Abstract
Age-related macular degeneration (AMD) is a major cause of blindness in developed countries, and the molecular pathogenesis of early events in AMD is poorly understood. Senescence-accelerated OXYS rats develop AMD-like retinopathy. The aim of this study was to explore the differences in retinal gene expression between OXYS and Wistar (control) rats at age 20 d and to identify the pathways of retinal cell death involved in the OXYS retinopathy initiation and progression. Retinal mRNA profiles of 20-day-old OXYS and Wistar rats were generated at the sequencing read depth 40 mln, in triplicate, using Illumina GAIIx. A terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick end labeling (TUNEL) assay was performed to measure the apoptosis level. GeneMANIA was used to construct interaction networks for differentially expressed (DE) apoptosis-related genes at ages 20 d and 3 and 18 months. Functional analysis was suggestive of a developmental process, signal transduction, and cell differentiation as the most enriched biological processes among 245 DE genes at age 20 d An increased level of apoptosis was observed in OXYS rats at age 20 d but not at advanced stages. We identified functional clusters in the constructed interaction networks and possible hub genes (Rasa1, cFLAR, Birc3, Cdk1, Hspa1b, Erbb3, and Ntf3). We also demonstrated the significance of the extrinsic apoptotic pathway at preclinical, early, and advanced stages of retinopathy development. Besides the cell death signaling pathways, immune system-related processes and lipid-metabolic processes showed overrepresentation in the clusters of all networks. These characteristics of the expression profile of the genes functionally associated with apoptosis may contribute to the pathogenesis of AMD-like retinopathy in senescence-accelerated OXYS rats.
Keywords: aging, apoptosis, age-related macular degeneration, cell death, OXYS rats, retinal transcriptome, RNA-Seq
Abbreviations
- AMD
age-related macular degeneration
- DE
differential expression
- DEGs
differentially expressed genes
- GO
gene ontology
- CNS
central nervous system
- RPE
retinal pigment epithelium.
Introduction
Age-related macular degeneration (AMD) is the leading cause of irreversible vision loss in the elderly in industrialized countries. AMD is classified into 2 clinical forms by international consensus: the wet type and the most prevalent dry atrophic type (˜90% of cases).1,2 The dry AMD results from atrophy of the retinal pigment epithelium (RPE) and the consequent death of photoreceptors. The etiology of AMD is multifactorial, involving besides age a complex interplay of genetic, environmental, metabolic, and functional factors.3 Local inflammation and oxidative stress appear to play fundamental roles in the pathogenesis of AMD. However, the molecular pathological changes and the genetic pathways underlying the initiation and progression of AMD have not yet been sufficiently studied.
The least information is available on the early stages of the disease: the mechanisms that turn normal age-related alterations into pathological processes. This is especially true for the regulated cell death, which is a physiological process that controls an organism's homeostasis. Activation of programmed cell death is one of the basic mechanisms of cell loss in the course of pathological changes in diseased organs and tissues. This phenomenon contributes to the development of structural and functional disturbances. Dysregulation of cell death can cause tissue damage and initiate the development of a number of human diseases.4 Functional studies in animal and cellular models have greatly expanded the understanding of the function of apoptosis in eye development and homeostasis,5 but the molecular genetic background and the mode of death of photoreceptors in AMD are still unclear.6 Cell death in the retina is controlled by complex mechanisms, which are poorly understood at present; this state of affairs hinders the development of effective treatments of AMD.
High-throughput genomic studies integrated transcriptomic next-generation sequencing with bioinformatic analysis of molecular pathways. Today, this approach seems to be productive at elucidation of disease development.7 In recent years, the data on the retinal transcriptome have accumulated substantially,8 but the information on its changes with age and at various stages of AMD is scarce. Most of the transcriptomic studies in humans are focused only on late stages of the disease, neglecting the changes at the early stage and the molecular prerequisites of development of the disease.9-11 Preclinical stages of AMD cannot be studied in humans, therefore, adequate animal models are needed.
There is evidence that senescence-accelerated OXYS rats are a suitable experimental model of AMD.11-16 OXYS rats develop retinopathy similar to the dry form of human AMD according to the clinical signs, morphological features, and some molecular changes. In these rats, the clinical signs of retinopathy appear by the age of 3 months during a reduction in the transverse area of the RPE and impairment of choroidal microcirculation.13 Significant pathological changes in the RPE as well as clinical signs of advanced stages of retinopathy are evident in OXYS rats older than 12 months and manifest themselves as excessive accumulation of lipofuscin and amyloid in the RPE regions and whirling extensions of the basement membrane into the cytoplasm. Just as the dry form of human AMD, the initial alterations in the RPE cells later lead to atrophy of the choriocapillaris and the complete loss of photoreceptor cells in the OXYS rats' retinas by the age of 24 months.12,14,15 Previously, by means of RNA sequencing (RNA-Seq), we determined that the retinopathy in OXYS rats at the first stage (age 3 months) and second stage (age 18 months) develops simultaneously with changes in mRNA levels of hundreds of genes. Most of them are linked to immune responses, inflammation, the response to oxidative stress, Ca2+ homeostasis, and apoptosis.16 The main aim of this study was to identify the pathways of retinal cell death involved in the retinopathy initiation and progression. Further research on the gene expression changes involved in the regulation of apoptosis and identification of novel modes of cell death at different stages of the development of retinopathy in OXYS rats (particularly at the early preclinical stage) are expected to advance the understanding of the relevant pathogenesis. Therefore, in this work, we compared gene expression profiles in the retina of 20-day-old OXYS rats and control Wistar rats using RNA-Seq. Additionally, the interaction networks were constructed among differentially expressed (DE) genes involved in cell death in the retina at the preclinical stage of retinopathy and during active manifestation and progression of this pathology. We believe that the findings of our study may shed some light on the roles of apoptosis-related genes in the development of AMD-like retinopathy.
Results
Evaluation of apoptosis in the retina of OXYS and Wistar rats
The TUNEL assay was used for detection of apoptotic cells. Fluorescent images of TUNEL-positive cells in the retina are presented in Figure 1. At the age of 20 days, in the retina of both OXYS and Wistar rats, TUNEL-positive cells were detected in ganglionar, inner nuclear, and outer nuclear layers (Fig. 1A). At this age, the number of TUNEL-positive cells in OXYS rats was 1.5-fold greater (p < 0.05) than that in Wistar rats. By the age of three months, the number of TUNEL-positive cells in the retina of both OXYS and Wistar rats significantly decreased to r solitary cases and remained at the same level in the retina of 18-month-old animals, without any interstrain differences (Fig. 1B).
Figure 1.
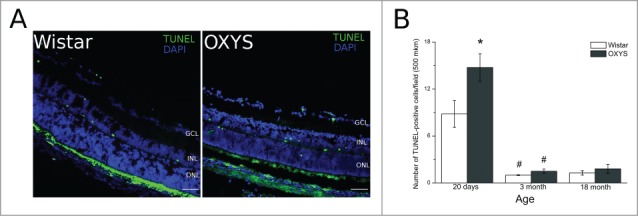
Apoptotic activity was analyzed by the terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick end labeling (TUNEL) assay. (A) Positive TUNEL straining (green) was observed under a fluorescence microscope. (B) Quantitative analysis. The number of apoptotic cells was calculated by averaging the number of positive TUNEL signals. *Significant interstrain differences, р < 0.05; #significant differences with the preceding age, р < 0.05. Bar 50 mkm.
Gene expression profiles in the retina of OXYS and Wistar
We used RNA-Seq to compare gene expression profiles of retinas of 20-day-old senescence-accelerated OXYS rats and age-matched control Wistar rats. Of 26,405 genes in the reference genome rno5, our reads were uniquely mapped to 14,640 genes, with at least 10 counts on average (Additional file 1). Differential expression of genes was evaluated using the DESeq software.17 We obtained a list of 245 DE genes (at padj <0.05). Of them, 160 were downregulated, and 85 were upregulated. When using the cutoff factor >2.0-fold, we obtained 82 genes total, all downregulated (Table 1). The most significantly downregulated genes in the OXYS retina were Tmem221, which is an integral component of the plasma membrane (transmembrane protein 221, fold change −87) and Gsta6 (similar to glutathione S-transferase A1, fold change −75), which regulates conjugation of reduced glutathione to a wide variety of exogenous and endogenous hydrophobic electrophilic molecules. We found significantly decreased expression of the genes encoding subunits of the mitochondrial respiratory chain: mt-ND4, mt-ND5, mt-ND4L (complex I, NADH/ubiquinone oxidoreductase), Atp5f1 (complex V, ATP synthase), and other mitochondrial proteins in the OXYS retina at the age of 20 d.
Table 1.
Differentially expressed genes (DEGs) with a fold change (FC) cutoff > 2.0 and adjusted p values < 0.05
| Gene title term | Gene ontology term | Gene Symbol | FC | Padj |
|---|---|---|---|---|
| transmembrane protein 221 | integral component of membrane | Tmem221 | −87.46 | 6.78E–52 |
| ENSRNOG00000045967 | unknown | unknown | −75.01 | 6.71E–14 |
| similar to Glutathione S-transferase A1 | Glutathione transferase activity | GSTA6 | −74.77 | 0.00104 |
| ENSRNOG00000045691 | unknown | unknown | −47.64 | 5.89E–05 |
| ENSRNOG00000017412 | unknown | unknown | −42.43 | 0.0058 |
| ribosomal protein L28-like | structural constituent of ribosome | RGD1565183 | −16.22 | 0.013 |
| ENSRNOG00000030351 | unknown | unknown | −14.96 | 2.5E–10 |
| ENSRNOG00000033517 | unknown | unknown | −13.24 | 0.021 |
| ENSRNOG00000030548 | Unknown | unknown | −10.41 | 5.59E–12 |
| nitric oxide synthase 2, inducible | arginine binding | NOS2 | −8.83 | 0.0067 |
| RT1 class I, locus CE4 | peptide antigen binding | RT1-CE4 | −8.73 | 0.0099 |
| ENSRNOG00000031667 | unknown | unknown | −8.57 | 0.021 |
| Osteopontin | extracellular matrix binding | Spp1 | −8.57 | 4.38E–13 |
| NLR family, apoptosis inhibitory protein 2 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process | Naip2 | −7.33 | 0.033 |
| ENSRNOG00000029389 | unknown | unknown | −6.89 | 0.0011 |
| SNRPN upstream reading frame protein-like | ubiquitin-protein transferase activity | Snurf | −6.86 | 1.48E–11 |
| neuropeptide VF precursor | receptor binding | Npvf | −6.6 | 0.0089 |
| ENSRNOG00000031454 | unknown | unknown | −6.56 | 0.02 |
| family with sequence similarity 3, member B | unknown | Fam3b | −6.31 | 0.035 |
| ENSRNOG00000032578 | unknown | unknown | −6.24 | 0.026 |
| patatin-like phospholipase domain containing 1 | lipid metabolic process | Pnpla1 | −5.75 | 0.0028 |
| coiled-coil domain containing 152 | unknown | Ccdc152 | −5.42 | 0.021 |
| Family with sequence similarity 70, member B | unknown | Tmem255b | −5.28 | 0.01 |
| thrombospondin 4 | calcium ion binding | Thbs4 | −5.11 | 0.034 |
| lumican | extracellular space | Lum | −4.74 | 0.01 |
| Cerebroside sulfotransferase | galactosylceramide sulfotransferase activity | Gal3st1 | −4.67 | 0.022 |
| phosphodiesterase 4D, cAMP-specific | 3′,5′-cyclic-AMP phosphodiesterase activity | Pde4d | −4.61 | 0.00055 |
| matrilin 3 | proteinaceous extracellular matrix | Matn3 | −4.48 | 0.01 |
| carbonic anhydrase 9 | carbonate dehydratase activity | Car9 | −4.14 | 0.011 |
| ENSRNOG00000033957 | unknown | unknown | −3.93 | 0.003 |
| ENSRNOG00000017661 | unknown | unknown | −3.86 | 0.003 |
| fibrinogen-like 2 | peptidase activity | Fgl2 | −3.83 | 4.23E–05 |
| Tp53rk binding protein | tRNA processing | TPRKB | −3.7 | 0.00098 |
| STEAP family member 4 | cupric reductase activity | Steap4 | −3.57 | 4.43E–05 |
| ENSRNOG00000029070 | unknown | unknown | −3.57 | 0.018 |
| ligase IV, DNA, ATP-dependen | DNA ligase (ATP) activity | Lig4 | −3.46 | 7.49E–19 |
| gremlin 1, DAN family BMP antagonist | transmembrane receptor protein tyrosine kinase activator activity | GREM1 | −3.44 | 0.0015 |
| myosin VIIb | ATP binding | Myo7b | −3.331 | 0.012 |
| family with sequence similarity 129, member A | negative regulation of protein phosphorylation | Fam129a | −3.26 | 0.036 |
| wingless-type MMTV integration site family, member 16 | frizzled binding | Wnt16 | −3.05 | 0.031 |
| interferon induced transmembrane protein 1 | integral component of membrane | Ifitm1 | −2.88 | 0.0033 |
| retinol saturase (all trans retinol 13,14 reductase) | all-trans-retinol 13,14-reductase activity | Retsat | −2.87 | 1.81E–11 |
| EGF-like module containing, mucin-like, hormone receptor-like 1 | calcium ion binding | Emr1 | −2.86 | 0.00076 |
| cellular retinoic acid binding protein 1 | retinal binding | Crabp1 | −2.85 | 5.2E–18 |
| ENSRNOG00000039025 | unknown | unknown | −2.84 | 0.012 |
| coiled-coil domain containing 146 | unknown | Ccdc146 | −2.8 | 9.9E–05 |
| BAI1-associated protein 2-like 1 | proline-rich region binding | Baiap2l1 | −2.71 | 8.3E–06 |
| sodium channel, voltage-gated, type VII, α | sodium channel activity | Scn7a | −2.64 | 0.031 |
| zinc finger protein 182-like | metal ion binding | Zfp943 | −2.62 | 7.96E–05 |
| family with sequence similarity 65, member C | unknown | Fam65c | −2.59 | 3.2E–11 |
| ENSRNOG00000032997 | unknown | unknown | −2.56 | 0.046 |
| ENSRNOG00000039790 | unknown | unknown | −2.56 | 0.00045 |
| YME1-like 1 (S. cerevisiae) | ATP binding | Yme1l1 | −2.54 | 5.51E–10 |
| solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 | amino acid transmembrane transporter activity | Slc7a11 | −2.53 | 0.0013 |
| Slc24a5 solute carrier family 24, member 5 | calcium, potassium:sodium antiporter activity | Slc24a5 | −2.53 | 0.0049 |
| Cd48 molecule | antigen binding | Cd48 | −2.51 | 0.037 |
| cubilin (intrinsic factor-cobalamin receptor) | calcium ion binding | Cubn | −2.49 | 0.0018 |
| coiled-coil domain containing 80 | heparin binding | Ccdc80 | −2.48 | 0.032 |
| mitochondrially encoded NADH 4L | NADH dehydrogenase (ubiquinone) activity | MT-ND4L | −2.46 | 7.58E–07 |
| phospholipase A2, group IIC | extracellular region | Pla2g2c | −2.37 | 0.023 |
| advillin | actin binding | Avil | −2.35 | 5.62E–05 |
| CGRP receptor component | DNA-directed RNA polymerase activity | Crcp | −2.35 | 9.8E–14 |
| asparaginase like 1 | asparaginase activity | Asrgl1 | −2.34 | 5.6E–06 |
| testis expressed 15 | fertilization | Tex15 | −2.33 | 4.23E–05 |
| zinc finger protein 420-like | metal ion binding | Zfp677 | −2.27 | 0.0012 |
| bone morphogenetic protein 6 | BMP receptor binding | Bmp6 | −2.25 | 0.0032 |
| disabled 2, mitogen-responsive phosphoprotein | phosphatidylinositol 3-kinase binding | Dab2 | −2.25 | 0.0098 |
| calcitonin receptor-like | adrenomedullin receptor activity | Calcrl | −2.23 | 0.035 |
| HAUS augmin-like complex, subunit 1 | cell division | Haus1 | −2.22 | 1.67E–07 |
| lysyl oxidase | carbohydrate binding | Lox | −2.21 | 0.0033 |
| sterile α motif domain containing 9-like | common myeloid progenitor cell proliferation | Samd9l | −2.2 | 0.0065 |
| hypothetical LOC300751 | unknown | RGD1311874 | −2.15 | 0.033 |
| zinc finger protein 873 | metal ion binding | Zfp873 | −2.16 | 0.013 |
| myosin IC | actin-dependent ATPase activity | Myo1c | −2.11 | 4.88E–08 |
| inositol (myo)-1(or 4)-monophosphatase 2 | inositol monophosphate 1-phosphatase activity | Impa2 | −2.10 | 0.0012 |
| cadherin 19, type 2 | calcium ion binding | Cdh19 | −2.09 | 0.007 |
| ENSRNOG00000048784 | unknown | unknown | −2.08 | 1.42E–06 |
| regulator of telomere elongation helicase 1 | ATP-dependent DNA helicase activit | Rtel1 | −2.07 | 2.19E–10 |
| diacylglycerol kinase, gamma | ATP binding | Dgkg | −2.06 | 0.035 |
| similar to Centromeric protein E (CENP-E protein) | unknown | Ccdc175 | −2.05 | 0.00989 |
| DNA primase, p49 subunit | DNA primase activity | Prim1 | −2.02 | 1.12E–06 |
| phosphoribosyl pyrophosphate synthetase 2 | ADP binding | Prps2 | −2.00 | 0.013 |
To identify biological functions associated (more than by chance) with the DE genes, we carried out gene annotation enrichment analysis in DAVID. The Gene Ontology (GOTERM_BP_ALL) terms of enriched biological processes are shown in Figure 2. DAVID revealed significant enrichment in Gene Ontology terms relevant to developmental processes, signal transduction, regulation of phosphorylation, enzyme-linked receptor protein signaling pathways, extracellular matrix organization, the response to a hormone stimulus, and oxidative phosphorylation. The enriched Gene Ontology terms (GOTERM_FAT) and results of functional annotation clustering are shown in the corresponding lists of Additional file 2.
Figure 2.
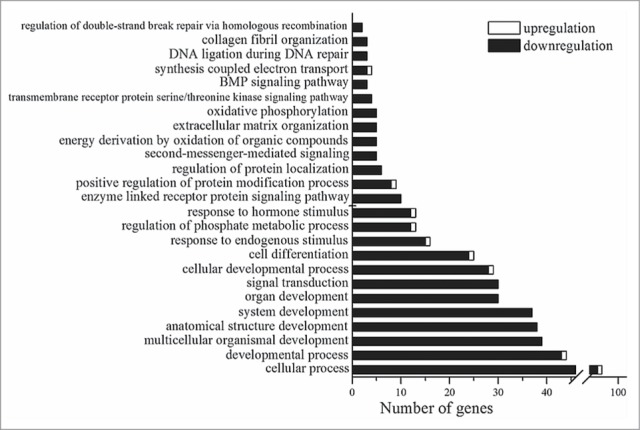
Statistically significant (p < 0.05) Gene Ontology terms that are related to the genes whose expression is changed in the retina of OXYS rats in comparison with Wistar rats at age 20 d.
We compared the data on transcriptomic analysis of the retina in 20-day-old animals with the results on 3- and 18-month-old OXYS and Wistar rats described earlier.16 Retinal expression of 64 genes was significantly different between OXYS and Wistar rats at both 20 d and 3 months of age. These genes participate in the processes regulating DNA repair (Lig4, Mdc1, RTEL1, and Hmg1l1) and metabolic processes related to DNA (Prim1, Lig4, Mdc1, and RTEL1), cell cycle (Cks2, Kif2c, Mdc1, Haus1, and Mapk12), and metabolic processes (n = 24) and catabolic processes associated with lipids (Acadsb, Pla2g2c, and Lipa). Comparison of 20-day-old and 18-month-old animals revealed 57 common DE genes involved in metabolism (n = 19) and cellular processes (n = 23), in particular in DNA repair (Lig4, Rtel1, and Hmg1l1) and regulation of the response to a hormonal stimulus (Rtel1, Il13ra1, Hmg1l1, and RT1-A1). The common gene ontologies, uniting the DE genes, were observed in 20-day-old and 3-month-old rats (a response to hormonal stimuli) and in 20-day-old and 18-month-old rats (organization of the extracellular matrix). Earlier to validate the RNA-Seq data, we performed quantitative RT-PCR for selected genes.16 Independent age-matched groups of animals were used for qPCR validation. RT-PCR confirmed that the chosen genes were significantly down/upregulated in the OXYS strain. Therefore, our RNA-Seq results must be reliable.16
In this study, in addition to gene expression profiles of 20-day-old animals, we used previously obtained RNA-Seq data on differences in gene expression between OXYS and Wistar retinas at ages of 3 and 18 months. According to DAVID, at the age of 20 days, 32 DE genes were found to be associated with apoptosis; of these, 26 were downregulated, and 6 were upregulated. At the age of 3 months, there were 48 DE genes associated with apoptosis. Of them, 42 were downregulated, and 6 were upregulated in OXYS rats. At the age of 18 months, of the 41 DE genes associated with apoptosis, mRNA expression of 4 genes was increased in OXYS rats and decreased for the other 37 genes, in comparison with Wistar rats. Figure 3 shows the Venn diagram visualizing the overlaps among the lists of DE genes associated with apoptosis in the 3 age groups. Interstrain differences in the expression level of the gene Lig4 were found at all 3 ages, and for the gene Aven between ages 20 d and 3 months. In addition, 9 common genes were found for ages 3 and 18 months (Bcl2l10, Cd24, Casp8, Casp7, LOC298795, C6, Cdk1, Alox12, and Myd88) and 4 common genes for ages 20 d and 18 months (Erbb3, Tgfa, Ctnna1, and Lig4).
Figure 3.
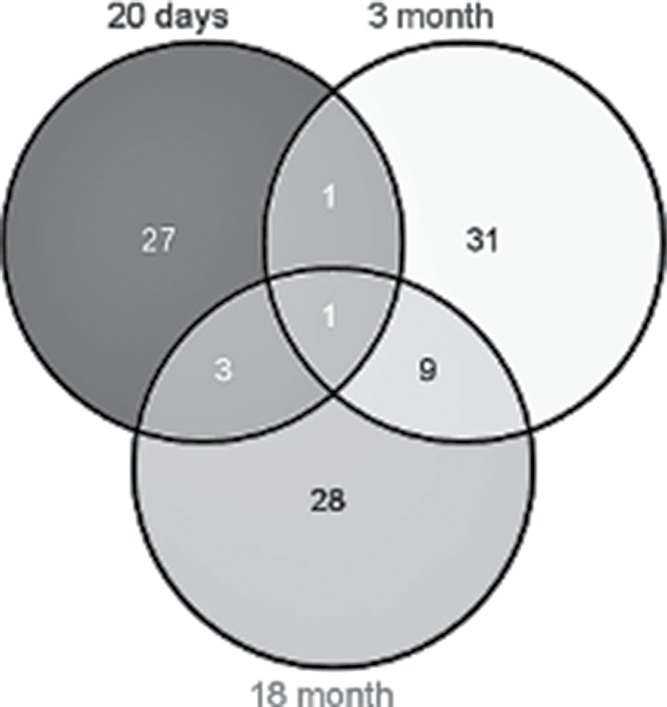
This Venn diagram shows overlapping sets of genes at the 3 ages.
Construction of the interaction network and identification of hub genes
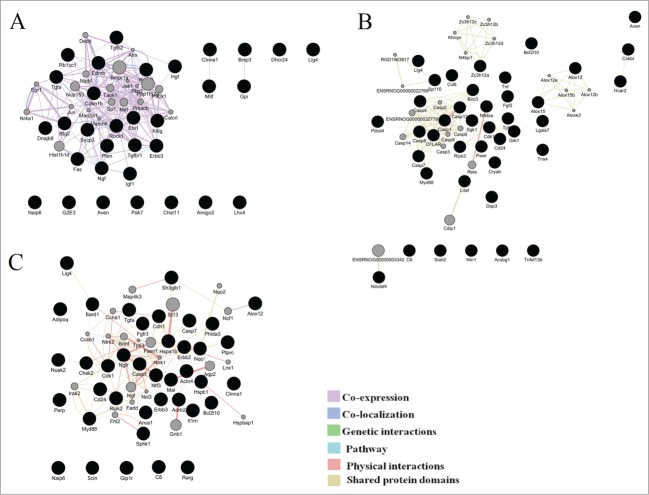
We used GeneMANIA to construct the interaction networks for the analysis of transcriptomic changes associated with functional categories of cell death during the retinopathy development. Figure 4 shows the resulting networks for ages 20 d, 3 months, and 18 months. It is evident that most of the genes are directly or indirectly related to one another.
Figure 4.
Illustration of the association networks of apoptosis in 20-day-old (A), 3-month-old (B), and 18-month-old rats (C). In each of these networks, black circles denote differentially expressed genes (DEG), whereas the GeneMANIA-predicted genes are shown in gray.
To describe the network architecture, we performed a topological analysis using the Network analyzer plugin of Cytoscape. The interaction networks were represented by 52 nodes and 13 multiedge node pairs for 20-day-old rats, 60 nodes and 23 multiedge node pairs for 3-month-old rats, and 58 nodes and 30 multiedge node pairs for 18-month-olds. The network diameter and the mean path length were found to be 4 and 1.9 units in 20-day-old rats, 5 and 2.5 units in 3-month-old rats, and 6 and 2.6 units in 18-mounth-old rats. These results were indicative of the “small-world” topology at all ages.
The node degree is the simplest and most important parameter: the higher the node degree, the greater the likelihood that the node is central or most important. We identified genes with the node degree >10 and analyzed other topological parameters for these genes such as the average length of the shortest path, closeness centrality, betweenness centrality, and the cluster coefficient (Table 2). The nodes that had a shorter average shortest path length, a greater clustering coefficient, greater closeness centrality, lower betweenness centrality, and higher node degrees were identified as highly connected nodes (hub genes). Therefore, at the age of 20 days, 8 hub genes were identified, of which only Rasa1 was a DE gene, and the remaining hubs were proposed by GeneMANIA. At the age of 3 months, 8 hub genes were identified, of which 7 were DE genes (Casp1, Casp12, Casp4, Cflar, Birc3, Casp7, and Nfkbia). At the age of 18 months among the 5 hub genes, 4 genes were DE between the 2 strains of rats (Cdk1, Hspa1b, Erbb3, and Ntf3).
Table 2.
Topological parameters of the nodes of association networks related to apoptosis in rats at ages 20 d and 3 and 18 months. The optimal parameters are in boldface
| Gene name | Degree | Average Shortest Path Length | Betweenness Centrality | Closeness Centrality | Clustering Coefficient |
|---|---|---|---|---|---|
| 20 days | |||||
| Cdkn1b | 15 | 1.68 | 0.064 | 0.59 | 0.27 |
| Rasa1 | 14 | 1.74 | 0.0095 | 0.58 | 0.64 |
| Kitlg | 14 | 1.74 | 0.031 | 0.58 | 0.56 |
| Ets1 | 14 | 1.71 | 0.08 | 0.59 | 0.41 |
| Tgfbr1 | 13 | 1.71 | 0.024 | 0.59 | 0.63 |
| Rock1 | 13 | 1.74 | 0.019 | 0.58 | 0.5 |
| Ednrb | 12 | 1.71 | 0.037 | 0.59 | 0.47 |
| Erbb3 | 12 | 1.84 | 0.012 | 0.54 | 0.56 |
| Nup153 | 11 | 1.87 | 0.034 | 0.54 | 0.51 |
| Ngf | 10 | 1.97 | 0.031 | 0.51 | 0.14 |
| Tgfa | 10 | 1.76 | 0.04 | 0.57 | 0.27 |
| Pten | 10 | 1.82 | 0.013 | 0.55 | 0.53 |
| 3 months | |||||
| Casp1 | 22 | 1.92 | 0.077 | 0.52 | 0.52 |
| Casp12 | 22 | 1.92 | 0.084 | 0.52 | 0.58 |
| Casp8 | 21 | 2.02 | 0.094 | 0.49 | 0.49 |
| Casp4 | 16 | 2.17 | 0.009 | 0.46 | 0.73 |
| CFLAR | 16 | 2.17 | 0.015 | 0.46 | 0.79 |
| Birc3 | 15 | 1.96 | 0.091 | 0.51 | 0.44 |
| Casp7 | 13 | 2.27 | 0.0 | 0.44 | 1.0 |
| Nfkbia | 13 | 1.88 | 0.189 | 0.53 | 0.2 |
| Zc3h12a | 13 | 2.04 | 0.142 | 0.49 | 0.32 |
| Casp14 | 11 | 2.27 | 0.0 | 0.44 | 1.0 |
| Alox15 | 10 | 2.42 | 0.127 | 0.41 | 0.39 |
| Ripk3 | 10 | 2.10 | 0.057 | 0.48 | 0.33 |
| Sgk1 | 10 | 2.17 | 0.030 | 0.46 | 0.31 |
| 18 months | |||||
| Cdk1 | 21 | 1.94 | 0.20 | 0.51 | 0.09 |
| Ngfr | 17 | 2.11 | 0.08 | 0.47 | 0.26 |
| Hspa1b | 17 | 1.93 | 0.13 | 0.52 | 0.19 |
| Erbb3 | 13 | 2.04 | 0.07 | 0.49 | 0.24 |
| Adrb2 | 13 | 2.13 | 0.08 | 0.47 | 0.18 |
| Ngf | 12 | 2.217 | 0.03 | 0.45 | 0.29 |
| Ntf3 | 12 | 2.26 | 0.03 | 0.44 | 0.38 |
| Mal | 12 | 2.11 | 0.05 | 0.47 | 0.24 |
| Erbb2 | 12 | 2.02 | 0.08 | 0.50 | 0.25 |
| Actn4 | 11 | 2.17 | 0.07 | 0.46 | 0.27 |
| Casp8 | 10 | 2.43 | 0.01 | 0.41 | 0.33 |
The GeneMANIA results on enrichment with Gene Ontology terms for the network's members are presented in Table 3. It is noteworthy that among the most enriched Gene Ontology terms were the extrinsic apoptotic signaling pathway (p < 3.53E–11) in the gene set of 20-day-old rats, a positive regulation of neuron apoptotic process (p < 1.82E–5) and a necroptotic process (p < 1.87E–5) in the gene set of 3-month-old rats, and a neuron apoptotic process (p < 6.2E–13) in the gene set of 18-month-olds.
Table 3.
Cell death gene ontology (GO) terms for differentially expressed genes (DEGs) and additional related genes according to GeneMANIA analysis for each category (20 days, 3 months, and 18 months)
| GO annotation | Genes/Total number of genes | p value |
|---|---|---|
| 20 days | ||
| GO:0097191 extrinsic apoptotic signaling pathway | 13/200 | 3.53e–11 |
| GO:0051402 neuron apoptotic process | 12/218 | 1.77e–9 |
| GO:0097192 extrinsic apoptotic signaling pathway in absence of ligand | 7/73 | 1.43e–6 |
| GO:0043154 negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | 6/75 | 3.21e–5 |
| GO:2001234 negative regulation of apoptotic signaling pathway | 7/151 | 6.94e–5 |
| GO:0008625 extrinsic apoptotic signaling pathway via death domain receptors | 5/56 | 1.7e–4 |
| GO:2001235 positive regulation of apoptotic signaling pathway | 5/123 | 2.93e–3 |
| GO:0008637 apoptotic mitochondrial changes | 4/90 | 1.14e–2 |
| GO:0090199 regulation of release of cytochrome c from mitochondria | 3/37 | 1.45e–2 |
| 3 months | ||
| GO:0043525 positive regulation of neuron apoptotic process | 6/52 | 1.82e–5 |
| GO:0070266 necroptotic process | 5/26 | 2.13e–5 |
| GO:0097193 intrinsic apoptotic signaling pathway | 8/215 | 1.26e–4 |
| GO:0097191 extrinsic apoptotic signaling pathway | 8/200 | 8.73e–5 |
| GO:0097194 execution phase of apoptosis | 4/60 | 7.24e–3 |
| GO:0097300 programmed necrotic cell death | 5/32 | 4.3e–5 |
| GO:0008637 apoptotic mitochondrial changes | 4/90 | 2.4e–2 |
| GO:0008625 extrinsic apoptotic signaling pathway via death domain receptors | 4/56 | 5.89e–3 |
| GO:1901214 regulation of neuron death | 7/217 | 1.19e–3 |
| GO:0043280 positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 9/179 | 1.02e–5 |
| 18 months | ||
| GO:0051402 neuron apoptotic process 15/218 | 6.2e–13 | |
| GO:0070997 neuron death | 15/253 | 1.45e–12 |
| GO:0097191 extrinsic apoptotic signaling pathway | 10/200 | 3.06e–7 |
| GO:0097194 execution phase of apoptosis | 3/60 | 3.64e–2 |
| GO:0008637 apoptotic mitochondrial changes | 4/90 | 1.09e–2 |
| GO:0043281 regulation of cysteine-type endopeptidase activity involved in apoptotic process | 5/179 | 1.25e–2 |
| GO:0070265 necrotic cell death | 3/39 | 1.44e–2 |
| GO:0072332 intrinsic apoptotic signaling pathway by p53 class mediator | 3/60 | 3.64e–2 |
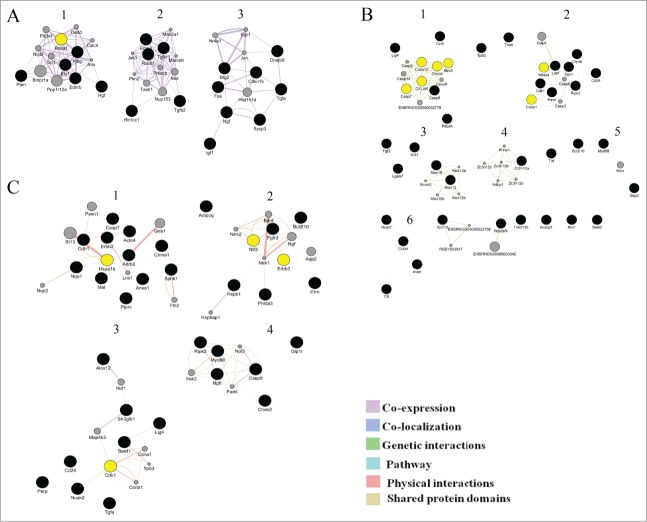
Cluster analysis: At the next step, in each network, we performed cluster analysis using the ClusterMarket plugin community cluster (GLay). The nodes nearest to one another in the network were combined into a single cluster. Each node could belong to only one cluster. Figure 5 shows the resulting network clusters for the ages 20 days, 3 months, and 18 months.
Figure 5.
Clusters in the schemes of gene networks for 20-day-old (A), 3-month-old (B), and 18-month-old rats (C). In each of these networks, black circles denote differentially expressed genes (DEG), whereas the GeneMANIA-predicted genes are shown in gray. The hub genes are yellow
In the gene network of 20-day-old rats, 3 clusters were identified. The cluster analyses show that the second and third clusters of genes participates in the regulation of apoptotic processes, but the first clusters of genes involved in the regulation of signaling of many factors (Table 4). In the gene network of 3-month-old rats, GLay identified 6 clusters, among which the largest number of genes that regulate apoptosis belonged to the first 2 clusters (Table 4).The third cluster included genes that regulate the metabolism of lipids and lipoproteins. The fourth cluster contained genes encoding proteins with CCCH-type zinc finger domains. This family of proteins regulates Toll-like receptor signaling and activation of macrophages participating in inflammatory diseases and host immunity.18 The fourth cluster also included the Tnf gene, an important inflammatory agent, and Diva (Bcl2l10), a member of the Bcl2 family of proteins playing an important role in the survival of cells via suppression of the mitochondrial pathway of apoptosis.19
Table 4.
Cluster analysis of gene networks for 20-day-old, 3-month-old and 18-month-old rats. Showing clusters with the number of nodes more than 3
| Cluster | Genes | Biological processes |
|---|---|---|
| 20 days | ||
| 1 | Pten, Rasa1, Hgf, Ets1, Kitlg, Ednrb | Regulation of signaling by interleukins (Il-2 and Il-7) and signal transduction EGFR, VEGF, FGFR, insulin receptor, NGF, PDGF, SCF-KIT, ERBB2, ERBB4, PIP3 activates AKT signaling, signaling by BMP, signaling by TGF-β receptor complex, signaling by GPCR; inositol phosphate metabolism and metabolism of lipids and lipoproteins and axon guidance. |
| 2 | Rock1, Erbb3, Tgfbr1, Tgfb2, Rb1cc1 | apoptotic execution phase, CaM pathway, calmodulin induced events, immune system and signal transduction. |
| 3 | Fas, Btg2, Dnajb9, Cdkn1b, Tgfa, Ngf, Sycp3, Igf1 | Caspase-8 activation by cleavage, FasL/CD95L signaling, regulation by c-FLIP, innate immune system. |
| 3 months | ||
| 1 | Lig4, Ccl6, Tns4, Tp63, Casp8, Pdcd4, Casp12, Casp4, Casp7, cFlar,Birc3 | TRAIL signaling, death receptor signaling, regulation by c-Flip, Caspase-8 activation by cleavage and dimerization of procaspase-8 |
| 2 | Casp1, Cdk1, Litaf, Cryab, Sgk1, Pawr, Ripk3, Cd24, Nfkbia | the role of DCC in the regulation of apoptosis and the execution phase of apoptosis |
| 3 | Fgf2, Lgals7, Grk1, Alox15, Alox12 | metabolism of lipids and lipoproteins (regulation of the metabolism of arachidonic acid) |
| 18 months | ||
| 1 | Hspa1b, Nqo1, Mal, Ptprc, Anxa1, Sphk1, Ctnna1, Adrb2, Cdh1, Erbb2, Actn4, Casp7 | FasL/CD95L signaling, TRAIL signaling, regulation of apoptosis by c-FLIP, in caspase 8 and procaspase 8 dimerization in the apoptotic pathways, and the regulation of the immune system |
| 2 | Hspb1, Phlda3, Il1rn, Erbb3, Ntf3, Fgfr3, Bcl2l10, Adipoq | apoptotic cleavage of cellular proteins (apoptotic execution phase), activation, myristolyation of BID and translocation to mitochondria, apoptotic factor-mediated response (cytochrome c-mediated apoptotic response and SMAC-mediated apoptotic response) in intrinsic pathway and caspase-8 activation by cleavage and dimerization of procaspase-8. |
| 3 | Alox12, Sh3glb1, Lig4, Bard1, Cdk1, Cd24, Perp, Nuak2, Tgfa | integration of energy metabolism, transmission across chemical synapses, potassium channels in neuronal system, signaling by NGF. |
| 4 | Ripk2, Myd88, Ngfr, Casp8, Chek2 | regulation of innate immune system and metabolism of lipids and lipoproteins. |
The fifth cluster consisted of the genes Myd88, Dap3, and Rela. Dap3 (death-associated protein 3) encodes a nucleotide-binding protein, which is a positive mediator of apoptosis. The protein product of Myd88 regulates Toll-like receptor signaling and determines the cell death pathway (apoptosis or necrosis) after UV irradiation.20
Analysis of the gene network in 18-month-old animals revealed the existence of 6 clusters: 2 large ones and 4 smaller ones. Only the first and the second clusters included genes involved in apoptotic processes (Table 4). Thus, the cluster analysis revealed a subsystem with distinct biological properties that is involved in the regulation of apoptosis.
Discussion
Aging-associated changes occurring at the molecular level contribute to cellular dysfunction and disease. It is important to determine the factors that affect normal aging in order to distinguish normal aging from aging affected by factors that are involved in a disease process, in our case, in AMD-like retinopathy in OXYS rats. It is also necessary to determine the molecular background of the disease at preclinical stages, which cannot be studied in humans. Our study includes a comparative RNA-Seq analysis of the retinal transcriptome between OXYS and Wistar rats at the age of 20 days, which is the period of completion of postnatal development of the organ of vision.
Although previous ophthalmological examination of OXYS rats showed no signs of retinal deterioration in 20-day-old animals,21 here we show that already by the age of 20 days, OXYS rats have substantial changes in the expression of 245 genes. The overwhelming majority of these genes was downregulated in OXYS rats in comparison with age-matched Wistar rats. OXYS rats may have an aberrant transcriptional response to a developmental process because all DE genes with the cutoff >2.0-fold were downregulated. This trend was also obvious at ages three and 18 months. We observed a lot of transcriptional alterations in genes associated with developmental processes and cell differentiation. Retinal development is a complex process involving glial and neuronal differentiation, establishment of specific neuronal pathways, formation of functional synapses and of vascular plexuses, and, eventually, the onset of vision.22 Thus, the identified early changes in gene expression in OXYS rats before the first clinical signs of retinopathy can be the foundation for the development and progression of retinopathy at a later age.
The main aim of this study was to identify the pathways of retinal cell death involved in the retinopathy progression. We found only stand-alone cases of TUNEL-positive cells in OXYS retinas at ages 3 and 18 months. This phenomenon can be explained as follows. Cell death may occur as an irregular process with undetectable peaks within a short period or simultaneously with chronic hypoxia caused by partial occlusion of blood vessels. Thus, there could have been a slow gradual cell loss. Another possible reason is that cell elimination is not accompanied by DNA fragmentation and therefore is not detected by the TUNEL assay.
Here we described the features of an apoptotic gene expression profile in OXYS rats at different stages of retinopathy development. There are several notable findings. First, most of the DE genes involved in cell death pathways showed a reduced level of mRNA in OXYS rats compared with age-matched Wistar rats regardless of age. Second, comparison of the RNA-Seq data from rats of the ages 20 d and 3 and 18 demonstrated that each stage was characterized by a different set of DE genes associated with cell death. In spite of the different sets of DE genes, we found that most of the DE genes that are linked to apoptosis in the OXYS retina at all ages are involved in the extrinsic apoptosis pathway and neuronal death. Furthermore, the enrichment with the Gene Ontology term extrinsic apoptotic signaling pathway points to the essential role of inflammatory processes in the initiation of cell death.
Cell death during healthy retinal development is necessary for formation of a functional vision organ. Apoptosis is the main mechanism of cellular homeostasis during the development of the retina: this process ensures the selection and elimination of extraneous neurons, reduces their number down to the physiological norm, promotes adequate neurogenesis, and ensures formation of well-defined neurons and selection of appropriate interneuronal connections.23,24 Disturbances in the signaling pathway of cell death at the early stages of development contribute to the onset of various diseases related to loss of vision (such as retinal dystrophy and retinopathy of prematurity).25 It is possible that the changes in the regulation of retinal apoptosis—that we uncovered here during the period of completion of retinal development in the early ontogenesis—contribute to the development of AMD-like retinopathy in OXYS rats by the age of 3 months. The same is probably true for suppression of mRNA expression of some genes involved in the processes of regulation of programmed cell death in OXYS rats.
It should be note, that our data show similar tendency to research of processes of wound healing – a marker for longevity or aging phenotype in the adult animals.26 For example, in old αMUPA mice the apoptosis are regulated after full wound closure and none is differentially expressed in the intact skin, whereas the WT mice exhibited significant age-related differences in gene expression of apoptosis both in intact animals and in the course of skin wound healing.27
Construction of association networks should help to identify crucial components of the pathophysiology. During construction of such networks by means of topological analysis, we detected central or most important nodes: the genes that probably play essential roles in the relevant biological systems.
It should be noted that the common gene among the 3 ages was Lig4, which is DNA ligase 4 participating in the repair of DNA double-strand breaks via nonhomologous end-joining (NHEJ).28 Expression of Lig4 mRNA is lowered at ages 20 d and 3 and 18 months in OXYS rats. It is known that changes in Lig4 expression lead to aberrant repair of DNA via NHEJ. If unrepaired double-strand breaks trigger apoptosis (via the ATM–ATR signaling pathway), then repair of such lesions is an important antiapoptotic mechanism. Nevertheless, the NHEJ pathway takes place mostly in the G1 phase of the cell cycle and is subject to errors; it is thought that this process is responsible for chromosomal rearrangements. At present, a hypothesis exists that DNA repair via NHEJ can touch off apoptosis.29 It is known that during aging, oxidative damage accumulates in DNA and the repair capacity of the cells is weakened; these changes may promote cell death via apoptosis.29 Suppression of the system of repair of single-strand and double-strand breaks in DNA is characteristic of many neurodegenerative diseases including AMD 30 and Alzheimer disease.27,31, There is evidence that mice with the defective NHEJ pathway undergo accelerated senescence.32 It is also known that mutations in the Lig4 gene are a cause of severe immunodeficiency.33
It should be noted that one of the DE genes in the retina of 3-month-old OXYS rats, Ripk3, is among key regulators of programmed necrosis. Mice homozygous for knockout alleles of Ripk3 show resistance to induced inflammatory responses.34 This finding is intriguing because OXYS rats show a weakened delayed hypersensitivity reaction and a decline of T cell-mediated immunity.35
We found that the most important node of the network in 20-day-old rats is Rasa1. The algorithm GeneMANIA offers for the hub gene Rasa1 the Gene Ontology annotation “neuronal apoptotic process.” KEGG annotated Rasa1 as a participant in the MAPK (Ras/mitogen-activated protein kinase) signaling. This pathway can trigger both programmed cell death and necrosis.36 In addition, the Rasa1 gene is a known negative regulator of the Ras pathway via enhancement of activity of the GAP proteins (GTPase-activating proteins).37 The strain of mice deficient in Rasa1 is characterized by the death of neuronal cells, disorganization of blood vessels, and high embryonic mortality.38
The most substantial changes in the expression of genes involved in the processes of regulation of cellular death were uncovered here in the retina of 3-month-old OXYS rats. Gene Ontology annotations of the relevant hub genes (Casp1, Casp12, Casp6, cFLAR, Birc3, Casp7, Nfkbia, and Casp14) were as follows: positive regulation of a neuronal apoptotic process, a necroptotic process, the execution phase of apoptosis, apoptotic mitochondrial changes, and the intrinsic apoptotic pathway. Cellular FLICE (FADD-like IL-1β-converting enzyme, also known as c-flip and cFLAR) is an inhibitory protein that controls activation of initiatory caspases and can suppress apoptosis that is induced by cytokines and chemotherapy.39 In addition, cFLAR performs an important function in necroptosis 40,41 and autophagy.42 Birc3 is a member of the family of inhibitors of apoptosis (IAPs) and performs various functions: it regulates apoptosis, innate immune responses and inflammation, and migration and proliferation of cells. Birc3 can bind to the effector caspases 3 and 7, causing their proteasomal degradation.43 There is evidence that Birc3 can activate via the Е3-dependent mode the NF-κB signaling pathway, which enhances the expression of prosurvival molecules.44 Besides, Birc3 protects the cells from death by regulating the activity of RIPK-1 and RIPK-3.45
At the age of 18 months, the hub genes are located in the first 3 clusters (Fig. 5C). KEGG analysis showed that the hub genes at age 18 months (Cdk1, Hspa1b, Erbb3, and Ntf3) participate in the MAPK and p53 signaling pathways. Gene Ontology annotations by GeneMANIA were as follows: “neuronal death” and “an extrinsic apoptosis pathway.” One of the hub genes, Cdk1 (cyclin-dependent kinase 1), is a key regulator of the mitotic transition. Activation of this kinase takes place at the beginning of mitosis and inactivation at the end.46 Studies also revealed the ability of the complex of Cdk1 with cyclin B1 to stimulate cell death via direct phosphorylation of the proteins Bcl-2, Bcl-XL, and Mcl1–1.47,48
The antiapoptotic effect of a member of the family of heat shock proteins Hspa1b (Hsp70.1) is predicated on its ability to protect cells from a number of proapoptotic stimuli and to regulate the induction of apoptosis by suppressing the release of SMAC, cytochrome c, and AIF from mitochondria, activation of caspases 3 and 9, and the release of cathepsins.49 Both survival and death of cells can be regulated by neurotrophin 3 (Ntf3), which in the retina mostly binds to the receptor TrkC, which is expressed by photoreceptors and Muller cells. The Trk receptors are mostly responsible for the maturation of neurotrophins and promote survival and differentiation of neurons as well as the synaptic function.50 It was reported recently that in the absence of its ligand, the TrkC receptor can induce apoptosis in various cell lines.51 Furthermore, in contrast to mature neurotrophins, their predecessors can drive the apoptotic processes via the complex of p75NTR and sortilin. These receptors are mostly located in Muller cells.52
In conclusion, in this work, we used systems biology to identify the DE genes associated with cell death and their functional characteristics. We also constructed 3 interaction networks. We identified functional clusters in these networks and likely hub genes (Rasa1, cFLAR, Birc3, Cdk1, Hspa1b, Erbb3, and Ntf3). In addition, we showed the significance of the extrinsic apoptotic pathway at preclinical, early, and advanced stages of retinopathy development. Aside from cell death signaling pathways, nonapoptotic immune system-related processes and lipid metabolic processes also show overrepresentation in the clusters of all networks. Changes in the expression of genes involved in the regulation of apoptosis in the retina of OXYS rats point to disturbances in the physiological processes of cell death that are needed for tissue homeostasis. Our findings may be useful for identification of biomarkers and possible therapeutic targets in retinal diseases.
Materials and Methods
Animals
Male senescence-accelerated OXYS rats (n = 12) and age-matched male Wistar rats (n = 12) at the age of 20 d and 3 and 18 months were obtained from the Breeding Experimental Animal Laboratory of the Institute of Cytology and Genetics, the Siberian Branch of the Russian Academy of Sciences (Novosibirsk, Russia). The rats were handled according to the Association for Research in Vision and Ophthalmology (ARVO) guidelines, and the study protocol was approved by the local ethics committee. At the age of 4 weeks, the pups were weaned and housed in groups of 5 animals per cage (57 × 36 × 20 cm) and kept under standard laboratory conditions (22 ± 2°C, 60+ relative humidity, and natural light). The rats were provided with standard rodent feed, PK-120–1, Ltd. (Laboratorsnab, Russia) and given water ad libitum.
RNA isolation
For RNA-Seq, we used OXYS (n = 3) and age-matched male Wistar rats (n = 3; as controls) at the age of 20 d The rats were euthanized using CO2 inhalation. After decapitation, the chorioretinal complex was excised rapidly, placed in RNAlater (Ambion, cat. # AM7020), frozen, and stored at −20°C prior to analysis. Frozen rat tissues were lysed with the TRIzol Reagent (Invitrogen, Cat. #15596–018), and total RNA was isolated according to the manufacturer's protocol. RNA quality and quantity were assessed using Agilent Bioanalyser (Agilent).
Illumina sequencing
More than 40 mln single-end reads 50 bp long were obtained for each sample of retinal RNA, using Illumina nonstranded sequencing on an Illumina GAIIx instrument at the Genoanalitika Lab, Moscow 53 in accordance with standard Illumina protocols (mRNA-Seq Sample Prep Kit, cat. # 1004816). Briefly, polyA-tailed mRNA was purified from total RNA using Sera-Mag Magnetic Oligo (dT) beads and then fragmented into small pieces by means of divalent cations and heating. Using a reverse transcriptase and random primers, we synthesized first- and second-strand cDNAs. The cDNA was processed in an end repair reaction with T4 DNA polymerase and Klenow DNA polymerase in order to blunt the termini. An “A” base was then added to the 3′ end of the blunt phosphorylated DNA fragments, and an Illumina adaptor with a single T overhang at its 3′ end was then ligated to the end of the DNA fragment, for hybridization in a single-read flow cell. After that, a size range of cDNA templates was selected, and these fragments were amplified on a cluster station using the Single-Read Cluster Generation Kit v2. Sequencing-by-synthesis (SBS) of 50-nucleotide length was performed using SBS v4 reagents on a Genome Analyzer IIx running the SCS2.8 software (Illumina, cat. #FC-940–4001).
Gene expression analysis: The sequencing data were preprocessed using the Cutadapt tool 53 to remove adapters and low-quality sequences. The resulting reads were mapped onto the Rnor_5.0 reference genome assembly in the TopHat2 software.54 The data were then converted into gene count tables using ENSEMBL gene annotation data. The resulting tables were subjected to the analysis of differential gene expression in the DESeq software. 55 The Benjamini–Hochberg correction for multiple testing was applied to the resulting p values, and the genes with an adjusted p value <0 .05 were selected as differentially expressed.
Construction and analysis of gene interaction networks
In addition to the gene expression profiles of 20-day-old animals, we used previously obtained RNA-Seq data on differences in gene expression between OXYS and Wistar retinas at ages 3 and 18 months.16 To construct gene interaction networks associated with cell death from lists of differentially expressed (DE) genes (raw p value <0.01) we selected genes that were in the following Gene Ontology categories: apoptosis, cell death, regulation of apoptosis, regulation of cell death, positive regulation of apoptosis, negative regulation of apoptosis, positive regulation of cell death, negative regulation of cell death, anti-apoptosis, induction of apoptosis according to DAVID (Database for Annotation, Visualization and Integrated Discovery, http://david.abcc.ncifcrf.gov/summary.jsp).
The gene interaction networks associated with cell death were identified by means of the GeneMANIA web server (http://www.genemania.org/) with default parameters. The topological properties of the networks were determined using the Network Analyzer plugin (version 2.6.1) of Cytoscape (ver. 2.8.2). Hub genes were identified by calculating the following topological properties:
Average shortest path length, i.e., the average number of steps along the shortest paths for all possible pairs of network nodes.
Betweenness centrality: the fraction of those shortest paths between all pairs of nodes that pass through one node. Betweenness centrality reflects the amount of control that this node exerts over the interactions of other nodes in the network.
Closeness centrality; it is defined as the reciprocal of the average shortest path length. It is a measure of how fast information spreads from a given node to other reachable nodes in the network.
The clustering coefficient, i.e., the normalized number of interactions among neighbors of each node.
The node degree: the average number of interactions per node.
The nodes that have shorter average shortest path length, a greater clustering coefficient, higher closeness centrality, lower betweenness centrality, and higher node degrees were identified as the highly connected nodes (hub genes).
Next, the functional clusters within the networks were detected by means of ClusterMarket plugin (ver. 1.11) community cluster (GLay) for Cytoscape (ver. 2.8.2). To identify the biological functions within each cluster, the detected clusters were subjected to functional analyses in Reactome (http://www.reactome.org/).
Functional analysis
To identify the Gene Ontology terms overrepresented in a DE gene list, the detected DE genes were subjected to functional enrichment analyses by means of the DAVID tool. Pathway analysis of the DE genes associated with cell death was conducted using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (http://www.genome.jp/kegg/).
Tissue dissection, fixation, and sectioning and the terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick end labeling (TUNEL) assay
Twenty-day-old and 3- and 18-month-old OXYS and Wistar (control) rats (3 per group) were euthanized using CO2 inhalation. The eyes were removed and fixed in fresh 4% paraformaldehyde in PBS for 2 h, washed 3 times in PBS, then cryopreserved in graded sucrose solutions (10%, 20%, and 30%). Posterior eyecups were embedded in Killik (Bio-Optica, cat. #05–9801), frozen, and stored at −70°C. Then, thin slices (14 μm thick) were made on a Microm HM-505 N cryostat (Microm, Germany) at −20°C, transferred onto Polysine glass slides (Menzel-Glaser, cat. #J1800AMNZ), and stored at −20°C. Apoptosis was analyzed by means of a TUNEL assay using the DeadEnd Fluorometric TUNEL System (Promega, cat. #G3250). The tissue slices were coverslipped with the Fluoroshield mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI; Abcam, cat. #ab104139). The positive TUNEL signals were counted under a microscope with a 20× objective lens (Axioskop 2 plus, Zeiss, Germany) and then averaged in each group of 4–5 slices per animal. The data were analyzed using the statistical software Statistica 8.0 (USA) with the Newman–Keuls post hoc test. The genotype and age were chosen as independent variables. The data were presented as mean ± SEM. Statistical significance was set to p < 0.05.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was supported by the Russian Foundation for Basic Research (project # 15-04-02195А) and by grants from the government of the Russian Federation ## 2012-220-03-435 and 14.B25.31.0033.
References
- 1.Horie-Inoue K, Inoue S. Genomic aspects of age-related macular degeneration. Biochem Biophys Res Commun 2014; 452:263-75; PMID:25111812; http://dx.doi.org/ 10.1016/j.bbrc.2014.08.013 [DOI] [PubMed] [Google Scholar]
- 2.Rickman CB, Farsiu S, Toth CA, Klingeborn M. Dry age-related macular degeneration: mechanisms, therapeutic targets, and imaging. Invest Ophthalmol Vis Sci 2013; 54:ORSF68-80; PMID:24335072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambati J, Fowler BJ. Mechanisms of age-related macular degeneration. Neuron 2012; 75:26-39; PMID:22794258; http://dx.doi.org/ 10.1016/j.neuron.2012.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Almagro MC, Vucic D. Necroptosis: Pathway diversity and characteristics. In Semin Cell Dev Biol 2015; 39:56-62; PMID:25683283; http://dx.doi.org/ 10.1016/j.semcdb.2015.02.002 [DOI] [PubMed] [Google Scholar]
- 5.Wright AF, Chakarova CF, El-Aziz MMA, Bhattacharya SS. Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat Rev Genet 2010; 11:273-84; PMID:20212494; http://dx.doi.org/ 10.1038/nrg2717 [DOI] [PubMed] [Google Scholar]
- 6.Murakami Y, Notomi S, Hisatomi T, Nakazawa T, Ishibashi T, Miller JW, Vavvas DG. Photoreceptor cell death and rescue in retinal detachment and degenerations. Prog Retin Eye Res 2013; 37:114-40; PMID:23994436; http://dx.doi.org/ 10.1016/j.preteyeres.2013.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian L, Kazmierkiewicz KL, Bowman AS, Li M, Curcio CA, Stambolian DE. Transcriptome of the human retina, retinal pigmented epithelium and choroid. Genomics 2015; 105:253-64; PMID:25645700; http://dx.doi.org/ 10.1016/j.ygeno.2015.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang H-J, Ratnapriya R, Cogliati T, Kim J-W, Swaroop A. Vision from next generation sequencing: Multi-dimensional genome-wide analysis for producing gene regulatory networks underlying retinal development, aging and disease. Prog Retin Eye Res 2015; 46:1-30; PMID:25668385; http://dx.doi.org/ 10.1016/j.preteyeres.2015.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newman AM, Gallo NB, Hancox LS, Miller NJ, Radeke CM, Maloney MA, Cooper JB, Hageman GS, Anderson DH, Johnson LV, Radeke MJ. Systems-level analysis of age-related macular degeneration reveals global biomarkers and phenotype-specific functional networks. Genome Med 2012; 4:16; PMID:22364233; http://dx.doi.org/ 10.1186/gm315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitmore SS, Mullins RF. Transcriptome changes in age-related macular degeneration. BMC Med 2012; 10:21; PMID:22369667; http://dx.doi.org/ 10.1186/1741-7015-10-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abu-Asab MS, Salazar J, Tuo J, Chan CC. Systems biology profiling of AMD on the basis of gene expression. J Ophthalmol 2013; 2013:453934; PMID:24349763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolosova NG, Muraleva NA, Zhdankina AA, Stefanova NA, Fursova AZ, Blagosklonny MV. Prevention of age-related macular degeneration–like retinopathy by rapamycin in Rats. Am J Pathol 2012; 181:472-77; PMID:22683466; http://dx.doi.org/ 10.1016/j.ajpath.2012.04.018 [DOI] [PubMed] [Google Scholar]
- 13.Zhdankina AA, Fursova AZ, Logvinov SV, Kolosova NG. Clinical and morphological characteristics of chorioretinal degeneration in early aging OXYS rats. Bull Exp Biol Med 2008; 146:455-58; PMID:19489319; http://dx.doi.org/ 10.1007/s10517-009-0298-4 [DOI] [PubMed] [Google Scholar]
- 14.Muraleva NA, Kozhevnikova OS, Zhdankina AA, Stefanova NA, Karamysheva TV, Fursova AZ, Kolosova NG. The mitochondria-targeted antioxidant SkQ1 restores α B-crystallin expression and protects against AMD-like retinopathy in OXYS rats. Cell Cycle 2014; 13:3499-505; PMID:25483086; http://dx.doi.org/ 10.4161/15384101.2014.958393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozhevnikova OS, Korbolina EE, Stefanova NA, Muraleva NA, Orlov YL, Kolosova NG. Association of AMD-like retinopathy development with an Alzheimer's disease metabolic pathway in OXYS rats. Biogerontology 2013; 14:753-62; PMID:23959258; http://dx.doi.org/ 10.1007/s10522-013-9439-2 [DOI] [PubMed] [Google Scholar]
- 16.Kozhevnikova OS, Korbolina EE, Ershov NI, Kolosova NG. Rat retinal transcriptome: effects of aging and AMD-like retinopathy. Cell Cycle 2013; 12:1745-61; PMID:23656783; http://dx.doi.org/ 10.4161/cc.24825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010; 11:R106; PMID:20979621; http://dx.doi.org/ 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minagawa K, Katayama Y, Matsui T. TFL, a hidden post-transcriptional modulator behind inflammation. Inflamm Cell Signal 2014; 1:1-7 [Google Scholar]
- 19.Liu NS, Du X, Lu J, He BP. Diva reduces cell death in response to oxidative stress and cytotoxicity. PloS One 2012; 7:e43180; PMID:22905226; http://dx.doi.org/ 10.1371/journal.pone.0043180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harberts E, Fishelevich R, Liu J, Atamas S, Gaspari A. MyD88 plays a role in deciding between apoptotic and necroptotic cell death after UV irradiation (P1238). Innate Immun 2013; 190:138-17 [Google Scholar]
- 21.Markovets AM, Saprunova VB, Zhdankina AA, Fursova AZ, Bakeeva LE, Kolosova N G. Alterations of retinal pigment epithelium cause AMD-like retinopathy in senescence-accelerated OXYS rats. Aging (Albany NY) 2011; 3:44; PMID:21191149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grün G. The development of the vertebrate retina: a comparative survey. Adv Anat Embryol Cell Biol 1982; 78:1-85; PMID:7158472; http://dx.doi.org/ 10.1007/978-3-642-68719-8_1 [DOI] [PubMed] [Google Scholar]
- 23.Valenciano AI, Boya P, de la Rosa EJ. Early neuronal cell death: numbers and cues from the developing neuroretina. Int J Dev Biol 2009; 53:1515-28; PMID:19247933; http://dx.doi.org/ 10.1387/ijdb.072446av [DOI] [PubMed] [Google Scholar]
- 24.Francisco-Morcillo J, Bejarano-Escobar R, Rodríguez-León J, Navascués J, Martín-Partido G. Ontogenetic cell death and phagocytosis in the visual system of vertebrates. Devl Dyn 2014; 243:1203-25; PMID:25130286; http://dx.doi.org/ 10.1002/dvdy.24174 [DOI] [PubMed] [Google Scholar]
- 25.Gregory-Evans CY, Wallace VA, Gregory-Evans K. Gene networks: dissecting pathways in retinal development and disease. Prog Retin Eye Res 2013; 33:40-66; PMID:23128416; http://dx.doi.org/ 10.1016/j.preteyeres.2012.10.003 [DOI] [PubMed] [Google Scholar]
- 26.Yanai H, Budovsky A, Tacutu R, Fraifeld V. Is rate of skin wound healing associated with aging or longevity phenotype? Biogerontology 2011; 12:591-97; PMID:21667230; http://dx.doi.org/ 10.1007/s10522-011-9343-6 [DOI] [PubMed] [Google Scholar]
- 27.Yanai H, Toren D, Vierlinger K, Hofner M, Nöhammer C, Chilosi M, Budovsky A, Fraifeld V. Wound healing and longevity: lessons from long-lived αMUPA mice. Aging (Albany NY) 2015; 7:167-76; PMID:25960543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeppesen DK, Bohr VA, Stevnsner T. DNA repair deficiency in neurodegeneration. Prog Neurobiol 2011; 94:166-200; PMID:21550379; http://dx.doi.org/ 10.1016/j.pneurobio.2011.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roos WP, Kaina B. DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett 2013; 332:237-48; PMID:22261329; http://dx.doi.org/ 10.1016/j.canlet.2012.01.007 [DOI] [PubMed] [Google Scholar]
- 30.Tokarz P, Kauppinen A, Kaarniranta KA, Blasiak J. Oxidative DNA damage and proteostasis in age-related macular degeneration. J Biochem Pharmacol Res 2013; 1:106-13 [Google Scholar]
- 31.Weissman L, de Souza-Pinto NC, Stevnsner T, Bohr VA. DNA repair, mitochondria, and neurodegeneration. Neuroscience 2007; 145:1318-29; PMID:17092652; http://dx.doi.org/ 10.1016/j.neuroscience.2006.08.061 [DOI] [PubMed] [Google Scholar]
- 32.Santos RX, Correia SC, Zhu X, Smith MA, Moreira PI, Castellani RJ, Nunomura A, Perry G. Mitochondrial DNA oxidative damage and repair in aging and Alzheimer's disease. Antioxid Redox Signal 2013; 18:2444-57; PMID:23216311; http://dx.doi.org/ 10.1089/ars.2012.5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh DK, Krishna S, Chandra S, Shameem M, Deshmukh AL, Banerjee D. Human DNA ligases: a comprehensive new look for cancer therapy. Med Res Rev 2014; 34:567-95; PMID:23959747; http://dx.doi.org/ 10.1002/med.21298 [DOI] [PubMed] [Google Scholar]
- 34.He S, Wang L, Miao L, Wang T, Du F, Zhao L, Wang X. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-α. Cell 2009; 137:1100-11; PMID:19524512; http://dx.doi.org/ 10.1016/j.cell.2009.05.021 [DOI] [PubMed] [Google Scholar]
- 35.Markova EV, Obukhova LA, Kolosova NG. Activity of cell immune response and open field behavior in Wistar and OXYS rats. Bull Exp Biol Med 2003; 136:377-9; PMID:14714088; http://dx.doi.org/ 10.1023/B:BEBM.0000010957.87077.ae [DOI] [PubMed] [Google Scholar]
- 36.Overmeyer JH, Maltese WA. Death pathways triggered by activated Ras in cancer cells. Front Biosci 2011; 16:1693-713; PMID:21196257; http://dx.doi.org/ 10.2741/3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawasaki J, Aegerter S, Fevurly RD, Mammoto A, Mammoto T, Sahin M, Mably JD, Fishman SJ, Chan J. RASA1 functions in EPHB4 signaling pathway to suppress endothelial mTORC1 activity. J Clin Invest 2014; 124:2774-84; PMID:24837431; http://dx.doi.org/ 10.1172/JCI67084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henkemeyer M, Rossi DJ, Holmyard DP, Puri MC, Mbamalu G, Harpal K, Shih TS, Jacks T, Pawson T. Vascular system defects and neuronal apoptosis in mice lacking ras GTPase-activating protein. Nature 1995; 377:695-701; PMID:7477259; http://dx.doi.org/ 10.1038/377695a0 [DOI] [PubMed] [Google Scholar]
- 39.Safa AR. Roles of c-FLIP in apoptosis, necroptosis, and autophagy. J Carcinog Mutagen 2013; Suppl 6:003; PMID:25379355; http://dx.doi:org/ 10.4172/2157-2518.S6-003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F, Zachariou A, Lopez J, MacFarlane M, Cain K, Meier P. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol Cell 2011; 43:432-48; PMID:21737329; http://dx.doi.org/ 10.1016/j.molcel.2011.06.006 [DOI] [PubMed] [Google Scholar]
- 41.Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M, Cain K, MacFarlane M, Häcker G, Leverkus M. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell 2011; 43:449-63; PMID:21737330; http://dx.doi.org/ 10.1016/j.molcel.2011.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee JS, Li Q, Lee JY, Lee SH, Jeong JH, Lee HR, Chang H, Zhou FC, Gao SJ, Liang C, Jung JU. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol 2009; 11:1355-62; PMID:19838173; http://dx.doi.org/ 10.1038/ncb1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi YE, Butterworth M, Malladi S, Duckett CS, Cohen GM, Bratton SB. The E3 ubiquitin ligase cIAP1 binds and ubiquitinates caspase-3 and -7 via unique mechanisms at distinct steps in their processing. J Biol Chem 2009; 284:12772-82; PMID:19258326; http://dx.doi.org/ 10.1074/jbc.M807550200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Estornes Y, Bertrand MJ. IAPs, regulators of innate immunity and inflammation. Semin Cell Dev Biology 2014; 39:1-9 [DOI] [PubMed] [Google Scholar]
- 45.Berthelet J, Dubrez L. Regulation of apoptosis by inhibitors of apoptosis (IAPs). Cells 2013; 2:163-87; PMID:24709650; http://dx.doi.org/ 10.3390/cells2010163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Potapova TA, Daum JR, Byrd KS, Gorbsky GJ. Fine tuning the cell cycle: activation of the Cdk1 inhibitory phosphorylation pathway during mitotic exit. Mol Bbiol Cell 2009; 20:1737-48; PMID:19158392; http://dx.doi.org/ 10.1091/mbc.E08-07-0771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harley ME, Allan LA, Sanderson HS, Clarke PR. Phosphorylation of Mcl-1 by CDK1–cyclin B1 initiates its Cdc20-dependent destruction during mitotic arrest. EMBO J 2010; 29:2407-20; PMID:20526282; http://dx.doi.org/ 10.1038/emboj.2010.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Terrano DT, Upreti M, Chambers TC. Cyclin-dependent kinase 1-mediated Bcl-xL/Bcl-2 phosphorylation acts as a functional link coupling mitotic arrest and apoptosis. Mol Cell Biol 2010; 30:640-56; PMID:19917720; http://dx.doi.org/ 10.1128/MCB.00882-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang B, Liang P, Deng G, Tu Z, Liu M, Xiao X. Increased stability of Bcl-2 in HSP70-mediated protection against apoptosis induced by oxidative stress. Cell Stress Chaperones 2011; 16:143-52; PMID:20890773; http://dx.doi.org/ 10.1007/s12192-010-0226-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friedman WJ. Proneurotrophins, seizures, and neuronal apoptosis. Neuroscientist 2010; 16:244-52; PMID:20360602; http://dx.doi.org/ 10.1177/1073858409349903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ichim G, Tauszig-Delamasure S, Mehlen P. Neurotrophins and cell death. Exp Cell Res 2012; 318:1221-28; PMID:22465479; http://dx.doi.org/ 10.1016/j.yexcr.2012.03.006 [DOI] [PubMed] [Google Scholar]
- 52.Shen W, Zhu L, Lee SR, Chung SH, Gillies MC. Involvement of NT3 and P75NTR in photoreceptor degeneration following selective Müller cell ablation. J Neuroinflammation 2013; 10:137; PMID:24224958; http://dx.doi.org/ 10.1186/1742-2094-10-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Genoanalitika Lab , Moscow. [http://www.genoanalytica.ru/]. [Google Scholar]
- 54.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMB Net J 2011; 17:10-12; http://dx.doi.org/ 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 55.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14 (4):R36; PMID:23618408; http://dx.doi.org/ 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.