The root apoplast secretome of Gossypium barbadense actively changes upon infection, and a thioredoxin plays an important role in apoplastic ROS balance.
Abstract
Examining the proteins that plants secrete into the apoplast in response to pathogen attack provides crucial information for understanding the molecular mechanisms underlying plant innate immunity. In this study, we analyzed the changes in the root apoplast secretome of the Verticillium wilt-resistant island cotton cv Hai 7124 (Gossypium barbadense) upon infection with Verticillium dahliae. Two-dimensional differential gel electrophoresis and matrix-assisted laser desorption/ionization tandem time-of-flight mass spectrometry analysis identified 68 significantly altered spots, corresponding to 49 different proteins. Gene ontology annotation indicated that most of these proteins function in reactive oxygen species (ROS) metabolism and defense response. Of the ROS-related proteins identified, we further characterized a thioredoxin, GbNRX1, which increased in abundance in response to V. dahliae challenge, finding that GbNRX1 functions in apoplastic ROS scavenging after the ROS burst that occurs upon recognition of V. dahliae. Silencing of GbNRX1 resulted in defective dissipation of apoplastic ROS, which led to higher ROS accumulation in protoplasts. As a result, the GbNRX1-silenced plants showed reduced wilt resistance, indicating that the initial defense response in the root apoplast requires the antioxidant activity of GbNRX1. Together, our results demonstrate that apoplastic ROS generation and scavenging occur in tandem in response to pathogen attack; also, the rapid balancing of redox to maintain homeostasis after the ROS burst, which involves GbNRX1, is critical for the apoplastic immune response.
Cotton (Gossypium spp.) is one of the most economically important crops worldwide and a number of pathogens affect the growth and development of cotton plants. The soil-borne pathogen Verticillium dahliae (V. dahliae) causes the destructive vascular disease Verticillium wilt, which results in devastating reductions in plant mass, lint yield, and fiber quality (Bolek et al., 2005; Cai et al., 2009). To date, Verticillium wilt has not been effectively controlled in the most common cultivated cotton species, upland cotton (Gossypium hirsutum), and cultivars with stably inherited resistance to this disease are currently unavailable (Aguado et al., 2008; Jiang et al., 2009; Zhang et al., 2012a). Unlike upland cotton, sea-island cotton (Gossypium barbadense), which is only cultivated on a small scale, possesses Verticillium wilt resistance. Exploring the molecular mechanisms involved in the defense responses against V. dahliae invasion in G. barbadense can provide useful information for generating wilt-resistant G. hirsutum species through molecular breeding.
During the past decades, progress has been made in studying the defense responses against V. dahliae infection in cotton. Global analyses have demonstrated that several signaling pathways, including those mediated by salicylic acid, ethylene, jasmonic acid, and brassinosteroids, activate distinct processes involved in V. dahliae defense (Bari and Jones, 2009; Grant and Jones, 2009; Gao et al., 2013a). Accumulating evidence indicates that many V. dahliae-responsive genes, such as GbWARKY1, GhSSN, GbERF, GhMLP28, GhNDR1, GhMKK2, and GhBAK1 (Qin et al., 2004; Gao et al., 2011, 2013b; Li et al., 2014a; Sun et al., 2014; Yang et al., 2015), play crucial roles in defense against Verticillium wilt. In addition, the biosynthesis of terpenoids, lignin, and gossypol also makes important contributions to V. dahliae resistance in cotton (Tan et al., 2000; Luo et al., 2001; Xu et al., 2011; Gao et al., 2013a). Together, these studies have greatly improved our understanding of the complex innate defense systems against V. dahliae infection in cotton.
The initial interaction between plants and pathogens takes place in the apoplast, the compartment of the plant cell outside the cell membrane, including the cell wall and intercellular space (Dietz, 1997). In response to pathogen colonization, the attacked plant cells undergo significant cellular and molecular changes, such as reinforcement of the cell wall and secretion of antimicrobial molecules into the apoplastic space (Bednarek et al., 2010). Thus, the apoplast serves as the first line of defense against microbe invasion, and apoplast immunity can be considered an important component of the plant immune response to pathogens.
Upon recognition of pathogen infection, rapid production of reactive oxygen species [the reactive oxygen species (ROS) burst] occurs in the apoplast (Lamb and Dixon, 1997; Torres et al., 2006; Torres, 2010). This ROS burst is regarded as a core component of the early plant immune response (Daudi et al., 2012; Doehlemann and Hemetsberger, 2013). During defense responses, apoplastic ROS can diffuse into the cytoplasm and serve as signals, interacting with other signaling processes such as phosphorylation cascades, calcium signaling, and hormone-mediated pathways (Kovtun et al., 2000; Mou et al., 2003). Apoplastic ROS can also directly strengthen the host cell walls by oxidative cross linking of glycoproteins (Bradley et al., 1992; Lamb and Dixon, 1997) or the precursors of lignin and suberin polymers (Hückelhoven, 2007). Moreover, apoplastic ROS can directly affect pathogens by degrading nucleic acids and peptides from microbes or causing lipid peroxidation and membrane damage in the microbe (Mehdy, 1994; Lamb and Dixon, 1997; Apel and Hirt, 2004; Montillet et al., 2005).
ROS levels in the apoplast increase rapidly in response to a variety of pathogens, but subsequently return to basal levels. The rapid production and dissipation of apoplastic ROS indicate that this process is finely regulated. Two classes of enzymes, NADPH oxidases and class III peroxidases, account for the rapid ROS burst in the apoplast (Bolwell et al., 1995; O’Brien et al., 2012). NADPH oxidases are directly phosphorylated by the receptor-like kinase BIK1 to enhance ROS generation (Li et al., 2014b). Also, due to the toxicity of high levels of ROS, plants have evolved enzymatic and nonenzymatic mechanisms to eliminate ROS, thereby preventing or reducing oxidative damage (Rahal et al., 2014; Torres et al., 2006). However, the molecular system responsible for the regulation of apoplastic ROS homeostasis during the immune response is not well understood.
In this study, we performed a comparative analysis of the apoplastic proteomes in control roots compared with V. dahliae-inoculated roots of Gossypium barbadense (wilt-resistant sea-island cotton) using the two-dimensional differential gel electrophoresis (2D-DIGE) technique. Among the differentially expressed apoplastic proteins, ROS-related proteins were found to be major components, including a thioredoxin, GbNRX1, which functions as an ROS scavenger in response to V. dahliae infection. Knock-down of GbNRX1 expression in cotton by virus-induced gene silencing (VIGS) resulted in reduced resistance to V. dahliae. Our results demonstrate that maintaining apoplastic ROS homeostasis is a crucial component of the apoplastic immune response and that GbNRX1 is an important regulator of this process.
RESULTS
Extracting Apoplastic Proteins from G. barbadense Roots
The root is the initial site of V. dahliae invasion. Thus, we identified V. dahliae-responsive apoplastic proteins in the root tissues of G. barbadense. We used the dip-inoculation method to infect cotton roots with V. dahliae spores, as previously described (Wang et al., 2011). To understand the process by which the pathogen colonizes root cells, we generated a V991-GFP strain, which enabled us to monitor the infection process of V. dahliae directly. Confocal observation showed that spores of the fungus appeared on the root surface randomly after infection with V991-GFP. At 0.5 d post-infection (dpi), some spores germinated on the surface of the root cells; at 1 dpi, most of the spores produced germ tubes and at 3 dpi, many hyphae appeared (Supplemental Fig. S1A). Examination of longitudinal cross sections showed that a small portion of the hyphae had penetrated into the inner layers of the root cells at 3 dpi (Supplemental Fig. S1B).
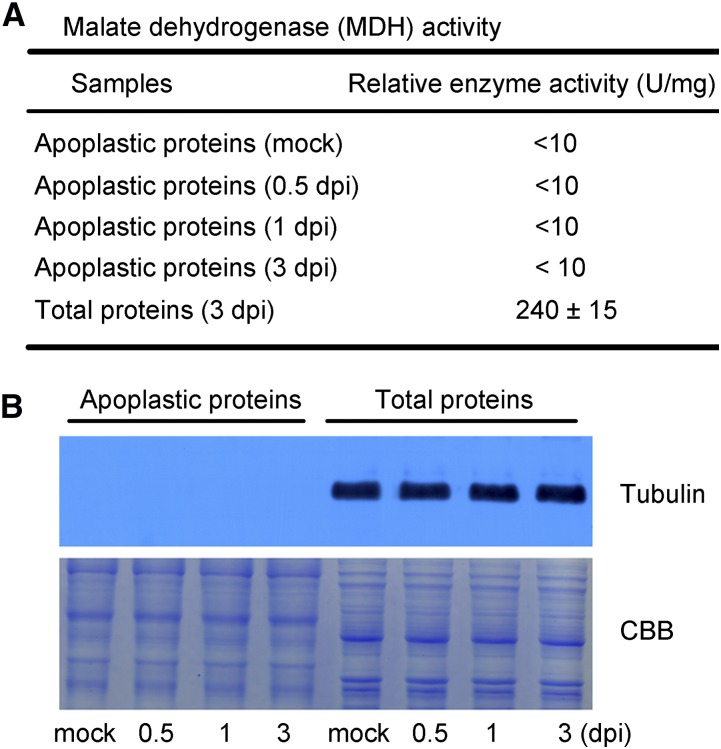
The vacuum infiltration-centrifugation method (Zhou et al., 2011) was applied to extract the apoplastic proteins from mock and infected cotton roots at 0.5, 1, and 3 dpi to investigate the early plant response to V. dahliae infection. We assayed the activity of the cytoplasmic enzyme malate dehydrogenase (MDH) to evaluate the purity of the apoplast extracts (Husted and Schjoerring, 1995). As shown in Figure 1A, MDH activity was low (<10 U/mg) in each of the apoplast extracts compared with the MDH activity in the total soluble protein fraction (240 ± 15 U/mg). To confirm the quality of the apoplast fractions, we also performed immunoblotting against the intracellular protein tubulin, as previously described (Zhang et al., 2009). As shown in Figure 1B, tubulin was not detected in the apoplast fractions but was clearly visible in the total protein fraction. These results indicate that there was little or no contaminating cytoplasmic protein in the apoplastic protein preparations.
Figure 1.
Assessment of purity of isolated apoplastic proteins. A, Measurement of MDH activity. Three independent apoplastic extracts of mock and infected cotton roots at 0.5, 1, and 3 dpi were used for MDH analysis. Total soluble proteins extracted from infected cotton roots at 3 dpi were used as a control to assess the purity of the isolated apoplastic proteins. Values are the means ± sd of three biological replicates. B, Immunoblot analysis of apoplastic and total proteins. Antibodies against tubulin were used for the test (top panel); 20 µg each of apoplastic and total proteins were used for CBB staining (bottom panel).
Proteomic Identification of V. dahliae-Responsive Apoplastic Proteins in Cotton Roots
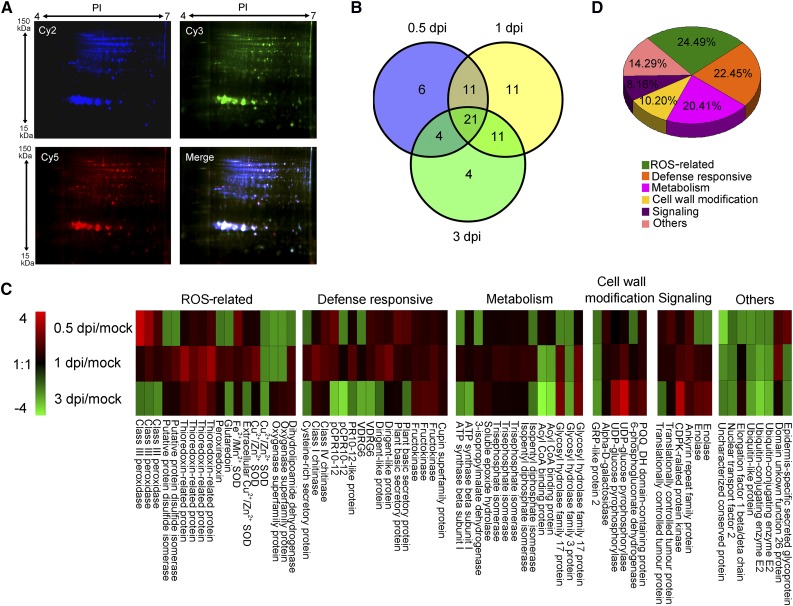
We analyzed the apoplastic proteins by 2D-DIGE. Representative expression profiles of apoplastic protein lysates isolated from mock-inoculated plants (A1, the first repeat of Mock) and plants at 0.5 dpi (B2, the second repeat of 0.5 dpi) are shown in Figure 2A. Spot analysis using DeCyder software (GE Healthcare Life Sciences) identified approximately 700 protein spots, 68 of which showed significant (P ≤ 0.05) differences in abundance (>1.5-fold) between samples. A Venn diagram analysis showed that 21 spots had different abundances at all three time points during V. dahliae infection (Fig. 2B); also, 42 spots had different abundances at 0.5 dpi, 54 spots differed at 1 dpi, and 40 spots differed at 3 dpi. Mass spectrometry (tandem matrix-assisted laser-desorption ionization time of flight) identified 49 unique proteins in the 68 spots. Expression profiles of these proteins are presented in Figure 2C and the detailed information is presented in Supplemental Table S1. As proteins extracted by vacuum infiltration-centrifugation may include fungal components, we also searched the mass spectrometry (MS) and tandem MS (MS/MS) results against the Verticillium database, finding that none of the identified proteins was of Verticillium origin.
Figure 2.
Analysis of V. dahliae-responsive apoplast secretome in cotton roots. A, DIGE images of apoplastic proteins isolated from mock-inoculated plants (A1, the first repeat of Mock) and plants at 0.5 dpi (B2, the second repeat of 0.5 dpi). Cy2 (blue) image of proteins represents an internal standard containing equal quantities of all samples. Cy3 (green) image of proteins from A1, and Cy5 (red) image of proteins from sample B2. The DIGE experimental design is shown in Supplemental Table S2. All images were observed with the Typhoon FLA9500 imager (GE Healthcare Life Sciences). Molecular masses (kDa) and pI are given on the left and top, respectively. B, Venn diagram of abundance changed protein spots in the cotton roots upon V. dahliae infection. The number of differentially expressed protein spots for each specific time point is shown in each different section. C, Expression profiling of differentially expressed protein spots based on their relative in-gel intensities. The average ratios of the fold changes are listed in Supplemental Table S1. The heat map was generated with Multi Experiment Viewer v. 4.9 (http://www.tm4.org/mev.html). Red represents upregulated protein spots; green represents down-regulated protein spots; black represents protein spots that remain the same under different conditions. D, Functional classification of differently expressed proteins using the Gene Ontology Tool (http://www.geneontology.org).
We then performed quantitative reverse transcription-PCR (qRT-PCR) to determine whether the changes in abundance of the identified proteins were consistent at the mRNA and protein levels. The results showed that the majority of the identified proteins exhibited similar changes at both transcriptional and translational levels, thus confirming the data from the 2D-DIGE analysis (Supplemental Figs. S2–S7 and Supplemental Table S1).
Gene ontology classification to analyze the functional categories of the differentially expressed apoplastic proteins classified these proteins into six categories, including ROS-related (24.49%), defense-responsive (22.45%), metabolism (20.41%), cell-wall modification (10.20%), signaling (8.16%), and others (14.29%; Fig. 2D).
To better understand the putative functions of these proteins and explore determinants for cotton defense against V. dahliae, we further analyzed and compared the transcript levels of the identified pathogen-responsive genes in G. barbadense with those in G. hirsutum. The results showed that the transcription of most of the ROS-related genes was up-regulated in both G. hirsutum and G. barbadense in response to pathogen attack, but the induction occurred more rapidly and to a higher extent in the G. barbadense plants (Supplemental Fig. S2), indicating that the ROS-mediated defense response was faster and stronger in the root apoplast of G. barbadense. We also found that transcription of some defense-responsive genes, such as PR1, PR4, and PR10, was markedly induced in G. barbadense, which likely contributes substantially to the Verticillium wilt resistance of these plants (Supplemental Fig. S3). Based on these data, it seems that the earlier and stronger ROS burst in G. barbadense may relate to the higher expression of the defense-related genes, thus leading to the disease resistance of the G. barbadense plants. Interestingly, we also observed that transcription of many genes involved in carbon metabolism was significantly more down-regulated in G. barbadense than those in G. hirsutum, suggesting that energy metabolism was actively reestablished in G. barbadense upon pathogen attack, which is likely beneficial for the plant to protect against V. dahliae infection (Supplemental Fig. S4).
Verifying the Apoplast Localization of the Identified Proteins
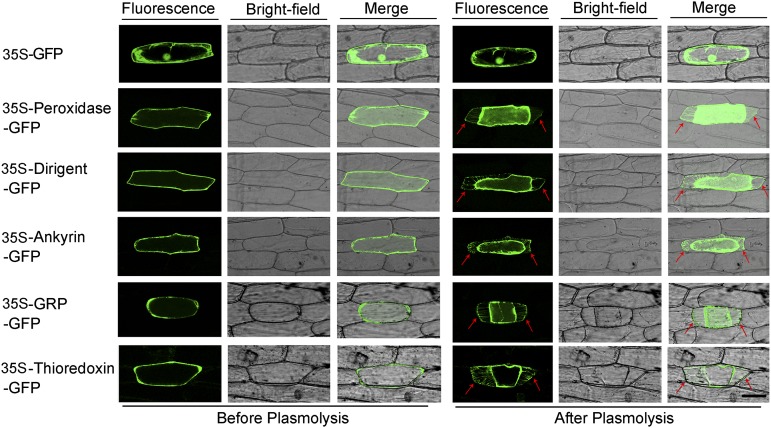
Two publicly available algorithms (SignalP and SecretomeP; Emanuelsson et al., 2000; Bendtsen et al., 2004) were used to predict the subcellular localizations of the identified proteins. As summarized in Supplemental Table S1, SignalP predicted that 15 out of the 49 identified proteins contain signal peptides, while SecretomeP analyses showed that 31 of these proteins were SecretomeP-positive. To gain direct evidence for their subcellular localization, we randomly selected five proteins, including three proteins containing a secretory signal peptide (peroxidase, thioredoxin, and dirigent-like protein) and two proteins lacking a secretory signal peptide (ankyrin and GRP-like protein 2). We examined their subcellular localizations after transiently expressing these proteins fused to GFP in onion epidermal cells. The vector pPZP111-GFP was used as a negative control, and the fluorescence was observed by confocal laser-scanning microscopy. As shown in Figure 3, fluorescence from the five GFP fusion proteins was detected in the cell wall or in the compartment between the cell membrane and the cell wall after Suc-induced plasmolysis, whereas the fluorescence of the GFP control was detected in the intracellular region and not in the extracellular region. Based on these results, it is possible that other proteins (both with and without secretory signal peptides) identified in our proteomics study also reside in the apoplast compartment. It may also validate that the vacuum infiltration-centrifugation method applied for extracting the apoplastic proteins from cotton roots was efficient.
Figure 3.
Subcellular localization of selected apoplastic proteins. Onion epidermal cells were transformed by particle bombardment. The top panels show the 35S-GFP fusion proteins, and the remaining panels show the 35S-Peroxidase-GFP, 35S-Dirigent-GFP, 35S-Ankyrin-GFP, 35S-GRP-GFP, and 35S-Thioredoxin-GFP fusion proteins, respectively. The onion epidermal cells were treated with 30 g/l Suc for plasmolysis. Arrows indicate the cell wall. Bar = 30 µm.
GbNRX1, a V. dahliae-Responsive Multidomain Thioredoxin Mainly Expressed in Vascular Tissue
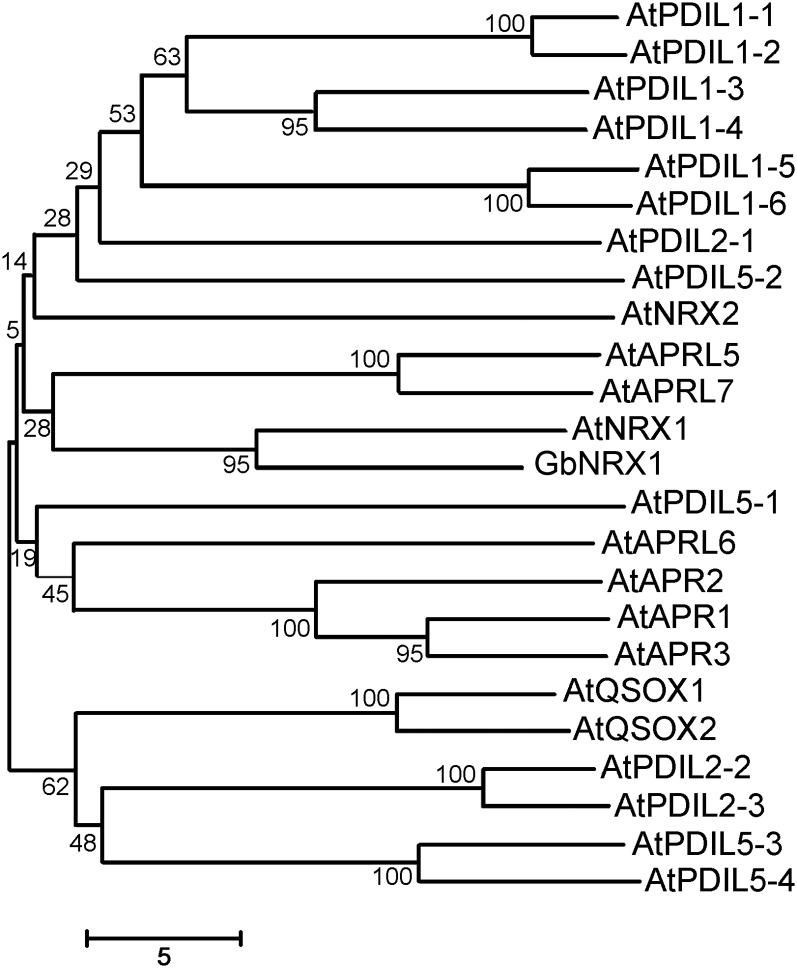
Of the ROS-related proteins we found, four spots (22, 24, 28, and 32) were identified as a thioredoxin. This relatively high frequency suggests that this thioredoxin protein plays important roles in the cotton–V. dahliae interaction. Thus, we used 5′ and 3′ rapid amplification of cDNA ends to amplify the corresponding full-length cDNA from a cDNA library derived from G. barbadense roots. The resulting cDNA was 1957 bp in length with an open reading frame of 1713 bp, which encodes a protein of 570 amino acids containing an N-terminal signal peptide, three typical thioredoxin domains, and a Cys-rich C-terminal domain. We examined the phylogenetic relationships of this protein with Arabidopsis TRX proteins containing multiple TRX domains, such as those encoding protein disulfide isomerase (PDI) and nucleoredoxin (NRX), revealing that the cotton protein is most closely related to AtNRX1 (Fig. 4). Hence, we designated this cotton thioredoxin as GbNRX1.
Figure 4.
Phylogenetic relationships among GbNRX1 and Arabidopsis multiple-TRX domain-proteins including PDI and NRX. The numbers on the nodes indicate the confidence values using 1000 replications.
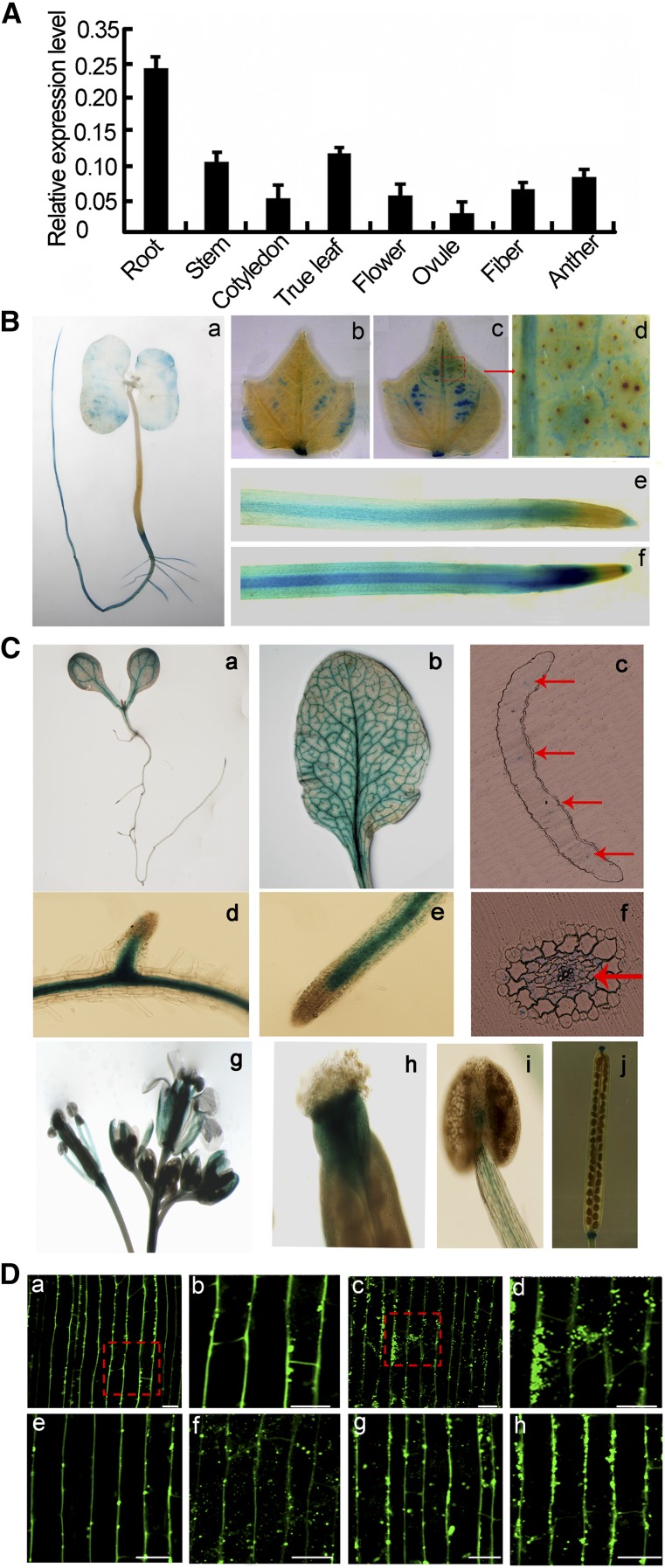
We conducted qRT-PCR analysis using total RNA extracted from various cotton organs with gene-specific primers to investigate the expression of GbNRX1. As shown in Figure 5A, GbNRX1 was expressed in all tissues examined, especially in roots and true leaves. The expression pattern of GbNRX1 was confirmed by GUS expression analysis in both cotton and transgenic Arabidopsis plants. We cloned a 1.3-kb upstream fragment of GbNRX1 by genomic walking and expressed the GUS cDNA under the control of this fragment in the plant expression vector pCAMBIA1301. As shown in Figure 5B a, GUS expression in cotton seedlings not infected by V. dahliae predominantly occurred in the roots. When these plants were treated with V. dahliae spores, GUS expression was strongly induced in roots and true leaves (Fig. 5B, b, c, e, and f), which confirms the finding that GbNRX1 is a V. dahlia-responsive gene. Furthermore, as shown in the enlarged image, GbNRX1 was mainly expressed in vascular tissue (Fig. 5B d). In transgenic Arabidopsis, expression of the GbNRX1pro::GUS fusion was detected in the roots and true leaves as well as other tissues, including petals, stigmas, stamen filaments, and seed pods (Fig. 5C).
Figure 5.
Tissue-specific expression of GbNRX1. A, qRT-PCR analysis of expression levels of GbNRX1 in cotton root, stem, cotyledon, true leaf, flower, ovule, fiber, and anther tissue. Error bars indicate sd from three technical replicates of one biological experiment. The experiments were repeated three times with similar results. B, Histochemical GUS staining of cotton plants transformed with the GbNRX1pro::GUS construct. (a) Young seedling; (b) true leaf without V. dahliae infection; (c) true leaf after V. dahliae infection for 48 h; (d) enlarged image of the section outlined by red dashed lines in (c); (e) root without V. dahliae infection; (f) root after V. dahliae infection for 48 h. C, Histochemical GUS staining of various organs from wild-type Arabidopsis plants transformed with GbNRX1pro::GUS. (a) Seedling; (b) true leaf; (c) vertical section of true leaf; (d) stem; (e) root; (f) cross section of root; (g) flower; (h) petals; (i) stigma stamen filaments; (j) seed pods. Arrows indicate vascular tissue. D, Immunofluorescence microscopy of GbNRX1 localized to the apoplast. GbNRX1 is mainly localized to the periphery of the cotton root cells without BFA treatment (a); aggregation of signals appears in BFA-treated cells (c); (b) and (d), enlarged images of the sections outlined in (a) and (c); localization of GbNRX1 in cotton root cells at 0, 0.5, 1, and 3 dpi, respectively (e–h). Bars = 50 µm.
To further validate the subcellular localization of GbNRX1 in cotton roots, we performed an immunofluorescence assay using GbNRX1 polyclonal antibodies. The results showed that GbNRX1 mainly localized in the peripheral regions of cotton root cells, whereas no signal was detected in the intracellular regions of these cells (Fig. 5D, a and b). To examine whether GbNRX1 moves from the cytoplasm to the extracellular regions, we treated root cells with Brefeldin A (BFA), an inhibitor of secretion (Staehelin and Driouich, 1997). As shown in Figure 5D, c and d, BFA treatment resulted in the aggregation of GbNRX1 signals within the cells, indicating that GbNRX1 is a secreted protein. In addition, immunofluorescence assays to investigate the localization of GbNRX1 during infection showed that GbNRX1 appeared both in the peripheral regions and inside the cotton root cells at 0.5 dpi. At 1 and 3 dpi, increasing amounts of GbNRX1 proteins accumulated at the peripheral regions of the cells (Fig. 5D, f–h).
GbNRX1 Possesses Trx Activity in Vitro
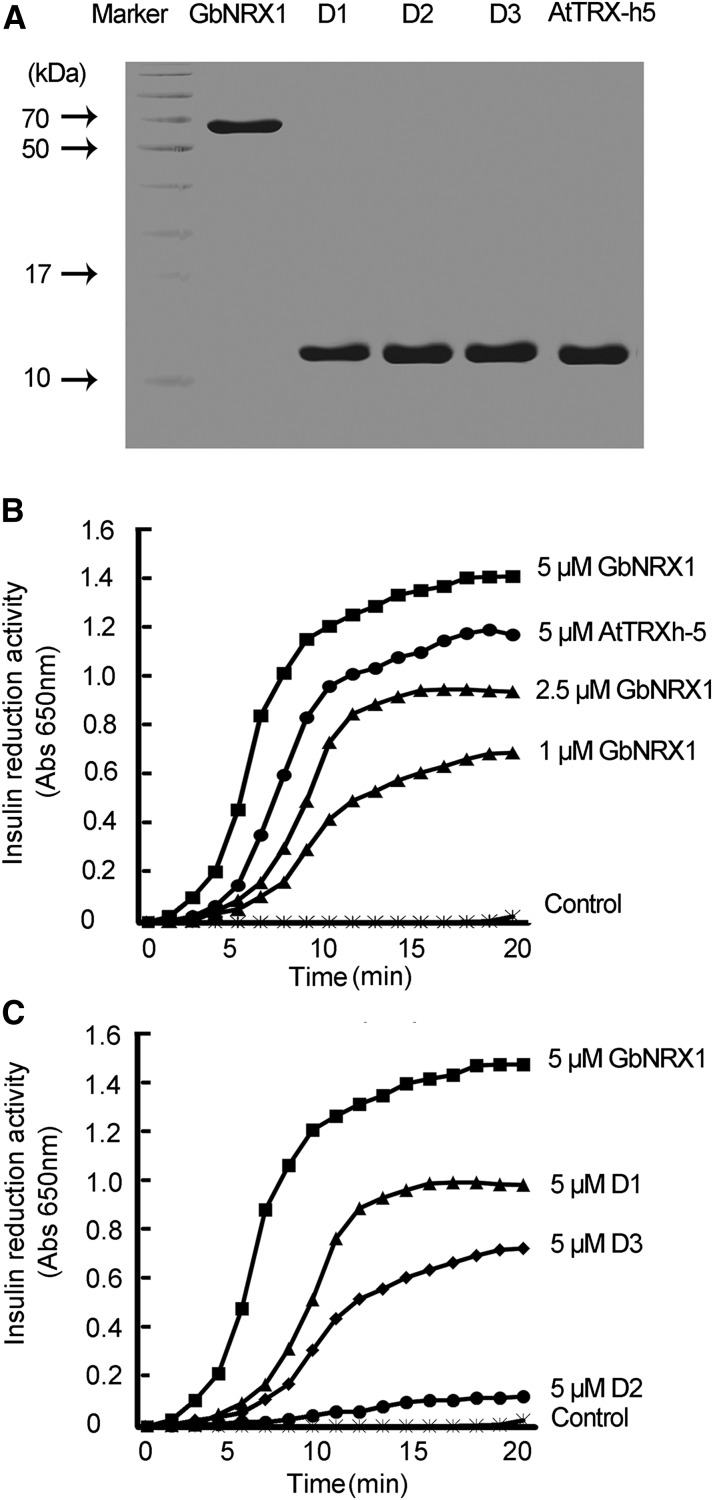
Trx proteins contain a conserved active site (WCGPC or WCCPC) that functions in the reduction of target molecules (Motohashi et al., 2001). Sequence alignment showed that GbNRX1 contains three thioredoxin domains (domain 1 [amino acids 29–158, D1] domain 2 [amino acids 189–317, D2], and domain 3 [amino acids 348–477, D3]). The first and third thioredoxin domains have typical active sites (D1 contains WCGPC and D3 contains WCCPC), whereas the second thioredoxin domain (D2) does not harbor such an active site. To investigate the redox activity of GbNRX1 and its three domains in vitro, we expressed His-tagged GbNRX1, the three domains of GbNRX1, and Arabidopsis TRX-h5 (AtTRX-h5) as a positive control [(Vellosillo et al., 2010); and see Fig. 6A]. We investigated the Trx activities of these recombinant proteins using the insulin reduction assay in vitro (Holmgren, 1989). Compared with AtTRX-h5, GbNRX1 demonstrated high insulin reduction activity in a dose-dependent manner (Fig. 6B). Among the three domains of GbNRX1, the reduction activity of D1 was more efficient than that of D3, while the reduction activity of D2 was much lower than that of D1 and D3, perhaps due to the lack of an active site. Based on these data, we ranked the insulin reduction activity of the three domains as follows: D1 > D3 > D2, and each of the three domains alone was less active than the native protein (Fig. 6C).
Figure 6.
Analysis of insulin disulfide reductase activity of GbNRX1. A, SDS-PAGE analysis of purified proteins. GbNRX1, D1 (domain 1), D2 (domain 2), D3 (domain 3), and AtTRX-h5 were expressed in E. coli, and the purified proteins were subjected to SDS-PAGE. Molecular masses (in kDa) are given on the left. B, Insulin reduction assay. Purified GbNRX1 (5, 2.5, and 1 µM) and AtTRX-h5 (5 µM) were used for the insulin reduction assay, and enzyme activity was quantified based on A650. C, Time course of insulin disulfide reductase activity. Five micromolars of intact GbNRX1, D1 (domain 1), D2 (domain 2), and D3 (domain 3) was used and reaction buffer was used as a negative control.
Silencing of NRX1 Leads to Reduced V. dahliae Resistance
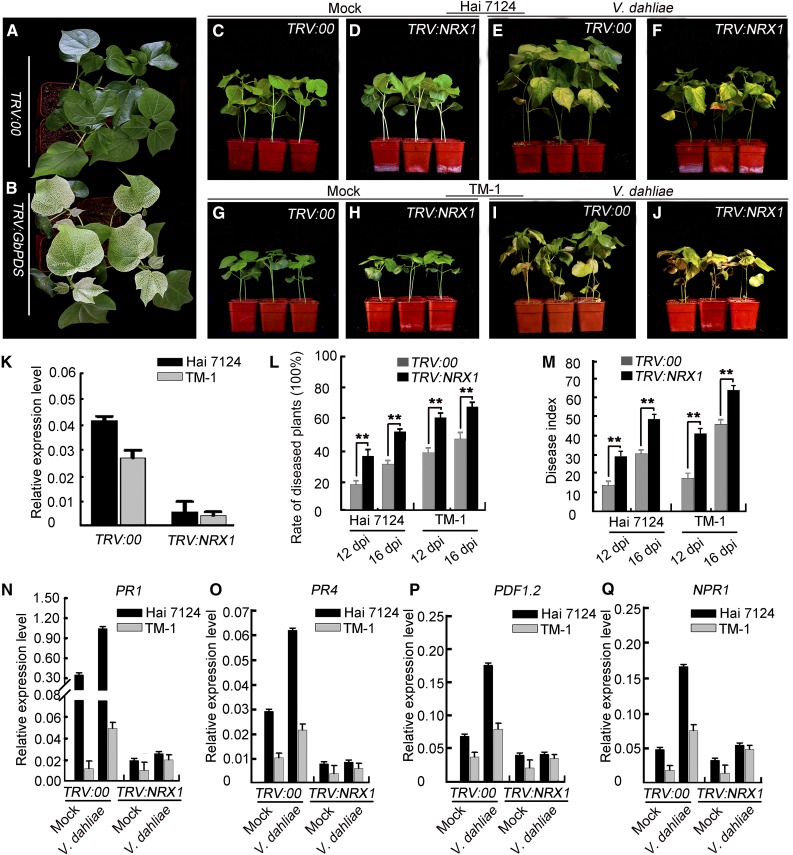
We employed VIGS to investigate the function of NRX1 in defense against V. dahliae infection. In a preliminary experiment, we silenced the expression of the phytoene desaturase gene (GbPDS), which is used as a visible marker to monitor the efficiency of VIGS (Pang et al., 2013). As shown in Figure 7B, the newly emerged true leaves of plants infiltrated with Agrobacterium carrying GbPDS exhibited an albino phenotype compared to the negative control (Fig. 7A), indicating that the VIGS system worked efficiently under our experimental conditions. Thus, we used this system to inhibit the expression of GbNRX1 in G. barbadense and its ortholog, GhNRX1, in G. hirsutum. The qRT-PCR analyses showed that the expression of both NRX1s was dramatically reduced in VIGS plants (Fig. 7K). Phenotypic analysis indicated that the growth of NRX1-silenced plants was comparable to that of the control plants under normal conditions (Fig. 7, C, D, G and H).
Figure 7.
NRX1-silenced cotton plants exhibit reduced resistance to V. dahliae infection. A and B, Preliminary assay of the efficiency of VIGS under our experimental conditions. Ten-d-old cotton plants were infiltrated with Agrobacterium carrying VIGS-control vector (TRV:00) and TRV:GbPDS. The photographs were taken at 2 weeks after infiltration. C, D, E, F, G, H, I, and J, Phenotypes of control and VIGS cotton plants. Ten-d-old cotton plants were infiltrated with Agrobacterium carrying VIGS-control vector (TRV:00) or TRV:NRX1 and then at 2 weeks after infiltration inoculated with a suspension of V. dahliae spores. Photographs were taken at 2 weeks after Agrobacterium infiltration (C and D; G and H) and at 2 weeks after V. dahliae infection (E and F; I and J). K, qRT-PCR analysis of NRX1 expression in cotton plants infiltrated with VIGS-control vector (TRV:00) and TRV:NRX1. Error bars indicate sd from three technical replicates of one biological experiment. The experiments were repeated three times with similar results. L and M, Rate of diseased plants and disease index of TRV:00 and TRV:NRX1 cotton plants. Error bars represent sd of three biological replicates (n ≥ 36), asterisks indicate statistically significant differences, as determined by the Student’s t test (**P < 0.01). N, O, P, and Q, qRT-PCR analysis of pathogenesis-related genes (PR1, PR4, PDF1.2, and NPR1) in TRV:00 and TRV:NRX1 cotton plants. Error bars represent sd of three technical replicates of one biological experiment. The experiments were repeated three times with similar results.
In response to challenge with V. dahliae, however, down-regulation of NRX1 expression resulted in reduced resistance to the pathogen (Fig. 7, E, F, I and J). The rate of diseased plants and the disease index of NRX1s-silenced plants clearly increased, compared with nonsilenced plants (Fig. 7, L and M). Also, the degree of susceptibility of GbNRX1-silenced G. barbadense plants and GhNRX1-silenced G. hirsutum plants was different; the G. hirsutum plants showed more severe disease symptoms than the G. barbadense plants after pathogen infection. Based on these observations, we believe that NRX1 participates in defense responses in both G. barbadense and G. hirsutum. In addition, the increased disease susceptibility of NRX1-silenced plants was further confirmed by culturing the fungus from the infected stems of G. barbadense plants. As shown in Supplemental Figure S8, V. dahliae propagation was better in GbNRX1-silenced plants compared to the control. We also conducted qRT-PCR analysis to examine the expression of pathogenesis-related genes in these plants. The results showed that PR1, PR5, PDF1.2, and NPR1 were up-regulated in control plants challenged with V. dahliae, whereas in NRX1s-silenced plants, these pathogenesis-related genes were either not activated or only slightly induced by the pathogen (Fig. 7, N–Q).
GbNRX1 Functions in Apoplastic ROS Dissipation in Response to V. dahliae Infection
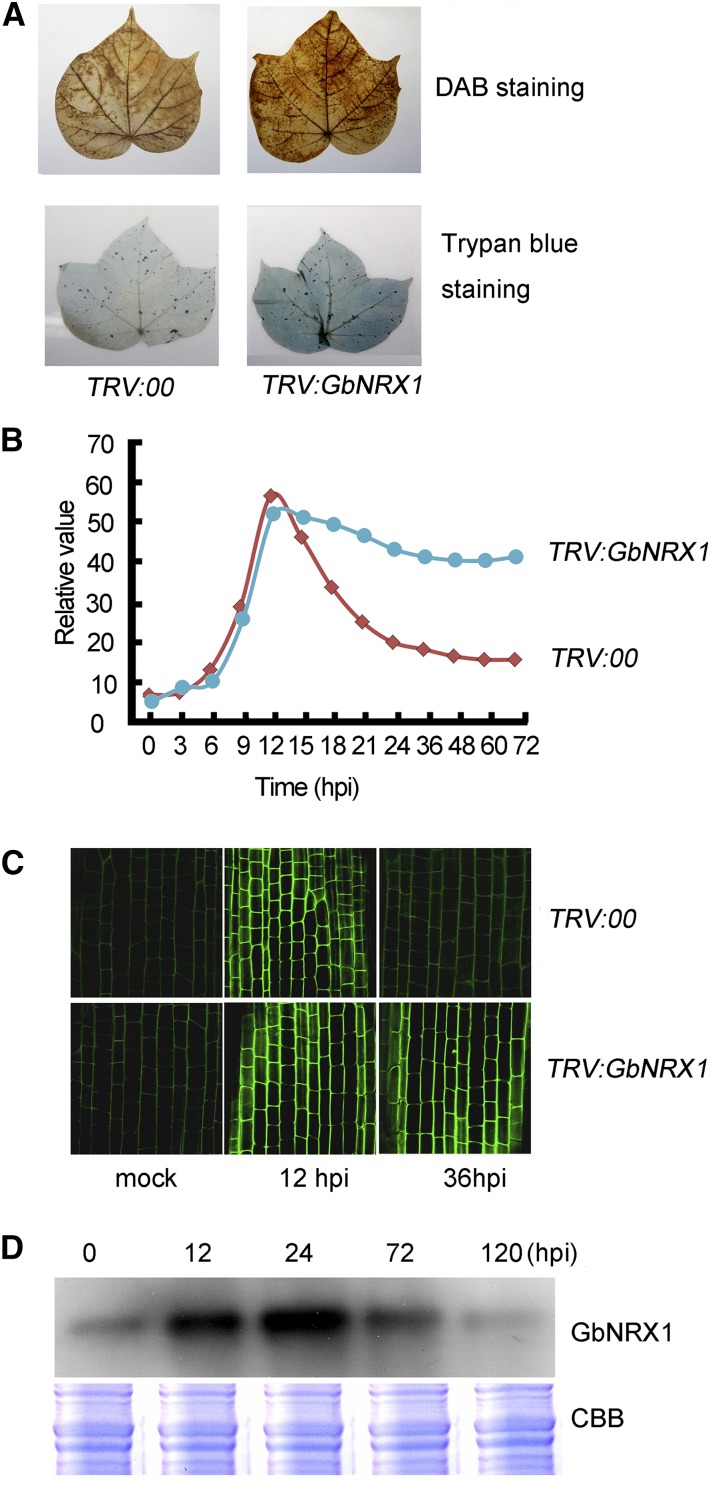
Given that GbNRX1 functions as an active thioredoxin in vitro, the reduced resistance of GbNRX1-silenced plants to V. dahliae infection may result from the effect of GbNRX1 on the redox status of infected cotton plants. To investigate this possibility, we first examined total ROS production in the leaves of control and GbNRX1-silenced plants challenged with V. dahliae infection. DAB (3,3′-diaminobenzidine tetrahydrochloride) staining showed that after V. dahliae infection, GbNRX1-silenced leaves accumulated more ROS than the control, and trypan blue staining demonstrated that silencing of GbNRX1 increased cell death in the infected leaves (Fig. 8A).
Figure 8.
Silencing of GbNRX1 results in defective apoplastic ROS elimination in response to V. dahliae. A, ROS levels and cell death analyses by DAB and trypan blue staining. Ten-d-old control (TRV:00) and GbNRX1-silenced (TRV:GbNRX1) cotton plants were inoculated with V. dahliae. Leaves were detached and stained with DAB (top panel) and trypan blue (bottom panel) at 12 dpi. B, ROS production in the cotton root cell apoplast at the indicated time points after inoculation with V. dahliae. C, Images showing apoplastic ROS in cotton root cells at 0, 12, and 36 h after inoculation with V. dahliae. The roots were initially processed with a cell-impermeable fluorogenic dye, and the fluorescence levels of apoplastic ROS were visualized by confocal microscopy. D, Immunoblot analysis of GbNRX1 levels in cotton roots at the indicated days post infection with V. dahliae.
GbNRX1 functions in apoplast ROS metabolism, so we next investigated the levels of plant apoplast ROS during the plant-pathogen interaction. We first examined the apoplast ROS dynamics by analyzing the levels of the ROS-related proteins characterized in our apoplast proteome. We found that the apoplast ROS production proteins (such as class III peroxidase) were strongly induced at 12 h post-infection (hpi). However, the ROS scavenging proteins identified in the proteome, including putative PDI, thioredoxin protein, peroxiredoxin, glutaredoxin, and oxygenase proteins, were mainly induced after 12 hpi (Fig. 2C). Accordingly, we next assessed ROS levels in the cotton root apoplast in response to V. dahliae infection. We performed a time-course experiment examining ROS production in the apoplast of control cotton root cells (summarized in Fig. 8B), which revealed an ROS peak at 12 hpi with V. dahliae spores, followed by a gradual decrease in ROS. Compared with the control, the GbNRX1-silenced plants showed relatively high apoplastic ROS levels after 12 hpi with V. dahliae spores. The apoplastic ROS status in response to V. dahliae was further confirmed using a cell-impermeable sensitive fluorogenic dye [OxyBURST Green H2HFF BSA; Molecular Probes, Eugene, OR; (Zhang et al., 2011)]. The results showed that the fluorescence intensity of apoplastic ROS at 12 hpi in the control root cells was higher than that at 36 hpi (Fig. 8C). In GbNRX1-silenced plants, an ROS burst occurred in the root apoplast at 12 hpi as it did in the control, but the ROS levels did not decrease at 36 hpi versus 12 hpi.
To obtain further evidence that GbNRX1 functions in the regulation of apoplastic ROS balance during the cotton–V. dahliae interaction, we performed immunoblot analysis to examine the relationship between GbNRX1 levels and apoplastic ROS abundance. The level of GbNRX1 in apoplast extracts from V. dahliae-infected cotton roots gradually increased, reaching a peak at 24 hpi, the time at which apoplastic ROS levels began to decrease (Fig. 8D). Together, these results indicate that GbNRX1 functions in ROS dissipation after the ROS burst that occurs in response to V. dahliae attack.
DISCUSSION
Changes In the Apoplastic Proteome in Response to V. dahliae Infection of Cotton Roots
Upon sensing invading pathogens, the plant secretes proteins into the apoplast, where the plant immune system recognizes conserved molecular patterns from the invading microbe and initiates a series of immune responses (Doehlemann and Hemetsberger, 2013). In this study, we identified 49 unique proteins that were secreted into the root apoplast upon V. dahliae infection. Moreover, we found that most of the proteins differentially expressed in response to pathogen infection act in the defense response and ROS metabolism. Previous apoplastic proteome studies found that pathogenesis-related proteins (PRs) were the major proteins secreted into the apoplast during the defense response. For example, secreted PR1 proteins act in the stress response and are associated with resistance to oomycete pathogens during tobacco development (Hugot et al., 2004). Also, chitinase (PR4) is an endogenous plant defense enzyme that also generates signaling molecules (elicitors) for further induction of defenses (van Loon et al., 2006) and PR10 can increase plant resistance to the oomycete pathogen Hyaloperonospora arabidopsidis (Choi et al., 2012). In addition, dirigent-like protein, FRK2, and Cupin superfamily protein also function in plant defense against pathogens (Ascencio-Ibánez et al., 2008). In this study, several PR proteins, such as Cys-rich secretory protein (PR1, spot 122), chitinase (PR4, spots 116 and 129), and PR10 family proteins (spots 11, 14, and 15), increased in abundance upon pathogen invasion. Similar expression patterns were also observed for dirigent-like protein (spots 27 and 34), FRK2 (spots 20, 45, and 88), and Cupin superfamily protein (spot 46; Supplemental Table S1). These results indicate that defense-responsive proteins act at the initial site of the cotton–V. dahliae interaction as the first line of defense, as also occurs in other plant–pathogen interactions (Floerl et al., 2008, 2012; Kim et al., 2014). These defense-responsive genes represent candidate genes for the improvement of Verticillium wilt resistance in G. hirsutum.
In addition, among the 49 proteins identified, only 15 contained a typical secretory signal, indicating that these proteins are secreted through an endoplasmic reticulum-Golgi-mediated classical secretion pathway (Palade, 1975; Rothman, 1994). However, 31 of the identified proteins are SecretomeP-positive, suggesting unconventional protein secretion (Nickel and Rabouille, 2009; Pinedo et al., 2012). Similar results were reported from other studies of apoplastic proteins (Fernández et al., 2012; Kim et al., 2013; Wang et al., 2013). Also, immunoblot and MDH enzyme assays indicated that the apoplast extract showed minimal contamination with intracellular components (Fig. 1). The subcellular localization experiments further demonstrated that such proteins reside in the apoplast (Fig. 3). These results indicated that the proteins we identified represent apoplastic proteins from the cotton roots.
Several proteomics investigations of proteins that are secreted during plant–pathogen interactions indicated that not only do plants secrete proteins during the defense response to fungal infection, but pathogens also secrete proteins (Kim et al., 2013, 2014). In this study, we tried to identify fungal proteins in the extracts, but found that none of the identified proteins originated from Verticillium. The failure to identify pathogen-derived proteins may be due, at least partially, to the following two reasons: 1. For preparation of apoplastic proteins, we used a gentle extraction method (1000 g centrifugation speed); however, such a low centrifugation speed may not be efficient enough to separate intracellular proteins from the pathogen cells. 2. Confocal observation showed that only a small portion of the V. dahliae hyphae could penetrate into the inner layers of the root cells (Supplemental Fig. S1). The amount of proteins secreted from the pathogen may be too low to be detected by MS. Similar result was reported in a previous study: when challenging Arabidopsis with Verticillium longisporum, no pathogen proteins were detected in the plant apoplastic proteome (Floerl et al., 2012; Gupta et al., 2015).
Properly Timed Expression of ROS-Generation and ROS-Scavenging Apoplastic Proteins Maintains ROS Homeostasis During the Early Immune Response Against V. dahliae
The accumulation of ROS is a core component of the early immune response in plants. ROS play multifaceted roles as signaling compounds that mediate the establishment of multiple responses (Doehlemann and Hemetsberger, 2013). However, ROS can also act as toxins. Therefore, after activating defense-related signaling, excess ROS must be scavenged to prevent cellular damage. In this study, we found that ROS-related proteins make up a large proportion (24.49%) of apoplastic proteins, which indicates that ROS metabolism in the apoplast is crucial during the cotton–V. dahliae interaction. We found that the apoplastic ROS-producing proteins (class III peroxidases) were strongly induced at 12 hpi and showed an expression peak consistent with the ROS burst (Fig. 2C, Supplemental Table S1), indicating that they function in apoplastic ROS generation. Compared to ROS-producing proteins, a set of ROS-scavenging proteins, including a putative PDI, thioredoxin protein, peroxiredoxin, glutaredoxin, and oxygenase proteins, exhibited peak expression at 24 or 72 hpi, i.e. after the initiation of the ROS burst (Fig. 2C, Supplemental Table S1). These results indicate that plant cells coordinately regulate the expression of ROS-producing and ROS-scavenging proteins and that these proteins are secreted into the apoplast in a timely manner in response to V. dahliae infection. Therefore, ROS levels must be balanced after the rapid ROS accumulation that occurs during the defense response against pathogen invasion. Consistent with this finding, more than 10% of pathogen-responsive proteins in the rice apoplast are antioxidant proteins (Kim et al., 2013). In addition, several other apoplastic proteomic studies have also revealed altered expression of ROS-related proteins in other plants in response to invasion by various pathogens (Wang et al., 2013; Kim et al., 2014).
GbNRX1 Is an Important Regulator of Apoplastic ROS Homeostasis
Of the many ROS-related proteins, our study identified thioredoxin at a relatively high frequency. Thioredoxin is a nucleoredoxin (NRX) family protein, as revealed by phylogenetic analysis (Fig. 4). The name “nucleoredoxin” was originally given to mouse thioredoxin due to its exclusively nuclear localization (Kurooka et al., 1997). However, some disagreed with this statement, finding that mouse NRX was mainly cytosolic and that two Arabidopsis NRXs localized to the cytosol or nucleus (Funato et al., 2006; Marchal et al., 2014). In this study, transient expression of GbNRX1-GFP in onion cells showed that GbNRX1 localizes in the apoplast (Fig. 3), as confirmed by immunofluorescence (Fig. 5D). In plants, a thioredoxin protein (thioredoxin-h5) plays important roles in defense against pathogens; thioredoxin-h5 regulates changes in the conformation of NPR1, the key regulator of local and systemic acquired resistance. NPR1 proteins normally occur in the cytosol as a thiol-bound oligomer. Upon pathogen challenge, thioredoxin-h5 reduces NPR1 oligomers, releasing monomers that translocate into the nucleus to regulate the expression of defense-related genes (Tada et al., 2008). Furthermore, thioredoxin-h5 plays a crucial role in plant defense against victorin-mediated cell death (Sweat and Wolpert, 2007; Lorang et al., 2012). In our study, we observed that the GbNRX1 proteins appeared both at the peripheral regions and inside the cotton root cells and silencing of GbNRX1 results in decreased expression of defense-related genes. Thus, like thioredoxin-h5, GbNRX1 may also function in regulation of the oligomer-to-monomer transition of NPR1 in plant defense responses.
In plants, apoplastic ROS are actively produced through the action of NADPH oxidases and class III peroxidases, but the biological significance and the mechanism by which these ROS are scavenged during the plant–pathogen interaction are not well understood. In this study, we found that GbNRX1 was secreted into the extracellular regions and that it reduced ROS levels in the apoplast after the ROS burst. We measured the apoplastic ROS levels in the control and GbNRX1-silenced plants after inoculation with V. dahliae. The results showed that V. dahliae caused an apoplastic ROS burst in cotton roots. Compared with the control plants, the GbNRX1-silenced plants also exhibited an ROS burst. However, more time was required for ROS scavenging in these plants, leading to longer periods of exposure to high levels of apoplastic ROS, thereby inducing more cell death (Fig. 8). Moreover, we detected more fungal propagation in GbNRX1-silenced plants compared to the control (Supplemental Fig. S8). The increased fungal colonization may have resulted from the high levels of extracellular ROS, which were reported to be important for the development and pathogenicity of some necrotrophic pathogens, such as Botrytis cinerea, Sclerotinia sclerotiorum, and Alternaria alternate (Govrin and Levine, 2000; Kim et al., 2011; Chung, 2012). These findings indicate that GbNRX1 functions as an important regulator of apoplastic ROS homeostasis after its rapid production in cotton in response to V. dahliae invasion.
Apoplastic TRX functions in the regulation of redox balance, which is important for salt stress tolerance in rice (Zhang et al., 2011). Here, we showed that GbNRX1 participates in the regulation of ROS homeostasis in response to pathogen invasion and that GbNRX1-silenced plants exhibited impaired disease resistance. Together, these findings reveal that TRX-family proteins play important roles in apoplastic redox regulation during both biotic and abiotic stress tolerance/resistance responses.
Redox Balance after the ROS Burst in the Apoplast Is Crucial for Downstream Defense Reactions
To determine whether GbNRX1 silencing affects the downstream defense process, we examined the expression of several defense-related marker genes including NPR1, PR1, PR4, and PDF1.2. The four genes examined were either not activated or only slightly induced after pathogen challenge compared to the control, indicating that the defense-related processes were perturbed in GbNRX1-silenced plants.
Upon recognition of pathogen infection, rapid production of reactive oxygen species (the ROS burst) occurs in the apoplast; this ROS burst is required for the early plant immune response (Lamb and Dixon, 1997; Durrant and Dong, 2004; Torres et al., 2006, 2010; Daudi et al., 2012; Doehlemann and Hemetsberger, 2013). However, when ROS are produced at an excessively high level, they can cause cellular injury and tissue damage (Govrin and Levine, 2000; Krishnamurthy and Rathinasabapathi, 2013; Vellosillo et al., 2010). This toxic effect of ROS could perturb the immune responses of the plants to microbes and ultimately result in the susceptibility of the plants to pathogen infection (Lorrain et al., 2003; Puertollano et al., 2011). In our study, an ROS burst was observed in the NRX1-silenced plants, indicating that knock-down of NRX1 did not affect apoplastic ROS generation and the rapid plant defense response was induced at an early stage of pathogen infection. Based on the literature, plants accumulating high levels of ROS often display increased resistance to pathogens, yet NRX1-silenced cotton plants in which ROS were sustained at high levels in the apoplast exhibited a V. dahliae-sensitive phenotype (Fig. 7 and 8). We speculate that this phenotype was due to the toxic effect of the prolonged presence of excess ROS.
In our study, we observed that the GhNRX1-silenced G. hirsutum plants had more-severe disease symptoms than the GbNRX1-silenced G. barbadense plants after pathogen infection. To understand the molecular basis for the different responses in the two species, we analyzed and compared the transcript levels of the genes encoding the identified pathogen-responsive proteins in G. barbadense with those in G. hirsutum. The results showed that, besides NRX1, the transcript levels of most of the ROS-related genes were up-regulated in both G. hirsutum and G. barbadense in response to the pathogen attack. However, the induction occurred more rapidly and to a higher extent in the G. barbadense plants (Supplemental Fig. S2), indicating that ROS-mediated defense response was faster and stronger in the root apoplast of G. barbadense, which may account for their disease resistance. Although the exact mechanism explaining how redox status regulates the defense system remains elusive, it is clear that redox balance after the ROS burst in the apoplast is required for a successful immune response in plants and that this task is fulfilled by a number of proteins, including GbNRX1, which are secreted into the apoplast in a timely manner.
MATERIALS AND METHODS
Cotton Growth and V. dahliae Culture
Seeds of Verticillium wilt-resistant island cotton cv Hai 7124 (G. barbadense) and susceptible upland cotton cv TM-1 (Gossypium hirsutum) were delinted with H2SO4 (98%) and sterilized in 70% ethanol for a few seconds, followed by three rinses in sterile water. Cotton plants were grown hydroponically as previously described (Qu et al., 2005) or cultured in a controlled environment chamber under a 16/8 h photoperiod and at 80% relative humidity.
The defoliating V. dahliae isolate V991 was used in this study (Zhang et al., 2012b). The fungal strain was grown on potato dextrose agar medium. For suspension spore production, mycelia growing on potato dextrose agar medium were collected and cultured in liquid Czapek’s medium (2 g NaNO3, 1g KH2PO4, 1 g MgSO4•7H2O, 1 g KCl, 2 mg FeSO4•7H2O, and 30 g Suc per l) at 150 rpm/min, 28°C for 3–5 d. For cotton inoculation, the concentration of spore suspension was adjusted to 1 × 106 conidia/ml with deionized water.
Infection of Cotton Seedlings and Sample Collection
The roots of cotton seedlings (10 d old) were dip-inoculated with V. dahliae conidia suspension (1 × 106 conidia/ml) as previously described (Wang et al., 2011). Control plants were mock-inoculated with sterile water. Roots were harvested at 0.5, 1, and 3 dpi and processed immediately for extraction of apoplastic proteins. For transcriptional analysis, roots were harvested at 0, 6, 12, 24, 36, 48, 60, and 72 hpi, frozen in liquid nitrogen, and stored at −80°C until use. The quantitative reverse-transcription PCR assays were performed using SYBR Green real-time PCR master mix (Toyobo, Osaka, Japan) and cotton Histon3 gene was used as an internal control. The primers used in this experiment are listed in Supplemental Table S3.
Generation of the V991-GFP V. dahliae Strain
The pSulPH expression vector was kindly provided by Dr. Chaozu He (Hainan University, Haikou, Hainan, China). The GFP open reading frame (ORF) was cloned into the pSulPH under the ToxA promoter and the recombinant plasmid was delivered into the V. dahliae V991 strain based on the method reported previously (Michielse et al., 2005).
Preparation of Apoplastic and Total Protein Extracts
To extract apoplastic proteins, a vacuum infiltration-centrifugation method (Zhou et al., 2011) was employed with minor modifications. Cotton roots were cut into segments of approximately 3 cm and washed four times with chilled deionized water for 5 min each time. Segments were blotted dry with filter paper and placed into a 20-ml disposable syringe, which was then submerged in the extraction buffer (100 mm Tris-HCl pH 7.5, 200 mm KCl, 1 mm PMSF) for 15 min at 4°C. The container was then exposed to a −70 kPa vacuum for up to 15 min in a vacuum chamber, after which the vacuum was slowly released for 5 min. These steps were repeated 1–2 times until the tissue was completely infiltrated. After the excess extraction buffer was dried under gravity, the syringe barrel was transferred to a 50-ml tube and centrifuged immediately at 1000 g at 4°C for 10 min. The flow-through was collected as the apoplast-enriched protein extract, which was concentrated with a Microcon YM-5 (Millipore, Billerica, MA) and subjected to the malate dehydrogenase enzyme assay or trichloroacetic acid precipitation for 2D electrophoresis. For total protein preparation, the roots were ground to powder in liquid nitrogen. The powder was transferred to new tubes containing 1 ml extraction buffer (100 mm Tris–HCl pH 7.5, 50 mm EDTA, 5 mm DTT, 200 mm KCl, 2 mm PMSF) and homogenized in a homogenizer for 3 min. Finally, the slurry was centrifuged at 15,000 g for 30 min at 4°C, and the supernatant was transferred to new tubes and stored at −80°C. Three biological replicates were performed per treatment.
Assessment of Purity of Apoplastic Protein Extracts
Malate dehydrogenase (MDH) activity measurements and immunoblotting were used to assess the purity of apoplastic protein extracts. The enzyme activity of MDH, a cytoplasmic biomarker, was assayed after 20 μg apoplast or total protein extract was added to a reaction mixture containing 0.17 mm oxalacetic acid and 0.094 mm β-NADH disodium salt in 100 mm Tris buffer, pH 7.5. The change in absorbance was monitored at 340 nm for 180 s in a spectrophotometer (model no. U-2001; Hitachi, Tokyo, Japan); the same reaction mixture containing only sample buffer was used as a blank. Antibodies specific to intracellular tubulin were used for immunoblot analysis.
Labeling of Proteins with Cy Dye Differential Gel Electrophoresis (DIGE) Fluors
After overnight precipitation in 10 volumes of 10% trichloroacetic acid in acetone at −20°C, apoplast extracts (three biological repeats) were centrifuged and subsequently resuspended in ice-cold rehydration buffer (7 m urea, 2 m thiourea, 4% CHAPS), and a 2-DE Clean-up kit (GE Healthcare Life Sciences, Pittsburgh, PA) was used to purify the proteins according to the instruction manual. Protein quantification was performed using the 2-DE Quant-kit (GE Healthcare Life Sciences). To allow effective labeling with Cy dye DIGE fluors, the protein concentrations were adjusted to 8.0 µg/µl with DIGE lysis buffer (7 m urea, 2 m thiourea, 2% CHAPS), pH 8.0, with 2 mm NaOH. Following the protocol in the Ettan DIGE User’s Manual (GE Healthcare Life Sciences), 50 µg of proteins from control or V. dahliae-infected cotton roots were separately labeled with 400 pmol of Cy3 or Cy5 dyes dissolved in dimethylformamide on ice for 30 min. Meanwhile, the pooled standard containing equal amounts of protein from all samples was labeled with 400 pmol of Cy2 dye as described above to serve as an internal standard. The labeling reaction was subsequently quenched by the addition of 10 mm Lys for 15 min. According to the principle of experimental design for biological replicates, each gel contained 50 µg of Cy3- and Cy5-stained samples supplemented with 50 µg of Cy2-stained internal standard; the sample distribution is shown in Supplemental Table S2. For preparatory gels, a total of 800 µg protein from pooled samples was used.
Two-Dimensional Electrophoresis, Image Acquisition, and Analysis of Protein Spots
According to standard guidelines, protein samples labeled with Cy2, Cy3, and Cy5 dyes to be separated in the same gel were mixed and the volume of each mixed sample was adjusted to 450 µl with DIGE lysis buffer (7 m urea, 2% CHAPS, 2 m thiourea, 20 mm DTT, 0.5% IPG buffer). For protein sample separation, samples to be loaded on IPG strips (24 cm, pH 4–7, linear; GE Healthcare Life Sciences) were first subjected to active rehydration at 30 V for 20 h on an Ettan IPGphor system (GE Healthcare Life Sciences). IEF was carried out at 100 V for 3 h, 300 V for 1 h, 500 V for 1 h, gradient to 1000 V for 1 h, gradient to 4000 V for 1 h, gradient to 8000 V for 12 h, and a final hold at 1000 V for 5 h. For the second dimension, the strips were incubated in equilibration buffer containing 15 mm DTT for 20 min in the first step, which was replaced with 2.5% iodoacetamide in the second step. The strips were transferred to the tops of 12.5% polyacrylamide gels and sealed with 0.5% agarose gel containing a trace amount of bromophenol blue. Electrophoresis was carried out at 1 W per gel for 1 h, followed by 8 W per gel in the dark until the dye front reached the bottom of the gels. The stained gels were scanned using a Typhoon FLA9500 Imager (GE Healthcare Life Sciences) at optimal excitation/emission wavelength. Image analysis was performed using DeCyder software v. 6.5 (GE Healthcare Life Sciences). According to the volume ratios of corresponding spots detected in the Cy2 image of the pooled-sample internal standard, the spots detected in individual gels were quantified and normalized using the differential in-gel analysis module. All normalized differential in-gel analysis data sets from all gels were collectively analyzed to acquire statistical data on average abundance for each protein spot among the DIGE gels produced in this study using the biological variation analysis module. ANOVA was applied to matched spots and the data were filtered to retain protein spots with P ≤ 0.05 (determined by one-way ANOVA) and a fold-change of ≥1.5.
The preparatory gel was run under the same conditions. The gel was then fixed in 40% ethanol and 10% acetic acid for 45 min, stained with colloidal Coomassie solution for 24–36 h, washed several times with Milli-Q water (Millipore) to remove the background stain, and scanned in a Typhoon scanner (GE Healthcare Life Sciences) using the Cy5 wavelength. After matching to the master gel, the differentially expressed spots in the preparatory gel image were manually excised and subjected to in-gel digestion using trypsin as the protease, followed by protein identification using a 4700 tandem matrix-assisted laser-desorption ionization time-of-flight mass spectrometer (Applied Biosystems, Framingham, MA). The combined mass spectrometry (MS) and tandem MS (MS/MS) peak lists were analyzed using ProteinPilot software (Applied Biosystems) with a Mascot search engine (MASCOT v. 2.2; www.matrixscience.com/mascot_support.html) and searched against Gossypium sequences in the NCBI (http://www.ncbi.nlm.nih.gov/) and Phytozome databases (http://www.phytozome.net/). In addition, the MS and MS/MS data were also searched against the V. dahliae VdLs.17 protein sequence database (http://www.broadinstitute.org/annotation/genome/verticillium_dahliae/MultiDownloads.html) to identify Verticillium proteins in cotton root apoplast extracts. Details of the procedures for in-gel digestion and mass spectrometric analysis referred to a previous work (Wang et al., 2011). Functional categories of the identified proteins were assigned using the Gene Ontology tool (http://www.geneontology.org).
Subcellular Localization Analysis of Identified Proteins
SignalP 3.0 software (for detecting the N-terminal signal peptide signature of classically secreted proteins; www.cbs.dtu.dk/services/SignalP/) and SecretomeP (for detecting the pathway-independent features of nonclassically secreted proteins; http://www.cbs.dtu.dk/services/SecretomeP/) were used to predict the conserved motifs in the identified proteins. To further detect the subcellular localization of these proteins, the empty vector pPZP111-GFP and recombinant plasmids fusing to GFP were transformed into onion epidermal cells by particle bombardment using the PDS-1000/He system (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions. After incubation on Murashige and Skoog medium in a growth chamber for 24–36 h, GFP fluorescence was visualized in the transformed epidermal cells under a confocal microscope (TCS SP8; Leica Microsystems, Wetzlar, Germany); 30 g/L Suc solution was used to initiate plasmolysis.
Virus-Induced Gene Silencing in Cotton and Pathogen Inoculation
Tobacco rattle virus (TRV)-based virus-induced gene silencing was performed as described previously (Liu et al., 2002). The pTRV1, pTRV2, and pTRV2 derivatives harboring the 5′-untranslated regions of the candidate genes amplified from G. barbadense or G. hirsutum cDNA were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. The primers used for fragment amplification are listed in Supplemental Table S3. Cotton gene silencing was performed by using a vacuum infiltration method (Qu et al., 2012). Ten-d-old seedlings were transfected with the mixture (1:1, v/v) of Agrobacterium cultures (OD600 = 1.5) harboring pTRV1 with pTRV2 or its derivative plasmids. After completion of agro-inoculation, the seedlings were washed three times with deionized water to remove excess agrobacterial inoculum and grown at 25°C under a 16 h/8 h light/dark cycle in a controlled environmental chamber. After two weeks of cultivation, the plants were inoculated with V. dahliae isolate V991 at a position approximately 1 cm below the cotyledons. The experiments were performed with at least 36 plants per treatment and repeated three times. The rate of diseased plants and the disease index were calculated as previously described (Gao et al., 2013a).
RNA Extraction and Cloning of Full-Length GbNRX1 cDNA by Rapid Amplification of cDNA Ends
Total RNA from cotton roots, stems, leaves, and flowers was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. For quantitative reverse-transcription PCR expression analysis, total RNA (2 μg) was reverse-transcribed using a PrimeScript Real-time PCR Kit (Takara Bio, Japan/Clontech Laboratories, Göteborg, Sweden). To clone the full-length GbNRX1 cDNA, first-strand cDNA was synthesized from RNA (1 μg) isolated from cotton roots using Reverse Transcriptase XL (Takara Bio), and nested PCR was performed with rapid-amplification-of-cDNA-ends primers and gene-specific primers (Supplemental Table S3). The nested PCR product was gel-purified, cloned into the pMD18-T vector (Takara Bio), and sequenced.
Phylogenetic Analysis
The neighbor-joining method was used to construct the phylogenetic tree of GbNRX1 and Arabidopsis TRX proteins containing multiple TRX domains using MEGA v. 5.1 [www.megasoftware.net; (Tamura et al., 2007)].
Promoter Isolation and GUS Staining
Using a DNA walker system, genomic DNA extracted from cotton roots was digested with Dra I, Ssp I, EcoR V, and Pvu II and ligated to adaptors to construct the GenomeWalker DNA Library (GenomeWalker Kit; Clontech Laboratories). PCR was performed with adaptor primers provided with the kit (Clontech Laboratories) and gene-specific primers, and an approximately 1.3-kb DNA fragment containing the start codon of GbNRX1 was amplified and sequenced. For tissue-specific expression analysis, the promoter fragment of GbNRX1 was fused upstream of the GUS gene in binary vector pCAMBIA1301. The recombinant plasmid was introduced into cotton and Arabidopsis Col-0, and the T1 transgenic plants were subjected to GUS staining (Jefferson et al., 1987).
Protein Expression, Purification, and Determination of Insulin Reduction Activity
The cDNA sequences encoding GbNRX1, domains of GbNRX1 (domain 1 [aa 29–158, D1], domain 2 [aa 189–317, D2], and domain3 [aa 348–477, D3]), and AtTRX-h5 were respectively cloned into the bacterial expression vector pET-28a and the resulting constructs were transformed into Escherichia coli strain BL21 (DE3). All His-tagged recombinant GbNRX1 and AtTRX-h5 proteins were purified using Ni-NTA resin following the manufacturer’s procedures (Qiagen, Hilden, Germany). To measure insulin reduction, His-GbNRX1, His-truncated GbNRX1, and His-AtTRX-h5 at the indicated concentrations were added to the reaction mixture containing 100 mm potassium phosphate buffer, pH 7.0, 0.1 mm EDTA, and 0.13 mm bovine insulin (Sigma I5500; Sigma-Aldrich, St. Louis, MO), and the reactions were initiated by the addition of 0.33 mm DTT. The reduction activity was determined based on the optical absorption of the solution at 650 nm at room temperature using a spectrophotometer (model no. U-2001; Hitachi).
Histochemical Assay
Total H2O2 accumulation was examined by staining with DAB (3,3′-diaminobenzidine hydrochloride). The leaves were incubated overnight in 1 mg/ml DAB-HCl (Sigma-Aldrich), pH 3.8 in the dark and destained with lactic acid/glycerol/ethanol (1:1:2). To identify dead plant cells, the leaves were stained with trypan blue and destained with saturated chloral hydrate solution (2.5 g/ml).
Apoplastic Reactive Oxygen Species (ROS) Detection
Apoplastic reactive oxygen species (ROS) production was measured as previously described (Roux et al., 2011; Schwessinger et al., 2011). Briefly, apoplastic fluids isolated from cotton roots were incubated in 96-well plates, followed by 200 µl of solution containing 10 µg/ml peroxidase (Sigma-Aldrich) and 20 µM luminol. Luminescence (expressed as relative light units) was measured and calculated using a Berthold Centro LB960 luminometer (Berthold Technologies, Zug, Switzerland). Visualization of apoplastic ROS was performed as previously described (Zhang et al., 2011). Cotton roots were incubated in OxyBURST Green H2HFF BSA (10 μg ml−1 O-13291; Molecular Probes, Eugene, OR) for 10 min in the dark, and the cells were examined under a confocal laser microscope (model no. TCS SP8; Leica Microsystems) in multitrack mode (at 0.75-μm steps with two-line averaging and one-frame averaging).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: GbNRX1 (KT372889), GbPDS (KC969206), GhNRX1 (CotAD_56209), AtTRXh5 (At1g45145), AtNRX1 (At1g60420), AtNRX2 (At4g31240), AtPDIL1-1 (At1g21750), AtPDIL1-2 (At1g77510), AtPDIL1-3 (At3g54960), AtPDIL1-4 (At5g60640), AtPDIL1-5 (At1g52260), AtPDIL1-6 (At3g16110), AtPDIL2-1 (At2g47470), AtPDIL2-2 (At1g04980), AtPDIL2-3 (At2g32920), AtPDIL5-1 (At1g07960), AtPDIL5-2 (At1g35620), AtPDIL5-3 (At3g20560), AtPDIL5-4 (At4g27080), AtQSOX1 (At1g15020), AtQSOX2 (At2g01270), AtAPR1 (At4g04610), AtAPR2 (At1g62180), AtAPR3 (At4g21990), AtAPRL5 (At3g03860), AtAPRL6 (At4g08930), and AtAPRL7 (At5g18120).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Confocal observation of the proliferation of V. dahliae in the cotton roots.
Supplemental Figure S2. qRT-PCR analysis of the genes encoding ROS-related proteins upon V. dahliae infection.
Supplemental Figure S3. qRT-PCR analysis of the genes encoding defense-responsive proteins upon V. dahliae infection.
Supplemental Figure S4. qRT-PCR analysis of the genes encoding metabolism-related proteins upon V. dahliae infection.
Supplemental Figure S5. qRT-PCR analysis of the genes encoding cell wall modification-related proteins upon V. dahliae infection.
Supplemental Figure S6. qRT-PCR analysis of the genes encoding signaling-related proteins upon V. dahliae infection.
Supplemental Figure S7. qRT-PCR analysis of the genes encoding other proteins upon V. dahliae infection.
Supplemental Figure S8. Proliferation of V. dahliae in cotton stem segments.
Supplemental Table S1. List of proteins with differential abundance in response to V. dahliae infection in G. barbadense identified by MS/MS.
Supplemental Table S2. Experimental design for DIGE analysis of apoplastic proteins extracted from cotton roots.
Supplemental Table S3. Primers used in this study.
Acknowledgments
We thank Dr. Yule Liu (Tsinghua University) and Dr. Chaozu He (Hainan University) for kindly providing us with VIGS vector and pSulPH expression vector, respectively.
Glossary
- DIGE
differential gel electrophoresis
- BFA
brefeldin A
- MDH
malate dehydrogenase
- NRX
nucleoredoxin
- ORF
open reading frame
- PDI
protein disulfide isomerase
- PR
pathogenesis-related
- ROS
reactive oxygen species
- TRV
tobacco rattle virus
- VIGS
virus-induced gene silencing
Footnotes
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDB11040600); National Science Foundation of China (grant no. 31401033) and China Postdoctoral Science Foundation (grant no. 2015M570168).
Articles can be viewed without a subscription.
References
- Aguado A, Santos BDL, Blanco C, Romero F (2008) Study of gene effects for cotton yield and Verticillium wilt tolerance in cotton plant (G. hirsutum). Field Crops Res 107: 78–86 [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Ascencio-Ibáñez JT, Sozzani R, Lee TJ, Chu TM, Wolfinger RD, Cella R, Hanley-Bowdoin L (2008) Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol 148: 436–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Jones JD (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69: 473–488 [DOI] [PubMed] [Google Scholar]
- Bednarek P, Kwon C, Schulze-Lefert P (2010) Not a peripheral issue: secretion in plant-microbe interactions. Curr Opin Plant Biol 13: 378–387 [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S (2004) Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel 17: 349–356 [DOI] [PubMed] [Google Scholar]
- Bolek Y, El-Zik KM, Pepper AE, Bell AA, Magill CW, Thaxton PM, Reddy OUK (2005) Mapping of verticillium wilt resistance genes in cotton. Plant Sci 168: 1581–1590 [Google Scholar]
- Bolwell GP, Butt VS, Davies DR, Zimmerlin A (1995) The origin of the oxidative burst in plants. Free Radic Res 23: 517–532 [DOI] [PubMed] [Google Scholar]
- Bradley DJ, Kjellbom P, Lamb CJ (1992) Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid defense response. Cell 70: 21–30 [DOI] [PubMed] [Google Scholar]
- Cai YF, He XH, Mo JC, Sun Q, Yang JP, Liu JG (2009) Molecular research and genetic engineering of resistance to Verticillium wilt in cotton. Afr J Biotechnol 8: 7363–7372 [Google Scholar]
- Choi DS, Hwang IS, Hwang BK (2012) Requirement of the cytosolic interaction between PATHOGENESIS-RELATED PROTEIN10 and LEUCINE-RICH REPEAT PROTEIN1 for cell death and defense signaling in pepper. Plant Cell 24: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KR. (2012) Stress response and pathogenicity of the necrotrophic fungal pathogen Alternaria alternata. Scientifica (Cairo) 2012: 635431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daudi A, Cheng Z, O’Brien JA, Mammarella N, Khan S, Ausubel FM, Bolwell GP (2012) The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell 24: 275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz K-J. (1997) Functions and responses of the leaf apoplast under stress. In Behnke HD, Lüttge U, Esser K, Kadereit J, Runge M, eds, Progress in Botany, Vol 58 Springer, Berlin, Germany, pp 221–254 [Google Scholar]
- Doehlemann G, Hemetsberger C (2013) Apoplastic immunity and its suppression by filamentous plant pathogens. New Phytol 198: 1001–1016 [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42: 185–209 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Fernández MB, Pagano MR, Daleo GR, Guevara MG (2012) Hydrophobic proteins secreted into the apoplast may contribute to resistance against Phytophthora infestans in potato. Plant Physiol Biochem 60: 59–66 [DOI] [PubMed] [Google Scholar]
- Floerl S, Druebert C, Majcherczyk A, Karlovsky P, Kües U, Polle A (2008) Defence reactions in the apoplastic proteome of oilseed rape (Brassica napus var. napus) attenuate Verticillium longisporum growth but not disease symptoms. BMC Plant Biol 8: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floerl S, Majcherczyk A, Possienke M, Feussner K, Tappe H, Gatz C, Feussner I, Kües U, Polle A (2012) Verticillium longisporum infection affects the leaf apoplastic proteome, metabolome, and cell wall properties in Arabidopsis thaliana. PLoS One 7: e31435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato Y, Michiue T, Asashima M, Miki H (2006) The thioredoxin-related redox-regulating protein nucleoredoxin inhibits Wnt-beta-catenin signalling through dishevelled. Nat Cell Biol 8: 501–508 [DOI] [PubMed] [Google Scholar]
- Gao W, Long L, Zhu LF, Xu L, Gao WH, Sun LQ, Liu LL, Zhang XL (2013a) Proteomic and virus-induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol Cell Proteomics 12: 3690–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Li F, Li M, Kianinejad AS, Dever JK, Wheeler TA, Li Z, He P, Shan L (2013b) Cotton GhBAK1 mediates Verticillium wilt resistance and cell death. J Integr Plant Biol 55: 586–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Wheeler T, Li Z, Kenerley CM, He P, Shan L (2011) Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J 66: 293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govrin EM, Levine A (2000) The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr Biol 10: 751–757 [DOI] [PubMed] [Google Scholar]
- Grant MR, Jones JDG (2009) Hormone (dis)harmony moulds plant health and disease. Science 324: 750–752 [DOI] [PubMed] [Google Scholar]
- Gupta R, Lee SE, Agrawal GK, Rakwal R, Park S, Wang Y, Kim ST (2015) Understanding the plant-pathogen interactions in the context of proteomics-generated apoplastic proteins inventory. Front Plant Sci 6: 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. (1989) Thioredoxin and glutaredoxin systems. J Biol Chem 264: 13963–13966 [PubMed] [Google Scholar]
- Hückelhoven R. (2007) Cell wall-associated mechanisms of disease resistance and susceptibility. Annu Rev Phytopathol 45: 101–127 [DOI] [PubMed] [Google Scholar]
- Hugot K, Rivière MP, Moreilhon C, Dayem MA, Cozzitorto J, Arbiol G, Barbry P, Weiss C, Galiana E (2004) Coordinated regulation of genes for secretion in tobacco at late developmental stages: association with resistance against oomycetes. Plant Physiol 134: 858–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husted S, Schjoerring JK (1995) Apoplastic pH and ammonium concentration in leaves of Brassica napus L. Plant Physiol 109: 1453–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Zhao J, Zhou L, Guo W, Zhang T (2009) Molecular mapping of Verticillium wilt resistance QTL clustered on chromosomes D7 and D9 in upland cotton. Sci China C Life Sci 52: 872–884 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Chen C, Kabbage M, Dickman MB (2011) Identification and characterization of Sclerotinia sclerotiorum NADPH oxidases. Appl Environ Microbiol 77: 7721–7729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Wu J, Kwon SJ, Oh H, Lee SE, Kim SG, Wang Y, Agrawal GK, Rakwal R, Kang KY, Ahn IP, Kim BG, et al. (2014) Proteomics of rice and Cochliobolus miyabeanus fungal interaction: insight into proteins at intracellular and extracellular spaces. Proteomics 14: 2307–2318 [DOI] [PubMed] [Google Scholar]
- Kim SG, Wang Y, Lee KH, Park ZY, Park J, Wu J, Kwon SJ, Lee YH, Agrawal GK, Rakwal R, Kim ST, Kang KY (2013) In-depth insight into in vivo apoplastic secretome of rice-Magnaporthe oryzae interaction. J Proteomics 78: 58–71 [DOI] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Tena G, Sheen J (2000) Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA 97: 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy A, Rathinasabapathi B (2013) Oxidative stress tolerance in plants: novel interplay between auxin and reactive oxygen species signaling. Plant Signal Behav 8: 4161–, 25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurooka H, Kato K, Minoguchi S, Takahashi Y, Ikeda J, Habu S, Osawa N, Buchberg AM, Moriwaki K, Shisa H, Honjo T (1997) Cloning and characterization of the nucleoredoxin gene that encodes a novel nuclear protein related to thioredoxin. Genomics 39: 331–339 [DOI] [PubMed] [Google Scholar]
- Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48: 251–275 [DOI] [PubMed] [Google Scholar]
- Li C, He X, Luo X, Xu L, Liu L, Min L, Jin L, Zhu L, Zhang X (2014a) Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM-DOMAIN1 expression. Plant Physiol 166: 2179–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y, Chen S, Zhou JM (2014b) The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15: 329–338 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Lorang J, Kidarsa T, Bradford CS, Gilbert B, Curtis M, Tzeng SC, Maier CS, Wolpert TJ (2012) Tricking the guard: exploiting plant defense for disease susceptibility. Science 338: 659–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Vailleau F, Balagué C, Roby D (2003) Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci 8: 263–271 [DOI] [PubMed] [Google Scholar]
- Luo P, Wang YH, Wang GD, Essenberg M, Chen XY (2001) Molecular cloning and functional identification of (+)-delta-cadinene-8-hydroxylase, a cytochrome P450 mono-oxygenase (CYP706B1) of cotton sesquiterpene biosynthesis. Plant J 28: 95–104 [DOI] [PubMed] [Google Scholar]
- Marchal C, Delorme-Hinoux V, Bariat L, Siala W, Belin C, Saez-Vasquez J, Riondet C, Reichheld JP (2014) NTR/NRX define a new thioredoxin system in the nucleus of Arabidopsis thaliana cells. Mol Plant 7: 30–44 [DOI] [PubMed] [Google Scholar]
- Mehdy MC. (1994) Active oxygen species in plant defense against pathogens. Plant Physiol 105: 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielse CB, Hooykaas PJ, van den Hondel CA, Ram AF (2005) Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr Genet 48: 1–17 [DOI] [PubMed] [Google Scholar]
- Montillet JL, Chamnongpol S, Rustérucci C, Dat J, van de Cotte B, Agnel JP, Battesti C, Inzé D, Van Breusegem F, Triantaphylidès C (2005) Fatty acid hydroperoxides and H2O2 in the execution of hypersensitive cell death in tobacco leaves. Plant Physiol 138: 1516–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K, Kondoh A, Stumpp MT, Hisabori T (2001) Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc Natl Acad Sci USA 98: 11224–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113: 935–944 [DOI] [PubMed] [Google Scholar]
- Nickel W, Rabouille C (2009) Mechanisms of regulated unconventional protein secretion. Nat Rev Mol Cell Biol 10: 148–155 [DOI] [PubMed] [Google Scholar]
- O’Brien JA, Daudi A, Butt VS, Bolwell GP (2012) Reactive oxygen species and their role in plant defence and cell wall metabolism. Planta 236: 765–779 [DOI] [PubMed] [Google Scholar]
- Palade G. (1975) Intracellular aspects of the process of protein synthesis. Science 189: 347–358 [DOI] [PubMed] [Google Scholar]
- Pang J, Zhu Y, Li Q, Liu J, Tian Y, Liu Y, Wu J (2013) Development of Agrobacterium-mediated virus-induced gene silencing and performance evaluation of four marker genes in Gossypium barbadense. PLoS One 8: e73211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinedo M, Regente M, Elizalde M, Quiroga IY, Pagnussat LA, Jorrin-Novo J, Maldonado A, de la Canal L (2012) Extracellular sunflower proteins: evidence on non-classical secretion of a jacalin-related lectin. Protein Pept Lett 19: 270–276 [DOI] [PubMed] [Google Scholar]
- Puertollano MA, Puertollano E, de Cienfuegos GA, de Pablo MA (2011) Dietary antioxidants: immunity and host defense. Curr Top Med Chem 11: 1752–1766 [DOI] [PubMed] [Google Scholar]
- Qin J, Zhao J, Zuo K, Cao Y, Ling H, Sun X, Tang K (2004) Isolation and characterization of an ERF-like gene from Gossypium barbadense. Plant Sci 167: 1383–1389 [Google Scholar]
- Qu J, Ye J, Geng YF, Sun YW, Gao SQ, Zhang BP, Chen W, Chua NH (2012) Dissecting functions of KATANIN and WRINKLED1 in cotton fiber development by virus-induced gene silencing. Plant Physiol 160: 738–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu ZL, Wang HY, Xia GX (2005) GhHb1: a nonsymbiotic hemoglobin gene of cotton responsive to infection by Verticillium dahliae. Biochim Biophys Acta 1730: 103–113 [DOI] [PubMed] [Google Scholar]
- Rahal A, Kumar A, Singh V, Yadav B, Tiwari R, Chakraborty S, Dhama K (2014) Oxidative stress, prooxidants, and antioxidants: the interplay. BioMed Res Int 2014: 761264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE. (1994) Mechanisms of intracellular protein transport. Nature 372: 55–63 [DOI] [PubMed] [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tör M, de Vries S, Zipfel C (2011) The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B, Roux M, Kadota Y, Ntoukakis V, Sklenar J, Jones A, Zipfel C (2011) Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet 7: e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin LA, Driouich A (1997) Brefeldin A effects in plants (are different Golgi responses caused by different sites of action?). Plant Physiol 114: 401–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zhu L, Xu L, Yuan D, Min L, Zhang X (2014) Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nat Commun 5: 5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweat TA, Wolpert TJ (2007) Thioredoxin h5 is required for victorin sensitivity mediated by a CC-NBS-LRR gene in Arabidopsis. Plant Cell 19: 673–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X (2008) Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tan XP, Liang WQ, Liu CJ, Luo P, Heinstein P, Chen XY (2000) Expression pattern of (+)-delta-cadinene synthase genes and biosynthesis of sesquiterpene aldehydes in plants of Gossypium arboreum L. Planta 210: 644–651 [DOI] [PubMed] [Google Scholar]
- Torres MA. (2010) ROS in biotic interactions. Physiol Plant 138: 414–429 [DOI] [PubMed] [Google Scholar]
- Torres MA, Jones JD, Dangl JL (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol 141: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CM (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44: 135–162 [DOI] [PubMed] [Google Scholar]
- Vellosillo T, Vicente J, Kulasekaran S, Hamberg M, Castresana C (2010) Emerging complexity in reactive oxygen species production and signaling during the response of plants to pathogens. Plant Physiol 154: 444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FX, Ma YP, Yang CL, Zhao PM, Yao Y, Jian GL, Luo YM, Xia GX (2011) Proteomic analysis of the sea-island cotton roots infected by wilt pathogen Verticillium dahliae. Proteomics 11: 4296–4309 [DOI] [PubMed] [Google Scholar]
- Wang Y, Kim SG, Wu J, Huh HH, Lee SJ, Rakwal R, Agrawal GK, Park ZY, Young Kang K, Kim ST (2013) Secretome analysis of the rice bacterium Xanthomonas oryzae (Xoo) using in vitro and in planta systems. Proteomics 13: 1901–1912 [DOI] [PubMed] [Google Scholar]
- Xu L, Zhu L, Tu L, Liu L, Yuan D, Jin L, Long L, Zhang X (2011) Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J Exp Bot 62: 5607–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CL, Liang S, Wang HY, Han LB, Wang FX, Cheng HQ, Wu XM, Qu ZL, Wu JH, Xia GX (2015) Cotton major latex protein 28 functions as a positive regulator of the ethylene responsive factor 6 in defense against Verticillium dahliae. Mol Plant 8: 399–411 [DOI] [PubMed] [Google Scholar]
- Zhang CJ, Zhao BC, Ge WN, Zhang YF, Song Y, Sun DY, Guo Y (2011) An apoplastic h-type thioredoxin is involved in the stress response through regulation of the apoplastic reactive oxygen species in rice. Plant Physiol 157: 1884–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sanogo S, Flynn R, Baral J, Bajaj S, Hughs SE, Percy R (2012a) Germplasm evaluation and transfer of Verticillium wilt resistance from Pima (Gossypium barbadense) to Upland cotton (G. hirsutum). Euphytica 187: 147–160 [Google Scholar]
- Zhang L, Tian LH, Zhao JF, Song Y, Zhang CJ, Guo Y (2009) Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol 149: 916–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WW, Jian GL, Jiang TF, Wang SZ, Qi FJ, Xu SC (2012b) Cotton gene expression profiles in resistant Gossypium hirsutum cv. Zhongzhimian KV1 responding to Verticillium dahliae strain V991 infection. Mol Biol Rep 39: 9765–9774 [DOI] [PubMed] [Google Scholar]
- Zhou L, Bokhari SA, Dong CJ, Liu JY (2011) Comparative proteomics analysis of the root apoplasts of rice seedlings in response to hydrogen peroxide. PLoS One 6: e16723. [DOI] [PMC free article] [PubMed] [Google Scholar]