Wounding induces expression of the NAC1 transcription factor that affects root tip emergence during de novo regeneration of adventitious root.
Abstract
Plants have powerful regenerative abilities that allow them to recover from damage and survive in nature. De novo organogenesis is one type of plant regeneration in which adventitious roots and shoots are produced from wounded and detached organs. By studying de novo root organogenesis using leaf explants of Arabidopsis (Arabidopsis thaliana), we previously suggested that wounding is the first event that provides signals to trigger the whole regenerative process. However, our knowledge of the role of wounding in regeneration remains limited. In this study, we show that wounding not only triggers the auxin-mediated fate transition of regeneration-competent cells, but also induces the NAC pathway for root tip emergence. The NAC1 transcription factor gene was specifically expressed in response to wounding in the leaf explant, but not in the wounded leaf residue of the source plant. Inhibition of the NAC1 pathway severely affected the emergence of adventitious root tips. However, the NAC1 pathway functioned independently of auxin-mediated cell fate transition and regulates expression of CEP genes, which encode proteins that might have a role in degradation of extensin proteins in the cell wall. Overall, our results suggest that wounding has multiple roles in de novo root organogenesis and that NAC1 acts as one downstream branch in regulating the cellular environment for organ emergence.
De novo organogenesis, through which detached or wounded plant organs regenerate adventitious roots and shoots, commonly occurs under natural conditions and allows plants to recover from damage (Duclercq et al., 2011; Xu and Huang, 2014). The ability of plants to undergo de novo organogenesis is also widely exploited in agricultural biotechnologies such as tissue culture and vegetative reproduction via cuttings (Sussex, 2008). In tissue culture, adventitious roots and shoots can regenerate from a group of pluripotent cell mass, termed callus. Callus is usually induced by a high level of auxin in the medium, and callus formation was shown to follow the rooting developmental pathway (Sugimoto et al., 2010; He et al., 2012; Liu et al., 2014).
By culturing leaf explants of the model plant Arabidopsis (Arabidopsis thaliana) on B5 medium without exogenous hormones, we established a simple method to study de novo root organogenesis mimicking natural conditions (Chen et al., 2014). In this system, adventitious roots can directly regenerate from the leaf explant under the control of endogenous hormones (Chen et al., 2014; Liu et al., 2014). The process of de novo root organogenesis from the leaf explant involves fate transition of regeneration-competent cells (i.e. procambium and vascular parenchyma cells) (Liu et al., 2014). The first fate-transition step from competent cells to root founder cells is controlled by the WUSCHEL-RELATED HOMEOBOX 11 (WOX11) transcription factor gene. The second fate-transition step is from root founder cells to root primordium cells and is marked by WOX5. The root apical meristem then differentiates from the root primordium and finally forms the adventitious root tip, which emerges from the leaf explant. Auxin was shown to play a critical role in promoting the fate transition of competent cells (Liu et al., 2014). Upon wounding, auxin is quickly produced in mesophyll cells of the detached leaf explant and is then polar-transported into competent cells near the wound to activate WOX11 expression, which triggers the first fate-transition step for competent cells. Therefore, activation of the hormone and gene expression networks can be traced back to the very beginning of the wounding event, in which the wound signals were proposed to be the initial molecules for regeneration (Liu et al., 2014; Xu and Huang, 2014). At present, the molecular nature of the wound signal(s) remains unclear.
The results of several studies have suggested that many physical events and chemical signals change quickly in response to wounding (León et al., 2001; Maffei et al., 2007). Other studies have characterized wound-responsive genes induced during plant regeneration, such as the WOUND INDUCED DEDIFFERENTIATION (WIND) family (Iwase et al., 2011a, 2011b). WIND genes are induced by wounding within several hours and are involved in cell dedifferentiation (Iwase et al., 2011a, 2011b). However, it is still unclear how wounding functions at the molecular level in plant de novo root organogenesis, and it is unknown whether wound signals are involved in processes other than the fate transition of competent cells.
In this study, we conducted a genome-wide transcriptome analysis of wounding in the leaf explant and identified that NAC1 (petunia NAM and Arabidopsis ATAF1, ATAF2, and CUC2), a member of the NAC family of transcription factor genes (Aida et al., 1997), is induced in response to wounding and functions in promotion of root rip emergence.
RESULTS
Identification of NAC Genes in Response to Explant-Specific Wounding
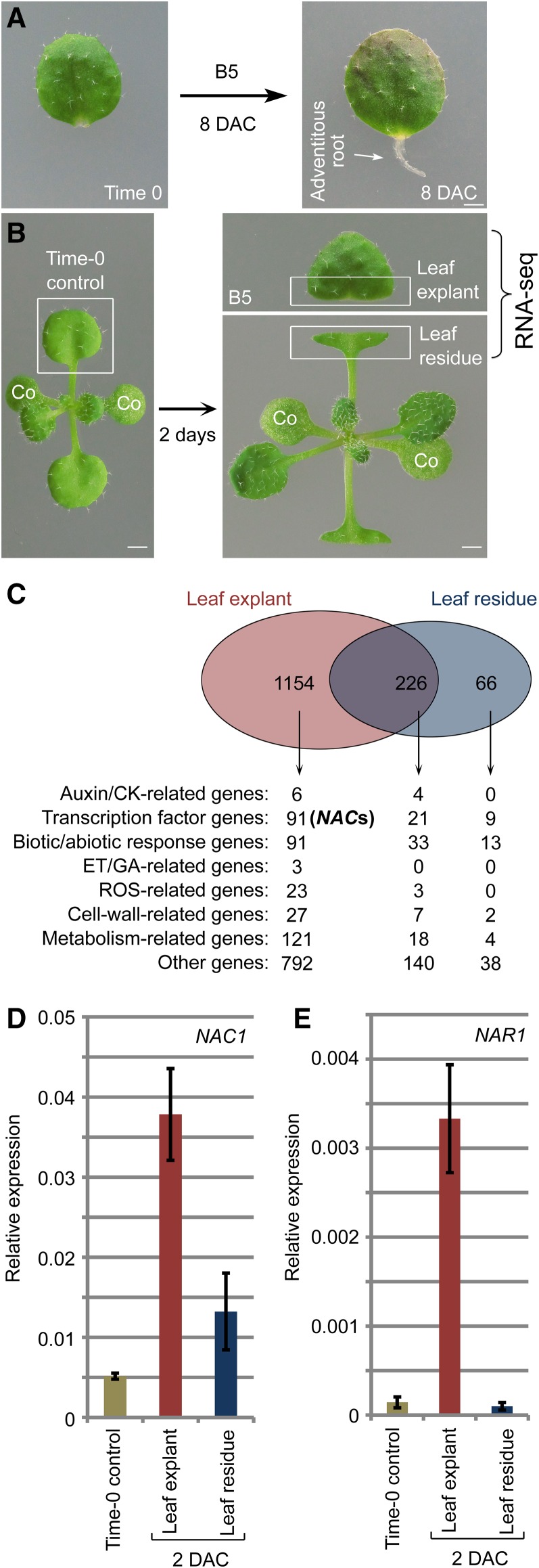
We previously established a de novo root organogenesis system using Arabidopsis leaf explants (Chen et al., 2014). In this system, detached leaf explants are cultured on B5 medium without exogenous hormones, and adventitious roots regenerate from the wounded region (Fig. 1A). Detachment of a leaf from a plant causes two wounds: one on the detached leaf explant and one on the leaf residue on the source plant. Adventitious roots usually regenerate from the detached leaf explant but not from the wounded leaf residue, suggesting that the two wounds have different molecular features. To study the effects of wounding on gene transcription during plant regeneration, we performed RNA-seq analyses on the two wounded tissues; the detached leaf explant cultured on B5 medium and the remaining wounded leaf residue at 2 d after culture (DAC). For both tissues, the RNA-seq data were compared with those obtained for the leaf prior to culture (time 0; Fig. 1B). All the tested materials were cultured under a 16-h-light/8-h-dark photoperiod.
Figure 1.
Identification of NAC genes induced in response to wounding. A, Regeneration system of de novo root organogenesis. B, Plant tissues used in RNA-seq analysis as indicated by the boxed regions. Leaf explant was cut and cultured on B5 medium without carbohydrates in light conditions. Wounded leaf residue was surrounded by air and did not touch the medium. co, cotyledon. C, RNA-seq data showing identification of NAC genes (also see Supplemental Table S1). D and E, qRT-PCR analysis of NAC1 (D) and NAR1 (E) transcript levels in leaf explants and leaf residue compared with those in time-0 leaf as indicated in B. Bars show sd from three PCR experiments. Results were confirmed in three biological repeats. Data from one repeat are shown. Bars = 1 mm in A and B.
The RNA-seq data revealed changes in gene expression patterns that were specific to the leaf explant, specific to the leaf residue, and common to both tissues (Fig. 1C; Supplemental Table S1). Because de novo root organogenesis usually occurs only in the leaf explant, we analyzed the leaf explant wounding-specific gene expression in more detail. Among many transcription factor genes showing changes in expression levels, we identified 12 NAC domain transcription factor genes (Ooka et al., 2003) that were significantly upregulated specifically in the leaf explant but not in the wounded leaf residue (Fig. 1C; Supplemental Table S1). We focused on the NAC family because this family is known to be involved in plant regeneration and lateral root formation (Xie et al., 2000; Asahina et al., 2011). To confirm the RNA-seq data, we performed quantitative RT-PCR (qRT-PCR) analyses of the closely related NAC genes, NAC1 and NAC1-RELATED1 (NAR1). The results showed that the two genes had relatively low transcript levels in time-0 leaf explants, but markedly higher transcript levels in 2-DAC leaf explants (Fig. 1, D and E). In addition, NAC1 was slightly upregulated in the 2-DAC leaf residue, while NAR1 was not (Fig. 1, D and E).
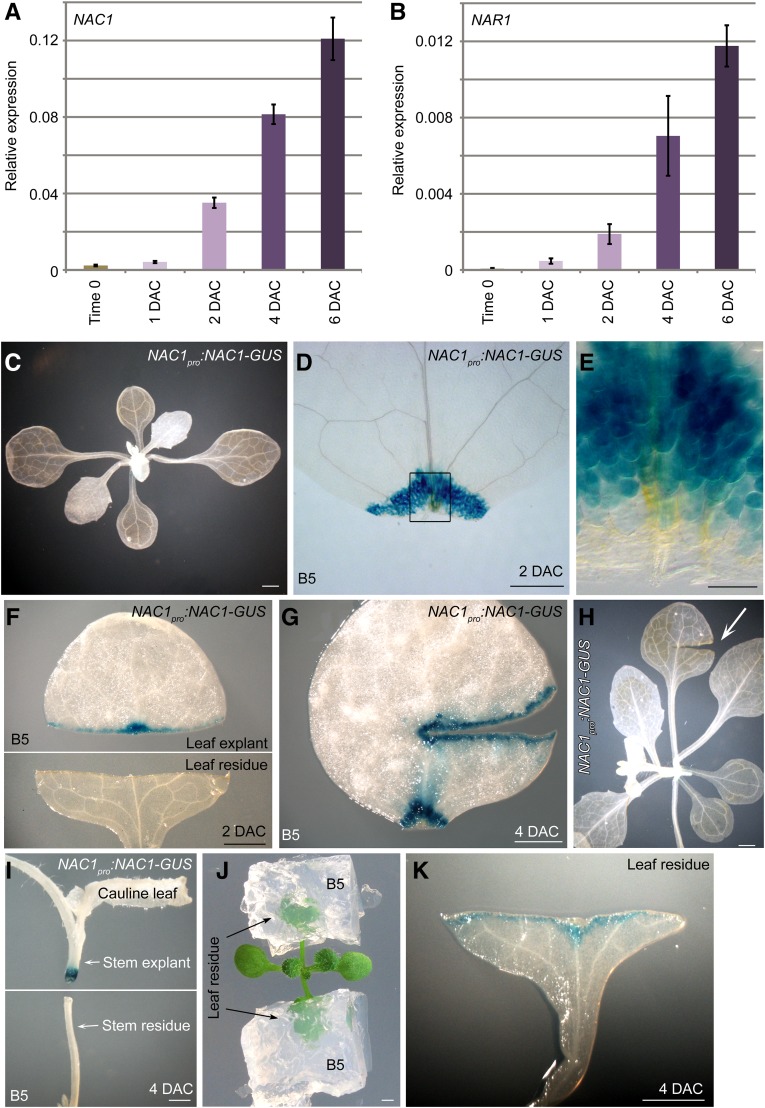
To analyze the expression patterns of NAC genes in more detail, we performed qRT-PCR analyses to detect NAC transcript levels during regeneration of leaf explants. The transcript levels of NAC1 and NAR1 were upregulated between 1 and 2 DAC and continuously upregulated afterward in leaf explants during de novo root organogenesis (Fig. 2, A and B). Next, we constructed the NAC1pro:NAC1-GUS reporter line. The GUS signal of the reporter line was barely detected in leaves at time 0 (Fig. 2C) but was strong and specific to the wounded region of leaf explants at 2 DAC (Fig. 2, D and E). The GUS signal was only observed in the wound of the leaf explant, and not in the wounded leaf residue on the source plant (Fig. 2F). Also, the GUS signal was present in multiple wounds in the detached leaf explant (Fig. 2G) but not in the wound on the attached leaf (Fig. 2H). To further examine the expression of NAC1 in the wounded region, we performed thin sectioning to show the GUS signals in the wounded region of the NAC1pro:NAC1-GUS leaf explant at 4 DAC. The result showed that the GUS signal was in both mesophyll cells and competent cells (Supplemental Fig. S1). In addition, the GUS signal was present in the wounded region of the detached stem explant but not in the wounded region of the stem from which it was derived (stem residue; Fig. 2I).
Figure 2.
Expression patterns of NAC genes. A and B, qRT-PCR analysis of NAC1 (A) and NAR1 (B) transcript levels in leaf explants from time 0 to 6 DAC. Bars show sd from three PCR experiments. Results were confirmed in two biological repeats. Data from one repeat are shown. C to E, GUS staining of NAC1pro:NAC1-GUS seedling (C) and 2-DAC leaf explant cultured on B5 medium (D and E). E, Enlargement of wounded region in D. F, GUS staining of NAC1pro:NAC1-GUS leaf explant (upper panel) and wounded leaf residue (lower panel) at 2 DAC. Cutting and culture times as indicated in Fig. 1B. G, Large wound on leaf explant from NAC1pro:NAC1-GUS cultured on B5 medium, showing GUS signal in wounded region. H, Large wound on seedling of NAC1pro:NAC1-GUS, showing no GUS signal in wounded region. I, Stem of NAC1pro:NAC1-GUS was cut, and stem explant (distal part) was cultured on B5 medium while wounded stem residue (proximal part) was still exposed to air. GUS signal was observed only in wounded region of stem explant. J and K, Wounded leaf residue was cultured in B5 medium (J), resulting in NAC1pro:NAC1-GUS signal at wounded region of leaf residue at 4 DAC (K). Bars = 1 mm in C and F to K, 500 μm in D, and 100 μm in E.
One of the differences between the detached leaf explant and the leaf residue was that the wound on the leaf explant touched the B5 medium, while that on the leaf residue did not touch B5 medium and was exposed to air. Therefore, we supplied B5 medium to the leaf residue (Fig. 2J) and observed that the wounded region of the leaf residue expressed NAC1 at 4 DAC (Fig. 2K). This result suggested that exposure of the wound to a wet environment could be very important for regeneration.
Overall, our data suggested that NAC1, together with NAR1 (Supplemental Fig. S2A), might be involved in an explant-specific wound-signaling pathway.
NAC1 Is Involved in de Novo Root Organogenesis
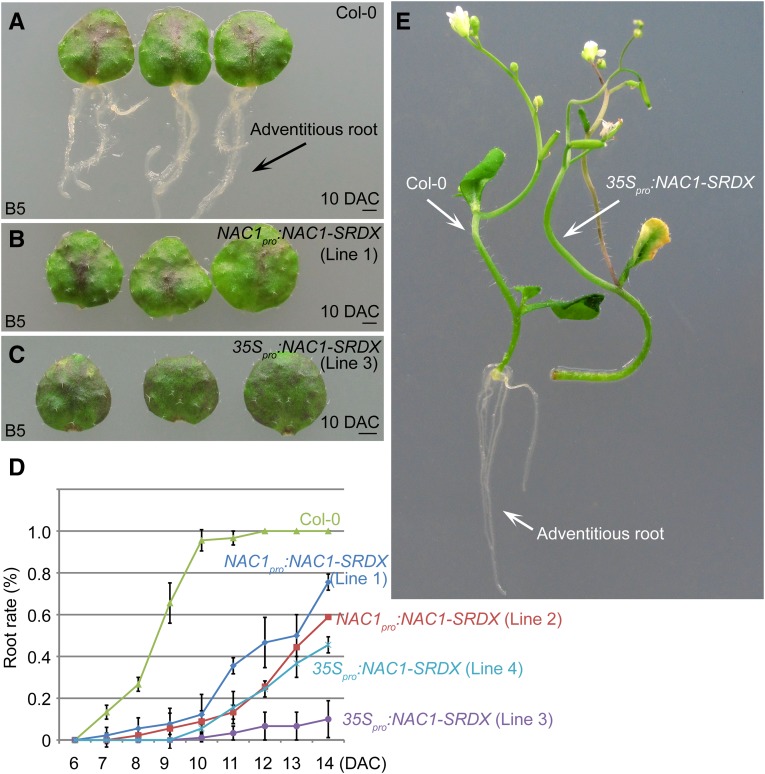
Because NAC genes were specifically induced in leaf explants, we explored their roles in de novo root organogenesis. The NAC transcription factors activate gene transcription (Xie et al., 2000). Therefore, we fused the NAC1 protein with the repression domain SRDX (Hiratsu et al., 2003) under the control of the NAC1 promoter to specifically suppress the expression of NAC1 target genes. This method also avoided redundancy among the NAC family genes. Compared with wild-type leaf explants that normally regenerate adventitious roots, leaf explants from the NAC1pro:NAC1-SRDX transgenic lines showed partially defective rooting (Fig. 3, A, B, and D). In the 35Spro:NAC1-SRDX lines harboring the chimeric NAC1-SRDX fusion under the control of the Cauliflower mosaic virus 35S promoter, rooting was more severely blocked (Fig. 3, C and D). In addition, regeneration of adventitious roots from stem explants was defective in 35Spro:NAC1-SRDX lines, compared with that in the wild type (Fig. 3E). Regeneration of adventitious roots from leaf explants of 35Spro:NAR1-SRDX lines was also defective (Supplemental Fig. S2B). These data suggested that the NAC pathway is involved in de novo root organogenesis in Arabidopsis.
Figure 3.
NAC1 is involved in de novo root organogenesis. A to C, Adventitious rooting from leaf explants of Col-0 (A), NAC1pro:NAC1-SRDX (B), and 35Spro:NAC1-SRDX (C) on B5 medium. Note that both transgenic lines showed defective rooting at 10 DAC. D, Rooting rate analyses of leaf explants from Col-0, NAC1pro:NAC1-SRDX, and 35Spro:NAC1-SRDX. Bars show sd from three biological repeats; n = 30 in each repeat. E, Adventitious rooting from stem explants of Col-0 (left) and 35Spro:NAC1-SRDX (right) cultured on B5 medium, showing rooting defect in 35Spro:NAC1-SRDX. Bars = 1 mm in A to C.
NAC1 Acts Independently of Auxin-Mediated Cell Fate Transition
The results of our previous studies suggested that auxin accumulation in competent cells in the vasculature near the wound initiates cell fate transition to WOX11-marked root founder cells. Root founder cells further undergo fate transition to become WOX5-marked root primordium cells via cell division (Liu et al., 2014). Therefore, we tested whether NAC1 is involved in this auxin-mediated cell fate transition pathway.
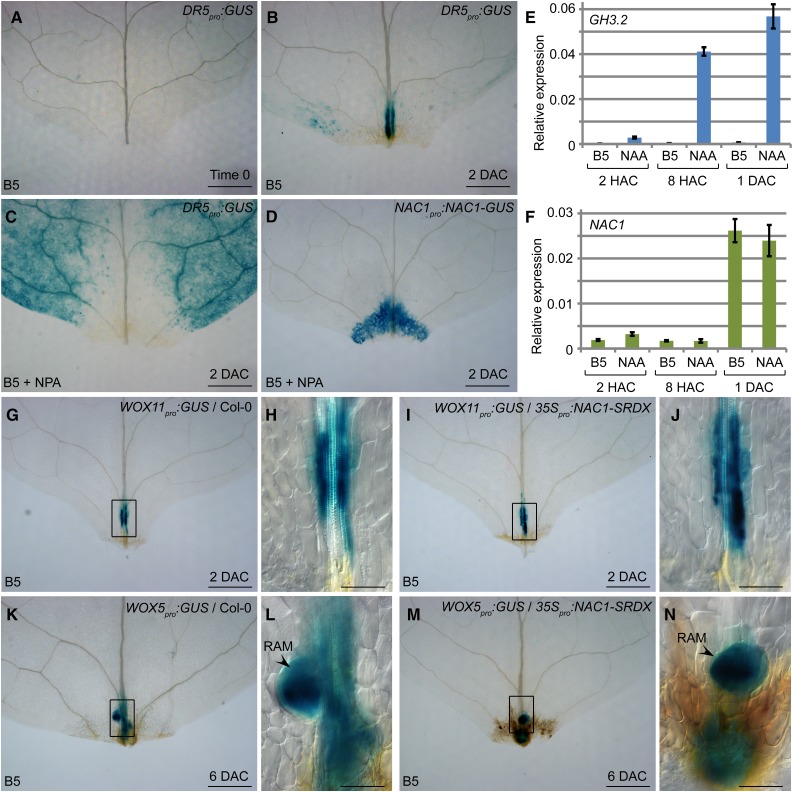
First, we tested whether NAC1 expression is controlled by auxin. Using the auxin reporter line DR5pro:GUS, we observed that auxin quickly accumulated in the mesophyll and vasculature in the wounded region at 2 DAC (Fig. 4, A and B). When napthylphthalamic acid (NPA; a polar auxin transport inhibitor) was supplied in the medium, auxin could not accumulate in the wounded region (Fig. 4C). Interestingly, NAC1 was expressed normally in the wounded region under NPA treatment (Fig. 4D). In addition, expression of NAR1 was also not affected by NPA treatment (Supplemental Fig. S2C). We conducted qRT-PCR analyses to test whether naphthylacetic acid (NAA; a type of auxin) could induce the transcription of the auxin response gene GH3.2 (Staswick et al., 2005; He et al., 2012) and NAC1. The results showed that the NAA treatment quickly induced GH3.2 transcription by 2 h after culture (Fig. 4E) but did not induce NAC1 (Fig. 4F). These data suggested that auxin does not control the expression of NAC1.
Figure 4.
NAC1 acts independently of auxin-mediated cell fate transition. A and B, GUS staining of DR5pro:GUS leaf explant cultured on B5 medium at time 0 (A) and 2 DAC (B). C, GUS staining of DR5pro:GUS leaf explant at 2 DAC on medium containing NPA. Note that the GUS signal did not accumulate in wounded region. D, GUS staining of NAC1pro:NAC1-GUS in leaf explant at 2 DAC cultured on B5 medium containing NPA. E and F, qRT-PCR analysis of GH3.2 (E) and NAC1 (F) transcript levels in leaf explants cultured on B5 medium without or with NAA. Bars show sd from three PCR experiments. Results were confirmed in three biological repeats. Data from one repeat are shown. G to J, GUS staining of WOX11pro:GUS (G and H) and WOX11pro:GUS/35Spro:NAC1-SRDX (I and J) leaf explants cultured on B5 medium at 2 DAC. K to N, GUS staining of WOX5pro:GUS (K and L) and WOX5pro:GUS/35Spro:NAC1-SRDX (M and N) leaf explant cultured on B5 medium at 6 DAC. RAM, root apical meristem. H, J, L, and N, Enlargements of boxed regions in G, I, K, and M, respectively. Bars = 500 μm in A to D, G, I, K, and M and 100 μm in H, J, L, and N.
Next, we tested whether NAC1 affects the competent cell fate transition in adventitious roots growing from leaf explants. We introduced the WOX11pro:GUS and WOX5pro:GUS reporter lines into the 35Spro:NAC1-SRDX transgenic lines to observe expression of the root founder cell marker WOX11 and the root primordium and root apical meristem marker WOX5. The results showed that, similar to their expression patterns in the wild-type background, WOX11 (Fig. 4, G–J) and WOX5 (Fig. 4, K–N) were expressed in the 35Spro:NAC1-SRDX transgenic lines. At 6 DAC, WOX5 expression began to be restricted toward the stem cell niche (Fig. 4, L and N), suggesting that the root apical meristem was forming at this stage in both the wild type and 35Spro:NAC1-SRDX. In addition, WOX11 and WOX5 were also expressed during regeneration of leaf explants from 35Spro:NAR1-SRDX (Supplemental Fig. S2D). These data suggested that NAC genes act independently of cell fate transition in de novo root organogenesis. Therefore, the block of rooting in 35Spro:NAC1-SRDX and 35Spro:NAR1-SRDX might be caused by the defect in root tip emergence.
NAC1 Promotes Expression of CEP Genes
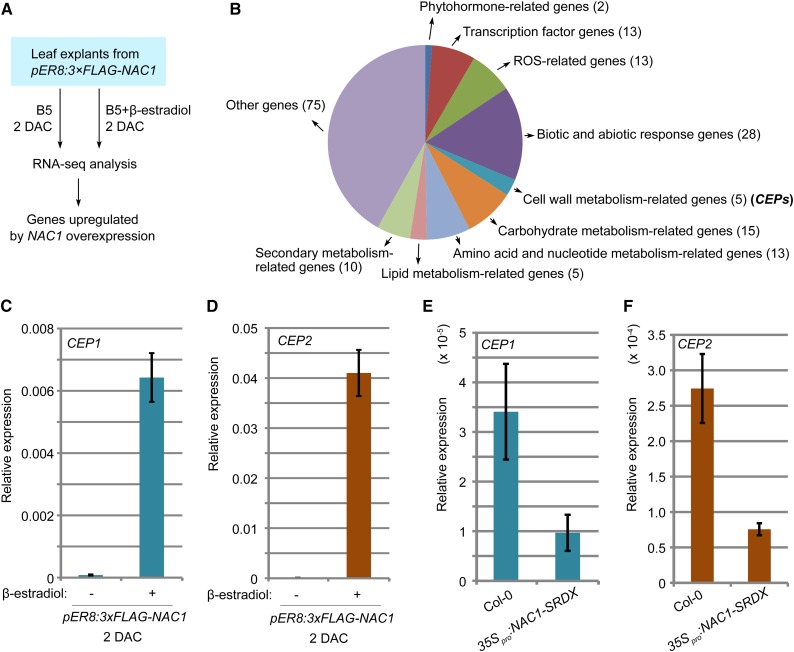
To analyze the possible downstream genes regulated by the NAC pathway, we constructed pER8:3×FLAG-NAC1 lines in which NAC1 was overexpressed under the control of the β-estradiol-inducible promoter (Zuo et al., 2000). We performed RNA-seq analyses using leaf explants of pER8:3×FLAG-NAC1 cultured on B5 medium containing β-estradiol for 2 d and leaf explants from the same line cultured on β-estradiol-free B5 medium and compared the data sets (Fig. 5A). The results showed that NAC1 overexpression induced many groups of genes, including metabolism-related genes (Fig. 5B; Supplemental Table S2).
Figure 5.
NAC1 promotes expression of CEP genes. A, RNA-seq analysis using 2-DAC leaf explants from pER8:3×FLAG-NAC1. Leaf explants were cultured on B5 medium containing β-estradiol/DMSO or DMSO only (control). B, Analysis of upregulated genes in RNA-seq data as indicated in A. Also see Supplemental Table S2. C and D, qRT-PCR analysis of CEP1 (C) and CEP2 (D) transcript levels in leaf explants of pER8:3×FLAG-NAC1 cultured on medium without (−) or with (+) 10 μm β-estradiol at 2 DAC. E and F, qRT-PCR analysis of CEP1 (E) and CEP2 (F) transcript levels in leaf explants of 35Spro:NAC1-SRDX at 4 DAC. Bars in C to F show sd from three PCR experiments. Results were confirmed in two biological repeats. Data from one biological repeat are shown.
We noticed that two KDEL-tailed Cys endopeptidase (CEP) genes, CEP1 and CEP2, were highly upregulated by NAC1 overexpression. In previous studies, CEPs were shown to be involved in programmed cell death and to be secreted to the cell wall for degradation of extensin (EXT) proteins (Greenwood et al., 2005; Helm et al., 2008; Hierl et al., 2012). EXT proteins are basic components of the cell wall, and genes encoding these proteins are induced by wounding (Hall and Cannon, 2002; Merkouropoulos and Shirsat, 2003; Cannon et al., 2008). EXT proteins are involved in cell proliferation (Hall and Cannon, 2002; Cannon et al., 2008; Xu et al., 2011) and wound healing (Bostock, 1989; Showalter, 1993; Tiré et al., 1994).
We performed qRT-PCR analyses to confirm the up-regulation of CEP1 and CEP2 in the pER8:3×FLAG-NAC1 background (Fig. 5, C and D). In addition, expression of CEP1 and CEP2 was partially inhibited by overexpression of NAC1-SRDX (Fig. 5, E and F). These data suggested that NAC1 may up-regulate CEP expression during de novo root organogenesis.
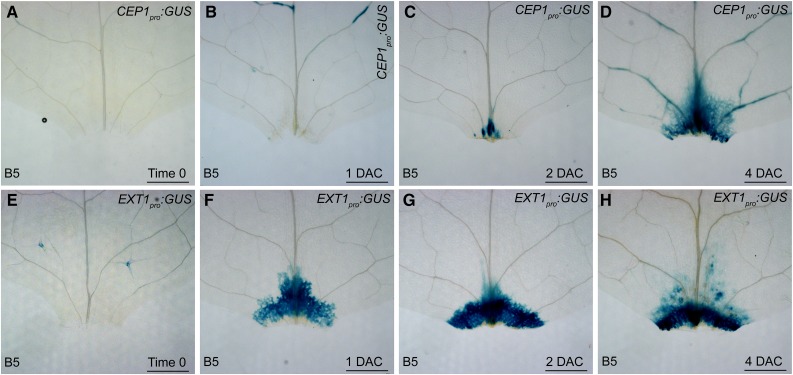
To determine whether CEP and EXT are involved in de novo root organogenesis, we analyzed the expression patterns of CEP1 and EXT1 using CEP1pro:GUS and EXT1pro:GUS reporter lines, respectively. The GUS signal from CEP1pro:GUS was barely detected at the wounded region in time-0 and 1-DAC leaf explants but began to accumulate in the wounded region at and after 2 DAC (Fig. 6, A–D). In addition, CEP1 expression was specifically induced in the wounded region of leaf explants but not in the leaf residue of the source plant (Supplemental Fig. S3). The GUS signal from EXT1pro:GUS was barely detected in time-0 leaf explants; however, its signal quickly became stronger at and after 1 DAC (Fig. 6, E–H), faster than that of CEP1pro:GUS. Therefore, expression of CEP1 and EXT1 was related to wounding during adventitious rooting from leaf explants, and EXT1 was expressed earlier than CEP1.
Figure 6.
Expression patterns of CEP1 and EXT1. A to D, GUS staining of CEP1pro:GUS leaf explant cultured on B5 medium at time 0 (A), 1 DAC (B), 2 DAC (C), and 4 DAC (D). E to H, GUS staining of EXT1pro:GUS leaf explant cultured on B5 medium at time 0 (E), 1 DAC (F), 2 DAC (G), and 4 DAC (H). Bars = 500 µm.
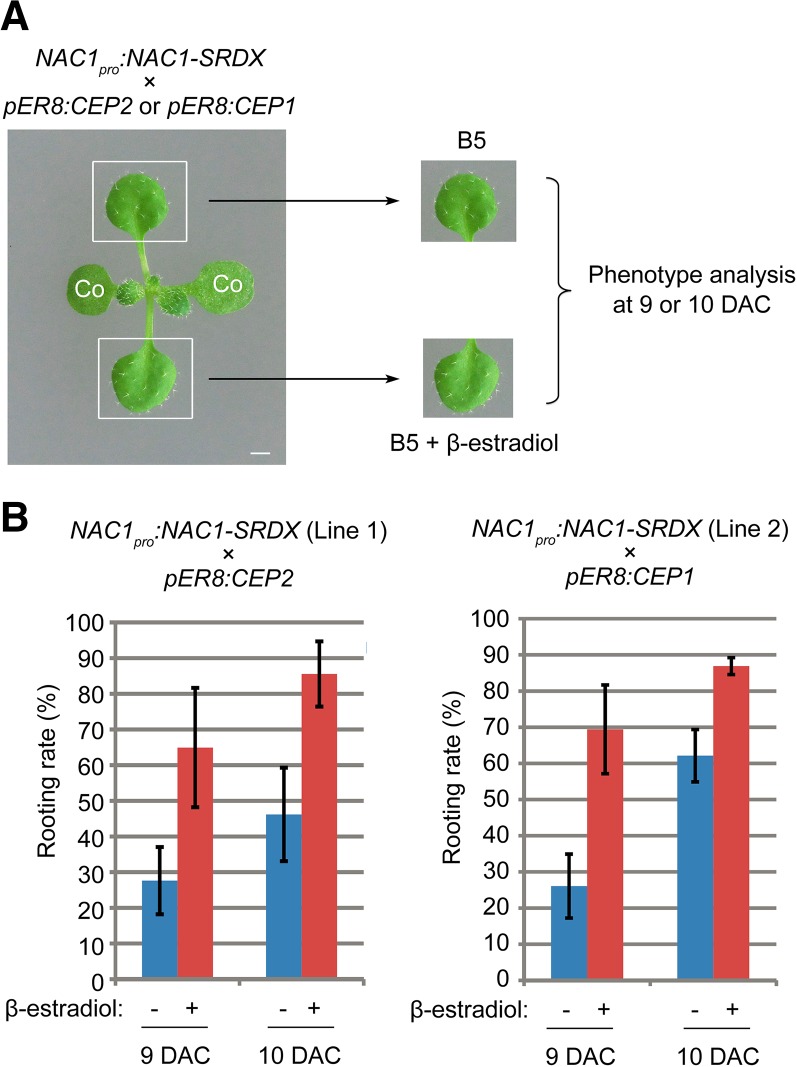
To test whether the CEP pathway contributes to the NAC1-mediated de novo root organogenesis, we overexpressed CEP1 or CEP2 in the NAC1-SRDX background by crossing NAC1pro:NAC1-SRDX with pER8:CEP1 or pER8:CEP2. Two independent NAC1pro:NAC1-SRDX lines were used in crossing. The F2 plants harboring both transgenic constructs were tested by placing one first-pair leaf on β-estradiol-free medium and the other on medium containing β-estradiol (Fig. 7A). CEP1 or CEP2 overexpression partially rescued the rooting defects in the NAC1pro:NAC1-SRDX background (Fig. 7B). These results suggested that the CEP pathway at least partly contributes to NAC1-mediated regeneration.
Figure 7.
Overexpression of CEP partially rescues rooting defect in NAC1pro:NAC1-SRDX. A, Design of rescue experiment. Each leaf explant from the same source plant was cultured on B5 medium containing β-estradiol/DMSO or DMSO only (control). co, Cotyledon. B, Rooting rate analyses of leaf explants from two independent crossed lines (NAC1pro:NAC1-SRDX/pER8:CEP2 in the left panel and NAC1pro:NAC1-SRDX/pER8:CEP1 in the right panel) cultured on medium without (−) or with (+) 10 μm β-estradiol at 9 and 10 DAC. Bars show sd with three biological repetitions. n ≥ 22 in each repetition.
DISCUSSION
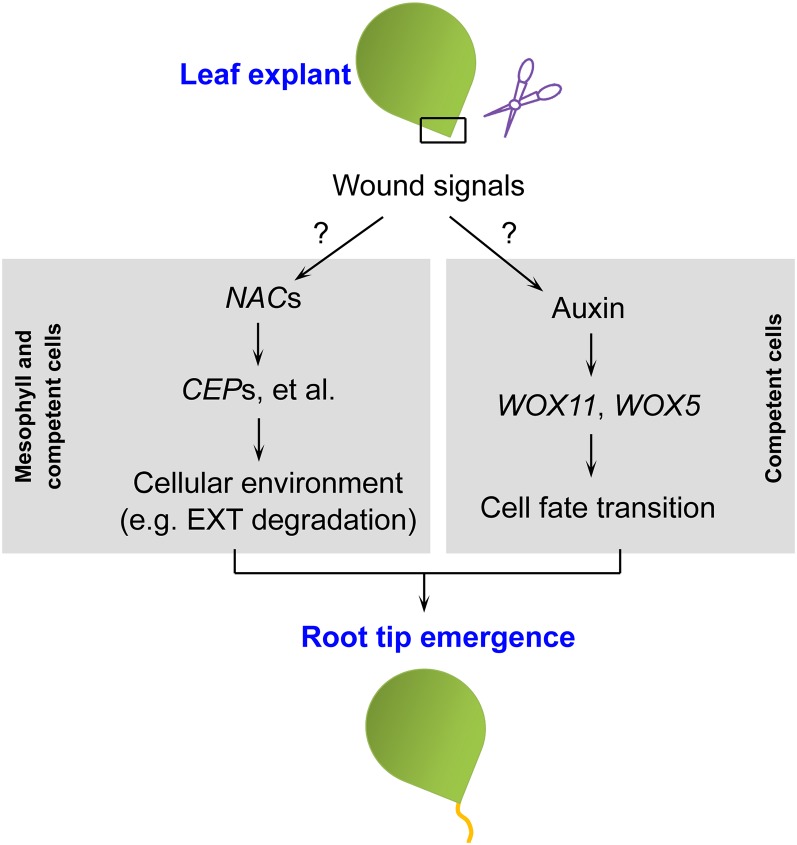
In this study, we revealed a wounding-induced NAC pathway that acts in parallel with auxin-mediated cell fate transition in de novo root organogenesis (see model in Fig. 8). We propose that wounding provides a complex mixture of signals that have multiple roles in controlling the direction of functionally independent molecular pathways. One of the core pathways is the auxin-mediated cell fate transition that occurs in the competent cells of the vasculature within the wounded region. Another pathway revealed in this study involves NAC transcription factors that likely function in regulating the cellular environment in both mesophyll and competent cells within the wounded region for promotion of root tip emergence.
Figure 8.
Model for wounding-induced de novo root organogenesis. Wound signals function in at least two separate pathways for de novo root organogenesis. The molecular features of wound signals remain unclear.
Many genes are activated in response to wounding. Previous studies suggested that WIND1 expression is induced by wounding in plant cell dedifferentiation (Iwase et al., 2011a, 2011b). However, the NAC pathway differs from WIND1. Expression of WIND1 is induced by wounding in both detached and attached tissues, whereas NACs expression is induced by wounding only in the detached explant. In addition, induction of WIND1 occurs rapidly, within hours (Iwase et al., 2011a), whereas up-regulation of NAC genes requires 1 to 2 d. Wounding also induces auxin production in de novo root organogenesis (Liu et al., 2014), and the NAC1 pathway is independent of the auxin-mediated WOX11 pathway. NAC1 expression is not induced by auxin, and NAC1 does not regulate WOX11 that controls cell fate transition. All these behaviors of NAC1 suggest that it functions in a novel wounding response pathway. The results of this study suggested that wounding controls many signals that act independently, but cooperatively, for regeneration. However, the molecular nature of wounding is still poorly understood. Further detailed studies on the physical/chemical events during and after wounding, and their molecular functions, will greatly improve our understanding of how regeneration begins.
NAC1 was previously identified as a regulator of lateral roots, and its expression was shown to be related to the auxin-signaling pathway during lateral root development (Xie et al., 2000). ANAC071, another NAC gene induced by auxin, was reported to be involved in tissue regeneration on the wounded stem (Asahina et al., 2011). In this study, we showed that NAC1 acts independently of auxin in de novo regeneration of adventitious roots from leaf explants. Therefore, NAC family genes may have divergent roles in organ formation and regeneration, and NAC1 may have different roles in lateral and adventitious root formation in response to different upstream regulators. Further analyses of how wounding regulates NAC1 expression and how NAC1 is regulated posttranscriptionally and posttranslationally (Xie et al., 2002; Mallory et al., 2004) will provide new insights into the role of NAC1 in plant regeneration.
The emergence of adventitious root tips requires cell wall degradation and assembly (Vidoz et al., 2010; da Costa et al., 2013). In grape (Vitis vinifera) stem cuttings, expression of the EXT protein was found to be induced during rooting (Thomas et al., 2003). Wounding has been shown to induce EXT1 expression, suggesting that cells at the wounded site accumulate EXT to strengthen cell walls during wound healing (Bostock, 1989; Showalter, 1993; Tiré et al., 1994; Merkouropoulos and Shirsat, 2003). The results of this study showed that NAC1 expression is also induced at the wounded site and promotes CEP expression. Up-regulation of CEPs might probably be related to the degradation of EXT. EXT promotes wound healing, and this might be a barrier for the emergence of regenerated root tips. Therefore, we hypothesize that the NAC1-CEP pathway antagonizes EXT-mediated wound healing, and this allows the emergence of regenerated root tips.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) Col-0 was used as the wild type in this study. To produce NAC1pro:NAC1-GUS and NAR1pro:NAR1-GUS transgenic plants, 5.0-kb NAC1 and 3.0-kb NAR1 genomic sequences including the promoters and coding regions were PCR amplified and inserted into the pBI101 vector (Clontech). CEP1pro:GUS and EXT1pro:GUS were constructed by inserting 1.6- and 5.1-kb CEP1 and EXT1 promoter sequences, respectively, into the pBI101 vector. NAC1pro:NAC1-SRDX was constructed by first fusing a sequence encoding the SRDX peptide (Hiratsu et al., 2003) with the 5.0-kb NAC1 genomic sequence including its promoter and coding region and then inserting the entire fusion construct into pCAMBIA1300 (Cambia). 35Spro:NAC1-SRDX and 35Spro:NAR1-SRDX were constructed by inserting the NAC1 and NAR1 cDNA-SRDX fusion constructs into the pMON530 vector, respectively. To construct pER8:CEP1, pER8:CEP2, or pER8:3×FLAG-NAC1, cDNA encoding the full-length CEP1, CEP2, or 3×FLAG-NAC1 was PCR amplified and inserted into the pER8 vector (Zuo et al., 2000). Transgenic plants were obtained by Agrobacterium tumefaciens-mediated transformation of each of these constructs into Col-0. WOX11pro:GUS, WOX5pro:GUS, and DR5pro:GUS transgenic plants were described previously (Ulmasov et al., 1997; He et al., 2012; Liu et al., 2014). The primers used for plasmid construction are listed in Supplemental Table S3.
Culture Conditions
Culture conditions were as described previously (Chen et al., 2014). Briefly, Arabidopsis seedlings were first cultured on 0.5× MS medium (Murashige and Skoog, 1962) at 22°C under a 16-h-light/8-h-dark photoperiod. Leaf explants from 12-d-old seedlings or stem explants from 26-d-old seedlings were cultured on B5 medium (Gamborg et al., 1968) without carbohydrates under the same photoperiod.
qRT-PCR and RNA-Seq Analyses
The RNA extractions and qRT-PCR analyses were performed as previously described (He et al., 2012) using gene-specific primers. The qRT-PCR results are shown as relative expression levels normalized against those produced using Actin-specific primers. The primers used for real-time PCR are listed in Supplemental Table S3.
For RNA-seq analyses, RNA was extracted using Trizol. Library construction and deep sequencing were carried out using the Illumina HiSeq 2000 platform following the manufacturer’s instructions by Genergy Biotechnology. The raw data comprised 100-bp paired-end sequences, and the cleaned reads were then mapped to Arabidopsis genome (TAIR10) using default settings of TopHat v2.0.8 (Kim et al., 2013). The duplicated reads were removed and alignments with MAPQ score > 20 were used for further analysis. RNA-seq alignments were processed using HTSeq-count (Anders et al., 2015), and differentially expressed genes were identified using DESeq (Anders and Huber, 2010) with |log2 fold change| > 3.5. The analyzed data are shown in Supplemental Tables S1 and S2.
Histology
We performed GUS staining and thin sectioning as previously described (Chen et al., 2014; Zeng et al., 2015). The differential interference contrast observations were performed using Nikon SMZ1500 and Nikon ECLIPSE 80i microscopes as previously described (Chen et al., 2014).
Accession Numbers
The RNA-seq data obtained in the wounding and pER8:3×FLAG-NAC1 analyses have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession numbers GSE74585 and GSE74584, respectively. Sequence data are listed in the Arabidopsis Genome Initiative under the following accession numbers: NAC1 (At1g56010) and NAR1 (At3g12977).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. NAC1 expression in the leaf explant.
Supplemental Figure S2. NAR1 in de novo root organogenesis.
Supplemental Figure S3. CEP1 expression responds to wounding of leaf explant.
Supplemental Table S1. RNA-seq analysis of genes expressed in response to wounding.
Supplemental Table S2. RNA-seq analysis of genes upregulated in pER8:3×FLAG-NAC1.
Supplemental Table S3. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank J. He for assistance in RNA-seq data analysis and T.J. Guilfoyle for Arabidopsis seeds.
Glossary
- DAC
days after culture
- NPA
napthylphthalamic acid
- NAA
naphthylacetic acid
Footnotes
This work was supported by grants from National Basic Research Program of China (973 Program, 2014CB943500/2012CB910503), the National Natural Science Foundation of China (91419302/31422005), and Youth Innovation Promotion Association CAS.
References
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahina M, Azuma K, Pitaksaringkarn W, Yamazaki T, Mitsuda N, Ohme-Takagi M, Yamaguchi S, Kamiya Y, Okada K, Nishimura T, et al. (2011) Spatially selective hormonal control of RAP2.6L and ANAC071 transcription factors involved in tissue reunion in Arabidopsis. Proc Natl Acad Sci USA 108: 16128–16132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock RM. (1989) Perspectives on wound healing in resistance to pathogens. Annu Rev Phytopathol 27: 343–371 [Google Scholar]
- Cannon MC, Terneus K, Hall Q, Tan L, Wang Y, Wegenhart BL, Chen L, Lamport DT, Chen Y, Kieliszewski MJ (2008) Self-assembly of the plant cell wall requires an extensin scaffold. Proc Natl Acad Sci USA 105: 2226–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Qu Y, Sheng L, Liu J, Huang H, Xu L (2014) A simple method suitable to study de novo root organogenesis. Front Plant Sci 5: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa CT, de Almeida MR, Ruedell CM, Schwambach J, Maraschin FS, Fett-Neto AG (2013) When stress and development go hand in hand: main hormonal controls of adventitious rooting in cuttings. Front Plant Sci 4: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclercq J, Sangwan-Norreel B, Catterou M, Sangwan RS (2011) De novo shoot organogenesis: from art to science. Trends Plant Sci 16: 597–606 [DOI] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50: 151–158 [DOI] [PubMed] [Google Scholar]
- Greenwood JS, Helm M, Gietl C (2005) Ricinosomes and endosperm transfer cell structure in programmed cell death of the nucellus during Ricinus seed development. Proc Natl Acad Sci USA 102: 2238–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall Q, Cannon MC (2002) The cell wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell 14: 1161–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Chen X, Huang H, Xu L (2012) Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet 8: e1002911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Schmid M, Hierl G, Terneus K, Tan L, Lottspeich F, Kieliszewski MJ, Gietl C (2008) KDEL-tailed cysteine endopeptidases involved in programmed cell death, intercalation of new cells, and dismantling of extensin scaffolds. Am J Bot 95: 1049–1062 [DOI] [PubMed] [Google Scholar]
- Hierl G, Vothknecht U, Gietl C (2012) Programmed cell death in Ricinus and Arabidopsis: the function of KDEL cysteine peptidases in development. Physiol Plant 145: 103–113 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Iwase A, Mitsuda N, Koyama T, Hiratsu K, Kojima M, Arai T, Inoue Y, Seki M, Sakakibara H, Sugimoto K, Ohme-Takagi M (2011a) The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol 21: 508–514 [DOI] [PubMed] [Google Scholar]
- Iwase A, Ohme-Takagi M, Sugimoto K (2011b) WIND1: a key molecular switch for plant cell dedifferentiation. Plant Signal Behav 6: 1943–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- León J, Rojo E, Sánchez-Serrano JJ (2001) Wound signalling in plants. J Exp Bot 52: 1–9 [DOI] [PubMed] [Google Scholar]
- Liu J, Sheng L, Xu Y, Li J, Yang Z, Huang H, Xu L (2014) WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26: 1081–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei ME, Mithöfer A, Boland W (2007) Before gene expression: early events in plant-insect interaction. Trends Plant Sci 12: 310–316 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Dugas DV, Bartel DP, Bartel B (2004) MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol 14: 1035–1046 [DOI] [PubMed] [Google Scholar]
- Merkouropoulos G, Shirsat AH (2003) The unusual Arabidopsis extensin gene atExt1 is expressed throughout plant development and is induced by a variety of biotic and abiotic stresses. Planta 217: 356–366 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 80: 662–668 [Google Scholar]
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, et al. (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10: 239–247 [DOI] [PubMed] [Google Scholar]
- Showalter AM. (1993) Structure and function of plant cell wall proteins. Plant Cell 5: 9–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Jiao Y, Meyerowitz EM (2010) Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev Cell 18: 463–471 [DOI] [PubMed] [Google Scholar]
- Sussex IM. (2008) The scientific roots of modern plant biotechnology. Plant Cell 20: 1189–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P, Lee MM, Schiefelbein J (2003) Molecular identification of proline-rich protein genes induced during root formation in grape (Vitis vinifera L.) stem cuttings. Plant Cell Environ 26: 1497–1504 [Google Scholar]
- Tiré C, De Rycke R, De Loose M, Inzé D, Van Montagu M, Engler G (1994) Extensin gene expression is induced by mechanical stimuli leading to local cell wall strengthening in Nicotiana plumbaginifolia. Planta 195: 175–181 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidoz ML, Loreti E, Mensuali A, Alpi A, Perata P (2010) Hormonal interplay during adventitious root formation in flooded tomato plants. Plant J 63: 551–562 [DOI] [PubMed] [Google Scholar]
- Xie Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14: 3024–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Guo HS, Dallman G, Fang S, Weissman AM, Chua NH (2002) SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419: 167–170 [DOI] [PubMed] [Google Scholar]
- Xu C, Takáč T, Burbach C, Menzel D, Samaj J (2011) Developmental localization and the role of hydroxyproline rich glycoproteins during somatic embryogenesis of banana (Musa spp. AAA). BMC Plant Biol 11: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Huang H (2014) Genetic and epigenetic controls of plant regeneration. Curr Top Dev Biol 108: 1–33 [DOI] [PubMed] [Google Scholar]
- Zeng M, Hu B, Li J, Zhang G, Ruan Y, Huang H, Wang H, Xu L (2015) Stem cell lineage in body layer specialization and vascular patterning of rice root and leaf. Sci Bull in press. DOI: 10.1007/s11434-015-0849-1 [Google Scholar]
- Zuo J, Niu QW, Chua NH (2000) Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.