Abstract
Seven-in-absentia homolog (SIAH) proteins are evolutionary conserved RING type E3 ubiquitin ligases responsible for the degradation of key molecules regulating DNA damage response, hypoxic adaptation, apoptosis, angiogenesis, and cell proliferation. Many studies suggest a tumorigenic role for SIAH2. In breast cancer patients SIAH2 expression levels correlate with cancer aggressiveness and overall patient survival. In addition, SIAH inhibition reduced metastasis in melanoma. The role of SIAH1 in breast cancer is still ambiguous; both tumorigenic and tumor suppressive functions have been reported. Other studies categorized SIAH ligases as either pro- or antimigratory, while the significance for metastasis is largely unknown. Here, we re-evaluated the effects of SIAH1 and SIAH2 depletion in breast cancer cell lines, focusing on migration and invasion. We successfully knocked down SIAH1 and SIAH2 in several breast cancer cell lines. In luminal type MCF7 cells, this led to stabilization of the SIAH substrate Prolyl Hydroxylase Domain protein 3 (PHD3) and reduced Hypoxia-Inducible Factor 1α (HIF1α) protein levels. Both the knockdown of SIAH1 or SIAH2 led to increased apoptosis and reduced proliferation, with comparable effects. These results point to a tumor promoting role for SIAH1 in breast cancer similar to SIAH2. In addition, depletion of SIAH1 or SIAH2 also led to decreased cell migration and invasion in breast cancer cells. SIAH knockdown also controlled microtubule dynamics by markedly decreasing the protein levels of stathmin, most likely via p27Kip1. Collectively, these results suggest that both SIAH ligases promote a migratory cancer cell phenotype and could contribute to metastasis in breast cancer.
Keywords: breast cancer, invasion, metastasis, migration, p27Kip1, SIAH ubiquitin ligases, seven-in-absentia, SIAH1, SIAH2, stathmin
Abbreviations
- BrdU
bromodeoxyuridine
- CAMKII
Ca2+/Calmodulin-dependent kinase II
- CDK
Cyclin-dependent Kinase
- DCC
deleted in colorectal cancer
- DOX
Doxorubicin
- EB3
Microtubule End-Binding protein 3
- ER
estrogen receptor
- HIF1α
Hypoxia-Inducible Factor 1α
- ERK
Extracellular Signal-Regulated Kinase
- HIPK2
Homeodomain-Interacting Protein Kinase 2
- MEFs
mouse embryonic fibroblasts
- N-CoR
nuclear receptor corepressor
- PHD
prolyl-hydroxylase domain protein
- PML
promyelocytic leukemia protein
- PP1
protein phosphatase 1
- Rb
retinoblastoma protein
- SIAH
seven-in-absentia homolog
- TRAF2
tumor necrosis factor receptor-associated factor 2
- VEGF
vascular endothelial growth factor
- VHL
von Hippel-Lindau protein.
Background
SIAH proteins are evolutionary conserved RING-type E3 ubiquitin ligases emerging as critical regulators in both normal development and cancer. SIAH proteins exert their primary functions by targeting selected proteins for proteasomal degradation by polyubiquitination.1 Whereas initial reports pointed to a role in tumor suppression,2 most recent studies indicate that SIAH proteins, especially SIAH2, are tumorigenic proteins and promote tumor cell proliferation in several tissues, although the molecular mechanisms have not yet been fully elucidated. Increased expression of SIAH has been reported in different human cancers such as prostate, lung and breast.3-5 Recently SIAH has also emerged as a tumor-specific biomarker in pancreatic cancer.6 The Cancer Genome Atlas lists the SIAH2 gene as amplified in many human tumors (e.g., in 30% of lung squamous cell carcinoma).7,8 Both SIAH1 and SIAH2 were reported to increase proliferation of liver cancer cells9,10 and SIAH2 expression was reported to promote tumor growth in head and neck tumors.11 SIAH inhibition approaches have produced promising results in preclinical mouse models of lung cancer,3 melanoma,12 and pancreatic cancer.13
Numerous substrates14 have been identified that mediate the tumor promoting effects of SIAH1 and SIAH2 in Ras, estrogen, DNA-damage, and hypoxia response pathways, as summarized in reference 15. Interestingly, SIAH proteins are central regulators of the hypoxic adaptation.16,17 The prolyl hydroxylase domain proteins PHD1, PHD2, and PHD3 mainly act to hydroxylate HIF1α, which is a prerequisite for its von Hippel-Lindau (VHL) protein-dependent degradation. SIAH targets the PHDs for degradation via ubiquitination, thus leading to stabilization of the transcription factor HIF1α, which in turn amplifies proangiogenic factors such as Vascular Endothelial Growth Factor A (VEGF-A) and thus induces tumor vascularization, tumor growth and metastatic potential.5,17,18 SIAH proteins also target the cell fate regulator Homeodomain-Interacting Protein Kinase 2 (HIPK2) for degradation, a critical event for full induction of hypoxia-induced genes that is also associated with decreased chemosensitivity of hypoxic cells.16,19,20 SIAH proteins have been reported to mediate polyubiquitination and degradation of more than a dozen of established tumor suppressors, including Numb,21 Promyelocytic Leukemia protein (PML),22 HIPK2,20 and Deleted in Colorectal Cancer (DCC).1
Many studies on the function of SIAH proteins in cancer have been conducted in breast cancer cell lines, patient samples, or mouse models.23 Whereas high SIAH expression in patient samples was predictive for the progression of ductal carcinoma in situ to invasive breast cancer,4 SIAH inhibition has been shown to reduce tumor growth in a murine breast cancer model.24 It was recently reported that SIAH1 and SIAH2 genes were amplified in samples from breast cancer patients by 17% and 10%, respectively.25 Similar to other cancer entities, SIAH2 primarily shows tumorigenic functions in breast cancer: SIAH2 knockout mice show delayed tumor onset and reduced infiltration in a spontaneous mouse breast cancer model,26 SIAH2 silencing reduced breast tumor growth in vivo,27 it is upregulated in basal-like breast cancer and its expression correlates with increased tumor aggressiveness.28 The role for SIAH1 in breast cancer remains less well described. In contrast to other cancer types, only few reports identify SIAH1 as a pro-tumorigenic protein in breast cancer similar to SIAH2,24,29 most point to a tumor suppressor role for SIAH1 in breast cancer.30-36
As high SIAH2 expression correlates with increased invasiveness and decreased overall patient survival in breast cancer,4,26,28 we aimed to determine if SIAH proteins play a role in breast cancer cell migration and metastasis. To date, the effects of SIAH inhibition, or silencing, on breast cancer metastasis or migration have not been reported; and also in other cancer types the general role of SIAH proteins in metastasis is not clear. For example, high SIAH2 expression correlated with metastasis in liver cancer,10 and SIAH inhibition strongly reduced metastasis in a syngeneic melanoma mouse model,18 yet anti-metastatic actions of SIAH1/2 have also been reported.37,38 Cancer cell migration and invasion are key components necessary for metastasis. Cell motility is mainly controlled by the Actin cytoskeleton, which provides the driving force at the leading edge of the cell, and the microtubule network that ensures rear retraction and controls protrusive and contractile forces. Actin and microtubule dynamics are highly cross-linked, regulating each other and being affected by adhesion and polarization.39,40 Both SIAH1 and SIAH2 were reported to promote migration of liver cancer cells,9,10 and SIAH1 silencing inhibited glioblastoma cell migration and invasion under hypoxia.41 Nevertheless, results from other groups indicate that SIAH1 exerts antimigratory activities in squamous cell carcinoma,42 neuronal cells,43 and mouse embryonic fibroblasts (MEFs).44
In this study we re-examined the role of SIAH1 and SIAH2 in breast cancer cell apoptosis, migration, and invasion. Our results support a tumor promoting role for both SIAH1 and SIAH2 in breast cancer cells.
Results
Regulation of hypoxic adaptation by SIAH1/2 in breast cancer cells
Initially, we determined the expression of SIAH1 and SIAH2 in various breast cancer cell lines. Although to different levels, SIAH1 and SIAH2 are expressed in MCF10A breast epithelial cells as well as MCF7, T47D, MDA-MB-231, and MDA-MB-468 breast cancer cells (Fig. 1A). Since MCF7 cells showed strong protein expression of both SIAH1 and SIAH2, we researched the effects of SIAH1 and SIAH2 depletion primarily in this cell model. To silence SIAH1 and SIAH2 in MCF7, we used siRNAs that had been previously published to work both potently and selectively.20,45-47 Knockdown of SIAH1 and SIAH2 with these siRNAs was confirmed both on mRNA level (Fig. S1A) and protein level (Fig. 1B).
Figure 1.
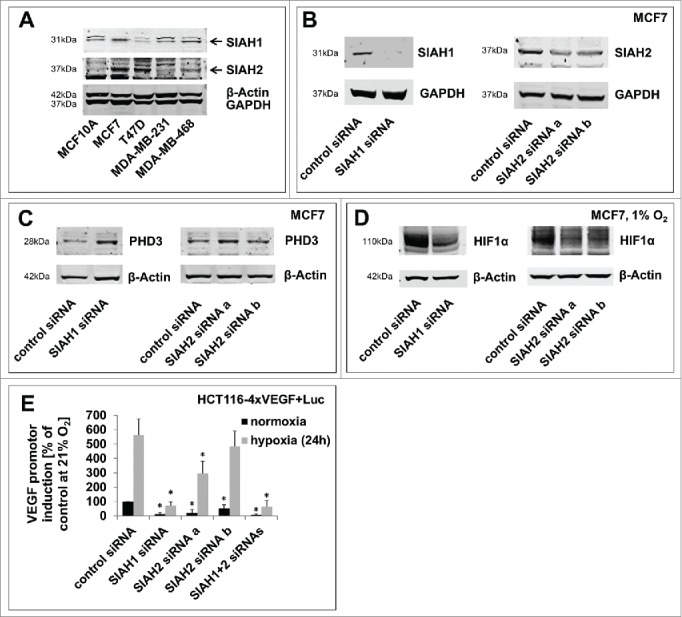
SIAH1/2 silencing reduces hypoxic adaptation in breast cancer cells. (A) Comparison of SIAH1 and SIAH2 expression levels in different breast cancer cell lines. Four breast cancer cell lines and MCF10A as a non-cancer control cell line were lysed and immunoblotted for SIAH1 expression. The membrane was reprobed for SIAH2. GAPDH and β-Actin serve as a loading control. (B) Efficient SIAH knockdown in MCF7 cells. MCF7 breast cancer cells were transfected with siRNAs targeting SIAH1 or SIAH2. After 48 h the cells were lysed and Western Blot was performed with specific antibodies. Instead of reprobing the membrane was cut in advance to allow clear visualization of the results. (C) Lysates of MCF7 cells silenced for expression of SIAH1 or SIAH2 were examined for expression of PHD3. (D) MCF7 cells were transfected with SIAH1 and SIAH2 siRNA and cultivated at 1% O2 for 48 h. Lysates were probed for HIF1α accumulation. (E) SIAH1 and SIAH2 were silenced in HCT116–4xVEGF+Luc cells for 24 h. The cells were then incubated at 21% O2 or 1% O2 for another 24 h, lysed, and a luciferase assay was performed to assess HIF target gene expression. Relative light units from the luciferase assay were normalized to the cell number measured with CellTiter-Blue Cell Viability assay. Bar graphs show mean values, error bars indicate SD. Data were analyzed using 1-way ANOVA followed by Dunnett's post-hoc test. n = 3; *, p<0.05.
To investigate the impact of SIAH ligase silencing on signaling in breast cancer cells, we looked at protein levels of known SIAH substrates. First, we determined the protein levels of the Tumor Necrosis Factor (TNF) receptor-associated factor 2 (TRAF2), a well-known SIAH substrate involved in TNF receptor signal transduction. TRAF2 was previously published to be a substrate of SIAH2, but not of SIAH1;48 consistently we found increased TRAF2 levels upon silencing of SIAH2, but not SIAH1 (Fig. S2A).
The pro-tumorigenic actions of SIAH1 and SIAH2 have been attributed to the regulation of PHD proteins, of which PHD3 is most strongly regulated by SIAH.17 PHD3 levels were increased after silencing of either SIAH1 or SIAH2 (Fig. 1C). Consistently, HIF1α accumulation under hypoxic conditions (1% O2) was decreased in SIAH1 and SIAH2 silenced cells (Fig. 1D). In this regard, SIAH1 and SIAH2 showed redundant function in MCF7 breast cancer cells, as previously reported in other cell lines.17,24,41,49 To investigate the effects of SIAH knockdown downstream of HIF1α, we employed a cell line which expresses luciferase under control of a VEGF-promotor (HCT116–4xVEGF+Luc50) to monitor HIF1α activity in a luciferase assay. It was confirmed that silencing of SIAH1 and SIAH2 had a comparable effect on the HIF downstream targets in this cell line as in MCF7 cells (Fig. S2B). Individual silencing of SIAH1, SIAH2, or in combination led to a significant decrease in VEGF promotor activation both under hypoxic and normoxic conditions (Fig. 1E).
Figure 2.
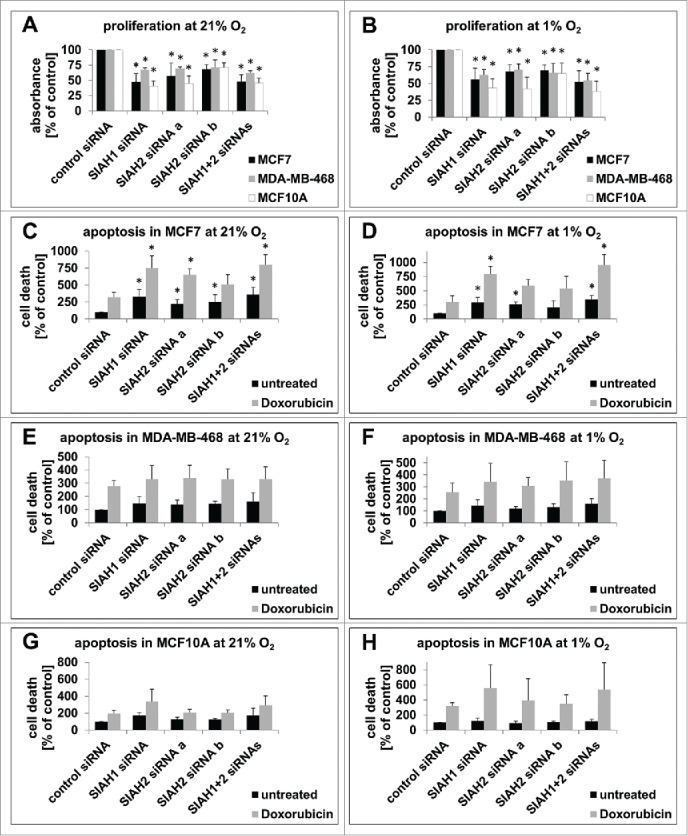
SIAH1/2 silencing inhibits proliferation and promotes apoptosis in breast cancer cells. (A, B) 48 h after transfection of MCF7, MDA-MB-468, and MCF10A cells with RNAi targeting SIAH1/2, cells were incubated in presence of BrdU for 2 h. BrdU incorporation into the DNA, signifying cell division, was quantified with an ELISA. Cells were either kept at 21% O2 (A) or 1% O2 (B) for the duration of the experiment. (C, D) 30 h post RNAi transfection, MCF7 cells were either left untreated or apoptosis was stimulated by adding the DNA-damage inducing chemotherapeutic drug Doxorubicin (1 µM) for 18 h. Then cells were lysed and apoptosis was measured with a TUNEL-based ELISA. The same experiments were performed with MDA-MB-468 (E, F) and MCF10A cells (G, H). Cells were either kept at 21% O2 (C, E, G) or 1% O2 (D, F, H) the whole time. Bar graphs show mean values, error bars indicate SD. Data were analyzed using 1-way ANOVA followed by Dunnett's post-hoc test. n = 3; *, p<0.05.
SIAH1 and SIAH2 control cell proliferation and cell death in breast cancer cells
Based on the results presented in Figure 1 we concluded that SIAH1 and SIAH2 have similar functions in MCF7 breast cancer cells. We thus hypothesized that SIAH1 may have a tumor promoting role in breast cancer cells similar to SIAH2. If this is the case, silencing of SIAH1 should have a similar effect on breast cancer cell proliferation and apoptosis as silencing of SIAH2. To compare the role of both SIAH ligases in different types of breast cancer, SIAH1 and SIAH2 functions were assessed in luminal-like, hormone receptor positive MCF7 breast cancer cells, basal-like, triple-negative MDA-MB-468 breast cancer cells, and MCF10A mammary epithelial cells as a non-cancer control cell line. In these additional cell lines, SIAH1 and SIAH2 were also successfully silenced by RNAi transfection (Fig. S1B, C, E, F). Cell proliferation was decreased in all 3 cell lines upon silencing of SIAH1, SIAH2, or both (Fig. 2A). Comparable effects could also be observed under hypoxic conditions (1% O2) (Fig. 2B). These results show that SIAH1 silencing inhibits proliferation in breast cancer cells. It had been previously observed in MEFs, that SIAH2 mRNA transcription was increased under hypoxia,17 which could also affect the efficiency of SIAH silencing in the cell proliferation assays. Therefore we monitored if SIAH protein expression was altered in cell culture under hypoxic conditions; however, this was not the case (Fig. S2C).
We determined spontaneous apoptotic cell death in SIAH-silenced cells in comparison to control siRNA-treated cells. Knockdown of SIAH1 and SIAH2 significantly increased the fraction of apoptotic cells in MCF7 (Fig. 2C). Similar results were observed in cells grown under hypoxic conditions (Fig. 2D). The influence of SIAH ligases on apoptosis has been linked to their role in DNA-damage response. Together with HIPK2, a substrate of both SIAH1 and SIAH2, SIAH ligases promote recovery from DNA-damage.16,20,45 We cultivated the cells in presence of the DNA-damaging drug Doxorubicin, a chemotherapeutic drug widely used in adjuvant breast cancer therapy.51 Doxorubicin treatment increased apoptosis in all cell lines tested (Fig. 2C-H). Cell death was even further increased in SIAH-silenced MCF7 cells. This could be detected when the cells were kept at 1% O2 as well as under normoxic conditions (Fig. 2C, D). These results indicate that SIAH1 and SIAH2 can inhibit apoptosis in MCF7 cells, as it has been shown before in other cancer cell types.52,53 However silencing of SIAH1 and SIAH2 did not yield a significant increase in apoptosis in MDA-MB-468 or MCF10A cells (Fig. 2E-H), similar to what has been described for SIAH-deficient mouse embryonic fibroblasts.54 Also the effect of Doxorubicin was not further increased by SIAH1 or SIAH2 silencing in these cell lines, neither at normoxic nor hypoxic conditions.
Role of ER receptor status in SIAH-controlled sensitivity of breast cancer cells
A possible explanation why MCF7 cells are more sensitive to the SIAH knockdown is their hormone receptor status. In contrast to MDA-MB-468 and MCF10A cells, the MCF7 cell line is estrogen receptor (ER)-positive.55 ER signaling is elicited by estradiol, which is present in the MCF7 culture medium, and is negatively regulated by the Nuclear receptor Corepressor (N-CoR).56-58 Estradiol can inhibit apoptosis in MCF7 cells59 and activate SIAH2.60 Both SIAH1 and SIAH2 promote N-CoR degradation,61,62 thereby in turn increasing estrogen signaling. Under these conditions, ER signaling and SIAH form a positive feedback loop, inducing high SIAH2 upregulation in ER-positive breast cancer.63 We determined N-CoR protein levels and confirmed increased N-CoR protein expression upon silencing of SIAH1 or SIAH2 (Fig. S3A). Silencing of N-CoR with siRNA (Fig. S3B) did not markedly change the apoptosis levels in MCF7, MDA-MB-468, or MCF10A cells (Fig. S3C). Yet, N-CoR silencing partly rescued the fraction of apoptotic cells in MCF7 silenced for expression of SIAH1, SIAH2, or both (Fig. S3D). This could also be observed after stimulation with Doxorubicin (Fig. S3E), suggesting that the N-CoR silencing partly counteracts the effects of the SIAH knockdown. These results indicate that SIAH silencing stabilizes N-CoR and thereby decreases antiapoptotic ER signaling. This effect contributes to the increased apoptosis observed upon SIAH silencing in ER-positive breast cancer cells.
To elaborate on the hypothesis that the ER receptor status is relevant, we repeated the cell death assays with another ER-positive breast cancer cell line, T47D.55 Also in these cells, SIAH1 and SIAH2 were silenced successfully (Fig. S1D, G). Similar to MCF7 cells, both baseline apoptosis, as well as Doxorubicin-induced apoptosis, were increased by silencing of SIAH2 both under normoxic and hypoxic conditions (Fig. S4A, B). The effect of SIAH1 silencing did not reach significance; it could be speculated that this might be connected to the lower endogenous SIAH1 expression level compared to MCF7 cells (Fig. 1A). Additional silencing of N-CoR partly rescued the fraction of apoptotic cells in T47D silenced for expression of SIAH1, SIAH2, or both (Fig. S4C, D), similar as in MCF7 cells.
Together the results show that knockdown of SIAH1 or SIAH2 led to increased apoptosis in ER-positive breast cancer cells, and reduced cell proliferation and hypoxic adaptation in both ER-positive and ER-negative breast cancer cells. These results point to a tumor promoting role for SIAH1 in breast cancer similar to SIAH2.
Silencing of both SIAH ligases inhibits breast cancer cell migration and invasion
As the observations in breast cancer patients point to a role for SIAH ubiquitin ligases in breast cancer metastasis,28 we hypothesized that SIAH ligases promote breast cancer cell migration. We induced a defined gap in a monolayer of MCF7 cells and calculated the cell migration speed from the gap closure after 24 h. When SIAH1, SIAH2, or both were silenced, gap closure was markedly decreased (Fig. 3A). Comparable effects were found in MDA-MB-468 and MCF10A cells. Quantification of the migration velocity shows that silencing of either SIAH ubiquitin ligase is sufficient to significantly inhibit cell migration (Fig. 3B). This was also observed under hypoxia to a comparable extent (Fig. 3C). To validate these results, cell migration was also assessed in a secondary assay. Instead of quantifying random cell motility, directed cell migration toward a chemical stimulus was measured in a modified Boyden chamber assay. This assay revealed a significant reduction in chemotaxis upon silencing of SIAH1 or SIAH2, in all 3 cell lines tested (Fig. 3D).
Figure 3.
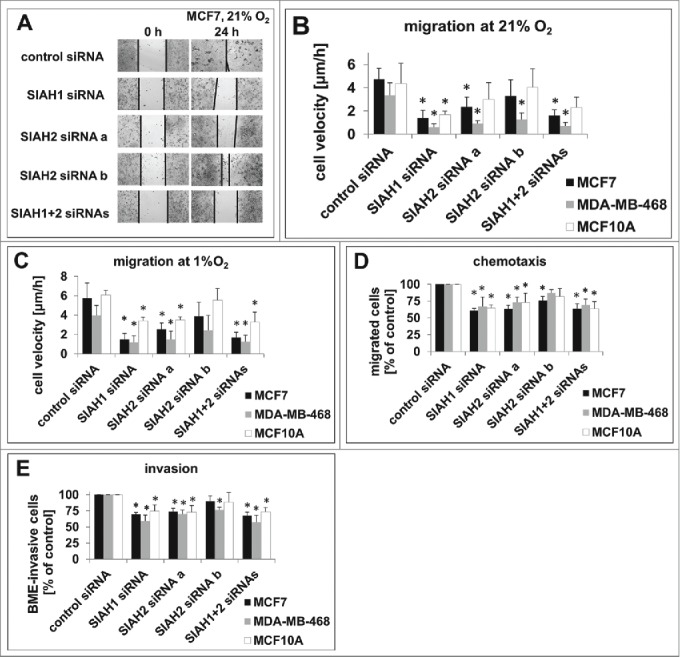
SIAH1/2 silencing inhibits migration and invasion in breast cancer cells. (A) Representative images of a wound healing assay in MCF7 cells silenced for SIAH1/2 expression. The gap was induced 48 h after RNAi transfection, cells were allowed to migrate for 24 h. Images were taken using a Zeiss Observer.Z1 microscope at 10-fold magnification. (B, C) Quantification of the cell migration speed in (A) employing 3 individual experiments with MCF7, MDA-MB-468, and MCF10A at 21% O2 (B) or 1% O2 (C). (D) Quantification of chemotaxis assays. MCF7, MDA-MB-468, and MCF10A cells were transfected with SIAH siRNAs and, after 48 h, seeded into the upper compartments of a modified Boyden chamber assay. The cells were allowed to migrate through a porous membrane toward the lower compartment filled with medium containing FCS for 24 h; the cells were then detached from the membrane undersurface, stained and quantified. (E) 48 h after RNAi transfection, the cells were seeded in a transwell coated with a rehydrated layer of basal membrane extract. Transmigrated cells were stained and quantified to assess the invasion capacity. Results are shown as means +SD. Data were analyzed using 1-way ANOVA followed by Dunnett's post-hoc test. n = 3; *, p<0.05.
Next, cell invasion was assessed in a transwell assay in which the cells migrated through a coating of basal membrane extract. The basal membrane is a continuous, sheet-like extracellular matrix between endothelial, epithelial, muscle, or neuronal cells and the adjacent stroma, and represents a first boundary for invading cancer cells.64 Silencing of SIAH1 or SIAH2 significantly decreased invasion of MCF7, MDA-MB-468, or MCF10A cells through the basal membrane extract (Fig. 3E). To confirm this result, we also performed a transendothelial migration assay, in which the cells invade through a monolayer of endothelial cells, mimicking cancer cell intravasation into blood vessels. Transmigration of breast cancer cells was markedly reduced when SIAH1 or SIAH2 was silenced (Fig. S5). Together these results indicate that silencing of both SIAH1 and SIAH2 inhibits breast cancer cell migration, adding to the effects on proliferation, apoptosis, and hypoxic adaptation.
SIAH ligases control the expression of the microtubule regulator stathmin
Unexpectedly, SIAH1 and SIAH2 significantly affect proliferation, apoptosis, and migration under both normoxic and hypoxic conditions to a comparable extent, suggesting that PHD proteins and the hypoxia-activated HIF1α signaling pathways are not the main drivers of the SIAH-mediated signaling effects. To this end, we analyzed potential links between SIAH and known migration regulators. Interestingly, SIAH2 induces the ubiquitination and degradation of Sprouty2, a negative regulator of Ras signaling.65 The small GTPase Ras activates many downstream signaling pathways, regulating cell migration by controlling Actin polymerization via Myosin Light Chain phosphorylation and proliferation through the Raf-MEK-MAPK pathway.66 SIAH ligases are also linked to microtubule dynamics via 2 substrates: p27Kip1 44 and the Microtubule Plus End-Binding protein 3 (EB3).67 p27Kip1 is an important regulator of cyclin-dependent kinases (CDKs)68 that also regulates microtubule polymerization.69 Increased p27Kip1 expression inhibits cancer cell migration by modifying microtubule dynamics in a complex with the central microtubule regulator stathmin,70 a phosphoprotein that regulates microtubule destabilization, cell cycle control,71 and cell migration.70 It has been proposed that p27 might also control stathmin expression:72 by inhibition of CDKs, p27Kip1 inhibits the phosphorylation of the Retinoblastoma (Rb) protein.73 Phosphorylated Rb is no longer able to inhibit the activity of the transcription factor E2F1,74 which drives stathmin expression.75,76
We detected Sprouty2 accumulation in MCF7 upon silencing of SIAH2, but not of SIAH1 (Fig. 4A), confirming previous reports in other cell lines.77 Consistently we found that phosphorylation of the Extracellular Signal-Regulated Kinase (ERK) 1/2 was reduced upon silencing of SIAH2, but not SIAH1, indicating decreased Ras activation. Therefore, this pathway might contribute to the effects observed, but it was not sufficient to explain why SIAH1 silencing also decreased cell migration and invasion.
Figure 4.
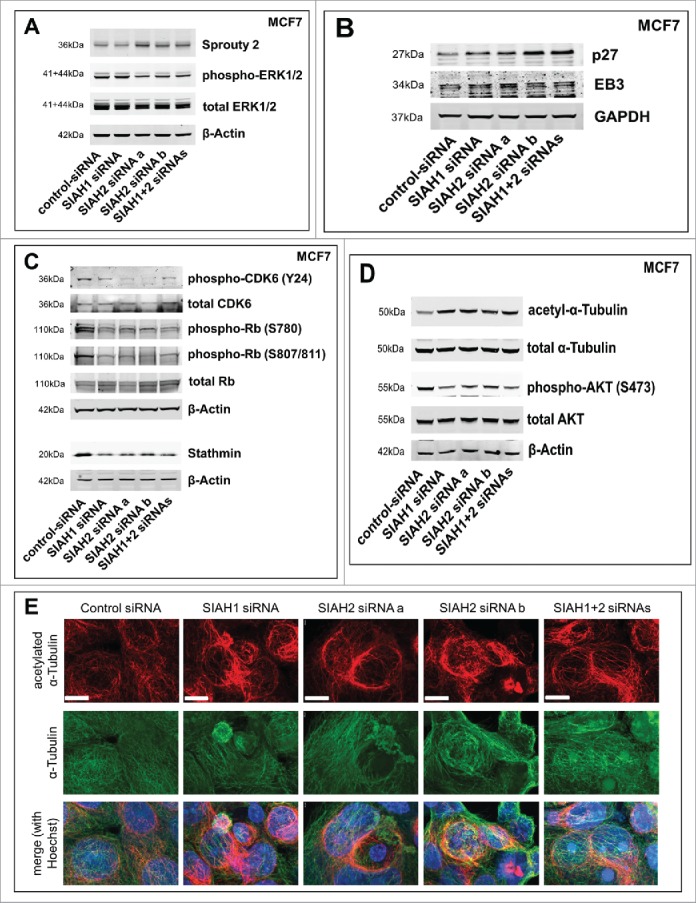
SIAH1/2 silencing affects expression of p27 and stathmin in breast cancer cells. (A) Following siRNA transfection (48 h), MCF7 cells were lysed. Western blot was performed to assess expression levels of the SIAH substrate Sprouty2 and activation status of ERK1/2. (B) Western Blot showing expression levels of the SIAH substrates p27Kip1 and EB3 after silencing of SIAH1 or SIAH2. (C) Western blot demonstrating the effect of SIAH1/2 silencing on total CDK6 levels and CDK6 phosphorylation at tyrosine 24, total Rb levels and Rb phosphorylation at serines 780 and 807/811, as well as on stathmin protein levels. (D) Western blot to determine the stability of microtubules (via acetylated α-Tubulin) as well as the activity of AKT (phosphorylation at serine 473) downstream of stathmin. (E) Immunofluorescent staining to determine microtubule stability after knockdown of SIAH1 and SIAH2. Following RNAi transfection (48 h), MCF7 cells were fixed and stained for α-Tubulin (DyLight 488, green) and acetylated α-Tubulin (Cy3, red). Cell nuclei were stained with Hoechst dye (blue). Scale bars, 10 µm.
Immunoblot analysis verified that both p27Kip1 and EB3 are affected by SIAH silencing in MCF7 breast cancer cells (Fig. 4B). Whereas EB3 expression was moderately increased by SIAH2 knockdown, p27Kip1 expression was markedly increased by silencing of either SIAH1 or SIAH2. These results indicate that p27Kip1 is not merely a SIAH1 substrate as previously shown,44 but presumably also one of SIAH2.
p27Kip1 is known to block the activating phosphorylation of several CDKs, one of which is CDK6.78 Downstream of p27Kip1, we determined the protein levels of total and phosphorylated CDK6 in MCF7 cells (Fig. 4C). CDK6 phosphorylation at tyrosine 24 was markedly decreased upon silencing of SIAH1 and SIAH2, while the total CDK levels remained unaltered. This indicates reduced CDK activity, which is consistent with increased p27Kip1 expression. Next, we determined the phosphorylation status of the CDK substrate Rb. Rb phosphorylation at serines 780, 807, and 811 inhibits sequestering of the E2F1 transcription factor.74 Our results show that Rb phosphorylation at these sites was prominently decreased upon SIAH1 or SIAH2 silencing. Total Rb protein levels remained largely unaltered. Downstream of Rb and E2F1, stathmin protein levels were markedly decreased after depletion of SIAH1 or SIAH2 (Fig. 4C), which was in line with the reduction in cell motility.
Stathmin phosphorylation can interfere with stathmin activity,71 and it has been suggested that this might be influenced by p27Kip1 as well.72 Analysis of stathmin phosphorylation revealed a complex regulation in response to SIAH silencing (Fig. S6A). The amount of serine 38-phosphorylated stathmin was not profoundly altered. In contrast, stathmin phosphorylation at serine 16 was decreased, pointing toward increased stathmin activity. Although the Ca2+/Calmodulin-dependent Kinases II and IV (CAMKII and IV) were proposed as the main kinases for stathmin activity regulation,79,80 4 additional kinases also phosphorylate Stathmin at serine 16: Aurora B kinase,81 cAMP-dependent protein kinase (PKA-C),82,83 p21-activated kinase 1 (PAK1),84 and Ribosomal protein S6 kinase α-3 (RSK2).85 We determined the activity of these kinases by testing the phosphorylation status. Exclusively phosphorylation of CAMKII β, which is responsible for 65% of the CAMKII activity,86 was markedly decreased (Fig. S6B-D), suggesting that stathmin phosphorylation at serine 16 was reduced due to decreased CAMKII activity upon SIAH silencing.
Next we determined protein levels of acetylated α-Tubulin as a measure of non-dynamic, stable microtubules.87,88 Elevated levels of acetylated α-Tubulin upon silencing of SIAH1 or SIAH2 in MCF7 cells (Fig. 4D) indicated reduced stathmin activity and increased microtubule polymerization and stability. In line with a previous report showing that stathmin activity promotes AKT activity,89 AKT phosphorylation was decreased upon SIAH silencing (Fig. 4D), further confirming that overall stathmin activity is negatively regulated by SIAH silencing. To investigate if CDK inhibition is sufficient to influence stathmin protein levels and thereby microtubule stability, we treated MCF7 cells with the pan-CDK inhibitor Roniciclib (BAY1000394). CDK inhibition resulted in decreased stathmin expression and increased α-Tubulin acetylation (Fig. S6E), which supports the hypothesis that SIAH ligases control stathmin expression and cell migration via p27Kip1.
To determine if alterations in microtubule turnover are visible looking directly at microtubules, we stained MCF7 cells for α-Tubulin and acetylated α-Tubulin (Fig. 4E). While total microtubule amount and appearance remained virtually unchanged, silencing of either SIAH1 or SIAH2 led to increased tubulin acetylation also in this setting, indicating increased microtubule stability.
Collectively, our results suggest that SIAH ligases promote cancer cell migration and invasion, at least in part, by inducing the degradation of p27Kip1 to promote microtubule depolymerization by increased stathmin expression.
Discussion
Depletion of SIAH1 and SIAH2 affects hypoxic adaptation, proliferation, migration and cell death
This report shows that SIAH1 assumes a tumor promoting role in breast cancer cells similar to SIAH2 and that both ubiquitin ligases affect cell migration. We show that, like SIAH2, SIAH1 silencing reduces hypoxic adaptation, proliferation and migration, and increases apoptosis and susceptibility to DNA-damaging chemotherapeutics in MCF7 breast cancer cells. This is in line with previous studies, which report that SIAH1 reduced PHD3 protein levels similar to that observed with SIAH2 in breast cancer24 and promotes the breast cancer stem cell phenotype.29 However it is in conflict with other reports that found SIAH1 has proapoptotic32,34-36 and antiproliferative30,32 properties in breast cancer cells. This discrepancy cannot be fully resolved, however our results show that the effects of SIAH1 silencing, especially with regards to apoptosis, are cell-line dependent (Fig. 2C-F). Additional factors, such as the hormone receptor status of the cell line and estrogen concentration of the cell culture media seem to have major impact on the biological effects of SIAH silencing. Our results suggest that SIAH silencing promotes apoptosis in estrogen-dependent MCF7 and T47D cells in part due to the accumulation of the estrogen signaling repressor N-CoR, and this can be partially rescued by N-CoR silencing. Breast cancer cell lines also show marked variation regarding SIAH1 and SIAH2 expression levels (Fig. 1A), which might influence sensitivity to SIAH knockdown as well. At least for SIAH2, this might be linked to estrogen signaling promoting SIAH2 expression.38,60 In MCF7 cells cultured without estrogen, the SIAH2 protein levels were found to be below the expression levels of MDA-MB-468 and MDA-MB-231 cells.47
Our results indicate that silencing of SIAH1 and SIAH2 inhibits both random cell motility and chemotactic migration in MCF7 and MDA-MB-468 breast cancer cells. The promigratory actions of SIAH ligases contribute to their tumor promoting role such as revealed in studies on hepatocellular carcinoma9,10 and glioblastoma.41 Also the invasive potential of breast cancer cells was significantly decreased upon SIAH1 and SIAH2 silencing. Notably, MCF7 cells and MDA-MB-468 cells represent different types of breast cancer cells and also differ in the invasive behavior. MCF7 cells have a pure luminal phenotype and are characterized by weak invasive capacity, whereas MDA-MB-468 cells show more mesenchymal characteristics and a high invasive capacity.90
The tumor promoting effects of SIAH ligases do not require hypoxic conditions
The tumor promoting effects of SIAH ligases have often been attributed to their substrates of the PHD family and subsequent influence on HIF1α activity.5,11,17,24,27,41 However, our results indicate that hypoxia is not a prerequisite for the effect of SIAH silencing on proliferation, migration and apoptosis since breast cancer cells grown under normoxic conditions show comparable effects on SIAH depletion. Thus SIAH ligases relay the tumor promoting effects through regulating signaling pathways different from the SIAH-PHD-HIF1α signaling axis. Moreover it has been shown that PHD2, which is hardly affected by SIAH, mainly controls HIF1α stability under hypoxia, while PHD1 and 3, which are profoundly degraded after SIAH-dependent ubiquitination, have a stronger influence on HIF1α stability under mild or moderate hypoxia (5-10% O2).91,92
Analysis of cell migration reveals novel mechanistic insight into tumor promoting effects of SIAH ligases
This is the first report to show that silencing of SIAH ligases inhibits the expression of the central microtubule regulator stathmin. Stathmin, also called Oncoprotein 18, is highly over-expressed in different cancer entities,71,93,94 and its depletion slows down proliferation and increases apoptosis.94,95 It is likely that SIAH controls migration and invasion in part through its influence on stathmin and its interaction partner p27Kip1. Stathmin binds α/β-tubulin heterodimers to facilitate the depolymerization of microtubules and can also inhibit microtubule polymerization.71 Consistently, silencing/inhibition of stathmin leads to increased tubulin acetylation,94 which slows down cell motility,96 while high stathmin protein levels inhibit tubulin acetylation.93 Previous reports demonstrated that the SIAH substrate p27Kip1 inhibits stathmin, promoting tubulin stability and inhibiting migration,70,97 suggesting that SIAH ligases exert their effects on migration and invasion via this axis. Of note, stathmin might also regulate p27Kip1 protein levels, presumably through their common interaction partner Kinase-interacting stathmin (KIS).72,98,99
Similar to the SIAH ligases, there have been conflicting reports on the influence of stathmin on migration. Microtubule destabilization was described as promoter of cell motility and migration in several studies reporting that stathmin has promigratory100-102 and proinvasive activities70,97,99,103,104 and promotes chemotherapy resistance.99,105 However, the destabilization of microtubule plus ends at the cell cortex can also inhibit directional cell migration,106,107 suggesting that stathmin might also inhibit migration under certain conditions.108 Uncovering the roles of stathmin is further complicated by the observation that proliferation effects seem to depend on the phosphorylation status while effects on migration rely on protein expression levels only.109 Also the role of the p21-related p27Kip1 in migration is under debate, although the many reports point to an antimigratory activity.70,110-114 One might speculate that the effect of SIAH ligases on migration diverges in different cell lines because their downstream effectors p27Kip1 and stathmin fulfil alternative roles in different cell lines. Although SIAH silencing negatively regulated total stathmin levels, this was opposed by decreased phosphorylation at serine 16, a major target of CAMKII. Potentially, the observed reduction in CAMKII activity is a direct consequence of SIAH silencing. CAMKII activity is controlled by protein phosphatase 1 (PP1),115-117 and some of the PP1 subunits have been described as an interaction partners or substrates of SIAH2.118-120 Therefore, it is possible that SIAH ligases regulate CAMKII phosphorylation via ubiquitination and degradation of PP1 subunits. It also appears plausible that the observed reduction in stathmin phosphorylation is independent of SIAH and part of a compensatory cellular response to facilitate the balance of microtubule dynamics. However, the effect on stathmin phosphorylation cannot compensate for the decrease in stathmin expression by SIAH silencing, as evident by increased tubulin acetylation and decreased cell migration and invasion.
The connection between SIAH ligases and stathmin provides a link to the SIAH knockdown effects on migration, invasion, proliferation, and apoptosis. A summary of the pathway suggested by our results is depicted in Figure 5. Still we assume that this pathway is only one of several contributing effector pathways downstream of SIAH1 and SIAH2. Others have shown previously that a number of different SIAH substrates also play a major role regarding the tumor promoting effects of SIAH family members.14,15
Figure 5.
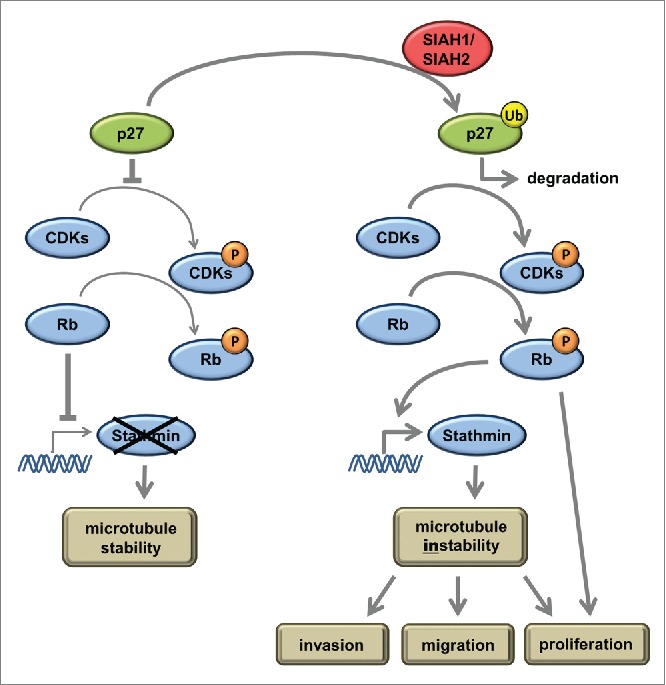
Summarizing model to explain the pathway how SIAH ubiquitin ligases influence migration, invasion, and proliferation according to our results. At low levels of active SIAH1 or SIAH2, p27 is present and inhibits phosphorylation of Rb via CDK inhibition. Non-phosphorylated Rb sequesters the E2F1 transcription factor and thereby inhibits stathmin expression. High levels of active SIAH1 and SIAH2 ligases induce ubiquitinylation of p27 and subsequent proteasomal degradation. Active CDKs phosphorylate Rb, inducing the release of E2F1 and stathmin expression. Stathmin depolymerizes microtubules, a prerequisite for increased migration, invasion, and proliferation.
In summary, our study supports previous findings that classify SIAH2 as a tumor promoting protein in breast cancer and, furthermore, strongly suggests that SIAH1 assumes a similar role. Our results show that both SIAH1 and SIAH2 promote breast cancer cell migration and invasion, 2 hallmarks of metastatic cancer subtypes. Along with previous findings that SIAH2 expression in breast cancer patients correlates with increased cancer invasiveness and decreased overall survival,4,26,28 and that SIAH inhibition reduced metastasis in a melanoma mouse model,18 these results indicate that SIAH ligases might very well promote breast cancer cell metastasis. The involvement of p27Kip1 and stathmin downstream of SIAH1/2 points to a novel component that is likely to contribute to the SIAH mechanism of action in metastasis. However, detailed future studies are required to further elucidate the role of SIAH1 and SIAH2 in breast cancer metastasis and to reveal additional regulation mechanisms downstream of SIAH.
As approximately 90% of all cancer-related mortalities are due to metastases,121 it could be beneficial to inhibit SIAH ligases in anticancer therapy. As SIAH1 and SIAH2 are similar in amino acid sequence and molecule structure,122 it would be difficult to find inhibitors selective for one isoform. However, this might not be required as our study shows that both human SIAH forms have similar roles in breast cancer. Further investigations are required to clarify whether SIAH ubiquitin ligases represent promising targets for anticancer therapy.
Materials & Methods
Cell culture
MCF7 cells (ATCC® HTB22™) were grown in RPMI 1640 (1x) medium without phenol red (Gibco, Cat.No. 32404-014) supplemented with 10% fetal bovine serum (FBS Superior; Biochrom, Cat.No. S0615), 1x10−7 M β-estradiol (Sigma, Cat.No. E2257), 10 µg/ml human recombinant insulin (Biochrom, Cat.No. K3620) and 5 ml 100x GlutaMAX™-I (Gibco, Cat.No. 35050-038). MDA-MB-468 cells (ATCC® HTB-132™) were grown in RPMI 1640 medium (1x) + GlutaMAX™-I (Gibco, Cat.No. 61870-010) supplemented with 10% FBS. MCF10A (ATCC® CRL10317™) were cultured in DMEM/F-12 (1:1) (1x) medium (Gibco, 11039-021) supplemented with 5% HI Horse Serum (Gibco, Cat.No. 26050-088), 9.5 µg/ml hydrocortisone (Biochrom, Cat.No. K3520), 20 ng/ml human recombinant EGF (Sigma, Cat.No. E9644), and 10 µg/ml human recombinant insulin. T47D (ATCC® HTB-133™) were cultured in RPMI 1640 (1x) medium + GlutaMAX™-I supplemented with 10% FBS, 1x10−7 M β-estradiol, and 10 µg/ml human recombinant insulin. HCT116-4xVEGF+Luc had previously been generated by stably transfecting HCT-116 cells (ATCC® CCL−247™) with pGL2-TK-HRE, containing the luciferase reporter gene under control of 4 copies of a HIF response element (HRE) derived from the human VEGF promoter.50 They were cultured in McCoy's 5a Medium Modified (1x) + GlutaMAX™-I (Gibco, Cat.No. 36600–021), supplemented with 10% FBS. Primary human umbilical vein endothelial cells (HUVEC) were obtained from Lonza (Cat.No. C2519AS) and cultured in endothelial cell growth medium (Lonza, Cat.no. CC-3162). They were used for assays only between passages 3 and 6.
All cell lines were maintained at 37°C. For experiments under normoxic conditions, cells were kept in Heracell 240i CO2 incubators (Thermo Scientific) at 21% O2 and 5% CO2. For hypoxia experiments, cells were kept in a Binder CB-160, type CB O2 incubator (Binder) at 1% O2 and 5% CO2 for the indicated times.
siRNA transfections were performed as reverse transfections according to the protocol of the manufacturer of the transfection reagent, employing Lipofectamine™ RNAiMAX (Invitrogen, Cat. No. 13778–075) and OptiMEM (Gibco, Cat.No. 31985). After 18 h the cells were transfected a second time (forward transfection) according to the protocol of the manufacturer. Assays were performed or cells were harvested 48 h after the second transfection unless indicated otherwise.
Where indicated, cells were treated with the pan-CDK inhibitor Roniciclib (BAY 1000394)123 (10nM or 30 nM) for 48 h.
siRNAs
The following previously published siRNAs were employed: SIAH1 siRNA (a), custom siRNA (Dharmacon, GATAGGAACACGCAAGCAA);20,45 SIAH2 siRNA a, On-Target plus SMARTpool (Dharmacon, Cat.No. L-006561-00);47 SIAH2 siRNA b, siGenome SMARTpool (Dharmacon, Cat.No. M-006561-02);46 AllStars negative Control siRNA (Qiagen, Cat.No. 1027280).
For the SIAH1/2 double knockdown, SIAH1 siRNA and SIAH2 siRNA a were employed.
Antibodies and immunoblots
Antibodies were obtained from the following sources: anti-β-Actin antibody [AC-74] (A5316), anti-Siah2 antibody [clone Siah2-369] (S7945) from Sigma-Aldrich; anti-Cdk6 (phospho Y24) antibody (ab131469), anti-EB3 antibody [KT36] (ab53360), anti-GAPDH antibody [6C5] (ab8245), anti-Sprouty 2 mouse antibody (ab60719) from abcam; acetyl-alpha-Tubulin (Lys40) (D20G3) antibody (#5335), phospho-Akt (Ser473) (D9E) antibody (#4060), AKT antibody (#9272), CDK6 (DCS83) Mouse mAb (#3136), phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) antibody (#9101), p44/42 MAPK (Erk1/2) antibody (#9102), p27 Kip1 (D69C12) XP® Rabbit mAb (#3686), Rb (4H1) Mouse mAb (#9309), phospho-Rb (Ser780) antibody (#9307), phospho-Rb (Ser807/811) antibody (#9308), Stathmin antibody (#3352) from Cell Signaling Technology; SIAH-1 antibody [N-15] (sc-5505) from Santa Cruz; purified mouse anti-human HIF-1α clone 54/HIF-1α (RUO) (610959) from BD Biosciences; PHD3/HIF Prolyl Hydroxylase 3 antibody (NB100-139) from Novus Biologicals.
Cells were harvested in Pierce® RIPA Buffer (Thermo Scientific, Cat.No. #89901) with protease inhibitor (cOmplete ULTRA Tablets, Roche, Cat.No. 04906837001) and phosphatase inhibitor (PhosSTOP, Roche, Cat.No. 05892791001). Protein concentration was assessed with a BCA protein assay kit (Novagen, Cat.No. 71285-3). Proteins were separated by electrophoresis on 4-12% Novex® NuPAGE 4-12% Bis-Tris Gel (Life Technologies, Cat.No. NP0335BOX) and transferred to nitrocellulose membranes (Novex® iBlot® Transfer Stacks, Life Technologies, Cat.No. IB301001) as stated in the manufacturer's protocol. Membranes were blocked with 5% skim milk in 0.5% Tween 20 in TBST and stained with primary antibodies at 4°C over night. The membrane was washed and incubated with IRDye®-conjugated secondary antibody (LI-COR Biosciences, Cat.Nos. 925-32214 (IRDye® 800CW Donkey anti-Goat IgG (H + L)); 925-32210 (IRDye® 800CW Goat anti-Mouse IgG (H + L)); 925-32211 (IRDye® 800CW Goat anti-Rabbit IgG (H + L)); 925-32219 (IRDye® 800CW Goat anti-Rat IgG (H + L)); 925-68070 (IRDye® 680RD Goat anti-Mouse IgG (H + L))) for 1 h at room temperature. Signal detection was done in an Odyssey Infrared Imager (LI-COR Biosciences).
Apoptosis assay
Apoptosis was quantified with the Cell Death Detection ELISA PLUS (Roche, Cat.No. 11774425001) according to the manufacturer's protocol. Briefly, following siRNA transfection (after 24 h) the cells were counted and 104 cells per well were seeded in a 96 well plate in growth medium as triplicates. After 4 h, 10 µl of growth medium with or without Doxorubicin was added (final concentrations 0, 0.1, or 1 µM). The cells were incubated for 18 h, then lyzed and centrifuged. The nucleosomes in the supernatant were then immediately quantified in the ELISA.
Luciferase assay
HIF target gene expression was monitored in a luciferase assay using the steadylite plus™ sytem (Perkin Elmer, Cat.No. 6066751) according to the manufacturer's protocol. Briefly, HCT116–4xVEGF+Luc cells were seeded and transfected in a 24 well plate. 48 h after transfection, the medium from all wells was pooled and a defined volume redistributed to the wells. The same volume steadylite plus reagent was added, the plate was protected from light and shaken gently for 15 min. From each well triplicates of 100 µl were transferred to a white 96 well plate (Nunc, Cat.No. 437796) and luminescence was measured in a Tecan infinite M200 PRO reader with Tecan i-control™ software.
To take into account that the different siRNAs have differential effects on cell viability, each plate was generated as a duplicate. The second plate was used for cell quantification with the CellTiter-Blue® Cell Viability assay (Promega, Cat.No. G8081) according to the manufacturer's protocol. Results from the luciferase assay were normalized to the cell viability.
Proliferation assay
Cell proliferation was determined by 5-bromodeoxyuridine (BrdU) incorporation (Cell Proliferation ELISA, Roche, Cat.No. 11647229001) according to the manufacturer's protocol. Epithelial cells were transfected with siRNAs. After 24 h the cells were counted and 7500 cells per well were seeded in a 96 well plate in growth medium as quintuplicates. After 24 h, BrdU was added for 2 h and newly synthesized DNA was detected with an enzyme-linked BrdU antibody.
Migration & Invasion assays
Cell motility was assessed with a wound healing assay as previously described124 in the presence of 10 μg/ml mitomycin C (Sigma, Cat.No. M4287) to block proliferation. μ-Dish 35 mm high Culture-Inserts ibiTreat (ibidi, Cat.No. 80209) were placed in a 24 well plate coated with rat tail collagen I (Life Technologies, Cat.No. A1048301). Following siRNA transfection (after 24 h), cells were trypsinized and 600,000 cells/ml were suspended in growth medium. Of this suspension, 100 μl were placed in each insert half. After 24 h, the inserts were removed and the gap width was measured at the indicated time points. The migration rate was calculated as the velocity of the moving cell front in μm/h.
Chemotaxis was determined with a modified Boyden chamber assay (CytoSelect 96-well Cell Migration Assay, Cell Biolabs Inc., Cat.No. CBA-106-CB). 24 h after the RNAi transfection, the cells were starved overnight in medium lacking FBS, counted, and seeded into the upper chambers in FBS-free medium containing 10 μg/ml mitomycin C. The lower chambers were filled with full medium containing 10% FBS as a stimulus and mitomycin C. Cells were allowed to migrate for 24 h, and then migratory cells were dissociated from the membrane, lysed and quantified with CyQuant® GR Fluorescent Dye.
Cell invasion was assessed in a transwell assay (CytoSelect 96-well Cell Invasion Assay, Cell Biolabs Inc., Cat.No. CBA-112-CB). 24 h after the RNAi transfection, the cells were starved overnight in medium lacking FBS. The transwells coated with basal membrane extract were rehydrated in serum-free medium for 1 h. The cells were counted and seeded into the transwell in FBS-free medium containing 10 μg/ml mitomycin C. The lower wells were filled with full medium containing 10% FBS as a stimulus and mitomycin C. Cells were allowed to invade through the basal membrane extract for 24 h, and then invasive cells were dissociated from the membrane, lysed and quantified with CyQuant® GR Fluorescent Dye.
Immunofluorescence staining
Cells were seeded on glass cover slips precoated with Poly-D-lysine hydrobromide (Sigma, Cat.No. P6407) 24 h after siRNA transfection. Following an incubation time of 24 h, the cells were fixed with 4% formaldehyde + 0.1% glutaraldehyde and permeabilized with 0.1% saponin. α-Tubulin and acetylated α-Tubulin were localized using α-Tubulin (DM1A) Mouse mAb (#3873, 1:4000), or acetyl-α-Tubulin (Lys40) (D20G3) antibody (#5335, 1:800; both from Cell Signaling Technology), respectively. Cy3-conjugated Goat Anti-Rabbit IgG (1:50, Jackson ImmunoResearch, Cat.No. 111–165–144) and Goat Anti-Mouse IgG (DyLight 488) preadsorbed (1:50, Thermo Scientific, Cat.No. 35502) were used as secondary antibodies. Nuclei were stained with Hoechst 33342 (1:1000, Molecular Probes, Cat.No. H1399) and slides were mounted with FluorSave Reagent (Calbiochem, Cat.No. 345789). Images were taken with a Zeiss laser confocal microscope (LSM700) at 63x magnification.
Statistical analysis
Results are expressed as means plus standard deviations unless stated otherwise. Comparisons between groups were analyzed by an Ordinary one-way ANOVA. Multiple comparisons were corrected with Dunnett's test. Probability values smaller than 0.05 were considered significant.
Disclosure of Potential Conflicts of Interest
S. Christian, H. Hess-Stumpp, A. Hägebarth, and C. Algire are full-time employees at Bayer Pharma.
Acknowledgments
We thank J. Pastorek & S. Pastoreková, Institute of Virology, Slovak Academy of Sciences, Bratislava, for providing the Carbonic Anhydrase IX monoclonal antibody, and Gerhard Siemeister, Bayer Pharma AG, Berlin, for providing the pan-CDK inhibitor. We thank the scientists of the GTRG Oncology II, GDD, Bayer Pharma AG, and the Research Review Committee of the German Cancer Research Center – Bayer HealthCare Alliance, especially Ruth Wellenreuther, for many fruitful discussions.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Hu G, Zhang S, Vidal M, Baer JL, Xu T, Fearon ER. Mammalian homologs of seven in absentia regulate DCC via the ubiquitin-proteasome pathway. Genes & Dev 1997; 11:2701-14; PMID:9334332; http://dx.doi.org/ 10.1101/gad.11.20.2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roperch JP, Lethrone F, Prieur S, Piouffre L, Israeli D, Tuynder M, Nemani M, Pasturaud P, Gendron MC, Dausset J, et al.. SIAH-1 promotes apoptosis and tumor suppression through a network involving the regulation of protein folding, unfolding, and trafficking: identification of common effectors with p53 and p21(Waf1). Proc Natl Acad Sci U S A 1999; 96:8070-3; PMID:10393949; http://dx.doi.org/ 10.1073/pnas.96.14.8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed AU, Schmidt RL, Park CH, Reed NR, Hesse SE, Thomas CF, Molina JR, Deschamps C, Yang P, Aubry MC, et al.. Effect of disrupting seven-in-absentia homolog 2 function on lung cancer cell growth. J Natl Cancer Inst 2008; 100:1606-29; PMID:19001609; http://dx.doi.org/ 10.1093/jnci/djn365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behling KC, Tang A, Freydin B, Chervoneva I, Kadakia S, Schwartz GF, Rui H, Witkiewicz AK. Increased SIAH expression predicts ductal carcinoma in situ (DCIS) progression to invasive carcinoma. Breast Cancer Res Treat 2011; 129:717-24; PMID:21088888; http://dx.doi.org/ 10.1007/s10549-010-1254-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi J, Nakayama K, Cardiff RD, Borowsky AD, Kaul K, Williams R, Krajewski S, Mercola D, Carpenter PM, Bowtell D, et al.. Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors. Cancer Cell 2010; 18:23-38; PMID:20609350; http://dx.doi.org/ 10.1016/j.ccr.2010.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin R, Smyrk TC, Reed NR, Schmidt RL, Schnelldorfer T, Chari ST, Petersen GM, Tang AH. Combining clinicopathological predictors and molecular biomarkers in the oncogenic K-RAS/Ki67/HIF-1alpha pathway to predict survival in resectable pancreatic cancer. Brit J Cancer 2015; 112:514-22; http://dx.doi.org/ 10.1038/bjc.2014.659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al.. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Disc 2012; 2:401-4; PMID:22588877; http://dx.doi.org/ 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al.. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signaling 2013; 6:pl1; PMID:23550210; http://dx.doi.org/ 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brauckhoff A, Malz M, Tschaharganeh D, Malek N, Weber A, Riener MO, Soll C, Samarin J, Bissinger M, Schmidt J, et al.. Nuclear expression of the ubiquitin ligase seven in absentia homolog (SIAH)-1 induces proliferation and migration of liver cancer cells. J Hepatol 2011; 55:1049-57; PMID:21356256; http://dx.doi.org/ 10.1016/j.jhep.2011.02.019 [DOI] [PubMed] [Google Scholar]
- 10.Malz M, Aulmann A, Samarin J, Bissinger M, Longerich T, Schmitt S, Schirmacher P, Breuhahn K. Nuclear accumulation of seven in absentia homologue-2 supports motility and proliferation of liver cancer cells. Int J Cancer J Int du Cancer 2012; 131:2016-26; PMID:22323152; http://dx.doi.org/ 10.1002/ijc.27473 [DOI] [PubMed] [Google Scholar]
- 11.Sun RC, Denko NC. Hypoxic regulation of glutamine metabolism through HIF1 and SIAH2 supports lipid synthesis that is necessary for tumor growth. Cell Metab 2014; 19:285-92; PMID:24506869; http://dx.doi.org/ 10.1016/j.cmet.2013.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah M, Stebbins JL, Dewing A, Qi J, Pellecchia M, Ronai ZA. Inhibition of Siah2 ubiquitin ligase by vitamin K3 (menadione) attenuates hypoxia and MAPK signaling and blocks melanoma tumorigenesis. Pigment Cell & Melanoma Res 2009; 22:799-808; PMID:19712206; http://dx.doi.org/ 10.1111/j.1755-148X.2009.00628.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt RL, Park CH, Ahmed AU, Gundelach JH, Reed NR, Cheng S, Knudsen BE, Tang AH. Inhibition of RAS-mediated transformation and tumorigenesis by targeting the downstream E3 ubiquitin ligase seven in absentia homologue. Cancer Res 2007; 67:11798-810; PMID:18089810; http://dx.doi.org/ 10.1158/0008-5472.CAN-06-4471 [DOI] [PubMed] [Google Scholar]
- 14.Qi J, Kim H, Scortegagna M, Ronai ZA. Regulators and effectors of Siah ubiquitin ligases. Cell Biochem Biophys 2013; 67:15-24; PMID:23700162; http://dx.doi.org/ 10.1007/s12013-013-9636-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.House CM, Moller A, Bowtell DD. Siah proteins: novel drug targets in the Ras and hypoxia pathways. Cancer Res 2009; 69:8835-8; PMID:19920190; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-1676 [DOI] [PubMed] [Google Scholar]
- 16.Calzado MA, de la Vega L, Moller A, Bowtell DD, Schmitz ML. An inducible autoregulatory loop between HIPK2 and Siah2 at the apex of the hypoxic response. Nat Cell Biol 2009; 11:85-91; PMID:19043406; http://dx.doi.org/ 10.1038/ncb1816 [DOI] [PubMed] [Google Scholar]
- 17.Nakayama K, Frew IJ, Hagensen M, Skals M, Habelhah H, Bhoumik A, Kadoya T, Erdjument-Bromage H, Tempst P, Frappell PB, et al.. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell 2004; 117:941-52; PMID:15210114; http://dx.doi.org/ 10.1016/j.cell.2004.06.001 [DOI] [PubMed] [Google Scholar]
- 18.Qi J, Nakayama K, Gaitonde S, Goydos JS, Krajewski S, Eroshkin A, Bar-Sagi D, Bowtell D, Ronai Z. The ubiquitin ligase Siah2 regulates tumorigenesis and metastasis by HIF-dependent and -independent pathways. Proc Natl Acad Sci U S A 2008; 105:16713-8; PMID:18946040; http://dx.doi.org/ 10.1073/pnas.0804063105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moehlenbrink J, Bitomsky N, Hofmann TG. Hypoxia suppresses chemotherapeutic drug-induced p53 Serine 46 phosphorylation by triggering HIPK2 degradation. Cancer Lett 2010; 292:119-24; PMID:20018442; http://dx.doi.org/ 10.1016/j.canlet.2009.11.016 [DOI] [PubMed] [Google Scholar]
- 20.Winter M, Sombroek D, Dauth I, Moehlenbrink J, Scheuermann K, Crone J, Hofmann TG. Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nat Cell Biol 2008; 10:812-24; PMID:18536714; http://dx.doi.org/ 10.1038/ncb1743 [DOI] [PubMed] [Google Scholar]
- 21.Susini L, Passer BJ, Amzallag-Elbaz N, Juven-Gershon T, Prieur S, Privat N, Tuynder M, Gendron MC, Israël A, Amson R, et al.. Siah-1 binds and regulates the function of Numb. Proc Natl Acad Sci U S A 2001; 98:15067-72; PMID:11752454; http://dx.doi.org/ 10.1073/pnas.261571998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fanelli M, Fantozzi A, De Luca P, Caprodossi S, Matsuzawa S, Lazar MA, Pelicci PG, Minucci S. The coiled-coil domain is the structural determinant for mammalian homologues of Drosophila Sina-mediated degradation of promyelocytic leukemia protein and other tripartite motif proteins by the proteasome. J Biol Chem 2004; 279:5374-9; PMID:14645235; http://dx.doi.org/ 10.1074/jbc.M306407200 [DOI] [PubMed] [Google Scholar]
- 23.Wong CS, Moller A. Siah: a promising anticancer target. Cancer Res 2013; 73:2400-6; PMID:23455005; http://dx.doi.org/ 10.1158/0008-5472.CAN-12-4348 [DOI] [PubMed] [Google Scholar]
- 24.Moller A, House CM, Wong CS, Scanlon DB, Liu MC, Ronai Z, Bowtell DD. Inhibition of Siah ubiquitin ligase function. Oncogene 2009; 28:289-96; PMID:18850011; http://dx.doi.org/ 10.1038/onc.2008.382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eirew P, Steif A, Khattra J, Ha G, Yap D, Farahani H, Gelmon K, Chia S, Mar C, Wan A, et al.. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 2015; 518:422-6; PMID:25470049; http://dx.doi.org/ 10.1038/nature13952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong CS, Sceneay J, House CM, Halse HM, Liu MC, George J, Hunnam TC, Parker BS, Haviv I, Ronai Z, Cullinane C, et al.. Vascular normalization by loss of Siah2 results in increased chemotherapeutic efficacy. Cancer Res 2012; 72:1694-704; PMID:22354750; http://dx.doi.org/ 10.1158/0008-5472.CAN-11-3310 [DOI] [PubMed] [Google Scholar]
- 27.Ma B, Chen Y, Chen L, Cheng H, Mu C, Li J, Gao R, Zhou C, Cao L, Liu J, et al.. Hypoxia regulates Hippo signalling through the SIAH2 ubiquitin E3 ligase. Nat Cell Biol 2015; 17:95-103; PMID:25438054; http://dx.doi.org/ 10.1038/ncb3073 [DOI] [PubMed] [Google Scholar]
- 28.Chan P, Moller A, Liu MC, Sceneay JE, Wong CS, Waddell N, Huang KT, Dobrovic A, Millar EK, O'Toole SA, et al.. The expression of the ubiquitin ligase SIAH2 (seven in absentia homolog 2) is mediated through gene copy number in breast cancer and is associated with a basal-like phenotype and p53 expression. Breast Cancer Res: BCR 2011; 13:R19; PMID:21306611; http://dx.doi.org/ 10.1186/bcr2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiang L, Gilkes DM, Hu H, Takano N, Luo W, Lu H, Bullen JW, Samanta D, Liang H, Semenza GL. Hypoxia-inducible factor 1 mediates TAZ expression and nuclear localization to induce the breast cancer stem cell phenotype. Oncotarget 2014; 5:12509-27; PMID:25587023; http://dx.doi.org/ 10.18632/oncotarget.2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruzzoni-Giovanelli H, Faille A, Linares-Cruz G, Nemani M, Le Deist F, Germani A, Chassoux D, Millot G, Roperch JP, Amson R, et al.. SIAH-1 inhibits cell growth by altering the mitotic process. Oncogene 1999; 18:7101-9; PMID:10597311; http://dx.doi.org/ 10.1038/sj.onc.1203187 [DOI] [PubMed] [Google Scholar]
- 31.Confalonieri S, Quarto M, Goisis G, Nuciforo P, Donzelli M, Jodice G, Pelosi G, Viale G, Pece S, Di Fiore PP. Alterations of ubiquitin ligases in human cancer and their association with the natural history of the tumor. Oncogene 2009; 28:2959-68; PMID:19543318; http://dx.doi.org/ 10.1038/onc.2009.156 [DOI] [PubMed] [Google Scholar]
- 32.He HT, Fokas E, You A, Engenhart-Cabillic R, An HX. Siah1 proteins enhance radiosensitivity of human breast cancer cells. BMC Cancer 2010; 10:403; PMID:20682032; http://dx.doi.org/ 10.1186/1471-2407-10-403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuynder M, Susini L, Prieur S, Besse S, Fiucci G, Amson R, Telerman A. Biological models and genes of tumor reversion: cellular reprogramming through tpt1/TCTP and SIAH-1. Proc Natl Acad Sci U S A 2002; 99:14976-81; PMID:12399545; http://dx.doi.org/ 10.1073/pnas.222470799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen YY, Yang ZQ, Song M, Li BL, Yao XH, Chen XL, Zhao J, Lu YY, Zhu JJ, Wang EH. The expression of SIAH1 is downregulated and associated with Bim and apoptosis in human breast cancer tissues and cells. Mol Carcinog 2010; 49:440-9; PMID:20082325 [DOI] [PubMed] [Google Scholar]
- 35.Wen YY, Yang ZQ, Song M, Li BL, Zhu JJ, Wang EH. SIAH1 induced apoptosis by activation of the JNK pathway and inhibited invasion by inactivation of the ERK pathway in breast cancer cells. Cancer Sci 2010; 101:73-9; PMID:19775288; http://dx.doi.org/ 10.1111/j.1349-7006.2009.01339.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L, Ma P, Sun LM, Han YC, Li BL, Mi XY, Wang EH, Song M. MiR-107 down-regulates SIAH1 expression in human breast cancer cells and silencing of miR-107 inhibits tumor growth in a nude mouse model of triple-negative breast cancer. Mol Carcinog 2015 [DOI] [PubMed] [Google Scholar]
- 37.Palmieri D, Fitzgerald D, Shreeve SM, Hua E, Bronder JL, Weil RJ, Weil RJ, Davis S, Stark AM, Merino MJ, et al.. Analyses of resected human brain metastases of breast cancer reveal the association between up-regulation of hexokinase 2 and poor prognosis. Mol Cancer Res: MCR 2009; 7:1438-45; PMID:19723875; http://dx.doi.org/ 10.1158/1541-7786.MCR-09-0234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jansen MP, Ruigrok-Ritstier K, Dorssers LC, van Staveren IL, Look MP, Meijer-van Gelder ME, Sieuwerts AM, Helleman J, Sleijfer S, Klijn JG, et al.. Downregulation of SIAH2, an ubiquitin E3 ligase, is associated with resistance to endocrine therapy in breast cancer. Breast Cancer Res Treat 2009; 116:263-71; PMID:18629630; http://dx.doi.org/ 10.1007/s10549-008-0125-z [DOI] [PubMed] [Google Scholar]
- 39.Etienne-Manneville S. Actin and microtubules in cell motility: which one is in control? Traffic 2004; 5:470-7; PMID:15180824; http://dx.doi.org/ 10.1111/j.1600-0854.2004.00196.x [DOI] [PubMed] [Google Scholar]
- 40.Wehrle-Haller B, Imhof BA. Actin, microtubules and focal adhesion dynamics during cell migration. Int J Biochem Cell Biol 2003; 35:39-50; PMID:12467646; http://dx.doi.org/ 10.1016/S1357-2725(02)00071-7 [DOI] [PubMed] [Google Scholar]
- 41.Shi H, Zheng B, Wu Y, Tang Y, Wang L, Gao Y, Gong H, Du J, Yu R. Ubiquitin ligase Siah1 promotes the migration and invasion of human glioma cells by regulating HIF-1alpha signaling under hypoxia. Oncol Rep 2015; 33:1185-90; PMID:25572001 [DOI] [PubMed] [Google Scholar]
- 42.Tanaka T, Iino M. Sec8 regulates cytokeratin8 phosphorylation and cell migration by controlling the ERK and p38 MAPK signalling pathways. Cell Signalling 2015; 27:1110-9; PMID:25725287; http://dx.doi.org/ 10.1016/j.cellsig.2015.02.015 [DOI] [PubMed] [Google Scholar]
- 43.Famulski JK, Trivedi N, Howell D, Yang Y, Tong Y, Gilbertson R, Solecki DJ. Siah regulation of Pard3A controls neuronal cell adhesion during germinal zone exit. Science 2010; 330:1834-8; PMID:21109632; http://dx.doi.org/ 10.1126/science.1198480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagano Y, Fukushima T, Okemoto K, Tanaka K, Bowtell DD, Ronai Z, Reed JC, Matsuzawa S. Siah1/SIP regulates p27(kip1) stability and cell migration under metabolic stress. Cell Cycle 2011; 10:2592-602; PMID:21734459; http://dx.doi.org/ 10.4161/cc.10.15.16912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crone J, Glas C, Schultheiss K, Moehlenbrink J, Krieghoff-Henning E, Hofmann TG. Zyxin is a critical regulator of the apoptotic HIPK2-p53 signaling axis. Cancer Res 2011; 71:2350-9; PMID:21248071; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-3486 [DOI] [PubMed] [Google Scholar]
- 46.Perez M, Garcia-Limones C, Zapico I, Marina A, Schmitz ML, Munoz E, Calzado MA. Mutual regulation between SIAH2 and DYRK2 controls hypoxic and genotoxic signaling pathways. J Mol Cell Biol 2012; 4:316-30; PMID:22878263; http://dx.doi.org/ 10.1093/jmcb/mjs047 [DOI] [PubMed] [Google Scholar]
- 47.Sarkar TR, Sharan S, Wang J, Pawar SA, Cantwell CA, Johnson PF, Morrison DK, Wang JM, Sterneck E. Identification of a Src tyrosine kinase/SIAH2 E3 ubiquitin ligase pathway that regulates C/EBPdelta expression and contributes to transformation of breast tumor cells. Mol Cell Biol 2012; 32:320-32; PMID:22037769; http://dx.doi.org/ 10.1128/MCB.05790-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Habelhah H, Frew IJ, Laine A, Janes PW, Relaix F, Sassoon D, Bowtell DD, Ronai Z. Stress-induced decrease in TRAF2 stability is mediated by Siah2. EMBO J 2002; 21:5756-65; PMID:12411493; http://dx.doi.org/ 10.1093/emboj/cdf576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gopalsamy A, Hagen T, Swaminathan K. Investigating the molecular basis of Siah1 and Siah2 E3 ubiquitin ligase substrate specificity. PloS One 2014; 9:e106547; PMID:25202994; http://dx.doi.org/ 10.1371/journal.pone.0106547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ellinghaus P, Heisler I, Unterschemmann K, Haerter M, Beck H, Greschat S, Ehrmann A, Summer H, Flamme I, Oehme F, et al.. BAY 87-2243, a highly potent and selective inhibitor of hypoxia-induced gene activation has antitumor activities by inhibition of mitochondrial complex I. Cancer Med 2013; 2:611-24; PMID:24403227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gluck S. Adjuvant chemotherapy for early breast cancer: optimal use of epirubicin. The Oncologist 2005; 10:780-91; PMID:16314288; http://dx.doi.org/ 10.1634/theoncologist.10-10-780 [DOI] [PubMed] [Google Scholar]
- 52.Christian PA, Fiandalo MV, Schwarze SR. Possible role of death receptor-mediated apoptosis by the E3 ubiquitin ligases Siah2 and POSH. Mol Cancer 2011; 10:57; PMID:21586138; http://dx.doi.org/ 10.1186/1476-4598-10-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsieh SC, Kuo SN, Zheng YH, Tsai MH, Lin YS, Lin JH. The E3 ubiquitin ligase SIAH2 is a prosurvival factor overexpressed in oral cancer. Anticancer Res 2013; 33:4965-73; PMID:24222137 [PubMed] [Google Scholar]
- 54.Frew IJ, Dickins RA, Cuddihy AR, Del Rosario M, Reinhard C, O'Connell MJ, Bowtell DD. Normal p53 function in primary cells deficient for Siah genes. Mol Cell Biol 2002; 22:8155-64; PMID:12417719; http://dx.doi.org/ 10.1128/MCB.22.23.8155-8164.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al.. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006; 10:515-27; PMID:17157791; http://dx.doi.org/ 10.1016/j.ccr.2006.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chien PY, Ito M, Park Y, Tagami T, Gehm BD, Jameson JL. A fusion protein of the estrogen receptor (ER) and nuclear receptor corepressor (NCoR) strongly inhibits estrogen-dependent responses in breast cancer cells. Mol Endocrinol 1999; 13:2122-36; PMID:10598586; http://dx.doi.org/ 10.1210/mend.13.12.0394 [DOI] [PubMed] [Google Scholar]
- 57.Kashima H, Horiuchi A, Uchikawa J, Miyamoto T, Suzuki A, Ashida T, Konishi I, Shiozawa T. Up-regulation of nuclear receptor corepressor (NCoR) in progestin-induced growth suppression of endometrial hyperplasia and carcinoma. Anticancer Res 2009; 29:1023-9; PMID:19414341 [PubMed] [Google Scholar]
- 58.Lavinsky RM, Jepsen K, Heinzel T, Torchia J, Mullen TM, Schiff R, Del-Rio AL, Ricote M, Ngo S, Gemsch J, et al.. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc Natl Acad Sci U S A 1998; 95:2920-5; PMID:9501191; http://dx.doi.org/ 10.1073/pnas.95.6.2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perillo B, Sasso A, Abbondanza C, Palumbo G. 17beta-estradiol inhibits apoptosis in MCF-7 cells, inducing bcl-2 expression via two estrogen-responsive elements present in the coding sequence. M Cell Biol 2000; 20:2890-901; PMID:10733592; http://dx.doi.org/ 10.1128/MCB.20.8.2890-2901.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frasor J, Danes JM, Funk CC, Katzenellenbogen BS. Estrogen down-regulation of the corepressor N-CoR: mechanism and implications for estrogen derepression of N-CoR-regulated genes. Proc Natl Acad Sci U S A 2005; 102:13153-7; PMID:16141343; http://dx.doi.org/ 10.1073/pnas.0502782102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, et al.. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol 2005; 7:665-74; PMID:15951807; http://dx.doi.org/ 10.1038/ncb1268 [DOI] [PubMed] [Google Scholar]
- 62.Zhang J, Guenther MG, Carthew RW, Lazar MA. Proteasomal regulation of nuclear receptor corepressor-mediated repression. Genes Dev 1998; 12:1775-80; PMID:9637679; http://dx.doi.org/ 10.1101/gad.12.12.1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Della NG, Senior PV, Bowtell DD. Isolation and characterisation of murine homologues of the Drosophila seven in absentia gene (sina). Development 1993; 117:1333-43; PMID:8404535 [DOI] [PubMed] [Google Scholar]
- 64.Kelley LC, Lohmer LL, Hagedorn EJ, Sherwood DR. Traversing the basement membrane in vivo: a diversity of strategies. J Cell Biol 2014; 204:291-302; PMID:24493586; http://dx.doi.org/ 10.1083/jcb.201311112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mason JM, Morrison DJ, Basson MA, Licht JD. Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol 2006; 16:45-54; PMID:16337795; http://dx.doi.org/ 10.1016/j.tcb.2005.11.004 [DOI] [PubMed] [Google Scholar]
- 66.Avruch J, Zhang XF, Kyriakis JM. Raf meets Ras: completing the framework of a signal transduction pathway. Trends Biochem Sci 1994; 19:279-83; PMID:8048167; http://dx.doi.org/ 10.1016/0968-0004(94)90005-1 [DOI] [PubMed] [Google Scholar]
- 67.Ban R, Matsuzaki H, Akashi T, Sakashita G, Taniguchi H, Park SY, Tanaka H, Furukawa K, Urano T. Mitotic regulation of the stability of microtubule plus-end tracking protein EB3 by ubiquitin ligase SIAH-1 and Aurora mitotic kinases. J Biol Chem 2009; 284:28367-81; PMID:19696028; http://dx.doi.org/ 10.1074/jbc.M109.000273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toyoshima H, Hunter T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell 1994; 78:67-74; PMID:8033213; http://dx.doi.org/ 10.1016/0092-8674(94)90573-8 [DOI] [PubMed] [Google Scholar]
- 69.Godin JD, Thomas N, Laguesse S, Malinouskaya L, Close P, Malaise O, Purnelle A, Raineteau O, Campbell K, Fero M, et al.. p27(Kip1) is a microtubule-associated protein that promotes microtubule polymerization during neuron migration. Dev Cell 2012; 23:729-44; PMID:23022035; http://dx.doi.org/ 10.1016/j.devcel.2012.08.006 [DOI] [PubMed] [Google Scholar]
- 70.Baldassarre G, Belletti B, Nicoloso MS, Schiappacassi M, Vecchione A, Spessotto P, Morrione A, Canzonieri V, Colombatti A. p27(Kip1)-stathmin interaction influences sarcoma cell migration and invasion. Cancer Cell 2005; 7:51-63; PMID:15652749; http://dx.doi.org/ 10.1016/j.ccr.2004.11.025 [DOI] [PubMed] [Google Scholar]
- 71.Cassimeris L. The oncoprotein 18/stathmin family of microtubule destabilizers. Curr Opin Cell Biol 2002; 14:18-24; PMID:11792540; http://dx.doi.org/ 10.1016/S0955-0674(01)00289-7 [DOI] [PubMed] [Google Scholar]
- 72.Iancu-Rubin C, Atweh GF. p27(Kip1) and stathmin share the stage for the first time. Trends Cell Biol 2005; 15:346-8; PMID:15951178; http://dx.doi.org/ 10.1016/j.tcb.2005.05.008 [DOI] [PubMed] [Google Scholar]
- 73.Harbour JW, Dean DC. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev 2000; 14:2393-409; PMID:11018009; http://dx.doi.org/ 10.1101/gad.813200 [DOI] [PubMed] [Google Scholar]
- 74.Knudsen ES, Wang JY. Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol Cell Biol 1997; 17:5771-83; PMID:9315635; http://dx.doi.org/ 10.1128/MCB.17.10.5771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen YL, Uen YH, Li CF, Horng KC, Chen LR, Wu WR, Tseng HY, Huang HY, Wu LC, Shiue YL. The E2F transcription factor 1 transactives stathmin 1 in hepatocellular carcinoma. Ann Surg Oncol 2013; 20:4041-54; PMID:22911364; http://dx.doi.org/ 10.1245/s10434-012-2519-8 [DOI] [PubMed] [Google Scholar]
- 76.Polzin RG, Benlhabib H, Trepel J, Herrera JE. E2F sites in the Op18 promoter are required for high level of expression in the human prostate carcinoma cell line PC-3-M. Gene 2004; 341:209-18; PMID:15474303; http://dx.doi.org/ 10.1016/j.gene.2004.06.052 [DOI] [PubMed] [Google Scholar]
- 77.Nadeau RJ, Toher JL, Yang X, Kovalenko D, Friesel R. Regulation of Sprouty2 stability by mammalian Seven-in-Absentia homolog 2. J Cell Biochem 2007; 100:151-60; PMID:16888801; http://dx.doi.org/ 10.1002/jcb.21040 [DOI] [PubMed] [Google Scholar]
- 78.Munoz-Alonso MJ, Acosta JC, Richard C, Delgado MD, Sedivy J, Leon J. p21Cip1 and p27Kip1 induce distinct cell cycle effects and differentiation programs in myeloid leukemia cells. J Biol Chem 2005; 280:18120-9; PMID:15746092; http://dx.doi.org/ 10.1074/jbc.M500758200 [DOI] [PubMed] [Google Scholar]
- 79.le Gouvello S, Manceau V, Sobel A. Serine 16 of stathmin as a cytosolic target for Ca2+/calmodulin-dependent kinase II after CD2 triggering of human T lymphocytes. J Immunol 1998; 161:1113-22 [PubMed] [Google Scholar]
- 80.Marklund U, Larsson N, Brattsand G, Osterman O, Chatila TA, Gullberg M. Serine 16 of oncoprotein 18 is a major cytosolic target for the Ca2+/calmodulin-dependent kinase-Gr. Euro J Biochem/FEBS 1994; 225:53-60; PMID:7925472; http://dx.doi.org/ 10.1111/j.1432-1033.1994.00053.x [DOI] [PubMed] [Google Scholar]
- 81.Gadea BB, Ruderman JV. Aurora B is required for mitotic chromatin-induced phosphorylation of Op18/Stathmin. Proc Natl Acad Sci U S A 2006; 103:4493-8; PMID:16537398; http://dx.doi.org/ 10.1073/pnas.0600702103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beretta L, Dobransky T, Sobel A. Multiple phosphorylation of stathmin. Identification of four sites phosphorylated in intact cells and in vitro by cyclic AMP-dependent protein kinase and p34cdc2. J Biol Chem 1993; 268:20076-84; PMID:8376365 [PubMed] [Google Scholar]
- 83.Gradin HM, Larsson N, Marklund U, Gullberg M. Regulation of microtubule dynamics by extracellular signals: cAMP-dependent protein kinase switches off the activity of oncoprotein 18 in intact cells. J Cell Biol 1998; 140:131-41; PMID:9425161; http://dx.doi.org/ 10.1083/jcb.140.1.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wittmann T, Bokoch GM, Waterman-Storer CM. Regulation of microtubule destabilizing activity of Op18/stathmin downstream of Rac1. J Biol Chem 2004; 279:6196-203; PMID:14645234; http://dx.doi.org/ 10.1074/jbc.M307261200 [DOI] [PubMed] [Google Scholar]
- 85.Alesi GN, Li D, Jin L, Chen GZ, Shin DM, Khuri F, Kang S. Abstract 3154: RSK2-mediated phosphorylation of stathmin promotes microtubule polymerization, providing a pro-invasive advantage to metastatic cancer cells. Poster presented at: AACR Annual Meeting 2014, April 5-9 2014. San Diego, CA, USA, 2014:(19 Suppl.). http://dx.doi.org/ 10.1158/1538-7445.AM2014-3154 [DOI] [Google Scholar]
- 86.Silva AJ, Stevens CF, Tonegawa S, Wang Y. Deficient hippocampal long-term potentiation in alpha-calcium-calmodulin kinase II mutant mice. Science 1992; 257:201-6; PMID:1378648; http://dx.doi.org/ 10.1126/science.1378648 [DOI] [PubMed] [Google Scholar]
- 87.Perdiz D, Mackeh R, Pous C, Baillet A. The ins and outs of tubulin acetylation: more than just a post-translational modification? Cell Signalling 2011; 23:763-71; PMID:20940043; http://dx.doi.org/ 10.1016/j.cellsig.2010.10.014 [DOI] [PubMed] [Google Scholar]
- 88.Schulze E, Asai DJ, Bulinski JC, Kirschner M. Posttranslational modification and microtubule stability. J Cell Biol 1987; 105:2167-77; PMID:3316248; http://dx.doi.org/ 10.1083/jcb.105.5.2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tamura K, Yoshie M, Miyajima E, Kano M, Tachikawa E. Stathmin regulates hypoxia-inducible factor-1alpha expression through the mammalian target of rapamycin pathway in ovarian clear cell adenocarcinoma. ISRN Pharmacol 2013; 2013:279593; PMID:23819061; http://dx.doi.org/ 10.1155/2013/279593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gordon LA, Mulligan KT, Maxwell-Jones H, Adams M, Walker RA, Jones JL. Breast cell invasive potential relates to the myoepithelial phenotype. Int J Cancer J Int du Cancer 2003; 106:8-16; PMID:12794751; http://dx.doi.org/ 10.1002/ijc.11172 [DOI] [PubMed] [Google Scholar]
- 91.Berra E, Benizri E, Ginouves A, Volmat V, Roux D, Pouyssegur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J 2003; 22:4082-90; PMID:12912907; http://dx.doi.org/ 10.1093/emboj/cdg392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Simon MC. Siah proteins, HIF prolyl hydroxylases, and the physiological response to hypoxia. Cell 2004; 117:851-3; PMID:15210106; http://dx.doi.org/ 10.1016/j.cell.2004.06.010 [DOI] [PubMed] [Google Scholar]
- 93.Belletti B, Nicoloso MS, Schiappacassi M, Berton S, Lovat F, Wolf K, Canzonieri V, D'Andrea S, Zucchetto A, Friedl P, et al.. Stathmin activity influences sarcoma cell shape, motility, and metastatic potential. Mol Biol Cell 2008; 19:2003-13; PMID:18305103; http://dx.doi.org/ 10.1091/mbc.E07-09-0894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Carney BK, Caruso Silva V, Cassimeris L. The microtubule cytoskeleton is required for a G2 cell cycle delay in cancer cells lacking stathmin and p53. Cytoskeleton 2012; 69:278-89; PMID:22407961; http://dx.doi.org/ 10.1002/cm.21024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alli E, Yang JM, Hait WN. Silencing of stathmin induces tumor-suppressor function in breast cancer cell lines harboring mutant p53. Oncogene 2007; 26:1003-12; PMID:16909102; http://dx.doi.org/ 10.1038/sj.onc.1209864 [DOI] [PubMed] [Google Scholar]
- 96.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature 2002; 417:455-8; PMID:12024216; http://dx.doi.org/ 10.1038/417455a [DOI] [PubMed] [Google Scholar]
- 97.Nadeem L, Brkic J, Chen YF, Bui T, Munir S, Peng C. Cytoplasmic mislocalization of p27 and CDK2 mediates the anti-migratory and anti-proliferative effects of Nodal in human trophoblast cells. J Cell Sci 2013; 126:445-53; PMID:23230143; http://dx.doi.org/ 10.1242/jcs.110197 [DOI] [PubMed] [Google Scholar]
- 98.Le XF, Pruefer F, Bast RC Jr. HER2-targeting antibodies modulate the cyclin-dependent kinase inhibitor p27Kip1 via multiple signaling pathways. Cell Cycle 2005; 4:87-95; PMID:15611642; http://dx.doi.org/ 10.4161/cc.4.1.1360 [DOI] [PubMed] [Google Scholar]
- 99.Watanabe A, Suzuki H, Yokobori T, Tsukagoshi M, Altan B, Kubo N, Suzuki S, Araki K, Wada S, Kashiwabara K, et al.. Stathmin1 regulates p27 expression, proliferation and drug resistance, resulting in poor clinical prognosis in cholangiocarcinoma. Cancer Sci 2014; 105:690-6; PMID:24708177; http://dx.doi.org/ 10.1111/cas.12417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Akhtar J, Wang Z, Zhang ZP, Bi MM. Lentiviral-mediated RNA interference targeting stathmin1 gene in human gastric cancer cells inhibits proliferation in vitro and tumor growth in vivo. J Trans Med 2013; 11:212; PMID:24040910; http://dx.doi.org/ 10.1186/1479-5876-11-212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Knight LM, Stakaityte G, Wood JJ, Abdul-Sada H, Griffiths DA, Howell GJ, Wheat R, Blair GE, Steven NM, Macdonald A, et al.. Merkel cell polyomavirus small T antigen mediates microtubule destabilization to promote cell motility and migration. J Virol 2015; 89:35-47; PMID:25320307; http://dx.doi.org/ 10.1128/JVI.02317-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li N, Jiang P, Du W, Wu Z, Li C, Qiao M, Yang X, Wu M. Siva1 suppresses epithelial-mesenchymal transition and metastasis of tumor cells by inhibiting stathmin and stabilizing microtubules. Proc Natl Acad Sci U S A 2011; 108:12851-6; PMID:21768358; http://dx.doi.org/ 10.1073/pnas.1017372108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Akhtar J, Wang Z, Yu C, Li CS, Shi YL, Liu HJ. STMN-1 is a potential marker of lymph node metastasis in distal esophageal adenocarcinomas and silencing its expression can reverse malignant phenotype of tumor cells. BMC Cancer 2014; 14:28; PMID:24433541; http://dx.doi.org/ 10.1186/1471-2407-14-28 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 104.Tan HT, Wu W, Ng YZ, Zhang X, Yan B, Ong CW, Tan S, Salto-Tellez M, Hooi SC, Chung MC. Proteomic analysis of colorectal cancer metastasis: stathmin-1 revealed as a player in cancer cell migration and prognostic marker. J Proteome Res 2012; 11:1433-45; PMID:22181002; http://dx.doi.org/ 10.1021/pr2010956 [DOI] [PubMed] [Google Scholar]
- 105.Lin X, Liao Y, Xie J, Liu S, Su L, Zou H. Op18/stathmin is involved in the resistance of taxol among different epithelial carcinoma cell lines. Cancer Biother Radiopharm 2014; 29:376-86; PMID:25379611; http://dx.doi.org/ 10.1089/cbr.2014.1649 [DOI] [PubMed] [Google Scholar]
- 106.Chiu CT, Liao CK, Shen CC, Tang TK, Jow GM, Wang HS, Wu JC. HYS-32-Induced Microtubule Catastrophes in Rat Astrocytes Involves the PI3K-GSK3beta Signaling Pathway. PloS One 2015; 10:e0126217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pagano A, Honore S, Mohan R, Berges R, Akhmanova A, Braguer D. Epothilone B inhibits migration of glioblastoma cells by inducing microtubule catastrophes and affecting EB1 accumulation at microtubule plus ends. Biochem Pharmacol 2012; 84:432-43; PMID:22634050; http://dx.doi.org/ 10.1016/j.bcp.2012.05.010 [DOI] [PubMed] [Google Scholar]
- 108.Ng DC, Lin BH, Lim CP, Huang G, Zhang T, Poli V, Cao X. Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J Cell Biol 2006; 172:245-57; PMID:16401721; http://dx.doi.org/ 10.1083/jcb.200503021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schmitt S, Safferling K, Westphal K, Hrabowski M, Muller U, Angel P, Wiechert L, Ehemann V, Müller B, Holland-Cunz S, et al.. Stathmin regulates keratinocyte proliferation and migration during cutaneous regeneration. PloS One 2013; 8:e75075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Daniel C, Pippin J, Shankland SJ, Hugo C. The rapamycin derivative RAD inhibits mesangial cell migration through the CDK-inhibitor p27KIP1. Lab Investigat; J Tech Methods Pathol 2004; 84:588-96; PMID:15064772; http://dx.doi.org/ 10.1038/labinvest.3700078 [DOI] [PubMed] [Google Scholar]
- 111.Goukassian D, Diez-Juan A, Asahara T, Schratzberger P, Silver M, Murayama T, Isner JM, Andrés V. Overexpression of p27(Kip1) by doxycycline-regulated adenoviral vectors inhibits endothelial cell proliferation and migration and impairs angiogenesis. FASEB J: Off Pub Federat Am Soc Exp Biol 2001; 15:1877-85; PMID:11532967; http://dx.doi.org/ 10.1096/fj.01-0065com [DOI] [PubMed] [Google Scholar]
- 112.Sun J, Marx SO, Chen HJ, Poon M, Marks AR, Rabbani LE. Role for p27(Kip1) in vascular smooth muscle cell migration. Circulation 2001; 103:2967-72; PMID:11413088; http://dx.doi.org/ 10.1161/01.CIR.103.24.2967 [DOI] [PubMed] [Google Scholar]
- 113.Zhang B, Ji LH, Liu W, Zhao G, Wu ZY. Skp2-RNAi suppresses proliferation and migration of gallbladder carcinoma cells by enhancing p27 expression. World J Gastroenterol: WJG 2013; 19:4917-24; PMID:23946596; http://dx.doi.org/ 10.3748/wjg.v19.i30.4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang D, Wang Y, Liang Y, Zhang M, Wei J, Zheng X, Li F, Meng Y, Zhu NW, Li J, et al.. Loss of p27 upregulates MnSOD in a STAT3-dependent manner, disrupts intracellular redox activity and enhances cell migration. J Cell Sci 2014; 127:2920-33; PMID:24727615; http://dx.doi.org/ 10.1242/jcs.148130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McCoy F, Darbandi R, Chen SI, Eckard L, Dodd K, Jones K, Baucum AJ 2nd, Gibbons JA, Lin SH, Colbran RJ, et al.. Metabolic regulation of CaMKII protein and caspases in Xenopus laevis egg extracts. J Biol Chem 2013; 288:8838-48; PMID:23400775; http://dx.doi.org/ 10.1074/jbc.M112.437186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schworer CM, Colbran RJ, Soderling TR. Reversible generation of a Ca2+-independent form of Ca2+(calmodulin)-dependent protein kinase II by an autophosphorylation mechanism. J Biol Chem 1986; 261:8581-4; PMID:3722161 [PubMed] [Google Scholar]
- 117.Strack S, Choi S, Lovinger DM, Colbran RJ. Translocation of autophosphorylated calcium/calmodulin-dependent protein kinase II to the postsynaptic density. J Biol Chem 1997; 272:13467-70; PMID:9153188; http://dx.doi.org/ 10.1074/jbc.272.21.13467 [DOI] [PubMed] [Google Scholar]
- 118.Cid C, Garcia-Bonilla L, Camafeita E, Burda J, Salinas M, Alcazar A. Proteomic characterization of protein phosphatase 1 complexes in ischemia-reperfusion and ischemic tolerance. Proteomics 2007; 7:3207-18; PMID:17683050; http://dx.doi.org/ 10.1002/pmic.200700214 [DOI] [PubMed] [Google Scholar]
- 119.Kim H, Claps G, Moller A, Bowtell D, Lu X, Ronai ZA. Siah2 regulates tight junction integrity and cell polarity through control of ASPP2 stability. Oncogene 2014; 33:2004-10; PMID:23644657; http://dx.doi.org/ 10.1038/onc.2013.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Twomey E, Li Y, Lei J, Sodja C, Ribecco-Lutkiewicz M, Smith B, Fang H, Bani-Yaghoub M, McKinnell I, Sikorska M. Regulation of MYPT1 stability by the E3 ubiquitin ligase SIAH2. Exp Cell Res 2010; 316:68-77; PMID:19744480; http://dx.doi.org/ 10.1016/j.yexcr.2009.09.001 [DOI] [PubMed] [Google Scholar]
- 121.Weigelt B, Peterse JL, van 't Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer 2005; 5:591-602; PMID:16056258; http://dx.doi.org/ 10.1038/nrc1670 [DOI] [PubMed] [Google Scholar]
- 122.Polekhina G, House CM, Traficante N, Mackay JP, Relaix F, Sassoon DA, Parker MW, Bowtell DD. Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-alpha signaling. Nat Struct Biol 2002; 9:68-75; PMID:11742346; http://dx.doi.org/ 10.1038/nsb743 [DOI] [PubMed] [Google Scholar]
- 123.Siemeister G, Lucking U, Wengner AM, Lienau P, Steinke W, Schatz C, Mumberg D, Ziegelbauer K. BAY 1000394, a novel cyclin-dependent kinase inhibitor, with potent antitumor activity in mono- and in combination treatment upon oral application. Mol Cancer Therap 2012; 11:2265-73; PMID:22821149; http://dx.doi.org/ 10.1158/1535-7163.MCT-12-0286 [DOI] [PubMed] [Google Scholar]
- 124.Adam MG, Berger C, Feldner A, Yang WJ, Wustehube-Lausch J, Herberich SE, Pinder M, Gesierich S, Hammes HP, Augustin HG, et al.. Synaptojanin-2 binding protein stabilizes the Notch ligands DLL1 and DLL4 and inhibits sprouting angiogenesis. Circ Res 2013; 113:1206-18; PMID:24025447; http://dx.doi.org/ 10.1161/CIRCRESAHA.113.301686 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


