Abstract
Although glutamine (Gln) is not an essential amino acid, it is considered a critical substrate in many key metabolic processes that control a variety of physiological functions and are involved in regulating early embryonic development. Thus, we investigated the effect of Gln on regulation of mouse embryonic stem cell (mESC) self-renewal and related signaling pathways. Gln deprivation decreased Oct4 expression as well as expression of cell cycle regulatory proteins. However, Gln treatment retained the expression of cell cycle regulatory proteins and the Oct4 in mESCs, which were blocked by compound 968 (a glutaminase inhibitor). In addition, Gln stimulated PI3K/Akt pathway, which subsequently elicited PKCε translocation to membrane without an influx of intracellular Ca2+. Inhibition of Akt and PKC blocked Gln-induced Oct4 expression and proliferation. Gln also stimulated mTOR phosphorylation in a time-dependent manner, which abolished by PKC inhibition. Furthermore, Gln increased the cellular population of both Oct4 and bromodeoxyuridine positive cells, suggesting that Gln regulates self-renewal ability of mESCs. Gln induced a decrease in HDAC1, but not in HDAC2, which were blocked by PKC inhibitors. Gln treatment resulted in an increase in global histone acetylation and methylation. In addition, Gln significantly reduced methylation of the Oct4 promoter region through decrease in DNMT1 and DNMT3a expression, which were blocked by PKC and HDAC inhibitors. In conclusion, Gln stimulates mESC proliferation and maintains mESC undifferentiation status through transcription regulation via the Akt, PKCε, and mTOR signaling pathways.
Keywords: glutamine, mouse embryonic stem cells, Oct4, proliferation, self-renewal
Abbreviations
- BrdU
Bromodeoxyuridine
- DNMT
DNA methyltransferase
- Gln
Glutamine
- HDAC
Histone deacetylases
- mESCs
Mouse embryonic stem cells
- mTOR
mammalian target of rapamycin
- Oct4
Octamer-binding transcription factor 4
- PKC
Protein kinase C
- PI3K
Phosphoinositide 3-kinase
- RFI
Relative fluorescence intensity
- ROD
Relative optical density
- Sox2
sex determining region Y-box 2
- TSA
Trichostatin A
Introduction
Various cellular microenvironment components such as hormones, cytokines, extracellular matrix, and nutrients can regulate stem cells pluripotency and somatic cells reprogramming. Recent studies demonstrating a relationship between stem cell metabolic pathways and physiological functions have led to an increased understanding of how metabolic pathways may affect stem cell pluripotency.1 Even though conventional culture media for stem cells contain various nutrients including amino acids, the effect of a specific amino acid on regulation of stem cell pluripotency is not fully described. Furthermore, compared with somatic cells, stem cells have distinct metabolic signatures and use different metabolic pathways. Moreover, several metabolic pathways can act as regulatory elements to maintain stem cell self-renewal by balancing energetic and biosynthetic requirements cooperatively.1 Amino acids are a source of energy, building blocks for nucleotide, and protein synthesis as well as being involved in the regulation of stem cells functions.2-3 In addition, amino acid requirements are precisely regulated depending on the developmental stage and they have a critical role in early embryogenesis. During embryo moves from the oviduct into the uterus, glucose concentration declined approximately 2-fold, which suggests that alternate energy sources are used in the 2 areas, and glutamine (Gln) is an obvious candidate energy source, because it has been reported that the addition of Gln significantly improves development.4-5 Although Gln is another source of metabolic energy for cells beside glucose,6-7 the physiological role of Gln in regulation of stem cell pluripotency is unclear. Gln synthetase, the only enzyme producing Gln de novo, is essential in early mouse embryogenesis due to its role in detoxification of ammonia production,8 which suggests that Gln is essential in early embryogenesis and embryo development.9 Furthermore, 2 unrelated human newborns with a congenital Gln synthetase deficiency presented with developmental disorders such as brain malformation, multiple organ failure, and infant death,9-10 which prompted us to examine the effects of Gln on regulation of embryonic stem cell (ESC) function and pluripotency. Thus, we investigated the role of Gln on the regulation of stem cell proliferation and the maintenance of stem cell self-renewal. Further studies are needed to identify the Gln signals that interact with epigenetic regulators to control cell fate, and such studies may result in novel therapeutic approaches.
Transcriptional activation or silencing of specific gene is needed for regulation of stem cell self-renewal and differentiation, which can be achieved via cross-talk between transcription factors and epigenetic modulators.11-12 Among the Yamanaka factors, Oct4 is irreplaceable and, when acting alone, has been reported to be sufficient for the reprogramming of somatic cells,13-14 indicating that Oct4 is the most important factor for maintenance of stem cells pluripotency. Transcription of Oct4 is regulated by a combination of transcription factors and epigenetic mechanisms involving DNA methylation and the remodeling of chromatin structures.15-17 Although the exact metabolic mechanisms contributing to epigenetic regulation in stem cells remain unclear, regulation of amino acid effects on ESC self-renewal and ESC functions via control of epigenetic modulators has been reported.2,18 In addition, the lack of glucose or Gln is reported to lead to the inhibition of cell cycle progression through gene expression reprogramming in hepatocellular carcinoma,19-20 suggesting that Gln may interact with an epigenetic modulator. However, the physiological responses of stem cells to Gln could be different in restricted progenitors and differentiated cells due to functional differences. Little is known about the relationship between Gln and epigenetic modification and its involvement in the maintenance of stem cell self-renewal.
Currently, ESCs, used frequently in developmental biology research, are reported to possess features that support infinite self-renewal; moreover, they have the capacity to differentiate into cellular derivates of 3 lineages, results that have attracted widespread interest in cell-based regenerative medicine.21 Elucidation of the precise mechanisms affecting the activity of Gln on stem cell self-renewal and maintenance is important in developing an understanding normal ESC development and pluripotency. Furthermore, such elucidation could be helpful in the development of novel strategies for cell-based regenerative medicine as well as the development of novel approaches to improving cellular reprogramming processes, either through alteration of culture media components or targeting the enzymes involved in Gln metabolism. In the present study, we investigated the role of Gln on the maintenance of mouse ESC (mESC) self-renewal and the molecular mechanisms underlying the Gln-mediated epigenetic control of Oct4 transcription in mESCs.
Results
Gln regulated maintenance of undifferentiation status in mESCs
To determine the role of Gln in the maintenance of undifferentiation status in mESCs, we cultured mESCs with Gln-free media for various periods (0–4 d) and observed the transcription levels of self-renewal marker genes and lineage specific differentiation marker genes by using qPCR. Provision of Gln-deprived media decreased mRNA expression levels of the self-renewal marker genes Oct4, Nanog, FoxD3, and Rex1, but mRNA expression of Sox2 was not affected (Fig. 1A). In addition, Gln deprivation significantly increased trophectoderm (Cdx2 and FGF4) and mesoderm (brachyury and MESP1) marker levels, but did not affect expression of endoderm and ectoderm marker genes (Fig. 1B). Consistenly, long-term culture without Gln up to 15 d increased MESP1 and Cdx2 expression levels in a time-dependent manner (Fig. 1C, D). Also, Gln deprivation elicited the atypical morphology of mESCs. The colonies were diffused and the cells were elongated and spread (Fig. 1E). Consistent with the decrease in mRNA expression levels, Oct4 and Nanog protein expression levels also decreased with Gln deprivation (Fig. 1F). After culturing mESCs with Gln-free media for 24 h, the addition of Gln (4 mM) restored Oct4 and Nanog mRNA and protein expression levels (Fig. 1G, H), which were inhibited by pretreatment with the glutaminase inhibitor compound 968 (Fig. 1I). When the cells were depleted with glucose and glutamine, glucose or Gln alone treatment increased Oct4 expression, and combined treatment of glucose and Gln synergistically increased Oct4 (Fig. S1). The populations of mESCs that maintained their undifferentiation status were examined by using a flowcytometer. Gln deprivation elicited a leftward shift in the Oct4- and Nanog-expressing cell population, but no such shift was observed in the Sox2-expressing cell population (Fig. 1J). In addition, the Gln-increased Oct4, Nanog, and SSEA1 expression levels were confirmed by immunocytochemistry (Fig. 1K). Taken together, the above results suggest that Gln has an important role in the maintenance of undifferentiation status in mESCs. Since Oct4 has been reported to be the most important regulator associated with maintenance of ESC pluripotency,22-23 we investigated the effect of Gln on Oct4 regulation in the next series of experiments.
Figure 1 (See previous page).
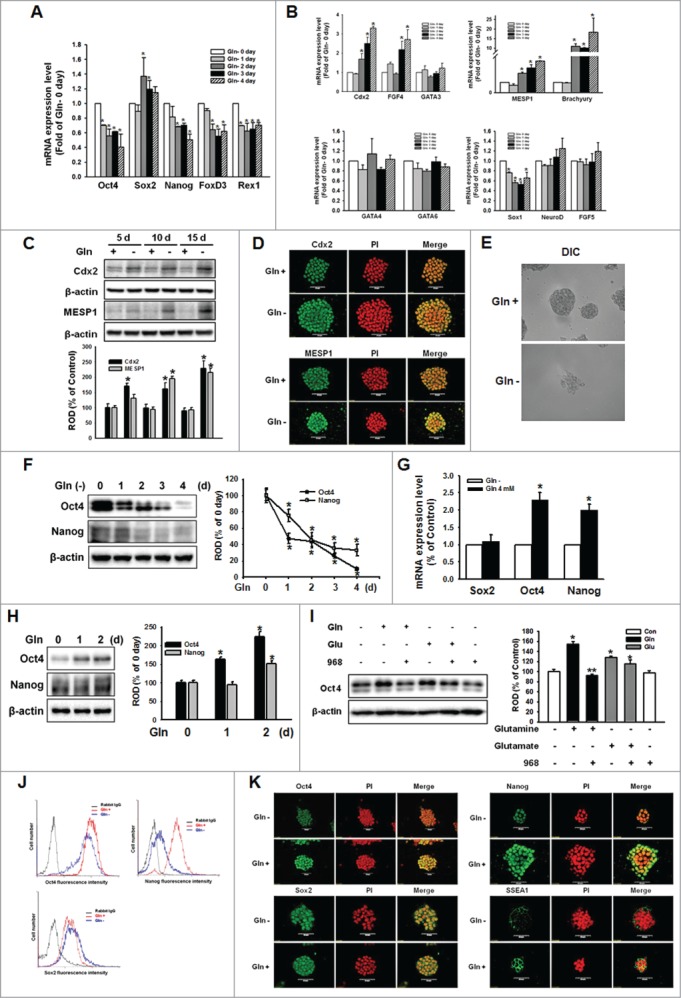
Glutamine (Gln) deprivation deteriorated self-renewal maintenance of mESCs. The mESCs were depleted of Gln for 4 d and the total RNA was extracted as described in Materials and Methods. (A, B) The mRNA of the self-renewal marker genes (Oct4, Sox2, Nanog, FOXD3, and Rex1), trophectoderm marker genes (Cdx2, FGF4, and GATA3), endoderm marker genes (GATA4 and GATA6), mesoderm marker genes (brachyury and MESP1), and ectoderm marker genes (Sox1, NeuroD, and FGF5) were detected. Error bars represent the mean ± SEM. *P < 0.05 vs. control (0 day of Gln deprivation). (C) mESCs were cultured with/without Gln up to 15 d. The cells were passaged every 5 d. After harvest the cells, the trophectoderm marker (Cdx2) and mesoderm marker (MESP1) proteins expression was detected by Western Blot analysis. The below panel depicted by bars denote mean ± SEM of 3 experiments for each condition determined by densitometry relative to β–actin. *P < 0.05 vs. control of each day. The cells were cultured with/without for 10 d. (D) The cells were immunostained with Cdx2 or MESP1. (E) The morphological examination was performed and DIC (differential interference contrast) were acquired at 400 × magnification. (F) Total lysates subjected to SDS-PAGE, and detected Oct4 and Nanog expression. The right panel of Fig. 1F depicted by bars denote mean ± SEM of 3 experiments for each condition determined by densitometry relative to β–actin. *P < 0.05 vs. 0 day of Gln deprivation. The cells were cultured with/without Gln (4 mM) for 2 d, and (G) Sox, Oct4, and Nanog mRNA expression level were detected using real-time PCR. Error bars of (G) represent the mean ± SEM. *P < 0.05 vs. control (0 day of Gln deprivation). (H) The Oct4 and Nanog proteins expression was detected by Western Blot analysis as described in Materials and Methods. (I) The cells were pretreated with compound 968 (glutaminase inhibitor; 1 μM) for 2 hours prior to incubation with Gln (4 mM) or glutamate (Glu; 4 mM) for 2 d. And Oct4 protein expression level was detected. The right panels of (H and I) depicted by bars denote mean ± SEM of 3 experiments for each condition determined by densitometry relative to β–actin. *P<0.05 vs. Gln 0 day or control, respectively. **P < 0.05 vs. Gln alone. (J) The cells were cultured with/without Gln for 2 d and then trypsinized and suspended in PBS. After labeling with anti-rabbit Oct4 IgG, anti-rabbit Sox2 IgG, or anti-rabbit Nanog IgG, the cells were incubated with secondary conjugated antibodies (goat-anti-rabbit IgG-conjugated FITC) and detected with flowcytometry. (K) The cells were double labeled with anti-Oct4, -Nanog, Sox2, or -SSEA1, and counter stained with PI. And then the cells were observed with a confocal microscope.
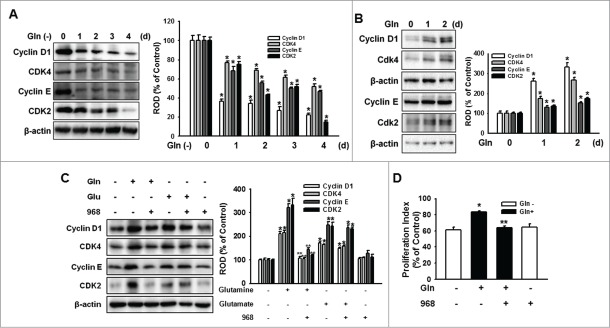
Gln increased cell cycle regulatory protein expression levels in mESCs
To determine the effect of Gln on mESC proliferation, mESCs were cultured with Gln-free media for various periods (0–4 d) after which the expression of cell cycle regulatory proteins were examined. Incubation with Gln-deprived culture media decreased cyclin D1/CDK4 and cyclin E/CDK2 (Fig. 2A). In contrast, Gln treatment significantly increased cell cycle regulatory protein expression levels (Fig. 2B). To investigate whether the restoration of mESC proliferation is dependent on Gln, mESCs were pretreated with compound 968 prior to incubation with Gln for 48 h. The Gln-increased cell cycle regulatory protein expressions were inhibited by the compound 968 pretreatment. In addition, glutamate also increased cell cycle regulatory protein expression but which were not affected by compound 968 (Fig. 2C). Consistently, Gln increased proliferation of mESC, which was inhibited by compound 968 (Fig. 2D). In addition, to elucidate the effect of Gln deprivation on cellular apoptosis, the cells were cultured with various times without Gln, and analyzed with annexin V/PI and MTT reduction assay. Gln deprivation did not induce cellular apoptosis and Gln treatment with/without compound 968 did not affect the cellular apoptosis. But, Gln increased MTT reduction level, which was blocked by compound 968 (Fig. S2). Moreover, to investigate the effect of energy depletion on mESCs proliferation, the cells were depleted with both glucose and Gln. Glucose or Gln alone treatment increased cell cycle regulatory proteins expression, and combined treatment of glucose and Gln synergistically increased cellular proliferation (Fig. S1). These results suggest that Gln and its metabolism play a critical role in the regulation of mESC proliferation. Next, we investigated the cell signaling pathways involved in Gln regulation of mESCs self-renewal.
Figure 2.
Gln restored proliferation of mESCs. (A) The mESCs were depleted of Gln up to 4 d and the total lysates subjected to SDS-PAGE and detected cell cycle regulatory proteins expression level. (B) The cell cycle regulatory proteins expression was detected by Western Blot analysis. The right panels of Fig. 1A and B depicted by bars denote mean ± SEM of 3 experiments for each condition determined by densitometry relative to β–actin. *P < 0.05 vs. Gln 0 d. (C) The cells were pretreated with compound 968 for 2 hours prior to incubation with Gln (4 mM) or Glu (4 mM) for 2 d. And cyclin D1 and cyclin E expression level was detected. The right part depicted by bars denotes mean ± SEM of 3 experiments for each condition determined by densitometry relative to β–actin. *P<0.05 vs. control. **P < 0.05 vs. Gln alone. (D) Cells were pretreated with compound 968 for 2 hours prior to culture with Gln for 2 d. The cells were washed with PBS, fixed, stained, and analyzed by flowcytometry. Gates were manually configured to determine the percentage of cells in S phase based on DNA content. The data were calculated using a proliferation index [(S+G2/M)/(G0/G1+S+G2/M)] and reported as the mean ± SEM of 3 different experiments, each conducted in triplicate. *P < 0.05 vs. control. **P < 0.05 vs. Gln alone.
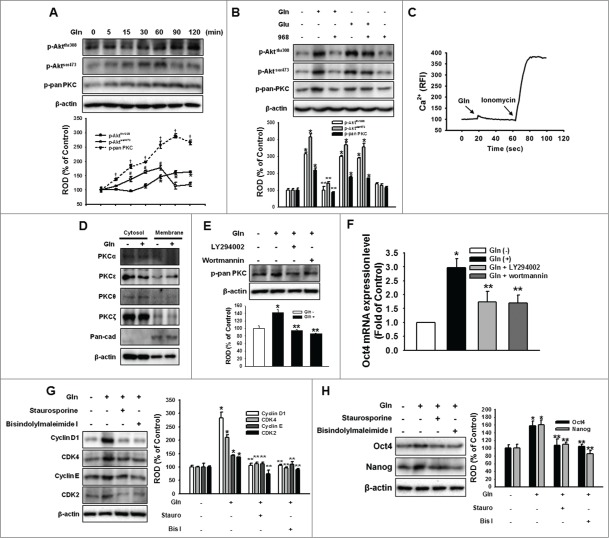
Akt and PKC activation is involved in Oct4 regulation and proliferation
To investigate the effect of Gln on PI3K/Akt and PKC activation, mESCs were treated with Gln (4 mM) for various periods (0–120 min). Treatment with Gln elicited phosphorylation of Aktthr308, Aktser473, and PKC in a time-dependent manner (Fig. 3A), and which could be inhibited by compound 968 pretreatment (Fig. 3B). In addition, the response of intracellular Ca2+ mobility to Gln treatment was examined in order to determine whether the regulation of mESC self-renewal ability by Gln was associated with an increase in cytosolic calcium ion concentration ([Ca2+]i). After staining mESCs with Fluo-3AM, they were treated with Gln and the resulting intracellular fluorescence levels were recorded. Ionomycin, which increases with [Ca2+]i, was used as a positive control. As shown in Fig. 3C, Gln did not affect [Ca2+]i. In addition, Gln stimulated the translocation of PKCε from cytosol to membrane (Fig. 3D). To examine the relationship between Akt and PKC, mESCs were pretreated with the PI3K/Akt inhibitors LY294002 and Wortmannin prior to culturing with Gln for 60 min. The Gln phosphorylated pan-PKC, which was inhibited by the PI3K/Akt inhibitors (Fig. 3E). These results suggest that the PI3K pathway lies upstream of PKC signaling in this Gln-induced signaling cascade. Consistent with those results, inhibition of PI3K with LY294002 and Wortmannin eliminated the Gln-induced increases in Oct4 mRNA expressions (Fig. 3F), and the inhibition of PKC with staurosporine and bisindolylmaleimide I, blocked the Gln-induced increases Oct4 protein and mESC cell cycle regulatory protein expressions (Fig. 3G, H). These results indicate that Gln-induced PI3K/PKC signaling is involved in maintenance of self-renewal in mESCs.
Figure 3.
Involvement of PI3K/Akt and PKC on Gln-induced maintenance of mESC self-renewal. (A) Cells were treated with 4 mM Gln with different time periods (0 to 120 minutes). Akt and pan-PKC phosphorylation was detected by Western blot analysis. The below panels of (A) depicted by line denote mean ± SEM of 3 experiments for each condition determined by densitometry relative to β–actin. *P < 0.05 vs. 0 minute control of p-Aktthr308. #P < 0.05 vs. 0 minute control of p-Aktser473. †P < 0.05 vs. 0 minute control of p-pan PKC. (B) The cells were pretreated with compound 968 for 2 hours prior to incubation with Gln (4 mM) or Glu (4 mM) for 60 min. And Akt and pan-PKC phosphorylation was detected. (C) Cells were loaded with 2 μM Fluo-3AM in serum-free medium for 40 minutes and treated with 4 mM Gln to examine the effects of Gln on intracellular calcium concentration ([Ca2+]i). Ionomycin (Ca2+ ionophore, 1 μM) was used as a positive control in order to validate the assay. The changes in [Ca2+]i were monitored using confocal microscopy and were expressed as relative fluorescence intensities (RFI). (D) Cells were treated with 4 mM Gln for 60 minutes, and cytosolic and membrane fractions were prepared. The PKC α, ε, θ, and ζ isoforms present in either cytosolic or membrane compartments were detected by Western blotting, as previously described. Pan-cadherin (pan-cad) was used as maker of membrane fraction. (E) The cells were pretreated with LY294002 (PI3 kinase inhibitor; 1 μM) and wortmannin (PI3 kinase inhibitor; 1 μM) prior to incubation with 4 mM Gln for 60 min, and pan-PKC phosphorylation was detected. (F) The cell were pretreated with LY294002 and wortmannin for 1 hour and treated with 4 mM Gln for 2 d. The Oct4 gene transcription level was detected. Error bars of (F) represent the mean ± SEM. *P < 0.05 vs. control. **P < 0.05 vs. Gln alone. (G, H) The cell were pretreated with staurosporine (protein kinase C inhibitor; 1 μM) and bisindolylmaleimide I (protein kinase C inhibitor; 1 μM) for 1 hour and treated with 4 mM Gln for 2 d. The cell cycle regulatory proteins, Oct4, and Nanog expression levels were detected. The below or right panels of (B, E, G, H) depicted by bars denote mean ± SEM of 3 experiments for each condition determined by densitometry relative to β–actin. *P < 0.05 vs. control. **P < 0.05 vs. Gln alone.
Crucial role of mTOR in Gln-induced proliferation
We investigated the phosphorylation of mTOR to determine whether the mTOR pathway participated in the Gln-induced maintenance of mESC self-renewal. As shown in Fig. 4A, mTOR, and p70S6K were phosphorylated in a time-dependent manner. To examine the relationship between PKC and mTOR, which was stimulated by Gln treatment, Gln-induced mTOR activation was inhibited by pretreatment with the compound 968 or PKC inhibitors staurosporine and bisindolylmaleimide I, respectively. The results (Fig. 4B, C) suggest that the mTOR pathway acts as a downstream signaling pathway of PKC. In addition, mTOR inhibition by rapamycin eliminated the effect of Gln on the expression of cell cycle regulatory proteins as well as proliferation (Fig. 4D, E). To confirm the involvement of mTOR in Gln-induced regulation of mESC self-renewal, Gln increased population of the cells that were double-labeled for Oct4 and bromodeoxyuridine (BrdU), which were inhibited by rapamycin (Fig. 4F, G). Furthermore, to test whether availability of Gln can influence pluripotent cell fate decisions, we performed colony formation assay. Gln treatment significantly increased the total number of colonies, which blocked by comound 968 or rapamycin (Fig. 4H). These results indicate that Gln mediated both mESC proliferation and maintenance of mESC self-renewal ability.
Figure 4.
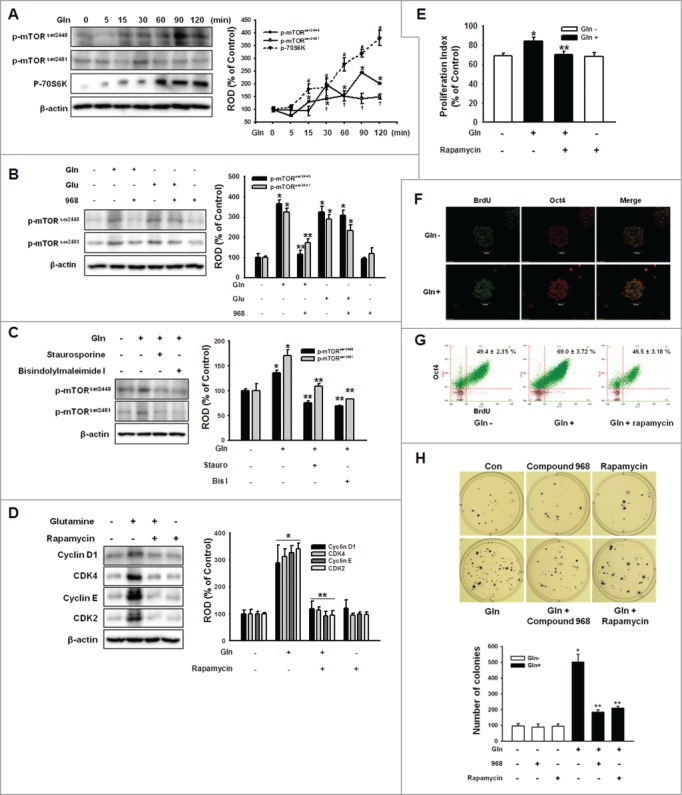
Involvement of mTOR on Gln-induced cell cycle regulatory protein expression. (A) Cells were treated with 4 mM Gln with different time periods (0 to 120 minutes). mTOR and 70S6K phosphorylation was detected by Western blot analysis. The right panels of (A) depicted by line denote mean ± SEM of 3 experiments for each condition determined by densitometry relative to β–actin. *P < 0.05 vs. 0 minute control of p-mTORser2448. †P < 0.05 vs. 0 minute control of p-mTORser2481. #P < 0.05 vs. 0 minute control of p-70S6K. (B, C) The cells were pretreated with compound 968, staurosporine, or bisindolylmaleimide I for 2 hours prior to incubation with Gln (4 mM) or Glu (4 mM) for 90 min. And mTOR phosphorylation was detected. The right panels of (B and C) depicted by bars denote mean ± SEM of 3 experiments for each condition determined by densitometry relative to β–actin. *P < 0.05 vs. control. **P < 0.05 vs. Gln alone. (D, E) The cells were pretreated with rapamycin (mTOR inhibitor; 10 nM) for 12 hours prior to incubation with Gln (4 mM) for 2 d. The cell cycle regulatory proteins and proliferation index were detected. The right panel of (D) depicted by bars denote mean ± SEM of 3 experiments. Error bars of (E) represent the mean ± SEM. *P < 0.05 vs. control. **P < 0.05 vs. Gln alone. (F) The mESCs were exposed to Gln (4 mM) after 24 hours incubation with Gln-free media for 2 d and double-labeled with Oct4 and BrdU antibodies simultaneously. (G) The mESCs were pretreated with rapamycin prior to Gln treatment for 2 d and dissociated in trypsin/EDTA. And then, double-labeled with Oct4 and BrdU antibodies and detected with flowcytometry. (H) The cells were plated at clonal density without Gln and media were changed twice a week. And the cells were treated with compound 968 or rapamycin. After 21 d, the cells were stained with nitro blue tetrazolium chloride for 12 hr and the colony numbers were counted. The below panel of (H) depicted by bars denote mean ± SEM of 3 experiments for each condition determined by colony number counting. *P < 0.05 vs. control. **P < 0.05 vs. Gln alone.
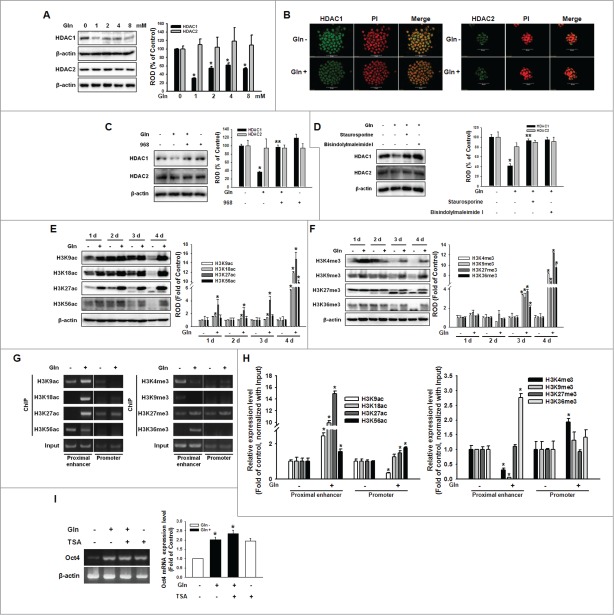
Histone acetylation and methylation regulated by Gln through PKC-dependent HDAC downregulation
To examine the effect of Gln on histone acetylation of mESCs, we investigated HDAC expression and changes in histone acetylation level by culturing mESCs with Gln. HDAC1 expression decreased with Gln treatment in a dose-dependent manner, whereas HDAC2 expression did not affect (Fig. 5A). Consistent with these results, Gln decreased HDAC1 expression compared with the expression levels in Gln-depleted cells (Fig. 5B). To examine whether the Gln-induced signaling cascade are involved in regulation of HDAC, mESCs were pretreated with compound 968, or a PKC inhibitor (staurosporine or bisindolylmaleimide I) prior to incubation with Gln for 48 h. As shown in Fig. 5C, D, the Gln-induced reduction of HDAC1 expression was inhibited by compound 968, staurosporine, and bisindolylmaleimide I, respectively. Consistent with the effect of Gln on HDAC1 expression, global histone acetylation and methylation levels were increased by adding Gln (Fig. 5E, F). In particular, H3K18ac, H3K27ac, and H3K56ac increased during the early period of Gln treatment (1–2 d), whereas histone methylation increased during a later period of Gln treatment (3–4 d). Furthermore, to clarify the histone acetylation and methylation involving Oct4 gene expression, ChIP assay were performed using primers of Oct4 proximal enhancer and promoter. Gln increased histone acetylation of proximal enhancer region (H3K9ac, H3K18ac and H3K27ac) and promoter region (H3K18ac and H3K27ac). In addition, Gln were involved in histone methylation of proximal enhancer (decrease of H3K4me and H3K9me, increase of H3K27me and H3K36me) (Fig. 5G, H). Furthermore, Gln-induced enhancement of Oct4 mRNA expression was blocked by pre-treatment of cells with TSA (Trichostatin A; a HDAC inhibitor) (Fig. 5I). Taken together, these results suggest that Gln could be involved in the epigenetic regulation of Oct4 gene. Next, we examined the relationship between histone acetylation and DNA methylation and the effect of Gln on Oct4 promoter region methylation.
Figure 5.
Involvement of HDAC on Gln-induced histone modification. (A) Cells were treated with various concentration of Gln (0–8 mM), and HDAC1 and HDAC2 expression level were detected with Western blot analysis. The right panel depicted by bars denotes mean ± SEM of 3 experiments for each condition determined by densitometry relative to β–actin. *P < 0.05 vs. control (0 mM of Gln). (B) The cells were incubated with Gln for 2 d, and were double labeled with anti-HDAC1, -HDAC2, and PtdIns and observed with a confocal microscope. (C, D) The mESCs were pretreated with compound 968, staurosporine, or bisindolylmaleimide I prior to Gln treatment, and HDAC1 and HDAC2 expression level were detected. The right panels of (C and D) depicted by bars denote mean ± SEM of 3 experiments for each condition determined by densitometry relative to β–actin. *P < 0.05 vs. control. **P < 0.05 vs. Gln alone. (E, F) Cells were cultured with/without Gln for 1–4 d. And histone acetylation and methylation level were detected. The right panels of (E and F) depicted by bars denote mean ± SEM of 3 experiments for each condition determined by densitometry relative to β–actin. *P < 0.05 vs. Gln deprivation group of each days. (G, H) Cells were incubated with/without Gln for 2 d, and the chromatin DNA was immunoprecipitated with antibodies against histone acetylation and methylation, the resulting samples were amplified with the primers for proximal enhancer and promoter of Oct4 gene. And then, the PCR products were verified by running on a 2% agarose gel or were analyzed with real-time PCR. (I) Cells were pretreated with TSA (trichostatin A; 100 nM) for 12 hours prior to incubation with Gln for 2 d, and Oct4 mRNA levels were detected. Error bars represent the mean ± SEM. *P < 0.05 vs. control. **P < 0.05 vs. Gln alone.
Transcriptional activity of Oct4 regulated by Gln through promoter region hypomethylation in a DNMT1/3a-dependent manner
When mESCs were cultured with/without Gln for 1–4 d or cultured with various concentration of Gln for 2 days, DNMT1 and DNMT3a expression levels decreased in dose-dependent manners, but DNMT3b expression was unaffected (Fig. 6A and B). In addition, pretreatment with PKC inhibitors (staurosporine and bisindolylmaleimide I) blocked Gln-induced reduction of DNMT1 andDNMT3, but a HDAC inhibitor (TSA) prior to incubation with Gln resulted in a synergistic effect on the decreases in DNMT1 and DNMT3a expression levels (Fig. 6C and D). To determine whether the Gln-induced decrease in DNMT1 and DNMT3a expression affected Oct4 promoter region methylation level, methylation-specific PCR was performed. As shown in Fig. 6E, Gln significantly reduced methylation of the Oct4 promoter region, which was synergistically increased by trichostatin A (TSA) pretreatment. Consistent with those results, pretreatment with TSA inhibited Gln-induced Oct4 expression levels (Fig. 6F). In order to examine the effect of Gln on Oct4 activity as a transcription factor, we performed Oct4 Cignal reporter assay. Gln treatment together or alone with TSA or 5-aza significantly increased Oct4 activity, but compound 968 or rapamycin inhibited Oct4 activity increasing by Gln (Fig. 6G). Consistently, Gln treatment increased mRNA expression of Oct4 downstream genes (Lefty1, Rif1, Foxo15, Otx2, and UTF1) (Fig. 6H). In addition, the colony forming ability was increased by Gln treatment with/without TSA or 5-aza (Fig. 6I). These results indicate that the Gln has an important role in controlling mESCs self-renewal ability through epigenetic regulation of the Oct4 genes.
Figure 6 (See previous page).
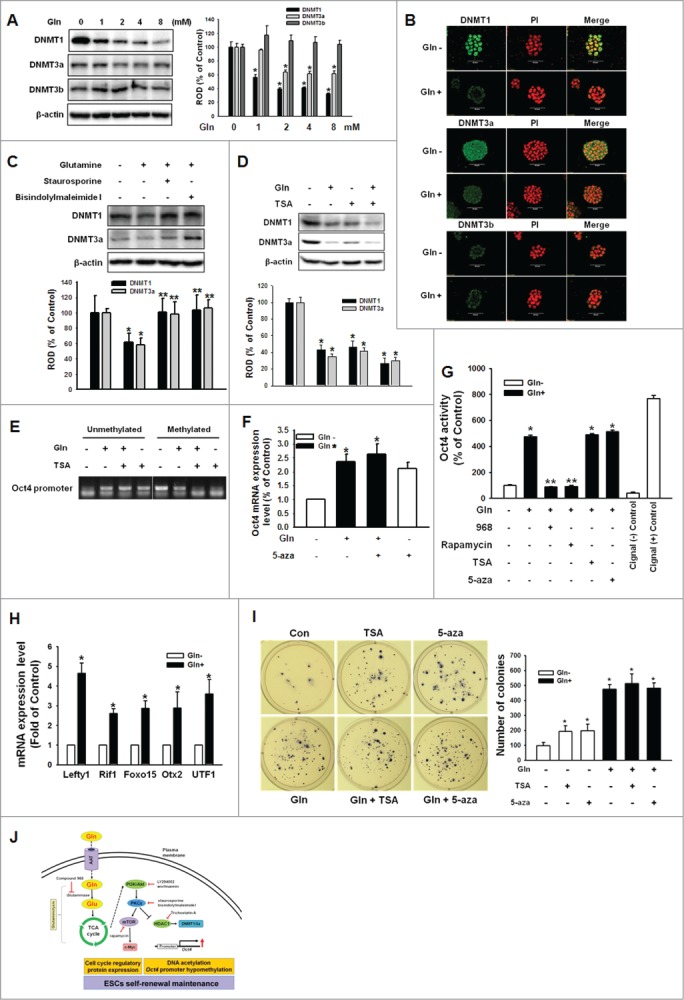
Involvement of DNMT1/3a on Gln-induced DNA methylation of Oct4 promoter region. (A) Cells were treated with various concentration of Gln (0–8 mM), and DNMT1, DNMT3a, and DNMT3b expression level were detected with Western blot analysis. The right panel depicted by bars denotes mean ± SEM of 3 experiments for each condition determined by densitometry relative to β–actin. *P < 0.05 vs. control (0 mM of Gln). (B) The cells were incubated with Gln for 2 d, and were double labeled with anti-DNMT1, -DNMT3a, DNMT3b and PI and observed with a confocal microscope. (C, D) The mESCs were pretreated with staurosporine, bisindolylmaleimide I, or TSA prior to Gln treatment, and DNMT1 and DNMT3a expression level were detected. The below parts of (C and D) depicted by bars denote mean ± SEM of 3 experiments for each condition determined by densitometry relative to β–actin. *P<0.05 vs. control. **P < 0.05 vs. Gln alone. (E) Cells were pretreated with TSA for 12 hours prior to treatment of Gln for 2 d. After harvest the cells, the total genomic DNA was extracted and bisulfate conversion was conducted as manufacture's instruction. Using a methylation and non-methylation specific primer for Oct4, methylation levels of Oct4 were detected. (F) The mESCs were pretreated with 5-aza-2'-deoxycytidine (5-aza; 100 nM) prior to incubation with Gln for 2 d, and Oct4 mRNA expression level was detected by real-time PCR. Error bars represent the mean ± SEM. *P < 0.05 vs. control. **P < 0.05 vs. Gln alone. (G) mESCs were transfected with Oct4 Cignal Reporter (100 ng), Cignal Negative Control (100 ng), Cignal Positive Control (100 ng) for 24 hr. The cells were pretreated with compound 968, rapamycin, TSA, or 5-aza prior to Gln treatment for 2d. Cell lysate were prepared using passive lysis buffer (PLB) and Dual-Luciferase Reporter Assay was performed. The Oct4 activity was measured by luminometer. Values are reported as the mean ± SEM of 3 independent experiments from triplicate dishes. *P < 0.05 vs. Gln free. **P < 0.05 vs. Gln alone. (H) The cells were incubated with/without Gln for 2d. The Oct4 downstream genes (Lefty1, Rif1, Foxo15, Otx2, and UTF1) mRNA expression levels were determined with real-time PCR. Error bars represent the mean ± SEM. *P < 0.05 vs. Gln free. (I) The cells were plated at clonal density without Gln and media were changed twice a week. And the cells were treated with TSA or 5-aza. After 21 d, the cells were stained with nitro blue tetrazolium chloride for 12 hr and the colony numbers were counted. The right panel of (I) depicted by bars denote mean ± SEM of 3 experiments for each condition determined by colony number counting. *P<0.05 vs. control. (J) The proposed model for the signaling pathways involved in Gln-induced maintenance of ESC self-renewal is shown. Gln metabolism through glutaminolysis activated PI3K/Akt, which subsequently stimulated phosphorylation and translocation of PKCε. Gln-induced PKC activation resulted in activation of mTOR signaling pathway, which elicited cell cycle regulatory proteins expression. In addition, Gln-induced PKC phosphorylation increased Oct4 gene transcription through reduction of HDAC1 and DNMT1/3a.
Discussion
In the present study, we show that the absence of Gln from culture media decreases mESC proliferation and undifferentiation status, whereas addition of Gln to the culture media downregulates HDAC1 and DNMT1/3a in PI3K-, PKC-, and mTOR-dependent manners, which subsequently increased the expression of cell cycle regulatory proteins and Oct4 (Fig. 6J). Gln has 2 nitrogen-containing side chains, amino and amido groups, and is the most abundant, naturally occurring, nonessential amino acid in the human body.24 Through its capacity to serve as a nitrogen or carbon donor, Gln is central to a variety of biochemical homeostasis, as well as to purine synthesis and the citric acid cycle. Gln is transported via ASCT2 as well as via SLC38 sodium-coupled neutral amino acid transporters (SNAT).25-26 The ASCT2 transporter plays a major role in HSCs (haematopoietic stem cells), and ASCT2 knockdown has resulted in a 40–75% decrease in Gln uptake.27 In the present study, Gln depletion significantly deteriorated mESC undifferentiation status and proliferation, suggesting that Gln, when present in culture media in vitro or plasma in vivo, is associated with mESC self-renewal. In addition, proline and threonine are involved in the control of ESC functions such as proliferation, motility, and teratoma formation.28-32 Moreover, L-proline positively or negatively regulates ESC differentiation, but the regulation depends on specific culture conditions,28 which suggests the possibility that amino acids can differentially regulate ESC functions depending on amino acid and cell line types. Consistently, the response to Gln deprivation was different in melanocyte and melanoma, suggesting possibility that the Gln metabolism could be differently regulated depending on cell type.33 Interestingly, the similarity between the effects of L-threonine and Gln on alteration of mESCs self-renewal markers (i.e., the decrease in undifferentiation markers and the increase in trophectoderm and mesoderm marker genes) suggests that these 2 amino acids may control mESC functions through common metabolic intermediates or signaling cascades.34 Gln is metabolized to pyruvate through glutaminolysis, which can contribute significantly to cellular metabolism under some conditions.6-7 Our results show that inhibition of glutaminolysis via a glutaminase inhibitor eliminates Gln-induced mESC proliferation, suggesting that Gln has an important role in the regulation of stem cell proliferation, which is mediated by Gln metabolites rather than by Gln itself. Consistent with our results, a deficiency of Gln has decreased the proliferation of adipose-derived stem cells without a concomitant increase in cell death.35 Our data show that Gln depletion significantly decreased mESCs proliferation and maintenance of their undifferentiation status, but both were restored by Gln treatment, which suggests that Gln is an essential factor in the maintenance of mESC self-renewal. These results indicate the possibility of using Gln for regulation of stem cell pluripotency and in the development of therapeutic strategies in the field of regenerative medicine. Our conceptual advance has important ramifications for understanding ESC stemness and for designing novel therapeutic treatments. However, determining the metabolic pathways involved and deciphering the underlying molecular mechanisms involved in ESC self-renewal are necessary for the advancement of stem cell–based therapies.
In stem cell proliferation, the PI3K pathway is stimulated by growth factors, cytokines, and nutrients such as glucose and amino acids.36 In addition, PI3K-Akt acts as an important regulator of stemness and proliferation, a result that is supported by the presence of substantial levels of active PI3K-Akt pathway in ESCs.37-39 In this study, we observed that the addition of Gln enhanced the phosphorylation of Akt at both Thr308 and Ser473, which supports previous study results showing that cellular amino acid deprivation reduces insulin-mediated phosphorylation of mTOR Ser2448 in an Akt-dependent manner.40 The activation of the PI3K pathway often indicates the activation of other intracellular signaling cascades such as the PKC pathway. The PtdIns-dependent protein kinases (PDKs) are involved in the PI3K/Akt pathway and lead to activation of PKC through phosphorylation at Thr410, a highly conserved motif in all PKC family members.41-43 In the present study, Gln enhanced PKCε activity in a glutaminase-dependent manner without changing the intracellular Ca2+ concentration, which suggests that Gln–induced Akt and PKC activation is significantly implicated in maintenance of mESC self-renewal. The evolutionarily conserved nutrient sensor mTOR directs cellular responses to nutrient status such as the availability of amino acids,44 and modulates stem cell maintenance.45-46 In addition, it has been suggested that mTOR acts as a convergence point for amino acid–mediated effects on translation initiation,47 which requires the activation of Akt and PKC.40,48 In this study, we investigated whether Gln elicits mTOR activation when mediated by PI3K/Akt and PKC. Our results showed that the PKC inhibition eliminated Gln-induced mTOR activation, suggesting that mTOR signaling activation is required for PKC activity. Consistent with those results, a novel PKC was reported to be involved in the control of mTOR activation and cap-dependent translation.49-50
The transcription factor Oct4 has critical roles in the establishment and maintenance of pluripotency during normal early embryonic development.51-54 In undifferentiated ESCs, the Oct4 promoter region is almost unmethylated, whereas that region in trophectoderm cells is heavily methylated,17 which indicates that Oct4 expression is precisely regulated via epigenetic modulation, such as that from histone modification and DNA methylation.55-57 In the present study, Gln–induced PKC activation in mESCs was associated with regulation of transcriptional activity through epigenetical modification. Relationships between signaling molecules and epigenetic regulators have been reported, and PKC signaling is involved in the regulation of promoter activity via detachment of HDAC1 from the promoter region or by the reduction of DNMT activity.58-60 In this study, we observed that a Gln-induced decrease in HDAC1 elicited global histone acetylation and methylation. Consistent with those results, a reduced condensation of the chromatin structure and the regulation of HDAC1 on the Oct4 gene have been reported in undifferentiated ESCs.61-62 However, those results are in contrast to previous reports indicating that the level of H3K9ac is important in somatic reprogramming of human and mouse cells.63-65 In this study, Gln-induced H3K9ac occurred at a relatively late time, which suggests the possibility that Gln is involved in a broad range of gene expressions and that intermediate signaling cascades may be involved in Gln-induced H3K9ac expression. Although further research is needed to distinguish the specific acetylation or methylation sites involving in pluripotency-related gene transcription, our results suggest that Gln is closely associated with chromatin remodeling via HDAC1. Interestingly, DNMT and HDAC can form a complex and can act as transcriptional repression factors for each other, suggesting that DNA methylation and histone modification, which is involved in chromatin silencing, are interdependent processes.66-68 Furthermore, HDAC1 is associated with DNMT1 stability through deacetylaltion of HDAC1, which protect HDAC1 from unbiquitination.69 We have shown that Gln hypomethylated the Oct4 promoter region through downregulation of DNMT1/3a activity in PKC–dependent manners. Consistent with our results, the methylation status of DNA has been linked to the chromatin structure of the upstream region of Oct4 in ESCs.17 Although we cannot exclude the possibility that Gln-induced DNMT downregulation is directly mediated by PKC and mTOR signaling, our results suggest that the Gln–induced reduction of DNMT1/3a depends on the loss of HDAC1, which is significantly implicated in Oct4 expression. Taken together, our results show a role of Gln in the regulation of self-renewal in mESCs. Moreover, the results can be used to provide a more complete description of the nature of stem cells as well as to provide a more comprehensive description of stem cell metabolism. However, additional work is needed to elucidate the metabolic intermediates of Gln that exert signaling activity and to determine how HDAC and DNMT can regulate specific chromatic structures or genes. In conclusion, Gln, through HDAC1 and DNMT1/3a inactivation via PI3K-PKC-mTOR signaling, is closely associated with proliferation and self-renewal maintenance of mESCs.
Materials and Methods
Materials
mESC line ES-E14TG2a was obtained from the American Type Culture Collection (Manassas, VA). Fetal bovine serum (FBS) was purchased from BioWhittaker (Walkersville, MD, USA). L-glutamine, glutamate, LY294002, wortmannin, staurosporine, bisindolylmaleimide I, rapamycin, 5-Bromo-2′-deoxyuridine (BrdU), ionomycin, and monoclonal anti-β-actin antibody were obtained from Sigma-Aldrich (St. Louis, MO, USA). Compound 968 and Akt inhibitor (1L6-hydroxymethyl-chiro-inositol-2-(R)-2-O-methyl-3-O-octadecyl-sn-lycerocarbonate) were purchased from Calbiochem (La Jolla, CA, USA). Phospho-pan PKC, phospho-mTORser2448, phospho-mTORser2481, phospho-70S6K, HDAC1, HDAC2, and histone acetylation (H3K9ac, H3K18ac, H3K27ac, and H3K56ac), and histone trimethylation (H3K4me3, H3K9me3, H3K27me3, and H3K36me3) antibodies were obtained from Cell Signaling Technology Inc.. (Beverly, MA, USA). The Oct4, Sox2, Nanog, DNMT1, DNMT3a, DNMT3b, and FITC-conjugated mouse anti-BrdU antibodies were purchased from Abcam (Cambridge, MA, US). Anti-SSEA1, anti-phospho-Aktser473, anti-phospho-Aktthr308, anti-PKCα, anti-PKCε, anti-PKCθ, anti-PKCζ anti-CDK4, anti-CDK2, anti-cyclin D1, and anti-cyclin E antibodies were purchased from Santa Cruz Biotechnology Inc.. (Santa Cruz, CA, USA). The goat anti-rabbit IgG was supplied by Jackson ImmunoResearch (West Grove, PA, USA). All other reagents were of the highest purity commercially available and were used as received.
mESC culture and glutamine deprivation
The mESCs (ES-E14TG2a) were cultured in Dulbecco's Modified Eagle's Media (DMEM) (Gibco-BRL; Gaithersburg, MD, USA) supplemented with 3.7 g/L sodium bicarbonate, 1% penicillin and streptomycin, 1.7 mM L-glutamine, 0.1 mM β–mercaptoethanol, 5 ng/ml mouse leukemia inhibitory factor (LIF), and 15% FBS, without a feeder layer. The cells were grown on 12-well plates or 60-mm culture dishes in an incubator maintained at 37°C in humidified atmosphere containing 5% CO2. After two to 3 d of culture, cells were washed twice with phosphate buffered saline (PBS) and maintained in serum- and glutamine-free DMEM, which contained all other supplements at the concentrations indicated above to derivates glutamine. After a 24 hours incubation period, the cells were washed twice with PBS and given fresh serum- and glutamine-free media containing the designated agents for the time periods indicated.
Real time polymerase chain reaction (Real time-PCR)
The mESCs were maintained in serum- and glutamine-free DMEM for 24 h. After refresh the culture media with serum- and glutamine-free DMEM, the cells were incubated up to 4 d with/without glutamine (4 mM) and the total RNA was extracted from the cells using RNA easy mini kit (Qiagen, Valencia, CA, USA). Reverse transcription was carried out using 1 μg of RNA using a MaximeTM RT-PCR PreMix kit (Intron Biotechnology, Seongnam, Korea) with the oligo(dT18) primers. One microliter of the reverse transcription (RT) products was then amplified using QuantiTect SYBR Green PCR Kits (Qiagen). Real-time quantification of RNA targets was performed in a Rotor-Gene 6000 real-time thermal cycling system (Corbett Research; NSW, Australia). The primers used are described in the Table S1. The reaction mixture (10 μl) contained 50 ng of total RNA, 0.5 μM of each primer, and appropriate amounts of enzymes and fluorescent dyes as recommended by the supplier. The PCR and real-time PCR were performed as follows: 30 minutes at 50°C for reverse transcription; 15 minutes at 95°C for DNA polymerase activation; 15 seconds at 95°C for denaturing; and 45 cycles of 15 seconds at 94°C, 30 seconds at 55°C, and 30 seconds at 72°C. Data collection was carried out during the extension step (30 seconds at 72°C) and analysis was performed using the software provided by the manufacturer. Following real-time PCR, melting curve analysis was performed to verify the specificity and identity of the PCR products.
Western blot analysis
Cells were harvested, washed twice with PBS, and lysed in lysis buffer [20 mM Tris (pH 7.5), 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 1 mg/ml aprotinin, 1 mM phenylmethylsulfonylfluoride (PMSF), and 0.5 mM sodium orthovanadate] for 30 minutes on ice. The lysates were cleared by centrifugation (30 minutes at 15,000 rpm, 4°C), and the protein concentration was determined using the Bradford method.70 Equal amounts of protein (20 µg) were resolved by 10% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were washed with TBST [10 mM Tris-HCl (pH 7.6), 150 mM NaCl, and 0.01% Tween-20], blocked with TBST containing 5% skim milk for 1 h, and incubated with the appropriate primary antibodies at the dilutions recommended by the suppliers. The membranes were then washed and the primary antibodies were detected with goat anti-rabbit IgG or goat anti-mouse IgG conjugated to horseradish peroxidase. Immunoreactive proteins were visualized by Immun-Star WesternC Chemiluminescence kit (Bio-Rad Laboratories, Inc.; Hercules, CA, USA). The results were measured optical density using Image J program (developed by Wayne Rasband, National Institutes of Health, Bethesda, MD; available at http://rsb.info.nih.gov/ij/index.html) and presented as means ± SEM.
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde for 20 min, permeabilized with 0.25% Triton X-100/PBS for 5 min, and blocked in 10% fetal bovine serum in 0.1% Triton X-100/PBS for 20 min. The cells were labeled with anti-Oct4, SSEA1, Nonog, Sox2, HDAC1, HDAC2, DNMT1, DNMT3a, or DNMT3b antibodies at a ratio of 1:100, followed by fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgM at 1:100, or Alexa Fluor® 555 goat anti-rabbit IgG at 1:400 for one hour each at room temperature. And the cells were counter stained with propidium iodide (PI; 250 μg/ml). Images were obtained using a FluoView™ 300 confocal microscope (Olympus; Tokyo, Japan).
Oct4 expression and BrdU incorporation
mESCs were cultured in serum- and glutamine-free media for 24 hours before stimulation with glutamine (4 mM). After the incubation period, 10 μM of BrdU was added to the cultures for one hour at 37°C. For flowcytometer analysis, BrdU-labeled cells were dissociated in trypsin/EDTA, pelleted by centrifugation, and resuspended at approximately 106 cells/ml in PBS containing 0.1% BSA. The cells were then fixed with 70% ice-cold ethanol for 30 minutes at 4°C, followed by incubation with rabbit anti-Oct4 (Abcam) at a ratio of 1:50, followed by Alexa Fluor® 555 goat anti-rabbit IgG at 1:100, or labeled with fluorescein isothiocyanate (FITC)-conjugated mouse anti-BrdU (Abcam) each for one hour at room temperature. And the Oct4 and BrdU positive cell population were analyzed with Cell Lab Quanta™ SC flowcytometer (Beckman Coulter Inc.; Brea, CA, USA).
Chromatin immunoprecipitation (ChIP)
ChIP was carried out using an EZ-ChIP™-Chromatin Immunoprecipitation Kit (EMD Millipore, Billerica, MA, USA) according to the manufacturer's instructions. Chromatin-protein complexes were immunoprecipitated using anti-histone acetylation (H3K9ac, H3K18ac, H3K27ac, and H3K56ac), and histone trimethylation (H3K4me3, H3K9me3, H3K27me3, and H3K36me3) antibodies. After overnight incubation, immune complexes were eluted with 200 μl (2 times of 100 μl each) elution buffer (1% SDS, 50 mM Tris-HCl, pH 7.5, and 10 mM EDTA), and then were incubated with RNase for 1 and 4 hours with proteinase K at 65°C. DNA was extracted and the PCR were performed using the Oct4 proximal enhancer primer or Oct4 promoter primer (Table S1) as previously described by Hattori et al.17. As inputs, we used products that corresponded to PCR reactions containing 1% of the total chromatin extract used in the immunoprecipitation reactions.
Preparation of cytosolic and total membrane fractions
The cytosolic and total membrane fractions were prepared using a modifications of the method reported by Markman et al.71. The cells were washed with ice-cold PBS and harvested by centrifugation and resuspended in buffer A [137 mM NaCl, 8.1 mM Na2HPO4, 2.7 mM KCl, 1.5 mM KH2PO4, 2.5 mM EDTA, 1 mM dithiothreitol, 0.1 mM PMSF, 10 g/ml leupeptin (pH 7.5)]. The resuspended cells were then mechanically lysed on ice by trituration with a 21.1-gauge needle. The lysates were first centrifuged at 1,000 × g for 10 minutes at 4°C. The cytosolic and total particulate fractions were prepared by centrifuging the supernatants at 100,000 × g for 1 hour at 4°C. The supernatants (cytosolic fraction) were then precipitated with 5 vol. of acetone, incubated for 5 minutes on ice and centrifuged at 20,000 × g for 20 minutes at 4°C. The pellets were resuspended in buffer A containing 1% (v/v) Triton X-100. The particulate fractions containing the membrane fraction were then washed and resuspended in buffer A containing 1% (v/v) Triton X-100.
Measurement of [Ca2+]i
Changes in [Ca2+]i were monitored using Fluo-3/AM dissolved in dimethylsulfoxide. Cells in 35 mm-diameter culture dishes were rinsed with a bath solution [140 mM NaCl, 5 mM KCl, 1 mM CaCl2, 0.5 mM MgCl2, 10 mM glucose, 5.5 mM HEPES (pH 7.4)] and were then incubated in a bath solution containing 3 mM Fluo-3/AM with 5% CO2–95% O2 at 37℃ for 40 min, rinsed, mounted on a perfusion chamber, and scanned at every seconds using Olympus FluoView 300 confocal microscope (Olympus, Hamburg, Germany) with 400X objective. The fluorescence was excited at 488 nm and the emitted light was read at 515 nm. Ionomycin (Ca2+ ionophore) was applied to the cells as a positive control in order to verify the assay. All [Ca2+]i analyses were processed at a single cell level and were expressed as the relative fluorescence intensity (RFI).
Proliferation assay using flowcytometer
Cells were detached with trypsin/EDTA, pelleted by centrifugation, and resuspended at ∼106 cells/ml in PBS containing 0.1% BSA. The cells were then fixed with 70% ice-cold ethanol for 1 hour at 4 °C, followed by incubation in a freshly prepared nuclei staining buffer consisting of 250 μg/ml propidium iodide (PI) and 100 μg/ml RNase for 30 minutes at 37 °C. Cell cycle histograms were generated after analyzing the PI-stained cells by FACS (Beckman Coulter). The samples were analyzed using Multicycle software (Phoenix Flow Systems Inc.; San Diego, CA) and the proliferation indices [(S+G2/M)/(G0/G1+S+G2/M)] calculated.
Colony formation assay
To evaluate the self-renewal ability of mESC, the colony formation assay was performed using modifications of the method reported by Borowicz et al.72 Six-well plates were coated with mixture of 0.5 % noble agar and DMEM with/without Gln. After 30 minutes incubation, the viable mESCs suspended at DMEM with/without Gln were mixed with same volume of 0.6 % noble agar, and seeded the 2000 cells/well. After cell/agar mixture solidify at room temperature for 30 minutes, the cells were incubated for 3 weeks at 37°C in humidified atmosphere containing 5% CO2. Media were changed twice a week. After incubation periods, the cells were stained with 1 mg/ml nitroblue tertrazolium cholide for 12 hours. Once colonies are stained, the total number of colonies were counted.
Measurement of Oct4 transcription factor activity
Oct4 transcription activity were detected using Cignal reporter assay kits for Oct4 procured from SABiosciences (cat# CCS-0025L, Qiagen). Transfections were performed as specified in the manufacturer's instruction sheet. Cells were collected and analyzed the Oct4 transcripiton activity using Dual Luciferase Reporter Assay system (cat# E1910, Promega) according to the manufacturer's protocol.
Methylation specific PCR
Extraction of genomic DNA and bisulfite conversion were conducted using the Blood & Cell Culture DNA Mini Kit (Qiagen, Valencia, CA, USA) and the EzWay DNA Methylation Detection Kit (Komabiotech, Seoul, Korea), respectively, according to the manufacturers' instructions. After bisulfate conversion, converted DNA was amplified by polymerase chain reaction (PCR) by using primers (Table S1) designed with MethPrimer (www.urogene.org/methprimer/index1.html).73 Two primer pairs were used for Oct4 promoter region methylation analysis, the first recognizes and anneals to methylated sequences only, whereas the second set amplifies unmethylated alleles. The PCR were performed using MaximeTM PCR PreMix Kiti-starTaq (Intron Biotechnology, Seongnam, Korea). The reaction mixture (20 μl) contained 1 μg of bisulfate converted DNA, 1 μM of each primer, and appropriate amounts of distilled water as recommended by the supplier. The cycling conditions were: 5 minutes at 95°C followed by 35 cycles of 15 seconds denaturation at 94°C, annealing for 30 seconds at 55°C, and elongation at 72°C for 30 sec.
Statistical analysis
Results are expressed as mean value ± standard error of mean (SEM). All experiments were analyzed by ANOVA, and some experiments which needed to compare with ≥3 groups were examined by comparing the treatment means to the control using a Bonferroni-Dunn test. A P value of <0.05 was considered statistically significant.
Funding
This research was supported by National R&D Program (2013M3A9B4076520) and Korea Mouse Phenotyping Project (2013M3A9D5072550) funded by the Ministry of Science, ICT & Future Planning hrough the National Research Foundation of Korea.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol 2014; 15:243-56; PMID:24651542; http://dx.doi.org/ 10.1038/nrm3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ochocki JD, Simon MC. Nutrient-sensing pathways and metabolic regulation in stem cells. J Cell Biol 2013; 203:23-33; PMID:24127214; http://dx.doi.org/ 10.1083/jcb.201303110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fafournoux P, Bruhat A, Jousse C. Amino acid regulation of gene expression. Biochem J 2000; 351:1-12; PMID:10998343; http://dx.doi.org/ 10.1042/bj3510001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris SE, Gopichandran N, Picton HM, Leese HJ, Orsi NM. Nutrient concentrations in murine follicular fluid and the female reproductive tract. Theriogenology 2005; 64:992-1006; PMID:16054501; http://dx.doi.org/ 10.1016/j.theriogenology.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Rezk Y, Huff C, Rizk B. Effect of glutamine on preimplantation mouse embryo development in vitro. Am J Obstet Gynecol 2004; 190:1450-4; PMID:15167866; http://dx.doi.org/ 10.1016/j.ajog.2004.02.025. [DOI] [PubMed] [Google Scholar]
- 6.Reitzer LJ, Wice BM, Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J Biol Chem 1979; 254:2669-76; PMID:429309. [PubMed] [Google Scholar]
- 7.Mazurek S, Eigenbrodt E, Failing K, Steinberg P. Alterations in the glycolytic and glutaminolytic pathways after malignant transformation of rat liver oval cells. J Cell Physiol 1999; 181:136-46; PMID:10457361; http://dx.doi.org/ 10.1002/(SICI)1097-4652(199910)181:1%3c136::AID-JCP14%3e3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 8.He Y, Hakvoort TB, Vermeulen JL, Lamers WH, Van Roon MA. Glutamine synthetase is essential in early mouse embryogenesis. Dev Dyn 2007; 236:1865-75; PMID:17557305; http://dx.doi.org/ 10.1002/dvdy.21185. [DOI] [PubMed] [Google Scholar]
- 9.Haberle J, Gorg B, Rutsch F, Schmidt E, Toutain A, Benoist JF, Gelot A, Suc AL, Hohne W, Schliess F, et al. Congenital glutamine deficiency with glutamine synthetase mutations. N Engl J Med 2005; 353:1926-33; PMID:16267323; http://dx.doi.org/ 10.1056/NEJMoa050456. [DOI] [PubMed] [Google Scholar]
- 10.Haberle J, Gorg B, Toutain A, Rutsch F, Benoist JF, Gelot A, Suc AL, Koch HG, Schliess F, Haussinger D. Inborn error of amino acid synthesis: human glutamine synthetase deficiency. J Inherit Metab Dis 2006; 29:352-8; PMID:16763901; http://dx.doi.org/ 10.1007/s10545-006-0256-5. [DOI] [PubMed] [Google Scholar]
- 11.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell 2007; 128:747-62; PMID:17320511; http://dx.doi.org/ 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 12.Bibikova M, Laurent LC, Ren B, Loring JF, Fan JB. Unraveling epigenetic regulation in embryonic stem cells. Cell Stem Cell 2008; 2:123-34; PMID:18371433; http://dx.doi.org/ 10.1016/j.stem.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Kim JB, Sebastiano V, Wu G, Arauzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D, et al. Oct4-induced pluripotency in adult neural stem cells. Cell 2009; 136:411-9; PMID:19203577; http://dx.doi.org/ 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 14.Tsai SY, Bouwman BA, Ang YS, Kim SJ, Lee DF, Lemischka IR, Rendl M. Single transcription factor reprogramming of hair follicle dermal papilla cells to induced pluripotent stem cells. Stem Cells 2011; 29:964-71; PMID:21563278; http://dx.doi.org/ 10.1002/stem.649. [DOI] [PubMed] [Google Scholar]
- 15.Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM, Choudhary J. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell 2010; 6:382-95; PMID:20362542; http://dx.doi.org/ 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Dai J. Concise review: isoforms of OCT4 contribute to the confusing diversity in stem cell biology. Stem Cells 2010; 28:885-93; PMID:20333750; http://dx.doi.org/ 10.1002/stem.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hattori N, Nishino K, Ko YG, Ohgane J, Tanaka S, Shiota K. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J Biol Chem 2004; 279:17063-9; PMID:14761969; http://dx.doi.org/ 10.1074/jbc.M309002200. [DOI] [PubMed] [Google Scholar]
- 18.Shan J, Hamazaki T, Tang TA, Terada N, Kilberg MS. Activation of the amino acid response modulates lineage specification during differentiation of murine embryonic stem cells. Am J Physiol Endocrinol Metab 2013; 305:E325-35; PMID:23736538; http://dx.doi.org/ 10.1152/ajpendo.00136.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuno T, Goto I. Glutaminase and glutamine synthetase activities in human cirrhotic liver and hepatocellular carcinoma. Cancer Res 1992; 52:1192-4; PMID:1346587. [PubMed] [Google Scholar]
- 20.Dang CV. Glutaminolysis: supplying carbon or nitrogen or both for cancer cells? Cell Cycle 2010; 9:3884-6; PMID:20948290; http://dx.doi.org/ 10.4161/cc.9.19.13302. [DOI] [PubMed] [Google Scholar]
- 21.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 2008; 132:661-80; PMID:18295582; http://dx.doi.org/ 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Kellner S, Kikyo N. Transcriptional regulation of the Oct4 gene, a master gene for pluripotency. Histol Histopathol 2010; 25:405-12; PMID:20054811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan GJ, Chang ZY, Scholer HR, Pei D. Stem cell pluripotency and transcription factor Oct4. Cell Res 2002; 12:321-9; PMID:12528890; http://dx.doi.org/ 10.1038/sj.cr.7290134. [DOI] [PubMed] [Google Scholar]
- 24.Bergstrom J, Furst P, Noree LO, Vinnars E. Intracellular free amino acid concentration in human muscle tissue. J Appl Physiol 1974; 36:693-7; PMID:4829908. [DOI] [PubMed] [Google Scholar]
- 25.Pollard M, Meredith D, McGivan JD. Identification of a plasma membrane glutamine transporter from the rat hepatoma cell line H4-IIE-C3. Biochem J 2002; 368:371-5; PMID:12171599; http://dx.doi.org/ 10.1042/bj20020982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mackenzie B, Erickson JD. Sodium-coupled neutral amino acid (System N/A) transporters of the SLC38 gene family. Pflugers Arch 2004; 447:784-95; PMID:12845534; http://dx.doi.org/ 10.1007/s00424-003-1117-9. [DOI] [PubMed] [Google Scholar]
- 27.Fuchs BC, Bode BP. Amino acid transporters ASCT2 and LAT1 in cancer: partners in crime? Semin Cancer Biol 2005; 15:254-66; PMID:15916903; http://dx.doi.org/ 10.1016/j.semcancer.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 28.Casalino L, Comes S, Lambazzi G, De Stefano B, Filosa S, De Falco S, De Cesare D, Minchiotti G, Patriarca EJ. Control of embryonic stem cell metastability by L-proline catabolism. J Mol Cell Biol 2011; 3:108-22; PMID:21307025; http://dx.doi.org/ 10.1093/jmcb/mjr001. [DOI] [PubMed] [Google Scholar]
- 29.Washington JM, Rathjen J, Felquer F, Lonic A, Bettess MD, Hamra N, Semendric L, Tan BS, Lake JA, Keough RA, et al. L-Proline induces differentiation of ES cells: a novel role for an amino acid in the regulation of pluripotent cells in culture. Am J Physiol Cell Physiol 2010; 298:C982-92; PMID:20164384; http://dx.doi.org/ 10.1152/ajpcell.00498.2009. [DOI] [PubMed] [Google Scholar]
- 30.Comes S, Gagliardi M, Laprano N, Fico A, Cimmino A, Palamidessi A, De Cesare D, De Falco S, Angelini C, Scita G, et al. L-Proline induces a mesenchymal-like invasive program in embryonic stem cells by remodeling H3K9 and H3K36 methylation. Stem Cell Reports 2013; 1:307-21; PMID:24319666; http://dx.doi.org/ 10.1016/j.stemcr.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 2013; 339:222-6; PMID:23118012; http://dx.doi.org/ 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Alexander P, Wu L, Hammer R, Cleaver O, McKnight SL. Dependence of mouse embryonic stem cells on threonine catabolism. Science 2009; 325:435-9; PMID:19589965; http://dx.doi.org/ 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ratnikov B, Aza-Blanc P, Ronai ZA, Smith JW, Osterman AL, Scott DA. Glutamate and asparagine cataplerosis underlie glutamine addiction in melanoma. Oncotarget 2015; 6:7379-89; PMID:25749035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryu JM, Han HJ. L-threonine regulates G1/S phase transition of mouse embryonic stem cells via PI3K/Akt, MAPKs, and mTORC pathways. J Biol Chem 2011; 286:23667-78; PMID:21550972; http://dx.doi.org/ 10.1074/jbc.M110.216283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Angel YC, Pichardo S, Salomir R, Petrusca L, Chapelon JY. Testing of a HIFU probe for the treatment of superficial venous insufficiency by using MRI. Conf Proc IEEE Eng Med Biol Soc 2006; 1:3533-6; PMID:17945783; http://dx.doi.org/ 10.1109/IEMBS.2006.260559. [DOI] [PubMed] [Google Scholar]
- 36.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene 2008; 27:5497-510; PMID:18794884; http://dx.doi.org/ 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Armstrong L, Hughes O, Yung S, Hyslop L, Stewart R, Wappler I, Peters H, Walter T, Stojkovic P, Evans J, et al. The role of PI3K/AKT, MAPK/ERK and NFκβ signalling in the maintenance of human embryonic stem cell pluripotency and viability highlighted by transcriptional profiling and functional analysis. Hum Mol Genet 2006; 15:1894-913; PMID:16644866; http://dx.doi.org/ 10.1093/hmg/ddl112. [DOI] [PubMed] [Google Scholar]
- 38.Singh AM, Reynolds D, Cliff T, Ohtsuka S, Mattheyses AL, Sun Y, Menendez L, Kulik M, Dalton S. Signaling network crosstalk in human pluripotent cells: a Smad2/3-regulated switch that controls the balance between self-renewal and differentiation. Cell Stem Cell 2012; 10:312-26; PMID:22385658; http://dx.doi.org/ 10.1016/j.stem.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe S, Umehara H, Murayama K, Okabe M, Kimura T, Nakano T. Activation of Akt signaling is sufficient to maintain pluripotency in mouse and primate embryonic stem cells. Oncogene 2006; 25:2697-707; PMID:16407845; http://dx.doi.org/ 10.1038/sj.onc.1209307. [DOI] [PubMed] [Google Scholar]
- 40.Nave BT, Ouwens M, Withers DJ, Alessi DR, Shepherd PR. Mammalian target of rapamycin is a direct target for protein kinase B: identification of a convergence point for opposing effects of insulin and amino-acid deficiency on protein translation. Biochem J 1999; 344 Pt 2:427-31; PMID:10567225; http://dx.doi.org/ 10.1042/bj3440427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chou MM, Hou W, Johnson J, Graham LK, Lee MH, Chen CS, Newton AC, Schaffhausen BS, Toker A. Regulation of protein kinase C ζ by PtdIns 3-kinase and PDK-1. Curr Biol 1998; 8:1069-77; PMID:9768361; http://dx.doi.org/ 10.1016/S0960-9822(98)70444-0. [DOI] [PubMed] [Google Scholar]
- 42.Dutil EM, Toker A, Newton AC. Regulation of conventional protein kinase C isozymes by phosphoinositide-dependent kinase 1 (PDK-1). Curr Biol 1998; 8:1366-75; PMID:9889098; http://dx.doi.org/ 10.1016/S0960-9822(98)00017-7. [DOI] [PubMed] [Google Scholar]
- 43.Le Good JA, Ziegler WH, Parekh DB, Alessi DR, Cohen P, Parker PJ. Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science 1998; 281:2042-5; PMID:9748166; http://dx.doi.org/ 10.1126/science.281.5385.2042. [DOI] [PubMed] [Google Scholar]
- 44.Dann SG, Selvaraj A, Thomas G. mTOR Complex1-S6K1 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol Med 2007; 13:252-9; PMID:17452018; http://dx.doi.org/ 10.1016/j.molmed.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 45.Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, Zack JA, Kornblum HI, Liu X, Wu H. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science 2001; 294:2186-9; PMID:11691952; http://dx.doi.org/ 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- 46.Zhou J, Shrikhande G, Xu J, McKay RM, Burns DK, Johnson JE, Parada LF. Tsc1 mutant neural stem/progenitor cells exhibit migration deficits and give rise to subependymal lesions in the lateral ventricle. Genes Dev 2011; 25:1595-600; PMID:21828270; http://dx.doi.org/ 10.1101/gad.16750211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anthony JC, Reiter AK, Anthony TG, Crozier SJ, Lang CH, MacLean DA, Kimball SR, Jefferson LS. Orally administered leucine enhances protein synthesis in skeletal muscle of diabetic rats in the absence of increases in 4E-BP1 or S6K1 phosphorylation. Diabetes 2002; 51:928-36; PMID:11916909; http://dx.doi.org/ 10.2337/diabetes.51.4.928. [DOI] [PubMed] [Google Scholar]
- 48.Minhajuddin M, Bijli KM, Fazal F, Sassano A, Nakayama KI, Hay N, Platanias LC, Rahman A. Protein kinase C-δ and phosphatidylinositol 3-kinase/Akt activate mammalian target of rapamycin to modulate NF-κB activation and intercellular adhesion molecule-1 (ICAM-1) expression in endothelial cells. J Biol Chem 2009; 284:4052-61; PMID:19074768; http://dx.doi.org/ 10.1074/jbc.M805032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar V, Pandey P, Sabatini D, Kumar M, Majumder PK, Bharti A, Carmichael G, Kufe D, Kharbanda S. Functional interaction between RAFT1/FRAP/mTOR and protein kinase cdelta in the regulation of cap-dependent initiation of translation. EMBO J 2000; 19:1087-97; PMID:10698949; http://dx.doi.org/ 10.1093/emboj/19.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parekh D, Ziegler W, Yonezawa K, Hara K, Parker PJ. Mammalian TOR controls one of two kinase pathways acting upon nPKCδ and nPKC. J Biol Chem 1999; 274:34758-64; PMID:10574945; http://dx.doi.org/ 10.1074/jbc.274.49.34758. [DOI] [PubMed] [Google Scholar]
- 51.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 1998; 95:379-91; PMID:9814708; http://dx.doi.org/ 10.1016/S0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 52.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 2000; 24:372-6; PMID:10742100; http://dx.doi.org/ 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 53.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 2003; 113:643-55; PMID:12787505; http://dx.doi.org/ 10.1016/S0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 54.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 2003; 113:631-42; PMID:12787504; http://dx.doi.org/ 10.1016/S0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 55.Farthing CR, Ficz G, Ng RK, Chan CF, Andrews S, Dean W, Hemberger M, Reik W. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet 2008; 4:e1000116; PMID:18584034; http://dx.doi.org/ 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell 2008; 2:160-9; PMID:18371437; http://dx.doi.org/ 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deb-Rinker P, Ly D, Jezierski A, Sikorska M, Walker PR. Sequential DNA methylation of the Nanog and Oct-4 upstream regions in human NT2 cells during neuronal differentiation. J Biol Chem 2005; 280:6257-60; PMID:15615706; http://dx.doi.org/ 10.1074/jbc.C400479200. [DOI] [PubMed] [Google Scholar]
- 58.Liao M, Zhang Y, Dufau ML. Protein kinase Cα-induced derepression of the human luteinizing hormone receptor gene transcription through ERK-mediated release of HDAC1/Sin3A repressor complex from Sp1 sites. Mol Endocrinol 2008; 22:1449-63; PMID:18372343; http://dx.doi.org/ 10.1210/me.2008-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lavoie G, Esteve PO, Laulan NB, Pradhan S, St-Pierre Y. PKC isoforms interact with and phosphorylate DNMT1. BMC Biol 2011; 9:31; PMID:21619587; http://dx.doi.org/ 10.1186/1741-7007-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hervouet E, Lalier L, Debien E, Cheray M, Geairon A, Rogniaux H, Loussouarn D, Martin SA, Vallette FM, Cartron PF. Disruption of Dnmt1/PCNA/UHRF1 interactions promotes tumorigenesis from human and mice glial cells. PLoS One 2010; 5:e11333; PMID:20613874; http://dx.doi.org/ 10.1371/journal.pone.0011333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kidder BL, Palmer S. HDAC1 regulates pluripotency and lineage specific transcriptional networks in embryonic and trophoblast stem cells. Nucleic Acids Res 2012; 40:2925-39; PMID:22156375; http://dx.doi.org/ 10.1093/nar/gkr1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dovey OM, Foster CT, Cowley SM. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc Natl Acad Sci U S A 2010; 107:8242-7; PMID:20404188; http://dx.doi.org/ 10.1073/pnas.1000478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krejci J, Uhlirova R, Galiova G, Kozubek S, Smigova J, Bartova E. Genome-wide reduction in H3K9 acetylation during human embryonic stem cell differentiation. J Cell Physiol 2009; 219:677-87; PMID:19202556; http://dx.doi.org/ 10.1002/jcp.21714. [DOI] [PubMed] [Google Scholar]
- 64.Liang G, Taranova O, Xia K, Zhang Y. Butyrate promotes induced pluripotent stem cell generation. J Biol Chem 2010; 285:25516-21; PMID:20554530; http://dx.doi.org/ 10.1074/jbc.M110.142059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mali P, Chou BK, Yen J, Ye Z, Zou J, Dowey S, Brodsky RA, Ohm JE, Yu W, Baylin SB, et al. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells 2010; 28:713-20; PMID:20201064; http://dx.doi.org/ 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 2003; 33 Suppl:245-54; PMID:12610534; http://dx.doi.org/ 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 67.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet 2000; 25:338-42; PMID:10888886; http://dx.doi.org/ 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 68.Zhou Q, Agoston AT, Atadja P, Nelson WG, Davidson NE. Inhibition of histone deacetylases promotes ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cells. Mol Cancer Res 2008; 6:873-83; PMID:18505931; http://dx.doi.org/ 10.1158/1541-7786.MCR-07-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hong Q, Shao ZM. Ubiquitination/deubiquitination and acetylation/deacetylation: making DNMT1 stability more coordinated. Acta Pharmacol Sin 2011; 32:139-40; PMID:21293465; http://dx.doi.org/ 10.1038/aps.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976; 72:248-54; PMID:942051; http://dx.doi.org/ 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 71.Mackman N, Brand K, Edgington TS. Lipopolysaccharide-mediated transcriptional activation of the human tissue factor gene in THP-1 monocytic cells requires both activator protein 1 and nuclear factor κB binding sites. J Exp Med 1991; 174:1517-26; PMID:1744583; http://dx.doi.org/ 10.1084/jem.174.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Borowicz S, Van Scoyk M, Avasarala S, Karuppusamy Rathinam MK, Tauler J, Bikkavilli RK, Winn RA. The soft agar colony formation assay. J Vis Exp 2014:e51998; PMID:25408172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 2002; 18:1427-31; PMID:12424112; http://dx.doi.org/ 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.