Although basic components required for the processes of DNA replication are conserved even between prokaryotes and eukaryotes, needs for coordinating it with other cell cycle events as well as with environmental perturbation render the eukaryotic replication machinery significantly more complex. Like many other chromosome reactions, DNA replication depends on sequential assembly of factors that ultimately constitute machinery, termed replisome, capable of continuously unwinding duplex DNA and synthesising DNA chains on both strands.1
The factors participating in eukaryotic DNA replication are surprisingly conserved from yeasts to human. The initial event for DNA replication is assembly of pre-RC (pre-Replicative Complex) composed of ORC, MCM and other accessory factors during G1 phase of cell cycle. At the G1/S boundary and probably throughout the S phase, the pre-RC matures into a more complex protein assembly through recruitment of other proteins including DNA polymerases and fork promoting- and -stabilizing factors. The phosphorylation mediated by 2 kinases, Cdc7 (DDK) and CDK, plays crucial roles in decision whether S phase can be initiated or not.
Earlier studies in yeasts and human indicate MCM subunits are key substrates of Cdc7 kinase. On the other hand, Sld2 and Sld3 are 2 critical substrates of Cdk in assembly of active initiation complex in budding yeast.2,3 In vivo analyses of assembly of replication complex in yeast led to identification of pre-LC (pre-Landing Complex) composed of CDK-phosphorylated Sld2, Dpb11, GINS and DNA polymerase ε that brings GINS to the origins. Cdc45 interacts with Sld3, which binds to origins through interaction with Mcm in a manner independent of CDK. Phosphorylation of Sld2 and Sld3 by CDK permits them to dock into the BRCT motifs of Dpb11, which would stabilize the complex.
In a recent issue of Cell Cycle, Im et al. reported their analyses on protein assembly for initiation of DNA replication in human cells.4 They have analyzed interactions among essential replication factors by combining coimmunoprecipitation assay techniques with the elegant split-GFP interaction assays. They have also carefully compared chromatin binding and origin binding activities of these factors. First, they have observed interactions between RecQL4 (Sld2), Mcm10 and Ctf4 (And-1) that are expressed in insect cells (Q10C complex). This interaction is mediated by Mcm10 which binds to both RecQL4 and Ctf4. In mammalian cells, RecQL4 binds to chromatin in a manner independent of pre-RC or phosphorylation events. Chromatin binding of Mcm10 and Ctf4 depends on RecQL4 but not on pre-RC. However, its binding to replication origins depends on pre-RC and Mcm10, which interacts with Mcm2-7 at the origins. Origin binding of RecQL4 depends also on Cdc7 and Cdk kinases. Thus, RecQL4, initially bound to chromatin at non-origin sequences, is recruited to origins through Mcm10 in a manner dependent on S phase phosphorylation events by Cdc7 and Cdk. Ctf4 binding to origins depend on RecQL4 and Mcm10, therefore, dependent on Cdc7 and Cdk. Thus, RecQL4 plays a crucial role in initial recruitment of Mcm10 and Ctf4 to chromatin and then recruitment of Ctf4 to origins. These authors also showed that assembly of the Q10C complex and their binding to origins is a target of DNA damage checkpoint.
The mechanism of replication factor assembly revealed in this manuscript shares some similarity to the replisome assembly pathway in budding yeast (Fig. 1). RecQL4, the likely homolog of Sld2, plays a crucial role for assembly of the Q10C complex on the chromatin, which is dependent on CDK. Pre-LC is assembled on CDK-phosphorylated Sld2 that recruits Dpb11 (TopBP1) and the polε-GINS complex. Thus, a central role of Sld2/RecQL4 in assembly of replisome appears to be conserved. However, there seem to be many differences as well. Whereas pre-LC delivers GINS to origins, RecQL4-Mcm10-Ctf4 is required for delivery of both GINS and Cdc45. In budding yeast, pre-formed Sld3-Cdc45 is recruited to pre-RC in G1 phase, and Cdk phosphorylates chromatin-bound Sld3, onto which pre-LC is delivered through Dpb11 to generate the CMG helicase complex. Treslin,5 a putative homolog of Sld3, interacts with TopBP1 (homolog of Dpb11), although the precise role of this interaction is unknown. Mcm10 in yeasts, on the other hand, is not required for origin association of the CMG components, but is required for its activation.
Figure 1.
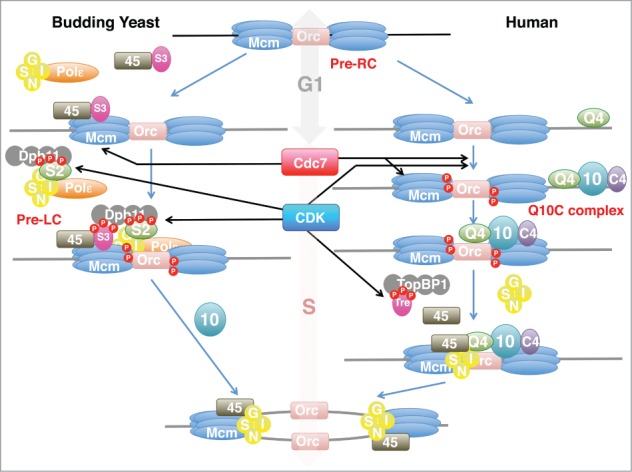
Assembly of a replisome in yeast and human cells. Cdk-dependent and pre-RC-independent formation of pre-LC or RecQL4-Mcm10-Ctf4(Q10C) complex may play an important role in assembly of replisome in yeast and human cells, respectively. See text for details. Only the relevant factors are depicted and other replisome factors including DNA polymerases, Mrc1(Claspin), Tof1-Csm3 (Timeless-Tipin), and FACT are omitted. S2, Sld2; S3, Sld3; 45, Cdc45; 10, Mcm10; Mcm, Mcm2-7; Q4, RecQL4; C4, Ctf4; Tre, Treslin.
It is interesting to note that the extent of conservation of the replisome assembly factors (Sld2, Sld3, and Dpb11 etc.) is low compared to that of the pre-RC factors including Orc, Cdc6 and Mcm2-7. This may suggest the functional divergence of the former factors, consistent with the mechanistic divergence of the replisome assembly pathway. Nevertheless, the assembly of replisome, critical for active duplex DNA unwinding, is regulated by Cdc7- and Cdk-mediated phosphorylation as well as by checkpoint pathway in both yeasts and higher eukaryotes. Recent establishment of an in vitro replication in yeast6,7 as well as future development of similar systems in mammalian cells will facilitate the detailed mechanistic studies and will disclose the common and unique mechanisms of replisome assembly in different species.
References
- 1.Masai H, et al.. Annu Rev Biochem 2010; 79:89-130; PMID:20373915; http://dx.doi.org/ 10.1146/annurev.biochem.052308.103205 [DOI] [PubMed] [Google Scholar]
- 2.Tanaka S, et al.. Nature 2007; 445:328-32; PMID:17167415 [DOI] [PubMed] [Google Scholar]
- 3.Zegerman P, Diffley JF. Nature 2007; 445:281-85; PMID:17167417 [DOI] [PubMed] [Google Scholar]
- 4.Im JS, et al.. Cell Cycle 2015; 14:1001-9; PMID:25602958; http://dx.doi.org/ 10.1080/15384101.2015.1007001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumagai A, et al.. Cell 2010; 140:349-59; PMID:20116089; http://dx.doi.org/ 10.1016/j.cell.2009.12.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heller RC, et al.. Cell 2011; 146:80–91; PMID:21729781; http://dx.doi.org/ 10.1016/j.cell.2011.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeeles JT, et al.. Nature 2015; 519:431-5; PMID:25739503; http://dx.doi.org/ 10.1038/nature14285 [DOI] [PMC free article] [PubMed] [Google Scholar]


