A xylan synthase engages an unusual regulatory mechanism linking ER complex assembly with forward trafficking.
Abstract
Xylans play an important role in plant cell wall integrity and have many industrial applications. Characterization of xylan synthase (XS) complexes responsible for the synthesis of these polymers is currently lacking. We recently purified XS activity from etiolated wheat (Triticum aestivum) seedlings. To further characterize this purified activity, we analyzed its protein composition and assembly. Proteomic analysis identified six main proteins: two glycosyltransferases (GTs) TaGT43-4 and TaGT47-13; two putative mutases (TaGT75-3 and TaGT75-4) and two non-GTs; a germin-like protein (TaGLP); and a vernalization related protein (TaVER2). Coexpression of TaGT43-4, TaGT47-13, TaGT75-3, and TaGT75-4 in Pichia pastoris confirmed that these proteins form a complex. Confocal microscopy showed that all these proteins interact in the endoplasmic reticulum (ER) but the complexes accumulate in Golgi, and TaGT43-4 acts as a scaffold protein that holds the other proteins. Furthermore, ER export of the complexes is dependent of the interaction between TaGT43-4 and TaGT47-13. Immunogold electron microscopy data support the conclusion that complex assembly occurs at specific areas of the ER before export to the Golgi. A di-Arg motif and a long sequence motif within the transmembrane domains were found conserved at the NH2-terminal ends of TaGT43-4 and homologous proteins from diverse taxa. These conserved motifs may control the forward trafficking of the complexes and their accumulation in the Golgi. Our findings indicate that xylan synthesis in grasses may involve a new regulatory mechanism linking complex assembly with forward trafficking and provide new insights that advance our understanding of xylan biosynthesis and regulation in plants.
It is believed that Golgi-localized, multiprotein complexes synthesize plant hemicellulosic polysaccharides, including xylans. Such complexes are not well characterized in plants (Zeng et al., 2010; Atmodjo et al., 2011; Chou et al., 2012), which is in sharp contrast with mammalian and yeast cells (Jungmann and Munro, 1998; McCormick et al., 2000; Giraudo et al., 2001). Xylans are the most abundant plant hemicellulosic polysaccharides on Earth and play an important role in the integrity of cell walls, which is a key factor in plant growth. Any mutations affecting xylan backbone biosynthesis seem to result in abnormal growth of plants due mostly to thinning and weakening of secondary xylem walls, described as the irregular xylem (irx) phenotype. Thus, characterizing the xylan synthase complex (XSC) would have an impact on plant improvement, as well as many industrial applications related to food, feed, and biofuel production (Yang and Wyman, 2004; Faik, 2010). Although the Arabidopsis (Arabidopsis thaliana) irx mutants have revealed the involvement of several glycosyltransferase (GT) gene families in xylan biosynthesis (Brown et al., 2007, 2009; Lee et al., 2007, 2010; Wu et al., 2009, 2010), no XSCs have been purified/isolated from Arabidopsis tissues, and we still do not know whether some of the identified Arabidopsis GTs can assemble into functional XSCs. Furthermore, if GTs do assemble into XSCs, we don’t know the mechanisms by which plant cells control their assembly and cellular trafficking. In contrast to dicots, xylan synthase activity was recently immunopurified from etiolated wheat (Triticum aestivum) microsomes (Zeng et al., 2010). This purified wheat XS activity was shown to catalyze three activities, xylan-glucuronosyltransferase (XGlcAT), xylan-xylosyltransferase (XXylT), and xylan-arabinofuranosyltranferase (XAT), which work synergistically to synthesize xylan-type polymers in vitro (Zeng et al., 2008, 2010). This work focuses on describing protein composition, assembly, and trafficking of this purified wheat XS activity.
In all eukaryotes, proteins of the secretory pathway (including GTs) are synthesized in the endoplasmic reticulum (ER) and modified as they go through the Golgi cisternae. Most proteins exit the ER from ER export sites (ERESs; Hanton et al., 2009) and use a signal-based sorting mechanism that allows them to be selectively recruited into vesicles coated by coat protein II complexes (Barlowe, 2003; Beck et al., 2008). For many Golgi-resident type II membrane proteins, di-Arg motifs, such as RR, RXR, and RRR located in their cytosolic NH2-terminal ends, have been shown to be required for their ER export (Giraudo et al., 2003; Czlapinski and Bertozzi, 2006; Schoberer et al., 2009; Tu and Banfield, 2010). Interestingly, di-Arg motifs located ∼40 amino acids from the membrane on the cytosolic side can also be used to retrieve some type II ER-resident proteins from cis-Golgi (Schutze et al., 1994; Hardt et al., 2003; Boulaflous et al., 2009). In contrast to the signal-based sorting mechanism involved in trafficking between the ER and Golgi, the steady-state localization/retention of proteins (including GTs) in the Golgi is thought to occur through vesicular cycling. Cycling is influenced by various mechanisms, including the length and composition of the transmembrane domain (TMD) of type II GTs (Bretscher and Munro, 1993; Colley, 1997; van Vliet et al., 2003; Sousa et al., 2003; Sharpe et al., 2010), and the oligomerization/aggregation of GTs (kin hypothesis), which suggests that formation of homo- or heterooligomers of GTs in the Golgi may prevent their recruitment into clathrin-coated vesicles (Machamer, 1991; Nilsson et al., 1993; Weisz et al., 1993; Cole et al., 1996). Some Golgi-resident GTs are predicted to have a cleavable NH2-terminal secretion signal peptide (SP) and would therefore exist as soluble proteins in the Golgi lumen. To maintain their proper Golgi localization, these processed GTs are likely part of multiprotein complexes anchored to integral membrane proteins. The fact that homologs of many of the trafficking proteins from mammalian and yeast cells are found in plants indicates that trafficking machineries of the plant secretory pathway are likely conserved (d’Enfert et al., 1992; Bar-Peled and Raikhel, 1997; Batoko et al., 2000; Pimpl et al., 2000; Phillipson et al., 2001; Hawes et al., 2008).
It is becoming increasingly evident that understanding the mechanisms controlling protein-protein interaction, sorting, and trafficking of polysaccharide synthases (including XSCs) will help elucidate how plants regulate cell wall synthesis and deposition during their development. To this end, we believe that the purified wheat XS activity (Zeng et al., 2010) is an excellent model for this type of study. In this work, proteomics was used to determine the protein composition of the purified XS activity. Confocal microscopy and immunogold transmission electron microscopy (TEM) were used to investigate the assembly and trafficking of the complex. Our proteomics data showed that the purified activity contains two GTs, TaGT43-4 and TaGT47-13, two putative mutases, TaGT75-3 and TaGT75-4, and two non-GT proteins: a germin-like protein (TaGLP) belonging to cupin superfamily and a protein specific to monocots annotated as wheat vernalization-related protein 2 (TaVER2). Microscopy analyses revealed that all these proteins interact in the ER, but the assembled complexes accumulate in the Golgi. Export of these complexes from the ER is controlled by the interaction between TaGT43-4 and TaGT47-13. Characterization of the wheat XSC and its trafficking furthers our understanding of xylan biosynthesis in grasses and helps elucidate how polysaccharide synthase complexes are assembled, sorted, and maintained in different compartments of the secretory pathway.
RESULTS
Proteomic Analysis of the Immunopurified Wheat XS Activity
Peptide sequences released by in-gel trypsin digestion of the previously immunopurified wheat XS activity (Zeng et al., 2010) were determined by Electrospray Ionization (ESI) tandem mass spectrometry (MS/MS) (ESI-MS/MS), resulting in a total of 153,586 peptide sequences. Initially, these peptide sequences were used to interrogate green plants (Viridiplantae) databases at NCBInr using the Mascot program. This search indicated that 18,122 peptide sequences matched 311 proteins, among which only 51 proteins satisfied our reliability criteria (e.g. protein score ≥60 and a minimum of two peptides having a -b or -y ion sequence tag of five residues or better; Table I). All 51 proteins, except four, were members of the GT43, GT47, or GT75 families of the CAZy database (Coutinho et al., 2003). Importantly, TaGT43-4 and TaGT47-13 were matched by the highest number of peptides and had the highest protein scores (Table I, Figure 1D). Phylogenetic analysis also showed that the identified proteins clustered with TaGT43-4 (homolog to Arabidopsis IRX14/IRX14-L; Figure 1A) and TaGT47-13 (homolog to Arabidopsis IRX10/IRX10-L; Figure 1B), but no proteins clustered with Arabidopsis IRX7 (Fig. 1B) or with IRX9/IRX9-L (Fig. 1A). This result is in agreement with western-blotting analysis in which purified anti-TaGT43-4 and anti-TaGT47-13 antibodies detected the presence of TaGT43-4 and TaGT47-13 in the purified wheat XS activity (Zeng et al., 2010). Nine members of the GT75 family were identified (Table I): EMT30339 (Aegilops tauschii), XP_003562308 (Brachypodium distachyon), XP_006856400 (Amborella trichopoda), XP_004488432 (Cicer arietinum), XP_008785739 (Phoenix dactylifera), XP_004490223 (C. arietinum), XP_003614188 (Medicago truncatula), XP_007138376 (Phaseolus vulgaris), and XP_006408433 (Eutrema salsugineum). EMT30339 and XP_003562308 were matched by the largest number of peptides and showed 96 and 99% identity with TaGT75-4, respectively, while XP_006856400, XP_008785739, and XP_004488432 showed 88 to 89% identity with TaGT75-3. TaGT75-3 and TaGT75-4 were not identified initially because their sequences were not present in NCBInr database at the time of the original search. Phylogenetic analysis clustered TaGT75-3 and TaGT75-4 with rice (Oryza sativa) UDP-Arap mutases 1 (UAM1) and UAM3, respectively (Fig. 1C), for which mutase activity has been confirmed experimentally (Konishi et al., 2007). Therefore, we concluded that TaGT75-3 and TaGT75-4 were most likely mutases and were also associated with the purified XS activity. This is also consistent with western-blotting data, in which antibodies against PsRGP1 (the pea protein homologous to TaGT75-3 and TaGT75-4) detected the presence of these proteins in the purified wheat XS activity (Zeng et al., 2010).
Table I. List of proteins identified in Mascot search along with their GI accession number in GenBank.
Only proteins with score higher than 60 and two peptides are shown. Proteins identified by the same peptides have the same Mascot protein score and were grouped together, which were classified in decreasing order for their Mascot protein scores.
| Accession No. in GenBank | Description | Prot. Score | No. of Pept. Matches | GT Family |
|---|---|---|---|---|
| gi|301072484 | Glycosyltransferase 47 [T. aestivum] (ADK56172) (TaGT47-13) | 25,516 | 17 | 47 |
| gi|475454583 | Exostosin-1 [A. tauschii] (F775_00782) | |||
| gi|194706890 | Unknown [Zea mays] (ACF87529) | |||
| gi|226529361 | LOC100281324 precursor [Z. mays] (NP_001147714) | 13,790 | 5 | 47 |
| gi|242088705 | Hypothetical protein SORBIDRAFT_09g027450 [Sorghum bicolor] | |||
| gi|63087754 | Glycosyltransferase [T. aestivum] (CAI93192) (TaGT43-4) | 13,749 | 15 | 43 |
| gi|63087744 | Glycosyltransferase [H. vulgare] (CAI93187) | 12,686 | 13 | 43 |
| gi|326487454 | Predicted protein [H. vulgare] (BAJ89711) | |||
| gi|475510687 | Galactosylgalactosylxylosylprotein 3-β-glucuronosyltransferase 1 [A. tauschii] (EMT05302) | 11,251 | 13 | 43 |
| gi|242088703 | Hypothetical protein SORBIDRAFT_09g027440 [S. bicolor] (EES18614) | |||
| gi|514747280 | Probable glucuronosyltransferase Os01g0926700-like [Setaria italica] (XP_004961328) | 8,793 | 5 | 47 |
| gi|514747276 | Probable glucuronosyltransferase Os01g0926600-like [S. italica] (XP_004961327) | 7,154 | 5 | 47 |
| gi|63087750 | Glycosyltransferase [Saccharum officinarum] (CAI93190) | 3,876 | 7 | 43 |
| gi|514767605 | Probable glucuronosyltransferase Os06g0687900-like [S. italica] (XP_004966284) | 3,664 | 6 | 43 |
| gi|16151819 | VER2 [T. aestivum] (BAA32786) | 3,627 | 11 | – |
| gi|475612348 | Hypothetical protein F775_25915 [A. tauschii] (EMT28373) | |||
| gi|242061766 | Hypothetical protein SORBIDRAFT_04g021180 [S. bicolor] (XP_002452172) | 2,856 | 3 | 47 |
| gi|357149222 | Probable glucuronosyltransferase Os02g0520750-like [B. distachyon] (XP_003575040) | 2,801 | 2 | 47 |
| gi|194701076 | Unknown [Z. mays] (ACF84622) | 2,535 | 2 | 43 |
| gi|413934620 | Glycosyltransferase [Z. mays] (AFW69171) | |||
| gi|29374017 | GLP-binding protein 1b [S. tuberosum] (AAO72974) | 1,857 | 2 | – |
| gi|53854259 | DING protein [S. tuberosum] (AAU95561) | 1,469 | 6 | – |
| gi|225456511 | Probable β-1,4-xylosyltransferase IRX10L [Vitis vinifera] (XP_002284685) | 1,266 | 2 | 47 |
| gi|75159221 | Probable glucuronosyltransferase Os01g0926700 [O. sativa Japonica Group] | |||
| gi|226503833 | Secondary cell wall-related glycosyltransferase family 47 precursor [Z. mays] (NP_001149319) | |||
| gi|242059743 | Hypothetical protein SORBIDRAFT_03g044530 [S. bicolor] (XP_002459017) | |||
| gi|357126610 | Probable glucuronosyltransferase Os01g0926700-like [B. distachyon] (XP_003564980) | 1,069 | 4 | 47 |
| gi|413951430 | Secondary cell wall glycosyltransferase family 47 [Z. mays] (AFW84079) | |||
| gi|514786644 | Probable glucuronosyltransferase Os01g0926700-like [S. italica] (XP_004971138) | |||
| gi|573915216 | Probable glucuronosyltransferase Os01g0926700-like [Oryza brachyantha] (XP_006645266) | |||
| gi|357121199 | UDP-arabinopyranose mutase 1-like [B. distachyon] (XP_003562308) | |||
| gi|475618083 | α-1,4-Glucan-protein synthase (UDP-forming) [A. tauschii] (EMT30339) | 796 | 5 | 75 |
| gi|672124692 | UDP-arabinopyranose mutase 3-like [P. dactylifera] (XP_008785739) | |||
| gi|502087115 | UDP-arabinopyranose mutase 3-like [C. arietinum] (XP_004488432) | |||
| gi|586769976 | Hypothetical protein AMTR_s00047p00209120 [A. trichopoda] (XP_006856400) | 691 | 3 | 75 |
| gi|502094462gi|593329899 | α-1,4-Glucan-protein synthase (UDP-forming)-like [C. arietinum] (XP_004490223) Hypothetical protein PHAVU_009G203300g [P. vulgaris] (XP_007138376) | 486 | 2 | 75 |
| gi|357487801 | α-1 4-Glucan-protein synthase [M. truncatula] (XP_003614188) | 486 | 2 | 75 |
| gi|698442876 | Probable β-1,4-xylosyltransferase IRX10L [Nicotiana sylvestris] (XP_009763728) | 405 | 2 | 47 |
| gi|460386806 | Probable β-1,4-xylosyltransferase IRX10L-like [Solanum lycopersicum] (XP_004239087) | |||
| gi|565364031 | Probable β-1,4-xylosyltransferase IRX10L-like [S. tuberosum] (XP_006348731) | 404 | 2 | 47 |
| gi|567204084 | Hypothetical protein EUTSA_v10021014mg [E. salsugineum] (XP_006408433) | 332 | 3 | 75 |
| gi|12231279 | Os06g0687900 UDP-glucuronyltransferase-l [O. sativa Japonica Group] (BAB20991) | 328 | 3 | 43 |
| gi|18642697 | Unknown protein [O. sativa Japonica Group] (AAL76189.1) | |||
| gi|21671946 | Unknown protein similar to exostosin-2 [O. sativa Japonica Group] (AAM74308.1) | |||
| gi|110288702 | Exostosin family protein, putative, expressed [O. sativa Japonica Group] (ABB46945.2) | 291 | 4 | 47 |
| gi|218184216 | Hypothetical protein OsI_32904 [O. sativa Indica Group] (EEC66643.1) | |||
| gi|115481310 | Os10g0180000 [O. sativa Japonica Group] | |||
| gi|212275360 | Uncharacterized protein LOC100191151 precursor [Z. mays] (NP_001130059) | 262 | 4 | 47 |
| gi|363543255 | Hypothetical protein precursor [Z. mays] (NP_001241842) | |||
| gi|5869973 | Germin-like protein (GLP) [T. aestivum] (CAB55558.1) | 60 | 6 | – |
| gi|5869975 | Germin-like protein [T. aestivum] (CAB55559.1) |
Figure 1.
Maximum likelihood phylogenetic analysis of proteins from GT47 (A), GT43 (B), and GT75 (C) families that were identified by proteomics (Table I). Wheat (Ta, in blue), Arabidopsis (At), and rice (Os) proteins are included for comparison. Trees were generated through the Phylogeny.fr platform using MUSCLE 3.7 (Edgar 2004) and PhyML 3.0 (Guindon et al., 2010) programs. Bootstrap values (based on 500 replicate) are indicated at the tree nodes. The scale measures evolutionary distance in substitution per amino acids. D, List of wheat proteins matched by the highest number of peptides and having the highest protein scores. Text in red indicates the amino acid sequences covered by the peptides. Protein coverage and the number of peptides matching each protein are indicated.
The four remaining proteins identified were non-GTs: one wheat protein (BAA32786) contained a dirigent domain and a jacalin lectin domain (Chong et al., 1998; Yong et al., 2003) and was annotated as vernalization-related gene 2 (TaVER2; Figure 1D); two wheat proteins (CAB55558 and CAB55558) contained a cupin domain and were annotated as wheat germin-like protein (TaGLP; Schweizer et al., 1999; Dunwell et al., 2000, 2004; Fig. 1D); and two potato (Solanum tuberosum) proteins annotated as DING (AAO72974) and GLP-binding proteins (AAU95561; Table I), which were matched by six peptides with protein coverage of 61%. DING proteins belong to a family of high-affinity phosphate-binding proteins having DINGGG motif at their NH2 termini (Berna et al., 2002, 2008). However, we were not able to identify homologs of potato DING and GLP-binding proteins in wheat, rice, or B. distachyon. Although the protein score of TaGLP was low (because peptides had low ion scores <20), the quality of the spectra was good. It is also known that germins and GLPs are recalcitrant to proteases (Dunwell et al., 2000), which may explain the difficulty in detecting TaGLP in this analysis.
No Members of the GT2, GT8, and GT61 Families Were Identified by Proteomic Analysis
It has been shown that members of the GT8 and GT61 families are xylan-α(1,2)GlcATs (Mortimer et al., 2010; Lee et al., 2012; Rennie et al., 2012) and xylan-α(1,3)AraT (Anders et al., 2012), respectively. Furthermore, several members of the GT2 family have been implicated in the synthesis of the backbone of some hemicellulosic β(1,4)glycans. Therefore, we searched for peptides that matched orthologs/homologs of these GTs using a small in-house database (∼600 sequences) consisting of protein sequences from the CAZy family GT2 (including CSL and CESA members), GT8, GT43, GT47, GT61, and GT75 of wheat, barley (Hordeum vulgare), rice, B. distachyon, and Arabidopsis. To overcome the fact that the wheat genome is not fully sequenced, we included sequences from the closest species to wheat, barley, for which the genome is fully sequenced (Mayer et al., 2012). There is usually a high rate of false discovery rate associated with the search of such small databases. Despite this high false discovery rate, the search resulted in the identification of only 100 proteins, and no members of the GT2, GT8, or GT61 families were among the hits. Among the 100 proteins, only 40 proteins passed our reliability criteria: 24 of these proteins belonged to the GT47 family, 11 were from the GT75 family, and five proteins were from the GT43 family. Again, the highest number of peptides matched TaGT43-4 (18 peptides), TaGT47-13 (28 peptides), TaGT75-4 (13 peptides), and TaGT75-3 (10 peptides). Protein coverage of these proteins is indicated in Figure 1D. Although TaGT75-4 and TaGT75-3 are 88% identical and 90% similar, they were specifically identified by at least two peptides unique to each of them. Therefore, we concluded that the immunopurified wheat XS activity is enriched mostly in TaGT43-4, TaGT47-13, TaGT75-3, TaGT75-4, TaVER2, and TaGLP proteins.
The NH2-Terminal Secretion SP of TaGT47-13 Is Functional
Sequence analysis indicated that TaGT75-3, TaGT75-4, and TaVER2 are predicted to be soluble proteins with no NH2-terminal secretion SP, while TaGT47-13 and TaGLP are predicted to have a cleavable SP. We tested whether the SP sequence of TaGT47-13 is functional by generating a construct encoding for a chimeric TaGT43-4 protein (SP-TaGT43-4) in which the first 71 amino acids, including TMD, were replaced with the first 31 amino acids of TaGT47-13. Expression of SP-TaGT43-4 alone in Pichia pastoris resulted in secretion of the protein in the culture medium (estimated size of ∼48 kD), but the presence of unprocessed SP-TaGT43-4 (SP not cleaved) was also detected in microsomal membranes (estimated size of ∼52 kD; Supplemental Fig. S1). Similarly, expression of TaGT47-13 alone in P. pastoris also resulted in its detection in both culture medium and microsomal membranes, with estimated sizes of ∼56 and ∼44 kD, respectively (Supplemental Fig. S1). These results confirmed that the first 31 amino acids of TaGT47-13 contain a cleavable SP, but the processing of this SP seems to be incomplete. It is unclear why unprocessed TaGT47-13 runs at much higher size (∼56 kD compared to the expected size of ∼47 kD) in SDS-PAGE. Interestingly, our previous data on purified wheat XSC suggest that wheat XSC contains only unprocessed TaGT47-13 (∼56 kD), since a band of smaller size (i.e. ∼44 kD) was never detected with anti-TaGT47-13 (Zeng et al., 2010).
TaGT43-4, TaGT47-13, TaGT75-3, and TaGT75-4 Can Form a Complex in Pichia pastoris
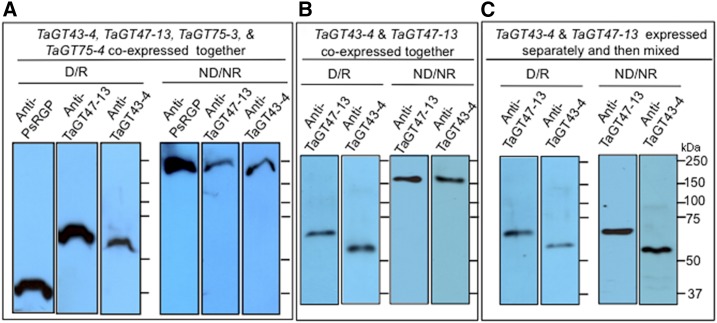
As a first step to test whether the proteins identified by proteomic analysis can form a complex, we expressed the four wheat proteins (TaGT43-4, TaGT47-13, TaGT75-3, and TaGT75-4) individually or in combinations in a system that does not have the capacity to synthesize xylans, P. pastoris. We have shown previously that, in nonreducing/nondenaturing gel electrophoresis, TaGT43-4, TaGT47-13, and TaGT75-3/TaGT75-4 comigrate as a complex that can be detected by purified anti-TaGT43-4, anti-TaGT47-13, and anti-PsRGP1 antibodies as a single band with a molecular mass around 240 kD (Zeng et al., 2010). Using this same immunoblot analysis, we monitored complex assembly in microsomes from transgenic P. pastoris cells coexpressing these wheat proteins. As shown in Figure 2A, immunoblot analysis of detergent-extracts of microsomes from transgenic yeast cells coexpressing the four wheat proteins revealed that, under reducing/denaturing (SDS-PAGE), anti-TaGT43-4, anti-TaGT47-13, and anti-PsRGP1 antibodies detected single bands corresponding to TaGT43-4, TaGT47-13, and TaGT75-3/-4 (TaGT75-4 and TaGT75-3 have a similar size; Supplemental Fig. S2). Under nonreducing/nondenaturing conditions, the three antibodies detected a same high molecular mass band around 240 kD. These data confirm that (1) the four GTs comigrated as a single complex, which mirrors our previous data obtained with microsomes from wheat seedlings (Zeng et al., 2010); and (2) only the unprocessed version of TaGT47-13 is engaged in a complex with TaGT43-4, as no band at ∼44 kD was observed (compare Fig. 2 and Supplemental Fig. S1). The estimated size of ∼240 kD of the complex is consistent with a presence of TaGT43-4, TaGT47-13, and TaGT75-3/-4 in a ratio of 2:1:2 (assuming that the estimated sizes of TaGT43-4, TaGT47-13, and TaGT75-3/-4 are ∼52, ∼56, and ∼40 kD, respectively).
Figure 2.
Immunoblot analysis of Triton extracts from microsomal membranes of transgenic P. pastoris cells. Triton extracts (20–25 μg proteins) were either analyzed under denaturing/reducing (D/R) conditions (1-D SDS-PAGE) or under nonreducing/nondenaturing (ND/NR) conditions (gels contain 0.05% cholate instead of SDS and no DTT) without boiling, as described by Zeng et al. (2010). A, Triton extracts from yeast cells coexpressing TaGT43-4, TaGT47-13, TaGT75-3, and TaGT75-4. B, Triton extracts from yeast cells coexpressing only TaGT43-4 and TaGT47-13. C, Triton extracts from yeast cells expressing TaGT43-4 or TaGT47-13 individually that were mixed (1:1 ratio) before analysis. Polyacrylamide gels were analyzed by immunoblot using anti-TaGT43-4, anti-TaGT47-13, and anti-PsRGP1 antibodies. Molecular weight markers (kD) are indicated.
A similar analysis was carried out using microsomes from transgenic P. pastoris cells coexpressing only TaGT43-4 and TaGT47-13 and showed that, under reducing/denaturing conditions, anti-TaGT43-4 and TaGT47-13 antibodies detected single bands corresponding to TaGT43-4 and TaGT47-13. Under nonreducing/nonreducing conditions, the two antibodies detected a high molecular mass band around 160 kD (Fig. 2B). These results confirm again that TaGT43-4 and TaGT47-13 comigrate as a complex that appears to contain TaGT43-4 and TaGT47-13 in a ratio of 2:1. Next, we asked whether mixing individual proteins (produced separately in P. pastoris cells) would assemble into a complex in vitro. To this end, detergent extracts of microsomes from transgenic yeast cell expressing TaGT43-4 or TaGT47-13 individually were mixed and subjected to the same immunoblot analysis. As shown in Figure 2C, gel electrophoresis under nonreducing/nondenaturing conditions did not result in a single high molecular mass band; instead, anti-TaGT43-4 and anti-TaGT47-13 antibodies detected single bands corresponding to TaGT43-4 and TaGT47-13 monomers. Taken together, these findings strongly suggest that for efficient assembly, TaGT43-4 and TaGT47-13 need to be in close proximity during their synthesis in the ER or may require specific membrane environments (or chaperones) that facilitate their interaction and assembly. In both cases, the assembly most likely occurs in the ER before export to the Golgi.
TaGT43-4, TaGT47-13, and TaGLP Assemble into a Complex in the ER, But Localize in the Trans-Golgi
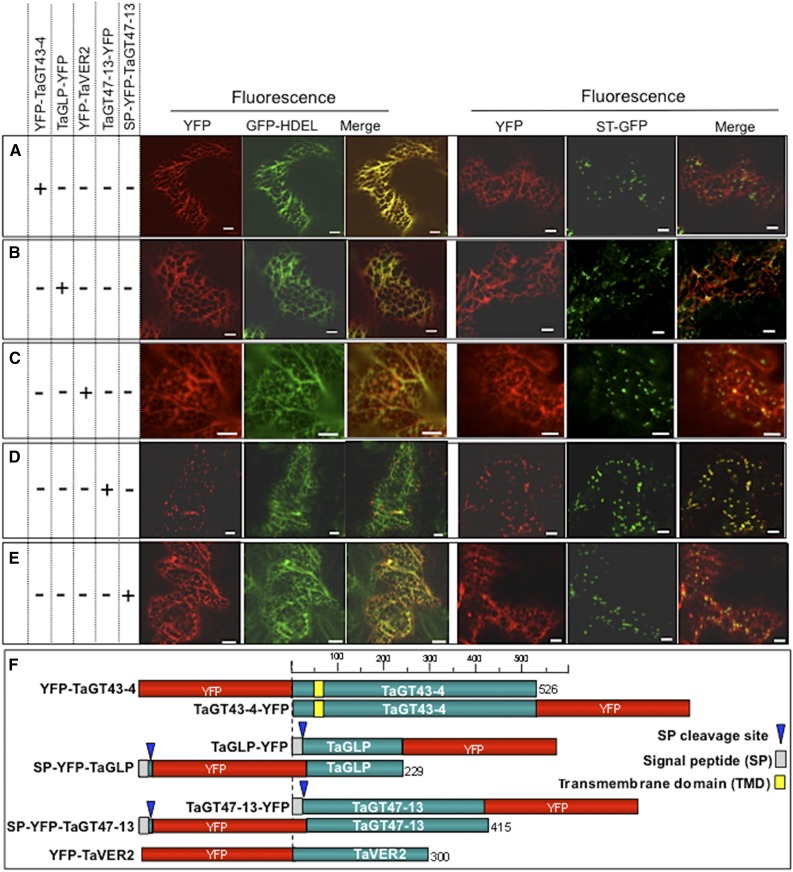
To test the hypothesis that wheat XSC assembles in the ER, we used confocal laser scanning microscopy to investigate subcellular localization of transiently expressed yellow fluorescent protein (YFP)-tagged proteins in live epidermal cells of tobacco (Nicotiana tabacum) leaves (Held et al., 2008). When YFP-TaGT43-4 construct was tested, YFP fluorescence colocalized with GFP-HDEL, an ER marker (Batoko et al., 2000), but not with α(2,6)-sialyltransferase transmembrane domain (STtmd) fused to GFP (referred to as ST-GFP), a trans-Golgi marker (Batoko et al., 2000; Fig. 3A). The same result was obtained with TaGT43-4 fused to YFP at its COOH-terminal end (TaGT43-4-YFP; Supplemental Fig. S3), indicating that ER localization was not due to YFP fusion to the NH2-terminal end of TaGT43-4. Similarly, when TaGLP-YFP and YFP-TaVER2 constructs were tested, YFP fluorescence was also colocalized with GFP-HDEL at the ER, but not with ST-GFP (Fig. 3, B and C, respectively). The same result was obtained using SP-YFP-TaGLP construct (YFP inserted downstream of the cleavage site for SP; Supplemental Fig. S3). This ER localization of YFP-tagged TaGT43-4, TaGLP, and TaVER2 was not an artifact of overproduction of the fusion proteins, as incubation time (<24 h or up to 4 d) after Agrobacterium tumefaciens infiltration or lowering the amount of infiltrated A. tumefaciens culture did not affect fluorescence localization. Also, it is unlikely that ER localization is due to the fact that tobacco, a dicot plant, was used as a system to express proteins from wheat, a monocot plant. Previous studies showed that transient expression of YFP-tagged rice xylosyl arabinosyl substitution of xylan 1 (XAX1, GT61) in onion epidermal cells (Chiniquy et al., 2012) and YFP-tagged wheat xylan arabinosyltransferase 2 (TaXAT2, GT61) in tobacco epidermal cells (Anders et al., 2012) both produced fluorescence that localized to the Golgi. Similarly, several CSL-F proteins (GT2 family) from various monocots including barley, rice, and B. distachyon have been shown to localize to the proper compartments (Golgi/plasma membrane) in tobacco and were functional (Jobling, 2015; Kim et al., 2015; Wilson et al., 2015). On the other hand, when transient expression of TaGT47-13-YFP construct alone was tested in tobacco leaves, YFP fluorescence colocalized with ST-GFP in the trans-Golgi (Fig. 3D), while SP-YFP-TaGT47-13 construct (YFP inserted after SP sequence) produced fluorescence that stained the ER (Fig. 3E). Coexpressing SP-YFP-TaGT47-13 with untagged TaGT43-4 construct did not result in a shift of fluorescence to the trans-Golgi, which could be explained by the fact that YFP insertion could affect the assembly of a functional “TaGT47-13/TaGT43-4” complex that is retained in the ER. Alternatively, YFP could affect the folding of TaGT47-13 required for the assembly of a functional complex. Nonfunctional complexes are retained in the ER. Therefore, TaGT47-13 may play a more important role in both complex assembly and together, these findings suggest that, individually, all these proteins (with the exception of TaGT47-13) are most likely retained in the ER and their export from the ER may require interactions with each other. Retention of SP-YFP-TaGT47-13 in the ER could be explained by the fact that YFP insertion could affect the folding of TaGT47-13 required for the assembly of a functional complex. Nonfunctional complexes are retained in the ER. Therefore, TaGT47-13 may play a more important role in both complex assembly and ER export.
Figure 3.
Subcellular localization of YFP-tagged TaGT43-4, TaGT47-13, TaGLP, and TaVER2 in epidermal cells of tobacco leaves. ER marker, GFP-HDEL, and α(2,6)sialyltransferase fused to GFP (ST-GFP), trans-Golgi marker are used to show the colocalization with YFP-tagged proteins. GFP and YFP fluorescence are shown in green and red, respectively, and their colocalization appears in yellow. Tagged proteins were transiently expressed individually (indicated by a “+” sign on the left of the figure). YFP-TaGT43-4, TaGLP-YFP, and YFP-TaVER2 localize to the ER (A–C, respectively). While TaGT47-13-YFP localizes with ST-GFP (D), SP-YFP-TaGT47-13 localizes with GFP-HDEL (E). Bars = 10 μm. Schematic presentations of YFP-tagged constructs used in this study are shown in F.
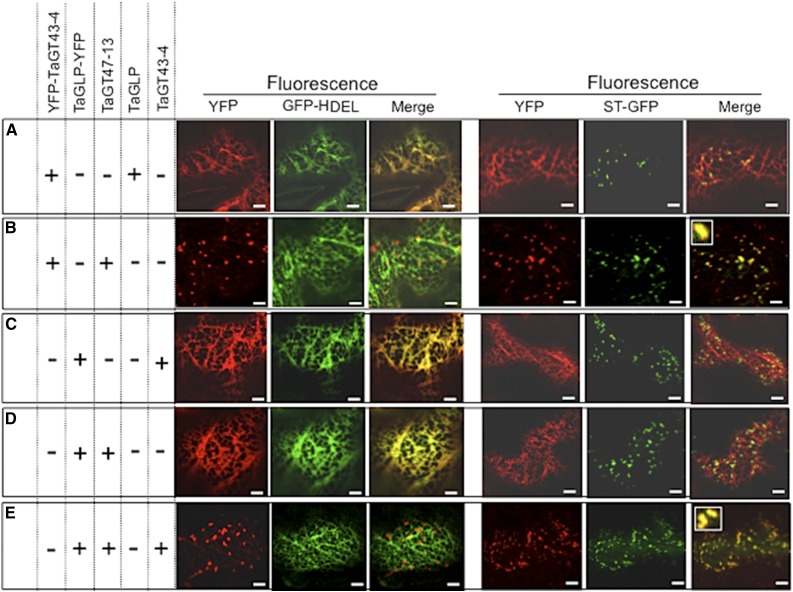
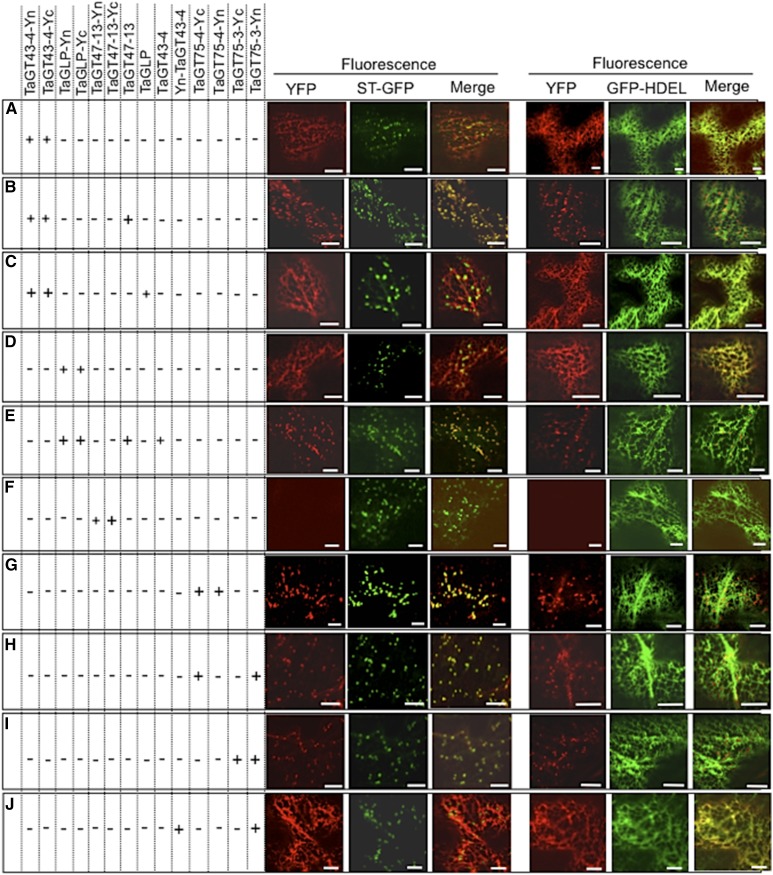
To test this hypothesis, we examined the coexpression of various combinations of TaGT43-4, TaGT47-13, and TaGLP. For example, YFP-TaGT43-4 was transiently coexpressed with either TaGLP or TaGT47-13 constructs (both untagged) and then simultaneously with both of them. The presence of TaGLP did not change ER localization of YFP-TaGT43-4 (Fig. 4A), whereas the presence of TaGT47-13 did result in a redistribution of the fluorescence to a trans-Golgi punctate pattern (Fig. 4B). Interestingly, coinfiltration of TaGLP-YFP with either TaGT43-4 or TaGT47-13 constructs (both untagged) did not change ER localization of YFP fluorescence (Fig. 4, C and 4D, respectively), but coinfiltration with both of these constructs resulted in a clear shift in YFP fluorescence to the trans-Golgi (Fig. 4E). These results indicate that for TaGT43-4 to exit the ER, the presence of TaGT47-13 is required, while for TaGLP to exit the ER, the presence of both TaGT43-4 and TaGT47-13 is needed. Therefore, we concluded that the three proteins can assemble into a complex in the ER and the interaction between TaGT43-4 and TaGT47-13 is the driver of ER export of the complex.
Figure 4.
Trafficking of YFP-TaGT43-4 and TaGLP-YFP when transiently expressed in combination with untagged TaGT47-13, TaGT43-4, and/or TaGLP in epidermal cells of tobacco leaves. Coinfiltrated constructs are indicated by the “+” at the left of the figure. ER marker (GFP-HDEL) and trans-Golgi marker (ST-GFP) were used to show the colocalization with YFP-tagged proteins. GFP and YFP fluorescence are shown in green and red, respectively, and their colocalization appears in yellow. Note that YFP-TaGT43-4 export from the ER relies on the presence of TaGT47-13 (B), while the presence of TaGLP does not affect its localization (A). TaGLP-YFP export from the ER requires the presence of both TaGT43-4 and TaGT47-13 (E), while the presence of TaGT43-4 alone does not affect its localization (C). The insets in merge pictures in B and E show the overlap between YFP and ST-GFP. Bars = 10 μm.
TaGT43-4 Interacts with Both TaGLP and TaGT47-13 in the ER Lumen
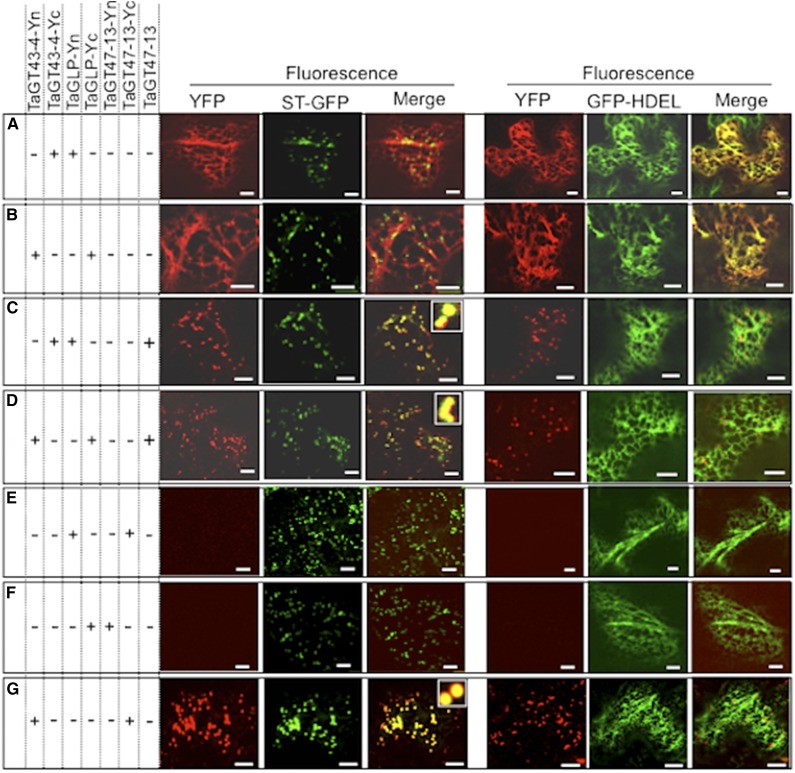
To investigate protein-protein interactions between TaGT43-4, TaGT47-13, and TaGLP, we used split-YFP bimolecular fluorescence complementation (BiFC; Bracha-Drori et al., 2004). We consistently observed YFP fluorescence in the ER, when [TaGT43-4-Yc and TaGLP-Yn] or [TaGT43-4-Yn and TaGLP-Yc] combinations were tested (Fig. 5, A and B), indicating that TaGT43-4 physically interacts with TaGLP and, as expected, both are retained in the ER. This fluorescence was redistributed to the trans-Golgi upon inclusion of untagged TaGT47-13 (Fig. 5, C and D). However, no YFP fluorescence was observed when [TaGT47-13-Yc and TaGLP-Yn] or [TaGT47-13-Yn and TaGLP-Yc] combinations were tested (Fig. 5, E and F), indicating that TaGT47-13 does not directly interact with TaGLP. Coinfiltration of TaGT43-4-Yn and TaGT47-13-Yc constructs resulted in YFP fluorescence that colocalizes with ST-GFP in the trans-Golgi (Fig. 5G), confirming that interaction between TaGT47-13 and TaGT43-4 regulates ER export of the complex. As expected, the controls consisting of coexpression of [Yn-TaGT43-4 and TaGLP-Yc] and [Yn-TaGT43-4 and TaGT47-13-Yc] combinations did not result in any fluorescence (Supplemental Fig. S4, A and B), as Yn and Yc ends are likely located on opposite sides of the membrane. Thus, we concluded that (1) TaGT47-13 interacts with TaGT43-4 but not with TaGLP, (2) TaGT43-4 interacts with both TaGT47-13 and TaGLP, suggesting that TaGT43-4 acts as a scaffold protein, and (3) the trafficking of the complex is dependent of the interaction between TaGT43-4 and TaGT47-13.
Figure 5.
TaGT43-4, TaGT47-13, and TaGLP assemble in the ER before export to the trans-Golgi. Protein-protein interactions were visualized via BiFC (split-YFP). Coinfiltrated constructs are indicated by the “+” at the left of the figure. ER marker (GFP-HDEL) and trans-Golgi marker (ST-GFP) were included to show the localization of the reconstituted YFP. GFP and YFP fluorescence are shown in green and red, respectively, and their colocalization (merge) appears in yellow. The insets in merge pictures in C, D, and G show the overlap between YFP and ST-GFP. Bars = 10 μm
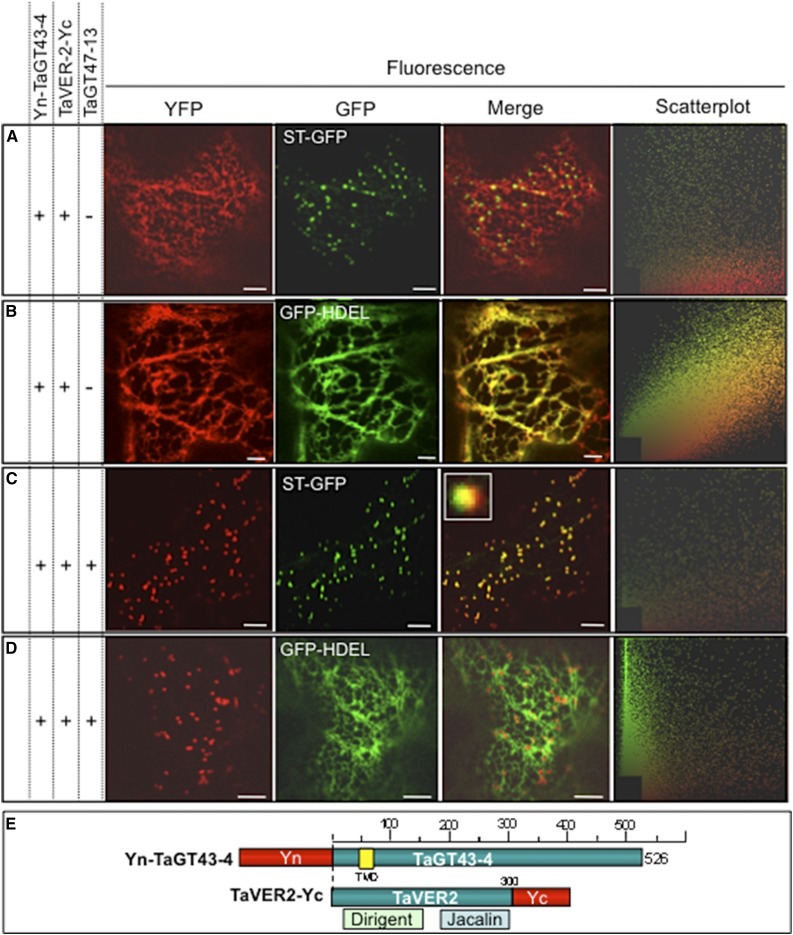
TaGT43-4 Interacts with TaVER2 and TaGT47-13 in the ER and the Assembled Complex Localizes in a Golgi Compartment Having Limited Overlap with Trans-Golgi
To investigate whether TaGT43-4 interacts with TaVER2 in the ER and whether TaGT47-13 is required for ER export of the complex, we used split YFP to monitor their protein-protein interactions and YFP fluorescence movements in live epidermal cells of tobacco leaves. Transient expression of [TaVER2-Yc and Yn-TaGT43-4] resulted in YFP fluorescence that localized in the ER (Fig. 6B) without any overlap with the trans-Golgi (Fig. 6A). However, transient coexpression of this combination with untagged TaGT47-13 resulted in the shift of the fluorescence to a Golgi compartment showing partial overlap with the trans-Golgi (scatterplot and inset in Fig. 6C) and no overlap with the ER (Fig. 6D). These findings suggest that TaVER2 may play a role in preventing the complex from reaching its proper and final localization within the Golgi. Additional experimental work is required to determine the exact physiological role of TaVER2 in trafficking of wheat XSC and xylan biosynthesis.
Figure 6.
TaGT43-4, TaVER2, and TaGT47-13 assemble in the ER before export to the Golgi. Protein-protein interactions were investigated using BiFC (split-YFP). The data show that TaGT43-4 interacts with TaVER2 in the ER to form a complex that is retained in the ER (A and B) until interaction with untagged TaGT47-13, which results in the export of the complex from the ER to a Golgi compartment that has partial overlap with trans-Golgi (C and D). ST-GFP and GFP-HDEL, and trans-Golgi and ER markers were used to show the colocalization of the assembled YFP. GFP and YFP fluorescence are shown in green and red, respectively, and their colocalization (merge) appears in yellow. Two-dimensional scatterplots on the right display the degree of overlap between the red and green in the images. The inset in merge picture in C shows the limited overlap between YFP and ST-GFP. Bars = 10 μm. Schematic presentations of YFP-tagged constructs used in this study are shown in E. TaVER2 is a soluble protein containing two domains: a dirigent domain at the NH2-terminal end and a lectin domain (Jacalin) at the COOH-terminal end.
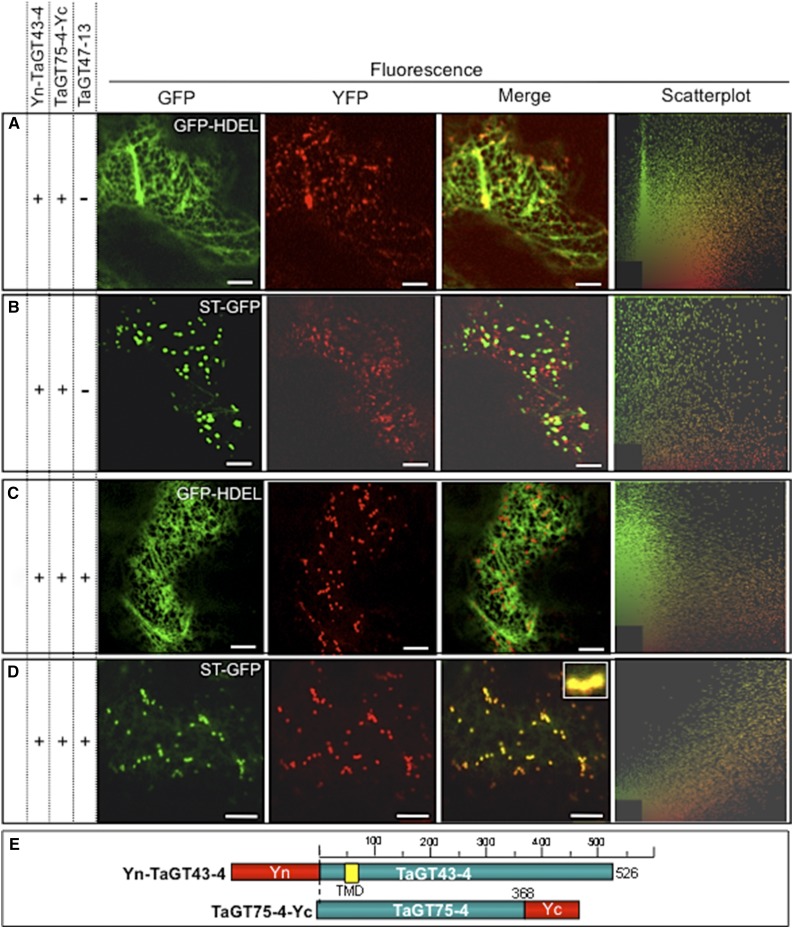
TaGT43-4 Interacts with TaGT75-4 and TaGT75-3 at Punctate Structures at the Surface of the ER
To test if TaGT43-4 acts as a scaffold protein, we determined whether it interacts with TaGT75-3 and TaGT75-4 on the cytosolic side of the membrane and whether this interaction also takes place in the ER. First, we determined the subcellular localization of TaGT75-3/-4 through transient expression of TaGT75-4-YFP, YFP-TaGT75-4, or TaGT75-3-YFP constructs individually. The resulting YFP fluorescence localizes mostly (80%) with ST-GFP, but ∼20% of the fluorescence localizes in certain areas at the surface of the ER seen as punctate structures that had no overlap with ST-GFP (indicated by arrows in Supplemental Fig. S5J). Using split-YFP BiFC, we next monitored YFP fluorescence reconstitution of [TaGT75-4-Yc and Yn-TaGT43-4] after coinfiltration into tobacco leaf epidermal cells. As demonstrated in Figure 7, the reconstituted YFP fluorescence stained the ER weakly and was mostly concentrated in certain areas of the ER seen as punctate structures that had limited overlap with the ER (GFP-HDEL; Fig. 7A) and no overlap with the trans-Golgi marker (ST-GFP; Figure 7B). Indeed, the fluorescence staining from GFP-HDEL was mostly excluded from these punctate structures (merge in Figure 7A) and appeared to be similar to the structures observed in Supplemental Figure S5J. Also, the mobility of YFP fluorescence in these structures was more similar to the mobility of the ER than trans-Golgi. As expected, the control [TaGT75-4-Yc and TaGT43-4-Yn], [TaGT75-4-Yc and TaGT47-13-Yn], or [TaGT75-4-Yn and TaGT47-13-Yc] combinations did not result in any fluorescence (Supplemental Fig. S4, C–E, respectively). Importantly, coinfiltration of TaGT75-4-Yc and Yn-TaGT43-4 constructs along with untagged TaGT47-13 resulted in a shift of the YFP fluorescence to the trans-Golgi, as judged by its overlap with ST-GFP (Fig. 7D). This result confirms that TaGT43-4 physically interacts with TaGT75-4 and the importance of TaGT47-13 in ER export of the complex. Furthermore, TaGT43-4 can also interact with TaGT75-3 (Fig. 8J). These findings further confirm the role of TaGT43-4 as a scaffold protein that can interact with at least five proteins (TaGT47-13, TaGLP, TaVER2, TaGT75-3, and TaGT75-4).
Figure 7.
TaGT43-4 interacts with TaGT75-4 at specific areas of the ER (A and B). The inclusion of untagged TaGT47-13 results in ER export of “TaGT43-4/TaGT75-4” complex to trans-Golgi (C and D). Protein-protein interaction between TaGT43-4 and TaGT75-4 was investigated using BiFC (split-YFP). ST-GFP and GFP-HDEL, and trans-Golgi and ER markers were used to show the colocalization of the assembled YFP. GFP and YFP fluorescence are shown in green and red, respectively, and their colocalization (merge) appears in yellow. Two-dimensional scatterplots on the right display the degree of overlap between the red and green in the images. The inset in merge picture in D shows the overlap between YFP and ST-GFP. Bars = 10 μm. Schematic presentations of YFP-tagged constructs used in this experiment are shown in E.
Figure 8.
Self-assembly of TaGT43-4, TaGT75-3, TaGT75-4, TaGT47-13, and TaGLP was investigated through BiFC (split-YFP). Both TaGT43-4 and TaGLP can form homodimers that localize to the ER (A and D, respectively). TaGT75-3 and TaGT75-4 can form homodimers mostly localize to the Golgi (G and I, respectively) and can also form heterodimers (H). TaGT47-13 cannot self-assemble (F). Interaction of TaGT43-4 homodimers with TaGT47-13 results in the shift of the fluorescence to the trans-Golgi (B), while TaGLP homodimers are retained in the ER until interaction with both TaGT43-4 and TaGT47-13 (E). ER marker (GFP-HDEL) and trans-Golgi marker (ST-GFP) were included to show the localization of the assembled YFP. GFP and YFP fluorescence are shown in green and red, respectively, and their colocalization (merge) appears in yellow. Bars = 10 μm.
Knowing from previous work that mutases are mostly associated with Golgi membranes but not with the ER membranes (Dhugga et al., 1997; Drakakaki et al., 2006), our results suggest that TaGT75-3 and TaGT75-4 are most likely added to the core wheat XSC (TaGT43-4/TaGT47-13 complex) in the last step, which takes place at these punctate structures that have less overlap with the ER. Our hypothesis is that these punctate structures could represent ERESs (daSilva et al., 2004; Hanton et al., 2009) where XSCs are assembled before delivery to the Golgi. Further work is needed to determine the exact nature of these punctate structures.
TaGT43-4, TaGT75-3, TaGT75-4, and TaGLP, But Not TaGT47-13, Can Self-Assemble
To determine the stoichiometry of the complex, we analyzed the homo-oligomerization of TaGT43-4, TaGT75-3/-4, TaGT47-13, and TaGLP using split-YFP. Transient coexpression of combinations [TaGT43-4-Yc and TaGT43-4-Yn] (Fig. 8A), [TaGLP-Yc and TaGLP-Yn] (Fig. 8D), [TaGT75-4-Yc and TaGT75-4-Yn] (Fig. 8G), or [TaGT75-3-Yc and TaGT75-3-Yn] (Fig. 8I) resulted in YFP fluorescence reconstitution, indicating that these proteins can form homodimers. TaGT47-13, on the other hand, did not seem to interact with itself, as no YFP fluorescence was observed with the infiltration of [TaGT47-13-Yc and TaGT47-13-Yn] combination despite many attempts (Fig. 8F). Heterodimerization was also observed with TaGT75-3 and TaGT75-4 ([TaGT75-3-Yc and TaGT75-4-Yn]; Fig. 8H) and with TaGT75-3 and TaGT43-4 ([TaGT75-3-Yc and Yn-TaGT43-4]; Fig. 8J). As expected, the control [Yn-TaGT43-4 and TaGT43-4-Yc] combination did not result in fluorescence reconstitution (Supplemental Fig. S4F). The fact that coexpression of untagged [TaGT43-4 and TaGT47-13] and [TaGT43-4, TaGT47-13, TaGT75-3, and TaGT75-4] combinations in P. pastoris cells resulted in the formation of complexes with molecular mass of ∼160 and ∼240 kD, respectively (Fig. 2, A and B), is consistent with the conclusion that TaGT43-4 and TaGT75-3/-4 form homo/heterodimers, while TaGT47-13 does not. Moreover, because immunoblot analysis under nonreducing/nondenaturing conditions of TaGT43-4 alone did not detect homodimers (Fig. 2C), the data suggest that no disulfide linkages are formed between the two TaGT43-4s in the homodimers and these homodimers are not stable to detergent solubilization in the absence of TaGT47-13.
Our data indicate that both TaGT43-4 and TaGLP homodimers are retained in the ER (Fig. 8, A and D). However, the fluorescence from TaGT43-4 homodimers clearly redistributes to the trans-Golgi in presence of untagged TaGT47-13 (Fig. 8B), but not in the presence of untagged TaGLP alone (Fig. 8C). On the other hand, and as expected, redistribution to the trans-Golgi of YFP fluorescence from TaGLP homodimers required the presence of both untagged TaGT43-4 and TaGT47-13 (compare Fig. 8, D and E). Taken together these findings are consistent with the conclusion that wheat XSC assembly initiates with the formation of TaGT43-4 homodimers (scaffold) in the ER to which other proteins (i.e. TaGLP and TaGT75s) are added with TaGT75-3/-4 added last.
Intracellular Localization of TaGT43-4, TaGT47-13, and TaGT75-3/-4 in Wheat Tissues
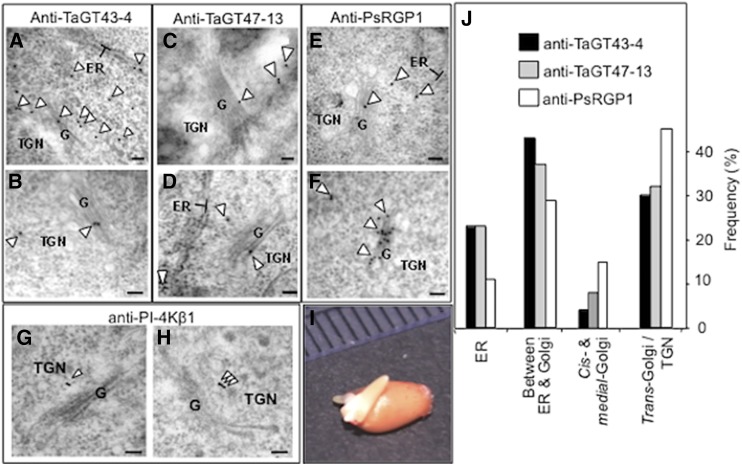
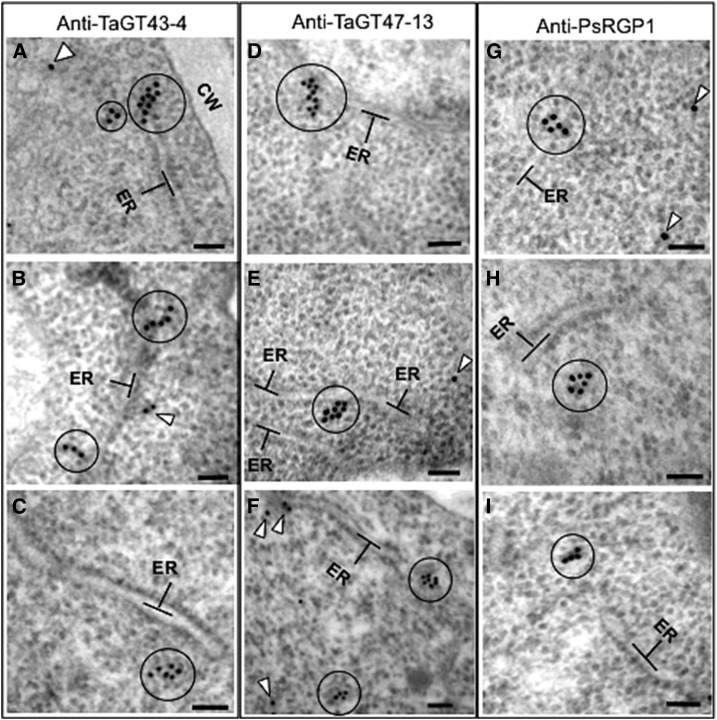
Having shown that TaGT43-4, TaGT47-13, TaGT75-3, and TaGT75-4 can form complexes in tobacco leaf epidermal and P. pastoris cells, we next sought to determine the intracellular localization of these proteins in their native tissues in young etiolated wheat seedlings (Fig. 9I) using purified anti-TaGT43-4 and anti-TaGT47-13 antibodies and immunogold labeling TEM. We used thin sections to increase epitope exposure and accessibility of antibodies to these epitopes. According to confocal microscopy data, TaGT43-4 and TaGT47-13 are expected to be localized to the ER and the Golgi but not in the plasma membrane. This turned out to be true, as the two antibodies gave very similar labeling patterns (Fig. 9, A–D). Specifically, they labeled mostly the ER and the area between the ER and cis-Golgi consistent with intense trafficking of these proteins between the ER and the cis-Golgi. The next most abundant labeling was observed on trans-Golgi and TGN. No labels were detected at the plasma membrane or cell wall. To determine the location of TGN, we used anti-PI-4Kβ1 antibodies against the Arabidopsis RabA4b effector protein (phosphatidylinositol-4 kinase, PI-4Kβ1; Kang et al., 2011), which localizes to budding secretory vesicles in the TGN or en route to the cell surface (Fig. 9, G and H). Statistical counting of ∼50 cells (gold particles n = 558 for anti-TaGT43-4 and n = 188 for anti-TaGT47-13) indicated that 65% of the labeling from both antibodies was associated with the ER and the area between the ER and the cis-Golgi, ∼30% with trans-Golgi and TGN, and only ∼5% of the labeling on cis- and medial-Golgi compartments (Fig. 9J). When anti-PsRGP1, a well characterized antibody with high specificity (Dhugga et al., 1997), was used, a very similar labeling pattern as anti-TaGT43-4 and anti-TaGT47-13 antibodies was observed (Fig. 9, E and F). Statistical counting (∼50 cells, gold particles n = 451) also showed similar distribution, namely, ∼10% of the labeling was associated with the ER, ∼30% with the area between the ER and the cis-Golgi, ∼50% with trans-Golgi and TGN, and only ∼10% of the labeling on cis- and medial-Golgi compartments (Fig. 9J). This distribution of labeling with anti-TaGT43-4, anti-TaGT47-13, and anti-PsRGP1 is consistent with confocal microscopy data and is expected if a multiprotein complex must be assembled in the ER before transfer to the Golgi.
Figure 9.
Immunoelectron micrographs of anti-TaGT43-4 (A and B), anti-TaGT47-13 (C and D), anti-PsRGP1 (E and F), and anti-PI-4Kβ1 (G and H) in shoots of 3-d-old etiolated wheat seedlings (I). Particles are marked with arrowheads. Individual immunogold particles (marked with arrowhead) are seen at the ER surface, between the ER and the cis-Golgi, and on the trans-Golgi and TGN. Note that anti-PsRGP1 labels on ER membranes were exclusively associated with the clusters (see Fig. 10), as no individual labels on ER membranes were observed outside of these clusters. Frequency (the number of particles counted from ∼50 cells) of immunogold labeling localization is presented in J. Bars = 100 nm.
During this analysis, we consistently observed that the three antibodies produced clusters of labels (3 to 10 labels) located in structures seen as protrusions/extensions from the ER (circled in Fig. 10, A, B, D, and G) or detached clusters but close to the ER (circled in Fig. 10, C, F, H, and I). We saw this clustering in 5 to 15% of the cells observed (depending on the antibody), with anti-TaGT47-13 showing the lowest amount of clustering. It is unlikely that these clusters of labeling are due to clumping of antibodies, as gold particles are not in contact with each other and there is no problem with the fixation of the samples. Anti-PsRGP1 labels on ER membranes were exclusively associated with the clusters, as no individual labels on ER membranes were observed outside of the clusters. This is consistent with confocal microscopy data (Fig. 7; Supplemental Fig. S5), showing TaGT75-3/-4 interacting with TaGT43-4 at areas having limited overlap with the ER. These data again suggest that TaGT75-3 and TaGT75-4 may be added to the complex later in the assembly of the wheat XSC.
Figure 10.
Immunoelectron micrographs of anti-TaGT43-4 (A–C), anti-TaGT47-13 (D–F), and anti-PsRGP1 (G–I) in shoots of 3-d-old etiolated wheat seedlings. Circles indicate clusters of 3 to 10 immunogold particles seen as protrusions/extensions from the ER (A, B, D, and G) or as clusters detached but near the ER (C, F, H, and I). Arrowheads indicate individual immunogold particles. Bars = 100 nm.
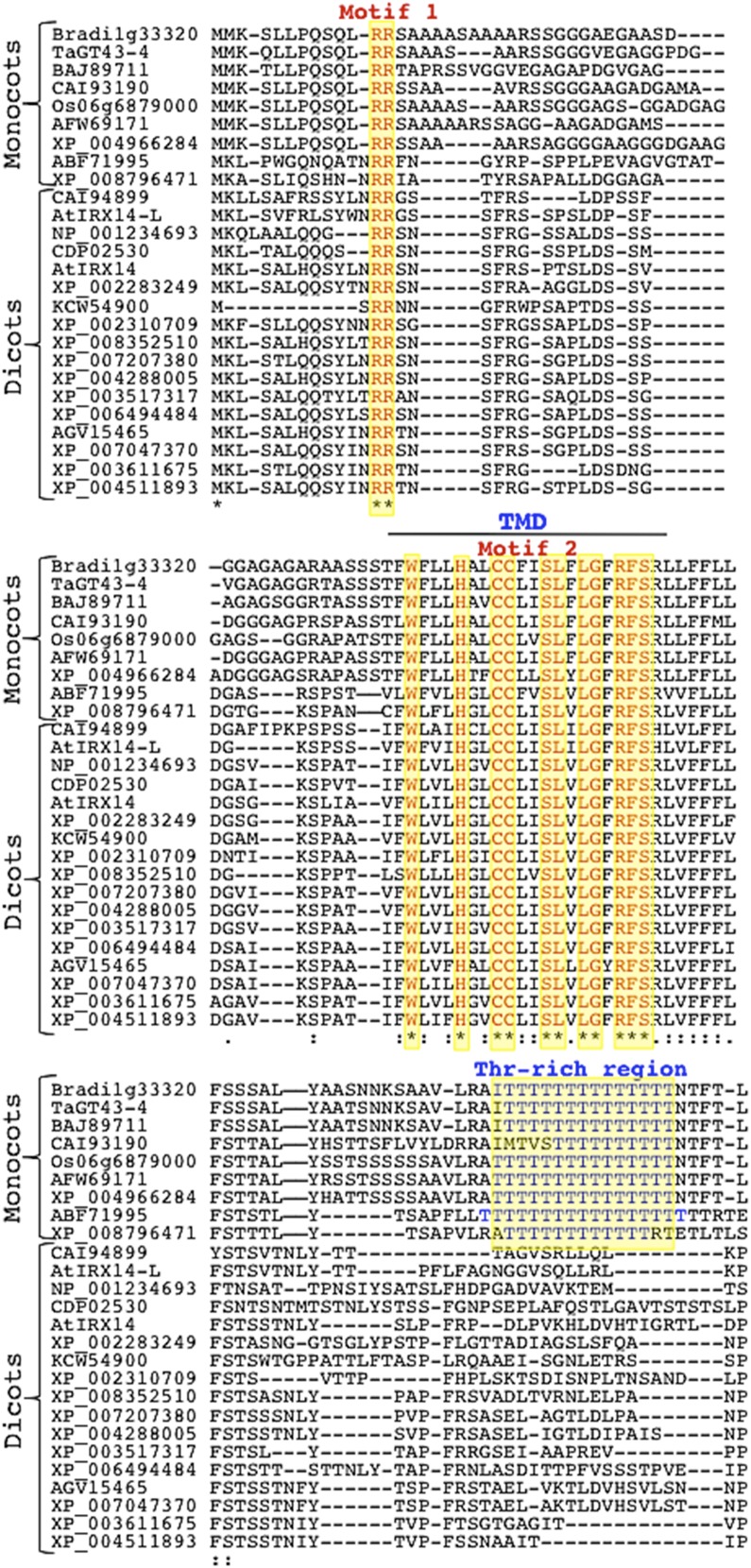
TaGT43-4 and Homologous Members from GT43 Family Have Poorly Conserved NH2-Terminal Regions, But Share Two Conserved Motifs
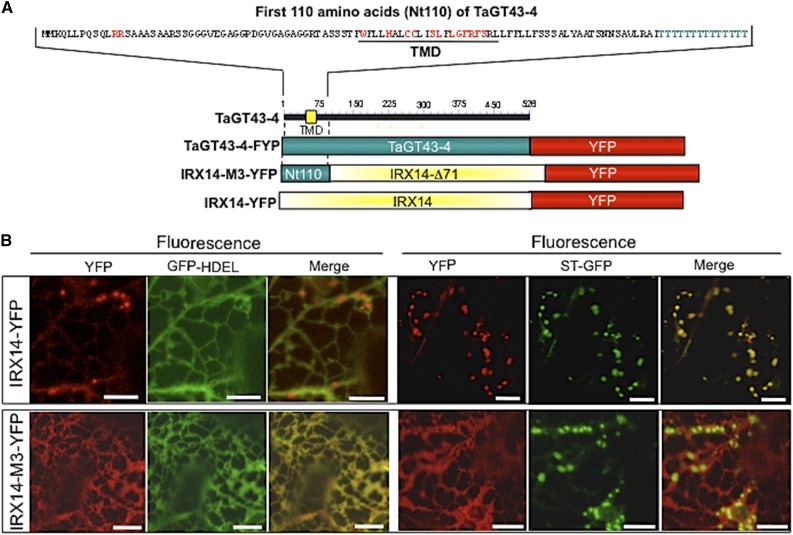
As mentioned above, TaGT43-4 is the only integral membrane protein (type II) identified by proteomics in the XSC. TaGT43-4 has a long NH2-terminal end (∼49 amino acids) on the cytosolic side of the ER and physically interacts with TaGT47-13, TaGT75s, and TaGLP (e.g. it acts as a scaffold protein). Thus, TaGT43-4 must contain the necessary sorting signal sequences that control trafficking and subcellular localization of the complex. This prompted us to analyze the sequences of TaGT43-4 and 25 homologous proteins of the GT43 family (including Arabidopsis IRX14/IRX14-L). Phylogenetic analysis clustered monocot and dicot proteins into two separate groups supported by strong bootstrap values (Supplemental Fig. S6). Interestingly, while the luminal regions (after the first 110 amino acids) of monocot and dicot proteins showed high sequence conservation (∼60% identity and ∼85% similarity), the NH2-terminal regions (the first 110 amino acids consisting of ∼49 amino acids on the cytosolic side, ∼20 amino acids of the predicted TMDs, and ∼40 amino acids on the lumen side) are poorly conserved (∼40% identity and ∼60% similarity). Despite this poor conservation, sequence alignment of the NH2-terminal regions revealed two conserved motifs, namely, a di-Arg (RR) motif located ∼10 amino acids from the NH2-terminal ends and a Wx3Hx2CCx2Sx2LGxRFS motif found within the predicted TMDs (Fig. 11). In addition, a Thr-rich region consisting of 12 to 14 Thr residues located ∼26 amino acids downstream of the TMDs on the luminal side (Fig. 11) seems to be specific to monocot homologs. Although the roles of the conserved motifs and Thr-rich regions are not known, we hypothesize that the NH2-terminal regions may contain different sorting signals specific to monocot and dicot proteins. To test this hypothesis, we generated a chimeric construct encoding for IRX14-M3-YFP fusion protein in which the first 71 amino acids of Arabidopsis IRX14 protein were replaced with the first 110 amino acids of TaGT43-4 (Fig. 12A). As shown in Figure 12B, expression of IRX14-YFP construct resulted in YFP fluorescence that colocalized mainly with ST-GFP in the trans-Golgi, while expression of the chimeric construct IRX14-M3-YFP resulted in YFP fluorescence that colocalized exclusively with GFP-HDEL in the ER. Trans-Golgi localization of IRX14-YFP contrasts ER localization of TaGT43-4-YFP (Figs. 3–5) but is in agreement with earlier work showing Golgi localization of Arabidopsis IRX14 when coexpressed with Arabidopsis fragile fiber8 (FRA8) from GT47 family (Lee et al., 2010). On the other hand, ER localization of IRX14-M3-YFP is reminiscent of ER localization of TaGT43-4-YFP and contrasts trans-Golgi localization of IRX14-YFP, which strongly supports the conclusion that NH2-terminal regions of TaGT43-4 and Arabidopsis IRX14 have different localization/sorting signals. The difference in sorting information along with the assembly-dependent trafficking may point to a major difference in the mechanisms that regulate/control xylan biosynthesis between monocots and dicots. This could explain why some monocot GT43 genes partially (or do not) complement Arabidopsis irx mutants (Lee et al., 2014). More analyses are underway to dissect the sorting mechanisms of these monocot proteins.
Figure 11.
Alignment of the NH2-terminal regions (first 100 amino acids) of TaGT43-4 and 25 homologous proteins of the GT43 family. Sequences were obtained from NCBI, Phytozome 10.3, or CAZy database. The alignment was generated through MUSCLE 3.7 program in the Phylogeny.fr platform. The two conserved motifs (RR and Wx3Hx2CCx2Sx2LGxRFS in red) as well as a Thr-rich region specific to monocot proteins (in blue) are highlighted in yellow. Monocot and dicot proteins with their accession numbers are indicated.
Figure 12.
Confocal images of epidermal cells of tobacco leaves transiently expressing Arabidopsis IRX14-YFP (B) or IRX14-M3-YFP, which encodes a protein having the first 71 amino acids replaced with the first 110 amino acids from TaGT43-4 (Nt110; C). Schematic presentations of these constructs are shown in A. ST-GFP and GFP-HDEL, and trans-Golgi and ER markers were used to show YFP localization. GFP and YFP fluorescence are shown in green and red, respectively, and their colocalization (merge) appears in yellow. NH2-terminal region of TaGT43-4 is sufficient to retain IRX14-M3-YFP in the ER. Bars = 10 μm.
DISCUSSION
Despite the fact that several GTs associated with xylan biosynthesis have been identified, isolation and characterization of multiprotein XSCs in plants is currently lacking. We previously immunopurified an active wheat XSC having XXylT, XAT, and XGlcAT activities working cooperatively to synthesize xylan-like polymers in vitro (Zeng et al., 2010). In this work, we describe proteomic analysis of this wheat XS activity, as well as its assembly and trafficking. Our data revealed that the purified activity is mostly enriched in six proteins: TaGT43-4, TaGT47-13, TaGT75-3, TaGT75-4, TaGLP, and TaVER2. We have no evidence that other GTs are associated with the purified XS activity, specifically, no wheat homologs of the previously described Arabidopsis GUX members of the GT8 family (Mortimer et al., 2010; Lee et al., 2012; Rennie et al., 2012) or the wheat α(1,3)XATs members of the GT61 family (Anders et al., 2012) were identified (Table I). Furthermore, the search of a smaller database containing protein sequences specifically from GT2, GT8, GT43, GT47, GT61, and GT75 families from wheat, barley, rice, B. distachyon, and Arabidopsis also failed to identify GT members other than GT43, GT47, and GT75. Therefore, we conclude that if members of the GT2, GT8, and GT61 families are part of wheat XS activity, they must interact weakly (or have unstable interactions) with the purified activity, which would explain why they weren’t detected in our proteomic analysis.
Implications of Proteomic Analysis in Xylan Biosynthesis
Earlier work showed that homologs of TaGT43-4 and TaGT47-13 in Arabidopsis (IRX14/IRX14-L and IRX10/IRX10-L, respectively) and other plant systems have XXylT activity responsible for xylan backbone synthesis (Lee et al., 2012a, 2014; Jensen et al., 2014; Urbanowicz et al., 2014). Thus, the presence of both TaGT43-4 and TaGT47-13 in wheat XSC is consistent with a role in xylan backbone synthesis and may suggest the need for two or more GTs cooperating in polymerizing Xyl residues into β(1,4)xylan chain, which may help overcome the fact that two consecutive Xyl residues need to be rotated 180° relative to each other. However, a recent site-directed mutagenesis study suggested that some members of the GT43 family might only have a structural role (Ren et al., 2014). TaGT43-4 in wheat XSC would fit this description, as it is the only integral membrane protein identified by proteomics, it is able to interact with five proteins (TaGT47-13, TaGT75s, TaVER2, and TaGLP), and its interaction with TaGT47-13 controls ER export of the complex. The fact that Ren et al. (2014) also showed that mutations in the conserved DXD motif in Arabidopsis IRX14 resulted in a protein that could not complement the irx14 mutant phenotype would suggest that TaGT43-4 might also have XXylT activity that contributes to xylan backbone synthesis. Thus, further work is needed to determine how several GTs work together to synthesize the xylan backbone. The identification of TaGT75-3 and TaGT75-4 (homologs to rice mutases UAM3 and UAM1, respectively) in wheat XSC is also in agreement with their role in arabinoxylan biosynthesis as putative mutases that catalyze the interconversion of UDP-Arap to UDP-Araf (Konishi et al., 2007; Rautengarten et al., 2011). Konishi et al. (2010) also observed a synergistic effect in their study on rice, where combining UAM2 with UAM1 or UAM3 resulted in an increase in mutase activity (up to 2-fold). The presence of two different mutases (TaGT75-3/-4) in wheat XSC could provide a similar positive synergistic effect. We previously showed that purified wheat XS activity is able to synthesize an arabinoxylan-like polymer in vitro (Zeng et al., 2010). However, it was surprising that no members of the GT61 families were identified in our proteomic analysis, as XAT is needed to account for the arabinosyl transfer activity observed in purified wheat XS activity. It was also puzzling that no retaining GTs (i.e. members of the GT8 family) were identified by our proteomic analysis to account for the XGlcAT activity also observed in purified wheat XS activity (Zeng et al., 2010). Although it might be true that the purified wheat XS activity may contain members of these two GTs that are present in nondetectable quantities, we cannot completely rule out the possibility that the wheat XSC, consisting only of TaGT43-4, TaGT47-13, TaGT75-3, TaGT75-4, TaGLP, and TaVER2, might be sufficient to catalyze the three transferase activities (XXylT, XAT, and XGlcAT). If this hypothesis is confirmed, it would mean that additional mechanisms for α-Ara and α-GlcA incorporation onto xylan backbone exist. More experimental work is needed to verify these possibilities and determine the mechanisms by which Xyl, Ara, and GlcA are cooperatively incorporated into xylan polymer by wheat XSC.
Another important implication from the proteomic analysis is the identification of two possible new players in xylan biosynthesis in grasses: TaGLP and TaVER2. Identification of TaGLP gives support to GLP’s gene ontology prediction: GO:0045492 (role in xylan biosynthesis). TaGLP belongs to the cupin superfamily, and cupin domain-containing proteins have previously been linked to polysaccharide synthesis in bacteria but not in plants. A bacterial protein involved in capsular polysaccharide biosynthesis (CapF) has been crystallized recently (2ZKL.pdb; http://www.rcsb.org/pdb/explore.do?structureId=2ZKL; Miyafusa et al., 2012). CapF contains a COOH-terminal cupin domain linked to an NH2-terminal Rossman-fold nucleotide-binding domain, common to GTs. The cupin domain stabilizes the CapF homodimer through protein-protein interaction and catalyzes C3 epimerization of UDP-4-hexulose (Miyafusa et al., 2012). Thus, cupin domain of TaGLP could act as a stabilizing protein for TaGT43-4 homodimers and/or TaGT43-4/TaGT47-13 complex. Since TaGLP can form homodimers (Fig. 8D), cupin domains could also simply function as stabilizers of TaGLP homodimers. Another possible function of TaGLP is to recruit other interacting proteins, such as GLP-binding proteins and DING proteins, which have been shown to associate with plant GLPs in vivo (Dunwell et al., 2000; Berna et al., 2002). DING proteins have a type 2 periplasmic binding fold (PBP2), which is also found in the substrate-binding domain of a cellulose-binding protein from Thermotoga maritima (TmCBP). TmCBP binds a variety of lengths of β(1,4)-linked Glc oligosaccharides (Cuneo et al., 2009). Therefore, it is possible that DING/GLP-binding proteins may act as polysaccharide chaperones that interact with the newly synthesized xylan chain to facilitate its rotation/movement through XSC.
TaVER2 was previously associated with wheat flowering, as knockdown of TaVER2 caused delayed flowering, while overexpressing wheat plants had accelerated-flowering phenotype (Yong et al., 2003; Xiao et al., 2014). However, TaVER2 has never been associated with cell wall polysaccharides biosynthesis. The fact that the assembled “TaGT43-4/TaVER2/TaGT47-13” complex showed limited overlap with trans-Golgi (Fig. 6C) suggests that the function of TaVER2 may be to control xylan synthesis by preventing XSC delivery to its proper localization (trans-Golgi), which could result in less activity. Because TaVER2 expression is up-regulated by jasmonic acid (Feng et al., 2009) and TaGLP has structural similarity to auxin-binding proteins, a possible function of these two proteins could be to modulate XS activity through the action of phytohormones (auxin and jasmonic acid). This modulating role is consistent with the observation that (1) cell growth in grass coleoptiles is induced by auxin and suppressed by jasmonic acid and abscisic acid, and (2) overexpression of OsJAC1 (rice homolog to TaVER2) suppresses coleoptile and stem elongation in rice (Jiang et al., 2007). As regulators, TaGLP and TaVER2 may not be required for XS activity and therefore may not be a permanent component of wheat XSC. Additional experimental work is required to determine the exact role of TaGLP and TaVER2 in xylan synthesis, and our hypotheses represent a starting framework for further experiments.
Implications of Assembly-Dependent Trafficking in Xylan Synthesis
We showed that the forward transport of TaGT43-4, TaGT47-13, and TaGLP proteins is coupled with their assembly into a complex in the ER. Such an assembly-dependent trafficking mechanism has been described in several membrane multiprotein complexes in mammalian and yeast cells and appears to involve a “quality control” step regulating the assembly of functional complexes before exiting the ER. For example, ATP-sensitive potassium channels in animal cells need to assemble into complexes containing specific numbers and correct subunits before exiting the ER (Zerangue et al., 1999), as do G-protein-coupled γ-aminobutyric acid receptors (Margeta-Mitrovic et al., 2000), invariant chain of the major histo-compatibility complex class II (Khalil et al., 2003), and IgG antibodies in animal blood cells (Baumal et al., 1971; Mains and Sibley, 1983). In all these cases, unassembled subunits or partial complexes are retained in the ER. This assembly-dependent trafficking mechanism has also been described in other membrane complexes involved in polysaccharide biosynthesis, such as heparan sulfate synthase (HA) in mammalian/yeast cells, which shares some biochemical properties with wheat XSC. Both wheat XSC and HA synthase are Golgi localized. Physical interaction between the two HA synthase subunits EXT1 and EXT2 is required for ER export (McCormick et al., 2000), and expression of EXT proteins individually results in their retention in the ER (McCormick et al., 2000), which is reminiscent of ER retention of TaGT43-4 and TaGLP. Similarly, TaGT43-4 and TaGT47-13 interaction is also required for ER export of the wheat XSC. Our immunogold TEM microscopy data suggest that the wheat XSC assembly process is sufficiently slow to allow the observation of ER sites where the complex is assembled before export, which are seen as protrusions/extensions from the ER labeled by clusters of immunogold labels (Fig. 10). These clusters of labels were also seen detached but close to the ER (Fig. 10). Our confocal microscopy data suggest that TaGT75-3 and TaGT75-4 are added to the complex in the last step at certain punctate structures in the ER before export from the ER (Fig. 7). These punctate structures are most likely the same ER areas labeled with clusters of immunogold particles in our TEM analysis.
Although the exact mechanism of assembly-dependent trafficking of wheat XSC assembly has not yet been fully elucidated, it is tempting to propose that the assembly of GTs into complexes, such as wheat XSC and HA synthase, occurs through a coupled folding and binding mechanism described for the assembly of IgG subclasses (Baumal et al., 1971; Dyson and Wright 2002), which involves chaperones to ensure that only specific combinations of heavy chains and two identical light chains are correctly associated and functional (Mains and Sibley, 1983). The assembly of IgGs starts with the formation of heavy chain homodimers that are actively retained in the ER unless they associate with light chains, which can exit the ER alone. In the case of wheat XSC, the assembly of the complex would start with the formation of TaGT43-4 homodimers. TaGT43-4 dimerization could possibly involve hydrogen bonds, hydrophobic and electrostatic interactions involving the COOH-terminal end of the globular domain of TaGT43-4, as was observed in the solved crystal structure of human glucuronosyltransferase, GlcAT-P (also belonging to the GT43 family), showing the COOH-terminal region forming a long loop that extends to the other molecule in the homodimer (Kakuda et al., 2004). The fact that the monomeric form of TaGT43-4 (∼52 kD) is the major species in nonreducing/nondenaturing gel electrophoresis (Fig. 2C) indicates that the two TaGT43-4s in a homodimer are not stable to detergent extraction and interaction with TaGT47-13 allows the formation of stable complexes. Partial complexes are retained in the ER (or ERES) unless they interact with TaGT47-13. Several close homologs of TaGT43-4, TaGT47-13, and TaGLP exist in wheat and other grasses, which can give rise to different complexes having various protein combinations and differences in biochemical characteristics. Therefore, a coupled folding and binding mechanism would ensure that only specific GT combinations are assembled into functional complexes, allowing a cell to assemble various complexes from closely related GTs (i.e. IRX9/IRX9-L versus IRX14/IRX14-L; IRX7/IRX7-L versus IRX10/IRX10-L, etc.), as was proposed earlier (Faik et al., 2013). The fact that the purified wheat XSC did not contain homologs to Arabidopsis IRX9/IRX9-L or IRX7/IRX7-L is also consistent with this idea and would also explain why IRX genes do not fully complement one another (Brown et al., 2007).
TaGT47-13 Might Play a More Important Role in the Assembly, Trafficking, and Subcellular Localization of Wheat XSC
It was surprising to observe that TaGT47-13-YFP alone can exit the ER and accumulate in the trans-Golgi (Fig. 3D). One explanation for this might be that the SP of TaGT47-13 is not cleaved from the mature protein and instead serves to anchor TaGT47-13 to the membrane and as a Golgi localization signal. In support of this hypothesis, expression of TaGT47-13 in P. pastoris cells showed both processed TaGT47-13 in the culture medium with the correct size (∼44 kD), as well as unprocessed TaGT47-13 (including SP) in microsomal fractions (∼56 kD; Supplemental Fig. S1). Furthermore, the XSC complex seems to contain only the unprocessed form of TaGT47-13, as no ∼44-kD band was observed in purified wheat XS activity (Zeng et al., 2010) or in XSC expressed in P. pastoris (Fig. 2, A and B). Therefore, unprocessed TaGT47-13 can exit the ER alone and accumulate in the Golgi (suggesting that the SP may in fact be a cryptic TMD and Golgi localization signal). Analysis of the SP sequence using the TMD organelle predictor program (Sharpe et al., 2010) predicts Golgi localization of unprocessed TaGT47-13 (with score 66), which is consistent with our data. The observation that SP-YFP-TaGT47-13 is retained in the ER may suggest that insertion of YFP after the SP cleavage site might induce defects in the folding of TaGT47-13 itself, which may interfere with the formation/assembly of a nonfunctional “SP-YFP-TaGT47-13/TaGT43-4” complex, resulting in a transport-incompetent complex that is retained in the ER. Therefore, our results suggest that the SP and near-luminal sequence of TaGT47-13 play important roles in the assembly of functional complexes as well as in the trafficking of these complexes. It is unlikely that TaGT47-13-YFP alone exits the ER because of an interaction with a tobacco homolog of TaGT43-4 because this should also apply to TaGT43-4, which is not observed. The data are also consistent with a coupled folding and binding mechanism involving a “quality control” step allowing only properly assembled and functional complexes to exit the ER. TaGT47-13 may not need interaction with other proteins to exit the ER, but its proper folding and assembly with the correct partners could be a prerequisite for ER export. Taken together, our data point to a more important regulatory role of TaGT47-13 in the assembly, trafficking, and subcellular localization of wheat XSC.
The Assembly and Trafficking of Wheat XSC Might Be Linked via Signal Sequences
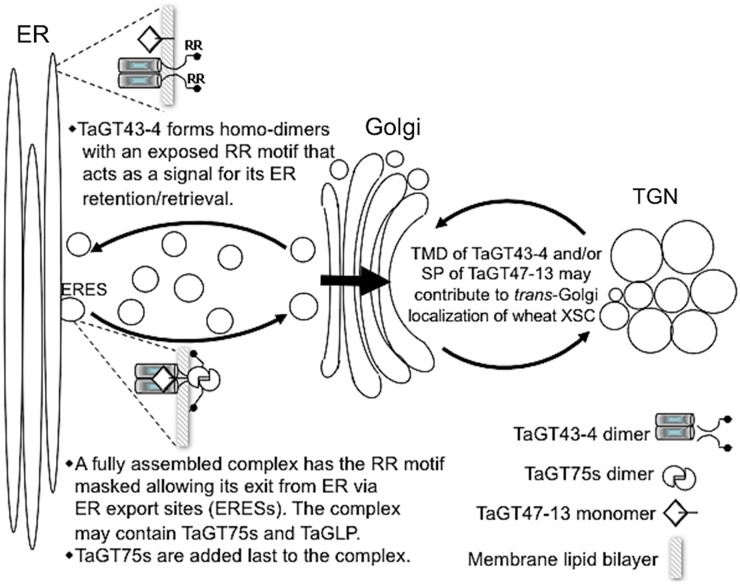
In the assembly-dependent trafficking process, di-basic motifs (i.e. RR, RXR, and RRR) have been involved in linking the assembly of the complexes to their trafficking through ER retrieval pathway (for review, see Michelsen et al., 2005). These di-basic motifs are usually located ∼40 amino acids from the TMDs in type II membrane proteins, such as GTs. Interestingly, one of the conserved motifs in the NH2-terminal region (the first 110 amino acids) of TaGT43-4 (and its homologs in GT43 family from several taxa) is a di-Arg (RR) motif located ∼36 amino acids from the TMD on the cytosolic side, which is consistent with a role in the ER retrieval pathway that retains unassembled proteins and partial/nonfunctional complexes in the ER. How might this RR motif link XSC assembly to its trafficking? It is conceivable that the RR motif is exposed in unassembled TaGT43-4 and partial/nonfunctional complexes, which would act as a “quality control” system to allow retention/retrieval of these partial complexes and ensure that only properly assembled and functional complexes with specific GTs exit the ER. This RR motif would become masked in fully assembled XSCs, allowing them to exit the ER. Conformation changes of TaGT43-4 in the fully assembled and functional XSCs could result in masking this RR motif through steric masking or other mechanisms, such as attachment of additional proteins or posttranslational modifications (i.e. phosphorylation, glycosylation, etc.). Once XSCs are fully assembled and released from the ER, they accumulate in specific subcompartments of the Golgi, which could be mediated by the second conserved motif (Wx3Hx2CCx2Sx2LGxRFS) located in the TMD (Fig. 11), as well as the SP sequence (acting as a TMD) of TaGT47-13. Indeed, TMDs have been shown to play an important role in Golgi retention and homodimerization of many GTs in mammalian, yeast, and plant systems (Aoki et al., 1992; Munro, 1995; Rayner and Pelham, 1997; Brandizzi et al., 2002a). Moreover, it has been shown previously that the Cys-29 and His-32 residues within the TMD of a mammalian galactosyltransferase are critical for its Golgi retention (Aoki et al., 1992). Therefore, it is possible that this second conserved motif in the NH2-terminal region of TaGT43-4, which also contains two Cys and one His residues, may play a role in Golgi retention/accumulation of fully assembled XSCs. A model for the assembly and trafficking of wheat XSC is presented in Figure 13. Work in our labs is underway to experimentally determine the role of the conserved motifs in targeting of XSC. Note that this model does not take in consideration the role of TaVER2 in this trafficking, which is speculative to this point.
Figure 13.
Model for the assembly and trafficking of wheat XSC. This model assumes that the central core of wheat XSC is formed of TaGT43-4 and TaGT47-13 but can include TaGT75s, TaGLP, and/or TaVER2. The assembly of this core complex occurs in the ER through a coupled folding and binding mechanism and starts with TaGT43-4 forming homodimers. However, the trafficking of the functionally assembled complex is signal mediated and involves a di-Arg (RR) motif at the NH2-terminal region of TaGT43-4. This RR motif is exposed in unassembled TaGT43-4 homodimers but is masked in a functional TaGT43-4/TaGT47-13 complex. Unmasked RR motif acts as an ER retention/retrieval signal, while masked RR motif allows the functional complex to escape this retention/retrieval process and exit the ER from ERESs. The accumulation of the complex in trans-Golgi is hypothesized to be under the control of the TMD of TaGT43-4 and the NH2-terminal secretion SP of TaGT47-13, acting as a cryptic TMD. This model assumes that TaGT75s are added to the complex at the ERESs before export to the Golgi.
In summary, the characterization of wheat XSC has revealed several new insights regarding the composition, assembly, and trafficking of wheat XSC. We believe that our work fills in some important gaps in our understanding of xylan and hemicellulose biosynthesis in plants and reconciles biochemical data with some of the genetic data. This work has also established the groundwork for future studies concerning the role of the NH2-terminal regions in controlling the trafficking/sorting of monocot and dicot XSC.
MATERIALS AND METHODS
Chemicals and Plant Materials
Triton X-100, BSA, dl-DTT, phenylmethylsulfonyl fluoride, acid-washed glass beads (425–600 μm), and other chemicals were from Sigma-Aldrich. Protease inhibitor cocktail tablets (cOmplete, EDTA-free) were from Roche, and protease inhibitor cocktail IV was from RPI. Anti-TaGT43-4 and anti-TaGT47-13 were prepared in our lab as described (Zeng et al., 2010). Anti-PsRGP1 and anti-PI-4Kβ1 were generous gifts from Dr. Kanwarpal Dhugga (Pioneer Hi-Bred) and Dr. Erik Nielsen (University of Michigan, Ann Arbor), respectively. Protein concentrations were determined by Bradford assay (Bio-Rad) using BSA as the standard. Plasmid miniprep and gel extraction kits were purchased from Qiagen.
Proteomic Analysis
Instrument and Conditions
Proteomics was carried out at the Mass Spectrometry and Proteomics Facility at the Campus Chemical Instrument Center of OH State University using capillary-liquid chromatography tandem mass spectrometry on a Thermo Scientific LTQ mass spectrometer equipped with a CaptiveSpray source (Bruker Daltonics) operated in positive ion mode. The liquid chromatography system was an UltiMate 3000 system from Thermo Scientific. Solvent A was water containing 50 mm acetic acid, and the solvent B was acetonitrile. Five microliters of each sample was first injected onto the μ-Precolumn Cartridge (Thermo Scientific) and washed with 50 mm acetic acid. The injector port was switched to inject and the peptides were eluted off of the trap onto the column. A 0.2 × 150 mm, 3u, 200A, Magic C18 (Bruker Daltonics) was used for chromatographic separations. Peptides were eluted directly off the column into the LTQ system using a gradient of 2 to 80% B over 45 min, with a flow rate of 2 μL/min. The total run time was 60 min. The mass spectra were acquired according to standard conditions established in the lab. Briefly, a CaptiveSpray source operated with a spray voltage of 3 kV and a capillary temperature of 200°C. The scan sequence of the mass spectrometer was based on the TopTen method; the analysis was programmed for a full scan recorded between 350 and 2000 D and a tandem mass spectrometry (MS/MS) scan to generate product ion spectra to determine amino acid sequence in consecutive instrument scans of the 10 most abundant peak in the spectrum. The AGC Target ion number was set at 30,000 ions for full scan and 10,000 ions for MSn mode. Maximum ion injection time was set at 20 ms for full scan and 300 ms for MSn mode. Micro-scan number was set at 1 for both full scan and MSn scan. The collision-induced dissociation (CID) fragmentation energy was set to 35%. Dynamic exclusion was enabled with a repeat count of 2 within 10 s, a mass list size of 200, an exclusion duration of 350 s, low mass width of 0.5, and high mass width of 1.5.
Sample Preparation and Analysis
Several immunopurified wheat (Triticum aestivum) XS activity fractions were combined (for detailed immunopurification procedures, see Zeng et al., 2010) and precipitated with TCA-acetone. Precipitated proteins (50 μg) were resuspended in 20 μL Invitrisol plus 1 μL of 10% ASB-14 detergent and 2 μL of 10× SDS-PAGE loading dye. After heating at 90°C for 7 min, proteins were separated by 1-D gel electrophoresis (SDS-PAGE) using precast gels (NuPAGE; Invitrogen). The gels were rinsed with water for 10 min to remove excess SDS and fixed by a nonmodifying, precipitation solution of ethanol-acetic acid (50% [v/v] and 10% [v/v], respectively), before staining with Coomassie Brilliant Blue solution (0.1% [w/v] Coomassie Brilliant Blue R350, 20% [v/v] methanol, and 10% [v/v] acetic acid) for 1 h and destained overnight in 50% (v/v) methanol in water with 10% (v/v) acetic acid. Next, the gel was sliced into 32 small pieces, and each piece was used for in-gel trypsin digestion (enzyme to substrate ratio of 1:10 [w/v]). Released peptides were then analyzed by MS/MS. Briefly, sequence information from the MS/MS data were processed by converting the raw data files into a merged file (.mgf) using an in-house program (RAW2MZXML_n_MGF_batch; merge.pl, a Perl script). The resulting .mgf files were searched against the full Viridiplantae (green plant) database at NCBInr (2,509,864 sequences) using Mascot Daemon by Matrix Science version 2.4.1. The mass accuracy of the precursor ions were set to 2.0 D given that the data were acquired on an ion trap mass analyzer and the fragment mass accuracy was set to 0.5 D. Considered variable modifications were Met oxidation and deamidated Asn (N) and Gln (Q) residues, with a fixed modification of carbamidomethyl Cys. Two missed cleavages for the enzyme were permitted. Peptides with a score of less than 50 were filtered. Protein identifications were checked manually, and proteins with a Mascot protein score of 60 or higher with a minimum of two unique peptides from one protein having a -b or -y ion sequence tag of five residues or better were accepted.
Transformation and Screening for Transgenic Pichia pastoris Cells
The P. pastoris X-33 strain was used for simultaneous transformation with multiple genes. Wheat genes (TaGT43-4, TaGT47-13, TaGT75-3, TaGT75-4, TaGLP, and TaVER2) were cloned by PCR from a wheat seed cDNA library (Zeng et al., 2010), using primers containing EcoRI restriction sites (except for TaGLP, where NotI sites were used). The amplified products were first ligated into pCR8 vector (Invitrogen) and then transferred to the pPICZ-A vector (Invitrogen) using EcoRI or NotI restriction enzymes. For TaGT43-4, forward primer was 5′-GACGAATTCATGATGAAGCAGCTGCTGCCG-3′ and reverse primer was 5′-GACGAATTCTCAGTTGTGGACGTCGCCC-3′. For TaGT47-13, forward primer was 5′-GACGAATTCATGGAGAAGCCGCGGGC-3′ and reverse primer was 5′-GACGAATTCCTACCATGGCTTCAGGTCGCC-3′. For TaGT75-3, forward primer was 5′-GACGAATTCATGGCCACCCCGACGAC-3′ and reverse primer was 5′-GACGAATTCTCACTTGCTCTTCAGAGCAGGGC-3′. For TaGT75-4, forward primer was 5′-GACGAATTCATGGCAGGGACGGTGACTG-3′ and reverse primer was 5′-GACGAATTCCTACTTGGCTGCTGCCTTGC-3′. For TaGLP, forward primer was 5′-GCGGCCGCATGGCATCACCCTCTTCCTTCC-3′ and reverse primer was 5′-GCGGCCGCTTAGTTGTGGTTGTTCTCCCAGAACTGA-3′. For TaVER2, forward primer was 5′-GACGAATTCATGGCCAAATTCCAGATTACACCGTTC-3′ and reverse primer was 5′-GACGAATTCTCAGACCGTGTAAACACCAAATGCG-3′. Equal amounts (∼5 μg) of linearized pPICZ-A plasmids containing individual wheat genes (a total of six plasmids) were mixed with 100 μL competent P. pastoris cells in a chilled electroporation cuvette (Fisher Scientific) and kept on ice. Competent cells were generated according to the manufacturer’s instructions (Invitrogen). The mixture pPICZ plasmids and competent P. pastoris cells were pulsed using a Gene Pulser II (Bio-Rad) according to the manufacturer’s recommendations (1.8 kV, 200Ω, at 25 μF). Immediately following electroporation, 1 mL of ice-cold 1 m sorbitol was added to the mixture and the yeast cells were kept at 30°C for 2 h (without shaking). After incubation, yeast cells were plated on YPD medium containing Zeocin (100 μg/mL) and allowed to grow at 28°C for 48 h. About 600 colonies of transgenic yeast cells were screened by PCR to determine which wheat genes were successfully inserted into the genome.
Protein Production in Transgenic P. pastoris Cells
Each transgenic P. pastoris cell line was cultured overnight at 28°C with shaking (200 rpm), in 15 mL BMGY medium (1% [w/w] yeast extract, 2% [w/w] peptone, 100 mm potassium phosphate, pH 6.0, 1.34% [w/w] yeast nitrogen base, 1% [v/v] glycerol, and 4 × 10−5% biotin) until the OD600 reached ∼2. Protein production was then initiated by transferring P. pastoris cells to 50 mL BMMY medium (1% [w/w] yeast extract, 2% [w/w] peptone, 100 mm potassium phosphate, pH 6.0, 1.34% [w/w] yeast nitrogen base, 4 × 10−5% biotin, and 0.5% [v/v] methanol) to an OD600 of 1.0 in a 250-mL baffled flask, and cultured at 28°C with shaking (250 rpm). Protein production was maintained by adding methanol (0.5% [v/v] final concentration) every 24 h. After 5 d of induction, cells were harvested by centrifugation at 4,000g for 30 min at 4°C and used directly for preparation of microsomal membranes.
Microsomal Membranes Preparation from P. pastoris Cells
Harvested P. pastoris cells were resuspended in 10 mL of extraction buffer (EB; 100 mm HEPES-KOH, pH 7.0, 0.4 m Suc, 1 mm DTT, 5 mm MgCl2, 5 mm MnCl2, and 5 mm phenylmethylsulfonyl fluoride), containing one Protease inhibitor cocktail tablet and 0.5 mL of protease inhibitor cocktail IV per 100 mL EB. P. pastoris cells were broken by vortexing with acid-washed glass beads (∼2 mL per 5 mL of P. pastoris cell suspension) for 8 × 40 s with 30s of cooling on ice between vortexing steps. After centrifugation (4,000g for 10 min at 4°C), supernatants were saved and laid carefully on top of 5 mL of cushion buffer (100 mm HEPES-KOH, pH 7.0, containing 1.8 m Suc and 1 mm DTT) in an ultracentrifugation tube. The tubes were centrifuged at 100,000g for 60 min at 4°C using a swinging rotor SW32 and an Optima l-80 XP ultracentrifuge (Beckman Coulter). Fractions at the interface between cushion buffer and EB were collected and transferred to a new ultracentrifuge tube and centrifuged again at 120,000g for 30 min at 4°C using a fixed angle rotor Ti70 (Beckman Coulter). The pellets (microsomal membranes) were resuspended in 500 μL EB and stored at −80°C until use. Protein concentration in these microsomal preparations was usually ∼5 μg/μL.
The presence of TaGT43-4, TaGT47-13, TaGT75-3, and TaGT75-4 was confirmed by immunoblot using anti-TaGT43-4, anti-TaGT47-13, and anti-PsRGP1, as described by Zeng et al. (2010).
PAGE Analysis
Microsomal membranes from transgenic P. pastoris cells were treated with 0.5% Triton X-100 and these Triton extracts (20–25 μg proteins/10–15 μL) were analyzed under reducing/denaturing conditions using 10% one-dimensional SDS-PAGE following standard methods. Triton extracts were mixed with loading buffer (50 mm Tris-HCl, pH 6.8, 10% [w/v] DTT, 2% [w/v] SDS, 0.1% [w/v] bromophenol blue, and 10% [v/v] glycerol) and boiled at 100°C for 10 min, and the mixtures were centrifuged briefly. The supernatants were loaded onto SDS-polyacrylamide gels. For analysis under nondenaturing/nonreducing conditions, polyacrylamide gels contained 0.05% [w/v] sodium cholate instead of SDS, and Triton extracts were mixed with loading buffer that contained 1% [w/v] sodium cholate instead of 1% [w/v] SDS and no DTT. The mixture was loaded directly (without boiling) onto nondenaturing/nonreducing polyacrylamide gels.
Immunoblotting
Proteins separated by polyacrylamide gels were transferred onto Immobilon membranes using the Mini-Protean Tetra system (Bio-Rad) according to the manufacturer’s recommendations. Immunoblot analysis was carried out using antisera to TaGT43-4 and TaGT47-13 (Zeng et al., 2010) and to PsRGP1 (Dhugga et al., 1997). Anti-TaGT43-4, anti-TaGT47-13, and anti-PsRGP1 were used as primary antibodies at dilutions varying between 1:2,000 and 1:10,000, and goat anti-rabbit antibody conjugated to peroxidase was used as the secondary antibody (at 1:5,000 dilution). Antibody binding was detected using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific).
Immunogold Electron Microscopy
Emerging shoots of 3-d-old etiolated wheat seedlings (Fig. 9I) were dissected and fixed in glutaraldehyde and OsO4 as described previously (Boudjeko et al., 2006), except fixation in glutaraldehyde was carried out overnight at 4°C. Samples were embedded in LR White resin (London Resin Company) or Epon resin (Electron Microscopy Sciences). Ultrathin sections (70–90 nm) were collected on 300 mesh Formvar-carbon film-coated gold grids (Electron Microscopy Sciences). Grids containing LR White sections and Epon sections pretreated with 0.5 m NaIO4 and 0.1N HCl as described (Follet-Gueye et al., 2003) were blocked for 1 h in 3% milk in PBS, pH 7.2. Grids were incubated in a primary antibody (anti-TaGT43-4 and anti-PsRGP1 used at 1:20 dilution, anti-TaGT47-13 at 1:15, and anti-PI-4Kβ1 at 1:5) in 3% milk in PBS for 2 h. After incubation, grids were washed with PBS containing 0.05% Tween to remove excess primary antibody before incubation for 1 h with 18-nm colloidal gold particles coupled to goat anti-rabbit IgG (Jackson ImmunoResearch) at 1:50 dilution in PBS containing 3% milk. Labeled grids were rinsed in PBS and then water before staining with 2% uranyl acetate for 25 min, followed by 10 min staining with Reynolds’ lead citrate diluted 10 times (Reynolds, 1963). Electron micrographs were obtained using a JEOL JEM 1010 electron microscope equipped with a Gatan model 791 Multiscan CCD camera and Gatan Digital Micrograph software. The number of gold particles present in specific cell compartments was quantified by direct visual examination of labeled sections from several blocks (∼50 cells from 30 sections were analyzed).
Confocal Laser Scanning Microscopy
The constructs and primer sequences are listed in Supplemental Table S1. All constructs were made in plasmid pVKH18-En6 (daSilva et al., 2004; kindly provided by Dr. Brandizzi, Michigan State University) containing full or halves (Yn or Yc) of YFP. Yn corresponds to the 1 to 154 residues of the NH2-terminal end, and Yc corresponds to the 155 to 239 residues of the COOH-terminal end. For the constructs producing fusion proteins with YFP or halves (Yn or Yc) at the COOH-terminal end of the target protein (i.e. TaGT43-4-YFP, TaGT43-4-Yn, TaGT43-4-Yc, TaGT47-13-YFP, TaGT47-13-Yn, TaGT47-13-Yc, TaGLP-YFP, TaGLP-Yn, and TaGLP-Yc), XbaI and SalI restriction sites were included in forward and reverse primers, respectively, while for constructs having YFP or halves (Yn or Yc) at the NH2-terminal of the target protein, BamHI and SacI restriction sites were included in forward and reverse primers, respectively. The coding regions of the target proteins were amplified by PCR and subcloned into pCR8 cloning vector (Invitrogen) and their sequences were checked for mistakes by DNA sequencing at OH University Genomic Facility before transfer to plasmid pVKH18-En6 using the listed restriction sites (XbaI/SalI or BamHI/SacI). If these restriction sites exist in the coding region of target proteins, an overlap PCR-based strategy was used to eliminate them. SP-YFP-TaGT47-13 and SP-YFP-TaGLP constructs were generated through PCR-based strategy using XhoI and HindIII sites to insert YFP after SP sequences (for TaGT47-13, YFP was inserted between amino acids 30 and 31; for TaGLP, YFP was inserted between amino acids 26 and 27). Untagged TaGT43-4, TaGT47-13, and TaGLP constructs were generated through PCR using primers containing XbaI and SalI restriction sites in the forward and reverse primers, respectively.
Transient expression of fluorescently tagged proteins using epidermal cell of tobacco (Nicotiana tabacum) leaves was performed as described (Brandizzi et al., 2002b). Leaves were infiltrated with Agrobacterium tumefaciens strain GV3101 transformed with plasmid pVKH18-En6 containing target protein-coding sequence fused to full YFP or its halves (Yn or Yc). Briefly, A. tumefaciens cultures (at an OD600 between 0.05 and 0.2) were infiltrated into the leaf epidermis of 4-week-old tobacco plants (grown at 22°C, 16 h light). Protein expression was under the control of enhanced Cauliflower mosaic virus 35S promoter and tobacco mosaic virus-Ω translation enhancer (Marusic et al., 2007). After 48 h of incubation, the infiltrated leaves were analyzed using an upright Zeiss 510 confocal laser scanning microscope (LSM510 META; Carl Zeiss Microscopy) with a Plan-NeoFluar 40×/1.3 oil immersion objective. For each experiment or construct, more than 20 individual cells were analyzed, and if 90 to 100% of the cells showed similar expression pattern, the result was accepted. Representative pictures from three to five leaves of three to five plants are shown in figures. YFP fluorescence was visualized with excitation of argon laser lines at 514 nm and emission at 565 nm. GFP fluorescence was visualized with excitation at 458 nm and emission at 500 nm. The line sequential scanning was selected in dual-channel observations to eliminate crossover between the two fluorophores. Time-lapse scanning was performed with Zeiss LSM 510 imaging system software. Postacquisition image processing was performed with ZEN 2012 software (Carl Zeiss Microscopy).
Accession Numbers
Sequence data in this article can be found in the GenBank/EMBL data libraries under the following accession numbers: TaGT43-4 (HM236487), TaGT47-13 (HM236485), TaGT75-4 (HM236489), TaGLP (CAB55558), TaVER2 (AB012103), and TaGT75-3 (KM670460).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. NH2-terminal secretion signal peptide of TaGT47-13 is functional.
Supplemental Figure S2. Immunoblotting using anti-PsRGP1 to detect the presence of TaGT75-3 and TaGT75-4 in microsomal membranes of transgenic Pichia cells.
Supplemental Figure S3. Confocal images of epidermal cells of N. tabacum leaves transiently expressing TaGT43-4-YFP or SP-YFP-TaGLP.
Supplemental Figure S4. Bimolecular fluorescence complementation (BiFC, split-YFP) assay of negative control combinations.
Supplemental Figure S5. Confocal images of epidermal cells of N. tabacum leaves transiently expressing TaGT75-3-YFP, TaGT75-4-YFP, or YFP-TaGT75-4.
Supplemental Figure S6. Maximum likelihood phylogenetic analysis of TaGT43-4 and 26 homologous proteins from GT43 family.
Supplemental Table S1. List of primers.
Supplementary Material
Acknowledgments
We thank Dr. Kanwarpal Dhugga (Dupont, Pioneer) for the gift of anti-PsRGP1 and Dr. Erik Nielson (University of Michigan) for the gift of anti-PI-4Kβ1. We also thank Dr. Christine Ondzighi-Assoume (University of Tennessee) and Dr. Robert Hikida (Ohio University) for the precious help with the immunogold TEM work. We thank Ms. Yuli Hu for her assistance with Figure 12.
Glossary
- XSC
xylan synthase complex
- GT
glycosyltransferase
- XGlcAT
xylan-glucuronosyltransferase
- XXylT
xylan-xylosyltransferase
- XAT
xylan-arabinofuranosyltranferase
- ER
endoplasmic reticulum
- TMD
transmembrane domain
- ERES
ER export site
- SP
signal peptide
- TEM
transmission electron microscopy
- BiFC
bimolecular fluorescence complementation
- HA
heparan sulfate synthase
- MS/MS
tandem mass spectrometry
- EB
extraction buffer
Footnotes
This work was supported by a National Science Foundation award (#IOS 1145887) and partly by a U.S. Department of Agriculture award (to A.F.).
Articles can be viewed without a subscription.
References
- Anders N, Wilkinson MD, Lovegrove A, Freeman J, Tryfona T, Pellny TK, Weimar T, Mortimer JC, Stott K, Baker JM, et al. (2012) Glycosyl transferases in family 61 mediate arabinofuranosyl transfer onto xylan in grasses. Proc Natl Acad Sci USA 109: 989–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki D, Lee N, Yamaguchi N, Dubois C, Fukuda MN (1992) Golgi retention of a trans-Golgi membrane protein, galactosyltransferase, requires cysteine and histidine residues within the membrane-anchoring domain. Proc Natl Acad Sci USA 89: 4319–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmodjo MA, Sakuragi Y, Zhu X, Burrell AJ, Mohanty SS, Atwood JA III, Orlando R, Scheller HV, Mohnen D (2011) Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc Natl Acad Sci USA 108: 20225–20230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled M, Raikhel NV (1997) Characterization of AtSEC12 and AtSAR1. Proteins likely involved in endoplasmic reticulum and Golgi transport. Plant Physiol 114: 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C. (2003) Signals for COPII-dependent export from the ER: what’s the ticket out? Trends Cell Biol 13: 295–300 [DOI] [PubMed] [Google Scholar]
- Batoko H, Zheng HQ, Hawes C, Moore I (2000) A rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12: 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumal R, Potter M, Scharff MD (1971) Synthesis, assembly, and secretion of gamma globulin by mouse myeloma cells. 3. Assembly of the three subclasses of IgG. J Exp Med 134: 1316–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck R, Sun Z, Adolf F, Rutz C, Bassler J, Wild K, Sinning I, Hurt E, Brügger B, Béthune J, Wieland F (2008) Membrane curvature induced by Arf1-GTP is essential for vesicle formation. Proc Natl Acad Sci USA 105: 11731–11736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berna A, Bernier F, Chabrière E, Perera T, Scott K (2008) DING proteins; novel members of a prokaryotic phosphate-binding protein superfamily which extends into the eukaryotic kingdom. Int J Biochem Cell Biol 40: 170–175 [DOI] [PubMed] [Google Scholar]
- Berna A, Bernier F, Scott K, Stuhlmüller B (2002) Ring up the curtain on DING proteins. FEBS Lett 524: 6–10 [DOI] [PubMed] [Google Scholar]
- Boudjeko T, Andème-Onzighi C, Vicré M, Balangé A, Ndoumou DO, Driouich A (2006) Loss of pectin is an early event during infection of cocyam roots by Pythium myriotyium. Planta 223: 271–282 [DOI] [PubMed] [Google Scholar]
- Boulaflous A, Saint-Jore-Dupas C, Herranz-Gordo MC, Pagny-Salehabadi S, Plasson C, Garidou F, Kiefer-Meyer MC, Ritzenthaler C, Faye L, Gomord V (2009) Cytosolic N-terminal arginine-based signals together with a luminal signal target a type II membrane protein to the plant ER. BMC Plant Biol 9: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha-Drori K, Shichrur K, Katz A, Oliva M, Angelovici R, Yalovsky S, Ohad N (2004) Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J 40: 419–427 [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Frangne N, Marc-Martin S, Hawes C, Neuhaus J-M, Paris N (2002a) The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell 14: 1077–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F, Snapp EL, Roberts AG, Lippincott-Schwartz J, Hawes C (2002b) Membrane protein transport between the endoplasmic reticulum and the Golgi in tobacco leaves is energy dependent but cytoskeleton independent: evidence from selective photobleaching. Plant Cell 14: 1293–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher MS, Munro S (1993) Cholesterol and the Golgi apparatus. Science 261: 1280–1281 [DOI] [PubMed] [Google Scholar]
- Brown DM, Goubet F, Wong VW, Goodacre R, Stephens E, Dupree P, Turner SR (2007) Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J 52: 1154–1168 [DOI] [PubMed] [Google Scholar]
- Brown DM, Zhang Z, Stephens E, Dupree P, Turner SR (2009) Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J 57: 732–746 [DOI] [PubMed] [Google Scholar]
- Chiniquy D, Sharma V, Schultink A, Baidoo EE, Rautengarten C, Cheng K, Carroll A, Ulvskov P, Harholt J, Keasling JD, et al. (2012) XAX1 from glycosyltransferase family 61 mediates xylosyltransfer to rice xylan. Proc Natl Acad Sci USA 109: 17117–17122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong K, Bao S-L, Xu T, Tan K-H, Liang T-B, Zeng J-Z, Huang H-L, Xu J, Xu Z-H (1998) Functional analysis of the VER gene using antisense transgenic wheat. Physiol Plant 102: 87–92 [DOI] [PubMed] [Google Scholar]
- Chou YH, Pogorelko G, Zabotina OA (2012) Xyloglucan xylosyltransferases XXT1, XXT2, and XXT5 and the glucan synthase CSLC4 form Golgi-localized multiprotein complexes. Plant Physiol 159: 1355–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B (2003) An evolving hierarchical family classification for glycosyltransferases. J Mol Biol 328: 307–317 [DOI] [PubMed] [Google Scholar]
- Cole NB, Smith CL, Sciaky N, Terasaki M, Edidin M, Lippincott-Schwartz J (1996) Diffusional mobility of Golgi proteins in membranes of living cells. Science 273: 797–801 [DOI] [PubMed] [Google Scholar]
- Colley KJ. (1997) Golgi localization of glycosyltransferases: more questions than answers. Glycobiology 7: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuneo MJ, Beese LS, Hellinga HW (2009) Structural analysis of semi-specific oligosaccharide recognition by a cellulose-binding protein of Thermotoga maritima reveals adaptations for functional diversification of the oligopeptide periplasmic binding protein fold. J Biol Chem 284: 33217–33223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czlapinski JL, Bertozzi CR (2006) Synthetic glycobiology: Exploits in the Golgi compartment. Curr Opin Chem Biol 10: 645–651 [DOI] [PubMed] [Google Scholar]
- daSilva LL, Snapp EL, Denecke J, Lippincott-Schwartz J, Hawes C, Brandizzi F (2004) Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. Plant Cell 16: 1753–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Enfert C, Gensse M, Gaillardin C (1992) Fission yeast and a plant have functional homologues of the Sar1 and Sec12 proteins involved in ER to Golgi traffic in budding yeast. EMBO J 11: 4205–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhugga KS, Tiwari SC, Ray PM (1997) A reversibly glycosylated polypeptide (RGP1) possibly involved in plant cell wall synthesis: purification, gene cloning, and trans-Golgi localization. Proc Natl Acad Sci USA 94: 7679–7684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G, Zabotina O, Delgado I, Robert S, Keegstra K, Raikhel N (2006) Arabidopsis reversibly glycosylated polypeptides 1 and 2 are essential for pollen development. Plant Physiol 142: 1480–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwell JM, Khuri S, Gane PJ (2000) Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol Mol Biol Rev 64: 153–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwell JM, Purvis A, Khuri S (2004) Cupins: the most functionally diverse protein superfamily? Phytochemistry 65: 7–17 [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE (2002) Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol 12: 54–60 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faik A. (2010) Xylan biosynthesis: news from the grass. Plant Physiol 153: 396–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faik A, Jiang N, Held M (2013) Xylan biosynthesis in plants, simply complex. In: NC Carpita, MS Buckeridge, MC McCann MC, eds, Plants and Bioenergy. Springer, New York, pp 153–181
- Feng H, Xu W-Z, Lin H-H, Chong K (2009) Transcriptional regulation of wheat VER2 promoter in rice in response to abscisic acid, jasmonate, and light. J Genet Genomics 36: 371–377 [DOI] [PubMed] [Google Scholar]
- Follet-Gueye ML, Pagny S, Faye L, Gomord V, Driouich A (2003) An improved chemical fixation method suitable for immunogold localization of green fluorescent protein in the Golgi apparatus of tobacco Bright Yellow (BY-2) cells. J Histochem Cytochem 51: 931–940 [DOI] [PubMed] [Google Scholar]
- Giraudo CG, Daniotti JL, Maccioni HJF (2001) Physical and functional association of glycolipid N-acetyl-galactosaminyl and galactosyl transferases in the Golgi apparatus. Proc Natl Acad Sci USA 98: 1625–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudo CG, Maccioni HJ (2003) Endoplasmic reticulum export of glycosyltransferases depends on interaction of a cytoplasmic dibasic motif with Sar1. Mol Biol Cell 14: 3753–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Hanton SL, Matheson LA, Chatre L, Brandizzi F (2009) Dynamic organization of COPII coat proteins at endoplasmic reticulum export sites in plant cells. Plant J 57: 963–974 [DOI] [PubMed] [Google Scholar]
- Hardt B, Kalz-Fuller B, Aparicio R, Volker C, Bause E (2003) (Arg)3 within the N-terminal domain of glucosidase I contains ER targeting information but is not required absolutely for ER localization. Glycobiology 13: 159–168 [DOI] [PubMed] [Google Scholar]
- Hawes C, Osterrieder A, Hummel E, Sparkes I (2008) The plant ER-Golgi interface. Traffic 9: 1571–1580 [DOI] [PubMed] [Google Scholar]
- Held MA, Boulaflous A, Brandizzi F (2008) Advances in fluorescent protein-based imaging for the analysis of plant endomembranes. Plant Physiol 147: 1469–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JK, Johnson NR, Wilkerson CG (2014) Arabidopsis thaliana IRX10 and two related proteins from psyllium and Physcomitrella patens are xylan xylosyltransferases. Plant J 80: 207–215 [DOI] [PubMed] [Google Scholar]
- Jiang J-F, Xu Y-Y, Chong K (2007) Overexpression of OsJAC1, a lectin gene, suppresses the coleoptile and stem elongation in rice. J Integr Plant Biol 49: 230–237 [Google Scholar]
- Jobling SA. (2015) Membrane pore architecture of the CslF6 protein controls (1-3,1-4)-β-glucan structure. Sci Adv 1: e1500069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J, Munro S (1998) Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with alpha-1,6-mannosyltransferase activity. EMBO J 17: 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuda S, Shiba T, Ishiguro M, Tagawa H, Oka S, Kajihara Y, Kawasaki T, Wakatsuki S, Kato R (2004) Structural basis for acceptor substrate recognition of a human glucuronyltransferase, GlcAT-P, an enzyme critical in the biosynthesis of the carbohydrate epitope HNK-1. J Biol Chem 279: 22693–22703 [DOI] [PubMed] [Google Scholar]
- Kang BH, Nielsen E, Preuss ML, Mastronarde D, Staehelin LA (2011) Electron tomography of RabA4b- and PI-4Kβ1-labeled trans Golgi network compartments in Arabidopsis. Traffic 12: 313–329 [DOI] [PubMed] [Google Scholar]
- Khalil H, Brunet A, Saba I, Terra R, Sékaly RP, Thibodeau J (2003) The MHC class II β chain cytoplasmic tail overcomes the invariant chain p35-encoded endoplasmic reticulum retention signal. Int Immunol 15: 1249–1263 [DOI] [PubMed] [Google Scholar]
- Kim SJ, Zemelis S, Keegstra K, Brandizzi F (2015) The cytoplasmic localization of the catalytic site of CSLF6 supports a channeling model for the biosynthesis of mixed-linkage glucan. Plant J 81: 537–547 [DOI] [PubMed] [Google Scholar]
- Konishi T, Miyazaki Y, Yamakawa S, Iwai H, Satoh S, Ishii T (2010) Purification and biochemical characterization of recombinant rice UDP-arabinopyranose mutase generated in insect cells. Biosci Biotechnol Biochem 74: 191–194 [DOI] [PubMed] [Google Scholar]
- Konishi T, Takeda T, Miyazaki Y, Ohnishi-Kameyama M, Hayashi T, O’Neill MA, Ishii T (2007) A plant mutase that interconverts UDP-arabinofuranose and UDP-arabinopyranose. Glycobiology 17: 345–354 [DOI] [PubMed] [Google Scholar]
- Lee C, O’Neill MA, Tsumuraya Y, Darvill AG, Ye Z-H (2007) The irregular xylem9 mutant is deficient in xylan xylosyltransferase activity. Plant Cell Physiol 48: 1624–1634 [DOI] [PubMed] [Google Scholar]
- Lee C, Teng Q, Huang W, Zhong R, Ye Z-H (2010) The Arabidopsis family GT43 glycosyltransferases form two functionally nonredundant groups essential for the elongation of glucuronoxylan backbone. Plant Physiol 153: 526–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Teng Q, Zhong R, Ye Z-H (2012) Arabidopsis GUX proteins are glucuronyltransferases responsible for the addition of glucuronic acid side chains onto xylan. Plant Cell Physiol 53: 1204–1216 [DOI] [PubMed] [Google Scholar]
- Lee C, Teng Q, Zhong R, Yuan Y, Ye Z-H (2014) Functional roles of rice glycosyltransferase family GT43 in xylan biosynthesis. Plant Signal Behav 9: e27809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Zhong R, Ye Z-H (2012a) Arabidopsis family GT43 members are xylan xylosyltransferases required for the elongation of the xylan backbone. Plant Cell Physiol 53: 135–143 [DOI] [PubMed] [Google Scholar]
- Machamer CE. (1991) Golgi retention signals: do membranes hold the key? Trends Cell Biol 1: 141–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mains PE, Sibley CH (1983) The requirement of light chain for the surface deposition of the heavy chain of immunoglobulin M. J Biol Chem 258: 5027–5033 [PubMed] [Google Scholar]
- Margeta-Mitrovic M, Jan YN, Jan LY (2000) A trafficking checkpoint controls GABA(B) receptor heterodimerization. Neuron 27: 97–106 [DOI] [PubMed] [Google Scholar]
- Marusic C, Nuttall J, Buriani G, Lico C, Lombardi R, Baschieri S, Benvenuto E, Frigerio L (2007) Expression, intracellular targeting and purification of HIV Nef variants in tobacco cells. BMC Biotechnol 7: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KF, Waugh R, Brown JW, Schulman A, Langridge P, Platzer M, Fincher GB, Muehlbauer GJ, Sato K, Close TJ, Wise RP, Stein N; International Barley Genome Sequencing Consortium (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491: 711–716 [DOI] [PubMed] [Google Scholar]
- McCormick C, Duncan G, Goutsos KT, Tufaro F (2000) The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proc Natl Acad Sci USA 97: 668–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelsen K, Yuan H, Schwappach B (2005) Hide and run. Arginine-based endoplasmic-reticulum-sorting motifs in the assembly of heteromultimeric membrane proteins. EMBO Rep 6: 717–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyafusa T, Caaveiro JMM, Tanaka Y, Tsumoto K (2012) Crystal structure of the enzyme CapF of Staphylococcus aureus reveals a unique architecture composed of two functional domains. Biochem J 443: 671–680 [DOI] [PubMed] [Google Scholar]
- Mortimer JC, Miles GP, Brown DM, Zhang Z, Segura MP, Weimar T, Yu X, Seffen KA, Stephens E, Turner SR, Dupree P (2010) Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. Proc Natl Acad Sci USA 107: 17409–17414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. (1995) A comparison of the transmembrane domains of Golgi and plasma membrane proteins. Biochem Soc Trans 23: 527–530 [DOI] [PubMed] [Google Scholar]
- Nilsson T, Slusarewicz P, Hoe MH, Warren G (1993) Kin recognition. A model for the retention of Golgi enzymes. FEBS Lett 330: 1–4 [DOI] [PubMed] [Google Scholar]
- Phillipson BA, Pimpl P, daSilva LL, Crofts AJ, Taylor JP, Movafeghi A, Robinson DG, Denecke J (2001) Secretory bulk flow of soluble proteins is efficient and COPII dependent. Plant Cell 13: 2005–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpl P, Movafeghi A, Coughlan S, Denecke J, Hillmer S, Robinson DG (2000) In situ localization and in vitro induction of plant COPI-coated vesicles. Plant Cell 12: 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C, Ebert B, Herter T, Petzold CJ, Ishii T, Mukhopadhyay A, Usadel B, Scheller HV (2011) The interconversion of UDP-arabinopyranose and UDP-arabinofuranose is indispensable for plant development in Arabidopsis. Plant Cell 23: 1373–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner JC, Pelham HRB (1997) Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO J 16: 1832–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Hansen SF, Ebert B, Lau J, Scheller HV (2014) Site-directed mutagenesis of IRX9, IRX9L and IRX14 proteins involved in xylan biosynthesis: glycosyltransferase activity is not required for IRX9 function in Arabidopsis. PLoS One 9: e105014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie EA, Hansen SF, Baidoo EEK, Hadi MZ, Keasling JD, Scheller HV (2012) Three members of the Arabidopsis glycosyltransferase family 8 are xylan glucuronosyltransferases. Plant Physiol 159: 1408–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds ES. (1963) The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol 17: 208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoberer J, Vavra U, Stadlmann J, Hawes C, Mach L, Steinkellner H, Strasser R (2009) Arginine/lysine residues in the cytoplasmic tail promote ER export of plant glycosylation enzymes. Traffic 10: 101–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer P, Christoffel A, Dudler R (1999) Transient expression of members of the germin-like gene family in epidermal cells of wheat confers disease resistance. Plant J 20: 541–552 [DOI] [PubMed] [Google Scholar]
- Schutze MP, Peterson PA, Jackson MR (1994) An N-terminal double-arginine motif maintains type II membrane proteins in the endoplasmic reticulum. EMBO J 13: 1696–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe HJ, Stevens TJ, Munro S (2010) A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell 142: 158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa VL, Brito C, Costa T, Lanoix J, Nilsson T, Costa J (2003) Importance of Cys, Gln, and Tyr from the transmembrane domain of human alpha 3/4 fucosyltransferase III for its localization and sorting in the Golgi of baby hamster kidney cells. J Biol Chem 278: 7624–7629 [DOI] [PubMed] [Google Scholar]
- Tu L, Banfield DK (2010) Localization of Golgi-resident glycosyltransferases. Cell Mol Life Sci 67: 29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowicz BR, Peña MJ, Moniz HA, Moremen KW, York WS (2014) Two Arabidopsis proteins synthesize acetylated xylan in vitro. Plant J 80: 197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet C, Thomas EC, Merino-Trigo A, Teasdale RD, Gleeson PA (2003) Intracellular sorting and transport of proteins. Prog Biophys Mol Biol 83: 1–45 [DOI] [PubMed] [Google Scholar]
- Weisz OA, Swift AM, Machamer CE (1993) Oligomerization of a membrane protein correlates with its retention in the Golgi complex. J Cell Biol 122: 1185–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SM, Ho YY, Lampugnani ER, Van de Meene AM, Bain MP, Bacic A, Doblin MS (2015) Determining the subcellular location of synthesis and assembly of the cell wall polysaccharide (1,3; 1,4)-β-d-glucan in grasses. Plant Cell 27: 754–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AM, Hörnblad E, Voxeur A, Gerber L, Rihouey C, Lerouge P, Marchant A (2010) Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L pairs of glycosyltransferase genes reveals critical contributions to biosynthesis of the hemicellulose glucuronoxylan. Plant Physiol 153: 542–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AM, Rihouey C, Seveno M, Hörnblad E, Singh SK, Matsunaga T, Ishii T, Lerouge P, Marchant A (2009) The Arabidopsis IRX10 and IRX10-LIKE glycosyltransferases are critical for glucuronoxylan biosynthesis during secondary cell wall formation. Plant J 57: 718–731 [DOI] [PubMed] [Google Scholar]
- Yang B, Wyman CE (2004) Effect of xylan and lignin removal by batch and flowthrough pretreatment on the enzymatic digestibility of corn stover cellulose. Biotechnol Bioeng 86: 88–95 [DOI] [PubMed] [Google Scholar]
- Yong W-D, Xu Y-Y, Xu W-Z, Wang X, Li N, Wu J-S, Liang T-B, Chong K, Xu Z-H, Tan K-H, Zhu Z-Q (2003) Vernalization-induced flowering in wheat is mediated by a lectin-like gene VER2. Planta 217: 261–270 [DOI] [PubMed] [Google Scholar]
- Xiao J, Xu S, Li C, Xu Y, Xing L, Niu Y, Huan Q, Tang Y, Zhao C, Wagner D, Gao C, Chong K (2014) O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat. Nat Commun 5: 4572. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zeng W, Chatterjee M, Faik A (2008) UDP-Xylose-stimulated glucuronyltransferase activity in wheat microsomal membranes: characterization and role in glucurono(arabino)xylan biosynthesis. Plant Physiol 147: 78–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Jiang N, Nadella R, Killen TL, Nadella V, Faik A (2010) A glucurono(arabino)xylan synthase complex from wheat contains members of the GT43, GT47, and GT75 families and functions cooperatively. Plant Physiol 154: 78–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerangue N, Schwappach B, Jan YN, Jan LY (1999) A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron 22: 537–548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.