Abstract
In recent years, genome engineering technology has provided unprecedented opportunities for site-specific modification of biological genomes. Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) 9 is one such means that can target a specific genome locus. It has been applied in human cells and many other organisms. Meanwhile, to efficiently enrich targeted cells, several surrogate systems have also been developed. However, very limited information exists on the application of CRISPR/Cas9 in chickens. In this study, we employed the CRISPR/Cas9 system to induce mutations in the peroxisome proliferator-activated receptor-γ (PPAR-γ), ATP synthase epsilon subunit (ATP5E), and ovalbumin (OVA) genes in chicken DF-1 cells. The results of T7E1 assays showed that the mutation rate at the three different loci was 0.75%, 0.5%, and 3.0%, respectively. In order to improve the mutation efficiency, we used the PuroR gene for efficient enrichment of genetically modified cells with the surrogate reporter system. The mutation rate, as assessed via the T7E1 assay, increased to 60.7%, 61.3%, and 47.3%, and subsequent sequence analysis showed that the mutation efficiency increased to 94.7%, 95%, and 95%, respectively. In addition, there were no detectable off-target mutations in three potential off-target sites using the T7E1 assay. As noted above, the CRISPR/Cas9 system is a robust tool for chicken genome editing.
Keywords: CRISPR/Cas9 system, surrogate reporter, chicken DF-1 cell, mutant efficiency, off-target, GenPred, Shared data resource, Genomic Selection
In recent years, genome engineering technologies have provided unprecedented opportunities for site-specific modification of biological genomes. These technologies could be used to investigate the function of coding genes, or regulatory elements, via gene editing (Li et al. 2015; Wanzel et al. 2016). The most important component of these technologies is a nuclease that can introduce double-strand breaks (DSBs) into specified regions of genomes. Representative examples of this technology, e.g., engineered zinc-finger nuclease (ZFN), transcription activator-like effector nuclease (TALEN), and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas)9, have developed rapidly in recent years.
CRISPR/Cas9 derives from bacterial and archaeal adaptive immune systems that defend against invasion by phages or foreign plasmids (Barrangou et al. 2007). The CRISPR/Cas9 system can target a specific genome locus by using a Cas9 protein and a guide RNA (gRNA), which includes a 20 nt sequence that binds to its DNA target by Watson-Crick base-pairing (Jinek et al. 2012). The target site must have a sequence motif, known as the protospacer adjacent motif (PAM), present just downstream of the 20 bp target sequence (Jinek et al. 2012). Unlike ZFN and TALEN, which require engineering of a new protein for each target sequence, the only required engineering in the CRISPR/Cas9 system is to match a 20 nt target-complementary gRNA with the target DNA sequence adjacent to the PAM. As such, these gRNAs can be rapidly constructed and are easy to use. After in vitro work showed the site-specific cleavage function (Jinek et al. 2012), the CRISPR/Cas9 system was promptly developed. To date, it has been applied in human cells, and in many other organisms (Mali et al. 2013a; Cong et al. 2013), such as zebra fish (Yin et al. 2015; Li et al. 2015), mice (Miura et al. 2015; Mou et al. 2015), rats (Guan et al. 2014; Li et al. 2013), pigs (Zhou et al. 2016; Ruan et al. 2015), and goats (Wang et al. 2015). However, there is little information about the application of this technology in chicken (Veron et al. 2015).
Not all designed nucleases are efficient enough to allow sufficient derivation of cells containing nuclease-driven mutations (Santiago et al. 2008; Kim et al. 2009). Laborious screening of many clones is often required to obtain enough gene-modified clones. Kim and colleagues devised several surrogate reporters that contained a nuclease target sequence to enrich gene-modified clones, and eliminate unmodified cells (Kim et al. 2011, 2009, 2013). In our previous work, we developed a dual reporter system for efficient enrichment of genetically modified cells (Ren et al. 2015). Here, we demonstrate efficient site-specific modification of the peroxisome proliferator-activated receptor-γ (PPAR-γ), ATP synthase epsilon subunit (ATP5E), and ovalbumin (OVA) genes in chicken DF-1 cells using the CRISPR/Cas9 system combined with a dual surrogate reporter system. The results indicate that this system is a robust tool for chicken genome editing.
Materials and Methods
Construction of the Cas9 nuclease expression vector
The CRISPR/Cas9 system used in this study was the engineered Streptococcus thermophilus CRISPR3-Cas (StCas9) system constructed by Xu et al. (2015). The human codon-optimized Cas9 gene and the gRNA were driven by the CMV and U6 promoters, respectively, and cloned into the pll3.7 vector. The targeting oligonucleotide sequences used for the respective gRNAs were: PPAR-γ, GCGAATGCCACAAGCGGAGA; ATP5E, GCCTCAGTACAAAGCTGAGG; and OVA, AGATGTTCTCATTG GCATGG.
Construction of the surrogate reporter
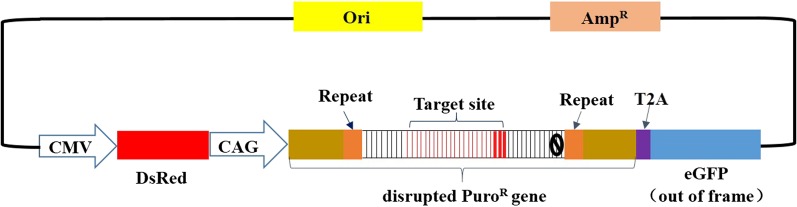
Three target sequences of approximately 170 bp in length for the PPAR-γ, ATP5E, and OVA genes were PCR-amplified using the chicken genome as a template using the primers shown in Table 1. Reference genomic sequences were extracted from GenBank (PPAR-γ, NC_006099; ATP5E, NC_006107; and OVA, NC_006089). Then, the target sequence was inserted into the parental DsRed-PuroR-eGFP (RPG) dual-reporter surrogate system based on the single strand annealing (SSA) repair pathway (SSA-RPG) (Ren et al. 2015) to generate corresponding reporter vectors. The surrogate SSA-RPG reporter vector included three reporter genes: DsRed, PuroR, and eGFP. DsRed was driven by a CMV promoter to measure transfection efficiency. The PuroR gene was driven by a CAG promoter, and fused with the eGFP gene via T2A as a dual-reporter. The CRISPR/Cas9 target sequence was flanked with 200 bp direct repeats, and inserted into the middle of the PuroR gene to interrupt the open reading frame (ORF) (Figure 1). When the CRISPR/Cas9 nuclease cleaves the target site of the surrogate reporter to introduce the DSBs, repair via SSA between the two direct repeats can result in correction of the ORF for the PuroR and eGFP reporter genes.
Table 1. Primer sequences for generating RPG reporter plasmid.
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| PPAR-γ | CCCgcggccgc TTCTTCAGCCATCAGGTTTGGG | CCCggatcc CCTTGGCTTTGGTCAGAGGG |
| ATP5E | CGCgcggccgc GTTTTCAGCTACATCCGGTACTC | CGCggatcc CCTTCTTGGTCTTCACAATCTTC |
| OVA | CCCgcggccgc AGAGTTCACCATGGGCTCCATC | CCCggatcc GTATACCATGGCTAGAGCTGAC |
Figure 1.
Schematic of the SSA-RPG reporter. The DsRed is driven by the CMV promoter. The PuroR gene is interrupted by a target sequence flanked with direct repeats as SSA arms. The disrupted PuroR and eGFP genes are fused by T2A, as a dual reporter, and driven by the CAG promoter.
Culture of DF1 cell line
The chicken DF-1 cell line was maintained in Dulbecco’s modified Eagle medium (DMEM, Gibco) containing 10% (v/v) fetal bovine serum (FBS, Sciencecell), 100 U/ml penicillin, and 100 μg/ml streptomycin in an incubator at 37° and 5% CO2. DF-1 cells were seeded into 24-well plates, and transfected 24 hr later.
Cell transfection
Cells were cotransfected with 1.6 μg plasmid DNA containing Cas9/gRNA, and the corresponding SSA-RPG reporter plasmid (molar ratio 1:1) using Sofast transfection reagent (Xiamen Sunma Biotechnology Co., Ltd. China), according to the manufacturer’s protocol. The Cas9 expression vector alone, with the SSA-RPG reporter, were used as controls. The cells were observed and photographed with a fluorescence microscope 2 d after transfection. Then, half of the cells were harvested for genome detection, and the other half for puromycin screening.
Puromycin selection
At 2 d after transfection, puromycin (2.5 μg/ml) (Sigma) was added to the culture medium and maintained for 4 d; the medium was changed daily. Then, the puromycin was removed, and the resistant cells were cultured sequentially until 90% confluence for subsequent genome detection.
Detection of nuclease-induced mutations
Genomic DNA was isolated from harvested DF-1 cells with the phenol-chloroform method. A T7E1 assay was performed as previously reported (Kim et al. 2009; 2011). PCR products were amplified using the primer pairs listed in Table 2, and purified by gel extraction. We denatured 100 ng purified product at 94°, annealed it to form heteroduplex DNA, subsequently treated it with 5 U of T7 nuclease I (New England Biolabs) for 30 min at 37°, and finally analyzed it using 2% agarose gel electrophoresis. Mutation frequencies were calculated as previously described based on the band intensities using ImageJ software and the following equation: mutation frequency (%) = 100 × [1 – (1 – F)1/2], where F represents the cleavage coefficient, which is the proportion of the total relative density of the cleavage bands to all of the relative densities of the cleavage bands and uncut bands (Guschin et al. 2010).
Table 2. Primers used in sequence analysis for detection of indels.
| Gene | Forward Primer | Reverse Primer | Product Size |
|---|---|---|---|
| PPAR-γ | AAGCGCTTCAGTAGTTTGCC | TTGTGGAAGATAACCTCTGG | 673 bp |
| ATP5E | CATGGTGGCGTACTGGCGGCAGGC | TGAGCTGCTCGCTGCATGTGCAGTG | 546 bp |
| OVA | TAGCCTACCATAGAGTACCCTG | CAACTGCTGGATGCAGAGCACTAGC | 697 bp |
To further confirm target locus mutations, PCR products were cloned into the pGEM-T Easy vector (Promega). For each sample, 19–20 random clones were sequenced to identify the mutation efficiency.
Off-target analysis in the DF-1 cell line
Three potential CRISPR/Cas9 off-target sites for the PPAR-γ, ATP5E, and OVA genes were selected for mutation analysis (Table 3). Off-target analysis was performed by T7E1 assay. Primers for the amplification of nine potential target fragments are listed in Table 3. Reference genomic sequences were obtained from GenBank (GNS, NC-006088; EHD3, NC-006090; ADCYAP1, NC-006089; KCNT2, NC-006095; DAD1, NC-006114; PITPNM2, NC-006102; ABCC9, NC-006088; TXNDC12, NC-006095; and LCP2, NC-006100).
Table 3. Primer sequences for detection of off-target analysis.
| Gene | Forward Primer | Reverse Primer | Product Size |
|---|---|---|---|
| GNS | AGCCCCACAGATAGCTGTC | TTCACGCCAGCCACTCCTTC | 544 bp |
| EHD3 | GAGGTTGTGACTCAGTCAGCAC | CACATACATTATCCGAGCCCG | 627 bp |
| ADCYAP1 | GAAATGGAGCAAACAGAGAG | CGTCCTTCATTTGTACTCAGG | 626 bp |
| KCNT2 | CCTCCCCAACAATCACCTCTTCCC | GCTGGGGAAGAAGCAGACA | 610 bp |
| DAD1 | CTCACACAAGGGCACCTCTG | GATAGCTACGGGGCTTCGTG | 636 bp |
| PITPNM2 | GCTGGATCTGTGCATACAAG | GGGTTCATACATGCCATGAC | 597 bp |
| ABCC9 | GGCTACTTGGGTACTGCACTC | CAGTTGCTGCAAAGATCACGC | 388 bp |
| TXNDC12 | CATGCTGACCCGGAAGTGAC | CCGTACACTGATTGATGTGGTG | 363 bp |
| LCP2 | ACCTGAGCCAAGCCTGACTC | TAGGGAGCTGGATTCATTTTCC | 387 bp |
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Detection of the CRISPR/Cas9 activity in DF-1 cells
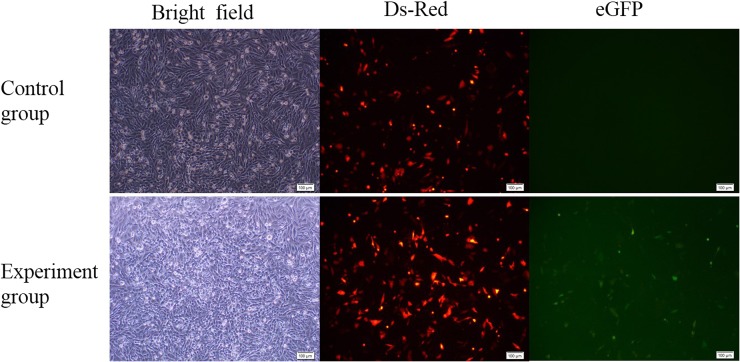
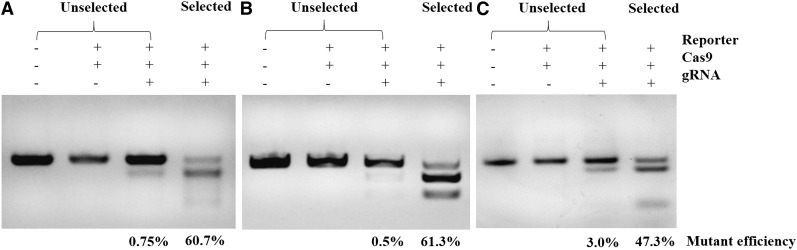
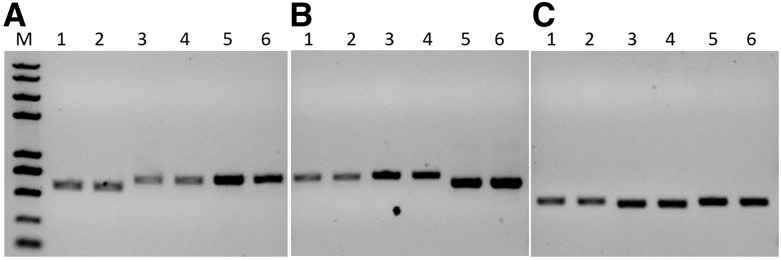
Three genes, PPAR-γ, ATP5E, and OVA, were selected as the target genes. To quickly test the activity of the CRISPR/Cas9 system, all Cas9 and gRNA plasmids, combined with the corresponding SSA-RPG reporter, were cotransfected into DF-1 cells in the experimental groups, with the Cas9 expression vector without gRNA being used in the control groups. At 48 hr after transfection, a considerable fraction of cells expressed Ds-Red, and a small portion of cells expressed eGFP in the experimental group, whereas cells in the control group expressed only Ds-Red. As expected, all of the eGFP-expressing cells also expressed Ds-Red in the experimental group (Figure 2). This indicated that the CRISPR/Cas9 system worked in DF-1 cells. The subsequent T7E1 assay demonstrated that the mutation frequencies within the PPAR-γ, ATP5E, and OVA loci were 0.75%, 0.5%, and 3.0%, respectively (Figure 3).
Figure 2.
Activity detection of the CRISPR/Cas9 system 48 hr after transfection. Visualization of DsRed and eGFP expression by fluorescence microscopy after transfection for 48 hr. Cells from the experimental group expressed DsRed and eGFP, but the control group expressed DsRed only.
Figure 3.
T7E1 assay for the indels induced by CRISPR/Cas9 within unselected and puromycin-selected cells. (A), (B), and (C) represent the indels induced by PPAR-γgRNA/Cas9, ATP5E.gRNA/Cas9, and OVA.gRNA/Cas9, respectively. The numbers at the bottom of the gel indicate the mutation frequency as measured by band relative density.
Mutant cell enrichment using puromycin screening
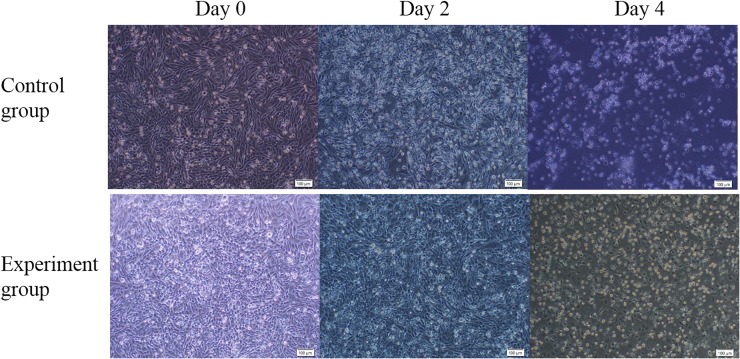
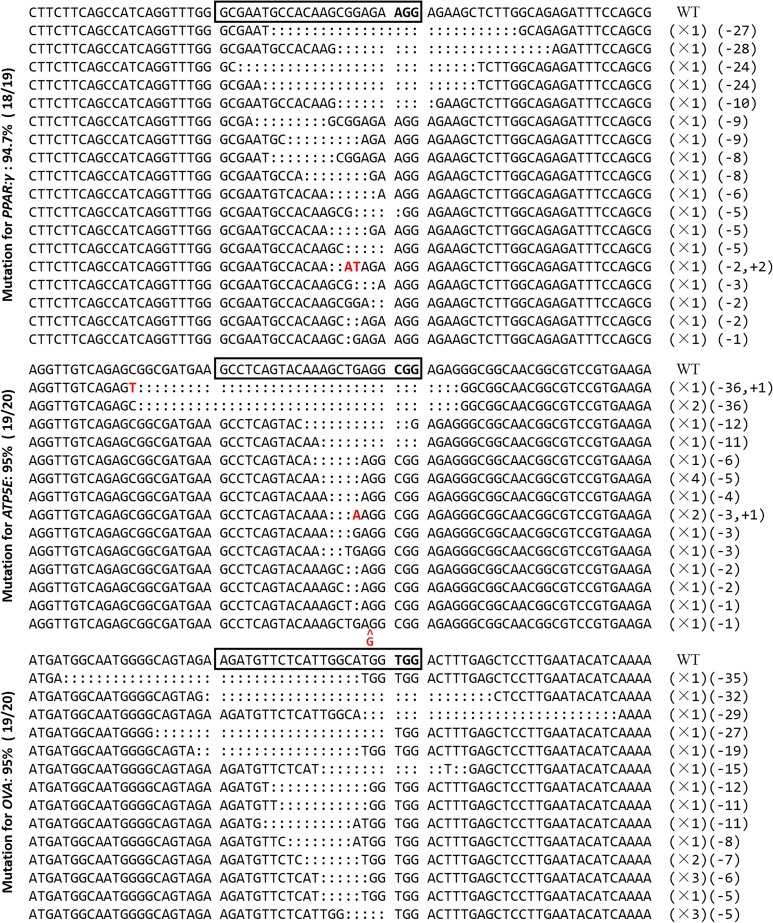
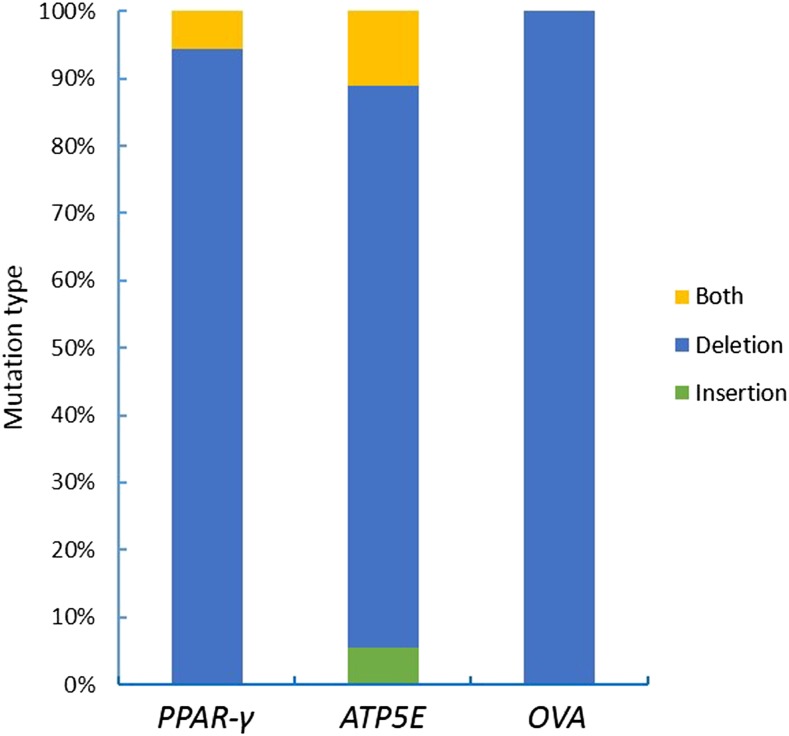
In order to obtain more mutant cells, we used the PuroR gene for fast and efficient enrichment of genetically modified cells. Puromycin was added to the medium of the experimental group and the control group; 4 d later, most of the cells in the control group were dead, and a fraction of the cells were alive in the experimental group (Figure 4). The remaining cells in the experimental group were harvested for sequencing analysis. T7E1 assay showed mutation frequencies within the PPAR-γ, ATP5E, and OVA loci for the screening cells of 60.7%, 61.3%, and 47.3%, respectively (Figure 4). However, since the T7E1 assay tends to underestimate fold enrichment (Kim et al. 2011), we subsequently sequenced the mutation site by cloning the PCR products in the T vector. The results showed different mutations at three target sites; efficiencies of up to 95% were observed for the three genes (Figure 5). This showed that the mutation efficiency was improved greatly by puromycin screening with the surrogate reporter. Most of the mutations were deletions, with only a few mutations being insertions, or both deletion and insertion (Figure 6). These data demonstrated that the selected gRNAs, combined with the surrogate reporter, worked efficiently in chicken genomes, and that deletion was the main type of mutation.
Figure 4.
Puromycin sensitivity after puromycin was added. Most of the cells from the control group died, and a portion of cells in the experimental group were alive after puromycin addition for 2 and 4 d. These results indicated that the CRISPR/Cas9 system worked in DF-1 cells. The control group shows cells transfected with Cas9 only and the corresponding RPG reporter. The experimental group refers to cells transfected with CRISPR/Cas9 and the corresponding RPG reporter.
Figure 5.
Sequencing results of mutations induced by CRISPR/Cas9 combined with an SSA-RPG reporter based on puromycin-screened cells. Boxes indicate target sites for CRISPR/Cas9 system. Dashes and red letters indicate deleted and inserted base pairs. X1, X2 ,X3, and X4 indicate the number of each clone.
Figure 6.
Mutation types in DF-1 cells induced by CRISPR/Cas9 combined with the SSA-RPG reporter.
Detection of an off-target effect
To test whether an off-target effect occurred in these puromycin-resistant cells, we predicted a total of nine potential off-target sites for PPAR-γ, ATP5E, and OVA. There were no detectable off-target mutations in these loci using the T7E1 assay (Figure 7).
Figure 7.
T7E1 assay of nine potential off-target effects. (A) Off-target effect for PPAR-γ.CRISPR/Cas9. One, 3, and 5 indicate the control group without puromycin screening for GNS, EHD3, and ADCYAP1, respectively. Two, 4, and 6 indicate the experimental group with puromycin screening for GNS, EHD3, and ADCYAP1, respectively. (B) Off-target effect for ATP5E.CRISPR/Cas9. One, 3, and 5 indicate the control group without puromycin screening for KCNT2, DAD1, and PITPNM2, respectively. Two, 4, and 6 indicate the experimental group with puromycin screening for KCNT2, DAD1, and PITPNM2, respectively. (C) Off-target effect for OVA.CRISPR/Cas9. One, 3, and 5 indicate the control group without puromycin screening for ABCC9, TXNDC12, and LCP2, respectively. Two, 4, and 6 indicate the experimental group with puromycin screening for ABCC9, TXNDC12, and LCP2, respectively (M, Plus2k marker).
Discussion
CRISPR/Cas9 depends on small RNAs for sequence cleavage, and only a programmable RNA is required to generate sequence specificity (Jinek et al. 2012). Therefore, CRISPR/Cas9-mediated genome engineering is easy to handle, highly specific, efficient, and multiplex (Mali et al. 2013a). It has been used widely in many organisms and cells since the demonstration of the site-specific cleavage function in vitro (Mali et al. 2013b; Cong et al. 2013). Here, we showed that the CRISPR/Cas9 system combined with a surrogate reporter can target chicken DF-1 cells efficiently.
The CRISPR/Cas9 system used in this study was from S. thermophilus (Xu et al. 2015), and the Cas9 gene in this system was codon humanized. The stCRISPR/Cas9 system provided the ability to cleave the target site effectively in chicken DF-1 cells. Véron and colleagues efficiently targeted the PAX7 gene in the chicken embryo using mammalian codon-optimized Cas9 (Véron et al. 2015), thus indicating the high similarity of codon usage between mammals and chickens. However, the mutation efficiency was very low according to T7E1 assay analysis. Therefore, we needed to remove the unwanted cells from the total cells so that we could enrich for the mutant cells. CRISPR/Cas9-modified cells can be enriched by selecting puromycin-resistant cells using a single vector expressing puromycin (Shalem et al. 2014; Yuen et al. 2015; Ran et al. 2013b). However, the activity of the nuclease cannot be assured using this single vector system, since this method enriches the transfected cells. In our surrogate system, the disrupted PuroR gene, fused with eGFP gene of the SSA-RPG reporter, would be corrected via the SSA repair mechanism when the CRISPR/Cas9 nuclease cleaves the target site of the surrogate reporter. So, the puromycin-resistant cells would be eGFP+ cells. A target sequence on a surrogate reporter could reflect nuclease activity in the same cell (Kim et al. 2011, 2014; Ramakrishna et al. 2014; Ren et al. 2015). Therefore, we could monitor CRISPR/Cas9 activity in live cells by fluorescent microscopy. Furthermore, enrichment can also be achieved with fluorescence activated cell sorting (FACS) by evaluating the fraction of eGFP+ cells alongside puromycin screening. The repeat length around the SSA-RPG surrogate reporter used in this reporter was 200 bp because the repair efficiency is much higher for the SSA-RPG reporter with a 200 bp repeat length (Ren et al. 2015). Surprisingly, it was found that the mutation efficiency increased to approximately 95% for these genes in the DF-1 cell line after puromycin screening. This was higher than the mutation efficiency observed in HEK293T cells (Ren et al. 2015). There are several probable reasons for the high efficiency using this system in DF-1 cells. First, DF-1 cells grow more slowly than HEK293T cells, so the Cas9 nuclease could work effectively in almost every CRISPR/Cas9 transfected DF-1 cell. Second, the SSA-mediated repair efficiency in the DF-1 cell line was probably higher than that in HEK293T cells, so we obtained more puromycin-resistant cells. Third, the large T-antigen of simian virus 40 (SV40) gene probably could influence the expression of CRISPR/Cas9 because HEK293T cells are stably transfected with the large T-antigen gene of SV40, while the DF-1 cell line was developed spontaneously from fibroblasts of chicken embryo (Himly et al. 1998). Among the mutations observed, deletion was the main type. These results are consistent with those found in transgenic chickens induced by TALEN (Park et al. 2014), and somatic cells induced by the CRISPR/Cas9 nuclease (Veron et al. 2015). However, the mechanisms require further study.
DNA damage can cause erroneous changes in the genetic code, leading to increased mutation load or wider-scale genome aberration that can threaten cell or organism viability (Jackson and Bartek 2009; Roos and Kaina 2013). So, off-target mutations remain a major concern for CRISPR/Cas9 (Cho et al. 2014; Tsai et al. 2015; Fu et al. 2013; Pattanayak et al. 2013; Veres et al. 2014; Zhang et al. 2015) because it is likely to cause unwanted chromosomal rearrangements (Cho et al. 2014). The potential off-target effect must not be ignored. Several methods could reduce off-target effects, such as considering the tolerance of mismatches and PAM mutations (Fu et al. 2013), controlling the dosage of CRISPR/Cas9 (Hsu et al. 2013; Pattanayak et al. 2013), and using paired Cas9 nickase (Shen et al. 2014; Ran et al. 2013a), or truncated gRNA (Fu et al. 2014). In this study, like most of the studies cited above, an off-target effect was not detected in the selected potential off-target sites by T7E1 assay. But T7E1 assay cannot detect off-target mutations that occur at frequencies < 1% due to its poor sensitivity (Kim et al. 2009). In order to detect off-target effect more reliably, several other methods, such as deep sequencing (Cho et al. 2014), high-throughput genomic translocation sequencing (HTGTS) (Frock et al. 2015), Genome-wide Unbiased Identification of DSBs Enabled by Sequencing (GUIDE-seq) (Tsai et al. 2015), etc. could be used.
In conclusion, we showed that the CRISPR/Cas9 system, combined with an SSA-mediated surrogate reporter, can efficiently target chicken DF-1 cells. This will help to study the function of chicken genes both efficiently and cheaply.
Acknowledgments
This work was supported by the National Science and Technology Major Project of China [2014ZX0801009B], and Fundamental Research Funds for the Central Universities (2452015150). The authors declare that no competing interests exist.
Footnotes
Communicating editor: R. A. Sclafani
Literature Cited
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., et al. , 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712. [DOI] [PubMed] [Google Scholar]
- Cho S. W., Kim S., Kim Y., Kweon J., Kim H. S., et al. , 2014. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 24: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frock R. L., Hu J., Meyers R. M., Ho Y. J., Kii E., et al. , 2015. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 33: 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Foden J. A., Khayter C., Maeder M. L., Reyon D., et al. , 2013. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31: 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Sander J. D., Reyon D., Cascio V. M., Joung J. K., 2014. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Shao Y., Li D., Liu M., 2014. Generation of site-specific mutations in the rat genome via CRISPR/Cas9. Methods Enzymol. 546: 297–317. [DOI] [PubMed] [Google Scholar]
- Guschin D. Y., Waite A. J., Katibah G. E., Miller J. C., Holmes M. C., et al. , 2010. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 649: 247–256. [DOI] [PubMed] [Google Scholar]
- Himly M., Foster D. N., Bottoli I., Iacovoni J. S., Vogt P. K., 1998. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248: 295–304. [DOI] [PubMed] [Google Scholar]
- Hsu P. D., Scott D. A., Weinstein J. A., Ran F. A., Konermann S., et al. , 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31: 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S. P., Bartek J., 2009. The DNA-damage response in human biology and disease. Nature 461: 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., et al. , 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Lee H. J., Kim H., Cho S. W., Kim J. S., 2009. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 19: 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Um E., Cho S. R., Jung C., Kim H., et al. , 2011. Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nat. Methods 8: 941–943. [DOI] [PubMed] [Google Scholar]
- Kim H., Kim M. S., Wee G., Lee C. I., Kim H., et al. , 2013. Magnetic separation and antibiotics selection enable enrichment of cells with ZFN/TALEN-induced mutations. PLoS One 8: e56476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. H., Ramakrishna S., Kim H., Kim J. S., 2014. Enrichment of cells with TALEN-induced mutations using surrogate reporters. Methods 69: 108–117. [DOI] [PubMed] [Google Scholar]
- Li J., Zhang B., Bu J., Du J., 2015. Intron-based genomic editing: a highly efficient method for generating knockin zebrafish. Oncotarget 6: 17891–17894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Teng F., Li T., Zhou Q., 2013. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat. Biotechnol. 31: 684–686. [DOI] [PubMed] [Google Scholar]
- Mali P., Esvelt K. M., Church G. M., 2013a Cas9 as a versatile tool for engineering biology. Nat. Methods 10: 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., et al. , 2013b RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura H., Gurumurthy C. B., Sato T., Sato M., Ohtsuka M., 2015. CRISPR/Cas9-based generation of knockdown mice by intronic insertion of artificial microRNA using longer single-stranded DNA. Sci. Rep. 5: 12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H., Kennedy Z., Anderson D. G., Yin H., Xue W., 2015. Precision cancer mouse models through genome editing with CRISPR-Cas9. Genome Med. 7: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T. S., Lee H. J., Kim K. H., Kim J. S., Han J. Y., 2014. Targeted gene knockout in chickens mediated by TALENs. Proc. Natl. Acad. Sci. USA 111: 12716–12721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V., Lin S., Guilinger J. P., Ma E., Doudna J. A., et al. , 2013. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31: 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna S., Cho S. W., Kim S., Song M., Gopalappa R., et al. , 2014. Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nuclease-induced mutations. Nat. Commun. 5: 3378. [DOI] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Lin C. Y., Gootenberg J. S., Konermann S., et al. , 2013a Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., et al. , 2013b Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C., Xu K., Liu Z., Shen J., Han F., et al. , 2015. Dual-reporter surrogate systems for efficient enrichment of genetically modified cells. Cell. Mol. Life Sci. 72: 2763–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos W. P., Kaina B., 2013. DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 332: 237–248. [DOI] [PubMed] [Google Scholar]
- Ruan J., Li H., Xu K., Wu T., Wei J., et al. , 2015. Highly efficient CRISPR/Cas9-mediated transgene knockin at the H11 locus in pigs. Sci. Rep. 5: 14253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago Y., Chan E., Liu P. Q., Orlando S., Zhang L., et al. , 2008. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 105: 5809–5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O., Sanjana N. E., Hartenian E., Shi X., Scott D. A., et al. , 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343: 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Zhang W., Zhang J., Zhou J., Wang J., et al. , 2014. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods 11: 399–402. [DOI] [PubMed] [Google Scholar]
- Tsai S. Q., Zheng Z., Nguyen N. T., Liebers M., Topkar V. V., et al. , 2015. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33: 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veres A., Gosis B. S., Ding Q., Collins R., Ragavendran A., et al. , 2014. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell 15: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veron N., Qu Z., Kipen P. A., Hirst C. E., Marcelle C., 2015. CRISPR mediated somatic cell genome engineering in the chicken. Dev. Biol. 407: 68–74. [DOI] [PubMed] [Google Scholar]
- Wang X., Yu H., Lei A., Zhou J., Zeng W., et al. , 2015. Generation of gene-modified goats targeting MSTN and FGF5 via zygote injection of CRISPR/Cas9 system. Sci. Rep. 5: 13878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanzel M., Vischedyk J. B., Gittler M. P., Gremke N., Seiz J. R., et al. , 2016. CRISPR-Cas9-based target validation for p53-reactivating model compounds. Nat. Chem. Biol. 12: 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Ren C., Liu Z., Zhang T., Zhang T., et al. , 2015. Efficient genome engineering in eukaryotes using Cas9 from Streptococcus thermophilus. Cell. Mol. Life Sci. 72: 383–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Jao L. E., Chen W., 2015. Generation of targeted mutations in zebrafish using the CRISPR/Cas system. Methods Mol. Biol. 1332: 205–217. [DOI] [PubMed] [Google Scholar]
- Yuen K. S., Chan C. P., Wong N. H., Ho C. H., Ho T. H., et al. , 2015. CRISPR/Cas9-mediated genome editing of Epstein-Barr virus in human cells. J. Gen. Virol. 96: 626–636. [DOI] [PubMed] [Google Scholar]
- Zhang X. H., Tee L. Y., Wang X. G., Huang Q. S., Yang S. H., 2015. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids 4: e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Wang L., Du Y., Xie F., Li L., et al. , 2016. Efficient generation of gene-modified pigs harboring precise orthologous human mutation via CRISPR/Cas9-induced homology-directed repair in zygotes. Hum. Mutat. 37: 110–118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.