Abstract
Ubiquitinated membrane proteins are sorted into intralumenal endosomal vesicles on their way for degradation in lysosomes. Here we summarize the discovery of the Cos proteins, which work to organize and segregate ubiquitinated cargo prior to its incorporation into intralumenal vesicles of the multivesicular body (MVB). Importantly, cargoes such as GPI-anchored proteins (GPI-APs) that cannot undergo ubiquitination, rely entirely on Cos proteins for sorting into intralumenal vesicles using the same pathway that depends on ESCRTs and ubiquitin ligases that typical polytopic membrane proteins do. Here we show Cos proteins provide functions as not only adaptor proteins for ubiquitin ligases, but also as cargo carriers that can physically usher a variety of other proteins into the MVB pathway. We then discuss the significance of this new sorting model and the broader implications for this cargo adaptor mechanism, whereby yeast Cos proteins, and their likely animal analogs, provide a ubiquitin sorting signal in trans to enable sorting of a membrane protein network into intralumenal vesicles.
Keywords: ubiquitin, multivesicular bodies, vacuole, lysosomes, Cos proteins, Tetraspanins, GPI-anchored proteins
The Cos Proteins
Canonical entry into the multivesicular body (MVB) pathway first involves ubiquitin conjugation of membrane proteins (Ub-cargo) followed by incorporation into intralumenal vesicles (ILVs) of the late endosome and delivery to the lysosome / vacuole for degradation.1,2 The Endosomal Sorting Required for Transport (ESCRT) machinery, which recognizes Ub-cargo and contributes to ILV formation,3 plays a key role in this pathway, but several aspects of the MVB sorting process are unaccounted for by the known function and interactions of the ESCRTs. One key question is how Ub-cargo is physically separated from other endosomal proteins during the MVB sorting process. Additionally, a wealth of data from different systems demonstrates sorting into MVBs occurs when the ESCRT machinery is severely compromised,4 so another key question is what other fundamental machineries work to effect sorting into the MVB pathway? Part of the answer in the yeast system comes in the form of Cos proteins, whose production is amplified during metabolic stress that results in broad downregulation of cell surface membrane proteins.5 The particular metabolite is NAD+, which is sensed through the NAD+-dependent deacetylase sirtuin, Sir2.5 One of the functions of Sir2 activity is to repress genes located in sub-telomeric regions.6 These subtelomeric regions house the conserved sequence (COS) genes, effectively placing them under nutrient control so that depletion of NAD+ stores allows for the further induction of COS genes and potentiation of their function. Conservation of duplicated genes can generally be explained by their ability to confer a selective advantage, either through increased protein levels or generation of divergent genes.7 Since COS genes are highly conserved at the nucleotide and amino acid level, the retention of so many COS genes is presumably due to their ability to collectively supply ample levels of Cos proteins. As a group, there are 11 Cos protein members (Cos1 - Cos10 & Cos12) whose overproduction under nutrient stress serves to relocate the steady-state distribution of a broad spectrum of cell surface proteins to the vacuole for degradation. This link between metabolism and protein trafficking serves not only to catabolize membrane proteins as a buffer against nutrient stress, by recycling of amino acids through the MVB pathway,8 but also to deplete a wide variety of plasma membrane transport functions to slow metabolic processes.
How Cos Proteins Function
While induction of Cos proteins (either experimentally or upon NAD+-depletion) increases sorting of membrane proteins into the MVB pathway, their absence dramatically decreases the efficiency of MVB sorting for a variety of ubiquitinated membrane proteins. A cosΔ strain, lacking all copies of the COS genes, is defective at sorting a variety of MVB cargoes even in nutrient replete conditions. This argues that basal Cos protein levels contribute to regular flux through the MVB pathway, and is supported by proteomic identification of ubiquitinated Cos proteins in such conditions, by our lab (unpublished) and others.9-11 More dramatically, Cos proteins are required for sorting of cargoes that do not require direct modification with Ub into the MVB pathway. One such example is a non-ubiquitinatable, lysine-less version of Sna3, a small membrane protein that has served as a paradigmatic example of Ub-independent cargo.12 More broadly, Cos proteins are essential for sorting Glycosylphosphatidylinositol - anchored proteins (GPI-APs), lipid anchored proteins that reside solely on the lumenal side of the membrane.13 So how do Cos proteins work to increase flux into the MVB pathway and to sort cargoes that are not ubiquitin-modified into the MVB pathway? There are several important biochemical features that enable this function of Cos proteins (Fig. 1).
Figure 1.
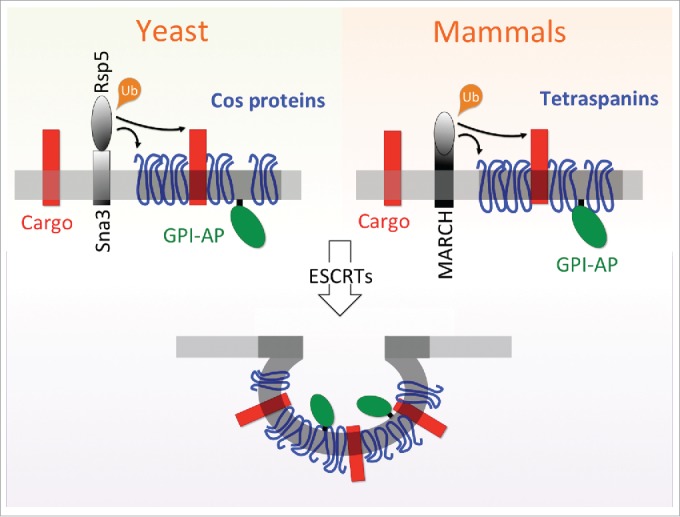
Model for Tetraspan protein function in the MVB pathway. A schematic model (left) for Cos protein function in yeast is depicted. Rsp5 mediated ubiquitination of conventional cargo, such as the methionine permease Mup1 (red), is required for sorting. The membrane bound Rsp5-Sna3 (and Rsp5-Bsd2) complex (black) ensures abundant ubiquitination of Cos proteins (blue). Cos proteins and Ub-cargo cluster in membrane subdomains composed of specific lipids (dark gray) prior to ESCRT recruitment. Proteins lacking a Ub-modification, including GPI-APs (green) attached to the membrane on the lumenal side, are also organized into these endosomal subdomains, before ESCRT mediated vesicle formation and cargo sorting to the vacuole. A proposed model for the analogous mechanism is mammalian cells (right) is shown, whereby Tspans, ubiquitinated by MARCH ligases, serve as cargo adaptors to drive sorting of GPI-APs, alongside canonical cargo like the Epidermal Growth Factor Receptor (EGFR).
Cos proteins are tetraspan (4 membrane-spanning domains) integral membrane proteins that are substantially ubiquitinated; as consequence, they are very efficiently sorted into the MVB pathway in an ESCRT-dependent manner. Their ability to drive the MVB sorting of other cargoes also depends on their ability to undergo ubiquitination. This robust ubiquitination is afforded in part by the curious abundance of acceptor lysine residues (>30) within Cos proteins, which can receive a Ub modification. Cos proteins can also directly associate with Rsp5, the Nedd4-family ubiquitin ligase that is chiefly responsible for ubiquitinating all known MVB membrane cargo proteins in yeast.14 Finally, Cos proteins functionally interact with Sna3 and Bsd2, which are small integral membrane proteins that also bind Rsp5 and serve as adaptor proteins that connect Rsp5 with different membrane protein substrates.12,15,16 Cos proteins interact with one another, both in vivo and in vitro, and localize strongly to late endosomes, where they cluster in distinct endosomal subdomains. These Cos-laden subdomains also contain other Ub-cargoes, which are spatially separated from other endosomal proteins that are ultimately targeted to other locales such as the limiting membrane of the vacuole or back to the cell surface. Importantly, Cos proteins themselves are required for this segregation, demonstrating their key role in contributing to a mechanism that physically sorts and separates one class or cargo from another. Cos proteins also work to retain cargo in endosomal subdomains even when that cargo undergoes deubiquitination. This aspect is important since removal of Ub from cargo by the Doa4 Ub-peptidase prior to incorporation into intralumenal vesicles serves the crucial function of replenishing ubiquitin levels and preventing excessive degradation of Ub in the vacuole.17
These data suggest that the pivotal function of Cos proteins is to serve as a type of cargo carrier, which associates with other Ub-cargo and creates a protein and lipid environment conducive to sorting. Moreover, the fact that Cos proteins are so heavily ubiquitinated themselves provides a mechanism for how they can supply a MVB sorting-signal in trans to the proteins with which they are associated. This model also explains how GPI-APs, which have no capacity to undergo ubiquitination, are sorted into the MVBs in the same canonical Rsp5- and ESCRT-dependent manner as conventional Ub-cargoes. In this specific model, it is Rsp5 that ubiquitinates Cos proteins that in-turn work as the cargo carriers for their associated GPI-APs.
An alternate possibility is that Cos proteins work instead as more conventional ligase adaptors for Rsp5, allowing Rsp5 to ubiquitinate some other target protein that in turn sorts clientele such as GPI-APs. Indeed, Rsp5 has myriad substrates besides Cos proteins,18,19 any of which, arguably, could be responsible for the block in GPI-AP sorting when ligase function is attenuated. While certainly Cos proteins might fulfill the additional role of a ligase adaptor, on face this seems insufficient to explain all of the functions and behaviors of Cos proteins. For instance, Cos proteins themselves need to undergo ubiquitination to sort other proteins, if the sole function of Cos proteins was as a ligase adaptor, they would merely require to bridge the association of Rsp5 with a Cos-associated target protein and would not themselves need to be ubiquitinated. As a further test of these possibilities, we determined whether the critical Rsp5 substrates for sorting GPI-APs into the MVB were indeed Cos proteins. Here we took advantage of a dominant-negative version of Rsp5 wherein Rps5 is fused to the catalytic domain of a Ub-peptidase, which deubiquitinates endogenous Rsp5 substrates and blocks the MVB sorting of a wide variety of integral membrane proteins.18 Expression of Rsp5-DUb also blocks sorting of GPI-APs into the MVB pathway showing that their delivery is dependent on an Rsp5-mediated ubiquitination event (Fig. 2). That event turns out to be the ubiquitination of Cos proteins themselves because expression of a “pre-ubiquitinated” version of a Cos protein (Cos5-HA-Ub) was sufficient to overcome the block imposed by Rsp5-DUb on the MVB sorting of GPI-APs. Cos5-HA-Ub carries Ub as an in-frame fusion at the C-terminus of Cos5 where it cannot be removed by Ub-peptidases. These data show the relevant Rsp5 targets in the sorting GPI-APs are Cos proteins and supports the overall model whereby Cos proteins form a ubiquitinated “Cos corral” that traps a variety of MVB cargoes into endosomal subdomains and provides them with a Ub sorting signal in trans. This mechanism assists with the sorting of conventional integral membrane proteins and is essential for GPI-AP MVB sorting. We speculate that integral membrane proteins that have poor capacity for ubiquitination and other peripherally associated lumenal proteins depend chiefly on the Cos proteins to achieve their MVB sorting.
Figure 2.
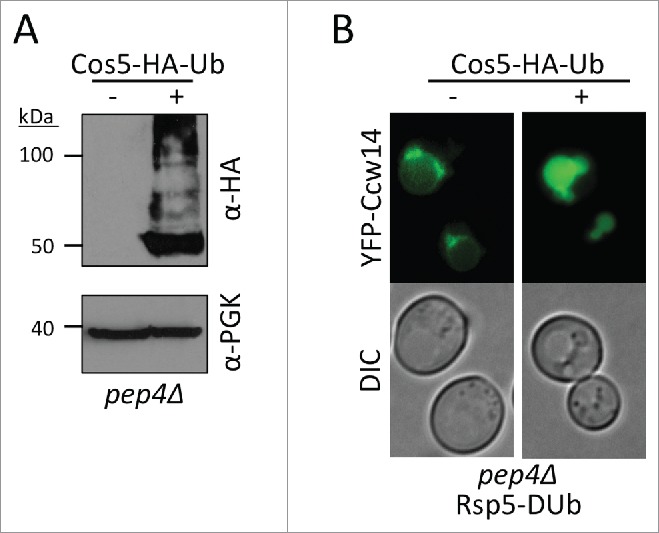
Cos ubiquitination is sufficient to drive GPI-AP sorting into the MVB pathway. (A) Lysates were generated from pep4Δ cells stably expressing an HA-tagged ubiquitin fusion of Cos5 (Cos5-HA-Ub) from the MET15 promoter (PLY4803), followed by immunblot analysis using anti-HA and anti-PGK antibodies. As a control, the parental strain (SEY6210 pep4Δ) was included. B) Vacuolar peptidase deficient mutant (pep4Δ) cells allow visualization of the ESCRT-dependent sorting of fluorescently tagged GPI-APs, such as YFP-Ccw14. Expression of Rsp5-DUb induces a complete block in delivery of YFP-Ccw14 to the lumen of the vacuole (upper). This block in sorting is suppressed in cells also expressing Cos5-HA-Ub, where YFP-Ccw14 instead is driven into the vacuolar lumen.
The Mechanics of Cos Proteins
The finding that ubiquitinated Cos proteins are sufficient to drive MVB sorting of GPI-APs (a diverse group of proteins with varied biochemical, structural and functional properties) suggests the cargo adaptor function of Cos proteins does not rely on conventional protein-protein interactions. We favor a model where Cos proteins organize lipids and Ub-cargo at sites primed for vesicle morphogenesis. Indeed, the regimented distribution of lipids throughout the endolysosomal pathway is known to contribute to cargo sorting, but the machinery that orchestrates such lipid organization is poorly understood.20,21 Cos proteins are promising candidates for lipid sorting at the late endosome, since they drive formation of the subdomain architecture that partitions Ub-cargo from vacuolar membrane proteins. We propose that the biophysical properties of the Cos protein trans-membrane domains (TMDs) are likely responsible for coordinating surrounding lipids in endosomal compartments. The TMDs of Cos proteins are unusual. When compared with all other members of the yeast TMD proteome, Cos TMDs have an extremely high frequency of Arg residues, in addition to abundant Trp, Cys and Pro residues (Fig. 3). Conversely, the TMDs of Cos proteins score as some of lowest across the TMD proteome for residues such as Ala and Thr. An attractive idea is that the buried charge residues within the endosomal membrane may induce conformational changes to recruit lipids predisposed to membrane deformation and ILV formation. Additionally, it has been proposed that functional lipid dependent sorting clusters GPI-APs.22,23 Since the Cos proteins are essential cargo adaptors for vacuolar trafficking of GPI-APs, it is likely that they can exert the necessary sorting of endosomal lipids to facilitate GPI-AP clustering, as again, physical interactions are unlikely to drive this process. Conceptually, lipid organization could initiate the sorting process upstream of ESCRTs, and a concentrated Ub-signal from cargo and Cos proteins in turn recruit ESCRTs to endosomes through their various Ub-binding domains and associations.24,25 Unlike ESCRTs, which are disassembled from ILVs prior to scission, Cos proteins are sacrificed to the vacuole for degradation. This presents the interesting possibility that the lipid composition of ILVs, interspersed with atypical Cos protein TMDs, might render them susceptible to vacuolar hydrolases. This would provide an explanation for how the limiting membrane of the vacuole, composed of lipids excluded by the “Cos corral” during endosomal partitioning, is largely protected from the resident lipases required for ILV degradation.
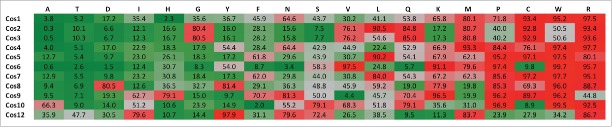
Figure 3.
Amino acid composition of Cos protein transmembrane domains. The frequency of each amino acid was calculated as a percentage of the total membrane spanning residues within each integral membrane protein. This frequency was used to rank all 652 TMD proteins in the S. cerevisiae proteome for abundance of particular amino acids. The percentile score of each Cos protein for all amino acids is denoted and colored based on frequency: low (green), high (red) and median (gray) across membrane spanning regions.
The Broader Role That Cos Function Fulfills
By sequence homology, Cos protein homologs are not readily found in the other eukaryotic kingdoms of plants and animals. Yet, sorting of GPI-APs into the MVB pathway has been documented in animal cells.26 Cos proteins execute this conserved function in yeast and serve as a simple explanation for how other non-ubiquitinatable cargoes can be sorted into the canonical Ub- and ESCRT- mediated MVB pathway. This conserved function in animal and plant cells is likely fulfilled by a functional analog of Cos proteins that may not share immediate sequence homology. Since it is not yet clear what biochemical features or signatures of Cos proteins drive their function, what proteins might execute their analogous function in animals is speculative. To begin to explore this, though, we used a supervised machine learning strategy to define compositional and positional features of residues within Cos protein TMDs that might reflect a signature found in mammalian homologs. A k-nearest neighbor algorithm27 was trained on a dataset of all S. cerevisiae transmembrane segments comprised of 33 Cos TMDs and 4,601 non-Cos-TMDs. During 10-fold cross validation, the algorithm predicted 30 yeast TMDs belonged to Cos proteins, 23 of which actually did (Table S1) and all S. cerevisiae Cos proteins were represented within those 23 TMDs except Cos12, which is predicted to have a unique topology among the Cos family.28 When this classification model was applied to the 20,756 member human TMD proteome, 132 of these (<0.65% total) were predicted to be a “Cos-like” neighbor (Table S2). STRING v.10 pathway analysis.29 revealed 2 of these proteins localize to the MVB, (the sortilin related receptor SORL1 and the tetraspanin (Tspan) CD63) and 2 others function as ubiquitin ligases, (RNF144B and MARCH6). Exactly what computed features of these proteins relate them to Cos proteins is not clear, but one implication is that proteins such as these might share some of the biophysical features and perhaps some of the functions of Cos proteins in yeast. Among the most intriguing possibilities are Tspans and MARCH ligases since they also share other biological parallels with Cos proteins.
Cargo sorting through the MVB pathway of mammalian cells in a ubiquitin- and/or ESCRT-independent manner has been proposed, including models that are driven by specific lipids.30-32 or accessory proteins that include CD63.33 Tspans such as CD63 organize Tspan enriched domains representing a meshwork of associated proteins together within a lipid subdomain enriched in sphingolipids and cholesterol.34 Because Cos proteins may also form and function in a similar organization, we propose that the best candidates for Cos analogs are Tspans. Like Cos proteins, Tspans have 4 TMDs that contain polar residues, have extracellular cysteines, and also interact as homo-multimers and cluster in membrane domains.35-37 The analogy extends further, as some Tspans contain lysosomal sorting motifs and directly interact with clathrin adaptor proteins.38,39 Most convincingly, Tspans can be ubiquitinated and have been implicated in MVB sorting of the EGFR, a prototypical Ub-cargo in animal cells,40-43 and Pmel17, a lumenal protein of melanosomes.33 More recently, the CD81 Tspan, which, like CD63, is also enriched in MVB internal vesicles,44 has been shown to play a role in mediating MVB sorting of the Ub-independent cargo, transferrin receptor 2 (TfR2). Although TfR2 is quickly degraded in lysosomes in an ESCRT-dependent manner, it does not itself undergo ubiquitination.45 Instead, its sorting is mediated by CD81, with which it associates.46
The key requirement for Cos protein function as cargo adaptors for GPI-APs is a strong Ub signal, indeed Cos proteins are chiefly responsible for Ub trafficking into the vacuole. To undergo maximal ubiquitination, Cos proteins partly rely on Ubiquitin-ligase membrane protein complexes defined by Rsp5 and the polytopic membrane adaptors Sna3 and Bsd2. Presumably the effective concentration of ligases and substrates resident in the same membrane contributes to the powerful Ub signal necessary to act as cargo adaptors. A similar scenario has been documented in mammalian cells through the Membrane associated RING-CH (MARCH) Ub-ligases, which have been proposed to ubiquitinate and downregulate Tspans.40,47 Our model would predict that ubiquitinated Tspans allow GPI-APs and other non-ubiquitinatable proteins to access ESCRT-dependent sorting to lysosomes in animal cells. The finding that MARCH ligases contribute to GPI-AP sorting serves to further validate this hypothetical model.48 It will be interesting and helpful to consider the cargo adaptor model proposed herein when analyzing future discoveries of MVB sorting and GPI-AP trafficking in animal cells.
Materials and Methods
Plasmids expressing HIS3 marked YFP-Ccw14.(pPL5707.5) and Rsp5-DUb,(pPL3742,18) and yeast strain SEY6210 pep4Δ.(PLY2463.24) have been described before. For this study, homologous recombination was used to create a MET15 marked plasmid that expresses YFP-Ccw14 (pPL5781) and also to integrate COS5-HA-Ub at the MET15 locus of strain PLY2463, using a methotrexate resistant marker,49 to create PLY4803. Yeast strains were grown at 30°C in synthetic defined (SD) media lacking appropriate amino acids for plasmid selection. Expression of Rsp5-DUb from the CUP1 promoter was induced by the addition of 50 µM copper chloride. Immunoblot analyses and microscopy techniques were used as described in MacDonald et al..5
Funding Statement
This work was funded by the American Heart Association (CM, 15POST-22980010) and the National Institutes of Health (RCP, NIHRO1-GM058202; DJK, NIHRO1-GM073024).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 2001; 106:145-55; PMID:11511343; http://dx.doi.org/ 10.1016/S0092-8674(01)00434-2 [DOI] [PubMed] [Google Scholar]
- 2.Urbanowski JL, Piper RC. Ubiquitin sorts proteins into the intralumenal degradative compartment of the late-endosome/vacuole. Traffic 2001; 2:622-30; PMID:11555416; http://dx.doi.org/ 10.1034/j.1600-0854.2001.20905.x [DOI] [PubMed] [Google Scholar]
- 3.Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol 2007; 23:519-47; PMID:17506697; http://dx.doi.org/ 10.1146/annurev.cellbio.23.090506.123319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hurley JH, Boura E, Carlson LA, Rozycki B. Membrane budding. Cell 2010; 143:875-87; PMID:21145455; http://dx.doi.org/ 10.1016/j.cell.2010.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacDonald C, Payne JA, Aboian M, Smith W, Katzmann DJ, Piper RC. A family of tetraspans organizes cargo for sorting into multivesicular bodies. Dev Cell 2015; 33:328-42; PMID:25942624; http://dx.doi.org/ 10.1016/j.devcel.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 1990; 63:751-62; PMID:2225075; http://dx.doi.org/ 10.1016/0092-8674(90)90141-Z [DOI] [PubMed] [Google Scholar]
- 7.Seoighe C, Wolfe KH. Yeast genome evolution in the post-genome era. Curr Opin Microbiol 1999; 2:548-54; PMID:10508730; http://dx.doi.org/ 10.1016/S1369-5274(99)00015-6 [DOI] [PubMed] [Google Scholar]
- 8.Jones CB, Ott EM, Keener JM, Curtiss M, Sandrin V, Babst M. Regulation of membrane protein degradation by starvation-response pathways. Traffic 2012; 13:468-82; PMID:22118530; http://dx.doi.org/ 10.1111/j.1600-0854.2011.01314.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hitchcock AL, Auld K, Gygi SP, Silver PA. A subset of membrane-associated proteins is ubiquitinated in response to mutations in the endoplasmic reticulum degradation machinery. Proc Natl Acad Sci U S A 2003; 100:12735-40; PMID:14557538; http://dx.doi.org/ 10.1073/pnas.2135500100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 2003; 21:921-6; PMID:12872131; http://dx.doi.org/ 10.1038/nbt849 [DOI] [PubMed] [Google Scholar]
- 11.Reggiori F, Pelham HR. Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J 2001; 20:5176-86; PMID:11566881; http://dx.doi.org/ 10.1093/emboj/20.18.5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacDonald C, Stringer DK, Piper RC. Sna3 Is an Rsp5 Adaptor Protein that Relies on Ubiquitination for Its MVB Sorting. Traffic 2012; 13(4):586-98; PMID:22212814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayor S, Riezman H. Sorting GPI-anchored proteins. Nat Rev Mol Cell Biol 2004; 5:110-20; PMID:15040444; http://dx.doi.org/ 10.1038/nrm1309 [DOI] [PubMed] [Google Scholar]
- 14.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol 2009; 10:398-409; PMID:19436320; http://dx.doi.org/ 10.1038/nrm2690 [DOI] [PubMed] [Google Scholar]
- 15.Hettema EH, Valdez-Taubas J, Pelham HR. Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J 2004; 23:1279-88; PMID:14988731; http://dx.doi.org/ 10.1038/sj.emboj.7600137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oestreich AJ, Aboian M, Lee J, Azmi I, Payne J, Issaka R, Davies BA, Katzmann DJ. Characterization of multiple multivesicular body sorting determinants within Sna3: a role for the ubiquitin ligase Rsp5. Mol Biol Cell 2007; 18:707-20; PMID:17182849; http://dx.doi.org/ 10.1091/mbc.E06-08-0680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amerik AY, Nowak J, Swaminathan S, Hochstrasser M. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol Biol Cell 2000; 11:3365-80; PMID:11029042; http://dx.doi.org/ 10.1091/mbc.11.10.3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stringer DK, Piper RC. A single ubiquitin is sufficient for cargo protein entry into MVBs in the absence of ESCRT ubiquitination. J Cell Biol 2011; 192:229-42; PMID:21242292; http://dx.doi.org/ 10.1083/jcb.201008121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta R, Kus B, Fladd C, Wasmuth J, Tonikian R, Sidhu S, Krogan NJ, Parkinson J, Rotin D. Ubiquitination screen using protein microarrays for comprehensive identification of Rsp5 substrates in yeast. Mol Sys Biol 2007; 3:116; PMID:17551511; http://dx.doi.org/ 10.1038/msb4100159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bissig C, Gruenberg J. Lipid sorting and multivesicular endosome biogenesis. Cold Spring Harb Perspect Biol 2013; 5:a016816; PMID:24086044; http://dx.doi.org/ 10.1101/cshperspect.a016816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hullin-Matsuda F, Taguchi T, Greimel P, Kobayashi T. Lipid compartmentalization in the endosome system. Semin Cell Dev Biol 2014; 31:48-56; PMID:24747366; http://dx.doi.org/ 10.1016/j.semcdb.2014.04.010 [DOI] [PubMed] [Google Scholar]
- 22.Sharma P, Varma R, Sarasij RC, Ira Gousset K, Krishnamoorthy G, Rao M, Mayor S. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell 2004; 116:577-89; PMID:14980224; http://dx.doi.org/ 10.1016/S0092-8674(04)00167-9 [DOI] [PubMed] [Google Scholar]
- 23.Raghupathy R, Anilkumar AA, Polley A, Singh PP, Yadav M, Johnson C, Suryawanshi S, Saikam V, Sawant SD, Panda A, et al.. Transbilayer lipid interactions mediate nanoclustering of lipid-anchored proteins. Cell 2015; 161:581-94; PMID:25910209; http://dx.doi.org/ 10.1016/j.cell.2015.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macdonald C, Buchkovich NJ, Stringer DK, Emr SD, Piper RC. Cargo ubiquitination is essential for multivesicular body intralumenal vesicle formation. EMBO Rep 2012; 13:331-8; PMID:22370727; http://dx.doi.org/ 10.1038/embor.2012.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shields SB, Piper RC. How ubiquitin functions with ESCRTs. Traffic 2011; 12:1306-17; PMID:21722280; http://dx.doi.org/ 10.1111/j.1600-0854.2011.01242.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satpute-Krishnan P, Ajinkya M, Bhat S, Itakura E, Hegde RS, Lippincott-Schwartz J. ER stress-induced clearance of misfolded GPI-anchored proteins via the secretory pathway. Cell 2014; 158:522-33; PMID:25083867; http://dx.doi.org/ 10.1016/j.cell.2014.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horton P, Nakai K. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc Int Conf Intell Syst Mol Biol 1997; 5:147-52 [PubMed] [Google Scholar]
- 28.Kim H, Melén K, Osterberg M, von Heijne G. A global topology map of the Saccharomyces cerevisiae membrane proteome. Proc Natl Acad Sci USA 2006; 103:11142-7; PMID:16847258; http://dx.doi.org/ 10.1073/pnas.0604075103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al.. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015; 43:D447-52; PMID:25352553; http://dx.doi.org/ 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falguieres T, Luyet PP, Bissig C, Scott CC, Velluz MC, Gruenberg J. In vitro budding of intralumenal vesicles into late endosomes is regulated by Alix and Tsg101. Mol Biol Cell 2008; 19:4942-55; PMID:18768755; http://dx.doi.org/ 10.1091/mbc.E08-03-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuo H, Chevallier J, Mayran N, Le Blanc I, Ferguson C, Faure J, Blanc NS, Matile S, Dubochet J, Sadoul R, et al.. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science 2004; 303:531-4; PMID:14739459; http://dx.doi.org/ 10.1126/science.1092425 [DOI] [PubMed] [Google Scholar]
- 32.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008; 319:1244-7; PMID:18309083; http://dx.doi.org/ 10.1126/science.1153124 [DOI] [PubMed] [Google Scholar]
- 33.van Niel G, Charrin S, Simoes S, Romao M, Rochin L, Saftig P, Marks MS, Rubinstein E, Raposo G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev Cell 2011; 21:708-21; PMID:21962903; http://dx.doi.org/ 10.1016/j.devcel.2011.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yáñez-Mó M, Barreiro O, Gordon-Alonso M, Sala-Valdés M, Sánchez-Madrid F. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol 2009; 19:434-46; http://dx.doi.org/ 10.1016/j.tcb.2009.06.004 [DOI] [PubMed] [Google Scholar]
- 35.Stipp CS, Kolesnikova TV, Hemler ME. Functional domains in tetraspanin proteins. Trends Biochem Sci 2003; 28:106-12; PMID:12575999; http://dx.doi.org/ 10.1016/S0968-0004(02)00014-2 [DOI] [PubMed] [Google Scholar]
- 36.Levy S, Shoham T. The tetraspanin web modulates immune-signalling complexes. Nat Rev Immunol 2005; 5:136-48; PMID:15688041; http://dx.doi.org/ 10.1038/nri1548 [DOI] [PubMed] [Google Scholar]
- 37.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol 2005; 6:801-11; PMID:16314869; http://dx.doi.org/ 10.1038/nrm1736 [DOI] [PubMed] [Google Scholar]
- 38.Berditchevski F, Odintsova E. Tetraspanins as regulators of protein trafficking. Traffic 2007; 8:89-96; PMID:17181773; http://dx.doi.org/ 10.1111/j.1600-0854.2006.00515.x [DOI] [PubMed] [Google Scholar]
- 39.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem 2003; 72:395-447; PMID:12651740; http://dx.doi.org/ 10.1146/annurev.biochem.72.121801.161800 [DOI] [PubMed] [Google Scholar]
- 40.Lineberry N, Su L, Soares L, Fathman CG. The single subunit transmembrane E3 ligase gene related to anergy in lymphocytes (GRAIL) captures and then ubiquitinates transmembrane proteins across the cell membrane. J Biol Chem 2008; 283:28497-505; PMID:18713730; http://dx.doi.org/ 10.1074/jbc.M805092200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Danglot L, Chaineau M, Dahan M, Gendron MC, Boggetto N, Perez F, Galli T. Role of TI-VAMP and CD82 in EGFR cell-surface dynamics and signaling. J Cell Sci 2010; 123:723-35; PMID:20144992; http://dx.doi.org/ 10.1242/jcs.062497 [DOI] [PubMed] [Google Scholar]
- 42.Odintsova E, Sugiura T, Berditchevski F. Attenuation of EGF receptor signaling by a metastasis suppressor, the tetraspanin CD82/KAI-1. Curr Biol: CB 2000; 10:1009-12; PMID:10985391; http://dx.doi.org/ 10.1016/S0960-9822(00)00652-7 [DOI] [PubMed] [Google Scholar]
- 43.Odintsova E, van Niel G, Conjeaud H, Raposo G, Iwamoto R, Mekada E, Berditchevski F. Metastasis suppressor tetraspanin CD82/KAI1 regulates ubiquitylation of epidermal growth factor receptor. J Biol Chem 2013; 288:26323-34; PMID:23897813; http://dx.doi.org/ 10.1074/jbc.M112.439380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wubbolts R, Leckie RS, Veenhuizen PT, Schwarzmann G, Mobius W, Hoernschemeyer J, Slot JW, Geuze HJ, Stoorvogel W. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J Biol Chem 2003; 278:10963-72; PMID:12519789; http://dx.doi.org/ 10.1074/jbc.M207550200 [DOI] [PubMed] [Google Scholar]
- 45.Chen J, Wang J, Meyers KR, Enns CA. Transferrin-directed internalization and cycling of transferrin receptor 2. Traffic 2009; 10:1488-501; PMID:19682329; http://dx.doi.org/ 10.1111/j.1600-0854.2009.00961.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J, Enns CA. CD81 promotes both the degradation of transferrin receptor 2 (TfR2) and the Tfr2-mediated maintenance of hepcidin expression. J Biol Chem 2015; 290:7841-50; PMID:25635054; http://dx.doi.org/ 10.1074/jbc.M114.632778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartee E, Eyster CA, Viswanathan K, Mansouri M, Donaldson JG, Früh K. Membrane-Associated RING-CH proteins associate with Bap31 and target CD81 and CD44 to lysosomes. PLoS One 2010; 5:e15132; PMID:21151997; http://dx.doi.org/ 10.1371/journal.pone.0015132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mansouri M, Viswanathan K, Douglas JL, Hines J, Gustin J, Moses AV, Fruh K. Molecular mechanism of BST2/tetherin downregulation by K5/MIR2 of Kaposi's sarcoma-associated herpesvirus. J Virol 2009; 83:9672-81; PMID:19605472; http://dx.doi.org/ 10.1128/JVI.00597-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacDonald C, Piper RC. Puromycin- and methotrexate-resistance cassettes and optimized Cre-recombinase expression plasmids for use in yeast. Yeast 2015; 32:423-38; PMID:25688547; http://dx.doi.org/ 10.1002/yea.3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.