Therapy with tyrosine kinase inhibitors (TKIs) is sufficient to prevent the progression to advanced chronic myeloid leukemia (CML) and inhibit recurrence. This has revolutionized the management of CML; but to maintain such “operational cure,” the Imatinib Mesylate (IM) is required indefinitely, incurring considerable cost and side effects due to the therapy.1
Based on these considerations, it would be beneficial to be able to identify patients who can stop taking the TKIs, remaining in treatment-free remission (TFR). In attempts to achieve this goal, STOP IM clinical trials (STIM, TWISTER, A-STIM) reported an average of 54% TFR at 2 y in patients stopping therapy after 2 y in remission based on mRNA qRT-PCR assays.2,3 The parameters of molecular response and molecular recurrence based on qRT-PCR results have been redefined during the trials, and the rate of TFR changed from 44% (41% STIM and 47% TWISTER) to 64% (A-STIM). The eligibility requirement for enrollment for STIM and TWISTER was undetectable minimal residual disease (UMRD) for at least 2 y by qRT-PCR (4.5 log of molecular response, MR4.5 or BCR-ABL1 ≤ 0.0032%). Molecular recurrence was defined by 2 consecutive samples that showed BCR-ABL1 positivity and a fold10- increase in the STIM, and by 2 consecutive positive samples independent of the fold-increase in the TWISTER The A-STIM study employed less stringent parameters: they included in the study patients with sporadic levels of BCR-ABL1 in the 2 y before therapy stopped, and the loss of major molecular response (MMR, BCR-ABL1>0,1%) in TFR. Based on the STIM definition, the rate of TFR at 2 y was 46%, and with the new standards was 64%.
Evidently the most decisive definition of molecular response and molecular recurrence is critical to plan safe discontinuation of TKI; but as shown, a definition based only on qRT-PCR can result in variable outcomes.
We proposed gDNA Q-PCR for the routine clinical monitoring of CML in addition to conventional qRT-PCR, thus redefining the concept of minimal residual disease (MRD) and potentially better identifying patients eligible for withdrawal of drug. gDNA Q-PCR is a patient-specific assay based on breakpoint sequence, and allows the calculation of the number of BCR-ABL1 positive cells independent of their transcriptional status.4,5
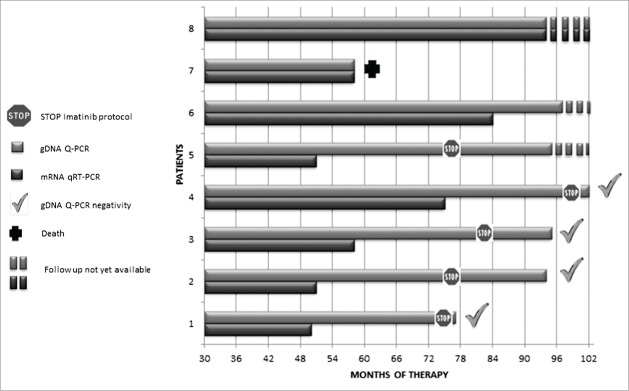
We monitored 8 chronic phase CML patients by both mRNA qRT-PCR and gDNA Q-PCR techniques for an average period of 90 months. In get information about the pathogenesis of CML we separated cell populations from bone marrow at the last follow-up. The analysis of cell sub-types could be very useful when we are at the limit of the sensitivity of the method, in order to enrich Ph-positive cells from the stem cell compartment.6
We confirmed the sensitivity of the gDNA Q-PCR technique, measuring positive levels of gDNA in all BCR-ABL1 mRNA positive samples. Notably, we detected Ph-positive cells by gDNA Q-PCR in 32.8% of samples that were negative for mRNA, and we never found any samples negative by gDNA Q-PCR that was positive for mRNA.7
According to the STIM protocol, we identified 5/8 patients with UMRD for at least 2 consecutive years as candidates eligible to withdrawal of therapy.
Furthermore, on the basis of gDNA Q-PCR, leukemic cells were present in all patients, and 4 out of 5 become negative by both techniques only an average of 34 months later. In addition, we confirmed the absence of leukemic clones by testing the CD34+ sub-fraction from bone marrow. A single sample showed gDNA positivity in all follow-ups, with BCR-ABL1 positivity in the CD34+ sorted cells. The CD34+ cells that scored positive could be a predictor of relapse, and thus in this case discontinuation of therapy is not advisable.
The gDNA technique is advantageous to monitor Ph-positive cells in the stem cells compartment because it is independent of their transcriptional status and can more conclusively indicate whether patients can safely stop the therapy. Its application is technically arduous because it requires the identification of the breakpoint and a customized patient-specific assay. However, the gDNA is not susceptible to degradation as is mRNA, facilitating the use of amplification and sequencing to infer the necessary sequence rapidly using standardized protocols.
In conclusion gDNA Q-PCR can identify residual leukemia in many patients who would be judged disease-free by the conventional method of mRNA qRT-PCR We thus propose to include gDNA negativity in peripheral blood and in bone marrow selected CD34+ cell as an additional criterion to enroll CML patient in STOP TKIs protocols.
References
- 1. Goldman J, Gordon M. Leuk Lymphoma 2006; 47: 1-7; PMID:16321820; http://dx.doi.org/ 10.1080/10428190500407996 [DOI] [PubMed] [Google Scholar]
- 2. Mahon FX, et al. . Lancet Oncol 2010; 11: 1029-35; PMID:20965785; http://dx.doi.org/ 10.1016/S1470-2045(10)70233-3 [DOI] [PubMed] [Google Scholar]
- 3. Ross DM, Hughes TP. Br J Haematol 2014; 166: 3-11; PMID:24754670; http://dx.doi.org/ 10.1111/bjh.12892 [DOI] [PubMed] [Google Scholar]
- 4. Mattarucchi E, et al. . Genes Chromosomes Cancer 2008; 47(7): 625-32; PMID:18398823; http://dx.doi.org/ 10.1002/gcc.20568 [DOI] [PubMed] [Google Scholar]
- 5. Mattarucchi E. et al J Mol Diagn 2009; 11(5): 482-7; PMID:19710400; http://dx.doi.org/ 10.2353/jmoldx.2009.080150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Michor F, et al. . Nature 2005; 30: 435(7046):1267-70; PMID:15988530; http://dx.doi.org/ 10.1038/nature03669 [DOI] [PubMed] [Google Scholar]
- 7. Pagani Ilaria S, et al. Oncoscience 2014; 1(7): 510–521; http://dx.doi.org/ 10.18632/oncoscience.65 [DOI] [PMC free article] [PubMed] [Google Scholar]