The E3 ubiquitin ligase PUB1 is a common negative regulator for both rhizobial and arbuscular mycorrhizal symbioses and interacts with a key receptor kinase.
Abstract
PUB1, an E3 ubiquitin ligase, which interacts with and is phosphorylated by the LYK3 symbiotic receptor kinase, negatively regulates rhizobial infection and nodulation during the nitrogen-fixing root nodule symbiosis in Medicago truncatula. In this study, we show that PUB1 also interacts with and is phosphorylated by DOES NOT MAKE INFECTIONS 2, the key symbiotic receptor kinase of the common symbiosis signaling pathway, required for both the rhizobial and the arbuscular mycorrhizal (AM) endosymbioses. We also show here that PUB1 expression is activated during successive stages of root colonization by Rhizophagus irregularis that is compatible with its interaction with DOES NOT MAKE INFECTIONS 2. Through characterization of a mutant, pub1-1, affected by the E3 ubiquitin ligase activity of PUB1, we have shown that the ubiquitination activity of PUB1 is required to negatively modulate successive stages of infection and development of rhizobial and AM symbioses. In conclusion, PUB1 represents, to our knowledge, a novel common component of symbiotic signaling integrating signal perception through interaction with and phosphorylation by two key symbiotic receptor kinases, and downstream signaling via its ubiquitination activity to fine-tune both rhizobial and AM root endosymbioses.
Many plant species establish associations with microorganisms of the rhizosphere that result in mutualistic symbioses. Interactions with arbuscular mycorrhizal (AM) fungi are widespread among land plants whereas rhizobial bacteria enter into symbiosis with a limited number of species, essentially with legume plants to form nitrogen-fixing nodules. Hyphae of AM fungi grow at root surfaces and form hyphopodia through which they penetrate into roots to reach the inner cortex, where they develop highly branched structures called arbuscules, through which nutrient exchange takes place (Gutjahr and Parniske, 2013). In legumes such as the model plants Medicago truncatula and Lotus japonicus, rhizobia enter into roots through infection threads (ITs) that develop within epidermal root hair cells and progress to inner root tissues. On reaching the induced nodule primordia, the bacteria are finally released from the ITs into the cytoplasm of nodule cells, where they differentiate to form nitrogen-fixing bacteroids (Oldroyd et al., 2011). To initiate mutual recognition and colonize host roots, rhizobia produce signal molecules identified as lipo-chitooligosaccharides (LCOs) called Nod factors (NFs; Lerouge et al., 1990), whereas AM fungi produce both LCOs (called Myc-LCOs; Maillet et al., 2011) and short-chain chitin oligomers (CO4-5, Genre et al., 2013).
Forward genetic approaches have identified lysin-motif receptor-like kinases (RLKs) as key components of NF perception and nodulation in a number of legume plants (Gough and Cullimore, 2011; Gust et al., 2012). In M. truncatula, NOD FACTOR PERCEPTION is required for all NF responses and rhizobial infection (Ben Amor et al., 2003), whereas LYSIN MOTIF RECEPTOR-LIKE KINASE 3 (LYK3) has been proposed to be an entry receptor controlling NF recognition for IT initiation and growth (Limpens et al., 2003; Smit et al., 2007). NF and Myc factor perception lead to activation of the common symbiosis signaling pathway (CSSP) that controls early root responses to both rhizobia and AM fungi, respectively (for reviews, see Nadal and Paszkowski, 2013; Oldroyd, 2013).
The Leucine-rich repeat (LRR) RLK, DOES NOT MAKE INFECTIONS 2 (DMI2 in M. truncatula, symbiosis receptor kinase (SYMRK) in L. japonicus), is an essential upstream component of the CSSP required to generate calcium oscillations that transmits the signal through the action of key transcription factors (Endre et al., 2002; Stracke et al., 2002; and for review, Oldroyd, 2013). The function of DMI2/SYMRK is critical for both rhizobial and AM early microbe recognition events at the root epidermis, and in later signaling events related to cortical cell infection and nodule primordia initiation (Catoira et al., 2000; Endre et al., 2002; Stracke et al., 2002; Limpens et al., 2005; Kosuta et al., 2011). DMI2/SYMRK presumably participates as part of receptor complexes in early signal perception, as recently suggested by the direct interaction of a truncated form of SYMRK with NFR5 (the L. japonicus NOD FACTOR PERCEPTION ortholog; Antolín-Llovera et al., 2014). Several other interacting partners of DMI2/SYMRK have been identified and most of them act as positive regulators of nodulation (Venkateshwaran et al., 2013). Some of these interactors are E3 ubiquitin ligases such as the SymRK-INTERACTING E3 ubiquitin ligase (SIE3) that binds to and ubiquitinates SYMRK (Yuan et al., 2012). Another E3 ubiquitin ligase, the SEVEN IN ABSENTIA 4 (SINA4), destabilizes SYMRK and negatively affects the rhizobial infection process (Den Herder et al., 2012).
E3 ubiquitin ligases are known to control the abundance or the activity of proteins through ubiquitination. Originally the functional role of ubiquitination was thought to be to target cytosolic proteins to the 26S proteasome for degradation (Hershko and Ciechanover, 1992). It is now clear that ubiquitination is also a major mechanism that regulates plasma membrane trafficking (Miranda and Sorkin, 2007; Barberon et al., 2011; Scheuring et al., 2012; Herberth et al., 2012), protein activity, localization, and protein-protein interactions (Komander and Rape, 2012). Ubiquitination involves the sequential action of an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme, and an E3 ubiquitin ligase to transfer ubiquitin (Ub) to a Lys residue on the targeted substrate (Kraft et al., 2005; Stone et al., 2005). As E3 ubiquitin ligases interact directly with the target proteins, they confer specificity to the ubiquitination process. E3 ubiquitin ligases are divided into families according to their mechanism of action and the presence of specific domains. One family, the Plant U-box E3 ubiquitin ligases (PUBs), is particularly expanded in plants indicating their importance in governing cellular processes specific to plants (Yee and Goring, 2009).
The M. truncatula plant U-box E3 Ubiquitin ligase 1 (PUB1) plays an important role during NF signaling and nodulation. PUB1 was identified in a yeast two-hybrid screen as an interactor of the intracellular region of the lysin-motif-RLK LYK3. PUB1 has a U-box-dependent E3 ubiquitin ligase activity and was shown to be phosphorylated by the kinase domain of LYK3. Functional studies indicate that PUB1 acts as a negative regulator of the LYK3 signaling pathway to control rhizobial infection and nodulation (Mbengue et al., 2010).
In this work, we identified the DMI2 receptor as a new interactor, to our knowledge, of the E3 ubiquitin ligase PUB1. As in nodulation, the expression of PUB1 is highly regulated at different steps of AM fungal colonization. We demonstrated that the ubiquitination activity of PUB1 plays a key role in the early control of M. truncatula root colonization by Rhizophagus irregularis and rhizobia. Thus, PUB1 interacts with two key symbiotic receptors and modulates the establishment of both rhizobial and AM fungal symbioses.
RESULTS
DMI2 Interacts with PUB1 in Yeast and in Planta
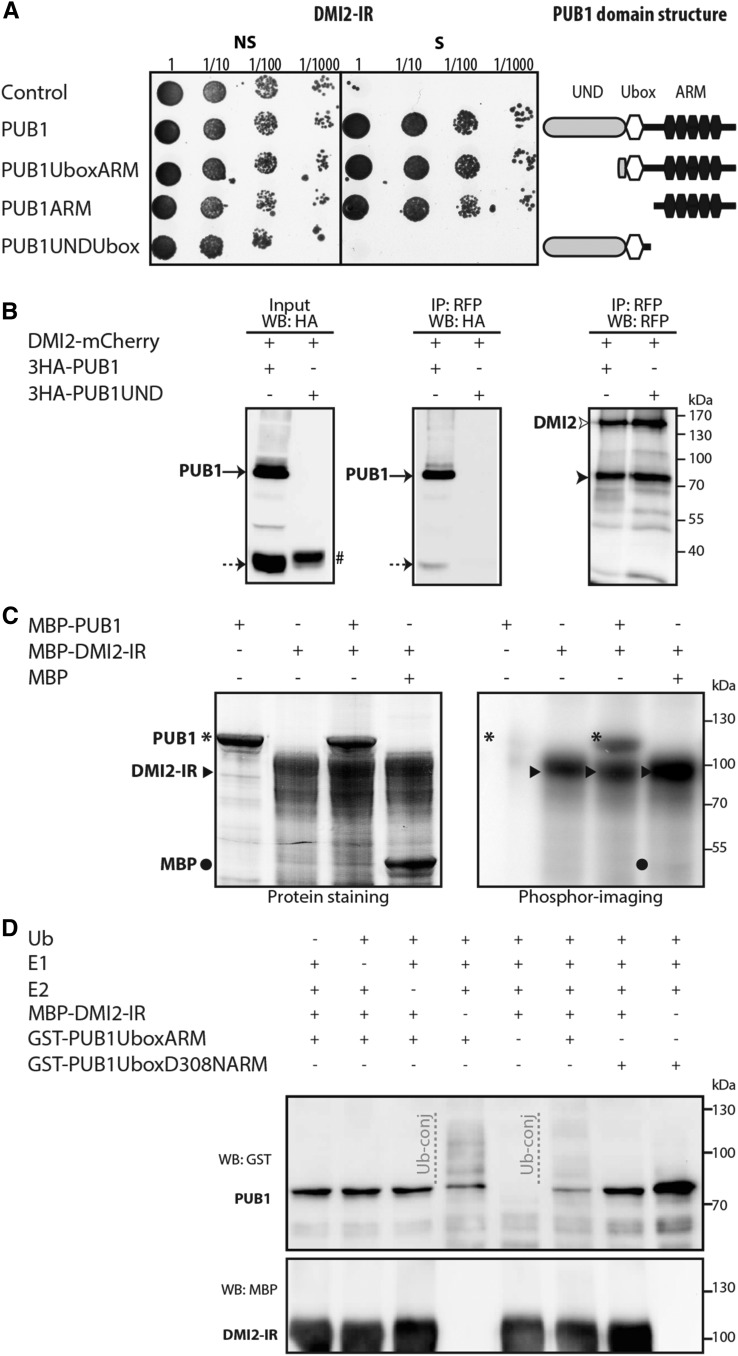
To identify new symbiotic NF signaling components we used the intracellular region (IR) of DMI2 (containing the kinase domain) as a bait to screen a GAL4-based yeast two-hybrid cDNA library prepared from root hair cells of M. truncatula roots treated with 10−8 M NFs (Andriankaja et al., 2007). Sixteen clones confirmed to interact with the DMI2 IR encoded either the full-length (8) or truncated versions (8) of the same previously described PUB1 protein (Mbengue et al., 2010; Fig. 1A). PUB1 is an E3 ubiquitin ligase containing three main domains: the N-terminal UND Domain (UND), the U-box Domain (U-box), and at least five ARMADILLO (ARM) repeats. Interacting-domain mapping revealed that the ARM repeats region is necessary and sufficient for the DMI2-IR/PUB1 interaction in yeast (Fig. 1A).
Figure 1.
PUB1 interacts with and is phosphorylated by DMI2. A, PUB1 interacts with the DMI2 IR in yeast. The negative control prey consists of the pGADT-T vector. A full-size PUB1 prey fusion and three deletion variants, PUB1UboxARM, PUB1ARM, and PUBUNDUbox, were tested for interaction with the DMI2 bait. Cotransformation of yeast cells is reported by yeast growth on nonselective medium lacking Trp and Leu (NS). Positive interactions are reported by yeast growth on selective (S) medium lacking Trp, Leu, and His and supplemented with 5 mm 3-amino-1,2,4-triazole. Serial dilutions 1–1:1000 are shown. The domain structure of PUB1 prey proteins is shown on the right-hand side. B, PUB1 coimmunopurifies with DMI2 in N. benthamiana. PUB1 or its deleted version PUB1UND N-terminally tagged with 3HA were coexpressed in N. benthamiana with DMI2-mCherry and solubilized complexes were affinity-purified with anti-RFP magnetic beads. Solubilized proteins (input) and immunopurified proteins (IP) were analyzed by western blot (WB) using anti-HA or anti-RFP antibodies (mCherry-tagged proteins). Black, dotted arrows and hashtag indicate 3HA-PUB1 full length (approximately 82 kD), cleaved/degraded (approximately 30 kD), or deleted 3HA-PUB1UND (approximately 35 kD), respectively. White and black arrowheads indicate DMI2-mCherry full length (>130 kD) and cleaved (approximately 75 kD), respectively. C, DMI2 transphosphorylates PUB1 in vitro. The auto- and transphosphorylation activities of MBP-DMI2-IR were assayed alone or by addition of MBP or MBP-PUB1. On the left panel, protein staining shows size and quantity of purified proteins. On the right panel, phosphor imaging reveals DMI2 kinase-dependent phosphorylation. Asterisks, black arrowheads, and black circles indicate MBP-PUB1 (approximately 117 kD), MBP-DMI2-IR (approximately 85 kD), and MBP (approximately 43 kD), respectively. D, DMI2 is not ubiquitinated in vitro by PUB1. The MBP-DMI2-IR protein was tested for ubiquitination in presence or absence of GST-PUB1UboxARM, GST-PUB1UboxD308NARM (an inactive form of PUB1; see Supplemental Fig. S2), yeast E1, Arabidopsis E2 (UBC8), and Ub. Anti-GST and anti-MBP antibodies were used to detect GST-PUB1 forms (approximately 72 kD) and MBP-DMI2-IR (approximately 85 kD), respectively. A range of proteins conjugated with ubiquitin (Ub-conj) was detected for GST-PUB1UboxARM (autoubiquitination) but not for GST-PUB1UboxD308NARM and MBP-DMI2-IR.
To evaluate whether DMI2 and PUB1 interact in planta, we performed coimmunopurification (co-IP) experiments with proteins produced in Nicotiana benthamiana leaves. PUB1 and its deleted version PUB1UND (1–296 amino acids), lacking the U-box and the ARM domains, were N-terminally fused with the 3× HUMAN INFLUENZA HEMAGGLUTININ (3HA) epitope (3HA-PUB1 and 3HA-PUB1UND) whereas DMI2 was C-terminally fused with a mCherry fluorescent protein (DMI2-mCherry). Western blot analysis revealed the presence of both full-length and cleaved and/or degraded forms of both 3HA-PUB1 and DMI2-mCherry proteins (Fig. 1B). 3HA-PUB1 coimmunopurified with DMI2-mCherry while 3HA-PUB1UND did not (Fig. 1B). Reciprocal co-IP of DMI2-mCherry and its C-terminal truncated form with 3HA-PUB1 produced similar outcomes (Supplemental Fig. S1A). To test the specificity of the PUB1/DMI2 interaction in planta, we performed co-IP of PUB1 with a nonsymbiotic LRR RLK of M. truncatula, LRRII.1 (Lefebvre et al., 2010), fused to the yellow fluorescent protein. PUB1 did not coimmunopurify with LRRII.1, thus suggesting specificity of the PUB1/DMI2 interaction in planta (Supplemental Fig. S1B).
DMI2 Phosphorylates PUB1 But Is Not a Direct Target of PUB1 Ubiquitin Ligase Activity
It was previously shown that SYMRK shows in vitro trans-phosphorylation activity (Chen et al., 2012). As we showed that PUB1 and DMI2 interact together, we tested whether the kinase domain of DMI2 could transphosphorylate PUB1. PUB1 and DMI2-IR were purified as MALTOSE BINDING PROTEIN (MBP) fusions and used in in vitro phosphorylation assays. MBP-DMI2-IR was autophosphorylated, as observed for SYMRK (Yoshida and Parniske, 2005), and able to transphosphorylate MBP-PUB1 but not the MBP domain alone (Fig. 1C). This result therefore indicates that PUB1 is a substrate of the in vitro kinase activity of DMI2.
To test whether DMI2 is a substrate for the ubiquitin ligase activity of PUB1, we performed in vitro ubiquitination assays using glutathione s-transferase (GST)-PUB1UboxARM and MBP-DMI2-IR proteins. Autoubiquitination of GST-PUB1UboxARM protein was observed as previously reported (Mbengue et al., 2010), but no higher bands corresponding to ubiquitinated forms of MBP-DMI2-IR were detected using anti-MBP antibodies (Fig. 1D), suggesting that DMI2-IR is not a substrate of PUB1 ubiquitin ligase activity in vitro. To explore whether the stability of the DMI2 protein could be dependent on PUB1 in planta, we coexpressed DMI2- and PUB1-tagged proteins in M. truncatula (Supplemental Fig. S1C) and N. benthamiana (Supplemental Fig. S1D). In both cases, overexpression of PUB1 did not significantly change DMI2 abundance and no additional higher band of DMI2 was detected. These results suggest that PUB1 does not affect the stability of DMI2, at least in the tested conditions.
PUB1 Is Expressed Throughout AM Fungal Colonization
As DMI2 is part of the CSSP and PUB1 is induced by Myc-LCOs (Camps et al., 2015), PUB1 could be involved in the AM symbiosis through its interaction with DMI2. To examine this hypothesis we analyzed the spatial expression patterns of DMI2 and PUB1 during AM fungal colonization. We analyzed the spatial expression pattern of DMI2 using a 3-kb upstream regulatory sequence of DMI2 fused to the GUS reporter gene (Bersoult et al., 2005) in Agrobacterium rhizogenes transgenic roots of M. truncatula. DMI2 was constitutively expressed in the entire root system of uninoculated composite plants and its expression did not change in response to AM fungal colonization, neither in intensity nor in location (Supplemental Fig. S3).
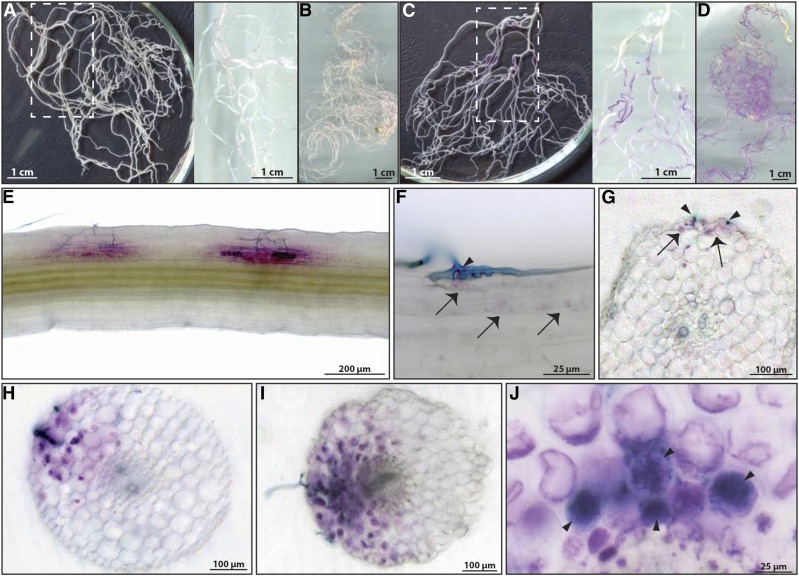
To examine the expression pattern of PUB1, we generated a stable transgenic line of M. truncatula expressing a 3-kb promoter region of PUB1 fused to the β-GLUCURONIDASE (GUS) reporter gene (ProPUB1-GUS). GUS activity was hardly detected in noninoculated roots; only a very faint staining was observed at 2 and 5 weeks (Fig. 2, A and B). In roots 2 weeks after inoculation (wpi) with R. irregularis, PUB1 expression was clearly detected in localized areas of the root system (Fig. 2C). Ink coloration of these GUS-stained areas revealed that these zones were always associated with the presence of the fungus (Fig. 2E). At 5 wpi, in a highly colonized root system, PUB1 expression was observed in almost the entire root system (Fig. 2D). Detailed analysis of the ProPUB1-GUS plants showed that PUB1 expression pattern was first detected in epidermal cells directly associated with visible hyphopodia forming on the root surface, and in a few restricted outer cortical cells in the immediate vicinity (Fig. 2, F and G). In roots with more advanced colonization, GUS activity was observed in infected outer cortical cells and in surrounding cells (Fig. 2, H and I). GUS activity was also detected both inside and in the vicinity of inner cortical cells containing arbuscules (Fig. 2J). In conclusion, these results show that the expression pattern of PUB1 is associated with all stages of AM fungal infection in epidermal and cortical infected cells, and in cells in the close vicinity.
Figure 2.
PUB1 expression is associated with all stages of symbiotic AM fungal infection. Transgenic lines expressing ProPUB1-GUS were analyzed in response to R. irregularis. Roots were stained for GUS activity (in magenta) and fungus was visualized by ink coloration (in blue). A and B, Noninoculated root systems of 2 (A) and 5 weeks (B). C and D, Inoculated root systems of 2 (C) and 5 wpi (D). White rectangles in (A) and (C) indicate the zone shown on the right of the panel on a white background to highlight the magenta GUS staining. E, GUS-stained areas are associated with the presence of the fungus (dark blue). F, G, Magnification (F) and transversal section (G) of the penetration zone showed expression of PUB1 in the epidermal cell directly in contact with a hyphopodium and in a few outer cortical cells in the immediate vicinity (arrows and arrowheads indicate the GUS staining and the fungus, respectively). H, I, Expression of PUB1 was detected in the cortex under the infection site in cortical cells directly associated with propagating internal hyphae or in cortical cells in the vicinity of the hyphae, but not necessarily in immediate contact. J, Magnification of a transversal section showing ProPUB1-GUS expression in cells containing arbuscules (arrowheads) and in adjacent cells without arbuscule.
PUB1 Negatively Regulates Fungal Colonization in the AM Symbiosis
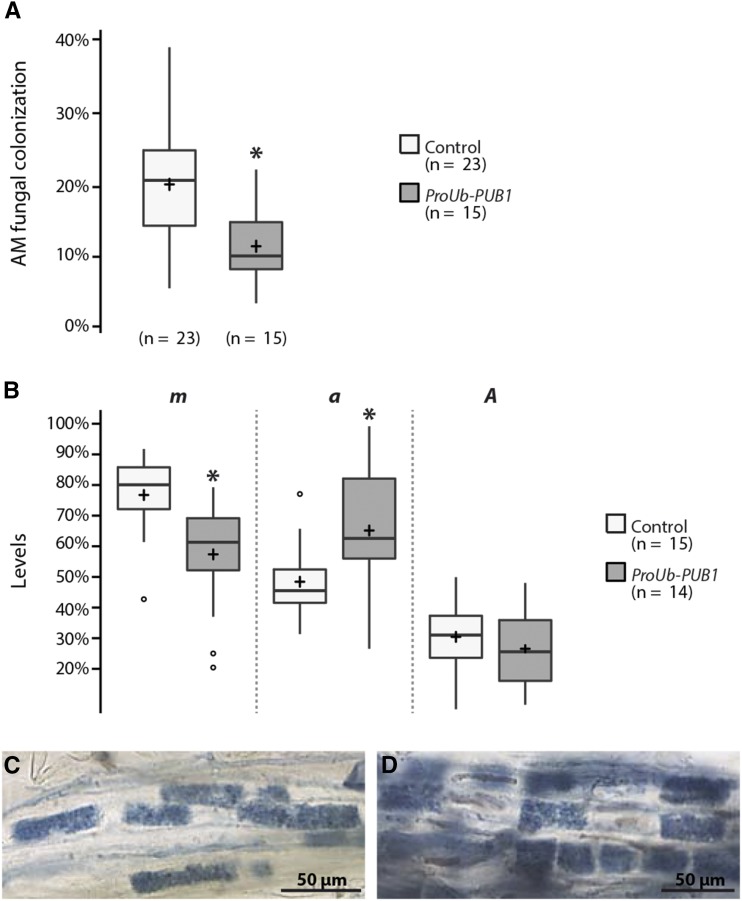
To investigate what role PUB1 plays during the AM symbiosis, we analyzed the level of colonization of M. truncatula roots overexpressing PUB1 by using the ubiquitin promoter of Lotus japonicus (ProUb-GFP-PUB1). Expression of GFP-PUB1 was detected by western blot in these roots (Supplemental Fig. S4). Three weeks after inoculation with R. irregularis, PUB1 overexpressing roots were about two times less colonized (−41%) by R. irregularis than control plants transformed with the empty vector (Fig. 3A). Analysis of these overexpressing PUB1 roots, using the Trouvelot method (Trouvelot et al., 1986), showed a decrease in the level of mycorrhizal colonization in root fragments (m) whereas arbuscule abundance in colonized root fragments was increased (a) compared to control roots (Fig. 3B). Estimated arbuscule abundance in the entire root system (A) was globally similar in the two types of samples (Fig. 3B) and no apparent difference in the development of arbuscules was observed (Fig. 3, C and D). These experiments indicate that overexpression of PUB1 negatively affects root colonization by R. irregularis hyphae without any major impact on cortical arbuscule development.
Figure 3.
PUB 1 overexpression reduces AM fungal colonization in wild-type M. truncatula roots. Wild-type roots were transformed with ProUb-GFP-PUB1 or an empty vector (control) and scored 24 d post-inoculation (dpi) with R. irregularis (1000 sp L−1) for total AM fungal colonization (A) and according to Trouvelot method (B). m, intensity of the mycorrhizal colonization in the colonized root fragments; a, arbuscule abundance in the colonized root fragments; A, arbuscule abundance in the root system. C and D, Control (C) and ProUb-GFP-PUB1 (D) roots colonized by R. irregularis at 24 dpi. No apparent difference in arbuscule development was observed. Center lines show the medians, crosses the means, and box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. Numbers of plants are indicated in brackets. Significant differences were detected by two-sample t tests (* P ≤ 0.05).
U-box Domain Integrity of PUB1 Is Critical for Its Negative Role in the AM Fungal Colonization
To investigate the importance of the ubiquitin ligase activity of PUB1 in root endosymbioses, we took advantage of a mutant line named pub1-1. This line contains a G to A substitution inducing a change of an Asp (D) to an Asn (N) at position 308 in a conserved residue of the U-box domain of PUB1 (mutation D308N, Supplemental Fig. S2A), which is predicted to affect the U-box activity of PUB1 (SIFT; http://sift.jcvi.org/). We tested whether the ubiquitination activity of the corresponding PUB1 protein was affected. The ubiquitin ligase activity of the recombinant protein lacking the UND domain and carrying the D308N (PUB1UboxD308NARM) fused to glutathione s-transferase (GST) at the N-terminal part was compared to the functional form of PUB1, also lacking the UND domain (GST-PUB1UboxARM) (Mbengue et al., 2010). Enzymatic activity was detected in in vitro ubiquitination assays with the wild-type U-box domain (GST-PUB1UboxARM), but not with the GST-PUB1UboxD308NARM mutant (Fig. 1D; Supplemental Fig. S2B). Thus, the PUB1D308N mutation in the U-box domain abolishes the ubiquitination activity of PUB1 in vitro, suggesting a lack of PUB1 E3 ligase activity in pub1-1 plants.
The pub1-1 plants showed a normal development compared to corresponding wild-type plants. A careful inspection of the root architecture of both non- and AM fungus-inoculated pub1-1 plants compared to controls did not reveal any major differences except a slight reduction (10%) of the total root length of pub1-1 plants at 2 weeks regardless of AM fungal colonization status (Supplemental Fig. S5A). Lateral root number, primary root length, and lateral root density were unchanged (Supplemental Fig. S5, B–D).
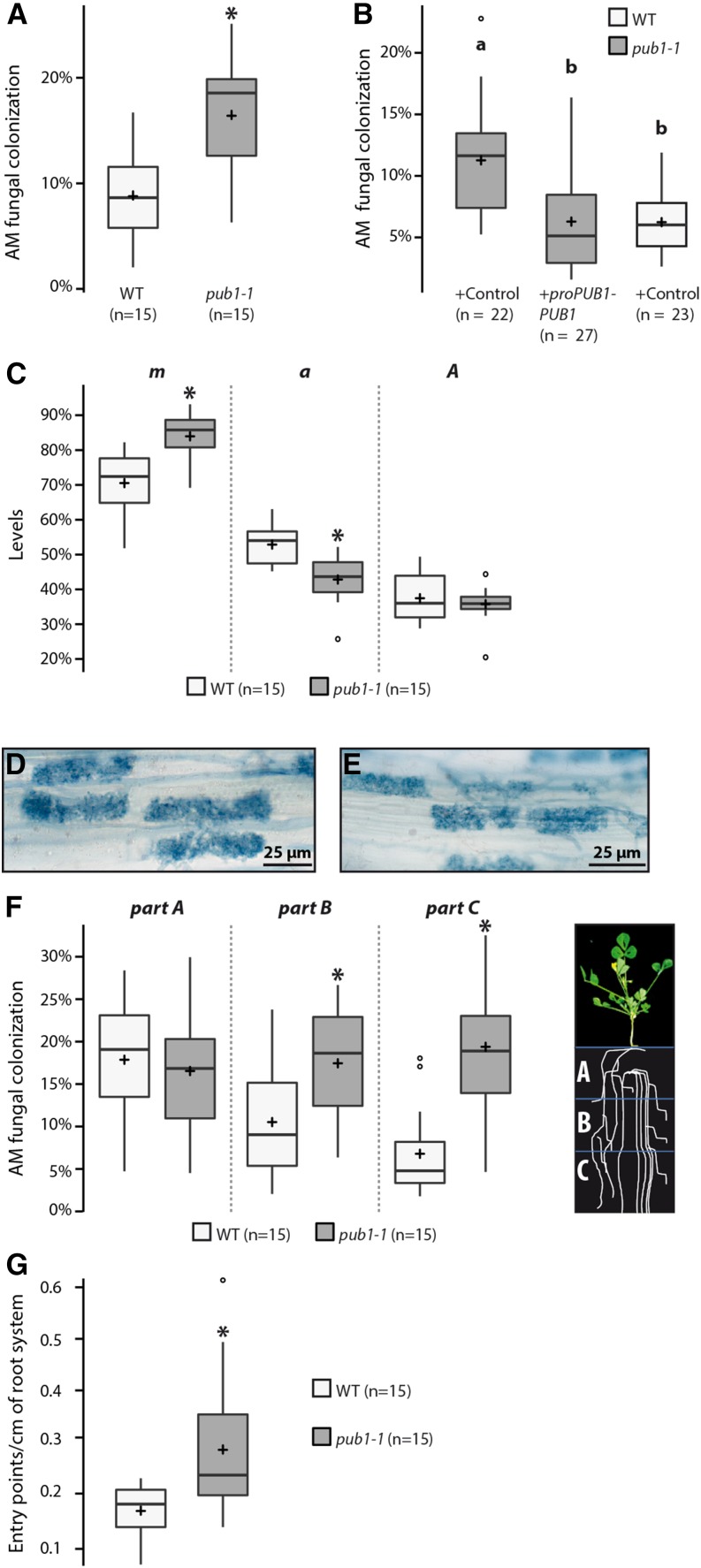
To test the implication of the ubiquitination function of PUB1 during AM symbiosis, pub1-1 and wild-type plants were compared for their fungal root colonization ability. Plants were first inoculated with a high number of spores of R. irregularis and colonization levels were quantified over time. By the gridline intersect method we found that the level of fungal colonization of pub1-1 plants was significantly increased (+42%) compared to wild type at the earliest time point (2 wpi), whereas no difference was observed in heavily colonized roots at later time points (Supplemental Fig. S6A). Recently, in M. truncatula and L. japonicus, use of a lower inoculum has provided evidence of a quantitative AM phenotype in several mutants where no visible phenotype was detected with a high inoculum (Maillet et al., 2011, Takeda et al., 2013; Delaux et al., 2013; Zhang et al., 2015a,b). By using a low inoculum, we found a significant 2-fold increase in fungal colonization in pub1-1 at 6 wpi (Fig. 4A). We complemented this phenotype by expressing the 3HA-PUB1 protein under its own promoter (ProPUB1-3HA-PUB1) or under the ubiquitin promoter (ProUb-GFP-PUB1) in pub1-1 roots (Fig. 4B; Supplemental Fig. S6B), confirming that the U-box domain of PUB1 is involved in the control of fungal colonization.
Figure 4.
PUB1 negatively regulates root AM symbiosis via its ubiquitination activity. A, Box plots show level of colonization per plant 6 weeks post-inoculation (wpi) with R. irregularis (350 sp L−1) on wild-type and pub1-1 roots. B, PUB1 complements pub1-1 AM root symbiotic phenotype. Wild-type and pub1-1 M. truncatula roots were transformed with A. rhizogenes strains containing ProPUB1-3HA-PUB1 or an empty vector (control). Transformed roots were inoculated with R. irregularis (1000 sp L−1) and scored at 25 dpi. Different letters indicate significant differences. C, Quantification of AM fungal colonization in wild-type and pub1-1 measured according to Trouvelot method at 6 wpi (350 sp L−1). m, intensity of the mycorrhizal colonization in the colonized root fragments; a, arbuscule abundance in the colonized root fragments; A, arbuscule abundance in the root system. D and E, wild-type (D) and pub1-1 (E) roots colonized by R. irregularis. No apparent difference in arbuscule development was observed. F, Each root system was transversally divided in three equal parts: part A (oldest part), part B (middle part), and part C (youngest part), as described on the cartoon. Levels of AM fungal colonization in wild-type and pub1-1 at 6 wpi with R. irregularis (350 sp L−1) are shown for each root part (A–C). G, Fungal entry points in wild type and pub1-1 at 2 wpi with R. irregularis (4000 sp L−1; normalized by the total root length). Significant differences (* P ≤ 0.05) were detected by two-sample t tests (C, G) and Wilcoxon rank sum tests (A, B, and F). For details, see box-plot descriptions on Figure 3. WT, wild type.
A detailed analysis of the colonized area of loss of function pub1-1 mutant roots using the Trouvelot method showed opposite results to those obtained by overexpression analyses (Fig. 3B): an increase in the intensity of mycorrhizal colonization in root fragments (m) and a decrease in arbuscule abundance in colonized root fragments (a). In addition, no differences in arbuscule abundance in the root system (A) (Fig. 4C) or in the development of arbuscules (Fig. 4, D and E) were observed. To analyze the spatial distribution of the colonized area on the root system, we transversally divided wild-type and pub1-1 inoculated root systems into three equal parts (Fig. 4F): part A (oldest part), part B (middle part), and part C (youngest part); each root part was scored individually. We found a significant increase in the number of colonized areas on the lower parts (B and C) of the pub1-1 root system in comparison to wild-type roots (Fig. 4F), indicating that an extended region of the pub1-1 root system was infected by R. irregularis.
Finally, we examined the first events of colonization, namely the capacity of R. irregularis to enter into the root. We observed a significant increase of fungal entry points in pub1-1 plants compared to the wild-type plants (+70%; Fig. 4G). As already observed for colonization (Fig. 4F), the entry points were localized over an extended region of the pub1-1 root system (Supplemental Fig. S7).
In conclusion, we showed that the ubiquitination function of PUB1 negatively regulates the AM fungal entry and modulates the intensity and spatial localization of AM fungal colonization.
U-box Domain Integrity of PUB1 Is Also Critical for Its Negative Role in the Rhizobial Symbiosis
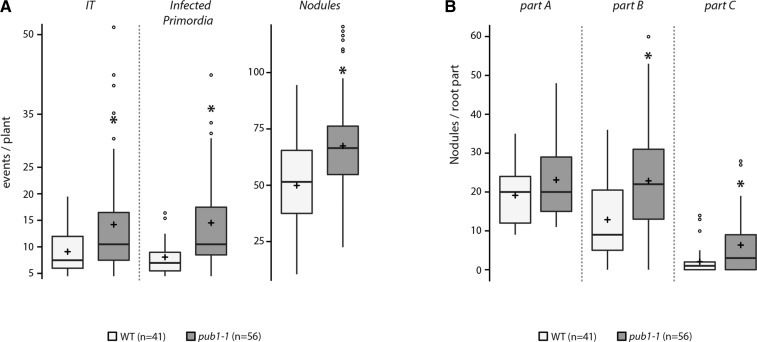
Using RNAi experiments, we previously reported that PUB1 plays a negative role during the rhizobial symbiosis (Mbengue et al., 2010). To investigate the importance of the ubiquitination activity of PUB1 during nodulation, we analyzed the phenotype of the loss of function pub1-1 line in response to inoculation by Sinorhizobium meliloti. At 4 wpi, pub1-1 plants produced significantly more nodules (increase of 35%), and 2-fold more infection threads (ITs) and infected nodule primordia compared to control plants (Fig. 5A). As for the AM symbiosis, we observed that the increase in nodulation was mainly localized on the lower parts of pub1-1 root systems (Fig. 5B), indicating an extended nodulation responsive zone in pub1-1. These results show that the negative role of PUB1 in nodulation requires its ubiquitination activity.
Figure 5.
PUB1 negatively regulates root nodule symbiosis via its ubiquitination activity. A, Box plots represent infection threads (ITs); infected primordia and nodules scored 28 days post-inoculation (dpi) with S. meliloti wild-type strain on wild-type and pub1-1 M. truncatula roots. B, Nodules observed on each root part (A–C). Each root system was transversally divided in three equal parts: part A (oldest part), part B (middle part), and part C (youngest part). Significant mean differences (*) were detected using Wilcoxon rank sum test (P ≤ 0.05). For details, see box-plot descriptions on Figure 3. WT, wild type.
DISCUSSION
Fine control of the rhizobial and AM symbioses is required to maintain mutually beneficial interactions between the plant and its microbial partners (Oldroyd, 2013). Here, we provide evidence that PUB1 is a common actor in both rhizobial and AM endosymbioses, by negatively regulating these processes via its ubiquitination activity. PUB1 can be phosphorylated by both LYK3 and DMI2 and thus its mode of action appears to occur through interaction with these two essential symbiotic receptors.
Domain interaction analyses in yeast and in planta showed that the intracellular regions of DMI2 and LYK3 interact with the same region of PUB1 containing the ARM repeats (this work; Mbengue et al., 2010). DMI2/SYMRK is required both for the nitrogen-fixing root nodule symbiosis with rhizobia to initiate nodule organogenesis and infection in legumes (Stracke et al., 2002; Endre et al., 2002), and for AM fungal colonization (Catoira et al., 2000; Endre et al., 2002; Stracke et al., 2002). LYK3 is essential for nodulation (Catoira et al., 2000; Limpens et al., 2003; Smit et al., 2007) and plays a weak role in the AM symbiosis (Zhang et al., 2015a). DMI2, LYK3, and PUB1 show overlapping expression patterns during nodulation (Bersoult et al., 2005; Mbengue et al., 2010; Supplemental Fig. S8), which are compatible with interactions between PUB1 and the two receptor-like kinases during different stages of rhizobial infection and nodule formation. The more general expression pattern of DMI2 in the AM symbiosis includes tissues in which PUB1 is induced (Fig. 2; Supplemental Fig. S3), which again is compatible with interaction of DMI2 and PUB1 during AM fungal colonization.
Phenotypic analyses of the pub1-1 mutant, impaired in PUB1 ubiquitination activity, show that this mutant is affected during the first steps of symbiotic infection with respective increases in the number of ITs in nodulation, and fungus entry points in the AM symbiosis. These phenotypes are in agreement with the spatial expression pattern of PUB1 and its early induction by NFs and by both sulfated and nonsulfated Myc-LCOs (Mbengue et al., 2010; Camps et al., 2015). The activation of early negative regulatory pathway(s) involving PUB1 suggests that rhizobial and AM signaling need to be down-regulated as early as the first steps of interaction between the symbiotic partners. In the same way, recent data suggest that NF signaling activates ethylene production, which attenuates NF signaling (Larrainzar et al., 2015).
In roots with established endosymbioses, we observed in the pub1-1 mutant an increase in the number of nodule primordia and nodules in the nitrogen fixing symbiosis, and an increase of hyphal colonization in the AM symbiosis. This quantitative AM phenotype is dependent on inoculum concentration. Because pub1-1 mutant contains a point mutation affecting PUB1 ubiquitin ligase activity, it could be possible that the phenotypes of pub1-1 are leaky due to additional function(s) of PUB1 unlinked to its ubiquitin ligase activity. However, overexpression of PUB1 in M. truncatula roots clearly reduced the level of AM colonization compared to control roots, and this was found to be through hyphal colonization without obvious impacts on the overall number and structure of arbuscules. Opposite reciprocal changes in the arbuscule abundance and the level of colonization were observed in colonized root fragments of the mutant and overexpressing plants. However, in both cases, no difference in arbuscule abundance in the whole root system was seen, as if the plant can offset the level of mycorrhizal colonization by the number of arbuscules developing in the colonized area. We therefore propose a role for PUB1 as a modulator of root endosymbiotic infection and nodulation/fungal colonization.
Autoregulatory mechanisms control the rhizobial and mycorrhizal symbioses. This is a phenomenon in which nodulation or root colonization by AM fungi inhibits the formation of subsequent nodules or root colonization in other parts of roots. This mechanism is mediated by a long-distance signal controlled by the shoot (Staehelin et al., 2011; Schaarschmidt et al., 2013; Huault et al., 2014). In addition, more locally, phytohormones are known to control establishment of rhizobial and mycorrhizal symbioses (Prayitno et al., 2006; Penmetsa et al., 2008; Foo et al., 2014). Given the observed extended zone responsive for colonization (in AM and nitrogen fixing symbioses) in the pub1-1 mutant, it is clear that PUB1 participates in a negative regulation aimed at limiting the overcolonization through a systemic or local control. Such a local control has already been proposed by Lauressergues et al. (2012), with a mechanism involving mi-RNA restricting colonization in root tips by AM fungi.
PUB1 expression is found associated with infected cells or those in close vicinity all along the symbiotic processes during both rhizobial and AM symbioses (this work and Mbengue et al., 2010; Roux et al., 2014). This suggests that PUB1 plays a role in modulating events in cells directly involved in or preparing for colonization. This could be to limit the cost to the plant and to spatially control colonization, two prerequisites essential for successful endosymbioses (Oldroyd, 2013). Plant cells actively prepare for intracellular accommodation of both rhizobial bacteria and AM fungi by the formation of preinfection structures (Genre et al., 2005, 2008). Thereafter, the development of plant-microbe interfaces involve an intense intracellular extension of the plasma membrane during IT or hyphae growth that involves common processes associated with extensive membrane dynamics controlled by the exocytotic machinery (Genre et al., 2012; Ivanov et al., 2012; Zhang et al., 2015b). The exocyst complex is part of the exocytotic machinery, and EXO70A1, a subunit of the exocyst complex, interacts with an ARM Repeat Containing 1 (ARC1), an UND-PUB-ARM protein, characterized in the self-incompatibility process in Brassica napus as a partner of the RLK s-locus receptor kinase (Gu et al., 1998; Samuel et al., 2009). Because of the similar domain structure and high sequence homology between ARC1 and PUB1, it is possible that PUB1 targets one or several members of the exocyst complex.
The U-box domain integrity is necessary for the biological function of PUB1. Based on the lack of in vitro ubiquitination activity of PUB1D308N, we conclude that the ubiquitination activity of PUB1 is essential for its symbiotic functions. However, we cannot exclude that this point mutation may also affect PUB1 stability, its interaction with DMI2 and/or LYK3, and/or its phosphorylation by the two receptors. Neither here nor in our previous studies (Mbengue et al., 2010) were we able to detect ubiquitination of LYK3-IR or DMI2-IR in vitro, and the stability of the symbiotic receptors was not obviously affected by the presence of PUB1 in planta. These data suggest that LYK3 and DMI2 downstream signaling could be down-regulated by a functional PUB1 during the rhizobial and AM symbioses, rather than PUB1 affecting the receptors themselves. In contrast, recent data show that two other E3 ubiquitin ligases (SIE3 and SINA4) control SYMRK stability (Den Herder et al., 2012; Yuan et al., 2012). PUB1 is structurally very different from SINA4 and SIE3 and may thus have a different mode of action. An open question is whether PUB1 has any functional relationship with SINA4 and SIE3 orthologs from M. truncatula to regulate DMI2 together.
Another hypothesis is that phosphorylation of PUB1 by DMI2 and LYK3 modulates the ubiquitination activity of PUB1 leading to changes in downstream signaling in both endosymbioses. It would be interesting to determine whether differential phosphorylation of PUB1 by DMI2 and LYK3 takes place in nodulation and AM symbioses, and whether this could participate in the specific regulation of mycorrhizal or rhizobial symbioses. Alternatively, equivalent phosphorylation of PUB1 by DMI2 and LYK3 could lead to a common mechanism of regulation of both endosymbioses. If DMI2 and/or LYK3 modulate the activity of PUB1 by phosphorylation during symbioses, we hypothesize that PUB1 could interact with and ubiquitinate downstream components in pathways leading to nodulation and/or AM fungal colonization. This ubiquitination could lead either to degradation of positive regulators or activation of negative regulators. In plant innate immunity, the RLK BAK1 directly phosphorylates the PUB1-like proteins PUB12 and PUB13 in Arabidopsis, and this phosphorylation is required for flg22-induced FLS2-PUB12/13 association (Lu et al., 2011). By analogy, phosphorylation of PUB1 by DMI2 and/or LYK3 could lead to interaction between PUB1 and other symbiotic components. In cell signaling regulation, interaction between ubiquitination and phosphorylation events is emerging as a common mechanism (Nguyen et al., 2013; Ryšlavá et al., 2013).
In conclusion, this work shows that, in addition to its role in nodulation, PUB1 is also an important regulator of the AM symbiosis. It negatively modulates early infection and colonization by both R. irregularis and rhizobium and interacts with the two symbiotic receptors DMI2 and LYK3 (Mbengue et al., 2010). PUB1 does not appear to affect the stability of LYK3 and DMI2, but the ubiquitination activity of PUB1 is crucial for its biological role. Understanding the molecular mechanisms by which PUB1 regulates both endosymbioses will help to decipher the commonalities of how rhizobial and AM fungal colonization are regulated. Identification of downstream partners of PUB1 should help in this way.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Medicago truncatula Jemalong A17 (wild type) and L416 (Journet et al., 2001) were used in this study. A homozygous line for Plant U-box E3 ubiquitin ligase 1 (PUB1) mutation was identified among a TILLING population of M. truncatula by the UMR1347 platform of INRA Dijon (Le Signor et al., 2009) and crossed with the L416 line. A homozygous line for both the PUB1 mutation and the ProMtENOD11-gusA gene was selected and used in this study (pub1-1). For all the experiments using the pub1-1 line, the control line (wild type) used was the L416 line.
Stable M. truncatula transgenic plants were obtained in the 2HA accession by transformation with Agrobacterium tumefaciens AGL1 as described in Chabaud et al. (2003). Ten independent stable plants containing a fusion of the 3-kb promoter region of PUB1 to the β-glucuronidase reporter gene (ProPUB1-GUS; Mbengue et al., 2010) were positively selected by PCR analyses. Three lines containing this construct, to our knowledge, never gave seeds. Five of these lines showed a β-glucuronidase reporter gene (GUS) expression in young nodule, similar to our previous observations in Agrobacterium rhizogenes-transformed roots expressing the same construct (Mbengue et al., 2010) but with different intensities in the response. Three of these lines were tested in AM symbiosis, and all showed a similar response. We have chosen the M66D8 line, which has a response of high intensity to perform contrasted pictures with the double-staining (magenta-Gluc substrate and ink). Three independent stable plants containing the ProDMI2-DMI2-3FLAG expressing the BRC1720 fragment described in Endre et al. (2002) with a 3 FLAG-tag at the 3′ end (A. Kereszt and G. Endre, unpublished) were obtained and only two produced seeds. The protein DOES NOT MAKE INFECTIONS 2 (DMI2)-3FLAG was expressed in these two lines. Because no good antibodies against the DMI2 protein of M. truncatula are available, the comparison with the expression of the endogenous protein is not possible by this method. However, comparison of total protein extracts from roots with or without nodules between our lines and the well-characterized stable line gDMI2:HAST published by Riely et al. (2013) showed similar results on western blot.
For root phenotyping, plants were grown individually in the Falcon tube system described by Maillet et al. (2011). Four independent replicates with at least 10 plants for each line were done for the experiments on noninoculated plants and two independent replicates with at least 20 plants for each line were done on inoculated plants. Root length was measured using a model no. 10000XL scanner (Epson, Suwa, Nagano, Japan) and the Winrhizo Scientific Software image analysis system (Regent Instruments, St. Foy, Quebec, Canada).
A. rhizogenes root transformations were done as described by Boisson-Dernier et al. (2005). Transformed roots were selected on kanamycin medium and/or by observation of DsRED fluorescence depending on the vectors used. All composite roots where then analyzed individually by western blot with corresponding antibodies.
Arbuscular Mycorrhizal Symbiosis Assays
Arbuscular mycorrhizal (AM) experiments were performed independently of nodulation experiments. Using sterile spores of Rhizophagus irregularis DAOM197198, at least three independent kinetics were performed at 2, 3, and 5 wpi on at least 10 plants of each genotype at each point with 4000 spores L−1 in the Falcon tube system described in Maillet et al. (2011). For low inoculum experiments, three independent repeats were performed with five plants of each genotype inoculated with 350 spores L−1 for 6 weeks as described by Lauressergues et al. (2012). Fungi were stained with Schaeffer black ink (Vierheilig et al., 1998). Colonization was estimated by the gridline intersect method on the entire root system (Giovannetti and Mosse, 1980) and then specified by the Trouvelot method (Trouvelot et al., 1986). Spatial localization was estimated on the root system divided in three transversal zones named A, B, and C, as symbolized in Figure 4F.
Nodulation Assays
Plant inoculations were done with Sinorhizobium meliloti GMI6526 (10 mL of an OD600 = 0.025) in small pots containing granules support (Oil Dry US Special; Brenntag, Mülheim, Germany). For phenotyping experiments, at least 20 plants were scored at 4 wpi for ITs, infected primordia, and nodules in two independent replicates. Spatial localization of nodules was estimated according to Figure 4F.
Complementation and Overexpression Assays
Three weeks after A. rhizogenes transformation, M. truncatula plants were transferred to granule support-containing small pots in propagator for nodulation analysis or in the Falcon system described by Maillet et al. (2011) for AM. Two independent replicates were performed with at least five plants per genotype per experiment. Plants were inoculated with 1000 spores L−1 and scored (as previously described) at 3.5 wpi. pCambia2200GG (Fliegmann et al., 2013) and pK7WG2-R-pUb (described below) were used as empty vectors, in pub1-1 complementation and other assays, respectively.
Histochemical Staining
AM colonization was performed on M66D8 plants as described in the AM symbiosis assays section and the noninoculated plants were grown in the same conditions. Three independent repeats were analyzed with at least five plants per condition per replicate. M66D8 plants and composite plants expressing the ProDMI2-GUS (Bersoult et al., 2005) were also inoculated with R. irregularis inoculum prepared as described in Journet et al. (2001) and roots were stained 10 d post-inoculation (dpi). GUS staining was performed as described by Mbengue et al. (2010). For plants inoculated with S. meliloti GMI6526, bacterial LacZ activity was performed as described by Bersoult et al., (2005). Observations were made with an Axioplan 2 microscope (Carl Zeiss, Jena, Germany) and images were taken with a digital camera and software (Leica, Wetzlar, Germany).
Yeast Two-Hybrid Assays
Sequence corresponding to the intracellular region (IR) of DMI2 (DMI2-IR), amino acids 544–924, was PCR-amplified; cloned by restriction in the pBD vector (Clontech Laboratories, Fitchburg, WI); and transformed into the yeast AH109. The screen was performed as described by Mbengue et al. (2010) using the M. truncatula Nod factor-elicited root hair cell cDNA library (Andriankaja et al., 2007). Domain mapping was carried out with PUB1U-box domain (Ubox) ARMADILLO (ARM; amino acids 271–694), PUB1ARM (amino acids 397–694), and PUB1UNDUbox (amino acids 1–387) as described by Mbengue et al. (2010).
Plasmid Construction
3× human influenza hemagglutinin (3HA)-PUB1, 3HA-PUB1UNDUbox, and 3HA-PUB1UND were cloned in pCambia2200GG under control of the cauliflower mosaic virus 35S promoter (2X35S) by golden gate strategy (Engler et al., 2009) and were introduced into A. tumefaciens LBA4404 and A. rhizogenes ARqua1 by electroporation. By similar technique, 3HA-PUB1 was cloned in pCambia2200GG under control of the PUB1 (3 Kb) promoter and introduced into A. rhizogenes ARqua1 strain. By using restriction sites, DNA corresponding to amino acids 544–924 of the DMI2-IR domain (EcoRI and SalI) and full-size PUB1 (SstI and SalI) were cloned in pMal-c2 to produce MALTOSE BINDING PROTEIN fusion proteins (BioLabs, Beverly, MA). Sequence corresponding to amino acids 271–694 of PUB1D308N was cloned using the Gateway Technology Cloning System (Thermo Fisher Scientific, Waltham, MA) into a pGEX vector to produce GST fusion proteins. Pro-35S-DMI2 (containing the first intron)-RFP from the lab of G. Endre has been subcloned in pBin. ProUb-GFP-PUB1 was created using the Gateway Technology Cloning System (Thermo Fisher Scientific) and the destination vector pK7WGF2-R-pUb. pK7WGF2-R-pUb derives from a pK7WG2-R (Ding et al., 2008) destination vector that was modified to add the GFP fusion tag from pK7WGF2 (Karimi et al., 2002) using SpeI and AatII restriction sites and replace the 35S promoter with pUb (pLjUb 553 bp from ATG (Maekawa et al., 2008) using SpeI-HindIII sites. Construct Leu-rich repeat II.1-YFP is described by Lefebvre et al., 2010; Pro-35SDMI2-mCherry has been subcloned into pBin+ 35S-mCherry with NheI and KpnI from the published CaMV 35S:DMI2-sYFP2 (Pietraszewska-Bogiel et al., 2013).
Transient Expression in Nicotiana benthamiana
Overnight bacterial cultures of A. tumefaciens LBA4404 were harvested by centrifugation. Cells were resuspended in induction buffer (10 mm MgCl2, 10 mm MES, pH5.6, and 50 μM acetosyringone) to an OD600 = 0.25 and coinoculated with p19 strain (Voinnet et al., 2003) diluted to OD600 = 0.1. Three days after infiltration, leaves were harvested.
Protein Extraction and Coimmunopurification
Total extract of N. benthamiana leaves were prepared as described in Mbengue et al. (2010). For M. truncatula, roots were ground in liquid nitrogen and proteins were solubilized directly by adding 2% SDS gel loading buffer and heating at 95°C for 5 min.
For coimmunopurification, protein complexes from tissues crushed in liquid nitrogen were solubilized 30 min at 4°C (gentle rotation) in IP buffer (25 mm HEPES pH7.5, 150 mm NaCl, 10% glycerol, 10 mm EDTA, 1 mm DTT, 100 μM iodoacetamide, protease inhibitor cocktail; see Mbengue et al., 2010) at a proportion of 1:4 (weight:volume) containing 0.2% of detergent n-dodecyl-β-d-maltoside (DDM; Enzo Life Sciences, Farmingdale, NY) and cocktail of inhibitor of phosphatase 1 mm Na4P2O7 and β-glycerol phosphate, 2 mm Na3VO4, 5 mm NaF. Samples were centrifuged (45 min at 47,000 g), and the supernatant (solubilized fraction) was diluted twice in IP buffer. For co-immunopurification, diluted supernatant was incubated either with 25 μL of washed magnetic beads coupled to anti-RFP or GFP (RFP-M and GFP-M; ChromoTek, Planegg, Germany) or with 20 μL of washed agarose anti-HA beads (Sigma-Aldrich, St. Louis, MO; 2 h at 4°C, gentle rotation) followed by magnetic separation or by centrifugation (30 s at 500 g). Beads were washed three times with IP buffer and proteins purified were eluted in 30 μL of 2× Laemmli buffer (95°C 5 min).
For analysis of DMI2 stability in M. truncatula, composite roots were harvested 17 dpi with S. meliloti (same growth conditions as in “Nodulation Assays”); total proteins were extracted from three biological replicates for each construct (with at least four plants pooled per replicate) and analyzed by western blot.
Expression and Purification of Escherichia coli Fusion Proteins
MBP-PUB1 and MBP-DMI2-IR proteins were purified from E. coli cell cultures DH5α and BL21, respectively, according to the manufacturer’s instructions (New England Biolabs, Ipswich, MA) with some modifications. After IPTG induction, cells were incubated at 16°C for 24 h, then collected, frozen at −20°C, and resuspended in ice-cold lysis buffer (10 mm of sodium phosphate buffer pH 7.2, 0.5 m NaCl, 10 mm β-mercaptoethanol, 10 mm EDTA), supplemented with protease inhibitors, and sonicated. The soluble fraction was incubated with prewashed amylose resin overnight at 4°C under agitation. Bound proteins on Amylose resin were washed with lysis buffer without EDTA. Proteins were eluted in wash buffer containing 20 mm of maltose. Fractions of elution were analyzed on SDS-PAGE gel and stained with InstantBlue (Expedeon, San Diego, CA). Fractions containing proteins were concentrated if necessary and buffer was changed (Tris 10 mm pH7.5, 1 mm DTT) using Amicon Ultra-2 K10 (Millipore, Billerica, MA). PUB1UboxARM- and PUB1UboxD308NARM glutathione s-transferase (GST)-fused proteins were expressed in E. coli strain DH5α and purifications were performed as described in Mbengue et al. (2010).
In Vitro Phosphorylation Assay
For transphosphorylation assays, about 1 μg of each purified protein was used. Proteins were incubated in 20 μL of 50 mm HEPES, pH7.4, 10 mm MgCl2, 10 mm MnCl2, 1 mm DTT, 1 μM ATP, and 10 μCi γ-P32 ATP at 30°C for 2 h. Reactions were stopped by adding Laemmli buffer and heating at 95°C for 5 min. Reactions were analyzed by SDS-PAGE stained with InstantBlue and gels were analyzed by phosphor imaging.
In Vitro Ubiquitination Assay
In vitro ubiquitination assays were performed as previously described in Mbengue et al. (2010). Quantification of proteins was done using ImageJ software (National Institutes of Health, Bethesda, MD).
Immunoblotting
Antibodies used for western blot are indicated in each figure with the following dilutions: Anti-HA-horseradish peroxidase (HRP) (1:1000; Roche, Basel, Switzerland); anti-glutathione s-transferase-HRP (1:10,000; GE Healthcare, Port Washington, NY); anti-FLAG-HRP (1:5000; Sigma-Aldrich); rabbit anti-GFP (1:7500; AMS Biotechnology, Abingdon, Oxfordshire, UK); rabbit anti-RFP (1:5000; Lefebvre et al., 2012); rabbit anti-MBP (1:7500; New England Biolabs); and anti-rabbit IgG-HRP (1:20,000; Millipore). HRP was detected using the SuperSignal West Dura Extended Duration Substrate (Thermo Scientific) and a digital camera (G-box; Syngene, Cambridge, UK).
Supplemental Data
The following materials are available in the on-line version of this article.
Supplemental Figure S1. PUB1 interacts specifically with DMI2 but does not affect DMI2 stability in planta.
Supplemental Figure S2. The D308N mutation in the U-box domain abolishes the ubiquitin ligase activity of the PUB1 protein.
Supplemental Figure S3. DMI2is expressed during AM symbiosis.
Supplemental Figure S4. Overexpression of PUB1 in Medicago roots.
Supplemental Figure S5. The pub1-1 root architecture.
Supplemental Figure S6. The ProUb-GFP-PUB1 construct complements the pub1-1 AM root colonization phenotype.
Supplemental Figure S7. PUB1 negatively regulates AM fungal entry and the root-responsive zone.
Supplemental Figure S8. The stable line ProPUB1-GUS shows PUB1 induction during nodulation.
Supplementary Material
Acknowledgments
From the Laboratoire des Interactions Plantes-Microorganismes, we thank Clare Gough and Benjamin Gourion for critical reading of the manuscript and for providing helpful comments; Charles Rosenberg for initiating golden gate cloning in our group and providing us with the pCambia2200 derivative vector; and Susana Rivas for providing UBC8-purified protein of Arabidopsis. We thank Gabriella Endre (Institute of Genetics, Hungry) and Anna Pietraszewska-Bogiel (University of Amsterdam, The Netherlands) for the ProDMI2-DMI2-3FLAG construct and CaMV 35S:DMI2-mCherry, respectively.
Glossary
- 3HA
3× human influenza hemagglutinin
- AM
arbuscular mycorrhizal
- ARC
ARM Repeat Containing 1
- ARM
ARMADILLO
- co-IP
coimmunopurification
- CSSP
common symbiosis signaling pathway
- DMI2
DOES NOT MAKE INFECTIONS 2
- GST
glutathione s-transferase
- GUS
β-glucuronidase reporter gene
- IR
intracellular region
- IT
infection thread
- LCO
lipo-chitooligosaccharide
- LRR
Leu-rich repeat
- LYK3
LYSIN MOTIF RECEPTOR-LIKE KINASE 3
- MBP
MALTOSE BINDING PROTEIN
- NF
Nod factor
- PUB
Plant U-box E3 ubiquitin ligases
- RLK
receptor-like kinase
- SIE3
SymRK-INTERACTING E3 ubiquitin ligase
- SINA4
SEVEN IN ABSENTIA 4
- SYMRK
symbiosis receptor kinase
- Ub
ubiquitin
- U-box, Ubox
U-box domain
- UND
UND domain
Footnotes
This work was funded by Agence Nationale Recherche projects Symnaling (no. ANR-12-BSV7-0001) and MycSignalling (no. ANR-09-BLAN-0241-01), and performed at the Laboratoire des Interactions Plantes Microorganismes, part of the Laboratoire d’Excellence program TULIP (no. ANR-10-LABX-41; no. ANR-11-IDEX-0002-02). The TILLING program was done at the Institut National de la Recherche Agronomique-UMR1347-Agroecologie and was supported by the Grain Legumes Integrated Project (no. FOOD-CT-2004-506223) of the European Commission FP6 Framework Program.
References
- Amor BB, Shaw SL, Oldroyd GED, Maillet F, Penmetsa RV, Cook D, Long SR, Dénarié J, Gough C (2003) The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J 34: 495–506 [DOI] [PubMed] [Google Scholar]
- Andriankaja A, Boisson-Dernier A, Frances L, Sauviac L, Jauneau A, Barker DG, de Carvalho-Niebel F (2007) AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19: 2866–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antolín-Llovera M, Ried MK, Parniske M (2014) Cleavage of the SYMBIOSIS RECEPTOR-LIKE KINASE ectodomain promotes complex formation with Nod factor receptor 5. Curr Biol 24: 422–427 [DOI] [PubMed] [Google Scholar]
- Barberon M, Zelazny E, Robert S, Conéjéro G, Curie C, Friml J, Vert G (2011) Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc Natl Acad Sci USA 108: E450–E458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersoult A, Camut S, Perhald A, Kereszt A, Kiss GB, Cullimore JV (2005) Expression of the Medicago truncatula DM12 gene suggests roles of the symbiotic nodulation receptor kinase in nodules and during early nodule development. Mol Plant Microbe Interact 18: 869–876 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Andriankaja A, Chabaud M, Niebel A, Journet EP, Barker DG, de Carvalho-Niebel F (2005) MtENOD11 gene activation during rhizobial infection and mycorrhizal arbuscule development requires a common AT-rich-containing regulatory sequence. Mol Plant Microbe Interact 18: 1269–1276 [DOI] [PubMed] [Google Scholar]
- Camps C, Jardinaud M-F, Rengel D, Carrère S, Hervé C, Debellé F, Gamas P, Bensmihen S, Gough C (2015) Combined genetic and transcriptomic analysis reveals three major signalling pathways activated by Myc-LCOs in Medicago truncatula. New Phytol 208: 224–240 [DOI] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Dénarié J (2000) Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell 12: 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabaud M, de Carvalho-Niebel F, Barker DG (2003) Efficient transformation of Medicago truncatula cv. Jemalong using the hypervirulent Agrobacterium tumefaciens strain AGL1. Plant Cell Rep 22: 46–51 [DOI] [PubMed] [Google Scholar]
- Chen T, Zhu H, Ke D, Cai K, Wang C, Gou H, Hong Z, Zhang Z (2012) A MAP kinase kinase interacts with SymRK and regulates nodule organogenesis in Lotus japonicus. Plant Cell 24: 823–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaux PM, Bécard G, Combier JP (2013) NSP1 is a component of the Myc signaling pathway. New Phytol 199: 59–65 [DOI] [PubMed] [Google Scholar]
- Den Herder G, Yoshida S, Antolín-Llovera M, Ried MK, Parniske M (2012) Lotus japonicus E3 ligase SEVEN IN ABSENTIA4 destabilizes the symbiosis receptor-like kinase SYMRK and negatively regulates rhizobial infection. Plant Cell 24: 1691–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Kalo P, Yendrek C, Sun J, Liang Y, Marsh JF, Harris JM, Oldroyd GED (2008) Abscisic acid coordinates Nod factor and cytokinin signaling during the regulation of nodulation in Medicago truncatula. Plant Cell 20: 2681–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kaló P, Kiss GB (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966 [DOI] [PubMed] [Google Scholar]
- Engler C, Gruetzner R, Kandzia R, Marillonnet S (2009) Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One 4: e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegmann J, Canova S, Lachaud C, Uhlenbroich S, Gasciolli V, Pichereaux C, Rossignol M, Rosenberg C, Cumener M, Pitorre D, Lefebvre B, Gough C, et al. (2013) Lipo-chitooligosaccharidic symbiotic signals are recognized by LysM receptor-like kinase LYR3 in the legume Medicago truncatula. ACS Chem Biol 8: 1900–1906 [DOI] [PubMed] [Google Scholar]
- Foo E, Ferguson BJ, Reid JB (2014) Common and divergent roles of plant hormones in nodulation and arbuscular mycorrhizal symbioses. Plant Signal Behav 9: e29593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Balzergue C, Puech-Pagès V, Novero M, Rey T, Fournier J, Rochange S, Bécard G, Bonfante P, Barker DG (2013) Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol 198: 190–202 [DOI] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Faccio A, Barker DG, Bonfante P (2008) Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell 20: 1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG (2005) Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 17: 3489–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A, Ivanov S, Fendrych M, Faccio A, Zársky V, Bisseling T, Bonfante P (2012) Multiple exocytotic markers accumulate at the sites of perifungal membrane biogenesis in arbuscular mycorrhizas. Plant Cell Physiol 53: 244–255 [DOI] [PubMed] [Google Scholar]
- Giovannetti M, Mosse B (1980) An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol 84: 489–500 [Google Scholar]
- Gough C, Cullimore J (2011) Lipo-chitooligosaccharide signaling in endosymbiotic plant-microbe interactions. Mol Plant Microbe Interact 24: 867–878 [DOI] [PubMed] [Google Scholar]
- Gu T, Mazzurco M, Sulaman W, Matias DD, Goring DR (1998) Binding of an arm repeat protein to the kinase domain of the s-locus receptor kinase. Proc Natl Acad Sci USA 95: 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust AA, Willmann R, Desaki Y, Grabherr HM, Nürnberger T (2012) Plant LysM proteins: modules mediating symbiosis and immunity. Trends Plant Sci 17: 495–502 [DOI] [PubMed] [Google Scholar]
- Gutjahr C, Parniske M (2013) Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu Rev Cell Dev Biol 29: 593–617 [DOI] [PubMed] [Google Scholar]
- Herberth S, Shahriari M, Bruderek M, Hessner F, Müller B, Hülskamp M, Schellmann S (2012) Artificial ubiquitylation is sufficient for sorting of a plasma membrane ATPase to the vacuolar lumen of Arabidopsis cells. Planta 236: 63–77 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1992) The ubiquitin system for protein degradation. Annu Rev Biochem 61: 761–807 [DOI] [PubMed] [Google Scholar]
- Huault E, Laffont C, Wen J, Mysore KS, Ratet P, Duc G, Frugier F (2014) Local and systemic regulation of plant root system architecture and symbiotic nodulation by a receptor-like kinase. PLoS Genet 10: e1004891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S, Fedorova EE, Limpens E, De Mita S, Genre A, Bonfante P, Bisseling T (2012) Rhizobium-legume symbiosis shares an exocytotic pathway required for arbuscule formation. Proc Natl Acad Sci USA 109: 8316–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet EP, El-Gachtouli N, Vernoud V, de Billy F, Pichon M, Dedieu A, Arnould C, Morandi D, Barker DG, Gianinazzi-Pearson V (2001) Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol Plant Microbe Interact 14: 737–748 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81: 203–229 [DOI] [PubMed] [Google Scholar]
- Kosuta S, Held M, Hossain MS, Morieri G, Macgillivary A, Johansen C, Antolín-Llovera M, Parniske M, Oldroyd GED, Downie AJ, Karas B, Szczyglowski K (2011) Lotus japonicus symRK-14 uncouples the cortical and epidermal symbiotic program. Plant J 67: 929–940 [DOI] [PubMed] [Google Scholar]
- Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau O-S, Deng X-W, Callis J (2005) Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol 139: 1597–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrainzar E, Riely BK, Kim SC, Carrasquilla-Garcia N, Yu HJ, Hwang HJ, Oh M, Kim GB, Surendrarao AK, Chasman D, Siahpirani AF, Penmetsa RV, et al. (2015) Deep sequencing of the Medicago truncatula root transcriptome reveals a massive and early interaction between nodulation factor and ethylene signals. Plant Physiol 169: 233–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauressergues D, Delaux P-M, Formey D, Lelandais-Brière C, Fort S, Cottaz S, Bécard G, Niebel A, Roux C, Combier J-P (2012) The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J 72: 512–522 [DOI] [PubMed] [Google Scholar]
- Lefebvre B, Klaus-Heisen D, Pietraszewska-Bogiel A, Hervé C, Camut S, Auriac M-C, Gasciolli V, Nurisso A, Gadella TWJ, Cullimore J (2012) Role of n-glycosylation sites and CXC motifs in trafficking of Medicago truncatula Nod factor perception protein to plasma membrane. J Biol Chem 287: 10812–10823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre B, Timmers T, Mbengue M, Moreau S, Hervé C, Tóth K, Bittencourt-Silvestre J, Klaus D, Deslandes L, Godiard L, Murray JD, Udvardi MK, et al. (2010) A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc Natl Acad Sci USA 107: 2343–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé JC, Dénarié J (1990) Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344: 781–784 [DOI] [PubMed] [Google Scholar]
- Le Signor C, Savois V, Aubert G, Verdier J, Nicolas M, Pagny G, Moussy F, Sanchez M, Baker D, Clarke J, Thompson R (2009) Optimizing TILLING populations for reverse genetics in Medicago truncatula. Plant Biotechnol J 7: 430–441 [DOI] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R (2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630–633 [DOI] [PubMed] [Google Scholar]
- Limpens E, Mirabella R, Fedorova E, Franken C, Franssen H, Bisseling T, Geurts R (2005) Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc Natl Acad Sci USA 102: 10375–10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Kusakabe M, Shimoda Y, Sato S, Tabata S, Murooka Y, Hayashi M (2008) Polyubiquitin promoter-based binary vectors for overexpression and gene silencing in Lotus japonicus. Mol Plant Microbe Interact 21: 375–382 [DOI] [PubMed] [Google Scholar]
- Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, Martinez EA, Driguez H, et al. (2011) Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63 [DOI] [PubMed] [Google Scholar]
- Mbengue M, Camut S, de Carvalho-Niebel F, Deslandes L, Froidure S, Klaus-Heisen D, Moreau S, Rivas S, Timmers T, Hervé C, Cullimore J, Lefebvre B (2010) The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell 22: 3474–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M, Sorkin A (2007) Regulation of receptors and transporters by ubiquitination: new insights into surprisingly similar mechanisms. Mol Interv 7: 157–167 [DOI] [PubMed] [Google Scholar]
- Nadal M, Paszkowski U (2013) Polyphony in the rhizosphere: presymbiotic communication in arbuscular mycorrhizal symbiosis. Curr Opin Plant Biol 16: 473–479 [DOI] [PubMed] [Google Scholar]
- Nguyen LK, Kolch W, Kholodenko BN (2013) When ubiquitination meets phosphorylation: a systems biology perspective of EGFR/MAPK signalling. Cell Commun Signal 11: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED. (2013) Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11: 252–263 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Murray JD, Poole PS, Downie JA (2011) The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45: 119–144 [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Uribe P, Anderson J, Lichtenzveig J, Gish JC, Nam YW, Engstrom E, Xu K, Sckisel G, Pereira M, Baek JM, Lopez-Meyer M, et al. (2008) The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations. Plant J 55: 580–595 [DOI] [PubMed] [Google Scholar]
- Pietraszewska-Bogiel A, Lefebvre B, Koini MA, Klaus-Heisen D, Takken FLW, Geurts R, Cullimore JV, Gadella TWJ (2013) Interaction of Medicago truncatula lysin motif receptor-like kinases, NFP and LYK3, produced in Nicotiana benthamiana induces defence-like responses. PLoS One 8: e65055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prayitno J, Rolfe BG, Mathesius U (2006) The ethylene-insensitive sickle mutant of Medicago truncatula shows altered auxin transport regulation during nodulation. Plant Physiol 142: 168–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riely BK, Larrainzar E, Haney CH, Mun JH, Gil-Quintana E, González EM, Yu HJ, Tricoli D, Ehrhardt DW, Long SR, Cook DR (2013) Development of tools for the biochemical characterization of the symbiotic receptor-like kinase DMI2. Mol Plant Microbe Interact 26: 216–226 [DOI] [PubMed] [Google Scholar]
- Roux B, Rodde N, Jardinaud MF, Timmers T, Sauviac L, Cottret L, Carrère S, Sallet E, Courcelle E, Moreau S, Debellé F, Capela D, et al. (2014) An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J 77: 817–837 [DOI] [PubMed] [Google Scholar]
- Ryšlavá H, Doubnerová V, Kavan D, Vaněk O (2013) Effect of posttranslational modifications on enzyme function and assembly. J Proteomics 92: 80–109 [DOI] [PubMed] [Google Scholar]
- Samuel MA, Chong YT, Haasen KE, Aldea-Brydges MG, Stone SL, Goring DR (2009) Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell 21: 2655–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaarschmidt S, Gresshoff PM, Hause B (2013) Analyzing the soybean transcriptome during autoregulation of mycorrhization identifies the transcription factors GmNF-YA1a/b as positive regulators of arbuscular mycorrhization. Genome Biol 14: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuring D, Künzl F, Viotti C, Yan MSW, Jiang L, Schellmann S, Robinson DG, Pimpl P (2012) Ubiquitin initiates sorting of Golgi and plasma membrane proteins into the vacuolar degradation pathway. BMC Plant Biol 12: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin C, Xie Z-P, Illana A, Vierheilig H (2011) Long-distance transport of signals during symbiosis: are nodule formation and mycorrhization autoregulated in a similar way? Plant Signal Behav 6: 372–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, Parniske M (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Takeda N, Tsuzuki S, Suzaki T, Parniske M, Kawaguchi M (2013) CERBERUS and NSP1 of Lotus japonicus are common symbiosis genes that modulate arbuscular mycorrhiza development. Plant Cell Physiol 54: 1711–1723 [DOI] [PubMed] [Google Scholar]
- Trouvelot A, Kough JL, Gianinazzi-Pearson V (1986). Mesure du taux de mycorhization VA d’un système radiculaire. Recherche de méthodes d’estimation ayant une signification fonctionnelle. In Physiological and General Aspects of Mycorrhizae, Gianinazzi-Pearson V. and Gianinazzi S., editors, INRA Press, Paris, France: 217–221 [Google Scholar]
- Venkateshwaran M, Volkening JD, Sussman MR, Ané J-M (2013) Symbiosis and the social network of higher plants. Curr Opin Plant Biol 16: 118–127 [DOI] [PubMed] [Google Scholar]
- Vierheilig H, Coughlan AP, Wyss U, Piche Y (1998) Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl Environ Microbiol 64: 5004–5007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Yee D, Goring DR (2009) The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J Exp Bot 60: 1109–1121 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Parniske M (2005) Regulation of plant symbiosis receptor kinase through serine and threonine phosphorylation. J Biol Chem 280: 9203–9209 [DOI] [PubMed] [Google Scholar]
- Yuan S, Zhu H, Gou H, Fu W, Liu L, Chen T, Ke D, Kang H, Xie Q, Hong Z, Zhang Z (2012) A ubiquitin ligase of symbiosis receptor kinase involved in nodule organogenesis. Plant Physiol 160: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Dong W, Sun J, Feng F, Deng Y, He Z, Oldroyd GED, Wang E (2015a) The receptor kinase CERK1 has dual functions in symbiosis and immunity signalling. Plant J 81: 258–267 [DOI] [PubMed] [Google Scholar]
- Zhang X, Pumplin N, Ivanov S, Harrison MJ (2015b) EXO70I is required for development of a sub-domain of the periarbuscular membrane during arbuscular mycorrhizal symbiosis. Curr Biol 25: 2189–2195 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.