The presence of a true quiescent state that is fundamentally distinct from slowly-growing cells in an extended G1 phase has been debated for decades.1 However, it is clear that the ability to remain in a reversible, nondividing state is essential to the survival of organisms. Such states are seen from budding yeast where nutritional cues promote a robust, stress-resistant cellular response through humans, where maintenance of stem cell populations hinges on controlled entry and exit from a proliferative program.2 The presence of an easily-separable, long-lived quiescent population in yeast has been discovered3 allowing for interrogation of properties and identification of mechanistic drivers of this conserved cellular state.4 However, strong evidence has brought into question the existence of a bona fide quiescent state in yeast, since nutrient limitation and starvation elicit similar responses while distinct nutrient challenges promote nutrient-specific quiescence programs.5,6
We recently described a large-scale repressive event that is detectable in purified quiescent yeast but not in cells undergoing diauxic shift or in cells starved and G1 arrested by rapamycin treatment.7 Using external controls to account for global shifts in transcriptional output, we showed that quiescent cells purified from stationary phase cultures demonstrate a ∼30-fold drop in mRNA levels compared to a ∼2-fold repression at the diauxic shift. This massive transcriptional shutoff is dependent on the conserved histone (or lysine) deacetylase (HDAC) Rpd3, which is specifically recruited to thousands of promoters in quiescent cells leading to global hypoacetylation of chromatin and gene repression. Deletion of RPD3 prevents progression from the diauxic shift chromatin state to the quiescence-specific chromatin state, and limits transcriptional shutoff to that observed in wild type cells undergoing diauxic shift. Thus we have identified a chromatin-based global mechanism required for stable repression of the transcriptome that is separable and distinct from the initial diauxic shift response to glucose consumption. We propose that acute responses to short term stresses such as nutrient elimination likely involve individual transcriptional programs for dealing with the specific environmental situation. Prolonged exposure to disparate stresses, however, may converge on a shared global transcriptional shutoff response such as that driven by Rpd3 after glucose consumption (Fig. 1). A distinct response to short-term stresses would be advantageous if the stress is truly short-lived, as cells would be poised to rapidly respond to a changing environment without committing resources to a global transcriptional shutoff. A common global response to prolonged stress is advantageous for conserving cellular resources under prolonged exposure to harsh environments.
Figure 1.
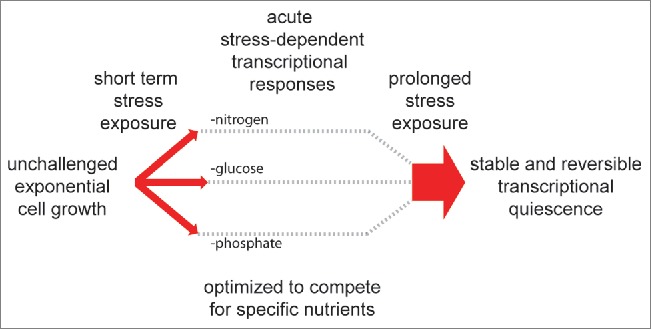
Cartoon diagram suggesting how different stresses may elicit distinct short-term transcriptional responses followed by a unified global quiescence response to endure prolonged stress exposure.
Our work has uncovered a unique and critical role for Rpd3 in establishing stable transcriptional quiescence, which has many potential implications for quiescence in yeast and beyond. We have identified global chromatin changes associated with yeast quiescence that are not seen in other cell cycle stages, thereby distinguishing quiescence as a truly separate cell cycle stage. It was unexpected to find such a widespread function for Rpd3 in yeast quiescence, as previous reports have demonstrated limited roles for Rpd3 in cycling cells. Many questions about the roles of Rpd3 and chromatin structure in transcriptional quiescence remain. What are the downstream events and cofactors responsible for transducing Rpd3-driven deacetylation to global transcriptional shutoff? What are the signals and pathways for “turning off” Rpd3 and reversing transcriptional quiescence? Are these mechanisms conserved in all eukaryotes? It is likely that many other yeast proteins function primarily during this under-characterized cell cycle stage and it will certainly be worthwhile reevaluating protein function in this unique context. Future research in yeast will likely uncover highly conserved, intricately-orchestrated molecular events that unify chromatin modification, genome reorganization, and massive transcriptional repression to promote the protective, reversible quiescent state.
References
- 1.Coller HA. Science 2011; 334:1074-5; PMID:22116874; http://dx.doi.org/ 10.1126/science.1216242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valcourt JR, et al. Cell cycle 2012; 11:1680-96; PMID:22510571; http://dx.doi.org/ 10.4161/cc.19879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen C, et al.. J Cell Biol 2006; 174:89-100; PMID:16818721; http://dx.doi.org/ 10.1083/jcb.200604072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L, et al. Mol Biol Cell 2013; 24:3697-709; PMID:24088570; http://dx.doi.org/ 10.1091/mbc.E13-05-0241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klosinska MM, et al. Gen Dev 2011; 25:336-49; PMID:21289062; http://dx.doi.org/ 10.1101/gad.2011311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brauer MJ, et al. Mol Biol Cell 2008; 19:352-67; PMID:17959824; http://dx.doi.org/ 10.1091/mbc.E07-08-0779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKnight JN, et al. Mol Cell 2015; 59:732-43; PMID:26300265; http://dx.doi.org/ 10.1016/j.molcel.2015.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]


