Abstract
The production of bacterial polyesters, polyhydroxyalkanoates (PHAs), has been improved by several rational approaches such as overexpression and/or engineering of the enzymes directly related to PHA biosynthetic pathways. In this study, a new approach at transcription level has been applied to a new category of the copolymer of lactate (LA) and 3-hydroxybutyrate (3HB), P(LA-co-3HB). When the 4 disrupting mutants of sigma factors in Escherichia coli, rpoN, rpoS, fliA, fecI, were used as platforms for production of P(LA-co-3HB), increases in the production level and LA fraction of the copolymer were observed for the mutant strain with rpoN disruption. These positive impacts on the polymer production were caused in an “indirect manner” via changes in the multiple genes governed by RpoN. A genome-wide engineering by sigma factors would be a versatile approach for the production of value-added products of interest and available for combination with the other beneficial tools.
Keywords: biobased plastic, Escherichia coli, polyhydroxyalkanoate, polyhydroxybutyrate, poly(lactic acid), sigma factor
Polyhydroxyalkanoate (PHA) is an attractive material that can be developed as a bio-based, biodegradable and biocompatible plastic applicable for environmental and medical applications.1 Compared to the fermentation management of natural PHA producers, PHA can be more efficiently synthesized in the recombinant microbial system by installing biosynthetic pathways for the production of both natural and tailor-made PHAs.2 The material properties of PHA should be governed by its monomeric composition, molecular weight and copolymer microstructure.3
The poly[lactate (LA)-co-3-hydroxybutyrate (3HB)], a new type of LA-based polymer, was synthesized by a microbial system carrying an LA-polymerizing enzyme (LPE).4,5 An interesting feature of the copolymer over PLA is that the variation of the LA/3HB ratio in P(LA-co-3HB) has demonstrated the generation of polymers with different properties. The increase of P(LA-co-3HB) production level and the control of LA fraction in copolymer is very important problem.
The system of bioplastics production using microorganisms has been evolved through the following process. First, researchers explored the optimization of cultivation conditions of bioplastic natural producing strains (first generation).6,7 Subsequently, the productivity of bioplastic was more increased by using a recombinant Escherichia coli system (second generation).8,9 Furthermore, the bioplastic production is enhanced by applying metabolic engineering and molecular alteration (enzyme evolution engineering) of the enzymes involved in the bioplastics production (third generation).10
So far, the P(LA-co-3HB) production was carried out by introducing bioplastics production enzymes in various E. coli species. However, the production level and the LA fraction of P(LA-co-3HB) varied depending on the species. This is most likely because of the difference of small genotype of various E. coli strain. In other words, it is likely that minor differences in the genes encoded by the E. coli genome have some effect on P(LA-co-3HB) production and LA fraction. These results show that the genes encoded by the E. coli genome, individually or in group, are involved in P(LA-co-3HB) productivity and LA fraction.
In the present study, we focused on the sigma factors that globally control the transcription of genes. It has been reported that E. coli possesses 4 non-essential sigma factors, RpoN, RpoS, FliA and FecI.11-13 RpoS is induced at stationary phase,14 while RpoN is activated under the nitrogen starvation conditions.15 The function of FliA is the transcriptional regulator of flagellar and chemotaxis genes.16 FecI is recognized as a member of the extracytoplasmic function subfamily of sigma factor.17 The disruption of these sigma factors was expectedly found to reduce or enhance the expression levels of a broad range of genes.13 Our plan is to observe the effects on the deletion of the sigma factor at P(LA-co-3HB) biosynthesis.
Result and Discussion
The sigma factor disrupted strains of E. coli, ΔrpoS, ΔrpoN, ΔfliA and ΔfecI, were obtained from the Keio collection and used in this study.18 The E. coli BW25113 was used as the parent strain. A plasmid pTV118NpctC1(STQK)AB which contains the P(LA-co-3HB) biosynthesis genes encoding propionyl-CoA transferase (PCT), phaC1 (STQK), phaA and phaB was introduced into these mutant strains. LB medium containing 2% glucose and 10 mM calcium panthothenate was used for P(LA-co-3HB) production. Cultivation was performed at 30°C for 48 hours with shaking at 180 rpm.19
Table 1 shows the production of P(LA-co-3HB) in the 4 sigma factor disrupted strains. In terms of the yield and LA fraction of P(LA-co-3HB), varied values were observed among the 4 mutant strains. It should be noted that the ΔrpoN showed the increases in both the yield and LA fraction of P(LA-co-3HB). The two mutants, the ΔrpoS and ΔfecI showed the increased P(LA-co-3HB) yield, although the LA fraction was decreased.
Table 1.
P(LA-co-3HB) production by E. coli BW25113 and sigma factor deleted strains
| Polymer production (g/l) |
|||||||
|---|---|---|---|---|---|---|---|
| Strain | Genotype | Cell dry weight (g/l) | Total | LA | 3HB | Polymer content (wt%) | LA fraction (mol%) |
| BW25113 | Parent | 9.1 ± 0.4 | 5.3 ± 0.2 | 0.9 ± 0.0 | 4.5 ± 0.2 | 58.3 ± 2.2 | 18.6 ± 0.9 |
| JW1907 | ΔfliA | 8.6 ± 1.3 | 4.9 ± 0.6 | 0.8 ± 0.1 | 4.2 ± 0.7 | 57.2 ± 2.0 | 18.4 ± 4.2 |
| JW3169 | ΔrpoN | 8.2 ± 0.8 | 6.2 ± 0.4 | 1.4 ± 0.1 | 4.8 ± 0.4 | 75.1 ± 3.3 | 26.2 ± 2.5 |
| JW4253 | ΔfecI | 10.7 ± 0.3 | 5.7 ± 0.2 | 0.2 ± 0.1 | 5.6 ± 0.2 | 53.5 ± 2.8 | 3.2 ± 1.4 |
| JW5437 | ΔrpoS | 10.0 ± 0.1 | 5.8 ± 0.1 | 0.6 ± 0.1 | 5.2 ± 0.0 | 57.6 ± 0.9 | 12.3 ± 1.4 |
| JW3169/ pCA24N a | ΔrpoN / Plac | 8.0 ± 0.2 | 6.2 ± 0.3 | 1.3 ± 0.1 | 4.9 ± 0.2 | 77.7 ± 2.2 | 24.4 ± 1.2 |
| JW3169/ pCA24N-rpoNa | ΔrpoN/ Plac::rpoN | 7.2 ± 0.5 | 5.0 ± 0.1 | 0.7 ± 0.1 | 4.3 ± 0.0 | 70.0 ± 4.3 | 15.8 ± 1.4 |
pCA24N: empty vector of ASKA clone.
JW3169 / pCA24N: empty vector introduced into JW3169 (rpoN deletion strain).
pCA24N-rpoN: rpoN gene cloned downstream of Plac in pCA24N.
JW3169 / pCA24N-rpoN: rpoN overexpression plasmid introduced into JW3169 (rpoN deletion strain).
100 μM of IPTG added.
To clarify the effect of rpoN disruption on P(LA-co-3HB) production, complementary experiment was performed. An expression vector of rpoN, pCA24N-rpoN which was extracted from JW3169 of ASKA clone collection,20 was introduced into the ΔrpoN strain. The P(LA-co-3HB) production level of the strain complemented with pCA24N-rpoN showed almost the same with that of the parent strain (Table 1). This result indicated the positive effect on P(LA-co-3HB) production by the rpoN disruption.
Next, we measured the time course profile of the glucose consumption and P(LA-co-3HB) production. As shown in Figure 1, an early stage of the cultivation at 12 h, the parent strain showed the higher P(LA-co-3HB) production than the ΔrpoN strain. At 18 h, both strain produced comparable level of P(LA-co-3HB) (Fig. 1A). Here, it should be noted that the LA fraction in P(LA-co-3HB) was higher than that of the parent strain. The accumulation of the LA unit into P(LA-co-3HB) was accelerated during the early stage of cultivation, while the accumulation of 3HB unit of P(LA-co-3HB) was increased rather than middle to late stage of cultivation. This P(LA-co-3HB) production profile was clearly different from the both strains. After 48 hours cultivation, glucose consumption was same level in both strains (Fig. 1B). The carbon yield (Ypol/C) in P(LA-co-3HB) synthesis from glucose was calculated to be 0.31 g/g for the ΔrpoN strain which is higher than that for the parent strain 0.26 g/g. The production of lactic acid concentration in the medium in the ΔrpoN strain was higher than that of the parent strain (Fig. 1C). The increased LA unit in P(LA-co-3HB) seemed to be correlated to the lactic acid production level.
Figure 1.
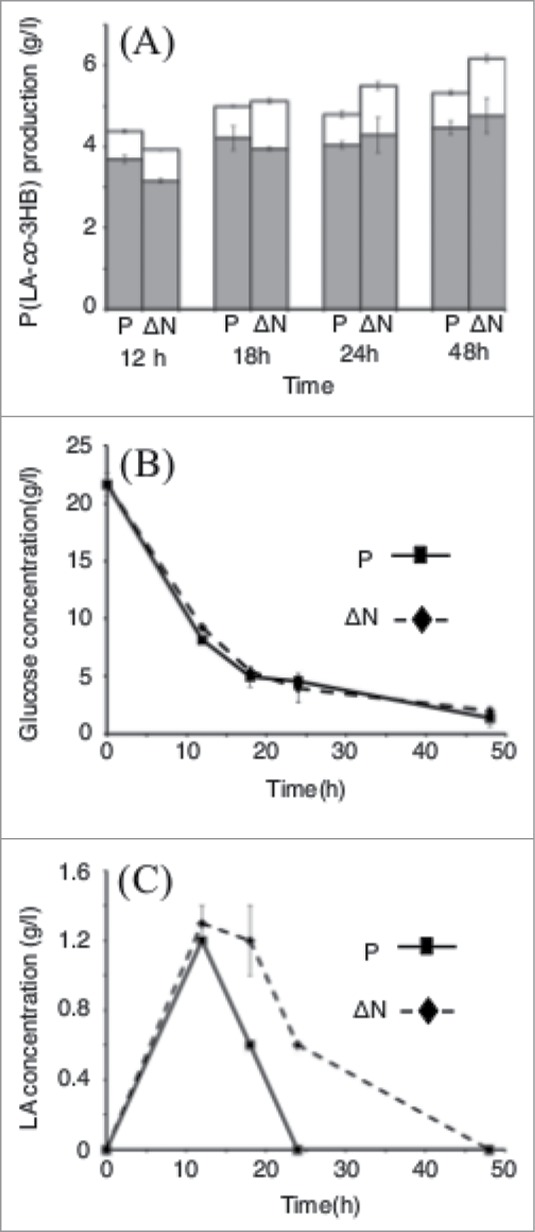
Time course profiles of the P(LA-co-3HB) production (A), amount for glucose (B) and lactic acid (C) in E. coli BW25113 (parent strain) and ΔrpoN (JW3169), respectively. (A) Gray, 3HB units in the copolymers; white, LA units in the copolymers. P: BW25113 (parent strain) and N: ΔrpoN (JW3169). (B) and (C) square and solid line, BW25113; diamond and broken line, ΔrpoN (JW3169).
To interpret the increase in lactic acid production by rpoN deletion, we searched a direct link between rpoN and lactic acid production. RpoN was proposed to regulated 74 genes based on the transcriptional assay of rpoN deleted and overexpressed strains.21 In addition, we added 45 genes, which are regulated in the same operon of the 74 genes based on the operon database “DOOR2”, to the candidates.22 As the result, no gene was found to be related to lactic acid production according to the COGs category (http://www.ncbi.nlm.nih.gov/COG/) (Table 2) and KEGG pathway (http://www.genome.jp/). Therefore, rpoN deletion presumably influenced lactic acid production in an indirect manner.
Table 2.
The number of genes controlled by RpoN
| COG category | Gene number |
|---|---|
| C: Energy production and conversion | 24 / 286 |
| E: Amino acid transport and metabolism | 23 / 362 |
| F: Nucleotide transport and metabolism | 2 / 98 |
| G: Carbohydrate transport and metabolism | 5 / 378 |
| H: Coenzyme metabolism | 1 / 157 |
| I: Lipid metabolism | 4 / 100 |
| J: Translation, ribosomal structure and biogenesis | 1 / 185 |
| K: Transcription | 8 / 311 |
| L: DNA replication, recombination and repair | 1 / 234 |
| M: Cell envelope biogenesis, outer membrane | 1 / 236 |
| N: Cell motility and secretion | 4 / 114 |
| O: Posttranslational modification, protein turnover, chaperones | 11 / 139 |
| P: Inorganic Ion Transport and Metabolism | 11 / 221 |
| Q: Secondary metabolites biosynthesis, transport and catabolism | 2 / 64 |
| R: General function prediction only | 11 / 408 |
| T: Signal transduction mechanisms | 2 / 180 |
| U: Intracellular trafficking and secretion | 1 / 130 |
| V: Defense mechanisms | 1 / 49 |
| S: Function unknown | 5 / 324 |
| No COG assignment | 12 / 614 |
Genes are classified into COG categories. The numbers represent the number of RpoN-controlled genes to the number of genes belonging to each of COG category.
In order to study the change of expression level of enzymes involved in bioplastic synthesis in the rpoN strain, intracellular protein levels of PCT, PhaA, PhaB and lactate dehydrogenase (LDH) in P(LA-co-3HB) production stage at 18 h were compared between the ΔrpoN strain and the parent strain by immunoblotting. The expression levels of these proteins were comparable in both strains (Fig. 2). These results indicated that the increased production level and LA fraction of the P(LA-co-3HB) in the ΔrpoN strain did not depend on the expression level of P(LA-co-3HB) biosynthetic enzymes.
Figure 2.
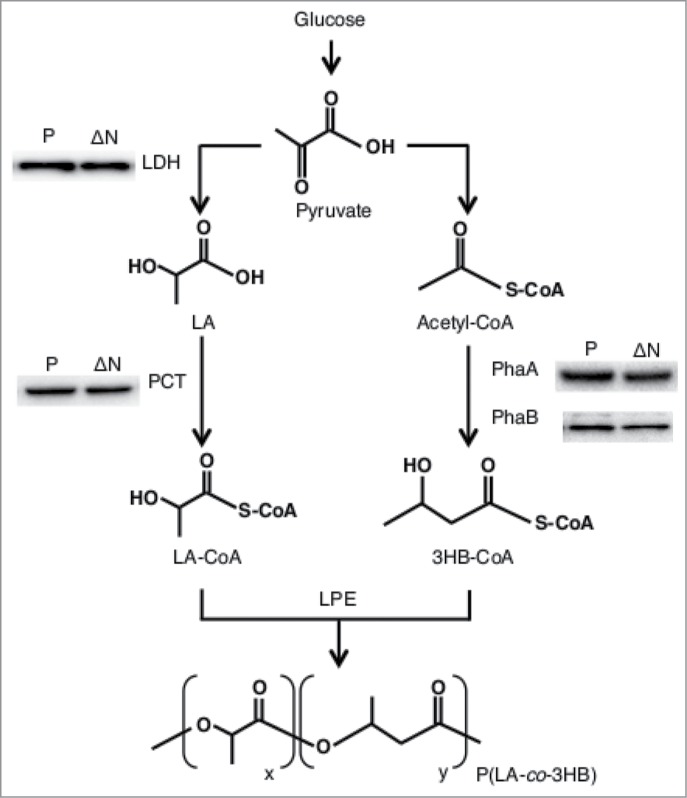
P(LA-co-3HB) synthesis pathway and immunoblot analysis of the polymer synthetic enzymes and LDH. Total protein was extracted from the BW25113 (parent strain) and ΔrpoN (JW3169) grown on 1.7 ml of LB media. Immnoblot analysis of the polymer biosynthesis enzymes (PCT, PhaA and PhaB) and LDH in the parent (P) and the ΔrpoN (ΔN) strain.
In other experiment, the overexpression of sigma factor SigE in Synechocystis sp. PCC 6803 is known to increase the production of P(3HB) under the condition of nitrogen starvation.23 The sigE overexpression elevates the levels of proteins implicated in glycogen catabolism, the oxidative pentose phosphate pathway, and polyhydroxybutyrate (PHB) biosynthesis. We think that the change of the transcriptional network by disruption of sigma factor RpoN has given a positive indirectly effect to the polymer production. The alterations in the expression of the genes governed by the RpoN seem to directly and/or indirectly modulate the carbon fluxes related to the polymer biosynthesis.
In our previous studies, xylose was found to contribute to the increased LA fraction compared to glucose as a carbon source.19,24 Therefore, in this study we attempted to examine the synergistic effect of xylose utilization also on the present mutant strain carrying ΔrpoN. The LA fraction of P(LA-co-3HB) in the ΔrpoN strain using xylose as a carbon source was 33.9 mol% (Table 3). This value is higher than that of obtained the LA fraction of copolymer using glucose cultivation (26.2 mol%). However, this value was same as the LA fraction of the parent strain using xylose cultivation (35.9 mol%). These results suggested the positive effect on the LA fraction of LA-co-3HB), but the synergistic effect of xylose utilization and rpoN disruption was not observed.
Table 3.
The LA fraction of P(LA-co-3HB) in the ΔrpoN strain and the parent strain using xylose or glucose as a carbon source
| Strain | Genotype | Carbon source | LA fraction of polymer (mol%) |
|---|---|---|---|
| JW3169 | ΔrpoN | Glucose | 26.2 |
| JW3169 | ΔrpoN | Xylose | 33.9 |
| BW25113 | parent | Glucose | 18.6 |
| BW25113 | parent | Xylose | 35.9 |
Conclusion and Further Perspective
Our experimental results suggested that the disruption of rpoN indirectly contributed to the increased in P(LA-co-3HB) production. The enhanced polymer production and LA fraction by the ΔrpoN strain may be attributed to the effect of the individual RpoN-regulated genes and/or their combinations. In order to resolve this issue, we plan to check the polymer production using the single gene deletion mutant collection (KEIO collection).
On the other hand, we have discovered that the gene relevant to high production of polymer exists besides rpoN disruptant. The deletion of mtgA may have some effect on the cell morphology and cell wall construction. Interestingly, the enlargement of the recombinant host cell took place in association with polymer accumulation. This changeable morphological property gives us good advantage in obtaining higher polymer productivity.25 The next issue is to study the possibility of producing more bioplastics by combining the enzyme evolution, metabolic engineering, mtgA deletion (cell morphological change) and rpoN deletion (to suppress the expression of multiple genes).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to the National BioResource Project, Japan, for providing the Keio collection strains and the rpoN-overexpression plasmid in ASKA clone.
Funding
This study was partly supported by the CREST, JST.
References
- 1.Misra SK, Valappil SP, Roy I, Boccaccini AR. Polyhydroxyalkanoate (PHA)/inorganic phase composites for tissue engineering applications. Biomacromolecules 2006; 7:2249-58; PMID:16903667; http://dx.doi.org/ 10.1021/bm060317c [DOI] [PubMed] [Google Scholar]
- 2.Matsumoto K, Taguchi S. Enzyme and metabolic engineering for the production of novel biopolymers: crossover of biological and chemical processes. Curr Opin Biotechnol 2013; 24:1054-60; PMID:23545442; http://dx.doi.org/ 10.1016/j.copbio.2013.02.021 [DOI] [PubMed] [Google Scholar]
- 3.Yamada M, Matsumoto K, Shimizu K, Uramoto S, Nakai T, Shozui F, Taguchi S. Adjustable mutations in lactate (LA)-polymerizing enzyme for the microbial production of LA-based polyesters with tailor-made monomer composition. Biomacromolecules 2010; 11:815-9; PMID:20166718; http://dx.doi.org/ 10.1021/bm901437z [DOI] [PubMed] [Google Scholar]
- 4.Taguchi S, Yamada M, Matsumoto K, Tajima K, Satoh Y, Munekata M, Ohno K, Kohda K, Shimamura T, Kambe H, et al.. A microbial factory for lactate-based polyesters using a lactate-polymerizing enzyme. Proc Natl Acad Sci U S A 2008; 105:17323-7; PMID:18978031; http://dx.doi.org/ 10.1073/pnas.0805653105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shozui F, Sun J, Song Y, Yamada M, Sakai K, Matsumoto K, Takase K, Taguchi S. A new beneficial mutation in pseudomonas sp. 61-3 polyhydroxyalkanoate (PHA) synthase for enhanced cellular content of 3-hydroxybutyrate-based PHA explored using its enzyme homolog as a mutation template. Biosci Biotechnol Biochem 2010; 74:1710-2; PMID:20699564; http://dx.doi.org/ 10.1271/bbb.100224 [DOI] [PubMed] [Google Scholar]
- 6.Anderson AJ, Dawes EA. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev 1990; 54:450-72; PMID:2087222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dawes EA, Senior PJ. The role and regulation of energy reserve polymers in micro-organisms. Adv Microb Physiol 1973; 10:135-266; PMID:4594739; http://dx.doi.org/ 10.1016/S0065-2911(08)60088-0 [DOI] [PubMed] [Google Scholar]
- 8.Peoples OP, Sinskey AJ. Poly-beta-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbC). J Biol Chem 1989; 264:15298-303; PMID:2670936 [PubMed] [Google Scholar]
- 9.Schubert P, Steinbuchel A, Schlegel HG. Cloning of the Alcaligenes eutrophus genes for synthesis of poly-beta-hydroxybutyric acid (PHB) and synthesis of PHB in Escherichia coli. J Bacteriol 1988; 170:5837-47; PMID:2848014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taguchi S, Doi Y. Evolution of polyhydroxyalkanoate (PHA) production system by “enzyme evolution”: successful case studies of directed evolution. Macromol Biosci 2004; 4:146-56; PMID:15468204; http://dx.doi.org/ 10.1002/mabi.200300111 [DOI] [PubMed] [Google Scholar]
- 11.Jishage M, Iwata A, Ueda S, Ishihama A. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J Bacteriol 1996; 178:5447-51; PMID:8808934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jishage M, Ishihama A. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of sigma 70 and sigma 38. J Bacteriol 1995; 177:6832-5; PMID:7592475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wösten MM. Eubacterial sigma-factors. FEMS Microbiol Rev 1998; 22:127-50; PMID:9818380; http://dx.doi.org/ 10.1111/j.1574-6976.1998.tb00364.x [DOI] [PubMed] [Google Scholar]
- 14.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol 1991; 5:49-59; PMID:1849609; http://dx.doi.org/ 10.1111/j.1365-2958.1991.tb01825.x [DOI] [PubMed] [Google Scholar]
- 15.Hirschman J, Wong PK, Sei K, Keener J, Kustu S. Products of nitrogen regulatory genes ntrA and ntrC of enteric bacteria activate glnA transcription in vitro: evidence that the ntrA product is a sigma factor. Proc Natl Acad Sci U S A 1985; 82:7525-9; PMID:2999766; http://dx.doi.org/ 10.1073/pnas.82.22.7525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnosti DN, Chamberlin MJ. Secondary sigma factor controls transcription of flagellar and chemotaxis genes in Escherichia coli. Proc Natl Acad Sci U S A 1989; 86:830-4; PMID:2644646; http://dx.doi.org/ 10.1073/pnas.86.3.830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braun V, Mahren S, Ogierman M. Regulation of the FecI-type ECF sigma factor by transmembrane signalling. Curr Opin Microbiol 2003; 6:173-80; PMID:12732308; http://dx.doi.org/ 10.1016/S1369-5274(03)00022-5 [DOI] [PubMed] [Google Scholar]
- 18.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2006; 2:2006 0008; PMID:16738554; http://dx.doi.org/ 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nduko JM, Matsumoto K, Ooi T, Taguchi S. Effectiveness of xylose utilization for high yield production of lactate-enriched P(lactate-co-3-hydroxybutyrate) using a lactate-overproducing strain of Escherichia coli and an evolved lactate-polymerizing enzyme. Metab Eng 2013; 15:159-66; PMID:23202750; http://dx.doi.org/ 10.1016/j.ymben.2012.11.007 [DOI] [PubMed] [Google Scholar]
- 20.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res 2005; 12:291-9; PMID:16769691; http://dx.doi.org/ 10.1093/dnares/dsi012 [DOI] [PubMed] [Google Scholar]
- 21.Zhao K, Liu M, Burgess RR. Promoter and regulon analysis of nitrogen assimilation factor, sigma54, reveal alternative strategy for E. coli MG1655 flagellar biosynthesis. Nucleic Acids Res 2010; 38:1273-83; PMID:19969540; http://dx.doi.org/ 10.1093/nar/gkp1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dam P, Olman V, Harris K, Su Z, Xu Y. Operon prediction using both genome-specific and general genomic information. Nucleic Acids Res 2007; 35:288-98; PMID:17170009; http://dx.doi.org/ 10.1093/nar/gkl1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osanai T, Numata K, Oikawa A, Kuwahara A, Iijima H, Doi Y, Tanaka K, Saito K, Hirai MY. Increased bioplastic production with an RNA polymerase sigma factor SigE during nitrogen starvation in Synechocystis sp. PCC 6803. DNA Res 2013; 20:525-35; PMID:23861321; http://dx.doi.org/ 10.1093/dnares/dst028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nduko JM, Matsumoto K, Ooi T, Taguchi S. Enhanced production of poly(lactate-co-3-hydroxybutyrate) from xylose in engineered Escherichia coli overexpressing a galactitol transporter. Appl Microbiol Biotechnol 2014; 98:2453-60; PMID:24337250; http://dx.doi.org/ 10.1007/s00253-013-5401-0 [DOI] [PubMed] [Google Scholar]
- 25.Kadoya R, Matsumoto K, Toshihiko O, Taguchi S. MtgA deletion-triggered cell enlargement of Escherichia coli for enhanced intracellular polyester accumulation. PLoS One 2015; 10:e0125163; PMID:26039058; http://dx.doi.org/ 10.1371/journal.pone.0125163 [DOI] [PMC free article] [PubMed] [Google Scholar]


