Abstract
Escherichia coli strain MRE600 was originally identified for its low RNase I activity and has therefore been widely adopted by the biomedical research community as a preferred source for the expression and purification of transfer RNAs and ribosomes. Despite its widespread use, surprisingly little information about its genome or genetic content exists. Here, we present the first de novo assembly and description of the MRE600 genome and epigenome. To provide context to these studies of MRE600, we include comparative analyses with E. coli K-12 MG1655 (K12). Pacific Biosciences Single Molecule, Real-Time sequencing reads were assembled into one large chromosome (4.83 Mb) and three smaller plasmids (89.1, 56.9, and 7.1 kb). Interestingly, the 7.1-kb plasmid possesses genes encoding a colicin E1 protein and its associated immunity protein. The MRE600 genome has a G + C content of 50.8% and contains a total of 5,181 genes, including 4,913 protein-encoding genes and 268 RNA genes. We identified 41,469 modified DNA bases (0.83% of total) and found that MRE600 lacks the gene for type I methyltransferase, EcoKI. Phylogenetic, taxonomic, and genetic analyses demonstrate that MRE600 is a divergent E. coli strain that displays features of the closely related genus, Shigella. Nevertheless, comparative analyses between MRE600 and E. coli K12 show that these two strains exhibit nearly identical ribosomal proteins, ribosomal RNAs, and highly homologous tRNA species. Substantiating prior suggestions that MRE600 lacks RNase I activity, the RNase I-encoding gene, rna, contains a single premature stop codon early in its open-reading frame.
Keywords: Escherichia coli, Shigella, MRE600, epigenetics, ribonuclease I, colicin
Introduction
Escherichia coli is a Gram-negative, nonsporulating, rod-shaped, facultative anaerobe that inhabits the intestines of warm-blooded animals and reptiles (Gordon and Cowling 2003). Escherichia coli is both a widespread gut commensal in vertebrates and a versatile and virulent pathogen that affects millions of humans each year (Kosek et al. 2003). Due to its ability to grow rapidly in chemically defined media, its metabolic versatility, and its ease of genetic manipulation, E. coli is also one of biology’s most important model organisms (Casali and Preston 2003). Escherichia coli has therefore become one of the most highly characterized organisms on Earth and, as an experimental model system, has been integral to our ability to investigate and understand many fundamental biological processes.
As a species, E. coli is exceptionally diverse and is comprised of innumerable strains that are differentiated by their genetic content and physiological properties. The first published E. coli genome assembly was of strain K-12 MG1655 (K12) which was chosen because it had been maintained in the lab with minimal genetic manipulation (Blattner et al. 1997). Subsequent analyses have revealed substantial genomic heterogeneity between different E. coli strains. For example, finished genomes listed in the Joint Genome Institute’s Integrated Microbial Genomes (IMG) database show that E. coli genome size can range from 3.98 Mb (strain K-12 subMDS42) to 5.86 Mb (strain O26:H11 11368), and can contain between 3,696 genes (strain K-12 subMDS42) and 5,919 genes (strain O157:H7 str. EDL933) (Markowitz et al. 2012). A recent study investigating the genomes of 20 E. coli strains identified a total of 17,838 distinct genes, with only 1,976 being common to all (Touchon et al. 2009). Such genomic variation contributes to each strain’s distinct physiological properties, such as their varied abilities to metabolize sugars, resistance to particular antibiotics, and growth rate-temperature profiles (Gordon 2004). The E. coli MRE600 strain has become a key workhorse for the RNA research community as the source for isolating RNA species such as mRNAs, tRNAs, and ribosomes due to its reported lack of RNAse I activity (Cammack and Wade 1965). The molecular basis of this distinction, however, has yet to be shown.
Although many of the details regarding the initial isolation of E. coli MRE600 (MRE600) (ATCC #29417, NCTC #8164, NCIB #10115, original strain reference C6) are not well documented, it is believed that this strain was derived from an environmental sample taken in 1950 by E. Windle Taylor of the Metropolitan Water Board of London (Public Health England). This strain was subsequently deposited into the Culture Collections of Public Health England, and in 1962, a personal communication written by A. Rogers described this strain as being RNase I deficient (Public Health England). Specifically, RNase I activity refers to a latent enzymatic degradation of the 30S ribosomal subunit upon exposure to denaturing conditions, such as urea, high salt, or ethylenediaminetetraacetic acid (Elson 1959). The first mention of MRE600 in the published literature came from the Microbial Research Establishment at Porton Down (UK). Cammack and Wade (1965) screened 13 bacterial strains for ribonuclease content using assays developed by Wade and Robinson (1963) and found that MRE600 exhibited negligible ribonuclease activity (Wade and Robinson 1963; Cammack and Wade 1965). Then, using a biochemical assay for ribonuclease activity, Wade and Robinson (1965) found that MRE600 lysate displayed a similar level of ribonuclease activity to known ribonuclease-deficient strains (Wade and Robinson 1965). As a result of these studies, MRE600 became the strain of choice for expressing and purifying stable RNA species and, as such, has played a key role in the field of translation biology.
In this report, we describe the 4.98 Mb E. coli MRE600 genome and epigenome for the first time. To provide context to the MRE600 genome, we provide comparative analyses with E. coli K12, a common lab strain, and perform additional phylogenetic and taxonomic studies to gain insight into its evolutionary history. Due to the importance of MRE600 to the translation biology field, we describe the genes encoding ribosomal components (ribosomal RNAs [rRNAs], ribosomal proteins [RPs]), while also highlighting other key characteristics of this organism. These key characteristics include the identification of a premature stop codon in the RNase I-encoding gene, rna, the possession of a colicin E1-expressing plasmid, which was previously predicted (Salaj-Smic 1978), and the lack of an hsdM gene, which encodes the type I methyltransferase, EcoKI.
Materials and Methods
Cell Growth Conditions and Genomic DNA Preparation
MRE600 was grown in lysogeny broth, Miller (EMD) at 37 °C in a rotating culture tube. Genomic DNA was extracted from cells using the QIAamp DNA Mini Kit (Qiagen) according to the manufacturer’s instructions. DNA quality and quantity were determined using an Agilent 2200 TapeStation and Qubit dsDNA BR Assay (Life Technologies), respectively.
Genome Sequencing, Assembly, and Epigenome Characterization
Single Molecule, Real-Time (SMRT) sequencing libraries were prepared using the SMRTbell Template Prep Kit 1.0 (Pacific Biosciences) and 20 kb Template Preparation Using BluePippin Size-Selection System protocol (Pacific Biosciences). Library quality and quantity were determined using an Agilent 2200 TapeStation and Qubit dsDNA BR Assay (Life Technologies), respectively. Sequencing was conducted using P5-C3 chemistry and a v3 SMRT Cell (Pacific Biosciences) at Weill Cornell Medical College. Genome assembly was conducted using the Hierarchical Genome Assembly Process 2.0 (HGAP 2.0) (Chin et al. 2013). Raw sequencing reads were filtered for length and quality such that the minimum polymerase read score was 0.8, the minimum subread length was 500 bp, and the minimum polymerase read score was greater than 100. The assembly was generated using CeleraAssembler v1 with the default parameters and was polished using the Quiver algorithm (Chin et al. 2013). To identify methylated DNA bases, we used the RS Modification and Motif Analysis module of Pacific Biosciences’ SMRT Analysis 2.3 with a Quality Value (QV) threshold of 30. This analysis uses an in silico kinetic reference and a t-test-based kinetic score to detect modified bases (Flusberg et al. 2010).
Genome Annotation
Genes were identified using Prodigal (Hyatt et al. 2010) as part of the Joint Genome Institute's genome annotation pipeline (Mavromatis et al. 2009). In this pipeline, predicted coding sequences are translated and used to search the National Center for Biotechnology Information nonredundant database, as well as the UniProt, TIGR-Fam, Pfam, PRIAM, KEGG, COG, and InterPro databases. RNA genes were identified using HMMer 3.0rc1 (Finn et al. 2011), tRNAscan-SE (Lowe and Eddy 1997), and INFERNAL 1.0.2 (Nawrocki et al. 2009). Further gene prediction analysis and annotation were performed within the Integrated Microbial Genomes—Expert Review (IMG-ER) platform (Markowitz et al. 2009).
Electron Microscopy
Cells were grown to mid-log phase before being prepared for electron microscopy. For negative staining, cells were placed on a formvar/carbon-coated copper 400-mesh grid and treated with 1.5% uranyl acetate solution. For transmission electron microscopy (TEM), cells were pelleted and fixed overnight at +4 °C in 0.1 M sodium cacodylate buffer (pH = 7.3) containing 2.5% glutaraldehyde and 4% paraformaldehyde. Cells were then washed with 0.1 M sodium cacodylate buffer (pH = 7.3) and postfixed in reduced osmium tetroxide (1% OsO4, 1.5% potassium ferrocyanide) for 1 h. Cells were washed again and then treated with 1.5% uranyl acetate solution for 1 h in the dark. Samples were dehydrated into ethanol, embedded in resin, and sectioned to 65–70 nm using a diamond knife (Diatome) on a Leica Ultracut S ultramicrotome. All imaging was performed using a JEOL JEM-1400 transmission electron microscope at Weill Cornell Medical College.
Results
The MRE600 Genome Includes a Colicin E1-Expressing Plasmid
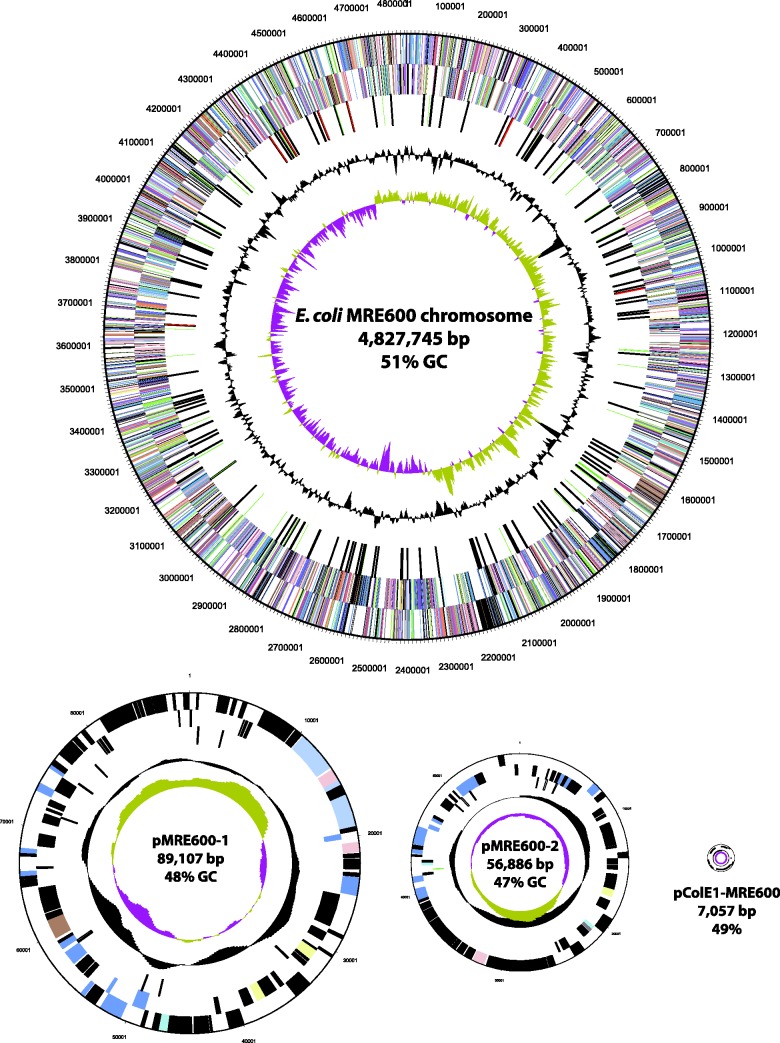
The genome sequence of MRE600 was determined at an average coverage of 184× and our assembly generated four contigs, including a 4.83 Mb chromosome and three plasmids of 89.1, 56.9, and 7.1 kb, respectively (fig. 1). The 7.1-kb plasmid, which we are calling pColE1-MRE600, contains a colicin E1 gene as well as the associated immunity protein. The identification of these genes was predicted by a previous study that showed MRE600 to possess colicinogenic activity (Salaj-Smic 1978). Our annotation identified a total of 5,181 genes, 4,913 of which are predicted to encode proteins, whereas 268 are classified as RNA genes (table 1). Of the identified genes, 3,566 (68.83%) could be categorized into Clusters of Orthologous Groups of proteins (COGs) (Tatusov et al. 2000); an overview of which can be seen in table 2.
Fig. 1.—
Graphical map of the E. coli MRE600 genome. Plasmids sizes are relative to pMRE600-1. The maps are annotated from bottom to top as follows: Genes on forward strand (colored by COG categories; Tatusov et al. 2000), genes on reverse strand (colored by COG categories; Tatusov et al. 2000), RNA genes (tRNAs = green, rRNAs = red, other RNAs = black), G + C content (black), G + C skew (purple/olive).
Table 1.
Comparing the General Genome Statistics of MRE600 and K12
| Attribute | MRE600 Value (% of total) | K12a Value (% of total) |
|---|---|---|
| Genome size (bp) | 4,980,795 (100) | 4,639,675 (100) |
| DNA coding (bp) | 4,357,764 (87.5) | 4,137,898 (89.2) |
| DNA G+C (bp) | 2,529,115 (50.8) | 2,356,477 (50.8) |
| DNA scaffolds | 4 | 1 |
| Total genes | 5,181 (100) | 4,497 (100) |
| Protein coding genes | 4,913 (94.8) | 4,321 (96.1) |
| RNA genes | 268 (5.2) | 179 (3.9) |
| rRNA genes | 22 (0.4) | 22 (0.5) |
| tRNA genes | 91 (1.8) | 89 (2.0) |
| Genes with function prediction | 4,254 (82.1) | 3,897 (86.7) |
| Genes assigned to COGsb | 3,566 (68.8) | 3,398 (75.6) |
| Genes with Pfam domainsc | 4,472 (86.3) | 3,932 (87.4) |
| Genes with signal peptides | 402 (7.8) | 426 (9.5) |
| Genes with transmembrane helices | 1,089 (21.0) | 1,038 (23.1) |
aIMG Genome ID: 646311926.
bAs described by Tatusov et al. (2000).
cAs described by Finn et al. (2010).
Table 2.
The Number of MRE600 and K12 Genes Associated with Clusters of Orthologous Groups of Proteins Functional Categories (Tatusov et al. 2000)
| Code | MRE600 Value (%) | K12a Value (%) | Description |
|---|---|---|---|
| J | 241 (6.08) | 236 (6.19) | Translation, ribosomal structure, and biogenesis |
| A | 2 (0.05) | 2 (0.05) | RNA processing and modification |
| K | 285 (7.18) | 294 (7.71) | Transcription |
| L | 149 (3.76) | 139 (3.64) | Replication, recombination, and repair |
| D | 40 (1.01) | 39 (1.02) | Cell cycle control, Cell division, chromosome partitioning |
| V | 87 (2.19) | 91 (2.39) | Defense mechanisms |
| T | 189 (4.76) | 191 (5.01) | Signal transduction mechanisms |
| M | 245 (6.18) | 242 (6.34) | Cell wall/membrane biogenesis |
| N | 133 (3.35) | 102 (2.67) | Cell motility |
| U | 60 (1.51) | 50 (1.31) | Intracellular trafficking and secretion |
| O | 157 (3.96) | 156 (4.09) | Posttranslational modification, protein turnover, chaperones |
| C | 265 (6.68) | 284 (7.44) | Energy production and conversion |
| G | 354 (8.92) | 381 (9.99) | Carbohydrate transport and metabolism |
| E | 341 (8.6) | 355 (9.31) | Amino acid transport and metabolism |
| F | 96 (2.42) | 107 (2.8) | Nucleotide transport and metabolism |
| H | 182 (4.59) | 179 (4.69) | Coenzyme transport and metabolism |
| I | 114 (2.87) | 121 (3.17) | Lipid transport and metabolism |
| P | 214 (5.39) | 223 (5.85) | Inorganic ion transport and metabolism |
| Q | 65 (1.64) | 68 (1.78) | Secondary metabolites biosynthesis, transport, and catabolism |
| R | 252 (6.35) | 261 (6.84) | General function prediction only |
| S | 202 (5.09) | 203 (5.32) | Function unknown |
| — | 1615 (31.17) | 1099 (24.44) | Not in COGs |
The percentage is relative to the total number of protein-coding genes.
aIMG Genome ID: 646311926.
The MRE600 Epigenome Reveals the Absence of EcoKI Type I Methyltransferase Activity
Our use of Pacific Biosciences SMRT sequencing also enabled the identification of 41,469 methylated DNA bases (0.83% of all bases) in the MRE600 genome. The majority of detected modifications were 6-methyladenine (m6A) at GATC motifs, but we were also able to detect putative 5-methylcytosine (m5C) modifications (table 3). Interestingly, we did not detect any m6A modifications at the well-known GCAC(N6)GTT motif in MRE600, although it was clearly identified in K12 (table 4). In bacteria, this particular modification is generated by the type I methyltransferase, EcoKI, which is encoded by the hsdM gene (Clark et al. 2013). Consistent with these epigenetic data, we were unable to identify the hsdM gene, or any close homologs, in our MRE600 assembly. Matching fragments were also absent in any of our sequencing reads, indicating that MRE600 is a knockout for the hsdM gene.
Table 3.
Summary of DNA Methylation Motifs Discovered across the Escherichia coli MRE600 Genome
| Motif | Modified Position | Modification Type | % Motifs Detected | No. Motifs Detected | No. of Motifs in the Genome | Mean Modification QV | Mean Motif Coverage |
|---|---|---|---|---|---|---|---|
| GATC | 2 | m6A | 99.65 | 39,329 | 39,468 | 143.35 | 91.11 |
| RCCGGCRYD | 2 | m5C* | 31.58 | 378 | 1,197 | 46.18 | 97.90 |
| CCAGGVDH | 2 | m5C* | 17.36 | 912 | 5,254 | 42.76 | 82.98 |
| RCCGGY | 3 | m5C* | 6.67 | 850 | 12,752 | 41.29 | 83.67 |
The modified base within each motif is shown underlined and in bold. The QV refers to the level of confidence that a particular base is methylated, and a score of >30 is considered significant. Modification types highlighted with an asterisk (*) were initially classified as either “unknown” or “m4C”; however, their low signal at this coverage suggests that they are more likely CCWGG, CCGG, or RCCGGY motifs containing the m5C modification.
Table 4.
Summary of DNA Methylation Motifs Discovered across the Escherichia coli K-12 MG1655 Genome
| Motif | Modified Position | Modification Type | % Motifs Detected | No. Motifs Detected | No. of Motifs in the Genome | Mean Modification QV | Mean Motif Coverage |
|---|---|---|---|---|---|---|---|
| GCAC(N6)GTT | 3 | m6A | 100 | 595 | 595 | 87.00 | 61.85 |
| AAC(N6)GTGC | 2 | m6A | 99.83 | 594 | 595 | 83.35 | 57.91 |
| GATC | 2 | m6A | 99.94 | 38,218 | 38,240 | 102.46 | 62.57 |
| CCAGGAVH | 2 | m5C* | 33.79 | 418 | 1,240 | 43.10 | 53.90 |
The modified base within each motif is shown underlined and in bold. The QV refers to the level of confidence that a particular base is methylated, and a score of >30 is considered significant. The GCAC(N6)GTT and AAC(N6)GTGC motifs are reverse complements of one another. The m5C modification (*) was initially called an m4C modification by the software; however, its low signal at this coverage suggests that it is more likely a CCWGG motif containing an m5C modification.
MRE600 Lacks Flagella and Exhibits Similarities to Shigella
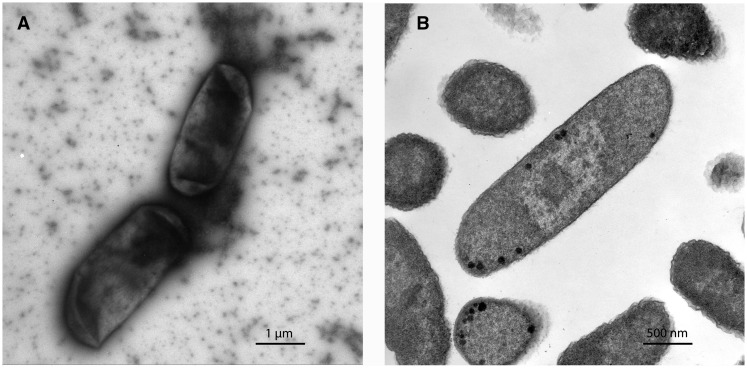
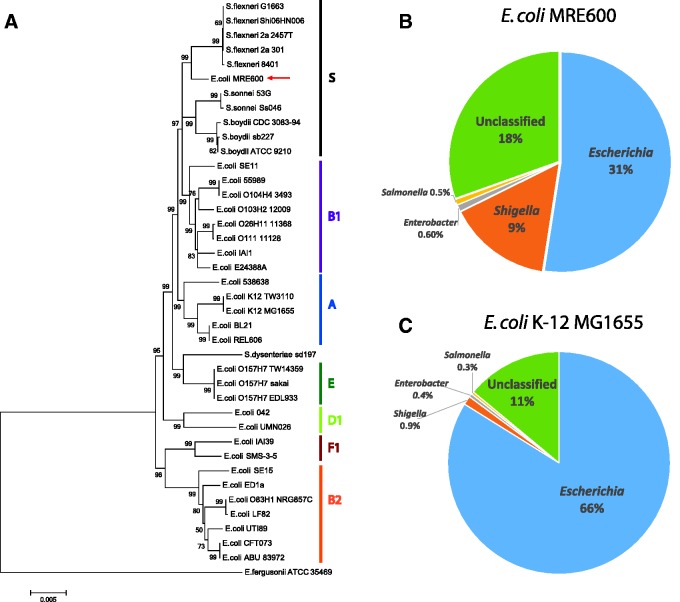
Images of MRE600 taken using TEM failed to identify flagella (fig. 2), which are commonly observed in E. coli. A lack of flagella, and consequently motility, is common, however, to species within the closely related genus, Shigella. To gain greater insight into the evolutionary relationship between MRE600, other strains of E. coli, and species within Shigella, we performed phylogenetic, taxonomic, and genetic analyses. First, we generated a phylogenetic tree using multilocus sequencing typing based on seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) (Wirth et al. 2006). This analysis found MRE600 to be more similar to species of Shigella than to other strains of E. coli (fig. 3A). We then performed a taxonomic analysis of MRE600 using Kraken (Wood and Salzberg 2014) and found that MRE600 exhibited 31% and 9% matches with Escherichia and Shigella, respectively (fig. 3B). As a comparison, we performed the same analysis using data from an E. coli K-12 MG1655 assembly (125× average coverage) that we generated. This showed a single major match of 66% with Escherichia followed by a minor match with Shigella (0.9%) (fig. 3C). These results prompted us to consider additional features that would help classify MRE600. To further investigate the appropriate taxonomic assignment of MRE600, we examined other documented genetic factors that differentiate shigellae from E. coli. Previously, it has been shown that shigellae differ from E. coli through certain genomic deletions that contain the cadA and ompT genes (Nakata et al. 1993; Maurelli et al. 1998). Neither of these genes was identified in MRE600, supporting its similarity to species of Shigella.
Fig. 2.—
Electron micrographs of E. coli MRE600 highlighting a lack of flagella. (A) MRE600 imaged using a negative stain to improve contrast. (B) MRE600 imaged using TEM. For more detail on the procedures used, see Materials and Methods.
Fig. 3.—
Phylogenetic and taxonomic analysis of E. coli MRE600. (A) Phylogenetic tree showing the relationship of E. coli MRE600 with various strains of E. coli and Shigella based on multilocus sequence typing using seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) (Wirth et al. 2006). The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei 1987). Escherichia coli phylogenetic groups (A, B1, B2, D1, E, F1) as well as a Shigella group (S) are indicated. Escherichia fergusonii ATCC 35469 was used as the outgroup. The optimal tree with the sum of branch length = 0.17149911 is shown. The confidence probability (multiplied by 100) that the interior branch length is greater than 0 was estimated using the bootstrap test (1,000 replicates) and is shown next to the branches (Dopazo 1994; Rzhetsky and Nei 1992). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al. 2004) and are in the units of the number of base substitutions per site. The analysis involved 40 sequences (9,015 positions each), and positions containing gaps and missing data were eliminated. All analyses were conducted in MEGA6 (Tamura et al. 2013). (B, C) Sequence classification of MRE600 and E. coli K12 using Kraken (Wood and Salzberg 2014). The filtered subreads used for this K12 analysis were from a 125× coverage assembly of E. coli K-12 MG1655, which we generated in the same manner as that for MRE600. Pie charts highlight the top four genera associated with the indicated strains. (B) MRE600 shows greater similarity to Escherichia (31% matches) than Shigella (9% matches). (C) Escherichia coli K12 also showed the greatest similarity to Escherichia (66%), but its match with Shigella was much lower (0.9%).
The Architecture of the MRE600 Genome Is Distinct from That of E. coli K12
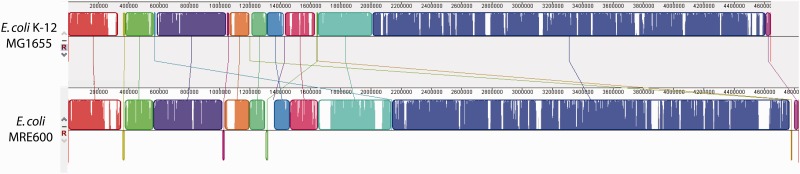
MRE600 has frequently been used as a model strain for understanding E. coli biology; however, our analysis suggests that it is distantly related to common lab strains. As genomes evolve, large-scale changes to genome architecture occur that are not easily resolvable by the comparison of short stretches of DNA. For this reason, we wanted to compare the genome structures of MRE600 and K12. To do this, we aligned the MRE600 and E. coli K12 chromosomes using MAUVE (Darling et al. 2010) to identify structural rearrangements, single nucleotide polymorphisms (SNPs), and gaps (fig. 4). We found 17 regions of high homology, called Local Collinear Blocks (LCBs), and identified a small number of structural rearrangements (fig. 4). According to our analysis, MRE600 and E. coli K12 have 63,880 SNPs and 1,850 gaps between their chromosomes. As a control, we performed the same comparative analysis on E. coli K12 and E. coli BL21 (accession number: NZ_CP010816.1), another strain in phylogenetic group A of E. coli (fig. 3) (Meier-Kolthoff et al. 2014). This analysis identified a single LCB, no rearrangements, 39,241 SNPs, and 978 gaps (data not shown). Collectively, these data show that E. coli K12 and E. coli BL21, which are two closely related strains, have similar genome structures, whereas MRE600 has a much different genome in terms of genomic variants and structure.
Fig. 4.—
Structural analysis of aligned E. coli MRE600 and E. coli K12a chromosomes. Sequences were aligned using the MAUVE aligner version 20150226 build 10 (Darling et al. 2010) and this figure was generated using the MAUVE viewer. LCBs are represented by blocks of different colors and the degree of similarity is indicated using white areas, that is, areas that are completely white within an LCB failed to align and likely contain genome-specific sequence elements. Rearrangements are highlighted using intergenome lines connecting similarly colored LCBs. This alignment identified a total of 63,880 SNPs and 1,850 gaps. The identified gaps had an average length of 722 ± 3,973 bp (SD). aAccession #U00096.3.
The MRE600 Translational Machinery Is Similar to That of K12
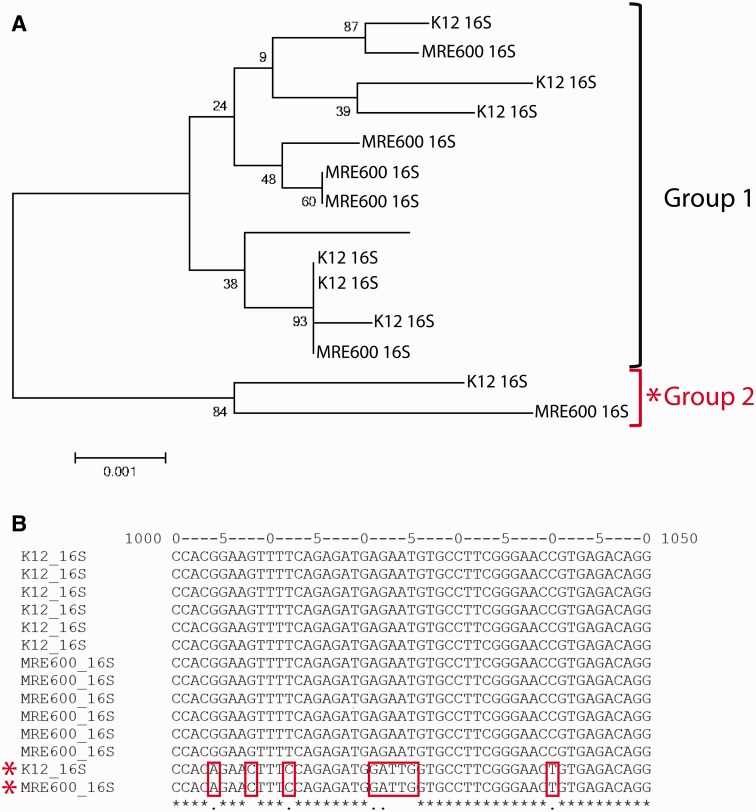
Given that MRE600 has been widely used in the field of translation biology, we used our genome assembly to compare the ribosome and tRNA genes of MRE600 and E. coli K12. First, we compared all of the 16S rRNA sequences from MRE600 with those in E. coli K12. Both strains have seven rRNA operons and all 16S rRNAs share greater than 99% identity (fig. 5A). We also found that MRE600 has the same 55 RP genes as E. coli K12 and that for all such proteins, their amino acid sequences share greater than 99% sequence identity (supplementary table S1, Supplementary Material online). Also, MRE600 is frequently used for the expression and purification of tRNAs. To compare the tRNAs of MRE600 with those of K12, we performed pairwise sequence alignments (supplementary figs. S1–S3, Supplementary Material online). Between MRE600 and K12, tRNAs containing the same anticodon generally showed very high sequence identity (>95%).
Fig. 5.—
The relationship between MRE600 and E. coli K12 16S rRNAs. (A) Phylogenetic tree showing the relationship between MRE600 and E. coli K12 16S rRNAs. Two groups of rRNAs were identified, which we named “Group 1” and “Group 2.” These relationships were inferred using the Neighbor-Joining method (Saitou and Nei 1987). The percentage of replicate trees in which the associated sequences clustered together in the bootstrap test (1,000 replicates) is shown next to the branches (Felsenstein 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al. 2004) and are in the units of the number of base substitutions per site. The analysis involved 14 sequences (1,543 positions each), and all positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA6 (Tamura et al. 2013). (B) The two groups of 16S rRNAs are identified by distinct sequences in the region encompassing nucleotides 1000–1050. This multiple sequence alignment was generated using MAFFT (Katoh and Standley 2013).
The RNase I Gene in MRE600 Bears a Premature Stop Codon
MRE600 became a widely used lab strain due to observations that it lacked ribonuclease I activity (Cammack and Wade 1965). We found that MRE600 possesses the RNase I-encoding rna gene, but that the gene’s fifth codon is a premature stop codon. Thus, as predicted, MRE600 should be unable to produce a full-length RNase I protein.
Discussion
Although E. coli MRE600 has served as a key strain for many laboratories focused on the isolation of stable RNAs prominent in translation biology, this study is the first to examine its genomic and epigenomic contents in detail. In addition to providing vital context to the myriad studies that have utilized this strain, we have identified MRE600 as a unique organism that possesses qualities similar to other strains of E. coli, but also to species of Shigella. Difficulties differentiating E. coli and Shigella have been well documented as numerous studies have suggested that Shigella should be regarded as a single species along with E. coli (Goullet 1980; Ochman et al. 1983; Whittam et al. 1983; Hartl and Dykhuizen 1984; Pupo et al. 1997). At this time, however, we refrain from suggesting that MRE600 should be reclassified, but note the intermediate location of this strain within the phylogeny (fig. 3).
One factor that prompted our investigation was the observation of endogenous plasmids within MRE600. The presence of plasmids is uncommon among typical lab strains of E. coli and we therefore rationalized that a genomic examination of this strain should be performed to assess the potential consequences of using MRE600 alongside other lab strains. Interestingly, we found a 7.1-kb plasmid that contains the genes for a colicin E1 protein as well as its associated immunity protein (fig. 1). The existence of a colicin-expressing plasmid in this organism was predicted by a previous study which identified colicinogenic activity in MRE600 (Salaj-Smic 1978). Colicin E1 expression is toxic to neighboring bacteria that do not express the appropriate immunity protein. Using BtuB as a receptor, colicin E1 proteins kill affected bacteria by forming depolarizing pores in their membranes (Cascales et al. 2007). Colicin-expressing strains are often found in the guts of animals, and although the ecological function of colicin expression is unclear, it is believed to play a role in anticompetition by enabling the invasion of a strain into an established ecological niche (Cascales et al. 2007). Alternatively, it has been proposed that colicin expression can play a defensive role in protecting an established ecological niche against invading strains (Cascales et al. 2007). At this time, we cannot speculate upon the function of colicin expression in MRE600 as we do not currently know the ecological niche in which it evolved.
Given the importance of this strain to the field of translation biology, our assembly and analysis is notable in that it predicts that the composition of E. coli K12 and MRE600 ribosomes is nearly identical at both the RNA and protein level (fig. 5 and supplementary table S1, Supplementary Material online). Similarities in the protein synthesis machineries of these two strains also extend to distinctions in the primary sequences of individual rRNA operons (fig. 5), including one operon in particular (the rrnH operon in E. coli K12). This operon, as well as a homologous operon in MRE600, contains a distinct and varied stretch of nucleotides between positions 1000–1050 within the helix 33 region of 16S rRNA (fig. 5B).
Substantiating reports that MRE600 lacks RNase I activity (Cammack and Wade 1965), our investigation identified a premature stop codon in the RNase I-encoding gene, rna. Escherichia coli RNase I is a nonspecific endoribonuclease that belongs to the T2 family (Meador and Kennell 1990). RNase I activity was first identified in ribonucleoprotein fractions from E. coli in 1958 (Elson 1958) and it was subsequently shown to be specifically associated with 30S ribosomal subunits (Elson and Tal 1959). Mechanistically, RNase I attacks the 3′ phosphates of RNA to yield ribonucleoside 3′-phosphates through a 2′, 3′-cyclic phosphate intermediate (Elson 1959; Spahr and Hollingworth 1961). The name RNase I was introduced in 1964 (Spahr 1964). We conclude that MRE600 does, in fact, lack the ability to express a functional RNase I protein due to an encoded termination site close to the N-terminus of the gene sequence.
Our epigenetic analysis identified a lack of EcoKI-mediated type I methyltransferase activity in MRE600. A subsequent genetic investigation failed to identify the EcoKI-encoding gene, hsdM, in MRE600. EcoKI methylates adenine residues within the 5′-GCAC(N6)GTT-3′ motif as well as its reverse complement 5′-AAC(N6)GTGC-3′ motif (modified adenines are underlined) (Clark et al. 2013). In bacteria, DNA base methylation has been shown to play important roles in a variety of biological processes, including virulence in E. coli (Rasko et al. 2011; Kozdon et al. 2013; Beaulaurier et al. 2015). Thus, as a natural EcoKI knockout strain, MRE600 may become an important model organism for further investigations of this particular modification.
Although the genome structure as well as epigenetic and genetic content of MRE600 shows that it is markedly distinct from common laboratory strains of E. coli, such as K12, targeted comparisons of the translational machinery from these two strains show that they are remarkable similar at the primary RNA and amino acid sequence level. These analyses validate the interchangeability of protein synthesis components derived from K12 and MRE600 strains for continued investigations of the translation mechanism.
Supplementary Material
Supplementary figures S1–S3 and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Lee Cohen-Gould and Juan Jimenez of the Weill Cornell Electron Microscopy and Histology Core Facility for their expert help with electron microscopy. They also thank Jonas Korlach for his insightful comments regarding their epigenetic analysis and to Ben Burnett, Justin Morse, and Greg Farber for their helpful comments on the manuscript. This work was supported by grants from the National Institute of Health (NIH) (R01GM079238) (to S.C.B.), the Tri-Institutional Stem Cell Initiative funded by the Starr foundation (to S.C.B. and C.T.V.), and the Sloan Foundation (2015-13964), the Bert L. and N. Kuggie Vallee Foundation, the Irma T. Hirschl and Monique Weill-Caulier Charitable Trusts, the WorldQuant Foundation, and the Pershing Square Sohn Prize (to C.E.M.).
Literature Cited
- Beaulaurier J, et al. 2015. Single molecule-level detection and long read-based phasing of epigenetic variations in bacterial methylomes. Nat Commun.. 6:7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. [DOI] [PubMed] [Google Scholar]
- Cammack KA, Wade HE. 1965. The sedimentation behaviour of ribonuclease-active and -inactive ribosomes from bacteria. Biochem J. 96:671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali N, Preston A. 2003. E. coli plasmid vectors: methods and applications. Totowa (NJ): Humana Press. [Google Scholar]
- Cascales E, et al. 2007. Colicin biology. Microbiol Mol Biol Rev. 71:158–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CS, et al. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 10:563–569. [DOI] [PubMed] [Google Scholar]
- Clark TA, et al. 2013. Enhanced 5-methylcytosine detection in single-molecule, real-time sequencing via Tet1 oxidation. BMC Biol. 11:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. 2010. Progressivemauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 5:e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopazo J. 1994. Estimating errors and confidence intervals for branch lengths in phylogenetic trees by a bootstrap approach. J Mol Evol. 38:300–304. [DOI] [PubMed] [Google Scholar]
- Elson D. 1959. Latent enzymic activity of a ribonucleoprotein isolated from Escherichia coli. Biochim Biophys Acta. 36:372–386. [DOI] [PubMed] [Google Scholar]
- Elson D. 1958. Latent ribonuclease activity in a ribonucleoprotein. Biochim Biophys Acta. 27:216–217. [DOI] [PubMed] [Google Scholar]
- Elson D, Tal M. 1959. Biochemical differences in ribonucleo-proteins. Biochim Biophys Acta. 36:281–282. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39:W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flusberg BA, et al. 2010. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 7:461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DM. 2004. The influence of ecological factors on the distribution and the genetic structure of Escherichia coli. EcoSal Plus; doi:19.1128/ecosalplus.6.4.1. [DOI] [PubMed] [Google Scholar]
- Gordon DM, Cowling A. 2003. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149:3575–3586. [DOI] [PubMed] [Google Scholar]
- Goullet P. 1980. Esterase electrophoretic pattern relatedness between Shigella species and Escherichia coli. J Gen Microbiol. 117:493–500. [DOI] [PubMed] [Google Scholar]
- Hartl DL, Dykhuizen DE. 1984. The population genetics of Escherichia coli. Annu Rev Genet. 18:31–68. [DOI] [PubMed] [Google Scholar]
- Hyatt D, et al. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosek M, Bern C, Guerrant RL. 2003. The global burden of diarrhoeal disease, as estimated from studies published between 1992 and 2000. Bull World Health Organ. 81:197–204. [PMC free article] [PubMed] [Google Scholar]
- Kozdon JB, et al. 2013. Global methylation state at base-pair resolution of the Caulobacter genome throughout the cell cycle. Proc Natl Acad Sci U S A. 110:E4658–E4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, et al. 2012. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 40:D115–D122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, et al. 2009. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25:2271–2278. [DOI] [PubMed] [Google Scholar]
- Maurelli AT, Fernandez RE, Bloch CA, Rode CK, Fasano A. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc Natl Acad Sci U S A. 95:3943–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavromatis K, et al. 2009. The DOE-JGI standard operating procedure for the annotations of microbial genomes. Stand Genomic Sci. 1:63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meador J, 3rd, Kennell D. 1990. Cloning and sequencing the gene encoding Escherichia coli ribonuclease I: exact physical mapping using the genome library. Gene 95:1–7. [DOI] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, et al. 2014. Complete genome sequence of DSM 30083(T), the type strain (U5/41(T)) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand Genomic Sci. 9:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata N, et al. 1993. The absence of a surface protease, OmpT, determines the intercellular spreading ability of Shigella: the relationship between the ompT and kcpA loci. Mol Microbiol. 9:459–468. [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR. 2009. Infernal 1.0: inference of RNA alignments. Bioinformatics 25:1335–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Whittam TS, Caugant DA, Selander RK. 1983. Enzyme polymorphism and genetic population structure in Escherichia coli and Shigella. J Gen Microbiol. 129:2715–2726. [DOI] [PubMed] [Google Scholar]
- Public Health England. Culture Collections: Bacteria Collection: Escherichia coli (NCTC 8164) [Internet] [accessed 2015 oct 20]. Available from: http://www.phe-culturecollections.org.uk/products/bacteria/detail.jsp?refId=NCTC+8164&collection=nctc
- Pupo GM, Karaolis DK, Lan R, Reeves PR. 1997. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun. 65:2685–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko DA, et al. 2011. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med. 365:709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzhetsky A, Nei M. 1992. Statistical properties of the ordinary least-squares, generalized least-squares, and minimum-evolution methods of phylogenetic inference. J Mol Evol. 35:367–375. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425. [DOI] [PubMed] [Google Scholar]
- Salaj-Smic E. 1978. Colicinogeny of Escherichia coli MRE 600. Antimicrob Agents Chemother. 14:797–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahr PF. 1964. Purification and properties of ribonuclease II from Escherichia coli. J Biol Chem. 239:3716–3726. [PubMed] [Google Scholar]
- Spahr PF, Hollingworth BR. 1961. Purification and mechanism of action of ribonuclease from Escherichia coli ribosomes. J Biol Chem. 236:823–831. [Google Scholar]
- Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 101:11030–11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, et al. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5:e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade HE, Robinson HK. 1963. Absence of ribonuclease from the ribosomes of Pseudomonas fluorescens. Nature 200:661–663. [DOI] [PubMed] [Google Scholar]
- Wade HE, Robinson HK. 1965. The distribution of ribosomal ribonucleic acids among subcellular fractions from bacteria and the adverse effect of the membrane fraction on the stability of ribosomes. Biochem J. 96:753–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittam TS, Ochman H, Selander RK. 1983. Multilocus genetic structure in natural populations of Escherichia coli. Proc Natl Acad Sci U S A. 80:1751–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T, et al. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 60:1136–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15:R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.