Abstract
Repressor activator protein 1 (Rap1) is essential for maintaining telomere length and structural integrity, but it also exerts other non-telomeric functions. The present study tested the hypothesis that Rap1 is released into the cytoplasm and induces production of pro-inflammatory cytokines via nuclear factor kappa B (NFκB) signaling in macrophages, a cell type involved in the development and progression of atherosclerotic lesions. Western blotting analysis confirmed that Rap1 was present in the cytoplasm of differentiated human monocytic leukemia cells (THP-1, a macrophage-like cell line). Co-immunoprecipitation assay revealed a direct interaction between Rap1 and I kappa B kinase (IKK). Knockdown of Rap1 suppressed lipopolysaccharide-mediated activation of NFκB, and phosphorylation of inhibitor of kappa B α (IκBα) and p65 in THP-1 macrophages. The reduction of NFκB activity was paralleled by a decreased production of NFκB-dependent pro-inflammatory cytokines and an increased expression of IκBα (native NFκB inhibitor) in various macrophage models with pro-inflammatory phenotype, including THP-1, mouse peritoneal macrophages and bone marrow-derived M1 macrophages. These changes were observed selectively in pro-inflammatory macrophages but not in bone marrow-derived M2 macrophages (with an anti-inflammatory phenotype), mouse lung endothelial cells, human umbilical vein endothelial cells or human aortic smooth muscle cells. Immunostaining revealed that Rap1 was localized mainly in macrophage-rich areas in human atherosclerotic plaques and that the presence of Rap1 was positively correlated with the advancement of the disease process. In pro-inflammatory macrophages, Rap1 promotes cytokine production via NFκB activation favoring a pro-inflammatory environment which may contribute to the development and progression of atherosclerosis.
Keywords: atherosclerosis; macrophages; NFκB; pro-inflammatory cytokines; repressor activator protein 1; signal transduction,; telomeric protein
Abbreviations
- GM-CSF
granulocyte macrophage colony-stimulating factor
- HASMCs
human aortic smooth muscle cells
- HUVECs
human umbilical vein endothelial cells
- ICAM-1
intercellular adhesion molecule-1
- IκB
inhibitor of kappa b
- IKK
I Kappa B kinase
- IL
interleukin
- LPS
lipopolysaccharide
- M-CSF
macrophage colony-stimulating factor
- MCP-1
monocyte chemotactic protein-1
- NFκB
nuclear factor kappa B
- PAECs
porcine aortic endothelial cells
- PCR
polymerase chain reaction
- PGC1α
peroxisome proliferator-activated receptor gamma coactivator 1 α
- PPAR
peroxisome proliferator-activated receptor
- Rap1
repressor activator protein 1
- siRNA
small interfering RNA
- RPMI
Roswell Park Memorial Institute
- TNFα
tumor necrosis factor α
- VCAM-1
vascular cell adhesion molecule-1
Introduction
Mammalian repressor activator protein 1 (Rap1) is a telomere-associated protein.1 It interacts with telomeric repeat-binding factor 2 which anchors Rap1 at the telomere.1,2 Rap1 protects telomeres against recombination and reduces their fragility by repressing homology-directed repair and preventing sister chromatid exchange.3–5
Besides the maintenance of telomere integrity, Rap1 also regulates gene transcription by binding to non-telomeric sites.4 It regulates genes involved in insulin secretion, peroxisome proliferator-activated receptor (PPAR) signaling and growth hormone pathways.4 In particular, Rap1 enhanced the expression of peroxisome proliferator-activated receptor gamma coactivator 1 α (PGC1α), an important transcriptional regulator of lipid metabolism.6 The transcriptional changes induced by Rap1 are distinct from its telomeric function.4
Phenotypic characterization of Rap1 deficient mice indicates that Rap1 is involved in signaling pathways connected to metabolism and contributes to body weight regulation.7,8 The levels of PPARα and PGC1α are reduced in the liver and gonadal white adipose tissue of Rap1 deficient mice.7 PPARα- and PGC1α-target genes (including carnitine palmitoyl transferase 1α, solute carrier family 27 member 2 and cluster of differentiation 36), which are involved in fatty acid oxidation and lipid uptake, are decreased in the liver of Rap1 deficient mice.7 Such metabolic alterations resulted in the development of hepatic steatosis in these mice, a phenomenon that is more severe in females than in males.7,8 Mice with genetic deletion of Rap1 also exhibit glucose intolerance, insulin resistance and became obese.7,8 Furthermore, Rap1 may potenitally contribute to chronic inflammation in age-assoicated diseases.9
Expression of Rap1 was initially believed to be restricted to the nucleus, however it was later found to be expressed in the cytoplasm of breast carcinoma BT476 and epithelial carcinoma HeLa S3 cell lines.10 In these human cancer cell lines, cytoplasmic Rap1 regulates the nuclear factor kappa B (NFκB) signaling cascade,10,11 a master transcription factor that plays fundamental roles in inflammatory, immune responses12 and other signaling cascades.13 Unlike in the nucleus, cytoplasmic Rap1 does not interact with telomeric repeat-binding factor 2, but rather constitutively binds to I Kappa B kinase (IKK),10 which is responsible for the phosphorylation and subsequent degradation of inhibitor of kappa b (IκB) protein,14 the endogenous inhibitor of NFκB.15 Cytoplasmic Rap1 ensures the efficient recruitment of IKKs and the phosphorylation of the p65 subunit of NFκB,10,16 an essential step for rendering NFκB transcriptionally competent.17 The presence of Rap1 has been positively correlated to the advancement of invasive human breast cancers.10 The ability of Rap1 to modify NFκB signaling is conserved in vivo, as heterozygous mice lacking one functional Rap1 allele do not develop proper immune responses.10 Collectively, the available experimental evidence suggests that Rap1 is not only a static structural component of the telomere but exerts other functions both within and outside the nucleus.
Inflammation plays a major role in all phases of atherosclerosis – from initiation through progression and even in its thrombotic complications.18–20 Being a major transcription factor regulating inflammatory responses, NFκB has been linked to the pathogenesis of atherosclerosis.12,21 Macrophages, endothelial cells and smooth muscle cells within atherosclerotic lesions exhibit enhanced NFκB activity.22 These cells cooperatively contribute to the pro-inflammatory environment within the atherosclerotic lesions by enhancing the expression of adhesion molecule and the release of chemokines and cytokines.22,23 Whether or not Rap1 exerts pro-atherogenic effects is unknown. Thus, the present study was designed to test the hypothesis that Rap1 modulates inflammatory processes via the NFκB signaling cascade in macrophages, endothelial and smooth muscle cells. Moreover, whether or not Rap1 abundance is associated with the advancement of human atherosclerotic lesions was examined.
Results
Establishing Rap1 knockdown in THP-1 macrophages
To demonstrate the involvement of Rap1 in controlling the expression of NFκB-dependent genes in macrophages, siRNA technology was applied to reduce intracellular Rap1 levels in THP-1 macrophages. A significant reduction of 73.1 ± 4.3% in Rap1 mRNA (Fig. 1A) and 84.7 ± 4.4% in Rap1 protein presence (Fig. 1B) was achieved in Rap1 knockdown cells, as compared to mock-transfected cells. To characterize the subcellular location of Rap1, whole cell lysates were separated into nuclear and cytosolic fractions. The lack of histone H3 within the cytosolic fraction confirmed the success of the fractionation (Fig. 1B).24 Endogenous Rap1 was present in both nuclear and cytosolic cell fractions, with its abundance being higher in the former. The introduction of Rap1 siRNA into THP-1 macrophages significantly suppressed both nuclear and cytosolic Rap1 protein levels by 55.1 ± 7.0% and 88.8 ± 9.7%, respectively (Fig. 1B).
Figure 1.
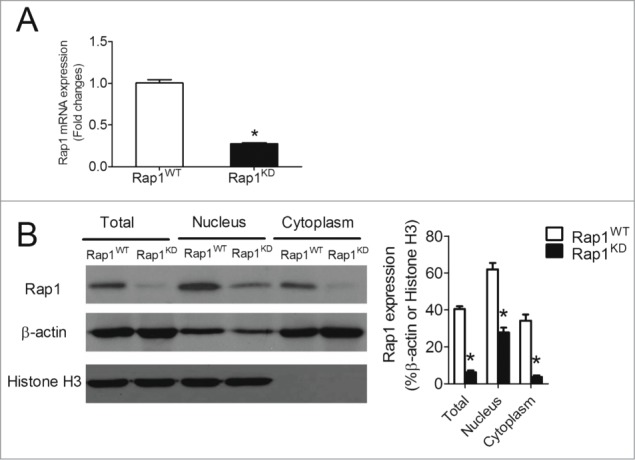
Knockdown of Rap1 in differentiated THP-1 macrophages. (A) mRNA expression of Rap1 in wild-type (Rap1WT) and Rap1 knockdown (Rap1KD) THP-1 macrophages, n = 6; (B) Protein levels of Rap1, β-actin and histone H3 in total, nuclear and cytoplasmic extracts of Rap1WT and Rap1KD THP-1 macrophages, n = 4. Data are shown as means ± SEM *P < 0.05 Rap1WT vs. Rap1KD.
Knockdown of Rap1 reduced NFκB activity and NFκB-dependent pro-inflammatory cytokines in macrophages
Bacterial endotoxins such as lipopolysaccharide (LPS) activate toll-like receptor 4 and aggravate the progression of atherosclerosis through multiple mechanisms, including increased production of reactive oxygen species, chemotactic and pro-inflammatory cytokines and other acute phase reactants, and augmented expression of adhesion molecules.25–28 Given the involvement of NFκB signaling in atherosclerosis, LPS was used to activate it and stimulate the production of NFκB-dependent pro-inflammatory cytokines.29,30 Indeed, LPS caused a sustained activation of p65 in THP-1 macrophages (Fig. 2A). Knockdown of Rap1 significantly reduced p65 activation by 39.0 ± 5.2%, 29.5 ± 4.7% and 32.7 ± 5.8% at 10, 60 and 240 minutes after exposure to LPS, respectively (Fig. 2A).
Figure 2.
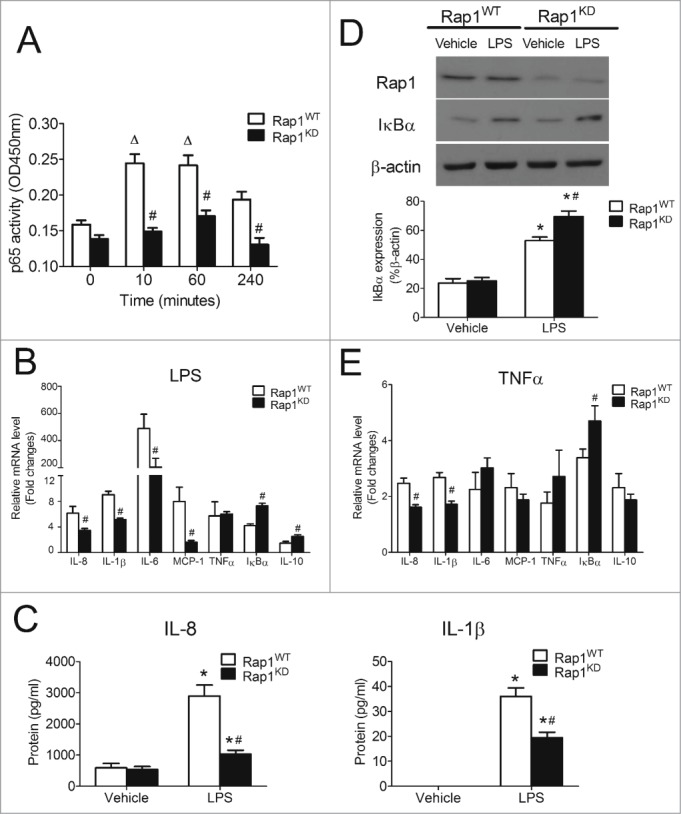
Knockdown of Rap1 reduced NFκB activity and NFκB-dependent pro-inflammatory cytokines in THP-1 macrophages. (A) p65 activity in Rap1WT and Rap1KD THP-1 macrophages stimulated with LPS (50ng/ml) at indicated time points; (B) mRNA expression of NFκB-dependent genes in Rap1WT and Rap1KD THP-1 macrophages stimulated with LPS (50ng/ml, 4 hours); (C) Protein concentrations of IL-8 and IL-1β released from Rap1WT and Rap1KD THP-1 macrophages stimulated with or without LPS (50ng/ml, 4 hours); (D) Protein levels of IκBα in Rap1WT and Rap1KD THP-1 macrophages stimulated with or without LPS (50ng/ml, 8 hours); (E) mRNA expression of NFκB-dependent genes in Rap1WT and Rap1KD cells stimulated with TNFα (100ng/ml, 4 hours). mRNA levels are expressed as fold changes against those mRNA expression in Rap1WT THP-1 macrophages with no stimulation. Data are shown as means ± SEM ΔP <0.05 vs. Rap1WT (time = 0); *P < 0.05 vehicle vs. LPS; #P < 0.05 Rap1WT vs. Rap1KD; n = 6.
Administration of LPS (50ng/ml for 4 hours) induced mRNA expression of interleukin (IL)-8, IL-1β, IL-6 and monocyte chemotactic protein-1 (MCP-1). Such induction was significantly attenuated in macrophages with Rap1 knockdown by 44, 43, 45 and 80%, respectively (Fig. 2B). The knockdown of Rap1 did not influence LPS-induced mRNA expression of tumor necrosis factor α (TNFα), a cytokine which is also under the transcription control of NFκB (Fig. 2B).31 LPS stimulated the expression of IκBα, the native inhibitor of NFκB and that of IL-10, an anti-inflammatory cytokine in macrophages.15 The knockdown of Rap1 further increased IκBα and IL-10 mRNA expression by 72.0 ± 10.2% and 69.8 ± 18.2%, respectively (Fig. 2B).
To determine whether or not these mRNA expression changes are related to protein presence, the protein levels of IL-8 and IL-1β in the supernatant were compared after 4 hours treatment with vehicle or with LPS. The knockdown of Rap1 did not influence endogenous protein levels of IL-8, but significantly reduced the LPS-stimulated protein level of the cytokine by 64.4 ± 4.4% (Fig. 2C). The protein level of IL-1β was undetectable in unstimulated macrophages, but became measurable after LPS stimulation. The knockdown of Rap1 significantly reduced the LPS-stimulated IL-1β protein level by 46.0 ± 6.1% (Fig. 2C). Likewise, in line with the mRNA changes, the protein level of IκBα in response to LPS was significantly enhanced by 31.5 ± 8.5% in macrophages with Rap1 knockdown (Fig. 2D). Taken in conjunction, these results indicate that the presence of Rap1 in macrophages favors a pro-inflammatory environment by selectively up-regulating certain (IL-8, IL-1β, IL-6 and MCP-1) but not all (TNFα) NFκB-dependent inflammatory genes and decreasing the expression of IκBα and IL-10.
NFκB signaling can also be activated by the pro-inflammatory cytokine TNFα.32 Thus, the effect of Rap1 knockdown on TNFα-induced responses was examined. Like LPS, TNFα (100ng/ml for 4 hours) significantly induced the mRNA expression of NFκB-dependent genes – including IL-8, IL-1β, IL-6, MCP-1, TNFα, IκBα and IL-10. Knockdown of Rap1 significantly suppressed TNFα-induced IL-8 and IL-1β mRNA expression (by 34.8 ± 3.8% and 35.9 ± 4.0%, respectively) and increased TNFα-induced IκBα mRNA expression (by 38.8 ± 16.0%; Fig. 2E). Knockdown of Rap1 had no significant effect on TNFα-induced increases in mRNA expression of IL-6, MCP-1, TNFα and IL-10 (Fig. 2E).
To circumvent potential off-target effects of siRNA, experiments were repeated using a second set of siRNA that targeted a different location within the Rap1 gene. A significant reduction of 68.5 ± 3.6% in Rap1 mRNA was achieved by the second set of siRNA and such knockdown consistently suppressed LPS- and TNFα-induced IL-8 and IL-1β mRNA expression (Fig. S1).
Positive feedback loop between Rap1 and NFκB
To evaluate if activation of NFκB up-regulates Rap1 levels in macrophages, Rap1 expression was quantified at various time points after stimulation with either LPS or TNFα. The mRNA expression of Rap1 was significantly increased in THP-1 macrophages in a time-dependent manner after LPS or TNFα stimulation. After 48 hours of LPS or TNFα treatment, Rap1 mRNA was increased by 123.5 ± 49.3% and 144.5 ± 38.9%, respectively, compared to unstimulated cells (Fig. 3A). Correspondingly, protein levels of Rap1 were increased by 49.5 ± 3.9% and 50.8 ± 7.0%, respectively, after 48 hours of LPS or TNFα stimulation (Fig. 3B). These results suggest the existence of a positive feedback loop between NFκB activation and Rap1 expression: once LPS or TNFα activate NFκB, the latter contributes to a sustained production of Rap1, which in turn triggers greater activation of NFκB in macrophages.
Figure 3.
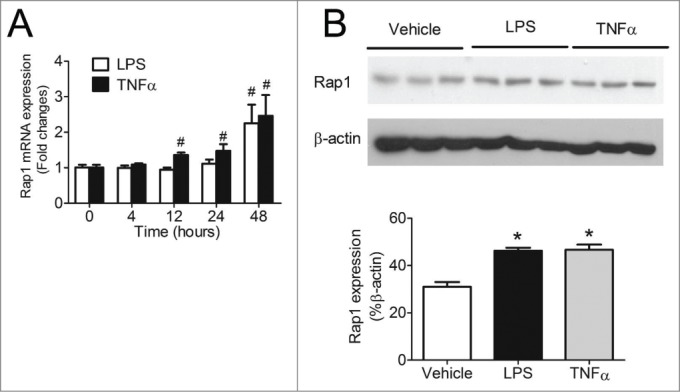
Activation of NFκB increased Rap1 levels. (A) mRNA expression of Rap1 in differentiated THP-1 macrophages after stimulation with LPS (50ng/ml), TNFα (100ng/ml) for 4, 12, 24 or 48 hours, n = 6; (B) Protein levels of Rap1 after 48 hours of LPS (50ng/ml) or TNFα (100ng/ml) stimulation in differentiated THP-1 macrophages, n = 3. Data are shown as means ± SEM *P < 0.05 vehicle vs. LPS or TNFα; #P < 0.05 vs. time = 0.
Rap1 mediate cytokine production predominately in macrophages with a pro-inflammatory phenotype
There are several caveats to the use of siRNA, including incompleteness or transient nature of the knockdown. To consolidate that Rap1 favors pro-inflammatory environment via NFκB, experiments were repeated in primary peritoneal macrophages and bone marrow-derived M1 and M2 macrophages isolated from Rap1 wild-type (Rap1+/+) and Rap1 knockout (Rap1−/−) mice. For peritoneal macrophages, as anticipated, deficiency of Rap1 significantly suppressed LPS-induced IL-8, IL-1β and MCP-1 mRNA expression (by 47.1 ± 2.7%, 38.4 ± 6.5% and 27.4 ± 3.7%, respectively; Fig. 4A) and increased IκBα mRNA expression (by 50.0 ± 17.3%; Fig. 4A) without any changes to LPS-induced IL-6, TNFα and IL-10 mRNA expression (Fig. 4A). Consistently, deficiency of Rap1 in bone marrow-derived M1 macrophages (with a pro-inflammatory phenotype) significantly suppressed LPS-induced IL-1β, MCP-1 and TNFα mRNA expression (by 34.7 ± 8.1%, 32.5 ± 4.6% and 29.0 ± 4.1%, respectively; Fig. 4B) and increased LPS-induced IκBα mRNA expression (by 33.6 ± 3.1%; Fig. 4B), but did not alter the mRNA expression of IL-8, IL-6 and IL-10 (Fig. 4B). By contrast, deficiency of Rap1 in bone marrow-derived M2 macrophages (with an anti-inflammatory phenotype) significantly increased LPS-induced IL-1β and IL-6 mRNA expression (by 103.9 ± 35.8% and 76.0 ± 25.2%, respectively; Fig. 4C). Yet, similar to macrophages with a pro-inflammatory phenotype, deficiency of Rap1 in M2 macrophages reduced LPS-induced MCP-1 (by 26.0 ± 5.6%; Fig. 4C) and enhanced IкBα (by 30.5 ± 6.5%; Fig. 4C).
Figure 4.
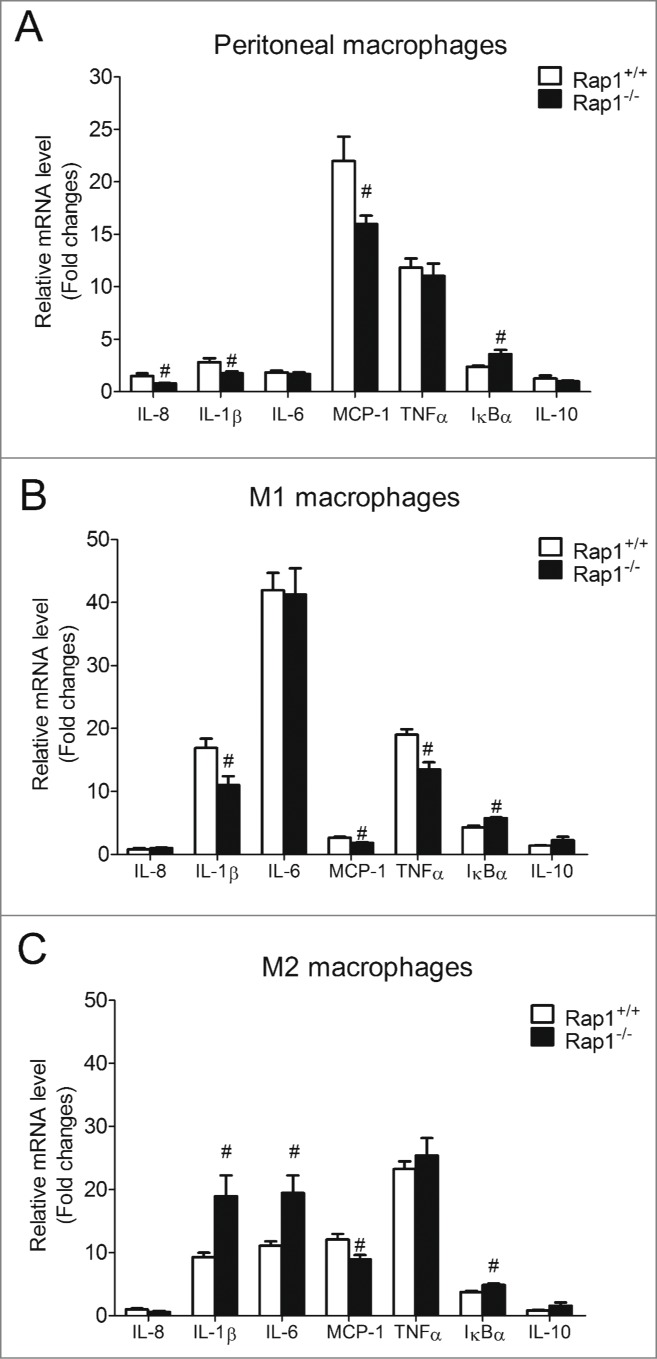
Rap1 knockout reduced NFκB-dependent cytokines in pro-inflammatory phenotypic macrophages. mRNA expression of LPS-induced (5ng/ml, 4 hours) NFκB-dependent genes in (A) peritoneal; (B) bone marrow-derived M1; (C) and bone marrow-derived M2 macrophages isolated from Rap1+/+ and Rap1−/− mice. mRNA levels are expressed as fold changes against those mRNA expression in Rap1+/+ macrophages with no stimulation. Data are shown as means ± SEM #P <0.05 Rap1+/+ vs. Rap1−/−; n = 6.
Rap1 induces phosphorylation of IκBα and p65
Given the observation that Rap1 knockdown impaired the transcription of NFκB targets in macrophages, key events in the NFκB signaling cascade were examined to assess which part of this pathway is regulated by Rap1.
To examine the effect of Rap1 on the activity of IKK, mock- and Rap1 siRNA-transfected macrophages were stimulated with LPS for 30 minutes and analyzed by Western blotting with antibodies specific for IKKα, IKKβ and phosphorylated IKK (S176/180). Rap1 knockdown did not affect the amount of IKKα or IKKβ, or the level of the phosphorylated form of IKK (S176/180) (Fig. 5A). Hence, the reduced transcription of NFκB target genes in Rap1 knockdown THP-1 macrophages is unlikely to be caused by the attenuation of IKK activity.
Figure 5.
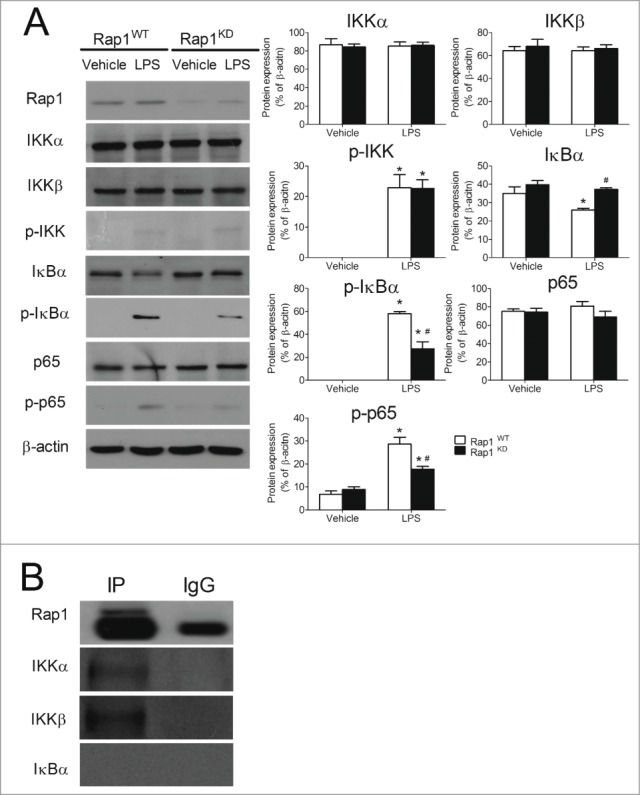
Knockdown of Rap1 suppressed the phosphorylation of IκBα and p65 in LPS-stimulated THP-1 macrophages. (A) Representative original Western blots of Rap1, IKKα, IKKβ, phosphorylated IKK (S176/180), IκBα, phosphorylated IκBα (S32/36), p65 and phosphorylated p65 (S536) in THP-1 macrophages and quantification of these proteins after normalization to β-actin. Protein presence of Rap1, IKKα, IKKβ and phosphorylated IKK was detected at 30 minutes post-treatment. Protein presence of IκBα and phosphorylated IκBα was detected 10 minutes and one hour after treatment, respectively. Protein presence of p65 and phosphorylated p65 was detected 4 hours after treatment; (B) Immunoprecipitation of endogenous Rap1 from differentiated THP-1 macrophages, followed by immunodetection for Rap1, IKKα, IKKβ and IκBα in immunoprecipitates (IP). Data are shown as means ± SEM *P < 0.05 vehicle vs. LPS; #P < 0.05 Rap1WT vs. Rap1KD (of the same treatment group); n = 4 to 5.
In order to investigate the effect of Rap1 on the phosphorylation and degradation of IκBα, mock- and Rap1 siRNA-transfected macrophages were stimulated with LPS for 10 minutes and subjected to immunoblotting using antibodies against non-phosphorylated or phosphorylated IκBα. Under LPS stimulation, knockdown of Rap1 significantly reduced the degradation of IκBα (by 43.0 ± 3.2%; Fig. 5A) and decreased the protein level of phosphorylated IκBα (S32/36) (by 52.7 ± 13.6%; Fig. 5A).
To investigate whether or not Rap1 affects p65 subunits and improves transcription initiation, mock- and Rap1 siRNA-transfected macrophages were stimulated with LPS for 4 hours and subjected to Western blotting with antibodies specific to p65 and phosphorylated p65(S536). Despite an absence of change in the expression of total p65, the protein level of phosphorylated p65 (S536) was decreased significantly in Rap1 knockdown macrophages after LPS stimulation (by 38.0 ± 7.5%; Fig. 5A). Therefore, the reduced phosphorylation on IκBα (S32/36) and that on p65 (S536) in Rap1 knockdown contributed to the alternation of LPS-induced transcription of NFκB target genes in THP-1 macrophages.
Endogenous interaction between Rap1 and the IKK complex
The observation that Rap1 stably resides in the cytosol of macrophages prompted to study its potential interaction with NFκB components. Rap1 was immunoprecipitated and its interaction with IKKα, IKKβ or IκBα was analyzed by immunoblotting. An interaction between Rap1 and the IKK complex (IKKα, IKKβ) was detected, as indicated by the stronger band intensity in the immunoprecipitated group than in IgG controls (Fig. 5B). Absence of IκBα band in both the immunoprecipitated and IgG groups, suggested the absence of interaction between Rap1 and IκBα (Fig. 5B). These data indicate that in macrophages the association of Rap1 to the IKK complex modulates its action and attenuates the subsequent phosphorylation of IκBα and p65.
Rap1 does not influence the expression of NFκB-dependent targets in endothelial and vascular smooth muscle cells
In human atheroma, endothelial and smooth muscle cells elaborate cytokines and contribute to the overall inflammatory environment.19 Therefore, the impact of Rap1 knockdown on the expression of NFκB-dependent targets was examined in these cells. Successful knockdown of Rap1 mRNA was achieved in both human umbilical vein endothelial cells (HUVECs) and human aortic smooth muscle cells (HASMCs) (Fig. 6A). The introduction of Rap1 siRNA into HUVECs and HASMCs significantly decreased Rap1 mRNA (by 66.7 ± 9.0% and 74.9 ± 5.2%, respectively, Fig. 6A) and Rap1 protein levels (by 65.6 ± 5.4% and 51.8 ± 10.5%, respectively, Fig. 6B). Endogenous Rap1 was found in both nuclear and cytosolic cell fractions of endothelial and smooth muscle cells. In HUVECs, transfection of Rap1 siRNA significantly suppressed both nuclear and cytosolic Rap1 protein levels by 44.0 ± 3.3% and 67.3 ± 3.4%, respectively (Fig. 6B). Similarly, introduction of Rap1 siRNA into HASMCs inhibited both nuclear and cytosolic Rap1 protein levels by 39.4 ± 8.0% and 62.6 ± 6.0%, respectively (Fig. 6B).
Figure 6.
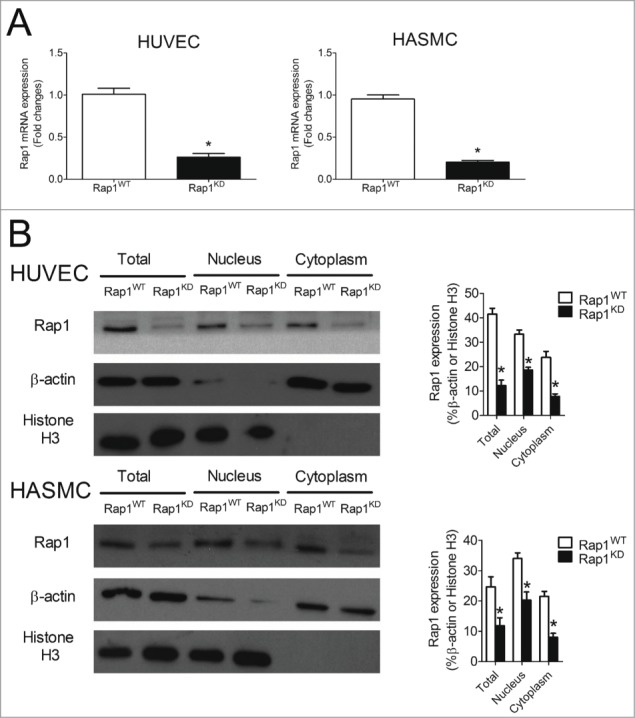
Knockdown efficiency of Rap1 in HUVECs and HASMCs. (A) mRNA expression of Rap1 in Rap1WT and Rap1KD HUVEC or HASMC cells, n = 6; (B) Protein levels of Rap1, β-actin and histone H3 in total, nuclear and cytoplasmic extracts of Rap1WT and Rap1KD HUVEC or HAMSC cells, n = 3 to 5; Data are shown as means ± SEM *P < 0.05 Rap1WT vs. Rap1KD.
Unlike THP-1 macrophages, stimulation with LPS (50ng/ml for 4 hours) did not induce expression of NFκB-dependent cytokines (IL-8, IL-1β) in HUVECs or HASMCs. A higher concentration of LPS (500ng/ml) or TNFα (100ng/ml) and a longer stimulation time (21 hours) were needed to stimulate NFκB-dependent cytokines (IL-8, IL-1β), adhesion molecules [intercellular adhesion molecule-1 (ICAM-1), P-selectin] in HUVECs and glycoproteins [macrophage colony-stimulating factor (M-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF)] in HASMCs. However, knockdown of Rap1 in either HUVECs or HASMCs did not significantly influence the mRNA expression of these NFκB-dependent genes (Figs. 7A and B).
Figure 7.
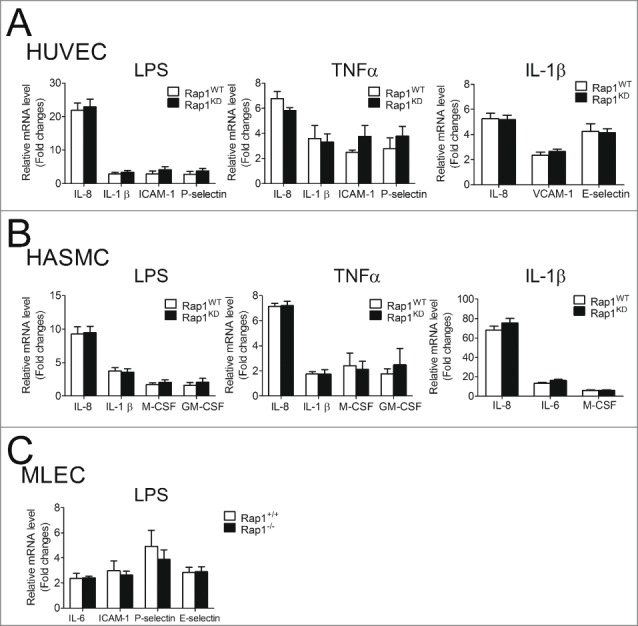
Knockdown or knockout of Rap1 did not alter LPS-, TNFα- or IL-1β-induced NFκB-dependent gene expression in endothelial or smooth muscle cells. (A) mRNA expression of NFκB-dependent genes in Rap1WT and Rap1KD HUVEC cells stimulated with LPS (500ng/ml, 21 hours), TNFα (100ng/ml, 21 hours) or IL-1β (10ng/ml, 4 hours), n = 6; (B) mRNA expression of NFκB-dependent genes in Rap1WT and Rap1KD HASMC cells stimulated with LPS (500ng/ml, 21 hours), TNFα (100ng/ml, 21 hours) or IL-1β (10ng/ml, 4 hours), n = 6; (C) mRNA expression of NFκB-dependent genes in primary lung endothelial cells (MLEC) from Rap1+/+ and Rap1−/− mice stimulated with LPS (500ng/ml, 21 hours), n = 6. mRNA levels are expressed against those in Rap1WT or Rap1+/+ with no stimulation. Data are shown as means ± SEM.
To consolidate the negative effect of Rap1 on NFκB-dependent target genes in endothelial and smooth muscle cells, experiments were repeated using a third stimulus (IL-1β) and further repeated using another cell type - primary mouse lung endothelial cells (MLEC) isolated from Rap1+/+ and Rap1−/− mice. Knockdown of Rap1 did not influence the mRNA expression of IL-1β-induced IL-8, vascular cell adhesion molecule-1 (VCAM-1), E-selectin in HUVEC and IL-8, IL-6, M-CSF in HASMC (Figs. 7A and B). Furthermore, deficiency of Rap1 did not alter LPS-induced mRNA expression of IL-1β, IL-6, ICAM-1 and P-selectin in mouse lung endothelial cells (Fig. 7C).
Rap1 is up-regulated in human atherosclerotic lesions
The expression of Rap1 in non-atherosclerotic arterial tissues and in various grades of human carotid atheroma was examined using a specific anti-Rap1 antibody. Minimal positive staining was detected in non-atherosclerotic arterial tissues (n=3). Faint positive staining was detected within fatty streak lesions (n=4) and significantly more staining was found within fibrous plaques (n=4). The most abundant Rap1 staining was found within atheromatous plaques (n=5). Omitting primary antibody for Rap1 gave negative results in these lesions. Hence, the abundance of Rap1 augmented with increasing grades of atherosclerosis (Fig. 8A). The results obtained by staining adjacent sections of human carotid atheroma with antibodies against CD68 (a marker of macrophages), CD31 (a marker of endothelial cells) and HHF35 (a marker of smooth muscle cells) indicated that Rap1 is localized predominately in macrophage-rich lesions rather than in endothelial or smooth muscle cell-rich areas (Fig. 8B). To validate that Rap1 localized predominately with macrophages, double immunofluorescence staining was performed. Merged images showed that Rap1 co-localized weakly with CD31 and HHF35 (endothelial and smooth muscle cell markers, respectively), but strong co-localization was found for Rap1 and CD68, the specific macrophage marker, in sections of human carotid atheroma (Fig. 8C).
Figure 8.
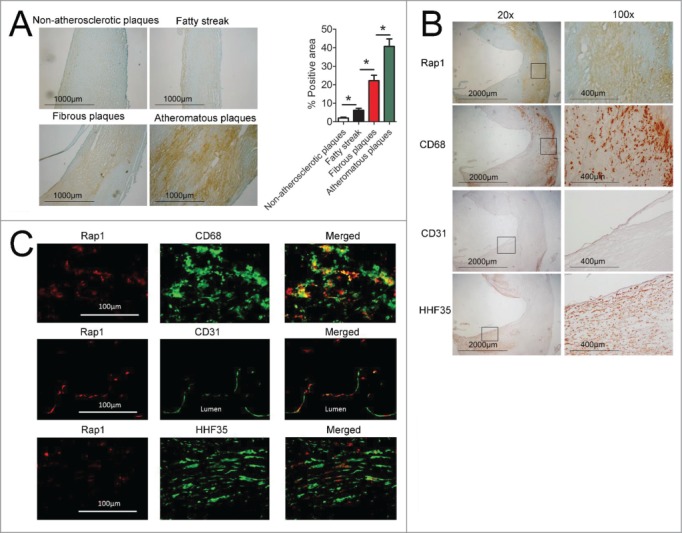
Staining of Rap1, CD68, CD31 and HHF35 in graded human atherosclerosis. (A) Left, Rap1 staining on representative samples from non-atherosclerotic plaques (n = 3), fatty streak (n = 4), fibrous plaques (n = 4), and atheromatous plaques (n = 5). Magnification×40, scale bars: 1000μm; Right, quantification of Rap1 expression levels in non-atherosclerotic arteries and atherosclerotic plaques, % positive area = positive staining area / entire intimal area × 100%. Data are shown as means ± SEM *indicates statistically significant differences between 2 groups (P < 0.05). (B) Adjacent sections of human atherosclerotic lesions were stained with anti-Rap1, anti-CD68, anti-CD31 and anti-HHF35. Left, Magnification×20, scale bars: 2000μm; square box indicate the chosen area for further magnification; Right, Magnification×100, scale bars: 400 μm. (C) Double immunofluorescence staining of Rap1 (detected by Alexa 555 red fluorescence) with cell-specific markers CD68, CD31 or HHF35 (detected by Alexa 488 green fluorescence) in the same section of human atherosclerotic lesions was merged into the overlapped image (Merged). Magnification×200, scale bars: 100 μm.
Discussion
Rap1 was originally believed to be a static protein found only within the nucleus.1,33 The present study confirms that Rap1 is not restricted to the nucleus, but that significant levels of this protein are found in the cytoplasm of macrophages. Such subcellular localization implies that Rap1 is likely to exert essential cytosolic functions in this cell type. Indeed, knockdown or knockout of Rap1 resulted in impaired LPS-induced NFκB activity and lowered expression of pro-inflammatory cytokines in THP-1 macrophages (including IL-8, IL-1β, IL-6 and MCP1) and in mouse peritoneal macrophages (including IL-8, IL-1β, and MCP1). This demonstrates that Rap1 facilitates the generation of NFκB-dependent cytokines in macrophages. In the present study, the stimulation of Rap1 knockdown THP-1 macrophages with LPS also enhanced the mRNA expression of IL-10, a pleiotropic anti-inflammatory cytokine.14 Therefore, the presence of Rap1 favors the production of pro-inflammatory cytokines and limits that of anti-inflammatory cytokines – overall promoting a pro-inflammatory environment. Knockdown or knockout of Rap1 did not influence LPS-induced TNFα expression in macrophages. Thus, the transcriptional modulatory effect of Rap1 is apparently selective for certain NFκB-target genes only. In response to TNFα, another NFκB-activating stimulus, knockdown of Rap1 selectively reduced the expression of IL-8 and IL-1β, but not that of IL-6 and MCP-1. These observations confirmed that endogenous Rap1 is a modulator of NFκB signaling in macrophages and revealed that the particular cohort of NFκB-target genes upregulated by Rap1 is stimulus-specific. Taken together, the data indicated that Rap1 is a potent activator of NFκB in macrophages.
The present study relied on work done on differentiated THP-1 cells and peritoneal macrophages, which are actually activated macrophages that exhibit a pro-inflammatory phenotype. To test whether or not a pro-inflammatory macrophage phenotype is necessary for Rap1 to release NFκB-dependent pro-inflammatory mediators, experiments were repeated using pro-inflammatory M1 and anti-inflammatory M2 macrophages. Rap1 deficiency in M1 macrophages lowered expression of pro-inflammatory cytokines (including IL-1β, MCP-1, TNFα) and increased IκBα expression. By contrast, Rap1 deficiency in M2 macrophages enhanced the expression of pro-inflammatory cytokines (including IL-1β, IL-6). However, similarly to what was observed in M1 macrophages, Rap1 deficiency in M2 macrophages reduced LPS-induced MCP-1 and enhanced IκBα expression. Taken in conjunction, these data demonstrate that Rap1 is a potent activator of NFκB-targeted genes predominately in macrophages with a pro-inflammatory phenotype, while, its effects on the anti-inflammatory M2 macrophages are less uniform as it potentiated some while inhibiting other NFκB-targeted genes.
In the unstimulated state, the nuclear localization sequence of p65 is masked by IκBα, sequestering the inactive NFκB in the cytoplasm.34,35 Upon stimulation by LPS, IKK complex activation is initiated which phosphorylates the inhibitory IκBα at serine 32 and 36.14 This phosphorylation marks IκBα for proteasome-mediated degradation.14 When IκBα dissociates from NFκB, the nuclear localization sequence is revealed and NFκB becomes activated and moves into the nucleus where it binds to specific DNA κB sites resulting in the transcription of specific subset of genes.14,36 Previous findings indicated that Rap1 directly binds to the IKK complex in Hela S3 carcinoma cells,10 but directly associates with IκBα in primary culture of porcine aortic endothelial cells (PAECs).37 The present results on macrophages are in line with the former observation that Rap1 binds directly to the IKK complex. Indeed, the present co-immunoprecipitation experiments revealed that Rap1 binds directly to both IKKα and IKKβ, but not to IκBα. Knowing that Rap1 binds to the IKK complex, it was logical to explore whether or not Rap1 influences its activation. However, knockdown of Rap1 did not affect IKKα/β phosphorylation at serine 176/180, but instead delayed the degradation of IκBα and reduced the phosphorylation of IκBα at serine 32 and serine 36. Hence, Rap1 does not influence the activation of the IKK complex but is vital for the initiation of IκBα degradation. Knockdown of Rap1 also reduced the phosphorylation of p65 at serine 536, indicating its ability to facilitate effective p65 translocation into the nucleus. Taken in conjunction, the present results indicate that Rap1 modulates transcription through NFκB. It facilitates the phosphorylation of IκBα, thus signaling it for degradation. This subsequently enables the phosphorylation of the p65 subunit of NFκB, which is essential for it to work as a competent transcription factor.
Prolonged activation of NFκB with either LPS or TNFα increased Rap1 levels, suggesting the existence of a positive feedback loop for Rap1 self-activation in macrophages. Indeed, the Rap1 promoter region contains NFκB binding sites.10 A positive regulation of Rap1 through NFκB signaling has been described in murine embryonic fibroblast cells.10 When NFκB activation was abrogated in these cells, total Rap1 expression was diminished.10 However, when the p65 subunit was over-expressed, more Rap1 was found in the total cellular extracts.10 Rap1 generated by self-activation ensures more cytokine release and further Rap1 production via NFκB – fueling, a vicious cycle that aggravates the chronic pro-inflammatory environment. This Rap1-NFκB feed-forward loop could be one possible mechanism by which NFκB activity is sustained throughout the lengthy process of atherosclerosis development and progression.12,18 This interpretation is in line with the observation that there is greater IκBα content upon LPS stimulation in Rap1 knockdown macrophages. The presence of Rap1 obviously represses the transcription of IκBα (the endogenous inhibitor of NFκB15), which may also facilitate sustained activation of NFκB.
Constitutive activation of NFκB in macrophages leading to aggravated release of cytokines has been implicated in the development and progression of atherosclerosis.17,38,39 In human atheroma, endothelial cells and smooth muscle cells also elaborate cytokines and contribute to the overall inflammatory state.40,41 Therefore, it was logical to test whether or not reduced Rap1 levels can diminish the release of NFκB-mediated pro-inflammatory mediators in endothelial and vascular smooth muscle cells. Modulation of Rap1 levels had no effect on IL-8, IL-1β, ICAM-1, VCAM-1, E-selectin and P-selectin expression in activated endothelial cells and did not affect IL-8, IL-1β, M-CSF and GM-CSF expression in vascular smooth muscle cells. These findings indicate that Rap1 exacerbates the release of pro-inflammatory mediators selectively in pro-inflammatory macrophages, but not in endothelial or smooth muscle cells. The cytoplasmic function of Rap1 in endothelial and smooth muscle cells remains to be elucidated.
Given the fundamental role of NFκB function in atherosclerosis,12 the presence of Rap1 was determined in diseased human arteries. When human atherosclerotic lesions of different severity were stained with anti-Rap1 antibody, the immunofluorescent analysis revealed greater presence of Rap1 in advanced complicated lesions as compared to early ones. By contrast, there was minimal Rap1 staining in non-atherosclerotic parts of the diseased arteries. These findings suggest the existence of a positive correlation between elevated Rap1 levels and increasing degrees of atherosclerotic lesions. The immunohistochemistry and immunofluorescence experiments revealed that Rap1 is predominately localized in macrophage-rich lesions. This finding is in line with the observations that Rap1 is a potent activator of NFkB in macrophages but not in human endothelial or vascular smooth muscle cells.
The amount of Rap1 in the nucleus is proportional to the length of the telomeres. Telomere attrition is accelerated as the cell ages or faces stress.42–44 Indeed, in senescent PAECs, the amount of Rap1 in the cytosol is elevated compared with young PAECs.37 Thus, it is tempting to speculate that as telomere length is reduced, the ratio between telomere-bound and “free” Rap1 (available for the cytoplasmic activation of the NFκB pathway) is affected. This hypothesis implies that Rap1 could be an important molecular switch that links aging to chronic inflammation.9 Telomere erosions may lead to the accumulation of cytoplasmic Rap1, which in turn could increasingly associate with IKK and enhance the phosphorylation of IκBα and p65, consequently leading to up-regulation of NFκB activity in pro-inflammatory macrophages – thereby driving age-associated inflammation that occurs with atherosclerosis.
In summary, the present study identified the existence of Rap1 in the cytoplasm of macrophages and revealed its role in increasing the production of NFκB-dependent pro-inflammatory mediators. Rap1 augmented the phosphorylation of IκBα and p65 and ultimately enhanced the binding of NFκB to specific DNA κB sites regulating the transcription of a subset of pro-inflammatory genes. The induction of NFκB-dependent pro-inflammatory mediators by Rap1 is specific to macrophages (and predominately in pro-inflammatory macrophages) as it does not occur in endothelial or smooth muscle cells. Extensive Rap1 expression is found in human atheroma with a positive correlation between elevated Rap1 levels and increasing severity of atherosclerotic lesions. By promoting the inflammatory process, Rap1 within pro-inflammatory macrophages may possibly contribute to the development and progression of human atherosclerosis. From a practical standpoint, further understanding on how Rap1 impacts inflammation may yield viable targets for the prevention of the atherosclerotic process.
Materials and Methods
Cell culture
THP-1 monocytic cells, HUVECs and HASMCs were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). THP-1 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Cat. no. 30–2001, ATCC) supplemented with 10% fetal bovine serum (FBS, Cat. no. 16000–044, Invitrogen, Carlsbad, CA, USA), 0.05 mM 2-mercaptoethanol (Cat. no. 21985, Invitrogen) and 1% penicillin/streptomycin (100U/ml, Cat. no. 15240–096, Invitrogen). Phorbol-12-myristate-13-acetate (100 ng/ml, 12 hours, Cat. no. P8139, Sigma, St. Louis, MO, USA) was used to induce the differentiation of THP-1 cells to macrophages. HUVECs were cultured in Ham's Kaighn's Modification F12K medium (Cat. no. 21127–022, Invitrogen) supplemented with 10% FBS, 1% penicillin/streptomycin (100 U/ml), heparin (0.1 mg/ml, Cat. no. 010040–03, LEO Pharma, Denmark) and endothelial cell growth supplement (0.3 mg/ml, Cat. no. 356006, BD Biosciences, San Jose, CA, USA). HASMCs were cultured in Ham's Kaighn's Modification F12K medium supplemented with 10% FBS, 1% penicillin/streptomycin (100U/ml) and 1% Insulin-Transferrin-Selenium (Cat. no. 41400–045, Invitrogen). All cells were incubated at 37°C in a room air atmosphere containing 5% CO2–95%O2.
Transfection and gene silencing
Small interfering RNA (siRNA) against Rap1 (Cat. no. TERF2IP_001) and scramble siRNA were synthesized by Ribobio (Guangzhou, Guangdong, China). Differentiated THP-1 macrophages, HUVECs and HASMCs were transfected with siRNA (50nM) using Lipofectamine 2000 (Cat. no. 11668–019, Invitrogen) for 48 hours according to the manufacturer's instructions and treated with vehicle or with LPS (50 ng/ml, 4 hours for THP-1 macrophages; 500 ng/ml, 21 hours for HUVECs and HASMCs, Cat. no. L2880, Sigma), TNFα (100 ng/ml, 4 hours for THP-1 macrophages; 100ng/ml, 21 hours for HUVECs and HASMCs, Cat. no. PHC3011, Invitrogen) or IL-1β (10 ng/ml, 4 hours for HUVECs and HASMCs, Cat. no. I9401, Sigma) before harvesting. The effect of transfection was validated by measuring the mRNA expression and protein presence of Rap1 through real-time polymerase chain reaction (PCR) and Western blotting, respectively. To ensure target specificity, results were confirmed with a second siRNA to the same target gene (50nM, Cat. no. TERF2IP_003, Ribobio).
Isolation and culture of mouse peritoneal macrophages, bone marrow-derived macrophages and lung endothelial cells
Rap1−/− mice were kindly provided by Dr. Tergaonkar from the Institute of Molecular and Cell Biology under the Agency for Science, Technology and Research (A*STAR) in Singapore.10 For peritoneal macrophages, 8 weeks old male Rap1+/+ and Rap1−/− mice were injected with 2ml of 4% thioglycolate broth (Cat. no. 90404, Sigma) into their peritoneal cavity 3 d prior to collection of the peritoneal exudate. Cells (3×105/ml/well) resuspended in Dulbecco's Modified Eagle medium (Cat. no. 11965–092, Invitrogen) supplemented with 10% FBS and 1% penicillin/streptomycin (100 U/ml) were seeded onto 24-wells plate. After four hours of incubation at 37°C, adhered macrophages were treated with LPS (5ng/ml, 4 hours) before harvesting. For bone marrow-derived M1 or M2 macrophages, bone marrow cells were harvested from the femur and tibia of male Rap1+/+ and Rap1−/− mice (8 weeks old) and cultured in RPMI1640 medium supplemented with 10% FBS, 0.05 mM 2-mercaptoethanol and 1% penicillin/streptomycin (100 U/ml). To induce differentiation into M1 or M2 macrophages, bone marrow cells were treated with GM-CSF (10ng/ml, Cat. no. 12343123, ImmunoTools, Friesoythe, Germany) or M-CSF (10 ng/ml, Cat. no. 12343113, ImmunoTools), respectively as described.45,46 The medium was changed every 2 d for 7 d. Harvested M1 and M2 macrophages (1×105/ml/well) were seeded onto 24-wells plate. To fully polarize M1 and M2 macrophages, GM-CSF-derived M1 macrophages were stimulated with LPS (10 ng/ml, 24 hours) and M-CSF-derived M2 macrophages were cultured with IL-4 (10 ng/ml, 24 hours, Cat. no. 12340043, ImmunoTools). These cells were further stimulated with vehicle or LPS (5 ng/ml, 4 hours) before harvesting. For mouse lung endothelial cells, lung tissues from male Rap1+/+ and Rap1−/− mice (8 weeks of age) were cut into pieces and digested in 20ml of collagenase A (2 mg/ml, Cat. no. C2674, Sigma) at 37°C for 45 minutes with occasional agitation. To isolate endothelial cells, the digested cells were incubated with Dynabeads (sheep anti-rat IgG, Cat. no. 11035, Invitrogen) pre-coated with CD144 (Cat. no. 550548, BD PharMingen, San Diego, CA, USA) and underwent magnetic separation (Dynal MPC-S, Invitrogen). Endothelial cells were plated onto fibronectin-coated cell culture dishes, cultured in Ham's Kaighn's Modification F12K medium supplemented with 20% FBS, 1% penicillin/streptomycin (100U/ml), heparin (0.1 mg/ml) and endothelial cell growth supplement (0.3 mg/ml) for 7 d and further purified with CD144-coated Dynabeads. Confluent endothelial cells (3×104/ml/well) were seeded onto 24-wells plate and treated with LPS (500 ng/ml) for 21 hours before harvesting. All cells were incubated at 37°C in the room air atmosphere. Animal studies were approved by The University of Hong Kong Committee on the Use of Live Animals for Teaching and Research.
Real-time polymerase chain reaction
Total RNA was isolated from cells cultured in 24-wells plates with Qiashredder (Qiagen, Valencia, CA, USA) and RNesay Mini Kit (Qiagen). Equal amounts were reverse-transcribed using the Omniscript RT kit (Qiagen), according to the manufacturer's instructions. Quantitative PCR was performed using SybrGreen Supermix (Cat. no. 170–8884AP, Bio-Rad, Hercules, CA, USA) in ABI 7000 Real-time PCR detection system (Applied Biosystems, Foster City, CA, USA). The conditions for amplification were 3 minutes at 95°C for denaturation, 40 cycles of 15 seconds at 95°C, 30 seconds at 55°C, and 30 seconds at 72°C. The primers sequences used are shown in Table 1. The mRNA levels of the different genes tested were normalized to those of β-actin (for THP-1 macrophages, mouse peritoneal macrophages and bone marrow-derived macrophages) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH, for HUVECs, HASMCs and mouse lung endothelial cells), used as reference genes in all experiments.
Table 1.
Primers used in quantitative real-time PCR on human (h) or mouse (m) cell lysates.
| Gene name a | Forward sequence 5′→3′ | Reverse sequence 5′→3′ |
|---|---|---|
| Rap1(h) | CAGAAGCTCAAGCGGAAGGCG’ | CCGGGTGGCTTCCACAAGC |
| IL-8(h) | AACTTCTCCACAACCCTCTG | TTGGCAGCCTTCCTGATTTC |
| IL-1β(h) | CCACGGCCACATTTGGTT | AGGGAAGCGGTTGCTCATC |
| IL-6(h) | GAAAAAGATGGATGCTTCCA | AACTGGATCAGGACTTTTGT |
| IκBα(h) | ACACTAGAAAACCTTCAGATGC | ACACAGTCATCATAGGGCAG |
| MCP-1(h) | AGACTAACCCAGAAACATCC | GACTGGGGCATTGATTGCATT |
| TNFα(h) | AAGGACACCATGAGCACTGA | AAGTGCAGCAGGCAGAAGAG |
| IL-10(h) | TTACCTGGGTTGCCAAGCCTT | CCTCAGCCTGAGGGTCTTCA |
| ICAM-1(h) | CGGAAATAACTGCAGCATTT | GCGCGTGATCCTTTATAGCG |
| VCAM-1(h) | GCTGCTCAGATTGGAGACTCA | CGCTCAGAGGGCTGTCTATC |
| P-selectin(h) | AGAAGTGGCAGCATGGACTT | CTGTAGTAGGGTAGGACCTT |
| E-selectin(h) | AATCCAGCCAATGGGTTCG | GCTCCCATTAGTTCAAATCCTTCT |
| M-CSF(h) | AGCAGGAGTATCACCGAGGA | CAACTGTTCCTGGTCTACAA |
| GM-CSF(h) | CACTGCTGCTGAGATGAATGAAA | GTCTGTAGGCAGGTCGGCTC |
| GAPDH(h) | CAATGACCCCTTCATTGACCTC | AGCATCGCCCCACTTGATT |
| β-actin(h) | GGACTTCGAGCAAGAGATGG | AGCACTGTGTTGGCGTACAG |
| IL-8(m) | AGAGATACCGCCACGTTCTG | GAGAGGCATCCGGTTCACAG |
| IL-1β(m) | TTGACGGACCCCAAAAGATG | AGAAGGTGCTCATGTCCTCAT |
| IL-6(m) | TGGAGTCACAGAAGGAGTGGCT | TCTGACCACAGTGAGGAATGTC |
| IκBα(m) | TGGAAGTCATTGGTCAGGTGAA | CAGAAGTGCCTCAGCAATTCCT |
| MCP-1(m) | GGCTGGAGAGCTACAAGAGG | TCTTGAGCTTGGTGACAAAAAC |
| TNFα(m) | ACGGCATGGATCTCAAAGAC | AGATAGCAAATCGGCTGACG |
| IL-10(m) | GCTCTTACTGACTGGCATGAG | CGCAGCTCTAGGAGCATGTG |
| ICAM-1(m) | GTGATGCTCAGGTATCCATCCA | CACAGTTCTCAAAGCACAGCG |
| VCAM-1(m) | AGTTGGGGATTCGGTTGTTCT | CCCCTCATTCCTTACCACCC |
| P-selectin(m) | TGAACTGAAGGGATCAAGAAGACT | GCCGAGGGACATCATCACAT |
| E-selectin(m) | ATGCCTCGCGCTTTCTCTC | GTAGTCCCGCTGACAGTATGC |
| GAPDH(m) | AGGTCGGTGTGAACGGATTTG | TGTAGACCATGTAGTTGAGGTCA |
| β-actin(m) | CCTGAGCGCAAGTACTCTGTGT | GCTGATCCACATCTGCTGGAA |
aAbbreviations: GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GM-CSF, granulocyte macrophage colony-stimulating factor; ICAM-1, intercellular adhesion molecule-1; IκBα, inhibitor of kappa B α; IL, interleukin; M-CSF, macrophage colony-stimulating factor; MCP-1, monocyte chemoattractant protein-1; Rap1, repressor activator protein 1; TNFα, tumor necrosis factor α; VCAM-1, vascular cell adhesion molecule-1
Interleukin-8 and interleukin-1β measurements
After LPS stimulation (4 hours), supernatants were collected for the quantification of IL-8 (Cat. no. DY208) and IL-1β (Cat. no. DY201) using enzyme linked immunosorbent assay kits according to manufacturer's instructions (R&D Systems, Minneapolis, MN, USA).
Cell lysis and separation of cellular fractions
Total cell lysates were prepared by lysing differentiated THP-1 macrophages with lysis buffer (20 mM Tris-HCl, 150mM NaCl, 1% Triton X-100, 1 mM ethylene glycol tetra-acetic acid, 1mM ethylenediaminetetraacetic acid, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate and 1mM sodium orthovanadate) supplemented with protease inhibitors [phenylmethylsulfonyl fluoride (1 mM, Cat. no. P7626, Sigma), dithiothreitol (1 mM, Cat. no. D5545, Sigma), aprotinin (1μg/ml, Cat. no. A1153, Sigma), leupeptin (1 μg/ml, Cat. no. L0649, Sigma) and pepstatin A (1μg/ml, Cat. no. P5318, Sigma)]. The cytoplasmic and nuclear fractions of differentiated THP-1 macrophages, HUVECs and HASMCs were separated using a commercially available nuclear extraction kit (Millipore, Billerica, MA, USA). The protein concentration of the samples was determined with the Bradford assay (Cat. no. 500–0006, Bio-Rad).
NFκB activation assay
Differentiated THP-1 macrophages were stimulated with or without LPS (50 ng/ml) for 10, 60 and 240 minutes.39 p65 activation within the nuclear extracts was quantified using the TransAM® NFκB p65 kit, according to the manufacturer's instructions (Cat. no. 40096, Active Motif, Carlsbad, CA, USA).
Immunoprecipitation
Differentiated THP-1 macrophages were lysed with nonidet P-40 lysis buffer (20 mM Tris-HCl, 300 mM NaCl, 2 mM EDTA, 1% nonidet P-40) supplemented with a protease inhibitor cocktail (Cat. no. 04693116001, Roche, Mannheim, Germany). The protein concentration of the samples was determined with the bicinchoninic acid assay (Cat. no. 23225, Thermo Fisher Scientific Inc., Rockford, IL, USA). Cell lysates were incubated overnight with primary Rap1 antibody (Cat. no. SC-28197, Santa Cruz Biotechnology, Dallas, TX, USA) or its respective isotype control (Cat. no. SC-2027, Santa Cruz Biotechnology, Santa Cruz, CA, USA), followed by incubation with Protein A agarose beads (Cat. no. 20333, Thermo Fisher Scientific Inc.) for 2 hours. Immunoprecipitated proteins were eluted with sodium dodecyl sulfate-nonidet P-40 buffer at 95°C for 10 minutes.
Western blotting
Protein lysates were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes for detection with appropriate antibodies. Primary antibodies against IKKα (1:1000, Cat. no. 2682S), IKKβ (1:1000, Cat. no. 2678S), phospho-IKK (1:1000, Cat. no. 2894S), IκBα (1:1000, Cat. no. 4814S), phospho-IκBα (1:1000, Cat. no. 9246S), p65 (1:2000, Cat. no. 4764S), phospho-p65 (1:1000, Cat. no. 3033S), Histone H3 (1:1000, Cat. no. 3638S) were purchased from Cell Signaling Technology (Danvers, MA, USA). Anti-Rap1 (1:500) and anti-β-actin (1:3000, Cat. no. A1978) were purchased from Santa Cruz Biotechnology and Sigma, respectively. Horseradish peroxidase-conjugated anti-mouse (Cat. no. NA931) or anti-rabbit (Cat. no. NA934) secondary antibodies (1:3000) were purchased from GE Healthcare (Boston, MA, USA). Blots were visualized with Amersham™ ECL™ Western Blotting Detection Reagent (Cat. no. RPN2106, GE Healthcare) and subsequently exposed to X-ray film (Fuji Super RX medical X-ray film; Fuji Photo Film, Dusseldorf, Germany). ImageJ software (National Institutes of Health, MD, USA) was used to analyze the optical densities of the immunoreactive bands. Protein presence was normalized to that of β-actin or Histone H3.
Immunohistochemistry and immunofluorescence
Atherosclerotic and non-atherosclerotic human carotid arteries were obtained during endarterectomy or from transplant donors, according to protocols preapproved by the Human Investigative Review Committee of Harvard Medical School. All patients gave informed consent. Serial cryostat sections (6μm) were prepared and stained with anti-Rap1 (1:200, Santa Cruz, CA, USA), anti-macrophage-specific CD68 (1:500, Cat. no. M0814, Dako, Carpinteria, CA, USA), anti-endothelium-specific CD31 (1:35, Cat. no. M0823, Dako) or anti-smooth muscle α-actin-specific HHF35 (1:40, Cat. no. C34931, Enzo Life Sciences, Syosset, NY, USA) antibodies. Antibody binding was visualized with 3,3′-diaminobenzidine (Cat. no. K3464, Dako) or 3-amino-9-ethyl carbazole (Cat. no. K3468, Dako). Images were captured with an Olympus BX41 microscope equipped with an Olympus DP72 digital camera (Olympus, Tokyo, Japan). Image analysis was performed with Image Pro Plus 5.0 (MediaCybernetics, Rockville, MD, USA). For the localization of Rap1 to respective cell types, double immunofluorescence staining was performed using rabbit anti-Rap1 antibody (Santa Cruz, CA, USA) mixed with cell selective monoclonal antibodies: mouse anti-CD68 (macrophages, Dako), or CD31 (endothelial cells, Dako), or HHF35 (smooth muscle cells, Enzo Life Sciences). Subsequently, a mixture of secondary antibodies goat anti-rabbit Alexa 555 (red, Cat. no. A-21428, Invitrogen) and goat anti-mouse Alexa 488 (green, Cat. no. A-11029, Invitrogen) was applied to visualize antigens. Images were captured by Inverted Nikon eclipse TE2000-U microscope.
Statistical Analysis
Data are expressed as means ± SEM. Comparisons between groups were carried out by 2-tailed non-parametric Mann-Whitney U test or unpaired Student's t-test, where appropriate, using the GraphPad Prism 5.0 software (San Diego, CA, USA). Differences were considered statistically significant when P was less than 0.05.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
We acknowledge Dr. Mary Y. K. Lee for helpful discussion of the isolation of mouse lung endothelial cells in this study.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work described in this paper is supported by the General Program of National Natural Science Foundation of China (Project No. 81270383).
References
- 1.Li B, Oestreich S, de Lange T. Identification of Human Rap1: implications for telomere evolution. Cell 2000; 101:471-83; PMID:10850490; http://dx.doi.org/ 10.1016/S0092-8674(00)80858-2 [DOI] [PubMed] [Google Scholar]
- 2.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 2005; 19:2100-10; PMID:16166375; http://dx.doi.org/ 10.1101/gad.1346005 [DOI] [PubMed] [Google Scholar]
- 3.Kabir S, Sfeir A, de Lange T. Taking apart Rap1: an adaptor protein with telomeric and non-telomeric functions. Cell Cycle 2010; 9:4061-7; PMID:20948311; http://dx.doi.org/ 10.4161/cc.9.20.13579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martínez P, Thanasoula M, Carlos AR, Gómez-López G, Tejera AM, Schoeftner S, Dominguez O, Pisano DG, Tarsounas M, Blasco MA. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat Cell Biol 2010; 12:768-80; PMID:20622869; http://dx.doi.org/20339076 10.1038/ncb2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. Loss of Rap1 Induces Telomere Recombination in the Absence of NHEJ or a DNA Damage Signal. Science 2010; 327:1657-61; PMID:20339076; http://dx.doi.org/ 10.1126/science.1185100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantó C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol 2009; 20:98-105; PMID:19276888; http://dx.doi.org/23791522 10.1097/MOL.0b013e328328d0a4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martínez P, Gómez-López G, García F, Mercken E, Mitchell S, Flores JM, de Cabo R, Blasco MA. Rap1 protects from obesity through Its extratelomeric role regulating gene expression. Cell Rep 2013; 3:2059-74; PMID:23791526; http://dx.doi.org/23791522 10.1016/j.celrep.2013.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeung F, Ramírez CM, Mateos-Gomez PA, Pinzaru A, Ceccarini G, Kabir S, Fernández-Hernando C, Sfeir A. Nontelomeric role for Rap1 in regulating metabolism and protecting against obesity. Cell Rep 2013; 3:1847-56; PMID:23791522; http://dx.doi.org/ 10.1016/j.celrep.2013.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh AS, Tergaonkar V. Telomeres and inflammation: Rap1 joins the ends? Cell Cycle 2010; 9:3834-5; PMID:20890110; http://dx.doi.org/ 10.4161/cc.9.19.13383 [DOI] [PubMed] [Google Scholar]
- 10.Teo H, Ghosh S, Luesch H, Ghosh A, Wong ET, Malik N, Orth A, de Jesus P, Perry AS, Oliver JD, et al.. Telomere-independent Rap1 is an IKK adaptor and regulates NF-kappa B-dependent gene expression. Nat Cell Biol 2010; 12:758-67; PMID:20622870; http://dx.doi.org/ 10.1038/ncb2080 [DOI] [PubMed] [Google Scholar]
- 11.Low KC, Tergaonkar V. Telomerase: central regulator of all of the hallmarks of cancer. Trends Biochem Sci 2013; 38:426-34; PMID:23932019; http://dx.doi.org/ 10.1016/j.tibs.2013.07.001 [DOI] [PubMed] [Google Scholar]
- 12.Xanthoulea S, Curfs DM, Hofker MH, de Winther MP. Nuclear factor kappa B signaling in macrophage function and atherogenesis. Curr Opin Lipidol 2005; 16:536-42; PMID:16148538; http://dx.doi.org/24877606 10.1097/01.mol.0000180167.15820.ae [DOI] [PubMed] [Google Scholar]
- 13.Tong L, Tergaonkar V. Rho protein GTPases and their interactions with NFκB: crossroads of inflammation and matrix biology. Biosci Rep 2014; 34:e00115; PMID:24877606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol 1998; 16:225-60; PMID:9597130; http://dx.doi.org/ 10.1146/annurev.immunol.16.1.225 [DOI] [PubMed] [Google Scholar]
- 15.Tedgui A, Mallat Z. Cytokines in atherosclerosis: pathogenic and regulatory pathways. Physiol Rev 2006; 86:515-81; PMID:16601268; http://dx.doi.org/ 10.1152/physrev.00024.2005 [DOI] [PubMed] [Google Scholar]
- 16.Lian S, Meng L, Liu C, Xing X, Song Q, Dong B, Han Y, Yang Y, Peng L, Qu L, Shou C. PRL-3 activates NF-κB signaling pathway by interacting with Rap1. Biochem Biophys Res Commun 2013; 430:196-201; PMID:23178297; http://dx.doi.org/ 10.1016/j.bbrc.2012.11.036 [DOI] [PubMed] [Google Scholar]
- 17.Monaco C, Paleolog E. Nuclear factor kappa B: a potential therapeutic target in atherosclerosis and thrombosis. Cardiovasc Res 2004; 61:671-82; PMID:14985064; http://dx.doi.org/ 10.1016/j.cardiores.2003.11.038 [DOI] [PubMed] [Google Scholar]
- 18.Libby P, Ridker PM, Hansson GK. Inflammation in atherosclerosis: from pathophysiology to practice. J Am Coll Cardiol 2009; 54:2129-38; PMID:19942084; http://dx.doi.org/ 10.1016/j.jacc.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Libby P, Ridker PM, Maseri A. Inflammation and atherosclerosis. Circulation 2002; 105:1135-43; PMID:11877368; http://dx.doi.org/ 10.1161/hc0902.104353 [DOI] [PubMed] [Google Scholar]
- 20.Ross R. Atherosclerosis — an inflammatory disease. N Engl J Med 1999; 340:115-26; PMID:9887164; http://dx.doi.org/ 10.1056/NEJM199901143400207 [DOI] [PubMed] [Google Scholar]
- 21.Brand K, Page S, Walli AK, Neumeier D, Baeuerle PA. Role of nuclear factor-kappa B in atherosclerosis. Exp Physiol 1997; 82:297-304; PMID:9129944; http://dx.doi.org/ 10.1113/expphysiol.1997.sp004025 [DOI] [PubMed] [Google Scholar]
- 22.Brand K, Page S, Rogler G, Bartsch A, Brandl R, Knuechel R, Page M, Kaltschmidt C, Baeuerle PA, Neumeier D. Activated transcription factor nuclear factor-kappa B is present in the atherosclerotic lesion. J Clin Invest 1996; 97:1715-22; PMID:8601637; http://dx.doi.org/ 10.1172/JCI118598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore KJ, Tabas I. Macrophages in the pathogenesis of atherosclerosis. Cell 2011; 145:341-55; PMID:21529710; http://dx.doi.org/ 10.1016/j.cell.2011.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manel N, Hogstad B, Wang Y, Levy DE, Unutmaz D, Littman DR. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature 2010; 467:214-7; PMID:20829794; http://dx.doi.org/ 10.1038/nature09337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pabst MJ, Johnston RB Jr. Increased production of superoxide anion by macrophages exposed in vitro to muramyl dipeptide or lipopolysaccharide. J Exp Med 1980; 151:101-14; PMID:7350246; http://dx.doi.org/ 10.1084/jem.151.1.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenstreich DL, McAdam KP. Lymphoid cells in endotoxin-induced production of the amyloid-related serum amyloid A protein. Infect Immun 1979; 23:181-3; PMID:370010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoll LL, Denning GM, Weintraub NL. Potential role of endotoxin as a pro-inflammatory mediator of atherosclerosis. Arterioscler Thromb Vasc Biol 2004; 24:2227-36; PMID:15472123; http://dx.doi.org/ 10.1161/01.ATV.0000147534.69062.dc [DOI] [PubMed] [Google Scholar]
- 28.Michelsen KS, Wong MH, Shah PK, Zhang W, Yano J, Doherty TM, Akira S, Rajavashisth TB, Arditi M. Lack of Toll-like receptor 4 or myeloid differentiation factor 88 reduces atherosclerosis and alters plaque phenotype in mice deficient in apolipoprotein E. Proc Natl Acad Sci 2004; 101:10679-84; PMID:15249654; http://dx.doi.org/ 10.1073/pnas.0403249101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawrence T. The nuclear factor NF-kappa B pathway in inflammation. Cold Spring Harb Perspect Biol 2009; 1:a001651; PMID:20457564; http://dx.doi.org/ 10.1101/cshperspect.a001651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghosh A, Saginc G, Leow SC, Khattar E, Shin EM, Yan TD, Wong M, Zhang Z, Li G, Sung WK, et al.. Telomerase directly regulates NF-κB-dependent transcription. Nat Cell Biol 2012; 14:1270-81; PMID:23159929; http://dx.doi.org/ 10.1038/ncb2621 [DOI] [PubMed] [Google Scholar]
- 31.Steer JH, Kroeger KM, Abraham LJ, Joyce DA. Glucocorticoids suppress tumor necrosis factor-alpha Expression by human monocytic THP-1 cells by suppressing transactivation through adjacent NF-kappa B and c-Jun-activating transcription factor-2 binding sites in the promoter. J Biol Chem 2000; 275:18432-40; PMID:10748079; http://dx.doi.org/ 10.1074/jbc.M906304199 [DOI] [PubMed] [Google Scholar]
- 32.McDonald PP, Bald A, Cassatella MA. Activation of the NF-kappa B pathway by inflammatory stimuli in human neutrophils. Blood 1997; 89:3421-33; PMID:912905017499040 [PubMed] [Google Scholar]
- 33.Bae NS, Baumann P. A Rap1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol Cell 2007; 26:323-34; PMID:17499040; http://dx.doi.org/ 10.1016/j.molcel.2007.03.023 [DOI] [PubMed] [Google Scholar]
- 34.Israël A. The IKK complex, a central regulator of NF-kappa B activation. Cold Spring Harb Perspect Biol 2009; 2:a000158; PMID:2030020315653325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viatour P, Merville MP, Bours V, Chariot A. Phosphorylation of NF-kappa B and Ikappa B proteins: implications in cancer and inflammation. Trends Biochem Sci 2005; 30:43-52; PMID:15653325; http://dx.doi.org/ 10.1016/j.tibs.2004.11.009 [DOI] [PubMed] [Google Scholar]
- 36.Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, Andersen JS, Mann M, Mercurio F, Ben-Neriah Y. Identification of the receptor component of the Ikappa B alpha-ubiquitin ligase. Nature 1998; 396:590-4; PMID:9859996; http://dx.doi.org/ 10.1038/25159 [DOI] [PubMed] [Google Scholar]
- 37.Bai B, Liang Y, Xu C, Lee MYK, Xu A, Wu D, Vanhoutte PM, Wang Y. Cyclin-dependent kinase 5–mediated hyperphosphorylation of sirtuin-1 contributes to the development of endothelial senescence and atherosclerosis. Circulation 2012; 126:729-40; PMID:22753194; http://dx.doi.org/ 10.1161/CIRCULATIONAHA.112.118778 [DOI] [PubMed] [Google Scholar]
- 38.Mallat Z, Tedgui A. NF-kappa B activation in atherosclerosis: a friend or a foe? Blood 2004; 103:754-5; http://dx.doi.org/ 10.1182/blood-2003-11-3818 [DOI] [Google Scholar]
- 39.Kanters E, Pasparakis M, Gijbels MJ, Vergouwe MN, Partouns-Hendriks I, Fijneman RJ, Clausen BE, Förster I, Kockx MM, Rajewsky K, et al.. Inhibition of NF-κappa B activation in macrophages increases atherosclerosis in LDL receptor–deficient mice. J Clin Invest 2003; 112:1176-85; PMID:14561702; http://dx.doi.org/ 10.1172/JCI200318580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doran AC, Meller N, McNamara CA. Role of smooth muscle cells in the initiation and early progression of atherosclerosis. Arterioscler Thromb Vasc Biol 2008; 28:812-9; PMID:18276911; http://dx.doi.org/ 10.1161/ATVBAHA.107.159327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gareus R, Kotsaki E, Xanthoulea S, van der Made I, Gijbels MJ, Kardakaris R, Polykratis A, Kollias G, de Winther MP, Pasparakis M. Endothelial cell-specific NF-kappa B inhibition protects mice from atherosclerosis. Cell metab 2008; 8:372-83; PMID:19046569; http://dx.doi.org/ 10.1016/j.cmet.2008.08.016 [DOI] [PubMed] [Google Scholar]
- 42.Fuster JJ, Andrés V. Telomere biology and cardiovascular disease. Circ Res 2006; 99:1167-80; PMID:17122447; http://dx.doi.org/ 10.1161/01.RES.0000251281.00845.18 [DOI] [PubMed] [Google Scholar]
- 43.Kurz DJ, Decary S, Hong Y, Trivier E, Akhmedov A, Erusalimsky JD. Chronic oxidative stress compromises telomere integrity and accelerates the onset of senescence in human endothelial cells. J Cell Sci 2004; 117:2417-26; PMID:15126641; http://dx.doi.org/ 10.1242/jcs.01097 [DOI] [PubMed] [Google Scholar]
- 44.Okuda K, Khan MY, Skurnick J, Kimura M, Aviv H, Aviv A. Telomere attrition of the human abdominal aorta: relationships with age and atherosclerosis. Atherosclerosis 2000; 152:391-8; PMID:10998467; http://dx.doi.org/ 10.1016/S0021-9150(99)00482-7 [DOI] [PubMed] [Google Scholar]
- 45.Zheng XF, Hong YX, Feng GJ, Zhang GF, Rogers H, Lewis MA, Williams DW, Xia ZF, Song B, Wei XQ. Lipopolysaccharide-induced M2 to M1 macrophage transformation for IL-12p70 production is blocked by Candida albicans mediated up-regulation of EBI3 expression. PLoS One 2012; 8:e63967; PMID:23724011; http://dx.doi.org/22547697 10.1371/journal.pone.0063967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lacey DC, Achuthan A, Fleetwood AJ, Dinh H, Roiniotis J, Scholz GM, Chang MW, Beckman SK, Cook AD, Hamilton JA. Defining GM-CSF- and macrophage-CSF-dependent macrophage responses by in vitro models. J Immunol 2012; 188:5752-65; PMID:22547697; http://dx.doi.org/ 10.4049/jimmunol.1103426 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


