These transcriptional modules were used in the ChipEnrich software package developed by Brady et al. (2007) and modified by Belmonte et al. (2013). ChipEnrich determines the significance of GO terms, metabolic processes, DNA motifs, and TFs using P values calculated from their hypergeometric distribution (Gadbury et al., 2009; Belmonte et al., 2013). For the hypergeometric distribution lists of GO terms, metabolic processes, DNA motifs, and TFs were used based on the Arabidopsis Gene Regulatory Information Server (http://arabidopsis.med.ohio-state.edu/AtTFDB/). An optimized ChipEnrich by Belmonte et al. (2013) was used to identify significantly enriched DNA motifs, associated TFs, and GO terms. Tables generated by ChipEnrich were imported into Cytoscape (version 2.8.2), and the transcriptional module networks were visualized using the yFiles organic layout.
Seed Germination, Seedling Establishment, and Root Growth Phenotype
AtHB20 mutant lines (athb20-1) were obtained from Barrero et al. (2010). Mutant plants from genotypes athb13-1, athb13-2, and athb13-1 + 35S::AtHB13 (lines 1 and 2) were described previously (Cabello et al., 2012; Ribone et al., 2015). Seeds of these mutants were sown on 5- × 5-cm Rockwool blocks in a climate cell (20°C day and 18°C night) with a photoperiod of 16 h of light and 8 h of dark. Each Rockwool block was watered with Hyponex solution (1 g L−1). Seeds were harvested in four replicates of at least three plants. In order to measure root growth, seeds were sterilized with commercial bleach (20%, v/v) and placed on solid medium containing 0.5× MS medium (Murashige and Skoog, 1962) without Suc. Thereafter, seeds were stratified for 72 h at 4°C to remove residual dormancy and transferred to a germination cabinet at 22°C with constant white light. Twenty-seven seeds were selected at the RP stage for Col-0 and for each of the mutants (athb13-1, athb13-2, athb13-1 + 35S::AtHB13 [lines 1 and 2], and athb20-1). These seeds were placed on new plates with 0.5× solid MS medium without Suc. Plates were placed vertically in a climate cell in the same conditions as described above. Root growth was scored 15 d later for each plant. Plates were scanned using an Epson document scanner, and root lengths were determined by SmartRoot 4.1 using ImageJ software.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Hierarchical clustering analysis of differentially expressed Arabidopsis seed-to-seedling transition transcripts.
Supplemental Figure S2. Overview of changes in transcript levels across seed-to-seedling developmental stages.
Supplemental Figure S3. Heat map showing the P value significance of GO enrichment terms for peak–transcript sets
Supplemental Figure S4. Transcriptional modules predicting regulation of the seed-to-seedling transition.
Supplemental Figure S5. Illustrative plates of wild type (Col-0), two AtHB13 mutant lines (athb13-1 and athb13-2) and two complemented lines (athb13-1+AtHB13 lines 1 and 2).
Supplemental Figure S6. An independent experiment of root growth phenotype of mutant lines (athb13-1 and athb20-1) and wild type (Col-0).
Supplemental Table S1. Validation of the seed-to-seedling transcriptome data set.
Supplemental Table S2. Transcript comparisons between previous and next stages, and their enriched GO terms, DNA motifs, and metabolic pathways.
Supplemental Table S3. Peak-transcripts and dominant pattern sets.
Supplemental Data Set S1. Abundance of all transcripts in the seed-to-seedling transition.
Supplemental Data Set S2. The seed-to-seedling network Cytoscape file.
Supplementary Material
Acknowledgments
We thank José M. Barrero for providing the athb20-1 mutants and Bas Dekkers for valuable discussions and critical reading of the article.
Glossary
- DP
dominant expression pattern
- Col-0
Columbia-0
- PCA
principal component analysis
- DS
dry seeds
- 6H
6-h imbibed
- TR
testa rupture
- RP
radicle protrusion
- RH
root hair
- GC
greening cotyledons
- OC
cotyledons fully opened
- GO
Gene Ontology
- TF
transcription factor
- ABA
abscisic acid
- MS
Murashige and Skoog
Footnotes
This work was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil.
Articles can be viewed without a subscription.
References
- Allen E, Moing A, Ebbels TM, Maucourt M, Tomos AD, Rolin D, Hooks MA (2010) Correlation network analysis reveals a sequential reorganization of metabolic and transcriptional states during germination and gene-metabolite relationships in developing seedlings of Arabidopsis. BMC Syst Biol 4: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amasino R. (2010) Seasonal and developmental timing of flowering. Plant J 61: 1001–1013 [DOI] [PubMed] [Google Scholar]
- Aoki K, Ogata Y, Shibata D (2007) Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol 48: 381–390 [DOI] [PubMed] [Google Scholar]
- Aryee MJ, Gutiérrez-Pabello JA, Kramnik I, Maiti T, Quackenbush J (2009) An improved empirical Bayes approach to estimating differential gene expression in microarray time-course data: BETR (Bayesian estimation of temporal regulation). BMC Bioinformatics 10: 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baima S, Forte V, Possenti M, Peñalosa A, Leoni G, Salvi S, Felici B, Ruberti I, Morelli G (2014) Negative feedback regulation of auxin signaling by ATHB8/ACL5-BUD2 transcription module. Mol Plant 7: 1006–1025 [DOI] [PubMed] [Google Scholar]
- Baima S, Nobili F, Sessa G, Lucchetti S, Ruberti I, Morelli G (1995) The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121: 4171–4182 [DOI] [PubMed] [Google Scholar]
- Barrero JM, Millar AA, Griffiths J, Czechowski T, Scheible WR, Udvardi M, Reid JB, Ross JJ, Jacobsen JV, Gubler F (2010) Gene expression profiling identifies two regulatory genes controlling dormancy and ABA sensitivity in Arabidopsis seeds. Plant J 61: 611–622 [DOI] [PubMed] [Google Scholar]
- Bassel GW, Fung P, Chow TF, Foong JA, Provart NJ, Cutler SR (2008) Elucidating the germination transcriptional program using small molecules. Plant Physiol 147: 143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Lan H, Glaab E, Gibbs DJ, Gerjets T, Krasnogor N, Bonner AJ, Holdsworth MJ, Provart NJ (2011) Genome-wide network model capturing seed germination reveals coordinated regulation of plant cellular phase transitions. Proc Natl Acad Sci USA 108: 9709–9714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, et al. (2013) Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci USA 110: E435–E444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Cabello JV, Arce AL, Chan RL (2012) The homologous HD-Zip I transcription factors HaHB1 and AtHB13 confer cold tolerance via the induction of pathogenesis-related and glucanase proteins. Plant J 69: 141–153 [DOI] [PubMed] [Google Scholar]
- Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE (2006) Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J 46: 805–822 [DOI] [PubMed] [Google Scholar]
- Carabelli M, Morelli G, Whitelam G, Ruberti I (1996) Twilight-zone and canopy shade induction of the Athb-2 homeobox gene in green plants. Proc Natl Acad Sci USA 93: 3530–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E, Holman T, Medhurst A, Peer W, Schmuths H, Footitt S, Theodoulou FL, Holdsworth MJ (2007) Gene expression profiling reveals defined functions of the ATP-binding cassette transporter COMATOSE late in phase II of germination. Plant Physiol 143: 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Jeong S, Kim DS, Na HJ, Ryu JS, Lee SS, Nam HG, Lim PO, Woo HR (2014) The homeodomain-leucine zipper ATHB23, a phytochrome B-interacting protein, is important for phytochrome B-mediated red light signaling. Physiol Plant 150: 308–320 [DOI] [PubMed] [Google Scholar]
- Dekkers BJW, Pearce S, van Bolderen-Veldkamp RP, Marshall A, Widera P, Gilbert J, Drost HG, Bassel GW, Müller K, King JR, et al. (2013) Transcriptional dynamics of two seed compartments with opposing roles in Arabidopsis seed germination. Plant Physiol 163: 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding YH, Liu NY, Tang ZS, Liu J, Yang WC (2006) Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. Plant Cell 18: 815–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait A, Angelovici R, Less H, Ohad I, Urbanczyk-Wochniak E, Fernie AR, Galili G (2006) Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol 142: 839–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng CZ, Chen Y, Wang C, Kong YH, Wu WH, Chen YF (2014) Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J 80: 654–668 [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Cadman CSC, Toorop PE, Lynn JR, Hilhorst HWM (2007) Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J 51: 60–78 [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Gampala SS, Lynch TJ, Thomas TL, Rock CD (2005) Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE (ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol Biol 59: 253–267 [DOI] [PubMed] [Google Scholar]
- Freeman TC, Goldovsky L, Brosch M, van Dongen S, Mazière P, Grocock RJ, Freilich S, Thornton J, Enright AJ (2007) Construction, visualisation, and clustering of transcription networks from microarray expression data. PLoS Comput Biol 3: 2032–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda M, Schnurr J, Abbadi A, Heinz E, Browse J (2004) Peroxisomal acyl-CoA synthetase activity is essential for seedling development in Arabidopsis thaliana. Plant Cell 16: 394–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadbury G, Garrett K, Allison D (2009) Challenges and approaches to statistical design and inference in high-dimensional investigations. In Belostotsky DA, ed, Plant Systems Biology. Humana Press, Totowa, NJ, pp 181–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, Smyth GK (2005) Limma linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer, New York, pp 397–420 [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C (2007) Combined networks regulating seed maturation. Trends Plant Sci 12: 294–300 [DOI] [PubMed] [Google Scholar]
- Hammani K, Gobert A, Small I, Giegé P (2011) A PPR protein involved in regulating nuclear genes encoding mitochondrial proteins? Plant Signal Behav 6: 748–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson E, Olsson ASB, Johannesson H, Johansson H, Hanson J, Engström P, Söderman E (2005) Homeodomain leucine zipper class I genes in Arabidopsis: expression patterns and phylogenetic relationships. Plant Physiol 139: 509–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJJ (2008a) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179: 33–54 [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Finch-Savage WE, Grappin P, Job D (2008b) Post-genomics dissection of seed dormancy and germination. Trends Plant Sci 13: 7–13 [DOI] [PubMed] [Google Scholar]
- Hu W, Franklin KA, Sharrock RA, Jones MA, Harmer SL, Lagarias JC (2013) Unanticipated regulatory roles for Arabidopsis phytochromes revealed by null mutant analysis. Proc Natl Acad Sci USA 110: 1542–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser P, Schmid M (2011) The control of developmental phase transitions in plants. Development 138: 4117–4129 [DOI] [PubMed] [Google Scholar]
- Huq E, Al-Sady B, Hudson M, Kim C, Apel K, Quail PH (2004) Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Liu Y, Sun H, Han Y, Li J, Li C, Guo W, Meng H, Li S, Fan Y, et al. (2013) The mitochondrial folylpolyglutamate synthetase gene is required for nitrogen utilization during early seedling development in Arabidopsis. Plant Physiol 161: 971–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannesson H, Wang Y, Hanson J, Engström P (2003) The Arabidopsis thaliana homeobox gene ATHB5 is a potential regulator of abscisic acid responsiveness in developing seedlings. Plant Mol Biol 51: 719–729 [DOI] [PubMed] [Google Scholar]
- Law SR, Narsai R, Taylor NL, Delannoy E, Carrie C, Giraud E, Millar AH, Small I, Whelan J (2012) Nucleotide and RNA metabolism prime translational initiation in the earliest events of mitochondrial biogenesis during Arabidopsis germination. Plant Physiol 158: 1610–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia J, Dekkers BJW, Provart NJ, Ligterink W, Hilhorst HWM (2011) The re-establishment of desiccation tolerance in germinated Arabidopsis thaliana seeds and its associated transcriptome. PLoS ONE 6: e29123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG (1996) The dynamics of seedling and cotyledon cell development in Arabidopsis thaliana during reserve mobilization. Int J Plant Sci 157: 280–295 [Google Scholar]
- Mazzella MA, Arana MV, Staneloni RJ, Perelman S, Rodriguez Batiller MJ, Muschietti J, Cerdán PD, Chen K, Sánchez RA, Zhu T, et al. (2005) Phytochrome control of the Arabidopsis transcriptome anticipates seedling exposure to light. Plant Cell 17: 2507–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Zhu L, Shen H, Huq E (2008) PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc Natl Acad Sci USA 105: 9433–9438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol Biol 60: 51–68 [DOI] [PubMed] [Google Scholar]
- Narsai R, Law SR, Carrie C, Xu L, Whelan J (2011) In-depth temporal transcriptome profiling reveals a crucial developmental switch with roles for RNA processing and organelle metabolism that are essential for germination in Arabidopsis. Plant Physiol 157: 1342–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando D, Brady S, Koch J, Dinneny J, Benfey P (2009b) Manipulating large-scale Arabidopsis microarray expression data: identifying dominant expression patterns and biological process enrichment. In Belostotsky DA, ed, Plant Systems Biology. Humana Press, Totowa, NJ, pp 57–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2011) R: A Language and Environment for Statistical Computing. Vienna, Austria : the R Foundation for Statistical Computing. http://www.R-project.org/.
- Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, Job D (2004) The effect of α-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol 134: 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribone PA, Capella M, Chan RL (2015) Functional characterization of the homeodomain leucine zipper I transcription factor AtHB13 reveals a crucial role in Arabidopsis development. J Exp Bot 66: 5929–5943 [DOI] [PubMed] [Google Scholar]
- Rougvie AE. (2005) Intrinsic and extrinsic regulators of developmental timing: from miRNAs to nutritional cues. Development 132: 3787–3798 [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Hwang YS, Quail PH (2006) phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J 48: 728–742 [DOI] [PubMed] [Google Scholar]
- Usadel B, Obayashi T, Mutwil M, Giorgi FM, Bassel GW, Tanimoto M, Chow A, Steinhauser D, Persson S, Provart NJ (2009) Co-expression tools for plant biology: opportunities for hypothesis generation and caveats. Plant Cell Environ 32: 1633–1651 [DOI] [PubMed] [Google Scholar]
- Vanstraelen M, Van Damme D, De Rycke R, Mylle E, Inze D, Geelen D (2006) Cell cycle-dependent targeting of a kinesin at the plasma membrane demarcates the division site in plant cells. Curr Biol 16: 308–314 [DOI] [PubMed] [Google Scholar]
- Verdier J, Lalanne D, Pelletier S, Torres-Jerez I, Righetti K, Bandyopadhyay K, Leprince O, Chatelain E, Vu BL, Gouzy J, et al. (2013) A regulatory network-based approach dissects late maturation processes related to the acquisition of desiccation tolerance and longevity of Medicago truncatula seeds. Plant Physiol 163: 757–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan CY, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223: 7–12 [DOI] [PubMed] [Google Scholar]
- Wang J, Wang Y, Wang Z, Liu L, Zhu XG, Ma X (2011) Synchronization of cytoplasmic and transferred mitochondrial ribosomal protein gene expression in land plants is linked to Telo-box motif enrichment. BMC Evol Biol 11: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.