Abstract
Antibiotic-resistant bacterial infections are a vexing global health problem and have rendered ineffective many previously-used antibiotics. Here we demonstrate that antibiotic-linkage to surface-functionalized silica nanoparticles (sNP) significantly enhances their effectiveness against Escherichia coli, and Staphylococcus aureus, and even methicillin-resistant S. aureus (MRSA) strains that are resistant to most antibiotics. The commonly-used antibiotic penicillin-G (PenG) was complexed to dye-labeled sNPs (15 nm diameter) containing carboxyl groups located as either surface-functional groups, or on polymer-chains extending from surfaces. Both sNPs configurations efficiently killed bacteria, including MRSA strains. This suggests that activities of currently-ineffective antibiotics can be restored by nanoparticle-complexation and used to avert certain forms of antibiotic-resistance.
The increasing prevalence, perseverance, and adaptability of bacterial resistance to antibiotics is a vexing healthcare problem; one which results in high morbidity/mortality and healthcare costs exceeding $20 billion annually.1–5 A wide range of infectious strains now exhibit antibiotic resistance. Common examples include MRSA (methicillin-resistant Staphylococcus aureus), Pseudomonas aeruginosa lung- and wound-infections, VREs (vancomycinresistant Enterococci), bacterial pneumonia strains, and urinary tract-infections (UTIs), as well as a host of infections that occur in association with human conditions such as AIDs, and intestinal/colon breaches.6–8 The frequency of community-acquired methicillin-resistant Staphylococcus aureus (MRSA) increased more than seven-fold from 1999 to 2006.9 Patients who acquire such infections are at increased risk for death and disease. Such patients can more than double inpatient hospital costs1 and account for increased outpatient treatment costs10 and spending on long-term care.
Many widely-used antibiotics (e.g., beta-lactam antibiotics) share similarities in molecular structure and modes of action.11 Since genetic mechanisms underlying antibiotic resistance can be readily exchanged among bacterial cells, a growing number of pathogenic strains now exhibit multiple antibiotic resistance (MAR).12 Overuse of antibiotics selects for the emergence and later persistence of a resistant infection following antibiotic treatment,13 a number that has been increasing over the past two decades. Consequently, many previously-used antibiotics (e.g. penicillin) have been rendered either less-effective or ineffective because of the preponderance of bacterial strains having genetically-transferable antibiotic resistance.14 In order to overcome the growing patterns of resistance, a more effective utilization of antibiotics against infections is required.
Nanoparticles (NP) and other nanotechnology-based approaches are now being investigated as therapeutic delivery-vehicles for antibiotics and other antimicrobial compounds to bacterial cells. While NPs have been used extensively for the delivery of anti-cancer drugs and other types of molecules to eukaryote cells,15–17 they have not been utilized to a great extent for delivery of antibiotics to bacterial cells.18 However, the size-dependent properties of NPs coupled to their high specific surface area and surface energies, potentially provide NPs with increased delivery capabilities when compared to dissolved molecules.19 NPs additionally offer emerging potential because their chemistry can be engineered with high-specificity to possess surfaces having different types and densities of chemical functional groups, charges and other properties.20–23 This can permit pre-determined quantities of antibiotic molecules to be complexed by a single nanoparticle, and subsequently used to kill infectious bacterial cells. The interiors or surfaces of nanoparticles similarly can be engineered with fluorophores to facilitate quantitative detection. These properties, when used in combination, also offer a unique and improved potential for probing bacterial infections with increasing resolution.
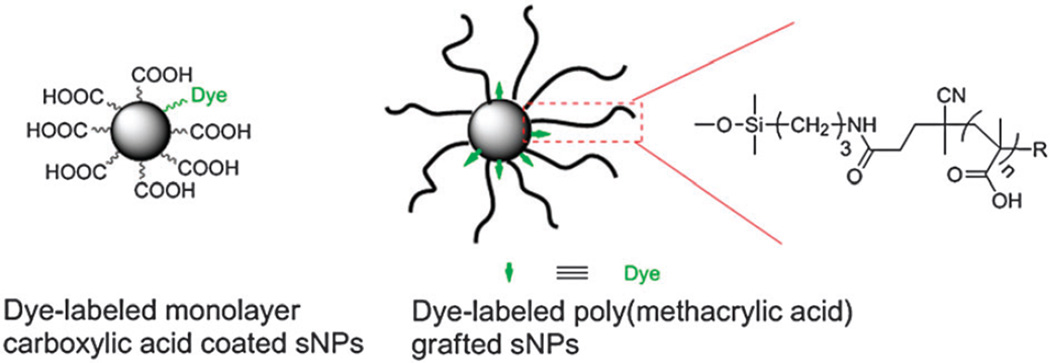
In the present study, we complexed the antibiotic penicillin-G using specifically-engineered nanoparticles, against bacterial pathogens, some of which were highly-resistant to the antibiotic. The activities of relatively non-effective antibiotics were significantly enhanced by nanoparticle-conjugation and avert certain forms of antibiotic resistance, especially if nanoparticles can specifically target bacteria. The antibiotic was complexed to engineered silica nanoparticles, the surfaces of which were functionalized with monolayer carboxylic acids, and poly(methacrylic acid) (PMAA) prepared by the RAFT polymerization technique (Fig. 1). Both forms of antibiotic–sNPs complexes demonstrated enhanced inhibition of the bacteria.
Fig. 1.
Structure of dye-labeled monolayer carboxylic acid and PMAA grafted silica nanoparticles (sNPs).
Our investigation studied ten bacterial pathogens (six Gram-negative and four Gram-positive bacteria), including methicillin-resistant Staphylococcus aureus (MRSA) strains. In disk-diffusion assays, treatment disks containing the nanoparticle complexes were tested on the same agar plates as the control (soluble PenG) disks to ensure that the dosage and growth stage of the bacterial lawn was comparable.
The carboxylic acid engineered sNPs were labeled with fluorescent dyes for monitoring the presence and movement of the sNPs using UV-vis spectroscopy and confocal scanning laser microscope (CSLM). A carboxylic acid monolayer was coated on sNPs with fluorescent tags in two steps (Fig. S1a, ESI†). First, colloidal silica nanoparticles were reacted with 3-aminopropyldimethylethoxysilane to generate amine-coated nanoparticles (with a predetermined amount of amine groups). Secondly, the amino particles were allowed to react with a small amount (<5 mol% relative to the amines) of 7-nitrobenzofurazan-(NBD)-based dye molecule. Subsequently, an excess of succinic anhydride was added to the reaction solution to generate fluorescent dye-labeled carboxylic acid-coated nanoparticles. Fig. S1c (ESI†) shows TEM images of amine and dye-labeled carboxylic acid functionalized nanoparticles, and exhibits an excellent dispersion of these particles. The IR spectra (Fig. S2, ESI†) of amine and carboxylic acid functionalized silica particles confirmed the successful conversion of amino groups to carboxylic acid due to the formation of a new peak at 1721 cm−1 ascribed to the carbonyl groups. The amount of dye molecules attached to the nanoparticle surface was determined by absorbance of functionalized nanoparticles (326 nm) and compared to a standard UV-vis absorption curve of free NBD-COOH (Fig. S1b, ESI†).
A series of dye- and carboxylic acid-functionalized sNPs at low-, medium-, and high-density were prepared, as shown in Table S1 (ESI†). Samples labeled ‘Entry 1’ and ‘Entry 5’ were used as control groups to test the toxicity of functionalized sNPs to bacteria. Bare sNP (entry 1, Table S1, ESI†) and 100% fluorescent dye-surface functionalized sNP (entry 5, Table S1, ESI†) demonstrated no detectable toxicity to either E. coli or S. typhimurium. Dynamic-light-scattering (DLS) (Fig. S3, ESI†) indicated the mean diameter of bare nanoparticles was 18.9 × 0.4 nm. The mean diameter of carboxylic acid coated-nanoparticles was 22.7 nm, with an increase of 3.8 nmafter surface modification.
Synthesis of PMAA grafted sNPs was based on previous work.21,22 Briefly, poly(tert-butyl methacrylate) grafted sNPs were prepared by RAFT polymerization of tert-butyl methacrylate mediated by 4-cyanopentanoic acid dithiobenzoate (CPDB) coated sNPs in THF employing azobisisobutyronitrile (AIBN) as the initiator at 60 °C. The poly(tert-butyl methacrylate) grafted sNPs with dithiobenzoate moieties were treated with excess of AIBN to remove the thiocarbonylthio groups, followed by deprotection of tert-butyl ester groups via trimethylsilyl iodide or trimethylsilyl bromide treatment. The extremely large abundance of carboxylic acid on the PMAA extending from the nanoparticles provides the capability for complexation of enormous numbers of antibiotic molecules to a single nanoparticle.
Fig. S4(a–c) (ESI†) shows TEM images of poly(tert-butyl methacrylate) and PMAA grafted sNPs, and reveals an excellent dispersion of nanoparticles after each step of chemical modifications. The diameter of individual PMAA grafted sNPs was approx. 30 nm diameter, and consistent with AFM-based measurements (Fig. S4d, ESI†). Fig. S5 (ESI†) shows thermogravimetric analyses (TGA) in nitrogen of PtBuMA, PtBuMA-grafted sNPs, and PMAAgrafted sNPs. The weight loss for each sample was consistent with the grafting density and chemical modifications across different grafting densities, and polymer molecular-weights investigated in this study.21
PenG was non-selectively complexed to monolayer carboxylic acid or PMAA grafted sNPs via physical interactions. Basically, PenG was added to the water solution of monolayer carboxylic acid or PMAA grafted nanoparticles for incubation at 28 °C with shaking for 3 hours. The PenG–nanoparticle complex was collected and separated from unbound PenG using Amicon Ultra Centrifuge Filters. As a result, UV-vis binding studies were used to determine PenG loadings (Fig. S6, ESI†). It was clearly observed that the conjugation of PenG was gradually increased until achieving saturation at a given concentration of PMAA grafted sNPs when increasing the loadings of PenG in the complex. In addition, the amount of unbound PenG was increased when the loading of PenG was higher than the saturation point (0.25 mM). Concurrently, the amount of particle-complexed PenG was constant, which is ascribed to the saturation conjugation.
Standardized agar disk-diffusion assays and broth dilution approaches were used to measure the antimicrobial activities of PenG and sNP–PenG complexes. The inhibition activity of nanoparticle-complexed antibiotic was verified by its presence in the inhibition zones of agar plates. Bare sNPs did not result in observable zones of inhibition. Free penicillin at a low dose (2.5 µg) had an activity of 0 in both assays, while the nanocomplexed form of penicillin with the same dose of PenG enhanced activities significantly (Fig. S7, ESI†).
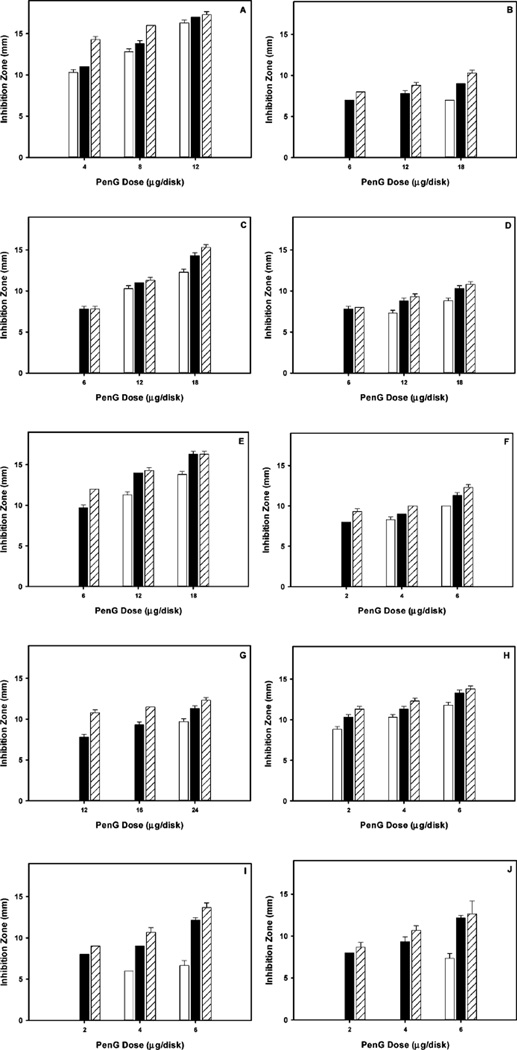
The results (Fig. 2) showed that in disk-diffusion assays using Gram-negative bacteria (B, D and I), both PenG-complexed to monolayer carboxylic acid and to PMAA grafted nanoparticles, exhibited much greater inhibition zones (>7 mm in B; >7.5 mm in D; >8 mm in I) while inhibition zones of free-PenG were effectively 0 mm when the PenG concentrations were low (B–G, I, J). Differences in observed inhibition zones between nanoparticlecomplexed PenG and free-PenG decreased when PenG concentrations were increased. The same trends were also observed in tests using Gram-positive bacteria and MRSA. The ability of PenG, when complexed to either monolayer carboxylic acid- or PMAAnanoparticles, to inhibit MRSA strains (Fig. 2F, G) illustrates the increased effectiveness of nanoparticle-complexed antibiotics. Minimum inhibitory concentrations (MIC) for a variety of bacteria were calculated using reported protocols by determination of the inhibition area (measuring the diameter and the depth of the agar).24,25 As shown in Table S2 (ESI†), MIC values indicated that PenG, when complexed to monolayer carboxylic acid- or poly(methacrylic acid) grafted-nanoparticles exhibited strong antimicrobial activities against both Gram-positive and Gram-negative bacteria, and even MRSA.
Fig. 2.
Inhibition activity of free-PenG (white), PenG-complexed to the monolayer carboxylic acids on sNPs (black), and PenG-complexed to carboxylated polymers on sNPs (hatched), as tested by disk-diffusion assays using B. cereus (A), P. aeruginosa (B), K. pneumoniae (C), P. vulgaris (D), E. aerogenes (E), S. typhimurium (F), CA-MRSA (G), HA-MRSA (H), E. coli (I), and S. aureus (J).
The disk-diffusion assays illustrate the high antimicrobial activity of the nanocomplexed antibiotics against both Gram-negative and Gram-positive bacteria, especially antibiotic-resistant MRSA. These results were surprising because PenG is not typically effective against MRSA. The question emerges from our results: why do nanoparticle-complexed antibiotics have increased antibacterial activity? In order to answer this question, control groups of dye-labeled monolayer carboxylic acid and PMAA grafted sNPs were used to conduct the same experiment as the sNP–PenG complex. It was found that these particles did not show any activities to the Gram-negative and Gram-positive bacteria, including MRSA. Thus, the real moiety which demonstrated the antimicrobial activity in the complex is the complexed PenG.
Beta-lactam antibiotics, such as PenG, work by interfering with the cross-linkage of peptidoglycan, a major component of the bacterial cell wall.26 Resistant bacterial strains often possess beta-lactamases, enzymes that specifically degrade the lactam structure and render the antibiotic ineffective. In order to verify the resistance mechanism, we tested the effects of nanoparticle–PenG complex on beta-lactamases by incubation of free PenG or nanoparticle-complexed PenG (both on monolayer carboxylic acids and PMAA coated sNPs) and E. coli (ATCC 25922) followed by the nitrocefin disk assays. Nitrocefin, containing a beta-lactam ring, is a chromogenic cephalosporin and often used to rapidly detect beta-lactamase.27 Usually, nitrocefin solution is yellow with a UV-vis absorption peak around 380 nm. Beta-lactamase hydrolyzes the beta-lactam ring of nitrocefin, resulting in a color change from yellow (negative) to red (positive), with a corresponding UV-vis absorption peak around 480 nm. Incubations with monolayer carboxylic acid coated sNPs remained a yellow color, which indicated no- or little hydrolysis; while incubations with PMAA resulted in a light yellow color, which reveal greater hydrolysis (Fig. S8, ESI†). However, compared to the control group of free PenG on nitrocefin disk (no sNPs) that resulted in a dark red color, incubations with the PenG–nanoparticle complexes generated much less beta-lactamase. This low concentration of beta-lactamase leads us to hypothesize that the binding of PenG on nanoparticle surfaces resulted in less recognition by the cell and a lower generation of beta-lactamase.
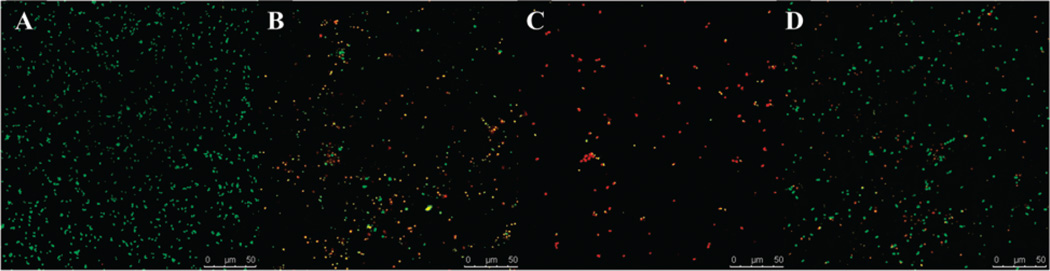
Confocal scanning laser microscopy (CSLM) confirmed that exposure to nanoparticle-complexed PenG resulted in cell death and low cell density. As shown in Fig. 3, cells exposed to nanoparticlecomplexed PenG and free PenG (B–D) were less dense and consisted of more dead (red fluorescent) cells than the control group (A), which consisted of mostly live (green) cells. Exposure to nanoparticle-complexed PenG (B and C) resulted in greater cell death than free PenG (D). Fig. 3(C) demonstrates that PenG-complexed to carboxylated polymers on sNPs resulted in greater cell death and lower cell density than PenG-complexed to monolayer carboxylic acids on sNPs (B). This result agrees well with previous disk diffusion assays.
Fig. 3.
Confocal scanning laser microscopy images of bacterial cells (A) S. aureus; (B) S. aureus exposed to complexed PenG on monolayer carboxylic acid coated sNPs; (C) S. aureus exposed to complexed PenG on carboxylated polymer grafted sNPs; (D) S. aureus exposed to free PenG (Scale bars: 50 µm). The Pn–nanoparticles complexes (B and C) have equal doses of PenG as the free PenG group (D).
We hypothesize that another reason for the high antimicrobial activities is the locally high concentrations of surface-attached antibiotic molecules on a nanoparticle. This effect has been reported by other groups.28–31 This effect may overwhelm a targeted bacterial cell’s ability to degrade the antibiotics using enzymes (i.e. beta-lactamases). In contrast, a typical antibiotic, once solubilized, will diffuse to form a rather homogeneous dispersion of molecules spread over bacterial cells, and results in a relatively lower concentration of antibiotic molecules reaching a single bacterium. Our studies suggest that when antibiotics are concentrated on a nanoparticle surface, they can supply a more locally-concentrated dose. The ability of PenG to prevent bacteria from building a peptidoglycan layer weakens the cell wall of the bacterium, which ultimately results in cell lysis.
In conclusion, we engineered carboxylic acid grafted silica nanoparticles (sNP) and examined their abilities to bind and deliver antibiotics to bacterial cells. We show that when a commonly-used antibiotic, penicillin-G, was linked to nanoparticles, their bacteriocidal efficiencies were increased significantly. Therefore, much lower concentrations of the antibiotic were needed to kill these bacteria under laboratory conditions. Additionally, when antibiotics are nano-linked, they become effective against antibiotic-resistant forms. And lastly, the type of linkage of the antibiotic to the nanoparticle and the density of antibiotic molecules linked to surface sites on the nanoparticle affect their efficacy. The beta-lactamase experiment indicates that the binding of PenG on nanoparticle surfaces resulted in reduced perception by the cell, lower generation of beta-lactamases, and thus a weakened defense. The high antimicrobial activity suggests that when high concentrations of antibiotic molecules are supplied to a bacterial cell (via a nanoparticle), the resistance mechanism(s) may be overwhelmed, resulting in destruction of the microbial cell. Through manipulation of their surface chemistry, nanoparticles can be used as efficient vehicles to allow antibiotics to more efficiently pervade cells in a concentrated dose, and accomplish a pre-determined task(s). A next major step lies in increasing the specificity of antibiotic-complexed nanoparticles for predetermined bacterial targets.
Acknowledgments
This work was supported by grants from the US National Science Foundation (BME-1032579) and the NanoCenter at the University of South Carolina. Dr Brian C. Benicewicz also acknowledges support from the SC SmartState Program.
Footnotes
Electronic supplementary information (ESI) available: Antibiotic–nanoparticle complexes and experimental procedures. See DOI: 10.1039/c4cc04936e
Notes and references
- 1.Evans AHL, Lefrak SN, Lyman J, Smith RL, Chong TW, McElearney ST, Schumlan AR, Hughes MG, Raymond DP, Pruett TL, Sawyer RG. Crit. Care Med. 2007;35:89–95. doi: 10.1097/01.CCM.0000251496.61520.75. [DOI] [PubMed] [Google Scholar]
- 2.Klevens RM, Edwards JR, Jr, Richards CL, Horan TC, Gaynes RP, Pollock DA, Cardo DM. Public Health Rep. 2002;122:160–165. doi: 10.1177/003335490712200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK. JAMA. 2007;15:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 4.Fischbach MA, Walsh CT. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greene SE, Reid A. Report from the American Academy of Microbiology. 1752 N Street, NW, Washington, 2012, DC 20036: p. 44. academy.asm.org. [Google Scholar]
- 6.Percival SL, Bowler PG, Russell D. J. Hosp. Infect. 2005;60:1–7. doi: 10.1016/j.jhin.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 7.Kim CK, Ghosh P, Pagliuca C, Zhu Z-J, Menichetti S, Rotella VM. J. Am. Chem. Soc. 2008;191:1960–1961. doi: 10.1021/ja808137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kallen AJ, Mu Y, Bulens S, Reingold A, Petit S, Gershman K, Ray SM, Harrison LH, Lynfield R, Dumyati G, Townes JM, Schaffner W, Patel PR, Fridkin SK. JAMA. 2010;6:641–647. doi: 10.1001/jama.2010.1115. [DOI] [PubMed] [Google Scholar]
- 9.Klein E, Smith DL, Laxminarayan R. Emerging Infect. Dis. 2007;13:1840–1846. doi: 10.3201/eid1312.070629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asche C, McAdam-Marx C, Seal B, Crookston B, Mullins CD. J. Antimicrob. Chemother. 2008;61:1162–1168. doi: 10.1093/jac/dkn073. [DOI] [PubMed] [Google Scholar]
- 11.Wright GD. Nat. Rev. Microbiol. 2007;5:175–186. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
- 12.Levy SB, Marshall B. Nat. Med. 2004;10:S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 13.Hannan S, Ready D, Jasni AS, Rogers M, Pratten J, Roberts AP. Med. Microbiol. 2010;59:345–349. doi: 10.1111/j.1574-695X.2010.00661.x. [DOI] [PubMed] [Google Scholar]
- 14.Nikaido H, Pages J-M. FEMS Microbiol. Rev. 2011;36:340–363. doi: 10.1111/j.1574-6976.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris JM, Chess RB. Nat. Rev. Drug Discovery. 2003;2:214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- 16.Steiniger SCJ, Kreuter J, Khalansky AS, Skidan IN, Bobruskin AI, Smirnova ZS, Severin SE, Uhl R, Kock M, Geiger KD, Gelperina SE. Int. J. Cancer. 2004;109:759–767. doi: 10.1002/ijc.20048. [DOI] [PubMed] [Google Scholar]
- 17.Liong M, Lu J, Kovochich M, Xia T, Ruehm SG, Nel AE, Tamanoi F, Zink JI. ACS Nano. 2008;2:889–896. doi: 10.1021/nn800072t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kell AJ, Stewart G, Ryan S, Peytavi R, Boissinot M, Huletsky A, Bergeron MG, Simard B. ACS Nano. 2008;2:1777–1788. doi: 10.1021/nn700183g. [DOI] [PubMed] [Google Scholar]
- 19.Mahapatra I, Clark J, Dobson PJ, Owenc R, Lead JR. Environ. Sci.: Processes Impacts. 2013;15:123–144. doi: 10.1039/c2em30640a. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Krentz TM, Wang L, Benicewicz BC, Schadler LS. ACS Appl. Mater. Interfaces. 2014;6:6005–6021. doi: 10.1021/am405332a. [DOI] [PubMed] [Google Scholar]
- 21.Cash BM, Wang L, Benicewicz BC. J. Polym. Sci., Part A: Polym. Chem. 2012;50:2533–2540. [Google Scholar]
- 22.Wang L, Benicewicz BC. ACS Macro Lett. 2013;2:173–176. doi: 10.1021/mz3006507. [DOI] [PubMed] [Google Scholar]
- 23.Li J, Wang L, Benicewicz BC. Langmuir. 2013;29:11547–11553. doi: 10.1021/la401990d. [DOI] [PubMed] [Google Scholar]
- 24.Bauer A, Kirby W, Sherris JC, Turck M. Am. J. Clin. Pathol. 1966;45:493. [PubMed] [Google Scholar]
- 25.Ganewatta MS, Chen YP, Wang J, Zhou J, Ebalunode J, Nagarkatti M, Decho AW, Tang C. Chem. Sci. 2014;5:2011–2016. [Google Scholar]
- 26.Tipper DJ, Strominger JL. Proc. Natl. Acad. Sci. U. S. A. 1965;54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ocallagh Ch, Shingler AH, Kirby SM, Morris A. Antimicrob. Agents Chemother. 1972;1:283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou C, Wang M, Zou K, Chen J, Zhu Y, Du J. ACS Macro Lett. 2013;2:1021–1025. doi: 10.1021/mz400480z. [DOI] [PubMed] [Google Scholar]
- 29.Yuan W, Wei J, Lu H, Fan L, Du J. Chem. Commun. 2012;48:6857–6859. doi: 10.1039/c2cc31529g. [DOI] [PubMed] [Google Scholar]
- 30.Zhu H, Geng Q, Chen W, Zhu Y, Chen J, Du J. J. Mater. Chem. B. 2013;1:5496–5504. doi: 10.1039/c3tb20713g. [DOI] [PubMed] [Google Scholar]
- 31.Zhang C, Zhu Y, Zhou C, Yuan W, Du J. Polym. Chem. 2013;4:255–259. [Google Scholar]