Abstract
Cytoplasmic aggregation of TDP-43, accompanied by its nuclear clearance, is a key common pathological hallmark of amyotrophic lateral sclerosis and frontotemporal dementia (ALS-FTD). However, a limited understanding of this RNA-binding protein (RBP) impedes the clarification of pathogenic mechanisms underlying TDP-43 proteinopathy. In contrast to RBPs that regulate splicing of conserved exons, we found that TDP-43 repressed the splicing of nonconserved cryptic exons, maintaining intron integrity. When TDP-43 was depleted from mouse embryonic stem cells, these cryptic exons were spliced into messenger RNAs, often disrupting their translation and promoting nonsense-mediated decay. Moreover, enforced repression of cryptic exons prevented cell death in TDP-43–deficient cells. Furthermore, repression of cryptic exons was impaired in ALS-FTD cases, suggesting that this splicing defect could potentially underlie TDP-43 proteinopathy.
Amyotrophic lateral sclerosis (ALS), a fatal adult-onset motor neuron disease characterized by progressive loss of upper and lower motor neurons, and frontotemporal dementia (FTD), a common form of dementia characterized by a gradual deterioration in behavior, personality, and/or language, share a common disease spectrum (1, 2). Transactivation response element DNA-binding protein 43 (TDP-43, TARDBP), is a heterogeneous nuclear ribonucleoprotein (hnRNP) thought to provide the neuropathological link to establish such a disease spectrum (1, 3). In sporadic ALS (~97% of all cases) and sporadic FTD (~45% of all cases), TDP-43 clears from the nucleus and forms ubiquitinated, cytoplasmic inclusions, termed TDP-43 proteinopathy (2). Missense mutations in TDP-43 are also linked to familial ALS, strongly supporting the idea that TDP-43 proteinopathy is central to the pathogenesis of sporadic disease (4, 5). Numerous genetic mutations associated with familial ALS-FTD—VCP, GRN, OPTN, ATXN2, SQSTM1, UBQLN2, PFN1, TBK1, and especially C9ORF72—result in TDP-43 proteinopathy, suggesting a convergent mechanism of neurodegeneration (6–9). Indeed, Tdp-43 is a tightly autoregulated (10), essential gene (11–13) that is required for neuronal physiology in mice (14), fruit flies (15), and zebrafish (16).
TDP-43's consensus binding motif has been identified as the “UG” dinucleotide repeat (17). Using a high-throughput sequencing approach combined with cross-linking immunoprecipitation (HITS-CLIP), direct targets of Tdp-43 have been identified (18, 19). Tdp-43 regulates alternative splicing of a subset of conserved exons by binding to UG-rich domains within proximal intronic regions (18, 19). However, the majority of Tdp-43 binds to repetitive elements within distal introns, locations where Tdp-43 has no clear function. To explore the RNA splicing role of Tdp-43, we assessed the transcriptome of cells lacking Tdp-43 with RNA-sequencing (RNA-seq) (12). These CreER-inducible Tdp-43 knockout mouse embryonic stem (mES) cells (iTDPKO) permit genetic deletion of Tdp-43 within 2 to 3 days of 4-hydroxytamoxifen (4HT) treatment. However, previous sequencing technologies were unable to provide robust transcriptome-wide splicing data. RNA-seq analyses performed on data with greater read length and coverage improved the splicing resolution and allowed us to identify cryptic exons in mES cells lacking Tdp-43 (Fig. 1).
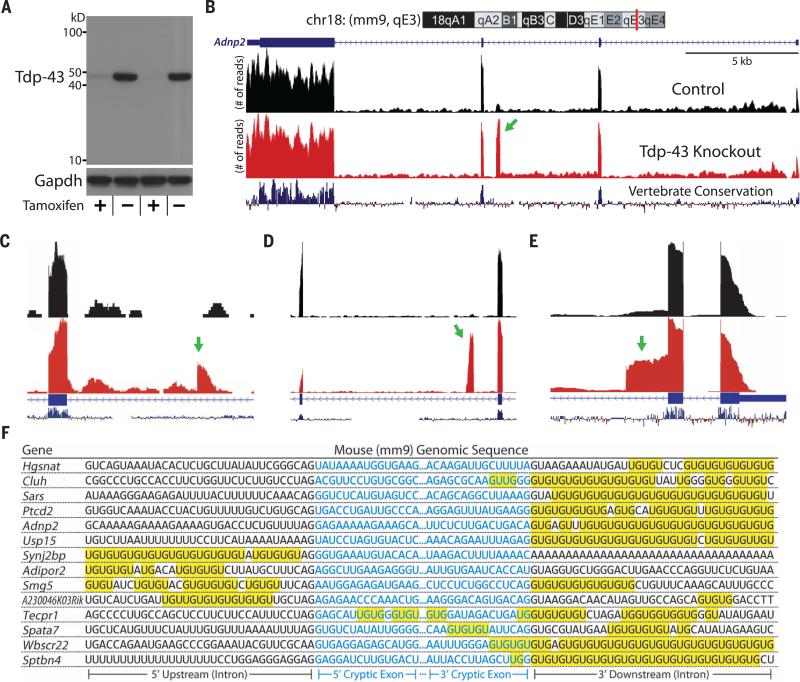
Fig. 1. Identification of Tdp-43–associated cryptic exons (Mus musculus).
(A) iTDPKO cells are depleted of Tdp-43 upon treatment with 4HT. (B) Visualization of the cryptic exon located in Adnp2 (red bar) of chromosome 18 (chr18) (top). Gene annotation is shown below, labeling exons (thick) and introns (thin). RNA-seq reads from iTDPKO untreated (“Control”) and 4HT treated (“Tdp-43 Knockout”) are aligned to the mm9 genome. The cryptic exon (green arrow) is not conserved (vertebrate conservation, bottom). Strand-specific analysis also confirms the incorporation of cryptic exons on the transcribing strand (fig. S11). (C to E) Most cryptic exons have standard 5′ and 3′ splice sites [Adipor2, (C)]. Occasionally, cryptic exons can present as transcriptional start sites [Sptbn4, (C)], polyadenylation sites [Synj2bp, (D)], or exon extensions [Wbscr22, (E)]. (F) Cryptic exons are flanked by UG tandem repeats that exist upstream, downstream, or internally.
These splicing variants have not been previously documented in any public databases and strictly appear in nonconserved regions of the genome. Traces of cryptic exons, however, can be detected from previously published RNA-seq data (fig. S1). Most cryptic exons introduce frameshifts and/or premature stop codons, characteristic of nonsense-mediated decay (NMD) (table S1). Additionally, we identified a subset of nonstandard cryptic exons that could be categorized as alternative transcriptional start sites, premature polyadenylation sites, or expansions of conserved canonical exons (Fig. 1, C to E).
To determine whether these cryptic exons were direct targets of Tdp-43 binding or missplicing events caused by indirect downstream effects, we looked for evidence of protein-RNA association. Tdp-43 has a very high affinity for UG repeats, with a dissociation constant (Kd) of 14 nM for only six repeats (20). As predicted, long UG dinucleotide repeats are adjacent to each cryptic exon (Fig. 1F). Tdp-43 localized to these flanking repeats could potentially act as a general splicing repressor. Further evidence of Tdp-43's role in repressing cryptic exons comes from previous HITS-CLIP data that mapped Tdp-43's direct RNA targets (19). As expected, clusters of HITS-CLIP reads were mapped onto cryptic exons, further supporting the hypothesis that Tdp-43 represses cryptic exons via direct interaction (fig. S2).
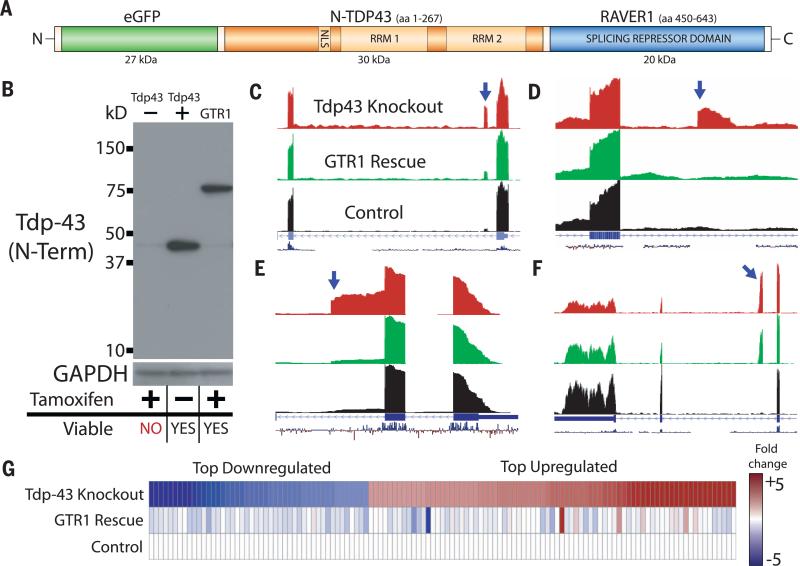
We next wanted to establish whether these cryptic exons were a main contributor to the cell death observed after genetic deletion of Tdp-43 (12). To this end, we designed a fusion protein by coupling TDP-43's N-terminal fragment (N-TDP) to a well-characterized splicing repressor domain, replacing TDP-43's C-terminal fragment (C-TDP) (Fig. 2A). N-TDP contains a dimerization domain, nuclear localization signal, and two UG binding RNA recognition motifs but is insufficient to act as a splicing repressor (21–23). In contrast, C-TDP is a glycine rich, prion-like domain that harbors the vast majority of mutations associated with ALS and mediates the protein-protein interac tions important for splicing (24). We replaced the C-TDP in our fusion protein with the minimal splicing repressor domain derived from ribonucleoprotein, PTB-binding 1 (RAVER1) (25, 26). This chimeric protein, termed GTR, translocated to the nucleus, bound to UG repeats, and repressed nearby exons (Fig. 2).
Fig. 2. Repression of cryptic exons is associated with the prevention of cell death induced by loss of Tdp-43.
(A) Diagram of the green fluorescent protein, N terminus TDP-43, RAVER1 splicing repressor domain chimeric construct (GTR). (B) Immunoblot using an antibody that recognizes mouse/human N terminus of Tdp-43. (C to F) Whereas iTDPKO cells are not viable when depleted of Tdp-43, GTR1 cells are fully viable. GTR represses a variety of cryptic exons; standard cassette [Adipor2, (C)], polyadenylation [Zfp809, (D)], and exon extensions [Wbscr22, (E)]. However, some cryptic exons are only partially rescued [Synj2bp, (F)]. (G) RNA-seq transcript expression levels for Tdp-43 knockout cells and GTR1 rescue cells were compared with untreated iTDPKO cells. The top 124 down-regulated/up-regulated genes in the Tdp-43 knockout cells were mostly restored to normal levels in the GTR1 rescued cells. Full transcript expression data are provided in the supplementary materials (data table S1).
The GTR construct and associated controls were then transiently transfected into 4HT-treated iTDPKO cells and screened for survivors by means of flow cytometry (fig. S3). After exposure to 4HT, the majority of Tdp-43–depleted iTDPKO cells underwent apoptosis. Whereas 4HT-treated iTDPKO cells transfected with control constructs also failed to survive, a portion of those transfected with the GTR construct remained viable.
To rule out the possibility that this rescue was only a transient effect of the GTR expression, we generated a stable transfected line of Tdp-43–deficient cells expressing GTR, termed GTR1 (Fig. 2B). The growth rate of GTR1 cells was almost identical to iTDPKO cells (fig. S4), and the majority of up-regulated and down-regulated transcripts in Tdp-43 knockout cells were also restored to normal values in GTR1 cells (Fig. 2G), including several transcripts markedly down-regulated because of nonsense-mediated decay (table S1). To verify that the rescue effect was correlated with repression of cryptic exons, we RNA-sequenced the GTR1 cells. Indeed, the majority of cryptic exons were highly repressed (Fig. 2, C to E, and table S2), with the remaining cryptic exons experiencing varying degrees of partial repression (Fig. 2F). Thus, a deficiency in cryptic exon repression could contribute to cell death.
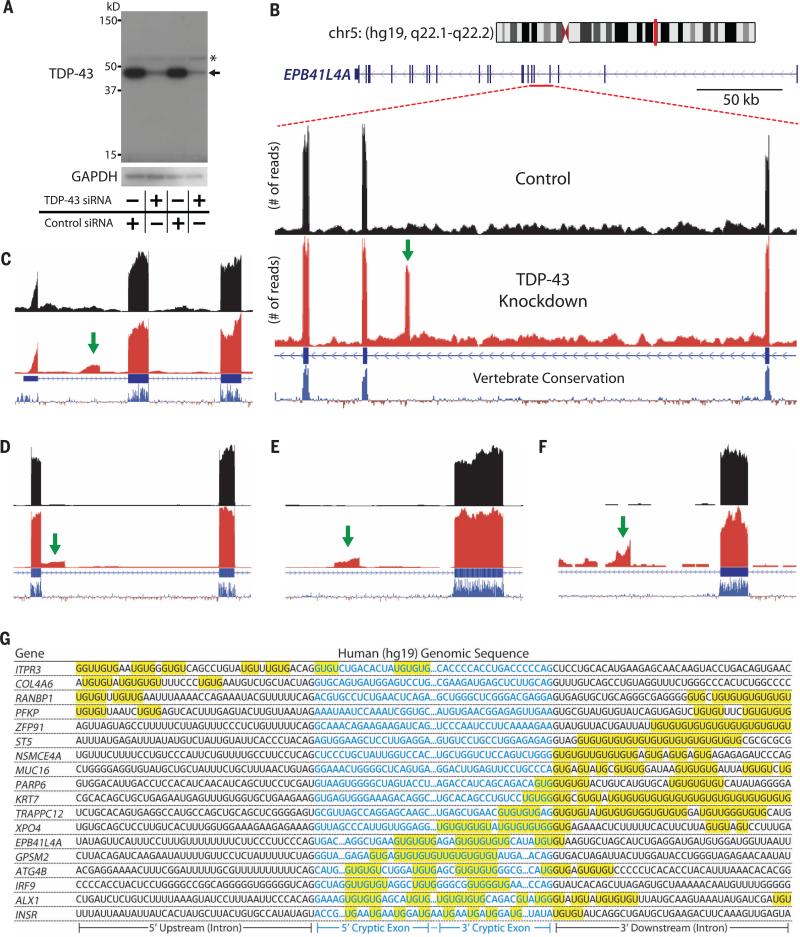
Because mouse cryptic exons are embedded within nonconserved sequences of the mouse genome, we predicted that human cryptic exons would be located in different regions of the human genome. HeLa cells treated with TDP-43 small interfering RNA (siRNA) (Fig. 3A) expressed numerous cryptic exons (Fig. 3, B to F, and table S3). These human-specific cryptic exons were flanked by UG tandem repeats (Fig. 3G) and were direct targets of TDP-43 (fig. S5). The human cryptic exons shared no overlap with mouse cryptic exons and instead influenced an entirely different set of genes (table S4).
Fig. 3. Identification of TDP-43–associated cryptic exons (Homo sapiens).
(A) TDP-43 protein levels are greatly reduced when HeLa cells are treated with TDP-43 siRNA (asterisk indicates nonspecific band). (B) Visualization of the cryptic exon located in EPB41L4A. Zoom-in of gene annotation demonstrates that the cryptic exon (green arrow) resides in a nonconserved region. Strand-specific analysis also verifies the incorporation of cryptic exons on the transcribing strand (fig. S12). (C to F) IRF9 contains a transcriptional start site (C), KRT7 contains an exon extension (D), GPSM2 contains a standard cryptic exon (E), and INSR contains a polyadenylation site (F). (G) UG repeats are also found adjacent to human cryptic exons.
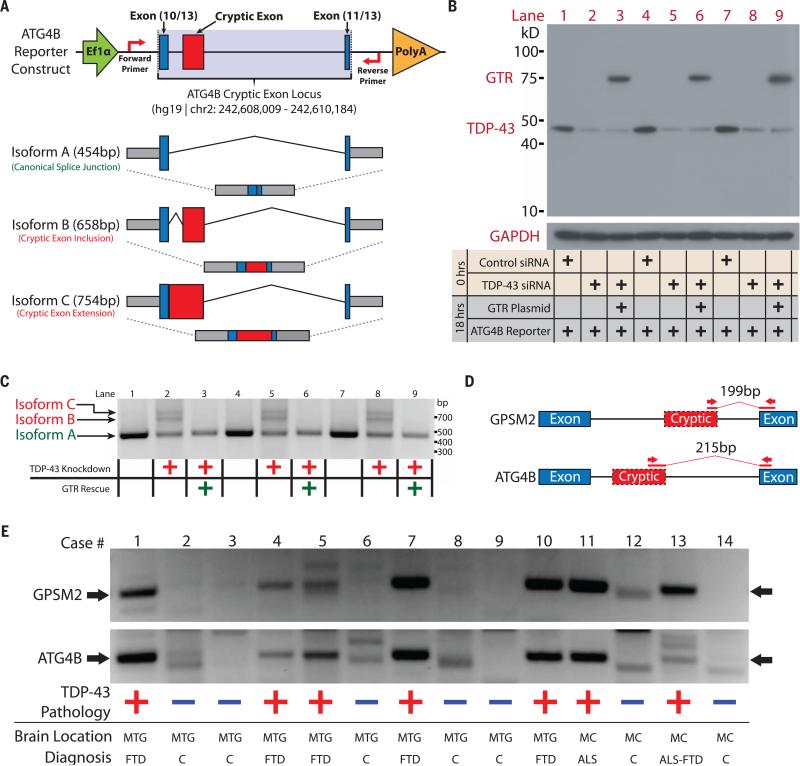
To further validate this model of cryptic exon repression, we engineered a mini-gene reporter construct encompassing a cryptic exon locus identified in ATG4B and transfected human HeLa cells lacking TDP-43 (Fig. 4). Under normal conditions, only isoform A was expressed (Fig. 4C, lanes 1, 4, and 7). After TDP-43 was depleted from the cell, cryptic isoforms B and C were no longer repressed (Fig. 4C, lanes 2, 5, and 8). Isoform C was not initially detected through RNA-seq owing to masking by an overlapping cryptic cassette exon; sequencing confirmed its identity as an exon extension (fig. S6). Isoforms B and C were repressed by GTR (Fig. 4C, lanes 3, 6, and 9), supporting the notion that TDP-43 may function as a general repressor whose specificity is determined by its affinity for UG repeats rather than its C-terminal domain.
Fig. 4. Validation of human cryptic exons and their detection in ALS-FTD brain tissue.
(A) Diagram of ATG4B reporter construct and potential splicing isoforms. (B) Immunoblots of HeLa transfections under normal or TDP-43–depleted conditions and (C) the associated electrophoretic gel analysis of splicing isoforms were performed in triplicate. Cryptic isoforms B and C only appeared under conditions of TDP-43 depletion and were rescued by the GTR protein. (D) Diagram of RT-PCR detection strategy. Primers were designed to amplify only the cryptic exon splice junction. (E) DNA fragments are detected at 199 base pairs (bp) (GPSM2) and at 215 bp (ATG4B) for all cases that display TDP-43 proteinopathy; control cases do not display these fragments. Full gel images and case demographics are provided in the supplementary materials (fig. S7 and table S5). Cases 1, 7, and 13 are positive for C9ORF72 expansions; the remaining cases are sporadic. MTG, middle temporal gyrus; MC, motor cortex.
Having identified a set of cryptic exons that TDP-43 regulates in humans, we then screened postmortem brain tissues from an ALS-FTD cohort (table S5) for the presence of cryptic exons. A reverse transcription polymerase chain reaction (RT-PCR) protocol was designed to amplify across the cryptic exon splice junctions of GPSM2 and ATG4B (Fig. 4D). Corresponding PCR products were readily observed in all ALS-FTD cases tested but not in controls (Fig. 4E) and validated by means of DNA sequencing (fig. S7). Thus, TDP-43 proteinopathy may be correlated with TDP-43 loss of function.
We have found that TDP-43 functions as a splicing repressor of nonconserved cryptic exons (fig. S8). A defect in this regulatory mechanism could be linked to TDP-43 proteinopathy in ALSFTD. Acting as an inhibitory hnRNP, TDP-43 may also regulate conserved exons (18, 19, 27). Analysis of mouse embryonic stem cell RNA-seq data suggests that some alternatively spliced conserved exons contain UG repeats and may be direct targets of TDP-43 (fig. S9). Further work will be required to determine TDP-43's role regarding splicing of conserved exons.
Although the subset of cryptic exons in mice (table S1) is entirely different from that of humans (table S3), TDP-43's cryptic exon repression function has been maintained across evolution. The protein sequence of TDP-43 is conserved and interchangeable across humans, mice, flies, and nematodes (17, 22), suggesting that TDP-43 cryptic exon repression could extend beyond mammals as well. From the perspective of human disease, however, we believe that studying genes specifically affected by cryptic exons in the human context could help clarify the development of TDP-43 proteinopathy. Two genes in particular—ATG4B and RANBP1—function in autophagy and nuclear import, respectively. We envision a feed-forward loop in which loss of TDP-43 function could undermine the cell's ability to restore TDP-43 to the nucleus, leading to further loss of function (fig. S10).
The discovery of TDP-43's role in repressing cryptic exons will advance our understanding of human diseases with TDP-43 proteinopathy and form the basis for previously unidentified bio-markers and therapeutic strategies.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Schaughency, S. Wheelan, and the rest of the Next Generation Sequencing Center [Johns Hopkins Medical Institution (JHMI)] staff for their RNA sequencing service. We also thank X. Zhang for flow cytometry assistance (Ross Flow Cytometry Core, JHMI), R. Roth for confocal imaging, and J. Bedont for editorial support. Last, we thank P. Rabin for encouragement and support of FTD studies and C. Onyike for clinical evaluations of cases of frontotemporal lobar degeneration (FTLD). This work was supported in part by The Robert Packard Center for ALS Research, Muscular Dystrophy Association, the Amyotrophic Lateral Sclerosis Association, Target ALS, the Johns Hopkins University Neuropathology Pelda fund, the Johns Hopkins Alzheimer's Disease Research Center (NIH P50AG05146), and the Samuel I. Newhouse Foundation. J.P.L. and P.C.W. have filed a patent application (number 62/180,988) in the United States that pertains to using incorporation of cryptic exons in RNA transcripts identified in human diseases exhibiting TDP-43 proteinopathy as the basis for biomarkers and therapeutic targets/strategies. Raw sequencing files (100 bp, paired-end) have been deposited at the National Center for Biotechnology Information Sequence Read Archive under SRP057819 and SRP057948. The authors declare no conflicts of interest. J.P.L. and P.C.W. designed, analyzed, and interpreted experiments. J.P.L. performed all studies. O.P. and J.C.T. evaluated brain tissues of ALS and FTLD cases and controls. J.P.L. and P.C.W. wrote the manuscript, and all authors discussed results and approved the manuscript.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/349/6248/650/suppl/DC1
Materials and Methods
Figs. S1 to S12
Tables S1 to S5 References (28–30)
Data Table S1
REFERENCES AND NOTES
- 1.Lee EB, Lee VM, Trojanowski JQ. Nat. Rev. Neurosci. 2012;13:38–50. doi: 10.1038/nrn3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ling S-C, Polymenidou M, Cleveland DW. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neumann M, et al. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 4.Sreedharan J, et al. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kabashi E, et al. Nat. Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 6.Janssens J, Van Broeckhoven C. Hum. Mol. Genet. 2013;22(R1):R77–R87. doi: 10.1093/hmg/ddt349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Renton AE, Chiò A, Traynor BJ. Nat. Neurosci. 2014;17:17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freischmidt A, et al. Nat. Neurosci. 2015;18:631–636. doi: 10.1038/nn.4000. [DOI] [PubMed] [Google Scholar]
- 9.Cirulli ET, et al. Science. 2015;347:1436–1441. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayala YM, et al. EMBO J. 2011;30:277–288. doi: 10.1038/emboj.2010.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sephton CF, et al. J. Biol. Chem. 2010;285:6826–6834. doi: 10.1074/jbc.M109.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiang P-M, et al. Proc. Natl. Acad. Sci. U.S.A. 2010;107:16320–16324. doi: 10.1073/pnas.1002176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraemer BC, et al. Acta Neuropathol. 2010;119:409–419. doi: 10.1007/s00401-010-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang C, et al. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E1121–E1129. doi: 10.1073/pnas.1322641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feiguin F, et al. FEBS Lett. 2009;583:1586–1592. doi: 10.1016/j.febslet.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 16.Schmid B, et al. Proc. Natl. Acad. Sci. U.S.A. 2013;110:4986–4991. doi: 10.1073/pnas.1218311110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukavsky PJ, et al. Nat. Struct. Mol. Biol. 2013;20:1443–1449. doi: 10.1038/nsmb.2698. [DOI] [PubMed] [Google Scholar]
- 18.Tollervey JR, et al. Nat. Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polymenidou M, et al. Nat. Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuo P-H, Doudeva LG, Wang Y-T, Shen C-KJ, Yuan HS. Nucleic Acids Res. 2009;37:1799–1808. doi: 10.1093/nar/gkp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y-J, et al. Hum. Mol. Genet. 2013;22:3112–3122. doi: 10.1093/hmg/ddt166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayala YM, et al. J. Mol. Biol. 2005;348:575–588. doi: 10.1016/j.jmb.2005.02.038. [DOI] [PubMed] [Google Scholar]
- 23.D'Ambrogio A, et al. Nucleic Acids Res. 2009;37:4116–4126. doi: 10.1093/nar/gkp342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nonaka T, et al. Cell Reports. 2013;4:124–134. doi: 10.1016/j.celrep.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 25.Gromak N, et al. EMBO J. 2003;22:6356–6364. doi: 10.1093/emboj/cdg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rideau AP, et al. Nat. Struct. Mol. Biol. 2006;13:839–848. doi: 10.1038/nsmb1137. [DOI] [PubMed] [Google Scholar]
- 27.Fu X-D, Ares Jr M. Nat. Rev. Genet. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.