Abstract
High-density lipoproteins (HDL) are a class of natural nanostructures found in the blood and are composed of lipids, proteins, and nucleic acids (e.g. microRNA). Their size, which appears to be well-suited for both tissue penetration/retention as well as payload delivery, long circulation half-life, avoidance of endosomal sequestration, and potential low toxicity are all excellent properties to model in a drug delivery vehicle. In this review, we consider high-density lipoproteins for therapeutic delivery systems. First we discuss the structure and function of natural HDL, describing in detail its biogenesis and transformation from immature, discoidal forms, to more mature, spherical forms. Next we consider features of HDL making them suitable vehicles for drug delivery. We then describe the use of natural HDL, discoidal HDL analogs, and spherical HDL analogs to deliver various classes of drugs, including small molecules, lipids, and oligonucleotides. We briefly consider the notion that the drug delivery vehicles themselves are therapeutic, constituting entities that exhibit “theralivery.” Finally, we discuss challenges and future directions in the field.
I. Introduction
High-density lipoproteins (HDL) represent a class of complex natural nanostructures appearing at high concentrations in human serum. Though HDLs serve multiple functions, they are most known for their roles in lipid transport and metabolism. For instance, HDLs play a critical role in reverse cholesterol transport, a process that results in the net transfer of cholesterol from peripheral tissues, such as cholesterol loaded macrophages in the arterial wall, to the liver for excretion.1, 2 HDLs interact with cells in a receptor-mediated fashion,3 and their natural cargo comprises a variety of lipids,4 proteins,5 and microRNAs.6 These observations motivate the use of HDL and HDL analogs for therapeutic delivery systems.
This highlight is composed of two sections. First, we review the biogenesis of natural HDL. We draw out features of natural HDL that make them attractive for drug delivery in their own right, as well as features that synthetic, nanotechnology-based approaches seek to mimic. From this background, we introduce the applications of natural HDL as therapeutic delivery systems, and the two major nanotechnology drug delivery platforms inspired by HDL: discoidal HDL biomimetics (also known as nanodisks, reconstituted HDL, and nanolipoparticles), and spherical HDL biomimetics. Finally we conclude by surveying future directions, and discussing challenges faced by the field in further advancing these concepts.
II. High-density Lipoprotein Structure and Function
Understanding the structure and function of endogenous HDL is critical to understanding the therapeutic delivery potential of exogenous HDL analogs. High density lipoproteins derive their name from the observation that among lipoproteins, HDL exhibit the highest density in classical ultracentrifugation experiments which first fractionated the lipoprotein components of serum.7 Accordingly, the density of HDL (by definition greater than 1.063 g/L) is higher than that of intermediate-density lipoproteins (IDL), low-density lipoproteins (LDL), very low-density lipoproteins (VLDL), and chylomicrons. The increased density of HDLs compared to other lipoproteins is due to their relatively high protein content, which ranges from approximately 30% to 60% protein by weight.8 Due to constant remodeling of HDL through interaction with cells, enzymes, and binding of various cargo molecules, natural HDL comprises a very heterogeneous class of nanoscale particles. HDL species exist along a continuum of size, ranging from nascent, discoidal species (called pre-β HDL) to more mature, spherical species, which again can be even further discriminated into a myriad of additional subspecies based on electrophoretic mobility.9 For instance, less mature, smaller (but denser) spherical HDL particles are termed HDL3, while larger, less dense particles are termed HDL2.7 Besides size, lipoprotein composition can also be used to distinguish different species of HDL. The main protein component of HDL is apolipoprotein A-I (apo A-I), a 28,000 kDa protein that represents approximately 70% of the total mass of protein on HDL species.2 However, a number of other apolipoproteins are also found on the surface of HDL. In a landmark proteomics study, Vaisal and colleagues detected approximately 60 different proteins on HDL, including proteins in the complement cascade and proteins involved in inflammatory modulation.5 Among the other apolipoproteins the best studied is apo A-II, which exists, chiefly, as a homodimer on HDL particles. Apo A-II is highly lipophilic; however, details of its function and mechanism are less well known.10 Apolipoprotein association with HDL is a dynamic process, as apolipoprotein spontaneously exchanges between its HDL-bound and lipid-free state.11 HDLs have also been found to carry microRNA, and the microRNA complement of HDL differs in patients with varying disease states.6 This rich structural and signaling diversity of high-density lipoproteins raises the possibility that there exists within HDL functionally specialized subpopulations with tailored combinations of biological macromolecules. The function, precise biochemical composition, and cell-specific interactions of HDL are best understood by first considering how high-density lipoproteins form, a process termed HDL biogenesis.
HDL Biogenesis and Function
HDL biogenesis (Figure 1) is initiated when free apo A-I protein, also called lipid-poor apo A-I, physically interacts with the cellular ATP-binding cassette transporter (ABCA1) on macrophages, leading to a unidirectional and ATP-dependent transfer of phospholipids and free cholesterol to the nascent HDL particle.12, 13 Termed pre-β HDL, these discoidal particles are thought to contain two or three apo A-I molecules, forming a belt around the hydrophobic acyl chains of the particle, thereby shielding them from the aqueous environment.14 15, 16 Pre-β HDL particles contain about 60-70% protein by weight, along with phospholipids and a small amount of unesterified cholesterol.8 While pre-β HDL continues to receive cholesterol from lipid-loaded macrophages in an ABCA1-dependendent manner, it can also mature to spherical HDL, predominantly through the action of lecithin-cholesterol acyltransferase (LCAT) enzyme found in serum. LCAT transfers an acyl group from a phospholipid donor to cholesterol to form cholesteryl ester (CE).17 CE leaves the surface of the discoidal HDL, intercalates within the hydrophobic environment of the bilayer, and progressively engenders the HDL particle with a spheroidal form. The stoichiometry of apo A-I on spherical HDL particles has not been defined, but a near-physiologic model system has shown the plausibility of three apo A-I molecules arranged in a trefoil pattern on the surface of an HDL.18
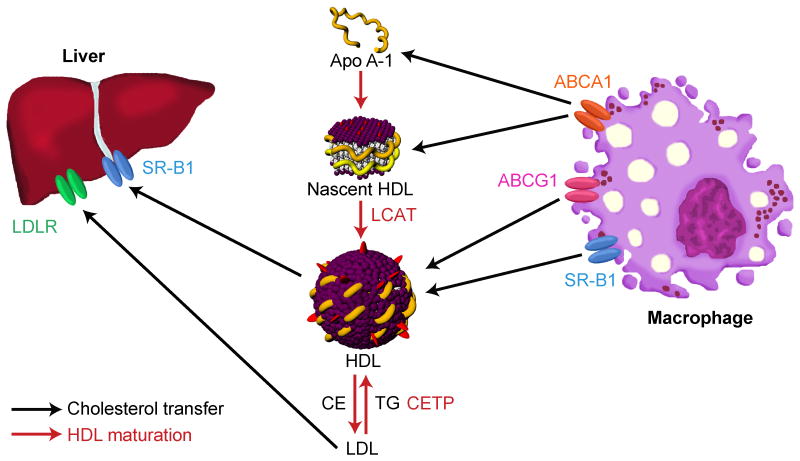
Figure 1. Maturation of High-Density Lipoproteins (HDL) and Lipoprotein Transport.
Through action of the ABCA1 transporter, macrophages efflux phospholipids (violet) and free cholesterol (red) to lipid-poor apo A-I, leading to the formation of nascent HDL, also called pre-β HDL. In this discoidal form of HDL, which is ≤ 8 nm in diameter, apo A-I is thought to form a “double belt” around the structure, protecting the hydrophobic acyl groups of the phospholipids from the aqueous environment. Through action of lecithin-cholesterol acyltransferase (LCAT), cholesterol is esterified. This renders cholesterol more hydrophobic, causing it to enter the core of the particle and create a spherical, more mature form of HDL. This mature, spherical HDL transfers its cholesterol cargo directly to the liver through the SR-B1 receptor. Alternatively, the cholesterol cargo of the HDL can be transferred to LDL by action of cholesterol ester transfer protein (CETP) in exchange for triglycerides (TG). LDL can, in turn, then be taken up by the liver through LDL receptor. For clarity, not shown is the distinction between the two main forms of spherical HDL: HDL3 particles, which are smaller (about 7.2-8.2 nm in diameter), and HDL2 particles, which are larger (about 8.8-12.9 nm in diameter).
Spherical HDL can undergo additional maturation by taking up phospholipids and cholesterol from cells through a variety of different mechanisms, including aqueous diffusion of lipids and cholesterol directly from the cell membrane14, 19 or through the membrane associated protein20 ATP-binding cassette G1 transporter (ABCG1). ABCG1 does not bind HDL particles, but may enhance efflux by promoting cholesterol localization on the plasma membrane. Spherical HDL also interacts with scavenger receptor B1 (SR-B1), an integral membrane glycoprotein21 that is expressed on a number of cell types, including macrophages,22 hepatocytes,23 and various cancer cell lines.24 In contrast to ABCA1 and ABCG1, SR-B1 can facilitate transfer of cholesterol both to and from HDL particles. SR-B1 also mediates the selective transfer of its CE payload to target cells. Highly relevant for the use of spherical HDL homologs for drug delivery, this so-called “selective lipid uptake” does not require lysosomal degradation or engulfment of the entire HDL particle.25, 26 Interaction between HDL and the receptor is most likely initiated by interaction of the receptor with amphipathic helices of apo A-I. Data demonstrate that multiple regions within apo A-I bind to the receptor with high affinity.27 Interestingly, SR-B1 binds to different HDL species with different affinities, depending on the shape and lipid composition of HDL, as well as the conformation of the presented apolipoprotein.28 For instance, bigger, less dense HDL2 species have a higher affinity to SR-B1 than smaller, denser HDL3 species.28 Selective cholesterol transport from HDL2 to the cell causes the particle to become smaller, which reduces the particles' affinity to the receptor and frees up cellular binding sites for other CE-rich HDL particles.28, 29 Lastly, by action of the enzyme cholesterol ester transfer protein (CETP), which transfers cholesteryl ester between lipoprotein species, cholesterol may be also transferred from high-density lipoproteins to low-density lipoproteins (LDL) in exchange for triglycerides (TG). LDL in turn may then be taken up by the liver through interaction with the LDL receptor.
Features of Natural HDL motivating adaptation for drug delivery
Nanotechnology-based biomimetics of natural HDL are an attractive platform for drug delivery for a variety of reasons (Table 1). HDL particles are highly stable structures30, 31 whose stability is both of thermodynamic as well as kinetic origin.11, 30-33 Further, their built-in receptor-mediated interactions29 allow for the potential for specific targeting.3 The concentration of HDL particles in the blood is quite high, approximately 30 μM, suggesting that HDL biomimetics could be tolerated at similar concentrations as well.34 HDL particles are typically <13 nm in diameter and have high surface area. This surface area is large enough to carry several small molecule drugs at once or even siRNA drugs,35 yet the overall size of the particle is small enough to penetrate tissue.36 Spherical HDL particles also appear to avoid endosomal sequestration,35, 37, 38 a feature which has potential to improve drug delivery. Finally, radiolabelled assays in humans have shown that certain protein components of HDL particles recirculate extensively.39 This aspect of HDL circulation potentially increases its bioavailability to target tissues. The above discussion delineates the role HDL play in human biology, and highlights features that make them desirable model nanostructures to emulate for therapeutic delivery systems. Below we discuss these systems in further detail.
Table 1. Advantages of HDL-Based Delivery Platforms.
HDL-based delivery platforms have several potential advantages, listed here alongside key references.
III. Natural HDL for Drug Delivery
Endogenous HDL has been explored as a vehicle for delivery of small interfering RNA (siRNA). In a landmark paper, Wolfrum et al. analyzed the mechanisms by which conjugation of siRNA to cholesterol and other lipophilic functional groups enhanced efficacy and targeting effects of the siRNA. As a model, the investigators used siRNA antisense to ApoB1, the gene responsible for the major protein on low-density lipoproteins (LDL). The authors demonstrated that lipophilic siRNA bind to HDL and LDL in the serum, and that modifying the hydrophobicity of the conjugate by varying the functional group altered the binding affinities. The authors then demonstrated that siRNA efficacy was enhanced by pre-incubating the lipophilic siRNA with natural HDL prior to injection in animals. Furthermore, the group showed that uptake of the siRNA-HDL conjugates was SR-B1 dependent through two lines of evidence. First, injection of the complex into mice lacking SR-B1 demonstrated reduced uptake into organs rich in SR-B1 expression, such as the liver. Second, the circulating half-life of the injected particles was approximately twice as long in SR-B1 -/- animals.40
The Yokota group has also explored the utility of natural HDL for conjugating siRNA. Kuwahara, et al. studied the effects of cholesterol-conjugated siRNA (chol-siRNA) in brain capillary endothelium. As a model target, the investigators selected organic anion transporter 3 (OAT3), a protein exclusively expressed on endothelial cells. They demonstrated by gel shift experiments that the chol-siRNA could be conjugated to HDL or LDL. After injection of the complex, siRNA was detected by Northern blot in the vascular compartment of brain tissue, and gene knockdown was achieved.41 Uno, et al. conjugated siRNA to the lipophilic molecule α-tocopherol (Vitamin E).42 HDL was isolated from mouse serum and incubated with the α-tocopherol-siRNA. Gel shift experiments showed conjugation to HDL, and RNA stability experiments demonstrated enhanced stability of the α-tocopherol-siRNA when conjugated to HDL. Knockdown of the target gene BACE1 after injection in mice was demonstrated by RT-PCR.
Chemical modification of natural HDL has also been explored as a method for conferring target organ specificity. In 1991, Bijsterbosch et al., noting that galactose uptake is quite specific to the liver, covalently conjugated galactose residues to isolated HDL and re-injected the radiolabelled particles. They noted rapid uptake in the liver over short time frames, and speculated that the approach might be useful for delivering lipophilic molecules.43
IV. Mimics of Discoidal HDL for Drug Delivery
Synthesis
In 1966, Scanu discovered that apo A-I and phospholipids isolated from healthy donors could be recombined in vitro and spontaneously assemble under physiologic conditions into complexes of definite protein:lipid stoichiometry. These complexes had similar density to nascent natural HDL and similar optical properties. Furthermore, Scanu demonstrated that these synthetic complexes were functional, activating the enzyme lipoprotein lipase, just as natural HDL.44 Subsequent studies using transmission electron microscopy demonstrated that various lipoprotein preparations using this technique were discoidal in nature and had a diameter of approximately 10 nm.45, 46 A limitation of this self-assembly approach was that unsaturated phospholipids and cholesterol could not be formulated. This limitation was overcome with the development of the cholate dialysis method, in which the detergent sodium cholate is used to facilitate assembly.47, 48 Using these approaches, and by varying the apolipoprotein and lipid components of the synthesis, a suite of discoidal nanostructures, termed recombinant HDL (rHDL) or nanodisks, have been formulated that range in diameter from approximately 9 to 30 nm.49 Others further extended this platform by deconstructing the biochemical properties of apolipoproteins allowing for assembly of these discoidal structures, and designing amphipathic peptides to control size.50
rHDL were initially developed to help investigate structure-function properties of naturally occurring HDL and have provided important insight into our understanding of the thermodynamics and biophysics of biological membranes.51-54 However, rHDL were quickly noted to have features useful for drug delivery as well. Due to their close structural similarity to natural HDL, many of these nanostructures are inherently biocompatible. The amphipathic nature of rHDL proves useful for formulating lipophilic drugs and improving their delivery. Being close mimics of natural HDL, they interact with cells through known receptor-ligand interactions, conferring some degree of inherent specificity. Furthermore, this inherent specificity can be further tailored by linking other targeting proteins to apo A-I.
Specific drugs and studies
A number of drugs have been successfully formulated using nanodisk approaches. Amphotericin B is a powerful antifungal drug with dose-limiting side effects. It is also a prototypical lipophilic drug. Amphotericin B has been loaded into nanodisks using a self-assembly strategy with a mixture of phospholipid:amphotericin B in a ratio such that amphotericin B was 20% of the total content of the structure, and likely nearly this high in the final product.55 These amphotericin B containing nanodisks have less toxicity as compared to conventional amphotericin B in a model cell line for liver cells, HepG2 cells, and for red blood cells. Data further showed that nanodisks with incorporated amphotericin B were effective in mice infected with Candida albicans. Amphotericin B nanodisks have also been used effectively in mouse models of leishmaniasis,56 a parasitic infection endemic in the tropics.
All trans retinoic acid (ATRA) is another lipophilic drug that has loaded in nanodisks.57, 58 This drug is a critical component of the treatment regimen for acute promyelocytic leukemia. Nanodisks were synthesized by self-assembly, with approximately 85% ATRA incorporated into the final nanodisk structures. These structures were noted to be relatively stable to long-term storage at 4°C. ATRA-containing nanodisks were tested against the conventional formulation in a cell culture model of lymphoma. Data demonstrate that the nanodisk formulation mediated an increase in cell death and cell cycle arrest.
Curcumin is a compound with antiproliferative effects, making it an attractive potential therapy for cancer. However, this compound has limited aqueous solubility. In a cell culture model, investigators demonstrated enhanced cell killing with the formulated version versus the unformulated drug.59 Using a modification of the cholate dialysis method, Lou and colleagues formulated aclacinomycin in rHDL, and demonstrated enhanced antitumor effect in a cell culture model of hepatocellular carcinoma.60 Thus both the direct self-assembly method and the cholate dialysis method have been used to load small molecules in nanodisks.
Simvastatin is a 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) inhibitor (statin) with potent cholesterol lowering and anti-inflammatory effects. However, this compound is largely taken up by the liver and inactivated, effectively decreasing the drug's bioavailability in target tissues such as in atherosclerotic plaques, where anti-inflammatory effects may be critical in reducing atherosclerosis.61 Duivenvoorden, et al. formulated simvastatin with reconstituted HDL ([S]-HDL) and studied its pharmacokinetics and therapeutic effects in the ApoE knockout mouse, an animal model of atherosclerosis.62 The investigators found that [S]-HDL labeled with gadolinium accumulated in the aortic wall of ApoE knockout mice at 24 hours after injection, indicating accumulation in areas prone to developing atherosclerosis. They showed that in vitro [S]-HDL decreased survival of macrophages, a key inflammatory cell type mediating atherosclerosis. Elegant in vivo studies showed a significant reduction in atherosclerotic burden in ApoE knockout mice on a high cholesterol diet. In a subsequent report, this group further dissected the mechanism of [S]-HDL effect in vivo, and demonstrated the importance of antiproliferative effects of [S]-HDL therapy on disease progression.63
Further modifications of nanodisks have been made in an attempt to enhance targeting. Iovannisci, et al. generated a chimeric protein comprising apo A-I and a single chain variable antibody directed to vimentin (scFv). Data demonstrated that the fusion protein did not interfere with the assembly of the nanodisk, and that the fusion protein could target vimentin. However no in vitro or in vivo testing of the constructs for targeting effect was conducted.64
V. Mimics of Spherical HDL for Drug Delivery
Spherical high-density lipoproteins represent the preponderance of HDL in the human body, yet relative to discoidal forms of HDL they remain understudied. This is in part because relative to the synthesis of analogs of discoidal HDL, such as rHDL, the development of spherical HDL analogs is somewhat more complicated. However, there are several rationales motivating the development of spherical HDL analogs as a drug delivery platform. First, spherical HDLs offer the opportunity for a greater variety of topologies of drug loading. In addition to drugs loadable on the membrane, or in the membrane, as is the case for discoidal HDL analogs, spherical HDL can also comprise drugs within the particle itself, providing a hydrophobic environment for payloads. Second, spherical forms of HDL engage the SR-B1 receptor,3 a receptor found in a variety of cell types, including tumor cells.65, 66 This may confer inherent target specificity to a class of cells of tremendous interest for therapeutic targeting. Emerging evidence supports the notion that SR-B1 is critical for maintaining cholesterol homeostasis, and that adequate cholesterol uptake is critical for supporting tumor cell growth. Third, mounting evidence demonstrates that spherical HDL has distinct roles in the body as compared to discoidal HDL, reinforcing the notion that for the purposes of drug delivery these two platforms function in distinct ways. For example, epidemiologic data suggest that patients with coronary heart disease may have lower concentrations of larger HDL species, and higher concentrations of smaller HDL species, highlighting the potential functional divergence between spherical and discoidal forms of HDL.67, 68
The discoidal rHDL can be biologically “matured” to spherical forms by the addition of low-density lipoproteins, which serve as a source of cholesterol, lipids, triglycerides, and LCAT69, 70 to convert cholesterol to a hydrophobic cholesteryl ester, which will partition to the potential space between the lipids in discoidal HDL, engendering a spherical form. Cholesterol ester transfer protein (CETP) may also be used.18, 71 However owing to the use of enzymes and complex macromolecular biologic structures such as LDL, these approaches offer limited synthetic control with regard to size and composition of the resulting nanostructure.
Spherical HDL for Small Molecule Drug Delivery
The cholate dialysis method has been used to generate spherical forms of HDL directly; however, by adding cholesteryl oleate (a cholesteryl ester) to the reaction mixture. This method was adapted by McConathy, et al. to formulate the chemotherapeutic paclitaxel (PTX) onto spherical HDL mimics.72 Apolipoprotein A-I, cholesterol, cholesteryl oleate, and phosphatidylcholine were mixed in a 1:5:1.3:115 ratio. Complexes were then isolated by preparative ultracentrifugation, followed by dialysis to further purify the particles and exchange them into buffer. The resulting particles were approximately 10% PTX by mass, and had approximately 25 PTX per particle. These particles had spherical geometry, with a mean diameter of approximately 11 nM, which compares favorably to the size of natural HDL particles. The IC50 of PTX formulated on the rHDL versus unformulated PTX were compared in breast, ovarian, and prostate cancer cell lines. Depending on the cell line, the formulated drug was 5 to 23 times more potent. As a marker of tolerability, the rHDL-formulated PTX induced less weight loss in treated mice as compared to unformulated drug. Using similar approaches, Sabnis, et al. have formulated the chemotherapeutic valrubicin into spherical rHDLs73, as well as fenretinide, a retinoid, potentially useful in neuroblastoma.74
The uptake mechanisms of rHDL-PTX were further studied by Mooberry et al.75 To investigate whether SR-B1 is involved in uptake of rHDL, ldl A7 cells overexpressing SR-B1 versus control cells were treated with rHDL-PTX. Approximately 3.5-fold higher uptake of rHDL-PTX was seen in the SR-B1 overexpressing cells. Furthermore, in a competition experiment, apo A-I alone, plasma-derived HDL, and rHDL without PTX could inhibit uptake of rHDL-PTX. These data provide compelling evidence that the spherical rHDL structure interacts with and is internalized by SR-B1.
The tailorability of targeting of rHDL was also assessed by Mooberry et al. Noting that folic acid receptor is overexpressed in the OVCAR-3 ovarian cancer cell line, the authors covalently linked folate to rHDL and conducted experiments to assess uptake of this species of rHDL. PTX uptake was 5-fold higher in OVCAR-3 cells treated with folate-rHDL-PTX, and 2-fold higher than cells treated with rHDL-PTX. Thus rHDL is not only inherently targeted through SR-B1 mediated interactions, but the platform may be modified to confer further enhanced targeting specificity.
Zhang et al. contributed important insights into the intracellular trafficking of spherical HDL analogs with fluorescently labeled compounds. The investigators generated a spherical HDL analog approximately 16 nm in diameter using an 18 amino acid apolipoprotein A-I peptide mimic, 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), and cholesteryl oleate to formulate the fluorescent dye 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindotricarbocyanine iodide bisoleate (DiR-BOA) as a model drug.76 Interestingly, while the loaded nanoparticle was spherical, the unloaded particle appeared to be discoidal. The DiR-BOA containing constructs trafficked to the cytosol and appeared to avoid sequestration in the endosomes. Zhang et al. also extended this work by linking endothelial growth factor (EGF) to the spherical HDL analogs and demonstrating that these EGF-containing particles had enhanced uptake in cells expressing EGF receptor. Injection of these constructs into mice demonstrated a long biologic half-life of 13.6 hours, demonstrating another desirable feature of HDL analogs with regard to drug delivery.
Modification of the apolipoprotein component of the construct to enhance particle uptake, targeting specificity, and drug delivery is an interesting approach that has been successfully employed by other investigators. Zhang et al. 77 studied the effects of using apolipoprotein A-IMilano as the protein component of rHDL. Apolipoprotein A-IMilano is a sequence variant of apo A-I that is naturally occurring and has been found in clinical studies to be associated with improved cardiovascular disease outcomes.78 Data demonstrated that a spherical rHDL generated using apolipoprotein A-IMilano incorporating the chemotherapeutic 10-hydroxycamptothecin was more potent than a spherical rHDL generated using conventional apolipoprotein A-I. In a very intriguing study, Dong et al.79 (vide infra), as part of their studies using HDL-like particles for siRNA-mediated knockdown, demonstrated that the addition of apolipoprotein E3 enhanced cellular uptake of their construct.
Spherical HDL for Nucleic Acid Drug Delivery
Using a gold nanoparticle as a template to control size and geometry, Thaxton et al. synthesized a spherical biomimetic of high-density lipoproteins.80 First, free apo A-I protein was added to the nanoparticle, and was permitted to self-assemble onto the nanostructure. Next, two lipids, the disulfide-containing 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[3-(2-pyridyldithio)propionate] and 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) were added to the gold nanoparticle-apo A-I complex to form the final spherical HDL-AuNP. These nanoparticles were approximately 18 nm in diameter, and contained approximately 3 apo A-I per particle. Building on observations that siRNA could be complexed to HDL for nucleic acid delivery, McMahon et al. studied the use of cholesterylated DNA for knockdown of intracellular targets.35 As a model system, cholesterylated DNA antisense to microRNA-210, a microRNA strongly upregulated by hypoxia, was investigated. Data demonstrated that HDL-NP complexed with cholesterylated DNA antisense to miR-210 could partially inhibit miR-210 induction by chemical hypoxia (Figure 2). Characterization of the particle showed that approximately 13 cholesteryl-DNA molecules were found per particle, that the addition of cholesterylated DNA to HDL-NP increased the diameter of the particles from 16 nm to 27 nm. Tripathy et al. then demonstrated that gold nanoparticle templated spherical HDL can formulate cholesterylated antisense RNA.81 In the system tested, antisense RNA to vascular endothelial growth factor receptor 2 (VEGFR2) could knockdown VEGFR2 mRNA levels and reduce endothelial survival and morphogenesis. Further data demonstrate that delivery of the HDL NP conjugates is dependent on expression of SR-B1. Thus, gold nanoparticle-templated HDL biomimetics are capable of formulating either DNA- or RNA-based antisense therapies, can knock down target gene expression, and demonstrate further downstream effects on the phenotypes of treated cells.
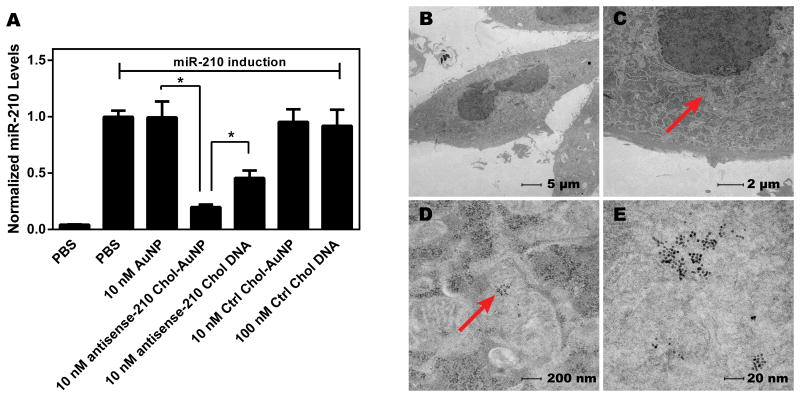
Figure 2. Gold-templated HDL-like Nanoparticles For Oligonucleotide Delivery.
(A) microRNA-210 (miR-210) is a small non-coding micro RNA induced by hypoxia, a defining feature of cancer. miR-210 in PC3 prostate cells can also be induced by exogenous addition of CoCl2. HDL-like NPs loaded with cholesterol-conjugated antisense miR-210 DNA (antisense-210 Chol-AuNP) effectively reduce CoCl2-induced miR-210 expression, both in comparison to the HDL-like NP alone (AuNP) and to an approximately equimolar dose of free cholesterol-conjugated antisense miR-210 DNA (antisense-210 Chol DNA). Scrambled DNA was used as a control. (B-E) Internalization of antisense-210 Chol-AuNP by PC3 cells 16 h after treatment was assessed by transmission electron microscopy (TEM). Arrows indicate AuNPs in the PC3 cells. Magnifications are (B) 890×, (C) 2,900×, (D) 23,000× and (E) 98,000×. Figure adapted from [35].
Yang et al. generated spherical rHDL mimetics using the cholate dialysis synthesis strategy, engendering a spherical shape to the final construct by inclusion of cholesteryl oleate into the synthesis.38 For the protein component, an apo A-I peptidomimetic was used. The resulting structure, termed HDL mimicking peptide-phospholipid scaffold (HPPS), was used to deliver siRNA antisense to Bcl-2. The construct had a radius of 25 nm and incorporated an average of 8 chol-siRNA per HPPS. Cytosolic delivery of payload was confirmed using fluorescent dye. Befitting the negative charge of the siRNA, loading of the particle with siRNA decreased the surface charge of the particle from -2.7±1.9 mV to -15.2±4.8 mV. The particles were found to be stable at 4 °C, able to knockdown Bcl-2 in KB cells, and capable of inducing apoptosis in treated cells.
Also using the cholate dialysis synthesis strategy, Shahzad et al. formulated and delivered siRNA. The investigators generated rHDL using a lipid mixture of cholesterol, cholesteryl ester, and phosphatidylcholine along with a protein component of apolipoprotein A-I.82 With this particle, siRNA antisense to STAT3 and FAK were formulated. The resulting complexes had a neutral surface charge and had spherical geometry. Data further demonstrate knockdown of the target STAT3 gene. Immunohistochemistry experiments done in a tumor model demonstrated the expected antitumor phenotype: less proliferation, less angiogenesis, and more apoptosis. Ding et al. used a similar approach to knockdown Pokemon by incorporating cholesterol-conjugated siRNA into a cholate dialysis based synthesis.83 Knockdown was achieved; characterization data show that the constructs generated were relatively large at approximately 90 nm in diameter.
Finally, Dong et al. in a related approach generated and tested a suite of 103 lipopeptide nanoparticles, so named because the key molecule in these self-assembling structures have lipid tails conjugated to amino acids, peptides, and polypeptide head groups.79 A lead material, cKK-E12 was found to be extremely potent and specific for effecting knockdown in hepatocytes. The addition of apoE3 greatly potentiated knockdown. Interestingly, the mechanism of uptake of these particles into cells appears to be through macropinocytosis.
VI. Inherently therapeutic HDL constructs: Theralivery
It is important to note that HDL mimicking nanostructures of all geometries, being biomimetics, have the potential for intrinsic therapeutic effect. Reconstituted HDL in discoidal form has been tested in animal as well as human models of atherosclerosis.84 Numata et al. demonstrated that nanodisks containing the pulmonary surfactant palmitoyloleoylphosphatidylglycerol (POPG) greatly reduced the infection rate of respiratory syncytial virus.85 Our laboratory has demonstrated that gold core HDL NP are capable of killing lymphoma cells, most likely through a mechanism involving gold core HDL NP binding to the SR-B1 receptor and abrogating normal cholesterol handling.86 This paradigm of drug delivery vehicles with intrinsic therapeutic effect might be termed “theralivery.”
VII. Challenges and Future Directions
HDL-like particles are not the only nanoparticles that have been employed for the purpose of drug delivery. A number of liposome-based drugs exist as well. HDL-like nanoparticles share many of the advantages liposomes exhibit with respect to drug delivery. Perhaps the most important benefits of using HDL-like nanoparticles are their inherent biocompatibility and active targeting properties.87
Since the development of rHDL - arguably the first nanotechnology platform - multiple advances have continued to drive the field of HDL-based drug delivery forward. As the preceding discussion demonstrates, important strides have been made in formulating particles of both discoidal and spherical geometries. Much progress has been made in controlling the size of these particles, a feature critically important for delivery of payload to the cytoplasm and avoidance of sequestration by the endosomal system. A wide variety of molecules has been bound to HDL - including small molecules, highly lipophilic drugs, and siRNA.
The major challenge facing the field is the ability to pilot multiple iterations of particles to find the best platform for a given application. Each synthetic HDL is composed of multiple molecules that may combine in a nearly limitless fashion. Finding the ideal particle to formulate the drug and achieve targeting may require iteration. A second major challenge to the field - and all drug delivery platforms in general - is how to confer specificity. Tailoring the composition of the nanoparticle would help; adding ligands for specific targeted receptors may be a further approach. A third major challenge is ensuring safety of these biomimetics to off target cells. A priori it would seem that as a biomimetic, HDL like drug delivery carriers would benefit from some degree of immunologic privilege. Data so far are promising regarding the safety of this material; ongoing studies will be needed to define the safety more precisely.
In summary, naturally occurring HDL, discoidal HDL biomimetics, and spherical HDL biomimetics have all been used to deliver drugs. With continued work in the field and further refinement of the techniques, HDL-based approaches hold great promise.
Table 2. Classes of HDL-Inspired Delivery Platforms and Potential Payloads.
HDL-inspired delivery platforms fall broadly into three categories: natural HDL itself, discoidal HDL analogs (termed reconstituted HDL or rHDL), and spheroidal HDL analogs. Payloads that have been tested include small molecules and oligonucleotides.
| Small Molecules | Oligonucleotides | |
|---|---|---|
| Natural HDL | 6, 40-42 | |
| Discoidal HDL | 55-63 | 99, 100 |
| Spherical HDL | 72, 73, 75-77 | 35, 38, 79, 81-83 |
Acknowledgments
For financial support we thank the Howard Hughes Medical Institute (HHMI) for a Physician Scientist Early Career Award, the Department of Defense/Air Force Office of Scientific Research (FA95501310192 and FA9550-11- 1-0275), the National Institutes of Health/National Cancer Institute (U54CA151880 and R01CA167041), and the Dixon Translational Grants Initiative through the Northwestern University Clinical and Translational Sciences Institute.
References
- 1.Camont L, Chapman MJ, Kontush A. Trends in molecular medicine. 2011;17:594–603. doi: 10.1016/j.molmed.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 2.Rosenson RS, Brewer HB, Jr, Davidson WS, Fayad ZA, Fuster V, Goldstein J, Hellerstein M, Jiang XC, Phillips MC, Rader DJ, Remaley AT, Rothblat GH, Tall AR, Yvan-Charvet L. Circulation. 2012;125:1905–1919. doi: 10.1161/CIRCULATIONAHA.111.066589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M. Science. 1996;271:518–520. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- 4.Rosso LG, Lhomme M, Merono T, Sorroche P, Catoggio L, Soriano E, Saucedo C, Malah V, Dauteuille C, Boero L, Lesnik P, Robillard P, Chapman M John, Brites F, Kontush A. Atherosclerosis. 2014;237:652–660. doi: 10.1016/j.atherosclerosis.2014.09.034. [DOI] [PubMed] [Google Scholar]
- 5.Vaisar T, Pennathur S, Green PS, Gharib SA, Hoofnagle AN, Cheung MC, Byun J, Vuletic S, Kassim S, Singh P, Chea H, Knopp RH, Brunzell J, Geary R, Chait A, Zhao XQ, Elkon K, Marcovina S, Ridker P, Oram JF, Heinecke JW. The Journal of clinical investigation. 2007;117:746–756. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. Nature cell biology. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenson RS, Brewer HB, Jr, Chapman MJ, Fazio S, Hussain MM, Kontush A, Krauss RM, Otvos JD, Remaley AT, Schaefer EJ. Clinical chemistry. 2011;57:392–410. doi: 10.1373/clinchem.2010.155333. [DOI] [PubMed] [Google Scholar]
- 8.Kontush A, Chantepie S, Chapman MJ. Arteriosclerosis, thrombosis, and vascular biology. 2003;23:1881–1888. doi: 10.1161/01.ATV.0000091338.93223.E8. [DOI] [PubMed] [Google Scholar]
- 9.Martin SS, Jones SR, Toth PP. Trends in endocrinology and metabolism: TEM. 2014;25:329–336. doi: 10.1016/j.tem.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Pownall HJ, Gillard BK, Gotto AM., Jr Clinical lipidology. 2013;8:551–560. doi: 10.2217/clp.13.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Handa D, Kimura H, Oka T, Takechi Y, Okuhira K, Phillips MC, Saito H. Biochemistry. 2015;54:1123–1131. doi: 10.1021/bi501345j. [DOI] [PubMed] [Google Scholar]
- 12.Smith JD, Le Goff W, Settle M, Brubaker G, Waelde C, Horwitz A, Oda MN. J Lipid Res. 2004;45:635–644. doi: 10.1194/jlr.M300336-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Gillotte KL, Davidson WS, Lund-Katz S, Rothblat GH, Phillips MC. J Lipid Res. 1998;39:1918–1928. [PubMed] [Google Scholar]
- 14.Phillips MC. The Journal of biological chemistry. 2014;289:24020–24029. doi: 10.1074/jbc.R114.583658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segrest JP, Jones MK, Klon AE, Sheldahl CJ, Hellinger M, De Loof H, Harvey SC. The Journal of biological chemistry. 1999;274:31755–31758. doi: 10.1074/jbc.274.45.31755. [DOI] [PubMed] [Google Scholar]
- 16.Davidson WS, Hilliard GM. The Journal of biological chemistry. 2003;278:27199–27207. doi: 10.1074/jbc.M302764200. [DOI] [PubMed] [Google Scholar]
- 17.Glomset JA. J Lipid Res. 1968;9:155–167. [PubMed] [Google Scholar]
- 18.Silva RA, Huang R, Morris J, Fang J, Gracheva EO, Ren G, Kontush A, Jerome WG, Rye KA, Davidson WS. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12176–12181. doi: 10.1073/pnas.0803626105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson WJ, Bamberger MJ, Latta RA, Rapp PE, Phillips MC, Rothblat GH. The Journal of biological chemistry. 1986;261:5766–5776. [PubMed] [Google Scholar]
- 20.Tarling EJ, Edwards PA. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19719–19724. doi: 10.1073/pnas.1113021108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu X, Kozarsky K, Krieger M. The Journal of biological chemistry. 2000;275:29993–30001. doi: 10.1074/jbc.275.39.29993. [DOI] [PubMed] [Google Scholar]
- 22.Ji A, Meyer JM, Cai L, Akinmusire A, de Beer MC, Webb NR, van der Westhuyzen DR. Atherosclerosis. 2011;217:106–112. doi: 10.1016/j.atherosclerosis.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pagler TA, Rhode S, Neuhofer A, Laggner H, Strobl W, Hinterndorfer C, Volf I, Pavelka M, Eckhardt ER, van der Westhuyzen DR, Schutz GJ, Stangl H. The Journal of biological chemistry. 2006;281:11193–11204. doi: 10.1074/jbc.M510261200. [DOI] [PubMed] [Google Scholar]
- 24.Cruz PM, Mo H, McConathy WJ, Sabnis N, Lacko AG. Frontiers in pharmacology. 2013;4:119. doi: 10.3389/fphar.2013.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Beer MC, Durbin DM, Cai L, Mirocha N, Jonas A, Webb NR, de Beer FC, van Der Westhuyzen DR. The Journal of biological chemistry. 2001;276:15832–15839. doi: 10.1074/jbc.M100228200. [DOI] [PubMed] [Google Scholar]
- 26.Nieland TJ, Ehrlich M, Krieger M, Kirchhausen T. Biochimica et biophysica acta. 2005;1734:44–51. doi: 10.1016/j.bbalip.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 27.Thuahnai ST, Lund-Katz S, Anantharamaiah GM, Williams DL, Phillips MC. Journal of lipid research. 2003;44:1132–1142. doi: 10.1194/jlr.M200429-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Liadaki KN, Liu T, Xu S, Ishida BY, Duchateaux PN, Krieger JP, Kane J, Krieger M, Zannis VI. The Journal of biological chemistry. 2000;275:21262–21271. doi: 10.1074/jbc.M002310200. [DOI] [PubMed] [Google Scholar]
- 29.de Beer MC, Durbin DM, Cai L, Jonas A, de Beer FC, van der Westhuyzen DR. Journal of lipid research. 2001;42:309–313. [PubMed] [Google Scholar]
- 30.Fukuda M, Nakano M, Miyazaki M, Handa T. The journal of physical chemistry B. 2010;114:8228–8234. doi: 10.1021/jp101071t. [DOI] [PubMed] [Google Scholar]
- 31.Jayaraman S, Cavigiolio G, Gursky O. The Biochemical journal. 2012;442:703–712. doi: 10.1042/BJ20111831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehta R, Gantz DL, Gursky O. Journal of molecular biology. 2003;328:183–192. doi: 10.1016/s0022-2836(03)00155-4. [DOI] [PubMed] [Google Scholar]
- 33.Cuhadar S, Koseoglu M, Atay A, Dirican A. Biochemia medica. 2013;23:70–77. doi: 10.11613/BM.2013.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mackey RH, Greenland P, Goff DC, Jr, Lloyd-Jones D, Sibley CT, Mora S. Journal of the American College of Cardiology. 2012;60:508–516. doi: 10.1016/j.jacc.2012.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMahon KM, Mutharasan RK, Tripathy S, Veliceasa D, Bobeica M, Shumaker DK, Luthi AJ, Helfand BT, Ardehali H, Mirkin CA, Volpert O, Thaxton CS. Nano letters. 2011;11:1208–1214. doi: 10.1021/nl1041947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perrault SD, Walkey C, Jennings T, Fischer HC, Chan WC. Nano letters. 2009;9:1909–1915. doi: 10.1021/nl900031y. [DOI] [PubMed] [Google Scholar]
- 37.Skajaa T, Zhao Y, van den Heuvel DJ, Gerritsen HC, Cormode DP, Koole R, van Schooneveld MM, Post JA, Fisher EA, Fayad ZA, de Mello Donega C, Meijerink A, Mulder WJ. Nano letters. 2010;10:5131–5138. doi: 10.1021/nl1037903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang M, Jin H, Chen J, Ding L, Ng KK, Lin Q, Lovell JF, Zhang Z, Zheng G. Small. 2011;7:568–573. doi: 10.1002/smll.201001589. [DOI] [PubMed] [Google Scholar]
- 39.Thompson PD, Cullinane EM, Sady SP, Flynn MM, Chenevert CB, Herbert PN. Circulation. 1991;84:140–152. doi: 10.1161/01.cir.84.1.140. [DOI] [PubMed] [Google Scholar]
- 40.Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, Rajeev KG, Nakayama T, Charrise K, Ndungo EM, Zimmermann T, Koteliansky V, Manoharan M, Stoffel M. Nature biotechnology. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 41.Kuwahara H, Nishina K, Yoshida K, Nishina T, Yamamoto M, Saito Y, Piao W, Yoshida M, Mizusawa H, Yokota T. Molecular therapy : the journal of the American Society of Gene Therapy. 2011;19:2213–2221. doi: 10.1038/mt.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uno Y, Piao W, Miyata K, Nishina K, Mizusawa H, Yokota T. Human gene therapy. 2011;22:711–719. doi: 10.1089/hum.2010.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bijsterbosch MK, Van Berkel TJ. Molecular pharmacology. 1992;41:404–411. [PubMed] [Google Scholar]
- 44.Scanu A. The Journal of biological chemistry. 1967;242:711–719. [PubMed] [Google Scholar]
- 45.Forte TM, Nichols AV, Gong EL, Levy RI, Lux S. Biochimica et biophysica acta. 1971;248:381–386. doi: 10.1016/0005-2760(71)90026-9. [DOI] [PubMed] [Google Scholar]
- 46.Hauser H, Henry R, Leslie RB, Stubbs J. European journal of biochemistry / FEBS. 1974;48:583–594. doi: 10.1111/j.1432-1033.1974.tb03801.x. [DOI] [PubMed] [Google Scholar]
- 47.Matz CE, Jonas A. The Journal of biological chemistry. 1982;257:4535–4540. [PubMed] [Google Scholar]
- 48.Jonas A. Methods in enzymology. 1986;128:553–582. doi: 10.1016/0076-6879(86)28092-1. [DOI] [PubMed] [Google Scholar]
- 49.Bricarello DA, Smilowitz JT, Zivkovic AM, German JB, Parikh AN. ACS nano. 2011;5:42–57. doi: 10.1021/nn103098m. [DOI] [PubMed] [Google Scholar]
- 50.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Journal of the American Chemical Society. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 51.Bayburt TH, Carlson JW, Sligar SG. Journal of structural biology. 1998;123:37–44. doi: 10.1006/jsbi.1998.4007. [DOI] [PubMed] [Google Scholar]
- 52.Carlson JW, Jonas A, Sligar SG. Biophysical journal. 1997;73:1184–1189. doi: 10.1016/S0006-3495(97)78150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grinkova YV, Denisov IG, Sligar SG. Protein engineering, design & selection : PEDS. 2010;23:843–848. doi: 10.1093/protein/gzq060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shih AY, Denisov IG, Phillips JC, Sligar SG, Schulten K. Biophysical journal. 2005;88:548–556. doi: 10.1529/biophysj.104.046896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oda MN, Hargreaves PL, Beckstead JA, Redmond KA, van Antwerpen R, Ryan RO. Journal of lipid research. 2006;47:260–267. doi: 10.1194/jlr.D500033-JLR200. [DOI] [PubMed] [Google Scholar]
- 56.Nelson KG, Bishop JV, Ryan RO, Titus R. Antimicrobial agents and chemotherapy. 2006;50:1238–1244. doi: 10.1128/AAC.50.4.1238-1244.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Redmond KA, Nguyen TS, Ryan RO. International journal of pharmaceutics. 2007;339:246–250. doi: 10.1016/j.ijpharm.2007.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh AT, Evens AM, Anderson RJ, Beckstead JA, Sankar N, Sassano A, Bhalla S, Yang S, Platanias LC, Forte TM, Ryan RO, Gordon LI. British journal of haematology. 2010;150:158–169. doi: 10.1111/j.1365-2141.2010.08209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh AT, Ghosh M, Forte TM, Ryan RO, Gordon LI. Leukemia & lymphoma. 2011;52:1537–1543. doi: 10.3109/10428194.2011.584253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lou B, Liao XL, Wu MP, Cheng PF, Yin CY, Fei Z. World journal of gastroenterology. 2005;11:954–959. doi: 10.3748/wjg.v11.i7.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bellosta S, Paoletti R, Corsini A. Circulation. 2004;109:III50–57. doi: 10.1161/01.CIR.0000131519.15067.1f. [DOI] [PubMed] [Google Scholar]
- 62.Duivenvoorden R, Tang J, Cormode DP, Mieszawska AJ, Izquierdo-Garcia D, Ozcan C, Otten MJ, Zaidi N, Lobatto ME, van Rijs SM, Priem B, Kuan EL, Martel C, Hewing B, Sager H, Nahrendorf M, Randolph GJ, Stroes ES, Fuster V, Fisher EA, Fayad ZA, Mulder WJ. Nature communications. 2014;5:3065. doi: 10.1038/ncomms4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang J, Lobatto ME, Hassing L, van der Staay S, van Rijs SM, Calcagno C, Braza MS, Baxter S, Fay F, Sanchez-Gaytan BL, Duivenvoorden R, Sager H, Astudillo YM, Leong W, Ramachandran S, Storm G, Perez-Medina C, Reiner T, Cormode DP, Strijkers GJ, Stroes ES, Swirski FK, Nahrendorf M, Fisher EA, Fayad ZA, Mulder WJ. Science advances. 2015;1 doi: 10.1126/sciadv.1400223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Iovannisci DM, Beckstead JA, Ryan RO. Biochemical and biophysical research communications. 2009;379:466–469. doi: 10.1016/j.bbrc.2008.12.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Danilo C, Gutierrez-Pajares JL, Mainieri MA, Mercier I, Lisanti MP, Frank PG. Breast cancer research : BCR. 2013;15:R87. doi: 10.1186/bcr3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Twiddy AL, Cox ME, Wasan KM. The Prostate. 2012;72:955–965. doi: 10.1002/pros.21499. [DOI] [PubMed] [Google Scholar]
- 67.Asztalos BF, Cupples LA, Demissie S, Horvath KV, Cox CE, Batista MC, Schaefer EJ. Arteriosclerosis, thrombosis, and vascular biology. 2004;24:2181–2187. doi: 10.1161/01.ATV.0000146325.93749.a8. [DOI] [PubMed] [Google Scholar]
- 68.Asztalos BF, Collins D, Cupples LA, Demissie S, Horvath KV, Bloomfield HE, Robins SJ, Schaefer EJ. Arteriosclerosis, thrombosis, and vascular biology. 2005;25:2185–2191. doi: 10.1161/01.ATV.0000183727.90611.4f. [DOI] [PubMed] [Google Scholar]
- 69.Rye KA, Barter PJ. The Journal of biological chemistry. 1994;269:10298–10303. [PubMed] [Google Scholar]
- 70.Jonas A, Kezdy KE, Williams MI, Rye KA. J Lipid Res. 1988;29:1349–1357. [PubMed] [Google Scholar]
- 71.Rye KA, Hime NJ, Barter PJ. The Journal of biological chemistry. 1995;270:189–196. doi: 10.1074/jbc.270.1.189. [DOI] [PubMed] [Google Scholar]
- 72.McConathy WJ, Nair MP, Paranjape S, Mooberry L, Lacko AG. Anti-cancer drugs. 2008;19:183–188. doi: 10.1097/CAD.0b013e3282f1da86. [DOI] [PubMed] [Google Scholar]
- 73.Sabnis N, Nair M, Israel M, McConathy WJ, Lacko AG. International journal of nanomedicine. 2012;7:975–983. doi: 10.2147/IJN.S28029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sabnis N, Pratap S, Akopova I, Bowman PW, Lacko AG. Frontiers in pediatrics. 2013;1:6. doi: 10.3389/fped.2013.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mooberry LK, Nair M, Paranjape S, McConathy WJ, Lacko AG. Journal of drug targeting. 2010;18:53–58. doi: 10.3109/10611860903156419. [DOI] [PubMed] [Google Scholar]
- 76.Zhang Z, Chen J, Ding L, Jin H, Lovell JF, Corbin IR, Cao W, Lo PC, Yang M, Tsao MS, Luo Q, Zheng G. Small. 2010;6:430–437. doi: 10.1002/smll.200901515. [DOI] [PubMed] [Google Scholar]
- 77.Zhang X, Chen B. Cancer letters. 2010;298:26–33. doi: 10.1016/j.canlet.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 78.Franceschini G, Sirtori CR, Capurso A, 2nd, Weisgraber KH, Mahley RW. The Journal of clinical investigation. 1980;66:892–900. doi: 10.1172/JCI109956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dong Y, Love KT, Dorkin JR, Sirirungruang S, Zhang Y, Chen D, Bogorad RL, Yin H, Chen Y, Vegas AJ, Alabi CA, Sahay G, Olejnik KT, Wang W, Schroeder A, Lytton-Jean AK, Siegwart DJ, Akinc A, Barnes C, Barros SA, Carioto M, Fitzgerald K, Hettinger J, Kumar V, Novobrantseva TI, Qin J, Querbes W, Koteliansky V, Langer R, Anderson DG. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:3955–3960. doi: 10.1073/pnas.1322937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thaxton CS, Daniel WL, Giljohann DA, Thomas AD, Mirkin CA. Journal of the American Chemical Society. 2009;131:1384–1385. doi: 10.1021/ja808856z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tripathy S, Vinokour E, McMahon KM, Volpert OV, Thaxton CS. Particle & particle systems characterization : measurement and description of particle properties and behavior in powders and other disperse systems. 2014;31:1141–1150. doi: 10.1002/ppsc.201400036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shahzad MM, Mangala LS, Han HD, Lu C, Bottsford-Miller J, Nishimura M, Mora EM, Lee JW, Stone RL, Pecot CV, Thanapprapasr D, Roh JW, Gaur P, Nair MP, Park YY, Sabnis N, Deavers MT, Lee JS, Ellis LM, Lopez-Berestein G, McConathy WJ, Prokai L, Lacko AG, Sood AK. Neoplasia. 2011;13:309–319. doi: 10.1593/neo.101372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ding Y, Wang W, Feng M, Wang Y, Zhou J, Ding X, Zhou X, Liu C, Wang R, Zhang Q. Biomaterials. 2012;33:8893–8905. doi: 10.1016/j.biomaterials.2012.08.057. [DOI] [PubMed] [Google Scholar]
- 84.Tardif JC, Gregoire J, L'Allier PL, Ibrahim R, Lesperance J, Heinonen TM, Kouz S, Berry C, Basser R, Lavoie MA, Guertin MC, Rodes-Cabau J. JAMA : the journal of the American Medical Association. 2007;297:1675–1682. doi: 10.1001/jama.297.15.jpc70004. [DOI] [PubMed] [Google Scholar]
- 85.Numata M, Grinkova YV, Mitchell JR, Chu HW, Sligar SG, Voelker DR. International journal of nanomedicine. 2013;8:1417–1427. doi: 10.2147/IJN.S39888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang S, Damiano MG, Zhang H, Tripathy S, Luthi AJ, Rink JS, Ugolkov AV, Singh AT, Dave SS, Gordon LI, Thaxton CS. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2511–2516. doi: 10.1073/pnas.1213657110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Akbarzadeh A, Rezaei-Sadabady R, Davaran S, Joo SW, Zarghami N, Hanifehpour Y, Samiei M, Kouhi M, Nejati-Koshki K. Nanoscale research letters. 2013;8:102. doi: 10.1186/1556-276X-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ng KK, Lovell JF, Zheng G. Accounts of chemical research. 2011;44:1105–1113. doi: 10.1021/ar200017e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Webb NR, Cai L, Ziemba KS, Yu J, Kindy MS, van der Westhuyzen DR, de Beer FC. Journal of lipid research. 2002;43:1890–1898. doi: 10.1194/jlr.m200173-jlr200. [DOI] [PubMed] [Google Scholar]
- 90.England CG, Priest T, Zhang G, Sun X, Patel DN, McNally LR, van Berkel V, Gobin AM, Frieboes HB. International journal of nanomedicine. 2013;8:3603–3617. doi: 10.2147/IJN.S51668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fischer NO, Weilhammer DR, Dunkle A, Thomas C, Hwang M, Corzett M, Lychak C, Mayer W, Urbin S, Collette N, Chiun Chang J, Loots GG, Rasley A, Blanchette CD. PloS one. 2014;9:e93342. doi: 10.1371/journal.pone.0093342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rui M, Tang H, Li Y, Wei X, Xu Y. Pharmaceutical research. 2013;30:1203–1214. doi: 10.1007/s11095-012-0957-4. [DOI] [PubMed] [Google Scholar]
- 93.Corbin IR, Chen J, Cao W, Li H, Lund-Katz S, Zheng G. The Journal of Biomedical Nanotechnology. 2007;3:367–376. [Google Scholar]
- 94.Matsumura Y, Maeda H. Cancer research. 1986;46:6387–6392. [PubMed] [Google Scholar]
- 95.Maeda H, Matsumura Y. Critical reviews in therapeutic drug carrier systems. 1989;6:193–210. [PubMed] [Google Scholar]
- 96.Sabnis N, Lacko AG. Therapeutic delivery. 2012;3:599–608. doi: 10.4155/tde.12.41. [DOI] [PubMed] [Google Scholar]
- 97.Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad OC. Advanced drug delivery reviews. 2014;66:2–25. doi: 10.1016/j.addr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huynh E, Zheng G. Wiley interdisciplinary reviews Nanomedicine and nanobiotechnology. 2013;5:250–265. doi: 10.1002/wnan.1217. [DOI] [PubMed] [Google Scholar]
- 99.Nakayama T, Butler JS, Sehgal A, Severgnini M, Racie T, Sharman J, Ding F, Morskaya SS, Brodsky J, Tchangov L, Kosovrasti V, Meys M, Nechev L, Wang G, Peng CG, Fang Y, Maier M, Rajeev KG, Li R, Hettinger J, Barros S, Clausen V, Zhang X, Wang Q, Hutabarat R, Dokholyan NV, Wolfrum C, Manoharan M, Kotelianski V, Stoffel M, Sah DW. Molecular therapy : the journal of the American Society of Gene Therapy. 2012;20:1582–1589. doi: 10.1038/mt.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ghosh M, Ren G, Simonsen JB, Ryan RO. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2014;92:200–205. doi: 10.1139/bcb-2014-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]