Abstract
Objective:
Chronic exposure to atrazine and other pesticides is reported to cause metabolic disorders, yet information on effects of atrazine on expression of genes relevant to mitochondrial function is largely missing. In the present study, therefore, we investigated the expression of a battery of nuclear- and mitochondrial-encoded genes involved in oxidative phosphorylation (OXPHOS) in human liver (HepG2) and rat muscle (L6) cell lines due to short-term atrazine exposure.
Materials and Methods:
We have determined the EC50 values of atrazine for cytotoxicity and mitochondrial toxicity (mitotoxicity) in terms of adenosine triphosphate (ATP) content in HepG2 and L6 cells. Further, the mRNA expression of nuclear- and mitochondrial-encoded genes was analyzed using quantitative real-time polymerase chain reaction.
Results:
The EC50 value of atrazine for mitotoxicity in HepG2 and L6 cells was found to be about 0.162 and 0.089 mM, respectively. Mitochondrial toxicity was indicated by reduction in ATP content following atrazine exposure. Atrazine exposure resulted in down-regulation of many OXPHOS subunits expression and affected biogenesis factors’ expression. Most prominently, superoxide dismutase (SOD) and sirtuin 3 (SIRT3) expressions were up-regulated in HepG2 cells, whereas SIRT3 expression was alleviated in L6 cells, without significant changes in SOD levels. Mitochondrial transcription factor A (TFAM) and SIRT1 expression were significantly down-regulated in both cell lines.
Conclusion:
Results suggest that TFAM and SIRT1 could be involved in atrazine-induced mitochondrial dysfunction, and further studies can be taken up to understand the mechanism of mitochondrial toxicity. Further study can also be taken up to explore the possibility of target genes as biomarkers of pesticide toxicity.
KEY WORDS: Gene expression, HepG2, L6, mitotoxicity
Introduction
Widespread use of pesticides in agriculture has always been a matter of concern, especially as India is the largest producer of pesticides in Asia.[1] Exposure to toxic compounds such as organochlorine, organophosphate, pyrethroids, and carbamate insecticides leads to widespread contamination of the ecosystem. Among many toxic pesticides, atrazine stands out for its more prevalent use in India although banned in many countries including the European Union.[2]
Atrazine belongs to the chloro-s-triazine family of herbicides, widely used for the control of broad-leaved grassy weeds.[3] Its chemical characteristics include lipophilicity, slow hydrolysis, low water solubility, and high solubility in organic solvents with high absorption by organic matter, clay, and fat tissues.[4] Atrazine has been reported to affect the reproductive system, causing decreased semen quality and birth defects in mammals.[5,6] It is also shown to act as disruptor of neuroendocrine axis and sexual development in male frogs, leading to complete sex reversal from male to female.[7]
In vitro atrazine exposure is reported to affect various cell types in humans; for example, increase in cellular proliferation in human intestinal epithelial cells,[8] decrease in growth of normal human fibroblasts,[9] and disruptions in the cell cycle regulation of immortalized human liver cells.[10] It has also decreased the natural killer cell-specific activity in peripheral blood lymphocytes[11] and cell viability in Chinese Hamster Ovary Cells.[12] One of the mechanisms which could be related to these cellular effects of atrazine may be its actions on mitochondrial functions in eukaryotes. Atrazine is reported to suppress mitochondrial oxidative phosphorylation (OXPHOS) in rats.[13] It was also able to cause apoptosis in grass carp cells (cell line ZC7901) and was involved in intracellular adenosine triphosphate (ATP) depletion.[14] Tadpoles of Xenopus laevis had difficulty maintaining energy homeostasis after 2 weeks of atrazine exposure.[15] Mitochondrial dysfunction is involved in many metabolic disorders such as diabetes, obesity, myopathies (cardiac and neuronal), and muscular dystrophy.[13,16] It has been hypothesized that atrazine binds to Complexes I and III of mitochondrial electron transport chain resulting into the suppression of OXPHOS.[13]
Taken together, these studies highlight the deleterious effects of atrazine on mitochondrial function in different mammalian cells. Studies into identifying the molecular targets of atrazine actions therefore hold the key to development of biomarkers and therapeutics. In the present study, we have used the human liver (HepG2) and rat muscle (L6) cell lines as models to study the gene targets of atrazine exposure. Cytotoxicity (EC50) and cellular ATP levels are determined for the mitochondrial toxicity using various concentrations at different time-points. Further, the expression of nuclear DNA- and mitochondrial DNA-encoded genes relevant to OXPHOS is examined.
Materials and Methods
Chemicals
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), and penicillin-streptomycin were purchased from Invitrogen (Barcelona, Spain). Atrazine (99% purity), glucose-free DMEM media, dimethyl sulfoxide (DMSO), sodium azide, and hydrogen peroxide were purchased from Sigma (USA). Hydrogen peroxide and sodium azide were used as a positive control for cell viability assay and mitochondrial toxicity assay, respectively.
Cell Culture
Human liver carcinoma (HepG2) and rat skeletal muscle (L6) cell lines were obtained from the National Centre for Cell Sciences (Pune, Maharashtra, India). Cells were grown in DMEM supplemented with 10% FBS, 0.2% sodium bicarbonate, and 1% antibiotic/antimycotic. The cells were maintained at 37°C in humidified atmosphere with 5% CO2. For mitochondrial toxicity assay, glucose- and serum-free DMEMs along with 10 mM galactose, 2 mM glutamine, and 1 mM sodium pyruvate were prepared.
Treatment of Cultured Cells with Test Compound
A stock solution of atrazine (1 mM; Sigma, USA) was prepared in DMSO. Cells were treated with various concentrations of atrazine (0.45, 0.30, 0.15, 0.1, 0.05, 0.01 mM) for cytotoxicity. The cells from the control group received vehicle (equal volume of DMSO).
Cell Viability Assay
The cells were seeded in a 96-well plate at a density of 1 × 104 cells/well. After 24 h of incubation, cells were treated with different concentrations of the atrazine (0.01–0.45 mM) for 6 h. At the end of the treatment, 20 μl of MTT solution (5 mg/ml in phosphate buffered saline) was added and cells were further incubated further for 3 h. For the analysis, the medium was removed and formazan crystals were dissolved in DMSO (200 μl). The absorbance was measured at 550 nm using a multimode plate reader (TeCAN infinite 200 PRO).
Assessment of Cellular Adenosine Triphosphate Content
Mitochondrial ToxGlo™ assay (Promega, USA) was used for the measurement of ATP levels. HepG2 and L6 cells were seeded at density 1 × 104 cells/well and 5 × 103 cells/well in a 96 well-plate, respectively and grown in glucose- and serum-free DMEMs at 37°C for 24 h. The cells were exposed to 0.05–0.2 mM atrazine dosage for HepG2 and 0.05–0.15 mM atrazine dosage for L6 and incubated for 3 h and 6 h, respectively. Cells for positive and vehicle controls were treated with sodium azide and DMSO, respectively. Cellular ATP concentrations were determined according to manufacturer protocol (ToxGlo™ assay; Promega, USA).
RNA Isolation and Quantitative Real-time Polymerase Chain Reaction
Effect of short-term atrazine exposure on gene expression in the HepG2 and L6 cells was analyzed using quantitative real-time polymerase chain reaction (qRT-PCR). HepG2 and L6 cells were treated with atrazine (HepG2 [0.1 mM and 0.2 mM] and L6 [0.05 mM and 0.1 mM]) for at 3 h, 6 h, and 9 h. Total RNA was extracted using Trizol reagent (Invitrogen, USA) and DNA was removed using the DNAse treatment. RNA was subjected to real-time PCR by using Power SYBR® Green RNA-to-CT™ 1-Step Kit (Applied Biosystems). The reaction conditions were 30 min at 48°C, 10 min at 95°C, 40 cycles of 15 s at 95°C, and 1 min at 60°C (annealing temperature). Applied Biosystem H7900 platform was used for the amplification. In addition to the dissociation curve analysis, the products were also analyzed on 1.2% agarose gel to confirm the single product amplification. The mRNA levels were normalized using 18S rRNA as an internal control. Primers used in this study are listed in Supplementary Table 1 (246.5KB, pdf) . Expression (mRNA) levels of 13 mitochondrial DNA (mtDNA)- and 10 nuclear DNA (ncDNA)-encoded OXPHOS genes and genes encoding 9 mitochondrial biogenesis controlling proteins were analyzed. For HepG2 cell line studies, primers were adopted from Koo et al. 2012, whereas primers were designed by primer select (DNASTAR) for the L6 cell line studies.
Statistical Evaluation
One-way analysis of variance was employed to test for differences among more than two groups followed by a post hoc comparison using Tukey's test. P <0.05 was considered statistically significant in all experiments.
Results
Cytotoxicity and Mitochondrial Toxicity in HepG2 and L6 Cell Lines
Cytotoxicity and mitotoxicity were determined as given in section 2.4 and 2.5. Approximately 50% reduction (n = 4;P < 0.01) in active mitochondria-dependent tetrazolium precipitation was observed in a dose-dependent manner. The cytotoxic concentration of atrazine for the liver (HepG2) and muscle (L6) cell lines was found to be more than 0.30 mM and 0.15 mM at 3 h, respectively [Figure 1], indicating that the muscle cell line was more sensitive to atrazine. These results were further utilized for the determination of the nontoxic range for the cell lines to perform the ToxGlo assay for the estimation of mitochondrial toxicity as well as cytotoxicity.
Figure 1.
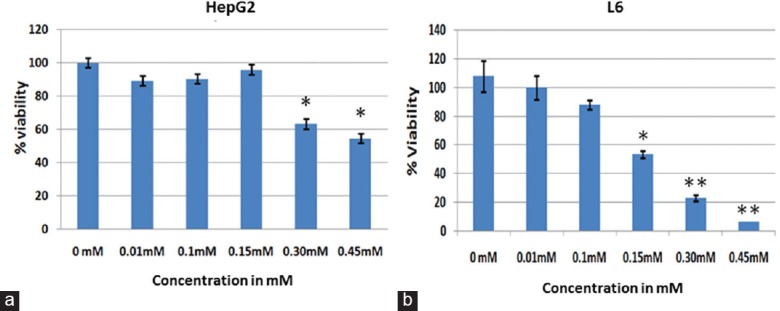
Graphs demonstrating the viability of HepG2 (a) and L6 (b) cell lines after atrazine exposure. Cytotoxicity study by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay in HepG2 and L6 cell lines after exposing to different concentrations of atrazine for 6 h revealed significant changes in viability of cells. Significantly different from other groups *P < 0.05, **P < 0.01
For the ToxGlo assay, as seen in Figure 1, 0.05–0.2 mM and 0.05–0.15 mM ranges were selected for HepG2 and L6 cell lines, respectively. The results of the ToxGlo assay demonstrated the effect of atrazine exposure on mitochondria differentiating between primary mitochondrial dysfunction and secondary cytotoxic events. As expected, the cytotoxicity was not observed at both the time-points using the subtoxic concentrations of atrazine. However, cellular ATP concentrations were found to be declining (n = 4;P < 0.05) after treatment with incremental atrazine concentrations suggesting the atrazine as a mitochondrial toxin. EC50 in the ToxGlo assay for HepG2 cell line was found to be 0.146 mM at 3 h exposure and 0.162 mM at 6 h exposure; whereas EC50 for L6 cell line was found to be 0.067 mM for 3 h exposure and 0.089 mM for 6 h exposure [Figure 2].
Figure 2.
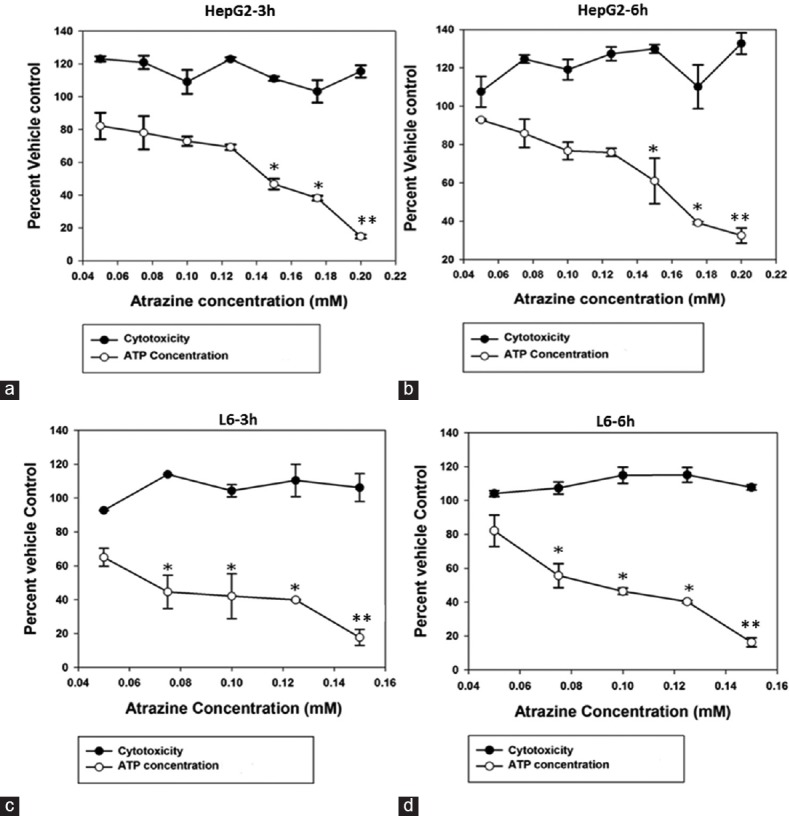
Graphs showing atrazine-induced mitochondrial toxicity in HepG2 (a and b) and L6 (c and d) cell lines at 3 h (a and c) and 6 h (b and d) exposure as studied by the ToxGlo assay. Similar to 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay, the nontoxic concentration of atrazine did not cause the cytotoxicity in both, the HepG2 and L6 cells. However, dose-response was observed in terms of atrazine-controlled reductions in adenosine triphosphate production by cell lines. Significantly different from other groups *P < 0.05, **P < 0.01
Gene Expression in Response to Mitochondrial Stress
Gene expression of all 13 subunits encoded by mtDNA and 10 selected ncDNA-encoded subunits were quantified by qRT-PCR to evaluate the effects of atrazine on mitochondrial function. Apart from this, expression of 9 mitochondrial biogenesis controlling genes namely mitochondrial transcription factor A (TFAM), manganese superoxide dismutase (SOD) 2 (SOD2, MnSOD), nuclear respiratory factor 1, peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), phosphoenolpyruvate carboxykinase 2, sirtuin 1 (SIRT1), sirtuin 3 (SIRT3), somatic cytochrome c, and uncoupling protein 2 were quantified.
Gene Expression Study with HepG2 Cell Line
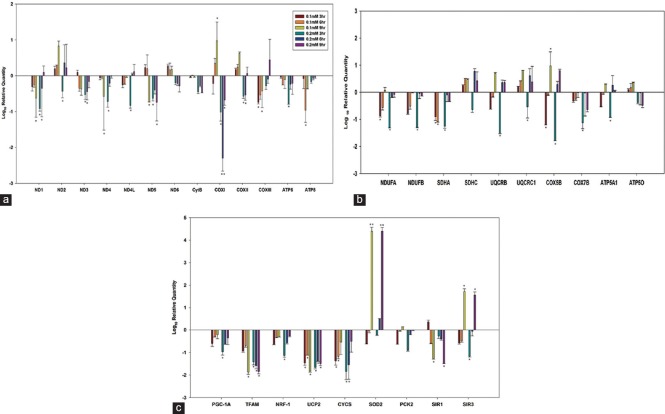
All mtDNA-encoded 13 subunits were significantly down-regulated (P < 0.05; n = 3) with 0.2 mM atrazine concentration at 3 h exposure time. However, three genes from the mitochondrially encoded NADH dehydrogenase group, i.e., ND1, ND2, and ND4L showed up-regulation when the exposure time was increased to 6 and 9 h. Genes associated with the mitochondrially encoded cytochrome C were down-regulated (P < 0.05), except for COXII and COXIII at 9 h exposure. COXI gene was most prominently affected as can be seen from maximum decline (P < 0.01). Variable response was observed with low concentration of atrazine, i.e., 0.1 mM. Both ATP synthase genes demonstrated down-regulation (P < 0.05, ATP6 - 0.2 mM at 3 h and ATP8 - 0.1 mM at 6 h) in all experimental conditions [Figure 3a].
Figure 3.
Gene expression studies with liver cell line demonstrating regulation of the oxidative phosphorylation system, (a) Mitochondrial DNA-encoded oxidative phosphorylation subunits. (b) Nuclear DNA-encoded oxidative phosphorylation subunits. (c) Mitochondrial biogenesis-related transcription factors. Statistically significantly different from other groups *P < 0.05, **P < 0.01
Gene expression of 10 nuclear DNA-encoded OXPHOS subunits with respect to atrazine exposure are represented in Figure 3b. Similar to the mitochondrial gene expression patterns, mRNA levels of all 10 nuclear DNA-encoded OXPHOS genes were also decreased (P < 0.05) after 3 h exposure of atrazine at 0.2 mM concentration. However, mRNA levels of three nuclear-encoded genes related to OXPHOS system, i.e., NDUFB, SDHA, and COX7B, were decreased in all experimental conditions. Formation of new mitochondria seems to be suppressed by the atrazine exposure [Figure 3c]. Cells treated with atrazine showed falling levels of expression of gene related to mitochondrial biogenesis (P < 0.05), except up-regulation of SOD and SIRT3 at 9 h exposure (P < 0.01).
Gene Expression Study with L6 Cell Line
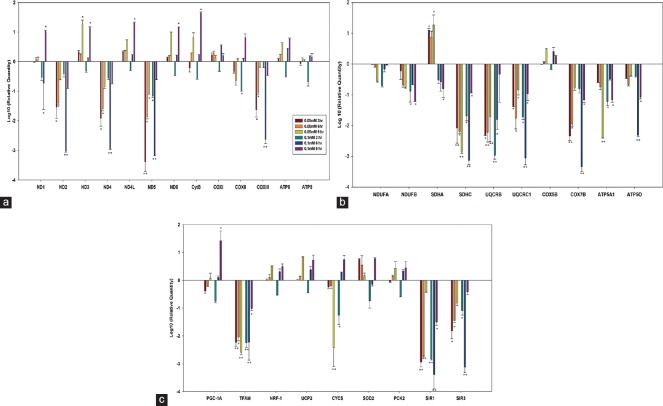
Atrazine exposure was found to be more toxic to muscle cell lines and hence, gene expression studies for L6 cell line were carried out at a concentration of 0.05 mM and 0.1 mM atrazine at 3, 6, and 9 h exposure [Figure 4]. Mitochondrial-encoded OXPHOS genes mainly demonstrated minimum of 2-fold down-regulation at 3 h exposure and 3-fold down-regulation at 6 h exposure. Genes from the NADH dehydrogenase group were especially expressed at low levels (P < 0.01) after the atrazine exposure, except ND3, ND4L, and ND6. No significant effect was observed at 9 h exposure [Figure 4a]. The mRNA levels of all ncDNA-encoded OXPHOS subunits, except SDHA and COX5B were decreased following atrazine exposure [P < 0.01; Figure 4b]. Three-fold down-regulation was observed with SDHC, UQCRC1, COX7B, and ATP5O in 0.1 mM at 6 h exposure. The mitochondrial biogenesis-related genes, i.e., TFAM, SIRT1, and SIRT3, were down-regulated in L6 cell lines in all experimental conditions [Figure 4c]. SIRT1 and SIRT3 demonstrated 3-fold down-regulation (P < 0.01).
Figure 4.
Gene expression studies with muscle cell lines demonstrating regulation of the oxidative phosphorylation system. (a) Mitochondrial DNA-encoded oxidative phosphorylation subunits. (b) Gene expression of nuclear DNA-encoded oxidative phosphorylation subunits. (c) Mitochondrial biogenesis-related transcription factors. Statistically significantly different from other groups *P < 0.05, **P < 0.01
Discussion
The toxic effects of pesticides especially, damage to mitochondria, have been shown to affect cellular functions leading to a number of health-related malfunctions. A number of pesticides have been reported to affect mitochondrial activity. For example, endosulfan reduces mitochondrial energy production in mice[17] and glyphosate-based herbicides affect mitochondrial activities and membrane integrity.[18] Atrazine has been reported to cause apoptosis in grass carp cells (cell line ZC7901) and was involved in mitochondrial membrane potential disruption and intracellular ATP depletion.[14] A study (Zaya et al. 2011) on X. laevis tadpoles showed significant increase in ADP: ATP ratios after 2 weeks of 200 μg/L atrazine exposure suggesting disturbed energy homeostasis. Low levels of ingested atrazine are bioaccumulated over a period in the liver which affected rat mitochondria.[13] However, a direct evidence of mitochondrial toxicity due to atrazine exposure in terms of gene expressions in rat and human cells is not available. The novel results of this study indicate the alterations in many mitochondrial-function-related genes in muscle and liver cell lines as a result of atrazine treatment. The data could be utilized toward the development of biomarkers.
OXPHOS is the first metabolic activity that may be affected by mitochondrial dysfunction. Energy released in OXPHOS is converted to ATP by five mitochondrial complexes, namely, (i) mitochondrial Complex I: NADH dehydrogenase (ubiquinone) subunits, (ii) mitochondrial Complex II: Succinate dehydrogenase subunits, (iii) mitochondrial Complex III: Ubiquinol-cytochrome c reductase complex subunits, (iv) mitochondrial Complex IV: Cytochrome c oxidase subunits, and (v) mitochondrial Complex V: ATP synthase subunits. Eighty-four subunits of these complexes are encoded by nuclear DNA and 13 are encoded by mitochondrial DNA.[19] We have observed the direct effect of atrazine on levels of expression of some of these genes as one of the possible mechanisms of the action of atrazine on mitochondrial function. Our results are consistent with the observations that low levels of nuclear-encoded PGC-1α and mitochondrial-encoded COX1 were associated with impaired mitochondrial function in insulin-resistant skeletal muscle cells.[20] Exposure to atrazine in muscle cell line demonstrated reduced levels of SOD, SIRT3, and UPC2 that indicated poor oxidative stress response. SOD activity was reported to decline on exposure to the pesticides endosulfan and chlorpyrifos.[21]
Mitochondrial stress leads to decreased ATP production. However, cellular ATP levels could also reduce to cytotoxic concentration resulting in false indication of mitochondrial toxicity.[14] Therefore, we have carried out the assays at the onset of the study which could help determine the atrazine concentrations for our gene expression studies related to mitochondrial toxicity. Moreover, atrazine exposure lead to reduced ATP levels, which was likely to be caused by the down-regulation of the OXPHOS system. Two genes were prominently down-regulated, i.e., TFAM and SIRT1, indicating mitochondrial dysfunction. While TFAM is essential for human mtDNA transcription and is a key regulator of mtDNA copy number,[22] SIRT1 can activate the expression of several genes related to mitochondrial oxidative functions.[23] It may be inferred that the down-regulation of TFAM and SIRT1 could be affecting the OXPHOS pathway leading to reduced ATP production. On the contrary, up-regulation of SOD and SIRT3 in the liver cell line may be a resultant stress response that was incurred by atrazine treatment. SIRT3 is reported in enhancing antioxidant activity and is associated with the development of age-associated diseases.[24,25] A dramatic up-regulation of SOD genes was seen in the liver cell line after 9 h exposure, unlike muscle cell line which showed moderate response [Figures 3c and 4c]. This difference in response indicates the higher sensitivity of L6 cells to atrazine exposure as opposed to muscle cells. Further studies are warranted to elucidate the underlying mechanisms of variable effects of atrazine on the expression of genes related to mitochondrial function.
Conclusion
The results of the present study clearly indicate that the mitochondrial toxicity after short-term exposure to water soluble range of atrazine in both HepG2 and L6 cells affects the OXPHOS system. Some of these effects may be attributed to the atrazine's actions on gene expression in muscle and liver cells. The present study provides information on effective concentrations of atrazine and downstream battery of gene targets in liver and muscle cells that are affected by the pesticide. Of these, two crucial genes in mitochondrial function, i.e., TFAM and SIRT1 emerge as possible targets for further studies and biomarker development since atrazine exposure down-regulated their expression at all-time points.
Financial Support and Sponsorship
Nil.
Conflicts of Interest
There are no conflicts of interest.
Acknowledgments
We acknowledge Senior Research Fellowship to Sneha Sagarkar by the Council of Scientific and Industrial Research (CSIR), India. The work was also supported by the funds from Department of Biotechnology, New Delhi, and CSIR-National Environmental Engineering Research Institute to Dr. Atya Kapley.
References
- 1.Gupta PK. Pesticide exposure – Indian scene. Toxicology. 2004;198:83–90. doi: 10.1016/j.tox.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 2.Jason Krutz L, Shaner DL, Weaver MA, Webb RM, Zablotowicz RM, Reddy KN, et al. Agronomic and environmental implications of enhanced s-triazine degradation. Pest Manag Sci. 2010;66:461–81. doi: 10.1002/ps.1909. [DOI] [PubMed] [Google Scholar]
- 3.Sagarkar S, Nousiainen A, Shaligram S, Björklöf K, Lindström K, Jørgensen KS, et al. Soil mesocosm studies on atrazine bioremediation. J Environ Manage. 2014;139:208–16. doi: 10.1016/j.jenvman.2014.02.016. [DOI] [PubMed] [Google Scholar]
- 4.Severi-Aguiar GD, Silva-Zacarin EC. Effects of Herbicide Atrazine in Experimental Animal Models, Herbicides – Properties, Synthesis and Control of Weeds. Dr. Mohammed Nagib Hasaneen (Ed.), ISBN: 978-953-307-803-8, InTech. DOI: 10.5772/32579;2012. Available from: http://www.intechopen.com/books/herbicides-properties-synthesis-and-control-of-weeds/effects-of-herbicideatrazine-in-experimental-animal-models .
- 5.Song Y, Jia ZC, Chen JY, Hu JX, Zhang LS. Toxic effects of atrazine on reproductive system of male rats. Biomed Environ Sci. 2014;27:281–8. doi: 10.3967/bes2014.050. [DOI] [PubMed] [Google Scholar]
- 6.Agopian AJ, Cai Y, Langlois PH, Canfield MA, Lupo PJ. Maternal residential atrazine exposure and risk for choanal atresia and stenosis in offspring. J Pediatr. 2013;162:581–6. doi: 10.1016/j.jpeds.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayes T, Haston K, Tsui M, Hoang A, Haeffele C, Vonk A. Herbicides: Feminization of male frogs in the wild. Nature. 2002;419:895–6. doi: 10.1038/419895a. [DOI] [PubMed] [Google Scholar]
- 8.Greenman SB, Rutten MJ, Fowler WM, Scheffler L, Shortridge LA, Brown B, et al. Herbicide/pesticide effects on intestinal epithelial growth. Environ Res. 1997;75:85–93. doi: 10.1006/enrs.1997.3766. [DOI] [PubMed] [Google Scholar]
- 9.Dhanwada KR, Deng Y, Clayton ME. Effects of the pesticides atrazine, metolachlor and diazinon and binary mixtures on proliferation of human fibroblasts. Int J Glob Health. 2003;2:21–36. [Google Scholar]
- 10.Powell ER, Faldladdin N, Rand AD, Pelzer D, Schrunk EM, Dhanwada KR. Atrazine exposure leads to altered growth of HepG2 cells. Toxicol In Vitro. 2011;25:644–51. doi: 10.1016/j.tiv.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Rowe AM, Brundage KM, Barnett JB. In vitro atrazine-exposure inhibits human natural killer cell lytic granule release. Toxicol Appl Pharmacol. 2007;221:179–88. doi: 10.1016/j.taap.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kmetic I, Gaurina Srcek V, Slivac I, Simic B, Kniewald Z, Kniewald J. Atrazine exposure decreases cell proliferation in Chinese hamster ovary (CHO-K1) cell line. Bull Environ Contam Toxicol. 2008;81:205–9. doi: 10.1007/s00128-008-9425-6. [DOI] [PubMed] [Google Scholar]
- 13.Lim S, Ahn SY, Song IC, Chung MH, Jang HC, Park KS, et al. Chronic exposure to the herbicide, atrazine, causes mitochondrial dysfunction and insulin resistance. PLoS One. 2009;4:e5186. doi: 10.1371/journal.pone.0005186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Li W, Piao X, Zhang J, Zhang D, Wei N, et al. Icariside II reduces testosterone production by inducing necrosis in rat Leydig cells. J Biochem Mol Toxicol. 2013;27:243–50. doi: 10.1002/jbt.21481. [DOI] [PubMed] [Google Scholar]
- 15.Zaya RM, Amini Z, Whitaker AS, Ide CF. Exposure to atrazine affects the expression of key genes in metabolic pathways integral to energy homeostasis in Xenopus laevis tadpoles. Aquat Toxicol. 2011;104:254–62. doi: 10.1016/j.aquatox.2011.04.022. [DOI] [PubMed] [Google Scholar]
- 16.Hirano M, Pavlakis SG. Mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes (MELAS): Current concepts. J Child Neurol. 1994;9:4–13. doi: 10.1177/088307389400900102. [DOI] [PubMed] [Google Scholar]
- 17.Kurutas EB, Doran F, Cıralık H. The effect of endosulfan on lactic dehydrogenase enzyme system in liver of Mus musculus: A histochemical study. Eur J Gen Med. 2006;3:148–51. [Google Scholar]
- 18.Mesnage R, Bernay B, Séralini GE. Ethoxylated adjuvants of glyphosate-based herbicides are active principles of human cell toxicity. Toxicology. 2013;313:122–8. doi: 10.1016/j.tox.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Leary SC, Sasarman F. Oxidative phosphorylation: Synthesis of mitochondrially encoded proteins and assembly of individual structural subunits into functional holoenzyme complexes. Methods Mol Biol. 2009;554:143–62. doi: 10.1007/978-1-59745-521-3_10. [DOI] [PubMed] [Google Scholar]
- 20.Heilbronn LK, Gan SK, Turner N, Campbell LV, Chisholm DJ. Markers of mitochondrial biogenesis and metabolism are lower in overweight and obese insulin-resistant subjects. J Clin Endocrinol Metab. 2007;92:1467–73. doi: 10.1210/jc.2006-2210. [DOI] [PubMed] [Google Scholar]
- 21.Saxena R, Garg P, Jain DK. In vitro anti-oxidant effect of Vitamin E on oxidative stress induced due to pesticides in rat erythrocytes. Toxicol Int. 2011;18:73–6. doi: 10.4103/0971-6580.75871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, et al. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–44. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 23.Liang F, Kume S, Koya D. SIRT1 and insulin resistance. Nat Rev Endocrinol. 2009;5:367–73. doi: 10.1038/nrendo.2009.101. [DOI] [PubMed] [Google Scholar]
- 24.Fu J, Jin J, Cichewicz RH, Hageman SA, Ellis TK, Xiang L, et al. Trans-(-)-ε-viniferin increases mitochondrial sirtuin 3 (SIRT3), activates AMP-activated protein kinase (AMPK), and protects cells in models of Huntington disease. J Biol Chem. 2012;287:24460–72. doi: 10.1074/jbc.M112.382226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sack MN, Finkel T. Mitochondrial metabolism, sirtuins, and aging. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a013102. pii: A013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.