Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
Dicer1 deletion in MKs alters platelet miRNA and mRNA profiles.
Dicer1-deficient platelets display increased integrins αIIb and β3 levels and enhanced in vitro and in vivo functional responses.
Abstract
Human platelets contain microRNAs (miRNAs) and miRNA processing machinery, but their contribution to platelet function remains incompletely understood. Here, we show that murine megakaryocyte (MK)-specific knockdown of Dicer1, the ribonuclease that cleaves miRNA precursors into mature miRNAs, reduces the level of the majority of miRNAs in platelets. This leads to altered platelet messenger RNA (mRNA) expression profiles and mild thrombocytopenia. Fibrinogen receptor subunits Itga2b (αIIb) and Itgb3 (β3) mRNAs were among the differentially expressed transcripts that are increased in platelets lacking Dicer1. Argonaute 2 (Ago2), a member of the miRNA silencing complex, co-immunoprecipitated with αIIb and β3 mRNAs in wild-type platelets. Furthermore, co-immunoprecipitation experiments suggested reduced αIIb/β3/Ago2 complexes in miRNA-deficient platelets. These results suggested that miRNAs regulate both integrin subunits. Subsequent 3′ untranslated region luciferase reporter assays confirmed that the translation of both αIIb and β3 mRNAs can be regulated by miRNAs miR-326, miR-128, miR-331, and miR-500. Consistent with these molecular changes, the deletion of Dicer1 resulted in increased surface expression of integrins αIIb and β3, and enhanced platelet binding to fibrinogen in vivo and in vitro. Heightened platelet reactivity, shortened tail-bleeding time, and reduced survival following collagen/epinephrine-induced pulmonary embolism were also observed in Dicer1-deficient animals. Combined Pf4-cre–mediated deletion of Drosha and Dicer1 did not significantly exacerbate phenotypes observed in single Dicer1 knockout mice. In summary, these findings indicate that Dicer1-dependent generation of mature miRNAs in late-stage MKs and platelets modulates the expression of target mRNAs important for the hemostatic and thrombotic function of platelets.
Introduction
MicroRNAs (miRNAs) are a class of short noncoding RNAs, ∼22 nucleotides in length.1-3 Generated by RNA polymerase II,4 primary miRNAs are conventionally processed into 70 bp hairpin miRNA precursors (pre-miRNA) in the nucleus by the enzyme Drosha.5 Dicer1 further processes pre-miRNA in the cytoplasm into mature miRNA duplexes.6,7 Guided by the miRNA effector protein Argonaute 2 (Ago2), miRNAs regulate posttranscriptional gene expression by inducing the degradation, sequestration, or translational repression of target messenger RNAs (mRNAs).8-10 miRNAs typically constrain gene expression by binding to the 3′ untranslated region (UTR) of mRNAs.11 They are often viewed as “rheostats” that tightly control and fine tune, rather than abolish, protein expression.
Several studies have described roles for miRNAs in megakaryocyte (MK) development.12,13 Their progeny, platelets, also possess miRNA processing machinery, and a diverse repertoire of miRNAs.14-16 Recent evidence suggests that miRNA expression patterns in platelets influence their reactivity.16,17 For example, increased expression of miR-96 regulates vesicle-associated membrane protein 8 expression, and a single nucleotide polymorphism in the vesicle-associated membrane protein 8 3′UTR, which lies within the miR-96 binding region, correlates with platelet reactivity.17 Expression of additional miRNA-mRNA pairs correlates with epinephrine-induced aggregation.16 Similarly, platelet miR-376c regulates phosphatidylcholine transfer protein levels, and its expression in platelets is inversely correlated with phosphatidylcholine transfer protein mRNA and protein, which regulates PAR4-mediated signals.18 The level of Dicer1, and several mature miRNAs, are significantly reduced in platelets from diabetic individuals and diabetic mice.19 Experiments using mice with a whole body knockout (KO) of miR-223, one of the miRNAs decreased in platelets from patients with diabetes, suggested that the reduction in miRNAs may alter platelet reactivity.19
Collectively, these studies suggest that miRNAs regulate the development and function of MKs and platelets. To directly test the functional requirement for miRNA expression and the effect of Dicer1 loss in the lineage, we generated mice with an MK-specific deletion of Dicer1.6,7,20-23 This resulted in mild thrombocytopenia, decreased level of mature miRNAs in platelets, and a corresponding increase in the level of several target mRNAs that regulate platelet function, including Itga2b (αIIb) and Itgb3 (β3) integrins. Dicer1-deficient platelets also exhibited increased fibrinogen binding and uptake, and enhanced activation of the αIIbβ3 receptor in response to PAR4 stimulation. In vivo, mice harboring Dicer1-deficient platelets demonstrated reduced bleeding time and blood loss after injury, and reduced survival following collagen/epinephrine-induced pulmonary embolism (PE). Thus, reducing miRNAs in platelets, by elimination of miRNA processing machinery, results in platelets with altered mRNA content that is associated with a lower activation threshold, and enhanced in vivo hemostasis and clot formation.
Materials and methods
Animals
All animal experiments complied with the regulatory standards of the University of Utah, the Walter and Eliza Hall Institute Animal Ethics Committee, or the Institutional Animal Care and Use Committee at Case Western Reserve University School of Medicine. Dicer1fl/fl (B6.Cg-Dicer1tm1Bdh/J), Droshafl/fl (B6.129P2-Droshatm1Litt/J), and platelet factor 4 (Pf4)-Cre (Tg[Pf4-cre]Q3Rsko/J) mice have been previously described.24-26 To control for background differences, experiments were performed on mixed backgrounds using gender-matched adult littermates.
Cell isolation
Platelets were purified using immune-magnetic negative selection to deplete red blood cells and leukocytes from platelet-rich plasma using previously published methods.27 Bone marrow (BM)-derived MKs were generated and enriched by density gradient separation, as previously described.28
Dicer1 activity assay
Supernatant of platelet lysates were centrifuged at 10 000 g for 10 minutes (S10 extract). S10 extracts were incubated with let-7a-3 pre-miRNA and processing into mature let-7a-3 miRNA was assessed as previously described.14
Platelet Ago2 αIIb or β3 mRNA complexes
Platelet-derived S10 extracts were cleared by centrifugation prior to immunoprecipitation using protein G-agarose beads (Roche Applied Science, Penzberg, Germany) conjugated with anti-Ago2 antibody (clone 2E12-1C9; Abnova) or isotype immunoglobulin G control (anti-FLAG; Sigma-Aldrich), as previously described.14,29 Ago2 and glyceraldehyde-3-phosphate dehydrogenase (Gapdh) protein levels were assessed by western blot. Ago2-associated αIIb or β3 mRNAs were isolated by phenol/chloroform extraction and ethanol precipitation, reverse transcribed with the miScript II Reverse Transcription Kit (Qiagen), and analyzed by real-time polymerase chain reaction (PCR) using specific oligonucleotides.
RNA preparation
For the RNA-seq and real-time PCR analysis of mRNA expression, washed mouse platelets were lysed in Trizol, and RNA was isolated and DNase treated. For miRNA array and real-time PCR analyses of small RNA, total RNA was extracted with mirVana Isolation Kit (Life Technologies). Samples were treated with DNase I, and RNA concentrated by ethanol precipitation followed by extensive ethanol washes.
miRNA array and RNA-seq analysis
An Agilent bio-analyzer was used to quality control and quantitate RNA. RNA integrity number scores were similar between all samples. Total RNA was equally loaded (100 ng) and arrays were hybridized simultaneously to Agilent miRNA arrays v2 (8X15K; 650 miRNAs with 20 probes per miRNA). One array per mouse for a total of 6 independent microarrays (3 per group) was used. Arrays were scanned on an Agilent G2565C scanner. Images were processed using Agilent Feature Extraction Software version 10.5. Of 650 miRNAs, a signal was detected for 267. Background corrected total gene signal estimation and filtering were done using in house software (http://healthcare.utah.edu/huntsmancancerinstitute/research/shared-resources/center-managed/bioinformatics/useful-links/agilent-filter.php#Launch_Agilen), and with the robust multi-array average algorithm in the AgiMicroRna R package.30 After default filtering, 160 miRNAs were detected above background. To focus on the more abundant miRNA, only those miRNAs above an arbitrary total signal threshold of 150 (123 miRNAs) were considered for further analysis. Global normalization strategies depend on the assumption that the majority of targets remain unchanged.31 Because we expected systemic miRNA changes, and arrays were equally loaded according to total RNA, fold changes and significance testing were performed on the background corrected, total gene signal without additional normalization. We confirmed this approach on selected candidates by real-time PCR using 18s ribosomal RNA (rRNA) as a reference gene. For visualization, simple scaling, clustering (Euclidean distance), and heat map generation were done in R.32,33
RNA-seq RNA from pooled platelets was prepared using TruSeq V2 with oligo-dT selection (Illumina, San Diego CA), and 36 bp-paired end reads sequenced on an Illumina GAIIx (1 sample sequenced for each group, with each containing platelets pooled from 3 mice per group). Reads were aligned to mouse genome build mm9 using NovoAlign. Reads were assigned to transcripts with DefinedRegionScanSeq in the USeq analysis package.34
Real-time PCR
For mRNA real-time PCR, superscript II and SYBR green was used. For small RNA, TaqMan miRNA RT and individual TaqMan assays were used. Relative mean fold changes and propagated standard errors were calculated according to the 2(−ΔΔCt) method.35 Gapdh, which was not altered in the RNA-seq dataset, was used as the reference for mRNA assays. For miRNA assays, references were sno-142 RNA in MKs or 18s rRNA in platelets (sno-142 was not detectable in mouse platelets). Prior to fold change estimation, single removal of outliers was performed using the modified Thompson’s τ method. For fold change plots, the calibrator group (wild-type [WT]) is set at 1, and the standard error range of the target (KO) group contains the combined error propagated from the calibrator and target.35 P values were calculated on the 2−ΔCt values.
Reporter gene activity assays
mRNA targets of miRNAs were predicted by using miRWalk 2.0 (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/) and the sequences aligned by using RNA hybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/). For reporter constructs, the 3′UTR sequence of αIIb and β3 mRNAs were cloned downstream of Renilla luciferase (Rluc) in the psiCHECK-2 vector (Promega). miRNA precursor constructs were created using the pGeneClip U1 Hairpin Cloning Systems (Promega). HEK293 cells were co-transfected with the reporter gene and miRNA expression vector constructs. Luciferase activity was measured at 48 hours using the Dual-Glo luciferase assay (Promega), and Firefly luciferase activity was normalized to Rluc. Relative luciferase activity was obtained by normalization of values with a nonrelevant short hairpin RNA expression vector.
Fibrinogen-based assays
To measure the in vivo uptake of exogenous fibrinogen, 488 labeled fibrinogen (150 μg; Life Technologies) was injected into the tail vein of mice. Blood was collected 48 hours later, and fluorescent fibrinogen intensity was determined by flow cytometry. Endogenous fibrinogen levels in circulating platelets were quantified by enzyme-linked immunosorbent assay (ELISA) (GenWay). Flow cytometry was used to assess ex vivo binding of fibrinogen to platelets stimulated with a PAR4 peptide (AYPGKF; Sigma-Aldrich). In brief, washed platelets were rested in M199 media for 30 minutes prior to dilution at 1 × 107/mL and incubation with 488 labeled fibrinogen in the presence of the PAR4 agonist peptide Ala-Tyr-Pro-Gly-Lys-Phe-NH2 (trifluoroacetate salt, A3227; Sigma-Aldrich). After 20 minutes, the platelets were fixed with BD lyse/fix buffer (Becton Dickinson) and analyzed by flow cytometry.
Flow cytometry analysis of surface integrin expression and platelet activation in whole blood
Whole blood from the carotid artery was diluted 1:50 in Hepes Tyrodes buffer. Agonists were added for 20 minutes prior to assessing JonA binding and surface P-selectin expression on CD41-positive platelets by using anti-JON/A (Emfret) and anti-CD62P (Emfret) antibodies, respectively. Surface integrins were assessed with anti-mouse CD61 (2c9.g3; eBioscience) and anti-mouse CD41 (mwreg30; eBioscience) antibodies.
Tail-bleeding assays
Tail-bleeding times were measured by clipping 3 mm from the tip of the tail of mice anesthetized with isoflurane. The tail was placed in pre-warmed saline at 37°C, and the time to cessation of bleeding was measured for 10 minutes. The quantity of blood loss was determined by lysing the red blood cells and measuring the absorbance of hemoglobin at 550 nm. The results were compared with a standard curve of known quantities of blood (1 to 200 μL).
Platelet counts and mean platelet volume (MPV)
Automated cell counts and MPV measurements were performed on blood collected from the retro-orbital plexus or facial vein into microtainer tubes containing EDTA (Sarstedt), using an Advia 2120 Hematology Analyzer (Siemens, Munich, Germany) and a Hemavet 950 (Drew Scientific, Waterbury, CT).
PE
PE was induced in anesthetized 8-week-old mice by injection into the plexus retro-orbital veins of a solution containing 60 mg/kg epinephrine and 150 mg/kg collagen, in phosphate-buffered saline (PBS) in a total volume of 200 μL, as previously described.36 Time to death was monitored for 30 minutes. Immediately post-expiration, the chest cavity was exposed and 500 μL of Evans blue dye in PBS was injected directly into the right ventricle of the heart. The heart and lungs were removed, rinsed briefly in PBS, and photographed. PE incidence was confirmed by dye exclusion from the lungs. As an additional control, a Pf4-Cre mouse was injected with PBS only and euthanized after 5 minutes; following Evans blue dye injection, the lungs stained blue, indicating no incidence of PE.
Statistical methods
For RNA-seq analysis, DefinedRegionScanSeq was used to estimate fold changes and rank/prioritize differentially expressed candidates. This software implements the binomial distribution to estimate significance (which was used rather as a ranking statistic) in samples without replicates according to a combination of read counts (abundant transcripts are given high priority) and fold change. Therefore, the term “top targets” refers to those given the lowest binomial P value, and represent abundantly expressed transcripts that are likely to have changed between groups. For microarray data, false discovery rate significance testing was performed using the “samr” library in R.37 A q-value < .05 was considered significant. Student t test, paired or unpaired as indicated, was used for hypothesis testing in 2 group comparisons. Analysis of variance and post hoc Students t tests were used for multiple comparisons. For survival analysis, a log-rank test was used. A value of P < .05 was considered significant (*). Significance at P value < .01 is also indicated (**).
Results
MK-specific deletion of Dicer1 reduces the level of mature miRNAs in platelets
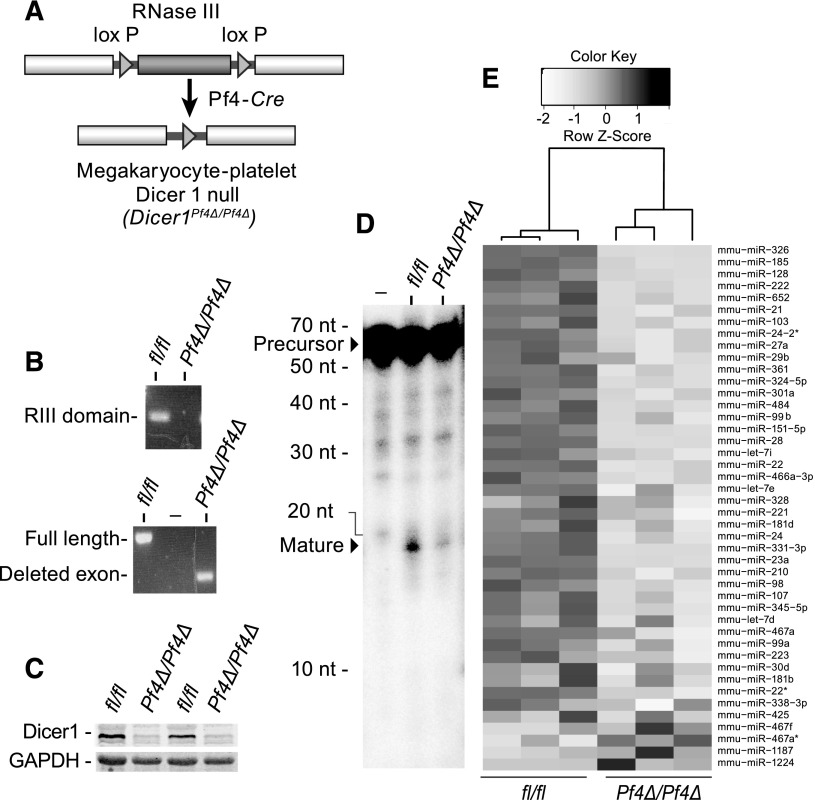
Dicer1 null mice die at E7.5.23 Therefore, to establish a causal role for miRNAs in platelet functional responses, we used a conditional deletion system to delete Dicer1 in MKs and platelets. Mice harboring a floxed allele of Dicer124 were crossed to animals expressing Cre recombinase controlled by the Pf4 promoter26 to generate MK-specific KO mice (Dicer1Pf4Δ/Pf4Δ mice). The Dicer1 conditional allele has loxP sites flanking an exon that encodes most of the second RNase III domain (Figure 1A), which catalyzes pre-miRNA processing into mature miRNA.23 Cre recombination effectively removed the RNase IIIb domain, resulting in a truncated mRNA for Dicer1 (Figure 1B) and a loss of Dicer1 protein in platelets (Figure 1C).
Figure 1.
Deletion of Dicer1 reduces mature miRNA level in platelets. (A) Schematic representing Pf4-cre recombinase-mediated deletion of Dicer1. LoxP sites flanking the second RNase IIIb domain of Dicer1 mediate its removal in the presence of Cre recombinase, which is under control of the MK and platelet-specific Pf4 promoter. (B) Real-time PCR amplification of platelet RNA from Dicer1Pf4Δ/Pf4Δ or Dicerfl/fl mice. The primers are designed to amplify within the Dicer RNase IIIb domain (top) or flank the Dicer RNase IIIb domain (bottom). Note that, in the Dicer1Pf4Δ/Pf4Δ mice, the transcript is present in platelets, but truncated. (C) Western blots for Dicer1 and Gapdh in platelets from Dicerfl/fl (lanes 1 and 3) or Dicer1Pf4Δ/Pf4Δ (lanes 2 and 4) mice. (D) Dicer activity assay to monitor the processing of 32P-labeled human pre–let-7a into mature let-7a using platelet lysates (or no lysate [–]; lane 1) from Dicer1Pf4Δ/Pf4Δ (lane 3) or Dicer1fl/fl littermate controls (lane 2). (E) Heat map depicting the clustering and relative fold changes of mature miRNAs, changed twofold or more, in platelets from Dicer1Pf4Δ/Pf4Δ compared with Dicer1fl/fl littermates (n = 3 mice per group. Each lane represents data from a single mouse/array for a total of 6 microarrays).
Next, we assessed Dicer1 activity in platelets by a pre-miRNA processing assay. Platelet-derived extracts from Dicer1fl/fl mice effectively processed let-7a-3 pre-miRNA into the mature miRNA form (Figure 1D). Processing of let-7a-3 pre-miRNA was lower when assessing platelet extracts isolated from Dicer1Pf4Δ/Pf4Δ mice (Figure 1D).
We next conducted a global screen of platelet miRNA expression by microarray analysis. As depicted in Figure 1E (also see supplemental Dataset 1, available on the Blood Web site), deletion of Dicer1 in platelets reduced the mean expression (up to ∼6.5-fold) of the majority (97 were reduced out of 123 analyzed) of miRNAs, and 49 of those miRNAs reached statistical significance (Q value < .05). No miRNAs significantly increased. The pattern and magnitude of changes observed by real-time PCR on selected candidates, using 18s rRNA as a reference gene (supplemental Figure 1A-B), matched the array results (Pearson correlation >.95, P < .001). Relative to WT counterparts, miRNA levels were also reduced in cultured Dicer1Pf4Δ/Pf4Δ MKs (supplemental Figure 1C).
To address whether miRNA levels could be reduced further by deletion of Drosha, which functions upstream of Dicer1 by processing primary miRNA into pre-miRNA in the nucleus,5 we generated DroshaPf4Δ/Pf4Δ single KO and (Dicer1 Drosha)Pf4Δ/Pf4Δ double KO mice. Similar to the ablation of Dicer1, the deletion of Drosha in MKs reduced, but did not eliminate, miRNA expression in platelets (supplemental Figure 2A). Combined loss of both Dicer1 and Drosha did not significantly exacerbate the changes in miRNA expression that were observed in either of the single KOs (supplemental Figure 2B).
Peripheral blood platelet counts were subtly, but significantly reduced in Dicer1Pf4Δ/Pf4Δ, DroshaPf4Δ/Pf4Δ, and (Dicer1 Drosha)Pf4Δ/Pf4Δ mice (supplemental Figure 3). No other developmental or age-related abnormalities were observed in each of the genotypic classes. Because miRNA levels were similarly reduced in all three cohorts and the cytosolic activity of Dicer1 is preserved in platelets (Figure 1D), we focused the remainder of the studies on Dicer1-deficient platelets.
Deletion of Dicer1 increases transcript levels, including αIIb and β3 in platelets
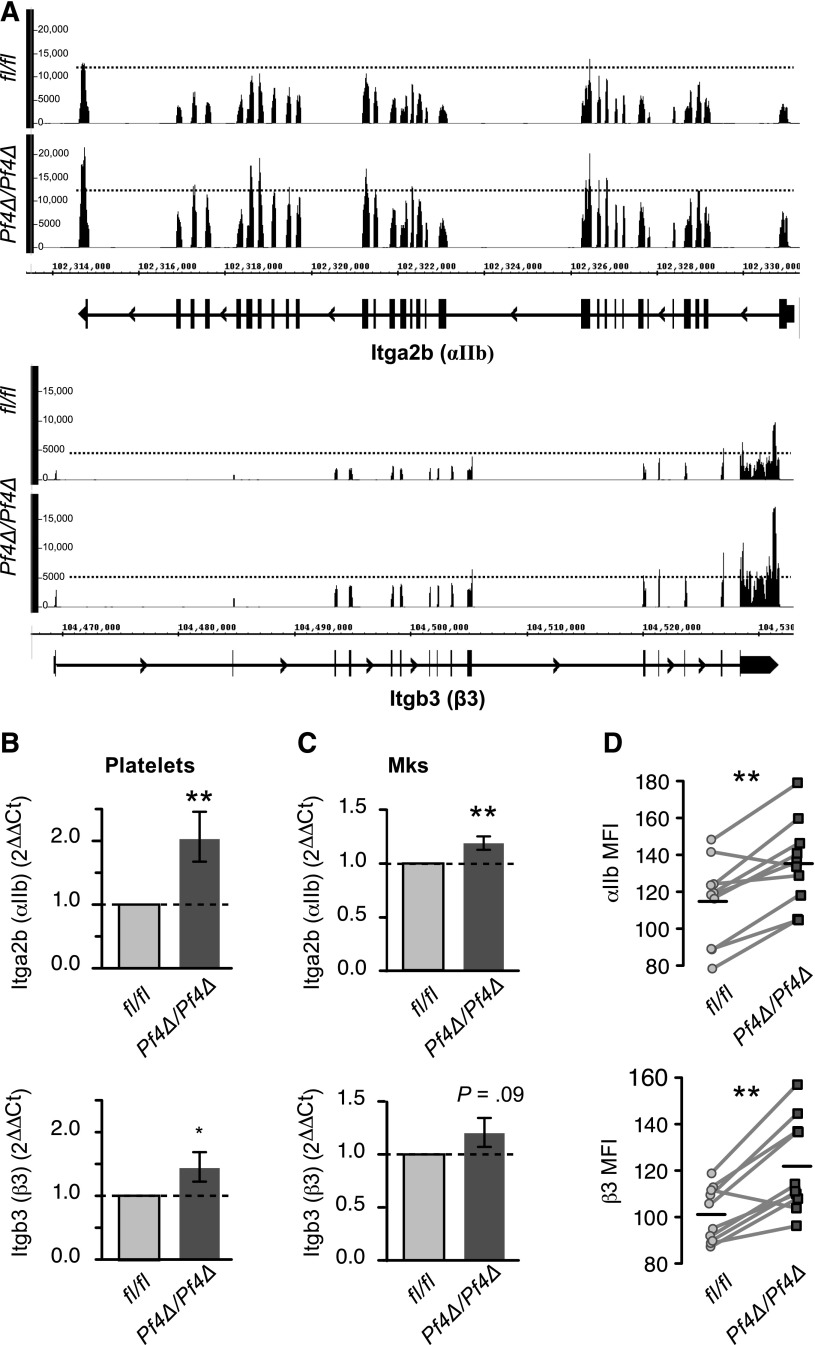
miRNAs regulate posttranscriptional gene expression by several mechanisms, including the destabilization of target mRNAs.8 To screen for target mRNAs potentially affected by reduced miRNA expression, we performed an RNA-seq analysis of poly-adenylated mRNA in platelets from Dicer1Pf4Δ/Pf4Δ and Dicer1fl/fl mice. Platelets of both genotypes expressed thousands of transcripts (>7000; supplemental Dataset 2). To identify potential direct targets of miRNA, we used a ranking strategy to prioritize targets according to magnitude of expression and fold change (see “Materials and methods”). We validated the utility of this strategy by real-time PCR on selected targets (supplemental Figure 4A-B and supplemental Dataset 2). As shown in supplemental Figure 4B-C, several of the selected targets altered in platelets were coordinately altered in BM-derived MKs. Of the targets identified as increased by RNA-seq, β3 and αIIb were ranked among the top differentially expressed candidates (see “Statistical methods”). Because of the critical role of αIIbβ3 integrin in homotypic aggregation, we focused on miRNA regulation of both αIIb and β3 in subsequent studies. As shown in Figure 2A, the numbers of sequence reads across αIIb and β3 mRNAs were increased by 1.4- and 1.6-fold, respectively, in Dicer1-deficient platelets. Real-time PCR analysis confirmed the increases in mRNA expression in additional platelet samples (Figure 2B). Increased expression of αIIb and β3 mRNA was also observed in cultured MKs, which was statistically significant for αIIb (Figure 2C). Consistent with the changes in transcript level, surface protein expression of αIIb and β3 was consistently higher on Dicer1Pf4Δ/Pf4Δ platelets relative to those purified from Dicer1fl/fl littermates (Figure 2D). Surface expression levels of Cd9 (a pan-leukocyte marker), Gp1bb, or Gp9 were not altered (not shown), even though RNA-seq suggested that Gp1bb RNA was considerably increased. The mean staining intensity of intracellular αIIb and β3 protein was higher in permeabilized platelets from Dicer1Pf4Δ/Pf4Δ mice relative to WT (αIIb geometric mean fluorescence intensity [gMFI]: 485 ± 3 vs 469 ± 3; β3 gMFI: 290 ± 14 vs 266 ± 22; mean ± standard error of the mean [SEM], n = 4 mice per group).
Figure 2.
αIIb and β3 expression is increased in Dicer1-deficient platelets. (A) RNA-seq reads from Dicer1Pf4Δ/Pf4Δ or Dicer1fl/fl platelets aligning to αIIb (top) or β3 (bottom) mRNAs. Refseq annotations are included underneath each tracing, where thick and thin lines represent exons and introns, respectively. In these depictions, reads are piled up at their alignment location, so that both the density and height of histogram peaks are indicative of relative abundance. RNA-seq was performed on a pool of 3 mice per group. (B-C) Real-time PCR expression analysis of αIIb or β3 mRNA in (B) platelets or (C) BM-derived MKs from Dicer1Pf4Δ/Pf4Δ vs Dicer1fl/fl mice. *P < .05; **P < .01; unpaired Student t test. n = 9 to 13 mice per group for (B) and n = 4 mice per group for (C). (D) Surface expression of αIIb and β3 protein. Platelets from Dicer1Pf4Δ/Pf4Δ or Dicer1fl/fl mice were isolated and stained in littermate pairs for flow cytometry analysis. The gMFI of surface staining of αIIb and β3 for each pair are shown. Light gray lines connect each matched littermate pair, and the horizontal lines represent the mean for each group (**P < .01; paired Student t test; n = 10 pairs).
Ago2 associates with αIIb and β3 transcripts in platelets
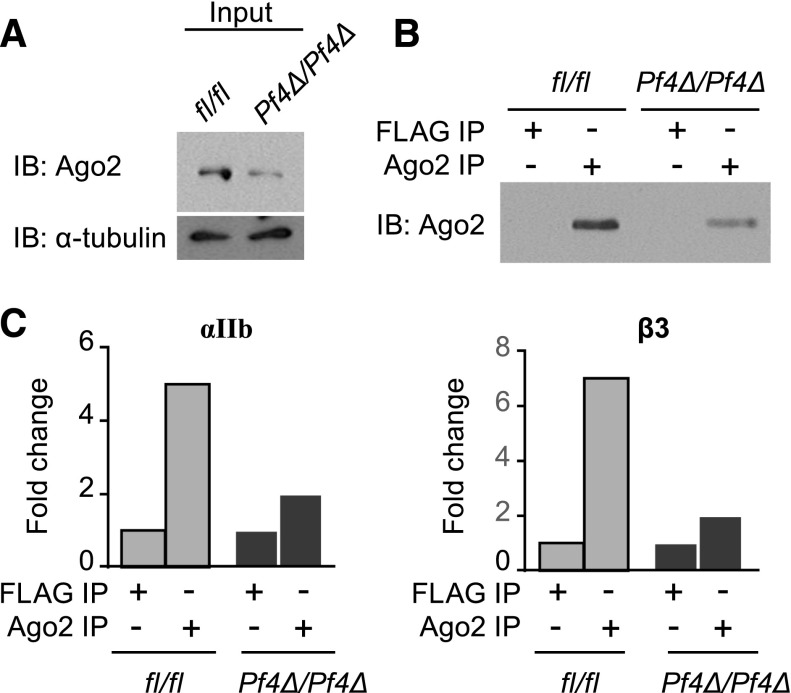
Ago2 binds and guides miRNAs to their target mRNA seed sites. Ago2 is expressed in human platelets.14 Western blotting demonstrated that Ago2 protein is also present in WT mouse platelets, but is reduced in Dicer1Pf4Δ/Pf4Δ platelets (Figure 3A). An antibody specific for Ago2, but not an isotype control antibody, is immunoprecipitated protein for Ago2 (Figure 3B). According to real-time PCR analysis of Ago2 immunoprecipitates (IP), Ago2 co-immunoprecipitated both αIIb and β3 mRNAs (Figure 3C), suggesting their association in platelets. Comparison of Ago2/mRNA IP from Dicer1fl/fl and Dicer1Pf4Δ/Pf4Δ platelets was suggestive of reduced αIIb and β3 in IP from Dicer1Pf4Δ/Pf4Δ platelets (Figure 3C), even though their total mRNA levels were higher (Figure 2B).
Figure 3.
Platelet αIIb and β3 mRNA associates with Ago2. Pooled lysates (5 to 7 mice per group) from Dicer1Pf4Δ/Pf4Δ or Dicer1fl/fl mouse platelets were immunoprecipitated with an antibody against Ago2 or against the isotype control FLAG followed by immunoblotting (IB) for the indicated protein. (A) Total Ago2 protein expression in platelets from Dicer1Pf4Δ/Pf4Δ or Dicer1fl/fl mice. Blot shown is representative of 2 independent experiments. (B) Immunoprecipitated Ago2 protein expression in platelets from Dicer1Pf4Δ/Pf4Δ or Dicer1fl/fl mice. n = 1 platelet pools (pooled from 5 mice per group). (C) Immunoprecipitated RNA was extracted from IP, and αIIb or β3 mRNA association to Ago2 was estimated by quantitative real-time PCR (in duplicates). The results are normalized with Gapdh mRNA to calculate a fold change vs FLAG IP. Results are representative of 2 independent experiments demonstrating enrichment of αIIb or β3 mRNA in Ago2 IP in pools (pooled from 5 to 7 mice per group) of Dicer1fl/fl platelet lysates, and n = 1 platelet pools per group (pooled from 5 mice per group) for the comparison with Dicer1Pf4Δ/Pf4Δ animals.
miRNA regulation of αIIb and β3 mRNA 3′UTR
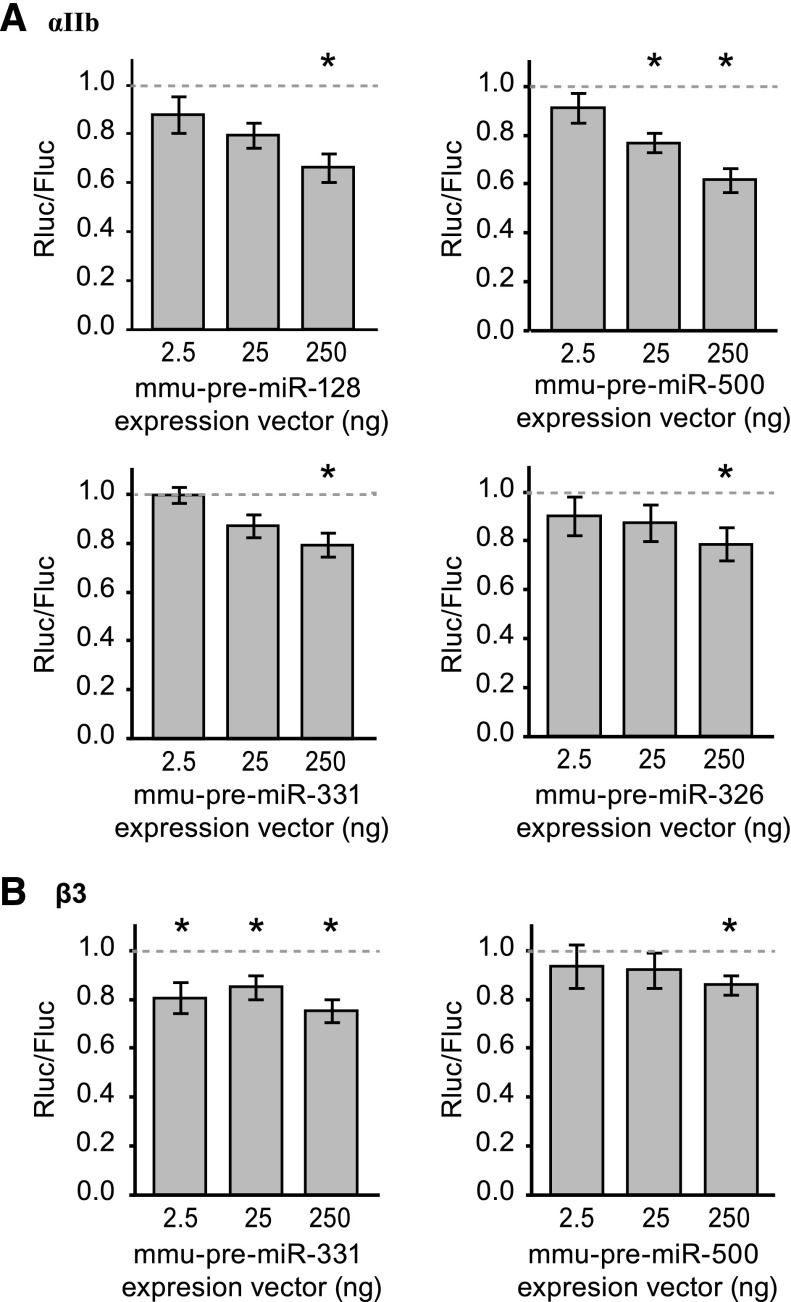
Next, we employed bioinformatics tools to identify miRNAs that potentially regulate αIIb or β3 expression using the following criteria where miRNAs needed to be: (1) expressed abundantly in platelets; (2) decreased in Dicer1Pf4Δ/Pf4Δ platelets relative to controls; and (3) predicted to bind the 3′UTR of αIIb or β3. We selected 4 miRNA’s that putatively target αIIb mRNA (miR-326, miR-128, miR-331, and miR-500), two of which also putatively target β3 mRNA (miR-331 and miR-500). As shown in Figure 4A, all 4 candidate miRNAs, as compared with a nonrelevant short hairpin control, downregulated reporter gene expression placed under the control of the αIIb mRNA 3′UTR. miR-331 and miR-500 also repressed reporter gene expression placed under the control of the β3 mRNA 3′UTR (Figure 4B).
Figure 4.
miRNAs that are decreased in Dicer1-deficient platelets regulate αIIb and β3 expression through their 3′UTR. HEK293 cells were transfected with a reporter gene construct that contains the 3′-UTR sequence of αIIb (A) or β3 (B), located downstream of the Rluc open reading frame, and vectors encoding pre–mmu-miR-128, pre–mmu-miR-500, pre–mmu-miR-331, pre–mmu-miR-326, or a nonrelevant short hairpin control. Rluc and Firefly luciferase signals were measured and normalized to the nonrelevant hairpin control vector, which is represented by the dashed line (set at 1). Shown are mean ± SEM. Analysis of variance: P < .05; *P < .05 vs control; unpaired Student t test. n = 5 to 6 experiments performed in duplicate.
Fibrinogen binding/uptake and platelet reactivity is increased in Dicer1-deficient mice
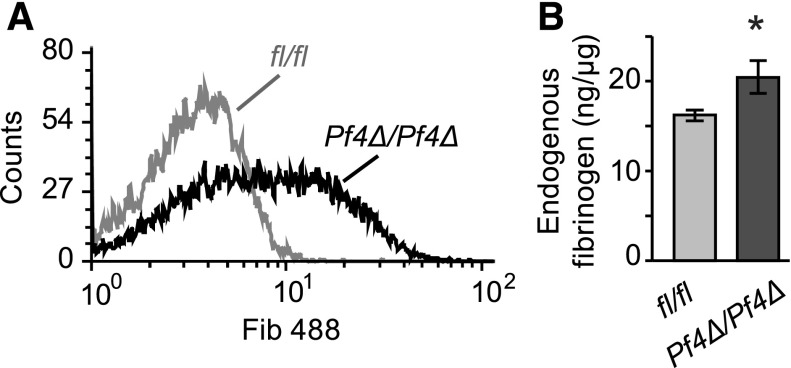
Circulating platelets take up fibrinogen in the absence of activation, and this uptake is dependent on integrin αIIbβ3.38 To assess fibrinogen uptake into Dicer1Pf4Δ/Pf4Δ platelets and controls, labeled fibrinogen was injected into mice and platelets were subsequently analyzed. After 48 hours, mean labeling intensity was significantly increased in platelets from Dicer1Pf4Δ/Pf4Δ compared with Dicer1fl/fl mice (Figure 5A). Circulating platelets from Dicer1Pf4Δ/Pf4Δ mice also contained more endogenous fibrinogen, as measured by ELISA (Figure 5B).
Figure 5.
In vivo fibrinogen uptake and endogenous fibrinogen levels are increased in Dicer1-deficient platelets. (A) Dicer1Pf4Δ/Pf4Δ or Dicer1fl/fl mice were IV injected with Alexa Fluor 488 labeled fibrinogen. After 48 hours, platelets were isolated, stained with an anti-CD41 antibody, and incorporation of the label was analyzed by flow cytometry. The histogram is representative of 6 independent samples isolated from Dicer1Pf4Δ/Pf4Δ and Dicer1fl/fl mice (difference in gMFI P < .05; unpaired Student t test; n = 6 samples per group). (B) Bar graph representing the amount of endogenous (not injected) fibrinogen in platelet lysates obtained from Dicer1Pf4Δ/Pf4Δ vs Dicer1fl/fl mice, as measured by ELISA (*P < .05; unpaired Student t test; n = 7 mice per group).
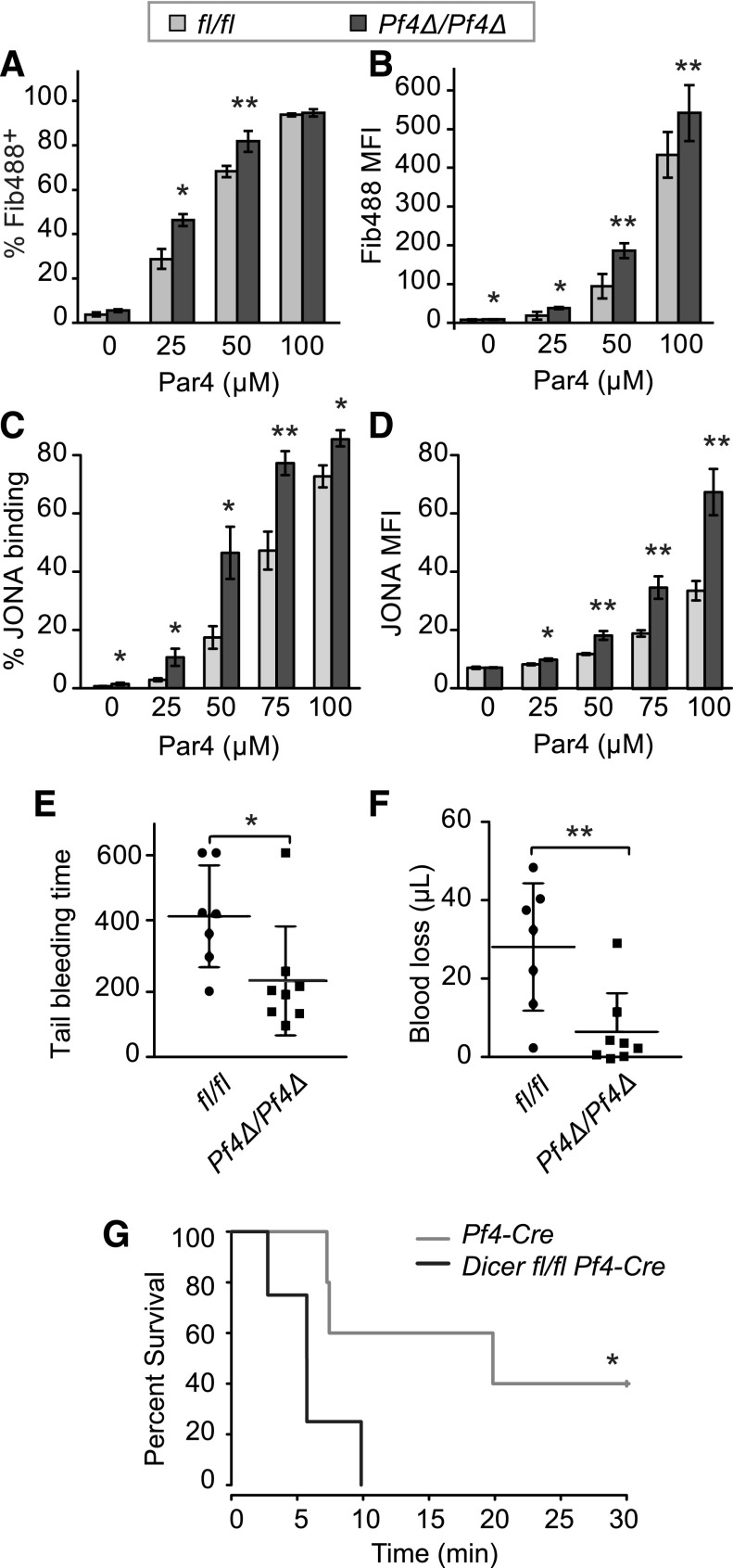
In response to ex vivo PAR4 stimulation, a greater proportion of washed platelets from Dicer1Pf4Δ/Pf4Δ mice bound fibrinogen at submaximal doses of PAR4 (Figure 6A). In addition, the percentage of Dicer1Pf4Δ/Pf4Δ platelets in diluted whole blood that bound JONA in response to PAR4 was increased (Figure 6C). There was no difference in JonA binding in Dicer1Pf4Δ/wt heterozygote mice compared with WT mice (supplemental Figure 5A). The level of fibrinogen and JONA binding per platelet was also higher in Dicer1Pf4Δ/Pf4Δ cells compared with Dicer1fl/fl counterparts (Figure 6B,D). Similar to JONA binding studies, subthreshold concentrations of PAR4 peptide induced a higher proportion of P-selectin–positive platelets from Dicer1Pf4Δ/Pf4Δ mice compared with Dicer1fl/fl WT or Dicer1Pf4Δ/wt heterozygote mice (supplemental Figure 5B). Activation with U46619, ADP, and convulxin also trended toward higher JONA binding in PAR4-stimulated platelets from Dicer1Pf4Δ/Pf4Δ compared with Dicer1fl/fl mice, but did not reach statistical significance (supplemental Figure 6). Consistent with heightened platelet reactivity, tail-bleeding time and blood loss following tail resection was reduced in Dicer1Pf4Δ/Pf4Δ mice compared with Dicer1fl/fl littermates (Figure 6E-F). Furthermore, survival was reduced in Dicer1Pf4Δ/Pf4Δ mice following collagen/epinephrine-induced PE (Figure 6G).
Figure 6.
Dicer1-deficient platelets are hyper reactive and prone to clot. (A-B) Flow cytometry analysis of fibrinogen binding to in vitro activated mouse platelets. Washed platelets from Dicer1Pf4Δ/Pf4Δ or Dicer1fl/fl mice were activated with increasing concentrations of a PAR4 agonist for 20 minutes in the presence of Alexa Fluor 488 labeled fibrinogen. (A) Mean ± SEM of the percentage of fibrinogen labeled (FIB488+) platelets or (B) intensity of fibrinogen 488 staining (gMFI). *P < .05; **P < .01; paired Student t test; n = 8 mice per group. (C-D) Flow cytometry analysis of JONA binding to platelets in whole blood in the presence of increasing concentrations of a PAR4 peptide. (C) Mean ± SEM of the percentage of JONA-labeled platelets or (D) the gMFI of JONA antibody staining (*P < .05; **P < .01; unpaired Student t test; n = 4 Dicer1fl/fl and n = 3 Dicer1Pf4Δ/Pf4Δ mice per group). (E-F) Analysis of tail-bleed time (E) and blood loss (F) after tail resection in Dicer1Pf4Δ/Pf4Δ vs Dicer1fl/fl mice (*P < .05; **P < .01; unpaired Student t test). (G) PE was induced in anesthetized mice of the indicated genotypes by IV injection of a solution of 60 mg/kg epinephrine and 150 mg/kg collagen. Time to death was monitored for 30 minutes. Shown is the Kaplan–Meier survival curve. *P < .05; log-rank test; n = 5 Pf4-Cre and n = 4 Dicer1Pf4Δ/Pf4Δ mice per group.
Discussion
Global ablation of Dicer1, the cytoplasmic enzyme that generates mature miRNAs, is embryonic lethal in mice.23 In contrast, we found that Dicer1Pf4Δ/Pf4Δ animals develop normally, are outwardly healthy, have a normal lifespan, and exhibit no major phenotypic abnormalities. Dicer1 deletion in MKs and platelets, however, resulted in a global reduction of miRNA expression levels, altered mRNA expression patterns, and redefined the activation threshold and hemostatic capacity of platelets.
As with human platelets,14 mature miRNAs were diverse and abundant in mouse platelets (supplemental Dataset 1), yet pre-miRNAs were nearly undetectable (cycle threshold, 32 to 36; data not shown). Loss of Dicer1 from MKs precipitated a global reduction in mature miRNA expression that varied in magnitude by up to 6.5-fold. These results are similar to other conditional deletion models of Dicer1 that result in reduction of miRNAs in target cells and tissue-specific phenotypes of varying intensity.24,39,40 On the other hand, several miRNAs did not decrease following Dicer1 deletion. These included miR-1224, a mirtron whose maturation is independent of Drosha and potentially Dicer141 and miR-451, which bypasses Dicer processing via direct cleavage by Ago2.42
Dicer1-deletion reduced but did not completely abolish miRNA expression, nor did the combined deletion of Dicer1 with Drosha. This suggests that the majority of platelet miRNAs are generated in MKs prior to PF4 activated loss of miRNA processing enzymes, and that the majority of MK lineage miRNAs are relatively stable in maturing MKs and, subsequently, circulating platelets.
miRNAs are predicted to regulate ∼60% of genes in humans,9 with each miRNA controlling hundreds of different targets.43 A major mechanism of miRNA regulation is degradation of target mRNAs.8 Our studies suggested systemic alterations in the platelet mRNA profile between Dicer1Pf4Δ/Pf4Δ and Dicer1fl/fl mice with some changes reflected in the MK.
Increased mRNA levels of αIIb and β3 in Dicer1-deficient platelets resulted in higher surface expression of αIIb and β3 proteins. Both primary and secondary effects of miRNA depletion could be influencing changes in αIIb and β3 expression. Studies have demonstrated a relationship between αIIb and platelet size,44 and others have reported an inverse relationship between MPV and platelet counts.45 Dicer1Pf4Δ/Pf4Δ mice exhibited a mild thrombocytopenia that also coincided with a higher MPV (supplemental Figure 7). Because other platelet markers did not increase, it is unlikely that the higher αIIb expression in Dicer1Pf4Δ/Pf4Δ platelets was simply related to their increased size. Furthermore, αIIb expression was higher in Dicer1Pf4Δ/Pf4Δ platelets, even when platelets were binned according to equivalent forward scatter intensity, which is proportional to cell surface area (supplemental Figure 8).
Regulation in human cell culture of β3 integrin subunit expression by miRNAs, including miR-30, miR-31, and let-7a has previously been documented.46-48 We identified 4 miRNAs (miR-326, miR-128, miR-331, and miR-500) that potentially directly regulate αIIb expression. The exact contribution of each of these, and of other miRNAs to surface αIIb and β3 levels, remains to be determined.
Ago2 protein is unstable in the absence of loaded miRNAs, and its abundance is reduced by posttranscriptional mechanisms in Dicer1- or DGCR8-deficient mouse embryonic stem cells.49 We found that Ago2 protein expression was reduced in Dicer1-deficient platelets suggesting that, as with other cells, the total miRNA load dictates Ago2 levels in MKs and/or platelets. Ago2 IP, but not isotype-matched controls, contained αIIb and β3 mRNAs. In contrast, our results suggested that αIIb and β3 was negligible in Ago2 IP from Dicer1-deficient platelets (Figure 3B), even though their total mRNA levels were higher (Figure 2B). The suggested loss of αIIb and β3 in Ago2 IP is consistent with the reduction of Ago2 leading to a general deficiency of gene knockdown, and with the reduction of specific miRNA that would normally direct the association of αIIb and β3 mRNAs with Ago2.
Thon and Devine50 have shown that stored human platelets synthesize αIIb and β3 protein. In preliminary studies, we have similarly found that mouse platelets constitutively synthesize protein for αIIb (data not shown). Therefore, it is likely that miRNAs continue to exert activity in circulating platelets.
αIIbβ3 is the major integrin complex in platelets that mediates homotypic aggregation, and fibrinogen is its major ligand. Circulating unstimulated platelets from Dicer1-deficient mice internalized more fibrinogen, and platelets activated in vitro bound more fibrinogen. Dicer1-deficient mice also have reduced tail-bleeding times, and reduced survival following induction of PE compared with their control counterparts. Considering the change in numbers of surface αIIbβ3, amplified by the sheer numbers of platelets interacting in the circulation, it is not improbable that this increase contributes to enhanced in vivo clot formation. Other pathways and targets of miRNA beyond αIIb and β3 are probably also contributing. For example, agonist-induced P-selectin translocation was also enhanced in Dicer1-deficient platelets. This could be a secondary effect of increased αIIb and β3 expression, but is more likely related to additional miRNA regulated targets. Both platelet-inhibitory and platelet-activating transcripts (supplemental Dataset 2) were increased, whereas others were decreased, probably contributing to and possibly balancing the phenotype.
It is interesting to note that a recent report associated Dicer1 reduction and miRNA suppression in platelets from patients with diabetes with enhanced platelet reactivity.17 They further suggested a role for miR-223, which is also reduced in our Dicer-deficient platelets. Whether the same miRNAs and targets are involved in the platelet phenotypes observed in diabetes-induced Dicer1 deficiency as in genetically induced Dicer1 deficiency will be an interesting avenue for future studies.
Based on studies primarily involving cultured megakaryocytic cells, many miRNAs have been implicated in MK development and platelet production.12,51 In one recent in vivo study by Chapnik et al,52 global knockdown in mice of miR-142 impaired MK maturation, polyploidization, and proplatelet formation. As a result, germline miR-142–deficient mice have pronounced thrombocytopenia and increased MPV. We also found that Dicer1Pf4Δ/Pf4Δ mice have reduced platelet counts and increased MPV compared with Dicer1fl/fl counterparts (supplemental Figure 8). Unlike the germline miR-142–deficient mice, however, thrombocytopenia was very mild (∼10%) in Dicer1Pf4Δ/Pf4Δ mice. In the Chapnik et al study, miR-142 deficiency affected the level of several actin cytoskeleton-related proteins in MKs.52 In our studies, several cytoskeletal-related genes ranked among the top differentially expressed candidates (see “Statistical methods”) increased by Dicer1 deletion. Gene set enrichment analysis ranked “Actin Filament Based Process” as the top enriched gene ontology category (nominal P < .05) according to P value. However, in a limited set of experimental conditions, we did not notice any overt differences in cytoskeletal morphology as assessed by immunocytochemistry (data not shown). The reasons for the mild thrombocytopenia and apparently unaltered cytoskeletal network in Dicer1Pf4Δ/Pf4Δ mice are not completely clear, but are likely due to the late onset of the Pf4-Cre expression.
Our studies demonstrate that disruption of the canonical miRNA-processing machinery in MKs modulates the miRNA and mRNA expression profile of platelets, which impacts platelet function. These results suggest that changes in miRNA-processing enzymes and/or miRNA expression may contribute to inter-individual variation in platelet responsiveness and reactivity in health and disease. Given the pleiotropic effects of miRNAs, future studies are required to dissect the role of specific miRNAs on platelets. Because miRNAs respond to various cellular stressors, future studies are needed to elucidate whether disease situations also alter platelet miRNA processing and function. These types of studies may shed considerable insight on the field, especially because recent studies suggest that Dicer1 is under translational control in platelets,53 and that diabetes is associated with decreased levels of Dicer1 in platelets.19
Acknowledgments
The authors are grateful for the contributions of Diana Lim in preparation of the figures, Jason Corbin and Patricia Landry for technical assistance, and Walter and Eliza Hall Institute Bioservices for outstanding animal husbandry.
This study was supported by grants from the National Institutes of Health (National Heart, Lung, and Blood Institute [NHLBI] grant R01: HL126547; Eunice Kennedy Shriver National Institute of Child Health and Human Development grant K08: HD049699; National Institute of General Medical Sciences grant K01: GM103806; and NHLBI grants T32: HL105321 and U54HL112311), the University of Utah Undergraduate Research Opportunities Program, the Australian National Health and Medical Research Council (fellowship 1016647, Program Grant 1016647, and an Independent Research Institutes Infrastructure Support Scheme grant [361646]), the Canadian Blood Services/Canadian Institutes of Health Research Blood Utilization and Conservation Initiative via Health Canada (286777), the Viertel Foundation (fellowship to B.T.K.), the Australian Cancer Research Foundation, and a Victorian State Government Operational Infrastructure Support grant. This study was also supported by the University of Utah Flow Cytometry Facility, in addition to the National Cancer Institute (5P30CA042014-24), and the American Heart Association (16GRNT27260319 to L.E.G). The views expressed herein do not necessarily represent the views of the Canadian government.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: J.W.R. performed experiments, data analysis, and drafting and preparation of the manuscript; S.C. and A.C. performed experiments and analyzed data; M.M.W.C. contributed reagents and experimental advice; R.C. performed experiments and contributed experimental advice; A.K., J.G.T.W., J.V.M., and B.K.M. performed experiments; M.M.M. and L.E.G. performed experiments and analyzed data; M.T.N. directed aspects of the study and prepared the manuscript; and B.T.K., P.P., and A.S.W. directed aspects of the study and prepared the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jesse W. Rowley, University of Utah School of Medicine, Eccles Institute of Human Genetics, Building 533, Room 4260, 15 North 2030 East, Salt Lake City, UT 84112; e-mail: jesse.rowley@u2m2.utah.edu.
References
- 1.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in caenorhabditis elegans. Science. 2001;294(5543):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Ambros V. An extensive class of small RNAs in caenorhabditis elegans. Science. 2001;294(5543):862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y, Ahn C, Han J, et al. The nuclear RNase III drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 6.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in caenorhabditis elegans. Science. 2001;293(5538):2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 8.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 11.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30(4):363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 12.Li H, Zhao H, Wang D, Yang R. microRNA regulation in megakaryocytopoiesis. Br J Haematol. 2011;155(3):298–307. doi: 10.1111/j.1365-2141.2011.08859.x. [DOI] [PubMed] [Google Scholar]
- 13.Edelstein LC, McKenzie SE, Shaw C, Holinstat MA, Kunapuli SP, Bray PF. MicroRNAs in platelet production and activation. J Thromb Haemost. 2013;11(suppl 1):340–350. doi: 10.1111/jth.12214. [DOI] [PubMed] [Google Scholar]
- 14.Landry P, Plante I, Ouellet DL, Perron MP, Rousseau G, Provost P. Existence of a microRNA pathway in anucleate platelets. Nat Struct Mol Biol. 2009;16(9):961–966. doi: 10.1038/nsmb.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruchova H, Merkerova M, Prchal JT. Aberrant expression of microRNA in polycythemia vera. Haematologica. 2008;93(7):1009–1016. doi: 10.3324/haematol.12706. [DOI] [PubMed] [Google Scholar]
- 16.Nagalla S, Shaw C, Kong X, et al. Platelet microRNA-mRNA coexpression profiles correlate with platelet reactivity. Blood. 2011;117(19):5189–5197. doi: 10.1182/blood-2010-09-299719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondkar AA, Bray MS, Leal SM, et al. VAMP8/endobrevin is overexpressed in hyperreactive human platelets: suggested role for platelet microRNA. J Thromb Haemost. 2010;8(2):369–378. doi: 10.1111/j.1538-7836.2009.03700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edelstein LC, Simon LM, Montoya RT, et al. Racial differences in human platelet PAR4 reactivity reflect expression of PCTP and miR-376c. Nat Med. 2013;19(12):1609–1616. doi: 10.1038/nm.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elgheznawy A, Shi L, Hu J, et al. Dicer cleavage by calpain determines platelet microRNA levels and function in diabetes. Circ Res. 2015;117(2):157–165. doi: 10.1161/CIRCRESAHA.117.305784. [DOI] [PubMed] [Google Scholar]
- 20.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15(20):2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grishok A, Pasquinelli AE, Conte D, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106(1):23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 22.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 23.Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nat Genet. 2003;35(3):215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 24.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102(31):10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chong MM, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease [published correction appears in J Exp Med. 2008;205(10):2449]. J Exp Med. 2008;205(9):2005–2017. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tiedt R, Schomber T, Hao-Shen H, Skoda RC. Pf4-Cre transgenic mice allow the generation of lineage-restricted gene knockouts for studying megakaryocyte and platelet function in vivo. Blood. 2007;109(4):1503–1506. doi: 10.1182/blood-2006-04-020362. [DOI] [PubMed] [Google Scholar]
- 27.Rowley JW, Oler AJ, Tolley ND, et al. Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes. Blood. 2011;118(14):e101–e111. doi: 10.1182/blood-2011-03-339705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi DS, Smith MCP, Campbell RA, et al. Proteasome function is required for platelet production. J Clin Invest. 2014;124(9):3757–3766. doi: 10.1172/JCI75247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perron MP, Landry P, Plante I, Provost P. Detection of human dicer and argonaute 2 catalytic activity. Methods Mol Biol. 2011;725:121–141. doi: 10.1007/978-1-61779-046-1_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.López-Romero P. Pre-processing and differential expression analysis of agilent microRNA arrays using the AgiMicroRna bioconductor library. BMC Genomics. 2011;12:64. doi: 10.1186/1471-2164-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pradervand S, Weber J, Thomas J, et al. Impact of normalization on miRNA microarray expression profiling. RNA. 2009;15(3):493–501. doi: 10.1261/rna.1295509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 33.Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller S, Schwartz M, Venables B. gplots: various R programming tools for plotting data. R package version 2.17.0. 2015. Available at: http://CRAN.R-project.org/package=gplots. Accessed January 25, 2016. [Google Scholar]
- 34.Nix DA, Courdy SJ, Boucher KM. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC Bioinformatics. 2008;9:523. doi: 10.1186/1471-2105-9-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Naik MU, Stalker TJ, Brass LF, Naik UP. JAM-A protects from thrombosis by suppressing integrin αIIbβ3-dependent outside-in signaling in platelets. Blood. 2012;119(14):3352–3360. doi: 10.1182/blood-2011-12-397398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tibshirani R, Chu G, Narasimhan B, Li J. samr: SAM: significance analysis of microarrays. R package version 2.0. 2011. Available at: http://CRAN.R-project.org/package=samr. Accessed January 25, 2016. [Google Scholar]
- 38.Handagama P, Scarborough RM, Shuman MA, Bainton DF. Endocytosis of fibrinogen into megakaryocyte and platelet alpha-granules is mediated by alpha IIb beta 3 (glycoprotein IIb-IIIa). Blood. 1993;82(1):135–138. [PubMed] [Google Scholar]
- 39.Cuellar TL, Davis TH, Nelson PT, et al. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc Natl Acad Sci USA. 2008;105(14):5614–5619. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L, Zhang B, Valdez JM, Wang F, Ittmann M, Xin L. Dicer ablation impairs prostate stem cell activity and causes prostate atrophy. Stem Cells. 2010;28(7):1260–1269. doi: 10.1002/stem.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sibley CR, Seow Y, Saayman S, et al. The biogenesis and characterization of mammalian microRNAs of mirtron origin. Nucleic Acids Res. 2012;40(1):438–448. doi: 10.1093/nar/gkr722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merkerova M, Belickova M, Bruchova H. Differential expression of microRNAs in hematopoietic cell lineages. Eur J Haematol. 2008;81(4):304–310. doi: 10.1111/j.1600-0609.2008.01111.x. [DOI] [PubMed] [Google Scholar]
- 43.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kunicki TJ, Williams SA, Nugent DJ, Yeager M. Mean platelet volume and integrin alleles correlate with levels of integrins α(IIb)β(3) and α(2)β(1) in acute coronary syndrome patients and normal subjects. Arterioscler Thromb Vasc Biol. 2012;32(1):147–152. doi: 10.1161/ATVBAHA.111.239392. [DOI] [PubMed] [Google Scholar]
- 45.Giles C. The platelet count and mean platelet volume. Br J Haematol. 1981;48(1):31–37. doi: 10.1111/j.1365-2141.1981.00031.x. [DOI] [PubMed] [Google Scholar]
- 46.Chen W, Harbeck MC, Zhang W, Jacobson JR. MicroRNA regulation of integrins. Transl Res. 2013;162(3):133–143. doi: 10.1016/j.trsl.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Augoff K, Das M, Bialkowska K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 is a broad regulator of β1-integrin expression and function in cancer cells. Mol Cancer Res. 2011;9(11):1500–1508. doi: 10.1158/1541-7786.MCR-11-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Müller DW, Bosserhoff A-K. Integrin beta 3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene. 2008;27(52):6698–6706. doi: 10.1038/onc.2008.282. [DOI] [PubMed] [Google Scholar]
- 49.Martinez NJ, Gregory RI. Argonaute2 expression is post-transcriptionally coupled to microRNA abundance. RNA. 2013;19(5):605–612. doi: 10.1261/rna.036434.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thon JN, Devine DV. Translation of glycoprotein IIIa in stored blood platelets. Transfusion. 2007;47(12):2260–2270. doi: 10.1111/j.1537-2995.2007.01455.x. [DOI] [PubMed] [Google Scholar]
- 51.Edelstein LC, Bray PF. MicroRNAs in platelet production and activation. Blood. 2011;117(20):5289–5296. doi: 10.1182/blood-2011-01-292011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chapnik E, Rivkin N, Mildner A, et al. miR-142 orchestrates a network of actin cytoskeleton regulators during megakaryopoiesis. ELife. 2014;3:e01964. doi: 10.7554/eLife.01964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cimmino G, Tarallo R, Nassa G, et al. Activating stimuli induce platelet microRNA modulation and proteome reorganisation. Thromb Haemost. 2015;114(1):96–108. doi: 10.1160/TH14-09-0726. [DOI] [PubMed] [Google Scholar]