Abstract
p53 protects cells from DNA damage by inducing cell-cycle arrest upon encountering genomic stress. Among other pathways, p53 elicits such an effect by inhibiting mammalian target of rapamycin complex 1 (mTORC1), the master regulator of cell proliferation and growth. Although recent studies have indicated roles for both p53 and mTORC1 in stem cell maintenance, it remains unclear whether the p53-mTORC1 pathway is conserved to mediate this process under normal physiological conditions. Spermatogenesis is a classic stem cell-dependent process in which undifferentiated spermatogonia undergo self-renewal and differentiation to maintain the lifelong production of spermatozoa. To better understand this process, we have developed a novel flow cytometry (FACS)-based approach that isolates spermatogonia at consecutive differentiation stages. By using this as a tool, we show that genetic loss of p53 augments mTORC1 activity during early spermatogonial differentiation. Functionally, loss of p53 drives spermatogonia out of the undifferentiated state and causes a consistent expansion of early differentiating spermatogonia until the stage of preleptotene (premeiotic) spermatocyte. The frequency of early meiotic spermatocytes is, however, dramatically decreased. Thus, these data suggest that p53-mTORC1 pathway plays a critical role in maintaining the homeostasis of early spermatogonial differentiation. Moreover, our FACS approach could be a valuable tool in understanding spermatogonial differentiation.
Keywords: differentiation, germline stem cell, mTORC1, p53, spermatogonia
Abbreviations
- mTORC1
mammalian target of rapamycin complex 1
- FACS
flow cytometry
- GDNF
glial cell line-derived neurotrophic
- Plzf
promyelocytic leukemia zinc finger
- hHLH
Basic helix-loop-helix
- Stra8
stimulated by the retinoic acid gene 8
- p-RPS6
phosphorylated ribosomal protein S6.
Introduction
Stem cells undergo both self-renewal and differentiation to maintain a lifelong production of mature cells for tissue regeneration. Imbalance between these processes results in the loss of tissue homeostasis that has been implicated in tissue degeneration, aging, and cancer. mTORC1 is a signaling complex that positively mediates cell growth, primarily through phosphorylation of components of the translation machinery, thus promoting protein translation.1,2 Previous studies suggest a critical role for mTORC1 signaling in determining the stem cell self-renewal and differentiation decision.3-5 Aberrant activation of mTORC1 leads to enhanced stem cell activity, activating the tumor suppressive mechanism in somatic stem cells and leading to cellular senescence or apoptosis.2,6 In germline stem cells, genetic deletion of an inhibitor of mTORC1 activity, e.g., Plzf or Tsc2, can lead to spermatogonial stem cell differentiation at the expense of self-renewal, and thereby results in a premature exhaustion of the spermatogonia pool and a testicular aging-like phenotype.7-10 Therefore, it is critical to understand how mTORC1 activity is precisely regulated, particularly under normal physiological conditions.
p53 has been extensively studied as the guardian of the genome for its tumor suppressor function.11 Now, it is well characterized that, under genotoxic stress, elevated level of p53 results in cell cycle arrest to allow time for DNA repair. If unsuccessful, p53 activation initiates cell death programming to eliminate cells with damaged or mutated DNA.12 Recently, mounting evidence suggests that p53 also controls the proliferation, self-renewal, and differentiation of embryonic and adult stem cells to maintain tissue or organ homeostasis.13 This raises the question of whether the molecular pathways that p53 employs under genotoxic stress might also function under normal physiological conditions. For example, upon DNA damage, p53 negatively regulates mTORC1 activity to limit metabolic support, prevent cell division, and cause cellular quiescence, thereby exerting its function as a tumor suppressor.14–16 Interestingly, under normal conditions, mTORC1 activity can be increased in tissue from p53-deficient mice such as the heart.17 However, it remains unclear whether the p53-mTORC1 pathway is conserved in regulating stem cell activity during tissue homeostasis.
Spermatogenesis is a classic stem cell dependent process by which the undifferentiated spermatogonia undergo both mitotic and meiotic divisions to produce spermatozoa.18 Morphological studies have classified spermatogonia into several subtypes.19 Specifically, spermatogonia in the single-cell state are known as A single (Asingle) spermatogonia, some of which are considered to be spermatogonial stem cells (SSCs). SSCs are traditionally regarded as the foundation of the spermatogonial differentiation hierarchy and give rise to paired A (Apaired) spermatogonia, which are connected by an intercellular bridge. Apaired cells then divide into chains of 4 – 32 aligned (Aaligned) spermatogonia. These three types of cells all maintain stem cell properties and are considered undifferentiated spermatogonia. After a differentiation step in which Aaligned spermatogonia are committed irreversibly to gamete production, the cells are classified as differentiating type A spermatogonia. This is followed by a series of divisions that culminate in B spermatogonia, which then divide to yield premeiotoic (preleptotene) spermatocytes that transition from the mitotic cell cycle to the meiotic cell cycle. While the transition from undifferentiated Aaligned spermatogonia to differentiating type A spermatogonia occurs every 8.6 d in mice, it takes an Asingle spermatogonia 35 d to fully differentiate into spermatozoa. In addition, recent studies have also characterized genes whose expressions are associated with specific stages during spermatogonial differentiation. For example, Gfra1, a component of the glial cell line-derived neurotrophic (GDNF) factor receptor, is predominately expressed in the Asingle spermatogonia.20 Functionally, GDNF stimulates the self-renewal of undifferentiated spermatogonia.21 The promyelocytic leukemia zinc finger (Plzf) is expressed in most of undifferentiated type A spermatogonia.7,8 Importantly, Plzf is essential for spermatogonia maintenance, as loss of Plzf promotes differentiation at the expense of self-renewal and results in the exhaustion of the spermatogonia. Plzf might mediate this effect by suppressing mTORC1 activity via transcriptionally activating Redd1.9 Basic helix-loop-helix (hHLH) protein Sohlh1 then appears in Aaligned spermatogonia and is essential for spermatogonial development.22,23 Preleptotene spermatocytes entering meiosis are characterized by their intense expression of stimulated by the retinoic acid gene 8 (Stra8), a protein indispensible for premeiotic DNA synthesis.24-26 Despite these progresses, the process of early spermatogonial differentiation is still not well understood.
Here, we report the development of a novel flow cytometry (FACS)-based approach that allows for separation and isolation of spermatogonia at consecutive differentiation stages. By using this as a tool, we show that loss of p53 amplifies mTORC1 signaling and promotes spermatogonial differentiation, suggesting a role for the p53-mTORC1 pathway in stem cell maintenance under tissue homeostasis.
Results
Quantitative analysis of spermatogonial differentiation
Meiosis can be viewed as a differentiation process unique to the germ line. Transcriptional activation of Stra8 that triggers meiotic initiation is perhaps the earliest event known to date during germline stem cell differentiation of both sexes.27 Therefore, we considered the possibility that Stra8 promoter activity might be useful in monitoring early germline stem cell differentiation. Toward this end, we have reported the successful development of a transgenic mouse line in which the 1.4-kb Stra8 promoter (−1400 to +11) drives the expression of the reporter gene, green fluorescence protein (GFP) (referred hereafter as the pStra8-GFP transgenic mouse line).28 The pStra8-GFP transgenic mice exhibit gonad-specific GFP expression in post-pubertal testes in males and sex-specific GFP expression in female embryonic ovaries during the developmental window of meiotic initiation. We combined the pStra8-GFP reporter with an established strategy of 2 additional cell surface markers to characterize spermatogonial differentiation:29 α6-integrin, which is a marker of undifferentiated spermatogonia with stem cell capacity, and c-Kit, which is turned on upon spermatogonial differentiation. These two cell surface markers allowed us to first identify the c-Kitnegative α6-integrinhigh, undifferentiated spermatogonia compartment and the c-Kitpositiveα6-integrinlow, differentiating spermatogonia compartment (Fig. 1A). EpCAM was not used as a pan-spermatogonia marker as previously reported,29 as recent studies as well as our own investigation suggest that EpCAM level is low in undifferentiated spermatogonia but increases along with early spermatogonial differentiation (data not shown).30
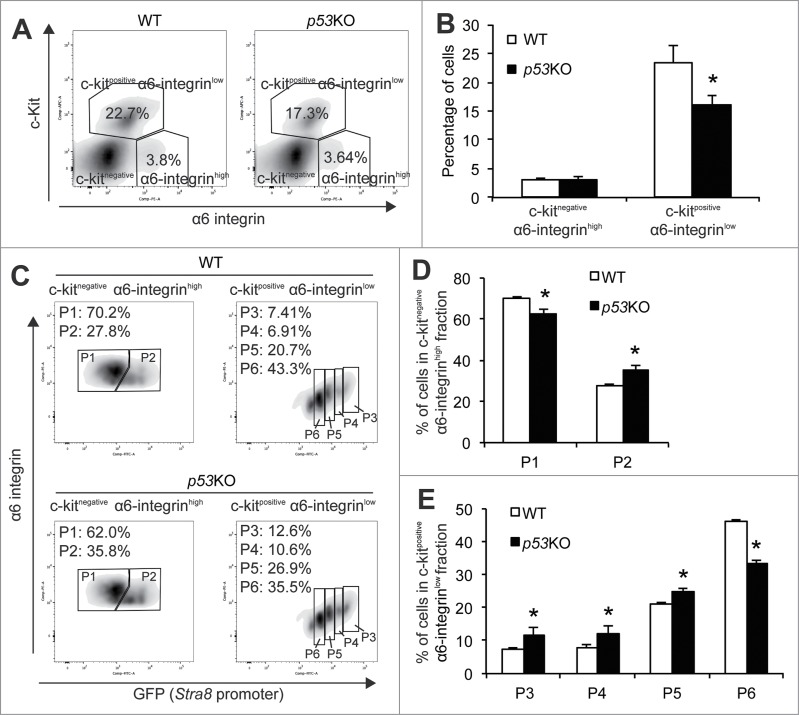
Figure 1.
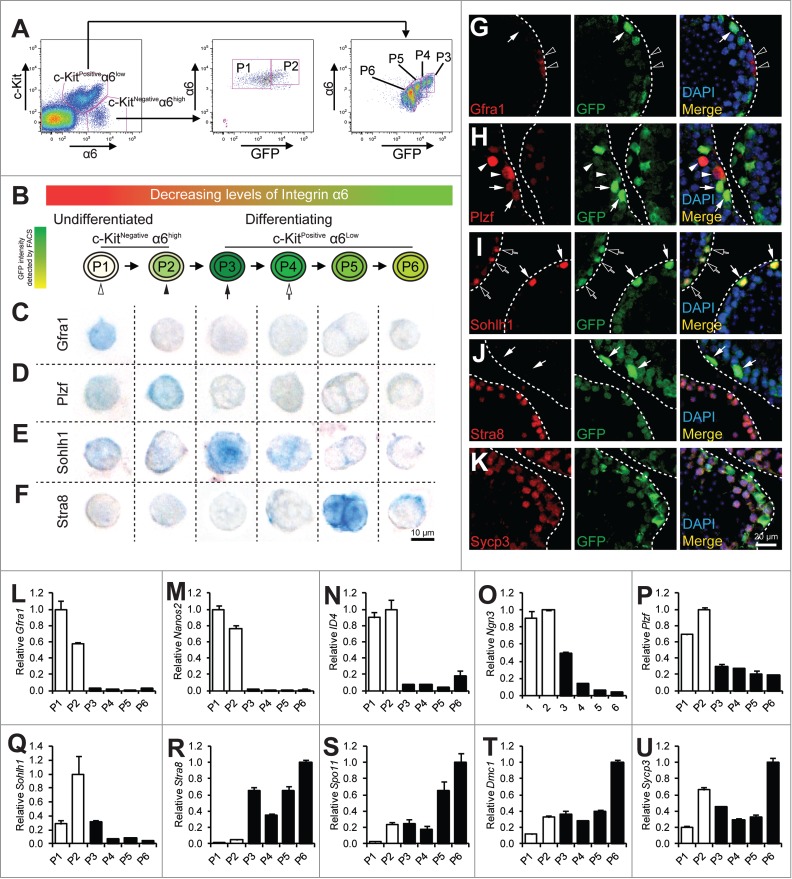
Analysis of spermatogonial differentiation by flow cytometry. (A) representative flow profile of separating spermatogonia differentiation into P1 to P6 by using a combination of markers for undifferentiated SSC (α6-integin), differentiating spermatogonia (c-Kit), and GFP driven by the Stra8 promoter. Note the concomitant decrease of α6-integrin level with GFP in the c-KitPos α6-integrinlow population. (B) schematic diagram showing progression of spermatogonial development from P1 to P6. C–F, immunohistochemistry staining of cells from P1 to P6 populations isolated from 4-weeks old juvenile pStra8-GFP mice. Expression of Gfra1 (C), Plzf (D), Sohlh1 (E), and Stra8 (F) was detected by alkaline phosphatase (AP). Cells shown are magnified views of a single cell. G–K, dual-immunofluorescence staining of GFP (green) with Gfra1 (G), Plzf (H), Sohlh1 (I), Stra8 (J), and Sycp3 (K) (red) in 4-weeks old juvenile mouse testes. L–U, qPCR analysis of relative mRNA expression of the indicated genes in P1 to P6. Twenty,000 cells were isolated from 4-weeks old juvenile pStra8-GFP mice by FACS as shown in panel A. mRNA levels are normalized to those of β-actin. Data are representative of 3 independent set of experiment. Graphs represent mean ± standard deviations from duplicate reactions.
Within the c-Kitnegative α6-integrinhigh compartment that has been shown to be composed mostly of undifferentiated spermatogonia, we were able to further identify a GFP-negative population (population 1 or P1) and a GFP-positive population (population 2 or P2) (Fig. 1A). Markers for undifferentiated spermatogonia (namely, Gfra1, Nanos2, Id4, Ngn3, and Plzf) were highly enriched in both P1 and P2 populations when compared with cells from the c-Kitpositiveα6-integrinlow, differentiating spermatogonia compartment (Fig. 1L-P).7,8,31-34 Since the Stra8 promoter activation may indicate commitment to meiotic initiation, we considered cells in the P1 population to be further enriched for the most primitive undifferentiated spermatogonia due to their complete lack of Stra8 promoter activity. We considered cells in the P2 population to represent the earliest differentiating spermatogonia within this undifferentiated spermatogonia compartment. This is because, while their c-Kit levels still remained negative, Stra8 promoter activity was already initiated (Fig. 1A). Expression of Gfra1 is present in earlier stages of spermatogonia development than that of Plzf. Indeed, we found cells in P1 expressed higher levels of Gfra1, while cells in P2 population expressed higher levels of Plzf mRNA (Fig. 1L and P). In addition, 80.3% (49 out of 61 cells examined) of cells from P1 expressed Gfra1 and this number dropped to 36.1% (26 out of 72 cells examined) in P2, while only 45.2% (42 out of 93) of cells from P1 expressed Plzf and this number increased to 90.6% (75 out of 83) in P2 (Fig. 1C and D). Moreover, to locate cells in P1 and P2 in juvenile testicular sections, we identified cells that expressed Gfra1 but not GFP (cells from P1) or expressed Plzf together with low levels of GFP (cells from P2) (Fig. G and H). Taken together, these molecular features suggest that, by using the Stra8 promoter-driven GFP as a reporter, we can further separate the previously reported undifferentiated spermatogonia population into the more primitive P1 population and the P2 population, which is on the verge of initial differentiation.
Within the c-Kitpositive α6-integrinlow differentiating spermatogonia compartment, we identified 4 separable populations based on their distinct levels of GFP expression (Fig. 1A). We refer to them hereafter as population 3 to population 6 (P3 to P6) based on their progressively decreasing levels of GFP expression with cells in P3 exhibiting the highest GFP and cells in P6 the lowest (Fig. 1A and B). We considered P3 to P6 populations to be differentiating spermatogonia that may include early spermatocytes at consecutive stages toward maturation. Interestingly, α6-integrin levels from P3 to P6 decrease concomitantly together with those of GFP expression (Fig. 1A). The differential GFP intensities are not due to various copies of transgenes, in that we “knocked” our pStra8-GFP transgenic cassette into the Hprt neutral locus on the X chromosome. Therefore, our male pStra8-GFP mice carry 1 copy of pStra8-GFP reporter per cell.28 Importantly, we identified P5 as the preleptotene spermatocytes, which were characterized by their intense Stra8 staining (80 out of 89 cells examined) and their doublet morphology, consistent with previous reports for preleptotene spermatocytes (Fig. 1F).35 Thus, cells in P3 and P4 populations are differentiating spermatogonia, an identification supported by their expression of Sohlh1 in cells from P3 populations (83 out of 93 cells examined) (Fig. 1E). Cells in P6 population are probably early meiotic spermatocytes. Consistently, cells from P3 to P6 population showed progressive features of meiosis initiation and progression by the expression of Stra8, Spo11, Dmc1, and Sycp3 mRNAs (Fig. 1R–U).24,36-38 We identified these cells in testicular sections as cells that exhibited decreasing levels of GFP expressions but showed Sohlh1 (P3), Stra8 (P5), and Sycp3 (P6) expression (Fig. 1J and K).
p53 deletion augments mTORC1 signaling during spermatogonial differentiation
p53 is expressed at low levels under normal physiological conditions and sometimes its activation can only be reflected by the activation of its promoter.39,40 To test whether p53 is an upstream suppressor of mTORC1 activity during spermatogonial differentiation, we compared the levels of the phosphorylated ribosomal protein S6 (p-RPS6) in wild-type (WT) and p53KO testes. We chose p-RPS6, because p-RPS6 is a downstream target of mTORC1 through ribosomal protein S6 Kinase 1 and has been previously used to monitor mTORC1 activity during spermatogonial differentiation.9,41,42 In western blotting analysis of whole testicular lysate, we found that p-RPS6 levels were increased in p53KO testes when compared to WT testes (Fig. 2A). Consistently, sections from p53KO testes exhibited a higher level of p-RPS6 staining than those from WT testes (Fig. 2B).
Figure 2.
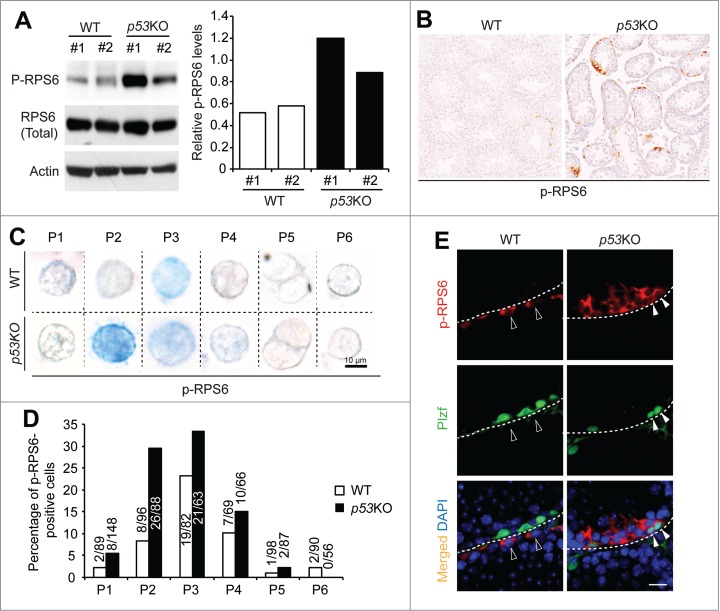
p53 loss amplifies mTORC1 activity in differentiating spermatogonia. (A) western blotting analysis of p-RPS6 from 2 mice per genotype (#1 and #2) (left). Quantification of the relative levels of p-RPS6 against total RPS6 (right). (B) immunohistochemistry staining of p-RPS6 in 4-weeks old juvenile WT and p53KO testis. Images shown are representative from 3 mice per genotype examined. (C) immunohistochemistry staining of p-RPS6 detected by alkaline phosphatase (AP) in cells from P1 to P6 populations isolated from 4-weeks old juvenile WT and p53KO pStra8-GFP mice. Cells shown are magnified views of a single cell. Number of positive cells detected and total number of cells examined are indicated. (D) comparison of the percentages of p-RPS6-positive cells in P1 to P6 populations. Graphs represent mean values ± SEM. n = 3 mice per genotype, *P < 0.05. (E) dual-immunofluorescence staining of p-RPS6 (red) and Plzf (green) in 4-weeks old juvenile WT and p53KO testis.
We next introduced the pStra8-GFP reporter allele into p53KO mice. WT littermates with pStra8-GFP reporter were used as controls. Since p-RPS6 has also been reported to be present not only in germ cells but also in the somatic Sertoli cells, we confirmed that p53 deletion-induced upregulation of mTORC1 activity in germ cells by isolating cells from P1 to P6 from WT and p53KO testes (Fig. 1A). Consistent with recent previous reports that mTORC1 activation defines a subset of spermatogonia committed to differentiation,10 p-RPS6 was mostly present during early spermatogonial differentiation at P3 stages in WT testes. However, in p53KO testes, there was a robust increase in the percentage of cells that expressed p-RPS6 at P2 stage of spermatogonial differentiation (Fig. 2C and D). Cells from P2 expressed high levels of Plzf (Fig. 1D), which has been reported to suppress mTORC1 activity to maintain self-renewal of spermatogonia.9 Consistently, we found that, in WT testes, Plzf-expressing cells usually did not show p-RPS6 expression (Fig. 2E). However, in p53KO testes, we identified cells that showed both strong Plzf and p-RPS6 expressions (Fig. 2E). Taken together, these data indicate that p53 suppresses mTORC1 activity during normal spermatogonia development and suggest that this pathway is independent of Plzf.
p53 deletion results in the loss of spermatogonia maintenance and meiotic differentiation
To examine the functional consequence of p53 loss on early spermatogonia differentiation, FACS analysis showed that, while p53 deletion had no appreciable effect on the frequency of c-Kitnegative α6-integrinhigh, undifferentiated spermatogonia compartment (Fig. 3A), the c-Kitpositiveα6-integrinlow, differentiating spermatogonia compartment was significantly decreased when compared with WT controls (Fig. 3B). This initial analysis seems to be opposite to our speculation that elevated mTORC1 activity would trigger spermatogonial differentiation and the previous reports that p53KO testes contain significantly higher numbers of differentiating spermatogonia.39
Figure 3.
Quantitative analysis of the impact of p53 loss on spermatogonial differentiation. (A) representative flow profiles of the c-Kitnegative α6-integrinhigh undifferentiated spermatogonia compartment and the c-Kitpositive α6-integrinlow differentiating spermatogonia compartment from 4-weeks old WT (left) and p53KO (right) testes. (B) quantification of frequency of c-Kitnegative α6-integrinhigh undifferentiated spermatogonia compartment and the c-Kitpositive α6-integrinlow differentiating spermatogonia compartment in testes of WT and p53KO mice at 4-weeks of age. Graphs represent mean values ± SEM. n = 4 mice per genotype, *P < 0.05. (C) representative flow profiles of P1 and P2 in the c-Kit-negative α6-integrin-high spermatogonia compartment (left panels) and P3 to P6 in the c-Kit-positive α6-integrin-low (right panels) from WT (upper panels) and p53KO (lower panels) testes. (D and E) quantification of frequency of P1 to P6 populations in testes of wild-type and p53KO mice at 4-weeks of age. Graphs represent mean values ± SEM. n = 4 mice per genotype, *P < 0.05.
We next examined the frequencies of P1 to P6 within these 2 compartments. We found that p53 deletion caused a 10.8% decrease in the percentage of P1 population but consistent increases in the percentages from P2 to P5 populations (27.5% in P2, 62.7% in P3, 52.9% in P4 and 17.2% in P5) (Fig. 3C–E). Thus, these data suggest that p53 deletion results in a phenotype resembling loss of the stem cell maintenance by driving spermatogonia out of the undifferentiated stage (P1) for differentiation (P2 to P5). The percentage of the P6 population, however, was dramatically decreased by 27.7% (Fig. 3C and E), which is consistent with the reported role for p53 in promoting meiotic recombination.40
We further confirmed these FACS-based observations through histological analysis. Comparing sections from the WT and p53KO testes, we observed a 39.4% increase in the numbers of cells that expressed intense levels of Plzf per tubule in the p53KO (Fig. 4A and B). Additionally, we found there was a 55.7% increase in the percentage of Stra8-expressing, stage VII seminiferous tubules (Fig. 4C and D). These observations are consistent with the increase in the P2 to P5 populations as shown through FACS. Furthermore, there was a 40.2% decrease in tubules containing Sycp3-expressing, meiotic spermatocytes (Fig. 4E and F), which matches the observed decrease in the P6 population shown through FACS (Fig. 3C and E).
Figure 4.
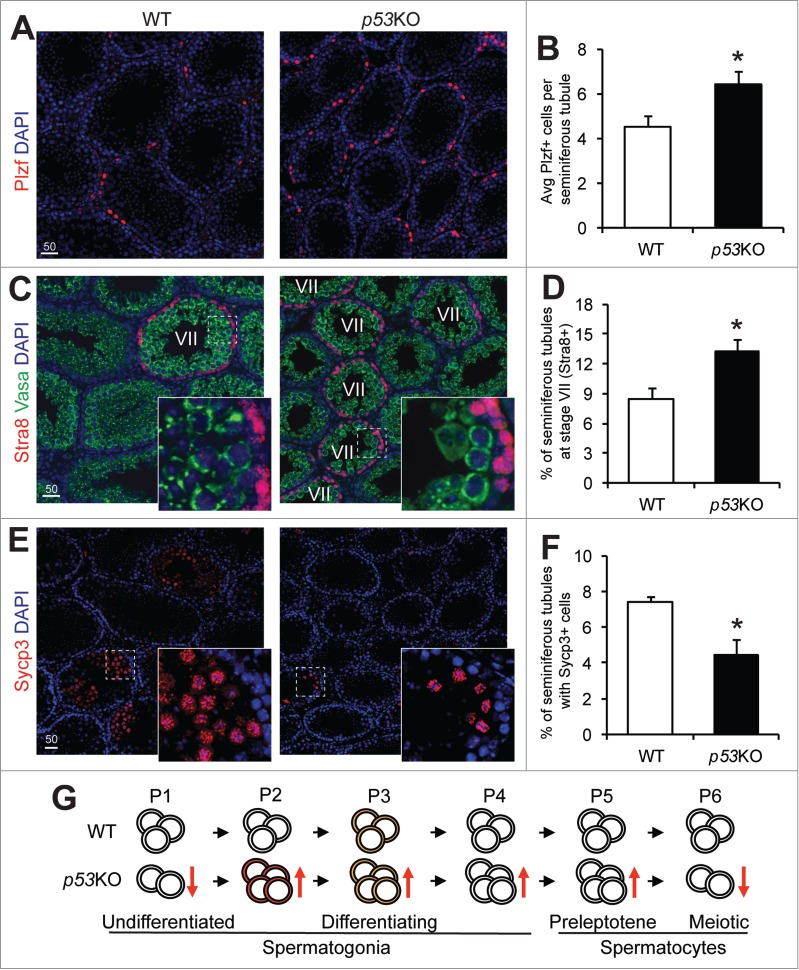
p53 loss expands the spermatogonia pool but causes a loss of meiotic spermatocytes. A) immunofluorescence staining of Plzf in 4-weeks old juvenile WT and p53KO testis. (B) quantification of cells showing strong positivity for Plzf in seminiferous tubules of testis from (A) representing early differentiating spermatogonia. Graphs represent mean values ± SEM. n = 4 mice per genotype, *P < 0.05. (C) dual-immunofluorescence staining of Stra8 (red) and Vasa (green) in 4-weeks old juvenile WT and p53KO testis. (D) quantification of percentage of stage VII seminiferous tubules in cross sections as shown in (C). Graphs represent mean values ± SEM. n = 4 mice per genotype, *P < 0.05. (E) immunofluorescence staining of Sycp3 in 4-weeks old juvenile WT and p53KO testis. (F) quantification of the percentage of seminiferous tubules containing meiotic spermatocytes showing strong positivity for Sycp3 from panel E. Graphs represent mean values ± SEM. n = 4 mice per genotype, *P < 0.05. (G) schematic diagram showing the effect of p53 deletion on spermatogonial differentiation. Orange and red colors in cells at P2 and P3 populations indicate mTORC1 activity.
Discussions
Similar to its role as the guardian of the genome in somatic cells, p53 function in germ cells of the testes primarily involves removing cells with DNA damage due to genotoxic stress through apoptosis.39 In addition, by using a traditional histological approach, a previous study shows that p53KO testes contain significantly higher numbers of differentiating spermatogonia.39 Here, by using a different strain of p53KO mice paired with a quantitative approach to analyze spermatogonial differentiation from undifferentiated spermatogonia to early meiotic spermatocytes, we further explored the impact of p53 deletion on the homeostasis of early spermatogonial differentiation. Specifically, we found that p53 deletion amplifies the differentiating spermatogonia compartment by depleting the undifferentiated spermatogonia compartment (Fig. 4G). This phenotype is associated with enhanced mTORC1 activity, which has been shown to promote spermatogonial differentiation at the expense of self-renewal.9 In addition, since mTORC1 activity is a primary determinant of cell size, enhanced mTORC1 may also contribute to the formation of the giant cells frequently observed in p53KO mice or mice with reduced levels of p53 protein.39,43 Taken together, these data suggest a critical role for the p53-mTORC1 pathway in regulating spermatogonial differentiation.
Age-related upregulation of p53 expression is frequently linked to cellular senescence and loss of tissue or organ function.44,45 Here, we also observed that p53 levels were significantly elevated in aged testes (Fig. S1). Since p53 is a negative regulator of mTORC1 activity, this raises the possibility that higher levels of p53 in aged testes may cause insufficient mTORC1 activity required for normal spermatogonial differentiation. In fact, lower levels of mTORC1 activity has been observed in germ cells from aged testes when compared with those from young testes.46
There has been a continuous interest to enrich undifferentiated spermatogonia by using genetic or cell surface markers. For example, the αv-integrinnegative Thy-1low c-Kitnegative fraction is reportedly enriched for spermatogonia with stem cell capacity.9 Thy-1 and Gfra1 have been used in previous studies to isolate undifferentiated spermatogonia.47 Recently, it is reported that cells transcriptionally active for Id4 defines a subset of the male germline stem cell population.48 In our current study, by combining the existing strategy of α6-integrin and c-Kit with the pStra8-GFP reporter, we were able to not only isolate an undifferentiated spermatogonia population (P1) but also a series of their progeny at consecutive differentiating stages toward maturation (P2 to P6). We consider cells in the P1 population to be enriched for more primitive undifferentiated spermatogonia than cells in the P2 population. This conclusion is supported by this population's complete lack of the Stra8 promoter activity, whose activation is among the earliest events known to date during meiotic initiation in germ cells of both sexes.24,25
This FACS-based approach may have many applications in the study of spermatogonial development or even provide insight into the general multi-stepped process during early stem cell differentiation. For example, transplantation of cells from P1 to P6 populations into chemotherapy-depleted recipient testes to test their repopulation capacity may be valuable in characterizing the progressive loss of stemness during spermatogonial differentiation as well as the molecular mechanisms responsible for this event. Moreover, since dedifferentiation has been reported to occur during early spermatogonial differentiation, transplantations of cells from P1 to P6 populations would be critical to elucidate the homeostasis. Since the Stra8 promoter activity is indicative of early spermatogonial differentiation, comparative gene expression analysis between P1 and P2 populations that share similar α6-integrinhigh c-Kitnegative profiles but show distinct Stra8 promoter activity may help reveal genes uniquely expressed in the most primitive undifferentiated spermatogonia or spermatogonial stem cells (P1). So far, most genes that have been reported to mark undifferentiated spermatogonia, e.g., Ngn3, Id4, Nanos2, are present in both P1 and P2 populations. So identification of genes uniquely expressed by cells in P1 population might help identify the most primitive undifferentiated spermatogonia that still remain elusive. On the other hand, genes uniquely enriched in P2 population may relate to differentiation commitment and therefore provide insight into the intrinsic cyclicity of spermatogenesis. In our initial analysis, we already identified certain discrete features for P1 and P2 populations. For example, P1 is highly enriched for p21 mRNA expression (data not shown), consistent with the role for p21 in keeping stem cells at a quiescence state, while cells in P2 are highly enriched for Sohlh1 mRNA expression (Fig. 1Q), a gene required for spermatogonial development (although Sohlh1 protein appears at P3 stage, highlighting a critical role for transcription/translation control during spermatogonial differentiation).22,49,50 In addition to identifying genes for the most primitive undifferentiated spermatogonia, comparative gene expression analysis between P4, the latest stage of differentiating spermatogonia, and P5, preleptotene premeiotic spermatocytes, will be helpful to identify genes involved in the process of mitosis to meiosis transition, the defining feature of germ cells. Therefore, an integrated transcriptome, proteome, and methylome analysis of cells from P1 to P6 populations may provide a comprehensive molecular landscape of spermatogonial differentiation.
Materials and Methods
Animals
p53KO were obtained from Taconic (TSG-p53).51 pStra8-GFP mice are generated as previously described.28 All procedures and care of animals were carried out according to the Massachusetts General Hospital Institutional Animal Care and Use Committee (IACUC).
Isolation of mouse testicular germ cells and FACS analysis
Dissociated testicular cells from wild-type or p53KO;pStra8-GFP mouse testes were generated by sequential digestion of dissected and minced seminiferous tubules with 1 mg/ml Type IV collagenase (Worthington) in the presence of 20 μg/ml DNAse I (Sigma) and 0.05% Trypsin (Gibco). The testes were dissected out and the capsules removed. Prior to mincing, tubules were washed twice with Hank's balanced salt solution (HBSS) to remove interstitial cells. After mincing, cells were resuspended in Type IV collagenase and DNase I solution and shaken vigorously at 37°C for 30 minutes. Cells were then passed through 20 G needles and washed with HBSS containing 0.2% FBS. An equivalent amount of Trypsin (to previous collagenase solution) was used to resuspend cells which were again shaken vigorously at 37°C for 30 minutes. Trypsin was neutralized by 10% fetal bovine serum (FBS) in HBSS and the cells were passed through a 70 μm strainer to remove clumps. Cells were collected by centrifugation and then resuspended in Cell Staining Buffer (Biolegend) for counting and subsequent staining. After blocking for 10 minutes with a 1:100 dilution of TruStain fcXTM anti-mouse CD16/32 (BioLegend), the following antibodies to cell surface markers were used: 1:40 dilution of APC-conjugated c-Kit antibody (CD117, Bioscience Catalog #553356) and 1:10 dilution of PE-conjugated α6-integrin (CD49f, Bioscience Catalog #555736). Cells were stained for 30 minutes, washed twice, and then resuspended with HBSS containing 0.2% FBS for sorting.
Immunocytochemistry and immunofluorescence
For frozen sections, testes were fixed in 4% paraformaldehyde for one hour at 4°C, dehydrated in 15% sucrose, and embedded in 7.5% gelatin. For paraffin sections, testes were fixed in 4% paraformaldehyde overnight at 4°C, embedded in paraffin. Tissues were then sectioned at 10 μm (frozen) or 6 μm (paraffin) for analysis. Antibodies used include: Gfra1 (AF560, R&D systems), Plzf (clone D-9, Santa Cruz Biotechnology Catalog# 28319), Sohlh1 (abcam), Stra8 (ab49602, Abcam), and Sycp3 (ab15093, Abcam). For immunofluorescence, detection was performed using Alexa fluor 488-conjugated donkey anti-mouse and Alexa fluor 546-conjugated donkey anti-rabbit secondary antibodies. Frozen sections stored in −80°C were dried at room temperature and then washed 3 times for 10 min each in PBS warmed to 37°C. Paraffin sections were hydrated 3 times in Xylene, 2 times in 100% Ethanol, 95% Ethanol, 80% Ethanol, 70% Ethanol, and H2O, all for 5 minutes each. Antigen retrieval was performed by boiling slides for 15 minutes in sodium citrate heated to 100°C. Slides were cooled to room temperature and then washed 3 times for 10 min each with TBST/PBST. Slides were blocked at room temperature for 1 hour with DAKO protein block and then incubated overnight at 4°C with primary antibodies in blocking solution (5% normal serum, 1% BSA, 0.3% Triton X-100 in PBST). After washing 3 times with PBST, slides were incubated at room temperature for 30 minutes with secondary antibodies in blocking solution. Slides were then washed, stained with DAPI, and mounted with ProLong(R) Gold antifade reagent (Life Technologies). For immunocytochemistry, detection was performed using Alkaline Phosphatase goat anti-rabbit, horse anti-goat, and horse anti-mouse IgG and the Alkaline Phosphatase Substrate Kit III from Vector Laboratories. Cells were sorted directly onto slides coated with poly-lysine-D, dried overnight at room temperature, and stored at 4°C. Prior to staining, slides were rinsed in PBS and fixed in 4% PFA in PBS for 15 minutes at room temperature. Slides were then washed 3 times in ice cold PBS and then permeabilized with 0.3% Triton X-100 for 10 minutes at room temperature. After washing, cells were incubated at room temperature for 1 hour with DAKO protein blocked and then incubated overnight at 4°C with primary antibody in blocking solution. After washing, slides were incubated at room temperature for 30 minutes with alkaline phosphatase-conjugated goat anti-rabbit, horse anti-mouse, or horse anti-goat secondary antibodies in blocking solution. Slides were washed again and developed with Vector Blue substrate working solution for 5–10 minutes at room temperature. Slides were rinsed and mounted in VectaMount AQ Aqeous Mounting Medium (Vector Laboratories).
Gene expression analysis
Total RNA from each sample was reverse transcribed by using SuperScript III from Invitrogen. qPCR was conducted by using SsoAdvanced™ Universal SYBR® Green Supermix (BioRad) with the following primers:
Plzf forward, 5′-AAACGGTTCCTGGACAGTTTGCGAC-3′
reverse, 5′-CCAGTATGGGTCTGTCTGTGTGTCTCC-3′
Stra8 forward, 5′-CCTGGTAGGGCTCTTCAACA-3′
reverse, 5′-GGCTTTCTTCCTGTTCCTGA-3′
Gfra1 forward, 5′-CACTCCTGGATTTGCTGATGT-3′
reverse, 5′-AGTGTGCGGTACTTGGTGC-3′
β-actin forward, 5′-CTGCCGCATCCTCTTCCTC-3′
reverse, 5′-GCCACAGGATTCCATACCCA-3′
Nanos2 forward, 5′-CTGCAAGCACAATGGGGAGT-3′
reverse, 5′-CGTCGGTAGAGAGACTGCTG-3′
ID4 forward, 5′-CAGTGCGATATGAACGACTGC-3′
reverse, 5′-GACTTTCTTGTTGGGCGGGAT-3′
Ngn3 forward, 5′-ACGACTTCCAGACGCAATTT-3′
reverse, 5′-CCATCCTAGTTCTCCCGACTC-3′
Sohlh1 forward, 5′-ACAGCTTGCAGACCTCATCACAAAG-3′
reverse, 5′-ACACAGACATCTCCAGGACAGATGC-3′
Spo11 forward, 5′-AACAATCAGATGGCTTGGTCTCCTC-3′
reverse, 5′-GGCTTGGATCTCTGCCTTCATCTTA-3′
Dmc1 forward, 5′-ATCGGGATTCCAAGATGATG-3′
reverse, 5′-TGCAGAGGGCTCTTCTTGTT-3′
Sycp3 forward, 5′-GGGGCCGGACTGTATTTACT-3′
reverse, 5′-GGAGCCTTTTCATCAGCAAC-3′
p53 forward, 5′-TGGAAGACTCCAGTGGGAAC-3′
reverse, 5′-TCTTCTGTACGGCGGTCTCT-3′
Oct4 forward, 5′-CACGAGTGGAAAGCAACTCA-3′
reverse, 5′-AGATGGTGGTCTGGCTGAAC-3′
CyclinD1 forward, 5′-AGAGGCGGATGAGAACAAGC-3′
reverse, 5′-CATGGAGGGTGGGTTGGAAA-3′
CyclinD2 forward, 5′-GCTGGAGTGGGAACTGGTAG-3′
reverse, 5′-CGGATCAGGGACAGCTTCTC-3′
p21 forward, 5′-CCTGGTGATGTCCGACCTG-3′
reverse, 5′-CCATGAGCGCATCGCAATC-3′
Western blotting analysis
Total protein was isolated in RIPA buffer supplemented with 1 mM PMSF (Sigma) and a protease inhibitor cocktail (Sigma P8340). Lysates were cleared by centrifugation at 14,000 × g for 10 min at 4°C, and protein concentrations in supernatants were determined (DC protein assay; BioRad). Equal amount of protein from each sample was mixed with LDS sample buffer (Invitrogen) plus sample reducing agent (Invitrogen), and denatured for 10 min at 70°C. Proteins were resolved in 4−12% Bis-Tris gels (Invitrogen), and transferred to PDVF membranes. Blots were probed with antibodies against p-RPS6 (Ser236/236, Cell Signaling Catalog#2211), total RPS6 (Cell Signaling Catalog#2217), or pan-Actin (NeoMarkers Catalog# MS-1295), washed and reacted with horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse IgG (BioRad). Detection was performed with the Clarity™ ECL Western Blotting Substrate (BioRad).
Data analysis and presentation
All experiments were independently replicated at least 3 times, using different mice, tissues or cells for each replicate. Where possible, assignment of mice to experimental groups was made randomly. Quantitative data from replicate experiments were pooled, analyzed by ANOVA or Student t-test in GraphPad Prism software, and are presented as the means ± SEM. Statistical analysis was performed. Representative images obtained from immunohistochemical and immunofluorescence assays are provided for qualitative assessment.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Dr. Jonathan L. Tilly for the permission to use pStra8-GFP mice and MGH HSCI flow cytometry core facility for expert assistance with flow cytometry analysis. N.W. is a recipient of an NIH Pathway-to-Independence Award from National Institute on Aging. Author contributions: M.X. and N.W. designed the experiments, analyzed data, and wrote the manuscript. M.X., I.F., O.Y. and N.W. conducted the experiments. N.W. supervised the study.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was supported by NIA (R00-AG-039512) and the Vincent Department of Obstetrics and Gynecology at Massachusetts General Hospital to N.W.
References
- 1.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 2009; 10:307-18; PMID:19339977; http://dx.doi.org/ 10.1038/nrm2672 [DOI] [PubMed] [Google Scholar]
- 2.Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature 2006; 441:475-82; PMID:16598206; http://dx.doi.org/ 10.1038/nature04703 [DOI] [PubMed] [Google Scholar]
- 3.Chen C, Liu Y, Liu R, Ikenoue T, Guan KL, Liu Y, Zheng P. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med 2008; 205:2397-408; PMID:18809716; http://dx.doi.org/ 10.1084/jem.20081297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magri L, Galli R. mTOR signaling in neural stem cells: from basic biology to disease. Cell Mol Life Sci 2013; 70:2887-98; PMID:23124271; http://dx.doi.org/ 10.1007/s00018-012-1196-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiang X, Zhao J, Xu G, Li Y, Zhang W. mTOR and the differentiation of mesenchymal stem cells. Acta Biochim Biophys Sin (Shanghai) 2011; 43:501-10; PMID:21642276; http://dx.doi.org/ 10.1093/abbs/gmr041 [DOI] [PubMed] [Google Scholar]
- 6.Castilho RM, Squarize CH, Chodosh LA, Williams BO, Gutkind JS. mTOR mediates Wnt-induced epidermal stem cell exhaustion and aging. Cell Stem Cell 2009; 5:279-89; PMID:19733540; http://dx.doi.org/ 10.1016/j.stem.2009.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet 2004; 36:647-52; PMID:15156142; http://dx.doi.org/ 10.1038/ng1366 [DOI] [PubMed] [Google Scholar]
- 8.Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet 2004; 36:653-9; PMID:15156143; http://dx.doi.org/ 10.1038/ng1367 [DOI] [PubMed] [Google Scholar]
- 9.Hobbs RM, Seandel M, Falciatori I, Rafii S, Pandolfi PP. Plzf regulates germline progenitor self-renewal by opposing mTORC1. Cell 2010; 142:468-79; PMID:20691905; http://dx.doi.org/ 10.1016/j.cell.2010.06.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hobbs RM, La HM, Makela JA, Kobayashi T, Noda T, Pandolfi PP. Distinct germline progenitor subsets defined through Tsc2-mTORC1 signaling. EMBO Rep 2015; 16:467-80; PMID:25700280; http://dx.doi.org/ 10.15252/embr.201439379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lane DP. Cancer. p53, guardian of the genome. Nature 1992; 358:15-6; PMID:1614522; http://dx.doi.org/ 10.1038/358015a0 [DOI] [PubMed] [Google Scholar]
- 12.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer 2014; 14:359-70; PMID:24739573; http://dx.doi.org/ 10.1038/nrc3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Solozobova V, Blattner C. p53 in stem cells. World J Biol Chem 2011; 2:202-14; PMID:21949570; http://dx.doi.org/ 10.4331/wjbc.v2.i9.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasty P, Sharp ZD, Curiel TJ, Campisi J. mTORC1 and p53: clash of the gods? Cell Cycle 2013; 12:20-5; PMID:23255104; http://dx.doi.org/ 10.4161/cc.22912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korotchkina LG, Leontieva OV, Bukreeva EI, Demidenko ZN, Gudkov AV, Blagosklonny MV. The choice between p53-induced senescence and quiescence is determined in part by the mTOR pathway. Aging (Albany NY) 2010; 2:344-52; PMID:20606252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levine AJ, Harris CR, Puzio-Kuter AM. The interfaces between signal transduction pathways: IGF-1/mTor, p53 and the Parkinson Disease pathway. Oncotarget 2012; 3:1301-7; PMID:23211569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leontieva OV, Novototskaya LR, Paszkiewicz GM, Komarova EA, Gudkov AV, Blagosklonny MV. Dysregulation of the mTOR pathway in p53-deficient mice. Cancer Biol Ther 2013; 14:1182-8; PMID:24184801; http://dx.doi.org/ 10.4161/cbt.26947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oatley JM, Brinster RL. Regulation of spermatogonial stem cell self-renewal in mammals. Annu Rev Cell Dev Biol 2008; 24:263-86; PMID:18588486; http://dx.doi.org/ 10.1146/annurev.cellbio.24.110707.175355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Rooij DG, Russell LD. All you wanted to know about spermatogonia but were afraid to ask. J Androl 2000; 21:776-98; PMID:11105904 [PubMed] [Google Scholar]
- 20.Buageaw A, Sukhwani M, Ben-Yehudah A, Ehmcke J, Rawe VY, Pholpramool C, Orwig KE, Schlatt S. GDNF family receptor alpha1 phenotype of spermatogonial stem cells in immature mouse testes. Biol Reprod 2005; 73:1011-6; PMID:16014811; http://dx.doi.org/ 10.1095/biolreprod.105.043810 [DOI] [PubMed] [Google Scholar]
- 21.Meng X, Lindahl M, Hyvonen ME, Parvinen M, de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H, Lakso M, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 2000; 287:1489-93; PMID:10688798; http://dx.doi.org/ 10.1126/science.287.5457.1489 [DOI] [PubMed] [Google Scholar]
- 22.Ballow D, Meistrich ML, Matzuk M, Rajkovic A. Sohlh1 is essential for spermatogonial differentiation. Dev Biol 2006; 294:161-7; PMID:16564520; http://dx.doi.org/ 10.1016/j.ydbio.2006.02.027 [DOI] [PubMed] [Google Scholar]
- 23.Suzuki H, Ahn HW, Chu T, Bowden W, Gassei K, Orwig K, Rajkovic A. SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev Biol 2012; 361:301-12; PMID:22056784; http://dx.doi.org/ 10.1016/j.ydbio.2011.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson EL, Baltus AE, Roepers-Gajadien HL, Hassold TJ, de Rooij DG, van Pelt AM, Page DC. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc Natl Acad Sci U S A 2008; 105:14976-80; PMID:18799751; http://dx.doi.org/ 10.1073/pnas.0807297105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baltus AE, Menke DB, Hu YC, Goodheart ML, Carpenter AE, de Rooij DG, Page DC. In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat Genet 2006; 38:1430-4; PMID:17115059; http://dx.doi.org/ 10.1038/ng1919 [DOI] [PubMed] [Google Scholar]
- 26.Zhou Q, Nie R, Li Y, Friel P, Mitchell D, Hess RA, Small C, Griswold MD. Expression of stimulated by retinoic acid gene 8 (Stra8) in spermatogenic cells induced by retinoic acid: an in vivo study in vitamin A-sufficient postnatal murine testes. Biol Reprod 2008; 79:35-42; PMID:18322276; http://dx.doi.org/ 10.1095/biolreprod.107.066795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menke DB, Koubova J, Page DC. Sexual differentiation of germ cells in XX mouse gonads occurs in an anterior-to-posterior wave. Dev Biol 2003; 262:303-12; PMID:14550793; http://dx.doi.org/ 10.1016/S0012-1606(03)00391-9 [DOI] [PubMed] [Google Scholar]
- 28.Imudia AN, Wang N, Tanaka Y, White YA, Woods DC, Tilly JL. Comparative gene expression profiling of adult mouse ovary-derived oogonial stem cells supports a distinct cellular identity. Fertil Steril 2013; 100(5):1451-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takubo K, Ohmura M, Azuma M, Nagamatsu G, Yamada W, Arai F, Hirao A, Suda T. Stem cell defects in ATM-deficient undifferentiated spermatogonia through DNA damage-induced cell-cycle arrest. Cell Stem Cell 2008; 2:170-82; PMID:18371438; http://dx.doi.org/ 10.1016/j.stem.2007.10.023 [DOI] [PubMed] [Google Scholar]
- 30.Valli H, Sukhwani M, Dovey SL, Peters KA, Donohue J, Castro CA, Chu T, Marshall GR, Orwig KE. Fluorescence- and magnetic-activated cell sorting strategies to isolate and enrich human spermatogonial stem cells. Fertil Steril 2014; 102:566-80 e7; PMID:24890267; http://dx.doi.org/ 10.1016/j.fertnstert.2014.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oatley MJ, Kaucher AV, Racicot KE, Oatley JM. Inhibitor of DNA binding 4 is expressed selectively by single spermatogonia in the male germline and regulates the self-renewal of spermatogonial stem cells in mice. Biol Reprod 2011; 85:347-56; PMID:21543770; http://dx.doi.org/ 10.1095/biolreprod.111.091330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida S, Sukeno M, Nakagawa T, Ohbo K, Nagamatsu G, Suda T, Nabeshima Y. The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development 2006; 133:1495-505; PMID:16540512; http://dx.doi.org/ 10.1242/dev.02316 [DOI] [PubMed] [Google Scholar]
- 33.Sada A, Suzuki A, Suzuki H, Saga Y. The RNA-binding protein NANOS2 is required to maintain murine spermatogonial stem cells. Science 2009; 325:1394-8; PMID:19745153; http://dx.doi.org/ 10.1126/science.1172645 [DOI] [PubMed] [Google Scholar]
- 34.Dettin L, Ravindranath N, Hofmann MC, Dym M. Morphological characterization of the spermatogonial subtypes in the neonatal mouse testis. Biol Reprod 2003; 69:1565-71; PMID:12855601; http://dx.doi.org/ 10.1095/biolreprod.103.016394 [DOI] [PubMed] [Google Scholar]
- 35.Boateng KA, Bellani MA, Gregoretti IV, Pratto F, Camerini-Otero RD. Homologous pairing preceding SPO11-mediated double-strand breaks in mice. Dev Cell 2013; 24:196-205; PMID:23318132; http://dx.doi.org/ 10.1016/j.devcel.2012.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chuma S, Nakatsuji N. Autonomous transition into meiosis of mouse fetal germ cells in vitro and its inhibition by gp130-mediated signaling. Dev Biol 2001; 229:468-79; PMID:11203703; http://dx.doi.org/ 10.1006/dbio.2000.9989 [DOI] [PubMed] [Google Scholar]
- 37.Di Carlo AD, Travia G, De Felici M. The meiotic specific synaptonemal complex protein SCP3 is expressed by female and male primordial germ cells of the mouse embryo. Int J Dev Biol 2000; 44:241-4; PMID:10794082 [PubMed] [Google Scholar]
- 38.Romanienko PJ, Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 2000; 6:975-87; PMID:11106738; http://dx.doi.org/ 10.1016/S1097-2765(00)00097-6 [DOI] [PubMed] [Google Scholar]
- 39.Beumer TL, Roepers-Gajadien HL, Gademan IS, van Buul PP, Gil-Gomez G, Rutgers DH, de Rooij DG. The role of the tumor suppressor p53 in spermatogenesis. Cell Death Differ 1998; 5:669-77; PMID:10200522; http://dx.doi.org/ 10.1038/sj.cdd.4400396 [DOI] [PubMed] [Google Scholar]
- 40.Lu WJ, Chapo J, Roig I, Abrams JM. Meiotic recombination provokes functional activation of the p53 regulatory network. Science 2010; 328:1278-81; PMID:20522776; http://dx.doi.org/ 10.1126/science.1185640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev 2002; 16:1472-87; PMID:12080086; http://dx.doi.org/ 10.1101/gad.995802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, Dor Y, Zisman P, Meyuhas O. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev 2005; 19:2199-211; PMID:16166381; http://dx.doi.org/ 10.1101/gad.351605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rotter V, Schwartz D, Almon E, Goldfinger N, Kapon A, Meshorer A, Donehower LA, Levine AJ. Mice with reduced levels of p53 protein exhibit the testicular giant-cell degenerative syndrome. Proc Natl Acad Sci U S A 1993; 90:9075-9; PMID:8415656; http://dx.doi.org/ 10.1073/pnas.90.19.9075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Keizer PL, Laberge RM, Campisi J. p53: Pro-aging or pro-longevity? Aging (Albany NY) 2010; 2:377-9; PMID:20657035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodier F, Campisi J, Bhaumik D. Two faces of p53: aging and tumor suppression. Nucleic Acids Res 2007; 35:7475-84; PMID:17942417; http://dx.doi.org/ 10.1093/nar/gkm744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferder IC, Wang N. Hypermaintenance and hypofunction of aged spermatogonia: insight from age-related increase of Plzf expression. Oncotarget 2015; PMID:25986924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kofman AE, McGraw MR, Payne CJ. Rapamycin increases oxidative stress response gene expression in adult stem cells. Aging (Albany NY) 2012; 4:279-89; PMID:22529334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan F, Oatley MJ, Kaucher AV, Yang QE, Bieberich CJ, Shashikant CS, Oatley JM. Functional and molecular features of the Id4+ germline stem cell population in mouse testes. Genes Dev 2014; 28:1351-62; PMID:24939937; http://dx.doi.org/ 10.1101/gad.240465.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kippin TE, Martens DJ, van der Kooy D. p21 loss compromises the relative quiescence of forebrain stem cell proliferation leading to exhaustion of their proliferation capacity. Genes Dev 2005; 19:756-67; PMID:15769947; http://dx.doi.org/ 10.1101/gad.1272305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes M, Scadden DT. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science 2000; 287:1804-8; PMID:10710306; http://dx.doi.org/ 10.1126/science.287.5459.1804 [DOI] [PubMed] [Google Scholar]
- 51.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr., Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 1992; 356:215-21; PMID:1552940; http://dx.doi.org/ 10.1038/356215a0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.