Proliferation, resistance to apoptosis and migration are hallmarks of cancer cells and the main focus of targeted therapies in recent years. The rationale behind targeted therapies is to attack cancer cells without damaging or affecting healthy cells consequently reducing the severe side effect associated with standard chemotherapy. Therefore, a thorough understanding of the underlying cellular pathways leading to proliferation, cell death and metastasis and their alterations in malignant cells is of most importance. In the last 20 years, profilin 1 (Pfn1), a small ubiquitously expressed cytoskeletal protein has been the subject of increased interest as a potential therapeutic target in the fight against several types of cancers including breast, pancreatic, hepatic and bladder cancer. Pfn1 is known to have a reduced expression in these cancer types and increased expression of Pfn1 in some breast cancer cells reduces the tumorigenicity of the cells. It has recently been shown that overexpression of Pfn1 in the human breast cancer cell line MDA-MB 231 induces alterations of the expression of a range of proteins involved in cell proliferation, motility and survival.1 However, the mechanisms underlying the reduced motility and proliferation and increased apoptosis obeserved in these MDA-MB 231 cells are still largely unknown. The study by Jiang et al.2 propose a novel pathway that link Pfn1 overexpression to the phosphorylation of p27 on threonine 198 (T198). This involves R-cadherin and activation of the LKB1-AMPK pathway as represented by the shaded boxes in Figure 1. Phosphorylation at T198 has been previously shown to induce p27 mislocalization from the nucleus to the cytoplasm, epithelial to mesenchymal cell transition and tumor metastasis.3 However, phosphorylation of p27 at T198 induced by increased expression of Pfn12 or by PKC activation4 is shown to both stabilize p27 in the nucleus and induce cell cycle arrest by inhibiting ubiquitination of p27 (Fig. 1). The mechanism responsible for the difference in p27 localization upon T198 phosphorylation is still unresolved although it might involve p27 phosphorylation on additional residues. Pfn1 can also be phosphorylated by PKC isoforms at serine 137 (S137). This phosphorylation disrupt the binding to proline rich domain (PRD) containing proteins and favors the localization of Pfn1 in the nucleus which appears to be necessary for its role in cell cycle arrest and proliferation.5 In the cytoplasm, increased Pfn1 level promotes the formation of actin cables following the interaction of profilin-actin with formins and Ena-Vasp rather than branched filament networks by Arp2/3 complex.6 This behavior is consistent with the stabilization of adherens junctions as proposed in Jiang et al. On the other hand, phosphorylation of Pfn1 at S137 would inhibit its interaction with formin and Ena-Vasp and prevent Pfn1 effect on adherens junction. It is therefore likely that the cell maintains pools of phosphorylated/unphosphorylated proteins which depending on either their cytoplasmic or nuclear localization, would have different but complementary functions. Future research will hopefully shed light on these questions. The paper by Jiang et al. adds another piece to the puzzle that is the mechanism responsible for the action of Pfn1 on cell proliferation, metastasis and apoptosis. An increased understanding of the role of Pfn1 in these processes and the effect of phosphorylation of the proteins involved in those pathways will provide the basis for exploring the development of new therapeutic approaches including drugs that could act as agonists of Pfn1. Such an approach based on supplementing cellular function has the potential to require low therapeutic doses and hence result in low toxicity.
Figure 1.
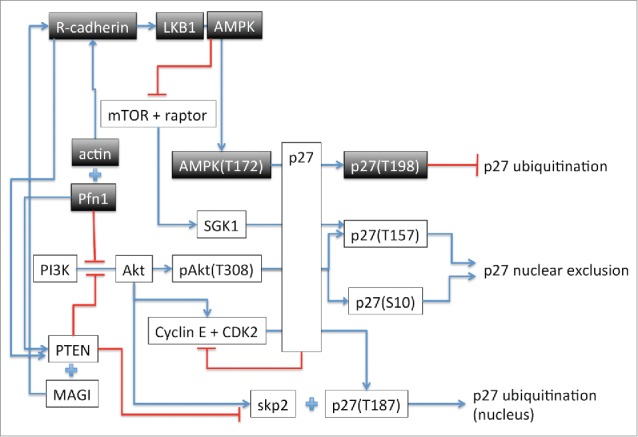
Schematic representation of the pathways implicated in Pfn1 effect on cell-cycle arrest when Pfn1 is overexpressed as described in Jiang et al. The arrows indicate activation or increased expression. The + sign indicates interaction between the proteins. The —| represents inhibition. The residue in () indicates the phosphorylated amino acid.
References
- 1.Coumans JV, et al.. Omics: a journal of integrative biology 2014; 18:778-91. PMID:25454514; http://dx.doi.org/ 10.1089/omi.2014.0075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang C, et al.. Cell cycle 2015; 14(18):2914-23; PMID:26176334; http://dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao D, et al.. Oncogene 2015. PMID:25684140; http://dx.doi.org/ 10.1038/onc.2014.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Vita F, et al.. Cell cycle 2012; 11:1583-92. PMID:22441823; http://dx.doi.org/ 10.4161/cc.20003 [DOI] [PubMed] [Google Scholar]
- 5.Diamond MI, et al.. J Biol Chem 2015; 290:9075-86. PMID:25681442; http://dx.doi.org/ 10.1074/jbc.M114.619874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henty-Ridilla JL, Goode BL. Dev Cell 2015; 32:5-6. PMID:25584793; http://dx.doi.org/ 10.1016/j.devcel.2014.12.022 [DOI] [PubMed] [Google Scholar]


