Abstract
Methods for cell-selective analysis of proteome dynamics will facilitate studies of biological processes in multicellular organisms. Here we describe a mutant murine methionyl-tRNA synthetase (designated L274GMmMetRS) that charges the noncanonical amino acid azidonorleucine (Anl) to elongator tRNAMet in hamster (CHO), monkey (COS7), and human (HeLa) cell lines. Proteins made in cells that express the synthetase can be labeled with Anl, tagged with dyes or affinity reagents, and enriched on affinity resin to facilitate identification by mass spectrometry. The method does not require expression of orthogonal tRNAs or depletion of canonical amino acids. Successful labeling of proteins with Anl in several mammalian cell lines demonstrates the utility of L274GMmMetRS as a tool for cell-selective analysis of mammalian protein synthesis.
Metabolic labeling of cellular proteins with noncanonical amino acids (ncAAs) has emerged as an important tool for the study of proteome dynamics.1−5 Bio-orthogonal noncanonical amino acid tagging (BONCAT)6 uses metabolic incorporation of ncAAs into newly made proteins to enable their enrichment and subsequent identification by mass spectrometry. Newly synthesized proteins can be detected in fixed cells or cell lysates by [3 + 2]-cycloaddition reactions between the azide side-chain of an ncAA and alkynyl probes7−9 (Scheme 1a) or tagged for affinity enrichment and identified by tandem mass spectrometry (Scheme 1b). Recent studies have combined BONCAT with SILAC (stable isotope labeling with amino acids in cell culture)10 to acquire quantitative information about protein synthesis in mammalian cell lines11−13 and to identify secreted proteins in serum-supplemented media.14 These studies illustrate the utility of the BONCAT method in studies of proteome dynamics in mammalian cells.
Scheme 1. Targeted Expression of Mutant Aminoacyl-tRNA Synthetases (aaRS) for Cell-Selective BONCAT in Mammalian Cells.
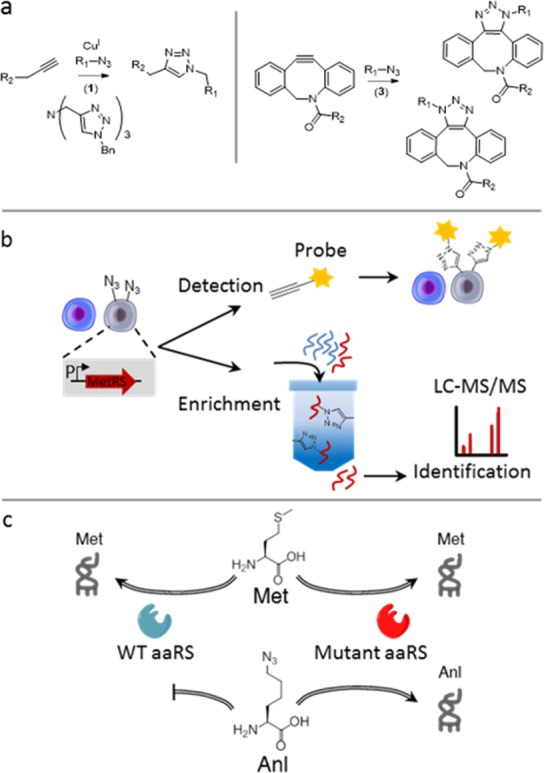
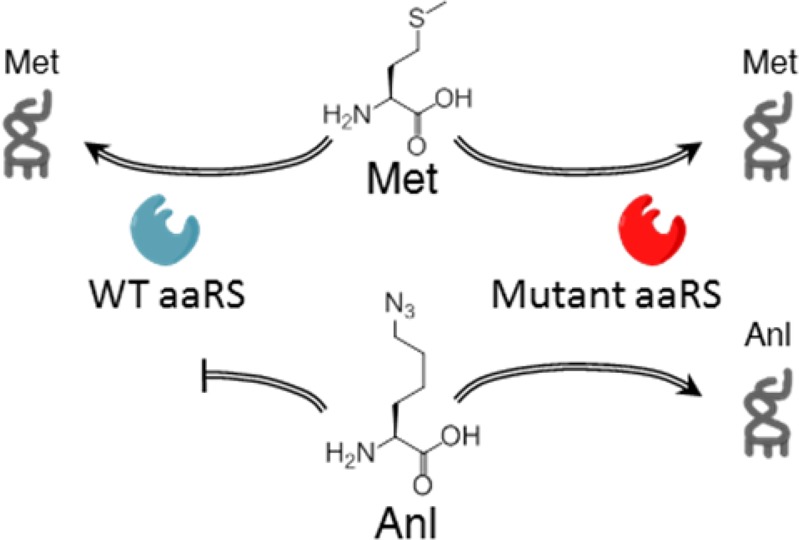
(a) The azide side chain of Anl in proteins (R1) is chemoselectively tagged through azide–alkyne cycloaddition reactions. (b) Controlled expression of mutant aaRS in cells of interest restricts labeling to those cells and enables the detection and identification of proteins in a cell-selective manner. (c) The endogenous mammalian aaRS charges Met but not Anl. A mutant aaRS is used to charge Anl to tRNAMet.
The temporal resolution provided by metabolic labeling methods can be complemented by cell-selectivity through the controlled expression of mutant aminoacyl-tRNA synthetases. Initial studies of this kind were implemented in bacterial systems15,16 and used to identify proteins injected into mammalian cells in the course of infection by the bacterial pathogen Yersinia enterocolitica.17 More recently, cell-selective methods have been developed for use in Caenorhabditis elegans(18) and Drosophila melanogaster.19 Extension of such methods to mammalian systems represents an attractive challenge.
Our cell-selective studies in bacterial systems used the L13N/Y260L/H301L mutant form of the E. coli methionyl-tRNA synthetase (which we designated NLL-MetRS) to activate the methionine surrogate l-azidonorleucine (Anl). Because wild-type MetRS does not activate Anl, BONCAT labeling occurs only in cells that express the NLL-MetRS. Although one might imagine using the NLL-MetRS for cell-selective Anl labeling of mammalian proteins (Scheme 1c), the E. coli enzyme charges only the initiator tRNA in mammalian cells; it does not charge elongator tRNAMet,20 and Anl labeling is restricted to N-terminal positions in mammalian proteins. This observation prompted us to develop new MetRS variants to tag mammalian cell proteins with Anl.
We investigated two complementary approaches to cell-selective incorporation of Anl into mammalian proteins. The first approach aimed to identify mutant mammalian aaRSs that would charge Anl to endogenous tRNAMet. The second approach involved coexpression of E. coli NLL-MetRS and E. coli elongator tRNAMet in mammalian cells. Reports by RajBhandary and co-workers showing successful concomitant expression of an active E. coli glutaminyl-tRNA synthetase and a functional amber suppressor tRNA that is not aminoacylated in vivo by any mammalian aaRS suggested to us that the mammalian translation machinery might co-opt an E. coli elongator tRNAMet that is charged by NLL-MetRS.21,22 To identify mutant mammalian aaRSs that charge Anl to endogenous tRNAMet, we made a small mutant aaRS library. We had previously observed that residues 13, 260, and 301 in the Met-binding pocket of E. coli MetRS23 can be mutated to enable activation of Anl.24 Because there are no reported structures of mammalian MetRSs, we used sequence alignment to determine the corresponding positions in the E. coli and eukaryotic MetRS enzymes (Figure 1a). Conservation of the residues that align with E. coli residues 13, 260, and 301 suggested that site-directed mutagenesis at the corresponding positions in the mouse MetRS (MmMetRS) would yield variants that charge Anl. Therefore, we constructed a small mutant library in which residues L274, Y527 and H562 of MmMetRS (“LYH”) were mutated to SLL, CLL, PLL, and NLL (Figure S1–S6). We also made an L274G mutant, reflecting our earlier observation that the corresponding L13G variant of the E. coli MetRS activates Anl.23 Finally, we overexpressed wild-type MmMetRS to determine whether overexpression of the wild-type enzyme causes observable levels of incorporation of Anl into cellular proteins.
Figure 1.
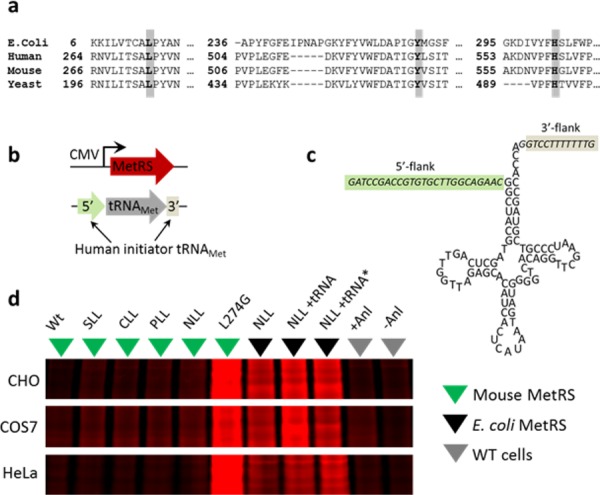
Identification of MetRS variants for metabolic labeling of mammalian proteins with Anl. (a) Sequence alignment of MetRS variants with positions 13, 260, and 301 of the E. coli MetRS highlighted. (b) Constructs for expression of MetRS and E. coli tRNAMet under control of the CMV promoter and an internal eukaryotic tRNA promoter, respectively (c) E. coli tRNAMet coding sequence carrying 5′- and 3′-flanking sequences derived from human initiator tRNAMet. (d) In-gel fluorescence image shows TAMRA labeling, which indicates Anl incorporation into cellular proteins. tRNA* carries a 3′-CCA tail in the tRNA coding sequence. The wild-type mouse MetRS is indicated by WT and the ±Anl conditions correspond to empty vector labeling controls. The same gels were stained with colloidal blue dye (Figure S11) for nonspecific labeling of all proteins.
MetRS variants were expressed under control of a CMV promoter (Figure 1b). To express the E. coli elongator tRNAMet, we adopted the approach of the RajBhandary group and introduced 5′- and 3′-flanking sequences derived from human initiator tRNA to enable proper RNA editing21 (Figure 1c). Notably, efficient transcription by mammalian RNA polymerase III requires a purine at position 9 of the tRNA coding sequence. In this case, adenine was used. This adenine lies in the A box of the internal promoter of eukaryotic tRNA genes.21 In mammalian cells, the 3′-CCA tail is added by a nucleotidyltransferase;25 therefore, we tested tRNA sequences with and without CCA tails (Figure S7–S9). We measured the activities of these constructs with respect to Anl incorporation by transiently transfecting several widely used mammalian cell lines, including Chinese hamster ovary (CHO) cells, monkey kidney cells (COS7), and an immortalized human cervical cancer cell line (HeLa). Cells were incubated in serum-containing medium supplemented with 1.5 mM Anl for 10 h and then lysed. Total cellular proteins were treated with alkyne-tetramethylrhodamine (TAMRA) (Figure S10) for labeling via copper-catalyzed azide–alkyne cycloaddition (reaction 1, Scheme 1a).7,26,27 After SDS-PAGE, TAMRA-labeled proteins were detected by in-gel fluorescence imaging (Figure 1d). As expected, Anl was not charged to any appreciable extent by the wild-type translational machinery. Anl incorporation was detected in cells carrying the E. coli NLL-MetRS, but we did not observe a significant increase in Anl incorporation as a function of tRNA expression. The variability of incorporation across different cell lines suggests that the expression levels of the tRNA and the synthetase may need to be optimized for each cell type, thereby limiting the general applicability of this approach. Nevertheless, variants of this strategy have already proven useful for cell-selective protein labeling: for example, Chin and co-workers have described successful proteome-wide labeling through the action of a genetically encoded pyrrolysyl-tRNA synthetase/tRNA pair in Drosophila.(28)
In contrast to our results with the E. coli NLL-MetRS/tRNAMet pair, expression of the L274G mouse MetRS, henceforth called L274GMmMetRS, resulted in robust incorporation of Anl across all three cell lines. Thus, a single mutation is sufficient to enable Anl incorporation into mammalian cell proteins. Surprisingly, we found no evidence of significant Anl labeling with any of the triple mutants we examined (Figure 1d).
An important determinant of the utility of a mutant synthetase in ncAA tagging is the selectivity of the synthetase with respect to activation of the ncAA and its canonical counterpart. To provide a practical measure of the selectivity of L274GMmMetRS, we examined protein labeling in CHO cells as a function of Anl concentration in serum-containing media. After Anl incorporation, proteins from cell lysates were labeled with alkyne-TAMRA (Figure 2a). We also performed Met competition assays by incubating CHO cells with 1.5 mM Anl in media supplemented with Met at different concentrations (Figure 2b). Assuming standard Michaelis–Menten kinetics for activation of both Met and Anl, we obtained the following expression (see Figure S12 for derivation):
where V is the rate of activation of Anl, KA and KM are the specificity constants (kcat/KM)) for Anl and Met, respectively, and C is a constant. Fitting this equation to the measured intensities of in-gel TAMRA fluorescence (taken as a measure of the rate of incorporation of Anl into cellular proteins) yielded a value of KA/KM of 0.25 (i.e., L274GMmMetRS activates Met 4-fold faster than Anl at equimolar concentrations of the two substrates).
Figure 2.
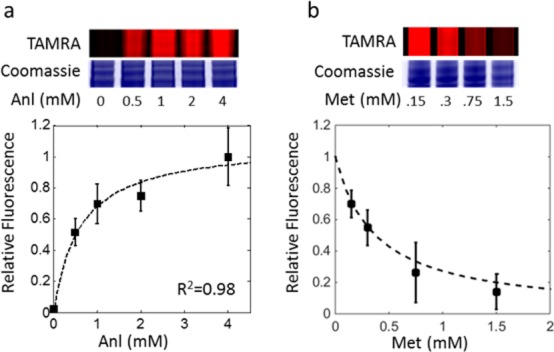
Selectivity of L274GMmMetRS. In-gel TAMRA fluorescence and Coomassie staining show Anl incorporation and total protein content, respectively. Fluorescence values were quantified and normalized to total protein levels. (a) Protein labeling in CHO media using the L274GMmMetRS at different Anl concentrations. (b) Protein labeling using both the L274GMmMetRS at increasing Met concentrations and 1.5 mM Anl. Plots on the bottom show Anl incorporation based on quantitated TAMRA fluorescence values. Error bars indicate standard deviation, and the dotted line represents line of best fit based on the equation above and was used to calculate selectivity.
The ability to restrict L274GMmMetRS expression to cells of interest should enable the cell-selective labeling of proteins for targeted analysis of protein synthesis. To validate this concept, we developed a reporter construct expressing both L274GMmMetRS and mCherry under control of a CMV promoter (pMaRSC). A T2A linker was used to express two disjointed proteins in the same open reading frame; we reasoned that fusing L274GMmMetRS and mCherry might alter the activity of the synthetase and confound protein labeling results (Figure 3a). As an additional control, we developed a similar expression construct without mCherry (pMaRS) (Figures S13 and S14). When transfected into CHO cells, both constructs enabled labeling of cellular proteins at comparable levels, indicating that L274GMmMetRS was functional in both contexts (Figures S14 and S15). We used the mCherry signal to track expression of L274GMmMetRS in pMaRSC-transfected cells. CHO cells transfected with pMaRSC and incubated with Anl (1.5 mM, 6 h) were fixed, permeabilized, and treated with alkyne-TAMRA. We observed TAMRA fluorescence exclusively in the mCherry-expressing cells, consistent with cell-selective incorporation of Anl into cellular proteins (Figure 3b).
Figure 3.
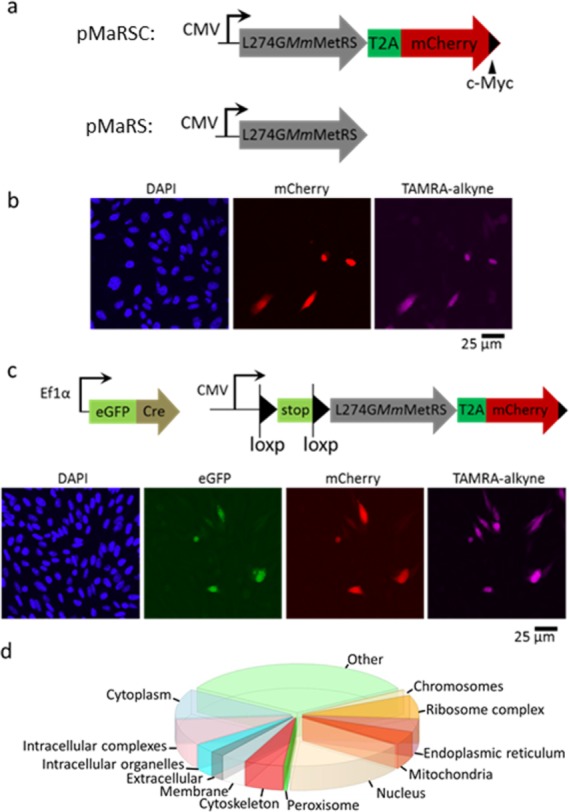
Cell-selective proteomic labeling and identification. (a) Design of pMaRSC and pMaRS constructs for the expression of L274GMmMetRS and a C-terminal Myc-tagged mCherry marker to track synthetase expression. (b) Fluorescence confocal microscopy image of CHO cells containing the pMaRSC construct. DAPI shows the position of cell nuclei. (c) Fluorescence confocal microscopy image of CHO cells with a LoxP-flanked L274GMmMetRS-T2A-mCherry sequence transiently transfected with an eGFP-Cre expression vector. The GFP signal identifies cells that express Cre; the mCherry signal marks cells in which L274GMmMetRS is expressed. All mCherry+ cells show TAMRA labeling. The TAMRA and mCherry signal intensities are linearly correlated (R2 = 0.7), but the mCherry intensity falls below the threshold for visual detection in some cells. (d) Identification of Anl-labeled proteins by tandem mass spectrometry and annotation in terms of cellular components using STRAP software.29 Proteins are listed in Table S1 and discussed more fully in Figure S16.
The CMV promoter drives constitutive expression of L274GMmMetRS. To enable conditional expression of the synthetase, we developed a stable CHO cell line containing an L274GMmMetRS-T2A-mCherry gene under control of a CMV promoter located just upstream of a loxP-flanked stop sequence (Figure 3c). These cells were transiently transfected with a second (activating) construct bearing a strong Ef1α promoter that drives expression of an eGFP-Cre fusion protein.
We used the eGFP signal to track expression of Cre and followed the mCherry signal to monitor recombination of the loxP sites. As expected, in the subset of cells that expressed Cre (as determined by the GFP signal), we observed both mCherry expression indicative of loxP recombination and TAMRA fluorescence from Anl-tagged cellular proteins, confirming that cell-selective proteomic labeling can be achieved through regulated transgene expression. These results suggest that targeted insertion of loxP-L274GMmMetRS cassettes into the ROSA26 locus of mouse embryonic stem (ES) cells30 should facilitate engineering of Cre-Lox mice in which tissue-selective proteomic studies can be performed.
To confirm the utility of L274GMmMetRS as a tool for proteomic analysis in mammalian systems, we used azadibenzocyclooctyne-functionalized beads to capture (reaction 2, Scheme 1a) and enrich Anl-labeled proteins from lysates of CHO cells that constitutively express the synthetase under CMV promoter control. After on-resin trypsinization of purified proteins, we analyzed the resulting tryptic peptides by tandem mass spectrometry (Table S1). A total of 996 proteins were identified in Anl-treated samples in short (120 min) analyses on a linear trap quadrupole (LTQ) Orbitrap mass spectrometer. Of these proteins, 786 were not found in Met-treated controls. Enrichment was confirmed by the acquisition of 19-fold more spectral counts in analyses of Anl-treated samples as compared to controls. We detected proteins associated with a wide variety of cellular components, including nuclear, cytoskeletal, mitochondrial, and membrane proteins (Figure 3d, S16). MS analysis of the proteome of HeLa cells expressing L274GMmMetRS identified 161 sites of Anl incorporation, 150 (93%) at internal Met positions (Figure S18, Table S2). This ratio mirrors the distribution of Met sites across the proteome.
In summary, we find that the L274GMmMetRS functions in a variety of mammalian cell lines and charges Anl at a rate sufficient for cell-selective proteomic analysis. Surprisingly, the single-site mutant is superior to all of the triple mutants we evaluated, including the murine orthologue of the E. coli NLL-MetRS used previously in cell-selective studies of protein synthesis.15 A variant of L274GMmMetRS has recently been used for cell-selective labeling of proteins in Drosophila,19 and we suggest that targeted expression of this enzyme will provide a broadly useful approach to proteome and secretome analysis in subpopulations of mammalian and other eukaryotic cells.
Acknowledgments
We are grateful for financial support by National Institutes of Health grant NIH R01 GM062523 and the Programmable Molecular Technology Initiative of the Gordon and Betty Moore Foundation, the Institute for Collaborative Biotechnologies through grant W911NF-09-0001 from U.S. Army Research Office. M.S. and S.H. were supported by the Gordon and Betty Moore Foundation through grant GMBF775 and NIH grant 1S10RR029594-01A1. A.M. was supported by a scholarship from the National Science and Engineering Research Council of Canada and by a postgraduate scholarship from the Donna and Benjamin M. Rosen Center for Bioengineering at Caltech. G.D.H. was supported by a postdoctoral fellowship from the National Science and Engineering Research Council of Canada.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.5b08980.
Author Present Address
⊥ Department of Chemistry, Princeton University, Princeton, New Jersey 08540, United States.
The authors declare no competing financial interest.
Supplementary Material
References
- Dieterich D. C.; Hodas J. J.; Gouzer G.; Shadrin I. Y.; Ngo J. T.; Triller A.; Tirrell D. A.; Schuman E. M. Nat. Neurosci. 2010, 13, 897. 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genheden M.; Kenney J. W.; Johnston H. E.; Manousopoulou A.; Garbis S. D.; Proud C. G. J. Neurosci. 2015, 35, 972. 10.1523/JNEUROSCI.2641-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinai L.; Rosenberg A.; Smith Y.; Segev E.; Ben-Yehuda S. Mol. Cell 2015, 57, 695. 10.1016/j.molcel.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammel M.; Zhang M. M.; Hang H. C. Angew. Chem., Int. Ed. 2010, 49, 5970. 10.1002/anie.201002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammel M.; Dossa P. D.; Taylor-Salmon E.; Hang H. C. Chem. Commun. 2012, 48, 1473. 10.1039/C1CC14939C. [DOI] [PubMed] [Google Scholar]
- Dieterich D. C.; Link A. J.; Graumann J.; Tirrell D. A.; Schuman E. M. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 9482. 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostovtsev V. V.; Green L. G.; Fokin V. V.; Sharpless K. B. Angew. Chem., Int. Ed. 2002, 41, 2596.. [DOI] [PubMed] [Google Scholar]
- Beatty K. E.; Fisk J. D.; Smart B. P.; Lu Y. Y.; Szychowski J.; Hangauer M. J.; Baskin J. M.; Bertozzi C. R.; Tirrell D. A. ChemBioChem 2010, 11, 2092. 10.1002/cbic.201000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y.; Mackey J. L.; Lopez S. A.; Liu F.; Houk K. N. J. Am. Chem. Soc. 2012, 134, 17904. 10.1021/ja309241e. [DOI] [PubMed] [Google Scholar]
- Ong S. E.; Blagoev B.; Kratchmarova I.; Kristensen D. B.; Steen H.; Pandey A.; Mann M. Mol. Cell. Proteomics 2002, 1, 376. 10.1074/mcp.M200025-MCP200. [DOI] [PubMed] [Google Scholar]
- Lu Y. Y.; Sweredoski M. J.; Huss D.; Lansford R.; Hess S.; Tirrell D. A. ACS Chem. Biol. 2014, 9, 334. 10.1021/cb400704n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagert J. D.; Xie Y. J.; Sweredoski M. J.; Qi Y.; Hess S.; Schuman E. M.; Tirrell D. A. Mol. Cell. Proteomics 2014, 13, 1352. 10.1074/mcp.M113.031914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden A. J.; Geoghegan V.; Katsch K.; Efstathiou G.; Bhushan B.; Boutureira O.; Thomas B.; Trudgian D. C.; Kessler B. M.; Dieterich D. C.; Davis B. G.; Acuto O. Nat. Methods 2013, 10, 343. 10.1038/nmeth.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichelbaum K.; Krijgsveld J. Methods Mol. Biol. 2014, 1174, 101. 10.1007/978-1-4939-0944-5_7. [DOI] [PubMed] [Google Scholar]
- Ngo J. T.; Champion J. A.; Mahdavi A.; Tanrikulu I. C.; Beatty K. E.; Connor R. E.; Yoo T. H.; Dieterich D. C.; Schuman E. M.; Tirrell D. A. Nat. Chem. Biol. 2009, 5, 715. 10.1038/nchembio.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdavi A.; Segall-Shapiro T. H.; Kou S.; Jindal G. A.; Hoff K. G.; Liu S.; Chitsaz M.; Ismagilov R. F.; Silberg J. J.; Tirrell D. A. J. Am. Chem. Soc. 2013, 135, 2979. 10.1021/ja400448f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdavi A.; Szychowski J.; Ngo J. T.; Sweredoski M. J.; Graham R. L.; Hess S.; Schneewind O.; Mazmanian S. K.; Tirrell D. A. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 433. 10.1073/pnas.1301740111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuet K. P.; Doma M. K.; Ngo J. T.; Sweredoski M. J.; Graham R. L.; Moradian A.; Hess S.; Schuman E. M.; Sternberg P. W.; Tirrell D. A. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 2705. 10.1073/pnas.1421567112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann I.; Marter K.; Kobler O.; Niehues S.; Abele J.; Muller A.; Bussmann J.; Storkebaum E.; Ziv T.; Thomas U.; Dieterich D. C. Nat. Commun. 2015, 6, 7521. 10.1038/ncomms8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo J. T.; Schuman E. M.; Tirrell D. A. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 4992. 10.1073/pnas.1216375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabkin H. J.; Park H. J.; RajBhandary U. L. Mol. Cell. Biol. 1996, 16, 907. 10.1128/MCB.16.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohrer C.; Sullivan E. L.; RajBhandary U. L. Nucleic Acids Res. 2004, 32, 6200. 10.1093/nar/gkh959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link A. J.; Vink M. K.; Agard N. J.; Prescher J. A.; Bertozzi C. R.; Tirrell D. A. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 10180. 10.1073/pnas.0601167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanrikulu I. C.; Schmitt E.; Mechulam Y.; Goddard W. A. 3rd; Tirrell D. A. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 15285. 10.1073/pnas.0905735106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H. F.; et al. Molecular Cell Biology, 4th ed.; W. H. Freeman: New York, 2000. [Google Scholar]
- Tornoe C. W.; Christensen C.; Meldal M. J. Org. Chem. 2002, 67, 3057. 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- Hong V.; Presolski S. I.; Ma C.; Finn M. G. Angew. Chem., Int. Ed. 2009, 48, 9879. 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott T. S.; Townsley F. M.; Bianco A.; Ernst R. J.; Sachdeva A.; Elsasser S. J.; Davis L.; Lang K.; Pisa R.; Greiss S.; Lilley K. S.; Chin J. W. Nat. Biotechnol. 2014, 32, 465. 10.1038/nbt.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia V. N.; Perlman D. H.; Costello C. E.; McComb M. E. Anal. Chem. 2009, 81, 9819. 10.1021/ac901335x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich G.; Soriano P. Genes Dev. 1991, 5, 1513. 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


