Abstract
This review focuses on the emerging role of site-specific mutagenesis and chimeragenesis for the functional improvement of proteins in areas where traditional protein engineering methods have been extensively used and practically exhausted. The novel path for the creation of the novel proteins has been created on the farther development of the new structure and sequence optimization algorithms for generating and designing the accurate structure models in result of x-ray crystallography studies of a lot of proteins and their mutant forms. Artificial genetic modifications aim to expand nature's repertoire of biomolecules. One of the most exciting potential results of mutagenesis or chimeragenesis finding could be design of effective diagnostics, bio-therapeutics and biocatalysts. A sampling of recent examples is listed below for the in vivo and in vitro genetically improvement of various binding protein and enzyme functions, with references for more in-depth study provided for the reader's benefit.
Keywords: activity, antibodies, enzymes, epitopes, hybrid proteins, lectins, molecular structure, site-specific mutagenesis, specificity
The idea of design and synthesis of proteins with altered properties through genetic modification was suggested by nature itself.1-8 Genes contain coded information that leads to the production of proteins. Proteins fold up to form 3D molecular architectures that are essential for biological functions. Although the natural proteins perform wide variety of tasks, they appear to use only a limited number of structural types. Based on perception that protein structures is more conserved than sequences, these are used over and over again, being altered through evolution to generate many different functions under environmental constraints.9,10,11,12 Mutations such as amino acid substitution, sequence deletion/insertion or DNA shuffling are selected depending on their impact on the chemical nature of catalytic residues, active site conformation, dynamical flexibility, local packing density, relative solvent accessibility, multimeric complexity, and the protein's ability to fold rapidly and stably.13-16 Site-specific evolutionary rates have been found to be mainly determined by side-chain packing.16 Moreover, the structurally disordered regions often have high conformational flexibility allowing to evolve structure and modulate function in recognition proteins and enzymes.11,17-19 The natural selection at the DNA and RNA level, including splicing and gene duplication or transfer, acting during the protein's production history can also affect protein evolutionary rates and amino acid choice that is as important as expression parameters.18,20,21
The analysis of the natural proteins and their mutants with X-ray crystallography allowed to develop the structure and sequence optimization algorithms for the new protein structure determination, prediction and design generating accurate structure models.22-27 Although the design principles used are learnt from natural proteins, some of the designed protein shapes are completely new and have not been observed in nature yet.28-32
Artificial genetic modifications aim to expand nature's repertoire of biomolecules. One of the most exciting potential results of mutagenesis or chimeragenesis finding could be design of effective diagnostics, bio-therapeutics and biocatalysts.24,28,33,34 Although certain wild type proteins are very effective pharmaceuticals or industrial agents, they generally suffer from heat/cold stability and activity, undesirable specificity/selectivity, foldability or utility problems.
This review focuses on the emerging role for site-specific mutagenesis and chimeragenesis for the functional improvement of proteins in areas where traditional protein engineering has been extensively used, such as chemical modification, modifying fluorescence, functional studies, biophysical analysis. A sampling of recent examples is listed below for the in vivo and in vitro genetically improvement of various binding protein and enzyme functions, with references for more in-depth study provided for the reader's benefit. A highlighting of in silico or experimentally obtained important structural details of the protein's functional domains demonstrates the power and potential of the protein genetic modifications for their application in pharmacology and industry.
Genetically Modified Proteins in Nature
There is sufficient data to study how proteins of living organism adapt to changes in the environment and how this is connected with cell metabolism and its regulation. Several levels of regulation such as transcription, translation, protein stability, enzyme regulation ensure that the response is optimal.35,36 In cells, proteins often occur together with other proteins in protein complexes to fulfill their function. However, at the early stage of evolution the protein's isoforms possessed a broad substrate specificity, high catalytic abilities and/or multifunctionality have evolved through mutation, duplication, and horizontal gene transfer from gene families diversified and promiscuous ancestral proteins under specific stress factors or emergence of new substrates.7,37,38 Instead of forming a post-genomic multi-enzyme complex, the rat peroxisomal multifunctional enzyme (rpMFE1) has 5 domains with different evolutionary histories connected via flexible linker or tightly associated, 3 catalytic activities and 2 active sites (Table 1).6 The N-terminal part of rpMFE1 (domains A and B) catalyzes both a hydratase and an isomerase reaction. The C-terminal part belongs to the 2 tightly associated domain HAD superfamily. The HAD fold consists of an NAD-binding Rossmann-like fold (domain C) plus a dimerization domain (domain D) that extended by one more domain (domain E), whose fold is topologically identical to the domain D. The C-terminal domains (C-E) form the second active site of (3S)-hydroxyacyl-CoA dehydrogenase. The two active sites are separated by a positively charged tunnel possibly facilitating an efficient channeling of substrate from the hydratase/isomerase catalytic site to the dehydrogenase catalytic site. Comparison of the structures of rpMFE1 with the monofunctional analogs such as crotonase and (3S)-hydroxyacyl-CoA dehydrogenase superfamily enzymes, and with the bacterial α2β2-fatty acid oxidation multienzyme complex, revealed that this tunnel could be important for substrate channeling.6
Table 1.
Genetically modified proteins in nature and engineered
| Protein | Modification | Structure characteristic | Function | Application |
|---|---|---|---|---|
| Rat peroxisomal enzyme rpMFE16 | Quinque-functional hybrid enzyme (Nature) | N-terminal active site: hydratase domain (A) – flexible linker – isomerase domain (B) – positively charged tunnel – C-terminal active site: (3S)-hydroxyacyl-CoA dehydrogenase HAD superfamily domains (C-E) fusion | Δ3,Δ2-Enoyl-CoA isomerase, Δ2-enoyl-CoA hydratase-1, (3S)-hydroxyacyl-CoA dehydrogenase activities | Enhancing metabolic efficiency in β-oxidation pathway of fatty acid degradation |
| Human intestinal bacterium Ruminococcus gvanus E1 α-galactosidase, AgaSK3 | Bifunctional hybrid enzyme (Nature) | N-terminal GH36 α-galactosidase domain (residues 1-720) – C-terminal nucleotide-binding motif (residues 721-935) | α-Galactosidase and sucrose kinase activities | Enhancing metabolic efficiency: melibiose and raffinose hydrolysis and phosphorylation of sucrose on the C6 position of glucose in the presence of ATP |
| Membrane protein, COX-2-10aa-mPGES-12 | Bifunctional hybrid enzyme (Nature) | C-terminus of cyclooxygenase isoform-2 (COX-2) - transmembrane linker – N-terminus of microsomal prostaglandin E2 synthase-1 (mPGES-1) | Cyclooxygenase and prostaglandin E2 synthase-1 activities | Acceleration of inflammation and cancers response: coupling reaction of converting arachidonic acid into prostaglandin E2 |
| Hepatitis C protein 3/4A, HCV NS3/4A5 | Bifunctional hybrid enzyme (Nature) | N-terminal chymotrypsin-like serine protease (residues 1-181) – linker residues 184-186 (regulatory switching domain–domain interface) – C-terminal superfamily 2 DExD-box helicase domain | Protease and 3′-5′ helicase activities | Enhancing viral propagation efficiency: duplex DNA and RNA unwinding, viral polyprotein cleavage for viral maturation, the cleavage of host innate immune response antiviral signaling protein (MAVS) and interferon-β (TRIF), autoregulation of the protease and helicase activities |
| Sponge Geodia cydonium galectin1 | Chimeric lectin (Nature) | Homologous sequences recombination within carbohydrate recognition domain and extension of galectin-3-like carbohydrate-binding subsite | Carbohydrate-binding activity | Extension of ligand specificity: accommodation to binding tetrasaccharide terminal GalNAcα1-3 moieties |
| Human dual-specificity phosphatase, DSP8 | Mutant (Nature) | Amino acid substitution L192A and noncatalytic C-terminal extension (residues 206-330) | Protein phosphatase and alkaline phosphatase activities | Enhancing signaling transduction efficiency: dephosphorylation of phosphoprotein monoesters and β-phosphate of RNA for cancer, diabetes, inflammation, and Alzheimer's disease prevention |
| Thermophilic bacterium G. stearothermophilus α-galactosidases, AgaA4 | Mutant (Nature) | Amino acid substitution A335E, displacement of Trp336 at catalytic subsite | α-Galactosidase and transglycosylation activities | Increasing yield of sucrose: raffinose hydrolysis and enzymatic synthesis of disaccharides |
| Bacteriophage lysine, Ply187N-V12C70 | Chimeric protein (Engineered) | Catalytic domain of Ply187 lysin; bacterial cell-binding domain of PlyV12 lysin (residues 146-314) | Lytic activity | Enhancing antimicrobial therapy efficiency: extension of lytic activity against human pathogens (staphylococci, streptococci, enterococci) |
| Bacillus megaterium steroid hydroxylase, CYP106A265 | Single mutant (Engineered) | Amino acid substitutions A395L or G397P | Cytochrome P450 activity | Enhancing steroid hydroxylation efficiency: extension of regiospecificity to 3-oxo-Δ4-steroids (progesterone, from 15 to 11-position; increase of 11α-, 9α- and 6α-hydroxylation activities up to fourfold |
| Candida antarctica lipaseB, CalB68 | Single, double, triple and quadruple mutants (Engineered) | Amino acid substitutions G39A, T103G, W104F, A141Q, I189Y, L278A | Lipase and amidase activities | Introducing and enhancing amidase activity up to 3-fold wild type activity for the preparation of optically active amines |
| Mannan-binding holothurian A. japonicus lectin, CmAP/MBL-AJ44 | Bifunctional hybrid protein, double mutant lectin (Engineered) | N-terminal alkaline phosphatase domain – flexible linker (G4S)3 – C-terminal mannan-binding lectin domain; amino acid substitutions A137N/F159K in the lectin active center | Alkaline phosphatase and carbohydrate-binding activities | Enhancing lectin activity by 25±5% and utility in enzyme linked lectin assay (ELLA) for early differentiated cervical cancer diagnosis |
| Trametes multicolor pyranose 2-oxidase, TmP2Ox66 | Double mutant (Engineered) | Amino acid substitutions V546C/E542K | D-glucose and D-galactose oxidase activity, mediated electron transfer (MET) | Enhancing catalysis efficiency: carbohydrate biotransformations in food applications (ketose sugar) or as the anodic bio-component in sensors and biofuel cells |
| Paenibacillus macerans cyclodextrin glycosyltransferase69 | Triple mutant (Engineered) | Amino acid substitutions Y260R/Q265K/Y195S | Transglycosidase activity | Enhancing maltodextrin specificity by 60% for production of vitamin C nonreducible highly antioxidant derivative AA-2G |
| Mucor hiemalis endo-β-N-acetylglucosaminidase, Endo-M67 | Single mutant (Engineered) | Amino acid substitution N175Q | Glycosid hydrolyse and transglycosidase activities | Enhancing glycosynthase activity with oxazoline and natural N-glycan for syntheses of glycopeptides/glycoproteins |
| Bacillus subtilis aminopeptidase, BSAP74 | Single mutants (Engineered) | Amino acid substitutions I387A, I387C and I387S | Aminopeptidase activities | Changing substrate specificity for ability to hydrolyze phenylalanine derivatives with large side chains |
| Guanylate kinase (GK)71 | Single mutant (Engineered) | Amino acid substitution S68P | Phosphotransferase ATP-GMP activity | Changing function from enzymatic to protein-binding (GKdom) of membrane associate guanylate kinases (MAGUK) in mitotic spindle orientation and cell adhesion |
| Transmembrane zinc metallopeptidase Neprilysin, NEP75 | Double mutant (Engineered) | Amino acid substitutions G399V/G714K | Peptidase activity | Changing protein cleavage site specificity for cleavage of amyloid β 1-40 (Ab1–40) at Phe20-Ala21 for the treatment of Alzheimer's disease |
| Plant Madagascar periwinkle flavin-dependent halogenase, RebH72 | Single mutant (Engineered) | Amino acid substitution Y455W | Tryptophan-specific halogenase activity | Changing substrate specificity to install chlorine preferentially onto tryptamine for production of halogenated alkaloid |
| Glucose 1-dehydrogenase IV, BmGlcDH-IV73 | Single mutant (Engineered) | Amino acid substitution G259A | D-glucose, D-xylose, D-mannose and D-galactose 1-dehydrogenase activities | Changing substrate specificity to only D-glucose for clinical assay to examine blood glucose levels |
| Bacillus subtilis phytase82 | Bifunctional hybrid enzyme (Engineered) | N-terminal β-1,4 endoglucanase domain – C-terminal phytase domain | β-1,4 endoglucanase and phytase activities | Enhancing nutrition uptake efficiency as a potential feed additive for monogastric animals |
| Xylanase Cel5A–XylT84 Glucanase XylT–Cel5A84 | Bifunctional hybrid enzymes (Engineered) | N-terminal xylonase domain – C-terminal glucanase domain; N-terminal glucanase domain – C-terminal xylonase domain | Heat-active endoglucanase and endoxylanase activities | Enhancing efficiency of β-glucan and beechwood xylan hydrolysis in biorefineries based on plant waste |
| α-Glucanohydrolase, mut-dexA85 | Bifunctional hybrid enzyme (Engineered) | N-terminal mutanase domain – C-terminal dextranase domain | α-1,3-glucanase and α-1,6-glucanase activity | Enhancing efficiency of α-glucans hydrolysis in the prevention of dental biofilm formation |
| Bacillus subtilis laccase, CotA-BglS83 | Bifunctional chimeric enzyme (Engineered) | Insertion of β-1,3;1,4-glucanase gene bglS (2–214 bp) into cotA gene of laccase (between 22–24 bp), forming inter-domain contacts in Gly319-Gly320, Asn354-Ala359 and Leu634-Ser638 loops of CotA-BglS | Benzenediol:oxygen oxidoreductase and β-1,3–1,4-glucanases activities | Enhancing efficiency of hydrolysis of plant cell wall β-glucans and oxidation of aromatic compounds for sugar release from milled sugarcane bagasse |
| Renilla reniformis luciferase, Aluc3043 | Bifunctional chimeric protein (Engineered) | Insertion of epitop sequences into N-terminal ALuc23 region (between residues Pro20-Val30) | Bioluminescence | Enhancing efficiency of optical intensity and expending utilities of luciferases for the broad use in bioassays in vivo and in vitro |
| Multivalent antibody scFv80 | Bifunctional hybrid antibody/ribonuclease (4D5 scFv-Bn) - ribonuclease inhibitor (Bs-UCNP) (Engineered) | Humanized monoclonal antibody 4D5 (scFv) (HER2-targeted module)- flexible linker - bacterial ribonuclease barnase (Bn):cysteine-free C40/82A mutant barstar (Bs) – upconversion nanophosphors (UCNPs) | Ribonuclease, HER2-binding, optical (photoluminescence) activities | Enhancing efficiency of malignant tumor detection deep in the living tissue for accurate molecular diagnosis and therapy of tumor diseases |
| Human lectin L-FCN/MBL79 | Chimeric protein (Engineered) | Lectin MBL carbohydrate recognition domain - L-ficolin (L-FCN) collagenous domain (simplified quaternary structure) | Ligand recognition and effector activities | Enhancing cooperativity between carbohydrate recognition domains and their cognate ligands, complement activation, and calreticulin binding dynamics for reducing infection by wild type Ebola Virus |
| Tumor-associated antigens MUC1 and HER292 | Biepitop hybrid antigen (Engineered) | MUC1 subunit – HER2 subunit | Binding antibodies | Enhancing efficiency of breast cancer detection in ELISA for detection of antibodies against MUC1 or HER2 in human serum |
| Enterotoxigenic E. coli colonization factor antigens (CFAs)93 | Multiepitop multifunctional hybrid protein (Engineered) | Seven E. coli CFAs – labile toxin subunit B – binding subunit of heat-labile toxoid (LT) – heat-stable toxoid (ST) | Recognition, binding and toxic activities | Enhancing efficiency of protection against enterotoxigenic E. coli (ETEC) strains in bacterial diarrhea therapy, inducion anti-CFA and antitoxin immunity |
Another example of the increase of metabolic efficiency by chimeragenesis is bifunctional enzyme AgaSK from human intestinal bacterium Ruminococcus gvanus E1 coupling α-galactosidase and sucrose kinase activities (Table 1).3 AgaSK is composed of 2 domains: the N-terminal one (residues 1–720) closely related to a-galactosidases of GH36 family enabling hydrolysis of melibiose and raffinose, and the C-terminal domain (residues 721–935) containing a nucleotide-binding motif for specifically phosphorylation of sucrose on the C6 position of glucose in the presence of ATP. The crystal structure of AgaSK highlighted an oligomeric state necessary for efficient substrate binding and suggesting a cross-talk between the galactose and kinase domains.3
For the improvement of catalytic efficiency in the conditions of the membrane topologies and structures, the C-terminus of cyclooxygenase isoform-2 (COX-2) was linked to the N-terminus of microsomal prostaglandin E2 synthase-1 (mPGES-1) through a transmembrane linker to form a hybrid enzyme, COX-2-10aa-mPGES-1 (Table 1).2 These inducible enzymes become up-regulated in inflammation and some cancers, and their coupling reaction of converting arachidonic acid (AA) into prostaglandin (PG) E2 (PGE2) is responsible for inflammation and cancers. The engineered hybrid enzyme expressed in HEK293 cells exhibited strong triple-catalytic functions in the continuous conversion of AA into PGG2 (catalytic-step 1), PGH2 (catalytic-step 2) and PGE2 (catalytic-step 3), a proinflammatory mediator. The hybrid enzyme retained similar Kd and Vmax values to that of the parent enzymes, suggesting that the configuration between COX-2 and mPGES-1 (through the transmembrane domain) could mimic the native conformation and membrane topologies of COX-2 and mPGES-1 in the cells. The results confirmed that the enzymes are localized near each other in a face-to-face orientation in nature, where the COX-2 C-terminus faces the mPGES-1 N-terminus in the ER membrane.2
Under the necessity of the regulatory mechanism improvement through chimeragenesis, a bifunctional enzyme of hepatitis C (HCV) protein 3/4A (NS3/4A) comprising 2 separate domains with protease and helicase activities evolved for the more effective viral propagation (Table 1).5 The functional analysis of the full-length protein and separately isolated catalytic domains suggested the existence of dynamic coupling through the interdomain communication. The full-length protein was more efficient in RNA unwinding than the isolated protease domain. However, substrate cleavage and DNA unwinding by the disrupted mutants were mostly enhanced compared with the wild-type protein. The HCV NS3/4A crystal structure and molecular dynamic simulations have shown that the hybrid enzyme adopts an “extended” catalytically active conformation, and interface formation acts as a switch to regulate activity modulating the protease and helicase activities.5
There are many examples of genetic modification of proteins in nature by recombination of sequences or domains between homologous proteins for obtaining chimeras with improved properties. The mammalian galectin-3 and galectin from the sponge, Geodia cydonium, and other members of this subfamily have chimera-type characteristic and sequence polymorphism within their carbohydrate recognition domains (Table 1).1 Additional sugar-binding amino acid residues and multiple substitutions were identified to involve in a putative extended carbohydrate-binding subsite conferring a galectin-3-like specificity, and the extended groove can accommodate a tetrasaccharide interacting with terminal GalNAcα1-3 moieties.1
Single amino acid substitution and sequence insertion/deletion were also found to alter appreciably specific activity/functionality in proteins. An important signaling enzyme, dual-specificity phosphatase (DSP), whose misregulation is linked to cancer, diabetes, inflammation, and Alzheimer's disease, possesses broad substrate specificity and dephosphorylates phosphoprotein monoesters and the β-phosphate of RNA (Table 1).8 A conservative mutation of L192 to arginine and noncatalytic C-terminal extension (residues 206−330) affected on enzymatic activity resulting in the appearence of diphosphatase activity in the monospecific ancestral protein phosphatase.8
Two α-galactosidases AgaA and AgaB from thermophilic bacterium Geobacillus stearothermophilus have 97% identity and differ from each other by only 22 amino acid residues (Table 1).4 This has drastically reflected on their different catalytic efficiency and themperature optimum. A-galactosidase AgaA is fully active at 338K against raffinose that can be used for increasing the yield of manufactured sucrose. AgaB has lower affinity for its natural substrates but is more efficient catalyst at 323K for the enzymatic synthesis of disaccharides by transglycosylation. It has been found that the A335E substitution in AgaA caused structural rearrangements resulting in a significant displacement of the invariant Trp336 at catalytic subsite that was not optimal position for ligand stacking. Hence, the active cleft of AgaA is narrowed in comparison with AgaB, and AgaA is more efficient hydrolase than AgaB against its natural substrates.4
Protein Engineering and Recombinant Production
Protein engineering includs different genomic and post-genomic strategies toward expansion of the functional ability and the synthesis, in vivo and in vitro, of proteins carrying novel chemical, physical and biological properties to use in many applications in basic research, biotechnology, material sciences and therapy.39,40 The strategies of modification proteins through genetic manipulations involve chimeragenesis, site-specific mutagenesis and de novo protein engineering. The synthesis of hybrid proteins by domain swapping or the ligation of chemically or biologically synthesized peptides are often used for biological probing.41-44 Combinatorial (random insertion, circular permutation, homologous and nonhomologous recombination) and non-combinatorial (rational) methods of chimeragenesis based on the original DNA shuffling or restriction-enzyme-based shuffling are also useful for the study of molecular structure/function at the specific subunit and/or domain levels.39,40,45 Directed evolution of DNA is one of the most important advances in biology today. Incorporation of specific amino acids into a target gene by site-directed mutagenesis allows to elucidate relationship between structure-function-biological roles of the natural proteins at the atomic level as well as to obtain the proteins with improved properties such as desired thermoactivity, specificity/selectivity, foldability and utility. The synthesis of proteins carrying a variety of unnatural amino acids are used to the study of receptors and channel proteins.30,39,43 Current computational methods of the structure prediction and design based on homology modeling or other structural principles adopted from natural proteins enable to predict or confirm significance either of the introduced protein modifications.23 Molecular modeling, docking with substrate/inhibitor, dynamic simulation can facilitate adoption of decision about the protein structure from a set of the mutants that should be realized through recombinant production to obtain the desired properties.26,29,44,46,47 Based on the information-driven combinatorial design, the directed evolution experiments consisted of mutation, recombination and screening processes identify interesting mutants. In vitro compartmentalization (IVC) is an in vitro gene screening system for directed evolution of proteins.48,49 Liposome-based IVC is characterized by in vitro protein synthesis from a single copy of a gene in a cell-sized unilamellar liposome and quantitative functional evaluation of the synthesized proteins.48 Today, monitoring and optimization of the recombinant protein production processes are also advantaged by data-based modeling or by the techniques that physically link individual members of diverse gene libraries to their translated proteins for the production of the water-insoluble proteins.49-52
However, the recombinant production of the protein that may have the properties of the desired valuable pharmaceutical product will often not have sufficient yield when adapted to the large-scale production due to low expression, solubility or stability.45,53 Therefore, there are now many examples of improving the recombinant production results including both genetic modification of the recombinant proteins and their producing cells.
Genetic Modification of Producing Strains
The high efficiency of the valuable proteins is crucial to the productivity of an industrial process. One of the expedients to enhance the protein specific activity or efficiency of interest is an archivement of the natural strain high productivity or expression selectivity. Some of the reports indicated the directed evolution in vivo by classical mutagenesis of the natural strain-producer with the use of physical/chemical methods or by recombination between the closely related strains.54-57 A random mutation in genetic materials by nucleotide substitution particularly by guanine alkylation improved the alkaline β-keratinase production by Brevibacillus sp. mutant strain AS-S10-II and it was succefully applied in the livestock feed formulation from the waste chicken-feathers.55 Psychrophilic Pseudoalteromonas haloplanktis TAC125 was engineered for the production of the aromatic oxidative activity encoded by toluene-oxylene monooxygenase from the mesophilic bacterium Pseudomonas sp. OX1 for its effective application in the decontamination of cold environments.54 The recombination of P. haloplanktis TAC125 genome was performed by insertional mutagenesis strategy, avoiding the introduction of any antibiotic resistance encoding gene followed by conversion of the inducible gene PhcopA into a constitutive one, substituting the copper inducible promoter located upstream the PhcopABCD cluster.54
However, the heterological expressions in Esherichia coli and the yeast strains such as Saccharomyces cerevisiae, Pichia pastoris are most often used for in vivo protein engineering.51,58-60 The commercially available DNA repair-deficient strains E. coli XL1-Red deficient in mutD, mutS, and mutT genes are transformed with a plasmid encoding the target DNA, and random mutations are accumulated at each round of DNA replication.55 Recently several new approaches for the genome-scale engineering of E. coli to enhance recombinant protein expression, including improvement of mRNA stability and translational efficiency, protein folding by chaperone co-expression, expression of disulfide-bonded proteins, acetylated and glycosylated protein production have been developed.51,61,62 Strains that confer improved protein expression can be engineered by screening libraries of chromosomal mutants as well as plasmid-encoded expression libraries of heterologous or native genes.51,58-60 The directed evolution based approach to increase protein production levels by the vector backbone optimization has also been reported.63 Randomly introducing mutations in the pET28a(+), pET22b(+) and pHY300PLK vector backbones allowed to increase cellulase, lipase and protease production from two- to fourfold, respectively.
Enhancing Specific Activity
To assess a potential role of each structural element in protein, the site-specific mutagenesis experiments under controlled laboratory conditions can apparent the relationship “structure-function” at the atomic level using X-ray crystallography or molecular modeling approaches.22-27 Generally, the significance of amino acid residue(s) for the exhibition of the protein specific activity is determined by disruption of inter- or intramolecular bonds that are formed by the taget residue(s).47,64 This is feasible by the substitution of the binding residue for Gly or Ala. The lost or decrease of a specific activity in the mutant variant allows to conclude about the level of participation of the substituted residues in the protein functionality. Thus, the point mutations of the residues Asp12, Asp273, Asp315, His316, Thr118, Glu268, and Trp274 to Gly in the marine bacterium alkaline phosphatase CmAP were predicted by in silico mutagenesis to bind ion Mg2+ maintaining the correct structure of the catalytic bimetal core.47 When the mutation interrupted a direct or indirect binding of the target residue with Mg2+, the enzymatic activity was completely lost, although the integrity of the molecular structure remained and only one or 2 hydrogen bounds were broken in the entire CmAP hydrogen bond network (Fig. 1). Moreover, it was predicted the key role of the substrate-binding residue Arg129 and a side residue Tyr441 protruded above the active site entrance for exhibiting the unexampled high specific activity among other known alkaline phosphatases by the computational pKa calculations at the high pH ≥ 10.0.47
Figure 1.
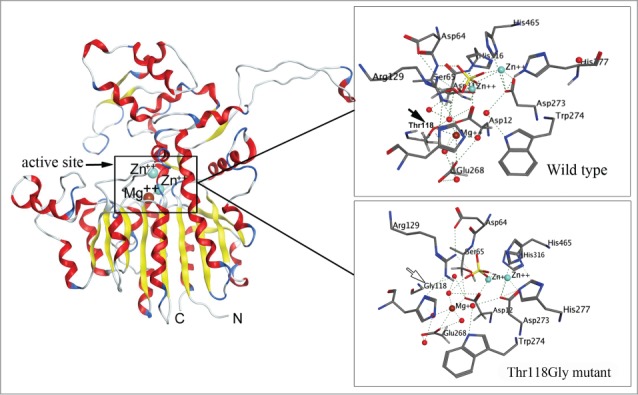
Molecular model of the alkaline phosphatase CmAP (left) and atomic structure of the wild type CmAP active center (above) and its mutant Tre118Gly (below) in silico generated with program MOE 2012.10.47 The coordinates of the metal ions produced by superposition model CmAP with crystal structure of VAP (PDB: 3E2D). Water is shown as red sphere. Metal ions Zn2+ and Mg2+ were shown as blue and brown spheres, respectively. Hydrogen bonds were shown as discrete lines. The residues Tre118 (above) and Gly118 (below) were shown as full and empty cursor, respectively.
By the steric effect of some amino acid residue substitutions, the significance of decrease in a specific activity of protein can be also explained. For example, the single substitution of Cys 170 in the conserved Cys-His-Asn catalytic triad for Arg or Ser decreased the ability of the chalcone synthase PpCHS to utilize hexanoyl-CoA as a starter molecule due to the smaller cavity volume of the mutant's active site.64
The Thr118Gly mutation resulted to the complete loss of activity due to the loss of hydrogen bond Thr118.OG1-Glu268.OE2 and hydrogen bonds with 3 water molecules in the wild type CmAP active site.47
At the same time, it is a challenging goal to change the recombinant protein specific activity or narrow/broad substrate specificity toward an increase their efficiency in any biotechnology or pharmacology process. The influence of the mutations on the catalytic or binding activity in many recombinant proteins of interest was theoretically and experimentally investigated to find the essential amino acid residues participated in the active site formation for designing of their more functionally usable mutants. In many pharmaceutically relevant enzymatic processes, it is desired to utilize the enzymes with enhanced specific activity and/or extended substrate specificity/selectivity.65-70
The bacteriophage lysin Ply187N-V12C was constructed by fusing the catalytic domain of the lysin Ply187 with the bacterial cell-binding domain (from 146 to 314 amino acid residues) of the lysin PlyV12 of another bacterial selectivity for extending the lytic activity spectrum toward pathogens from staphylococcal strains to streptococci and enterococci (Table 1).70
Five mutants in the substrate recognition site 6 (S394I, A395L, T396R, G397P and Q398S) of the bacterium Bacillus megaterium steroid hydroxylase (CYP106A2) related to cytochromes P450 were selected with the use of molecular docking calculations for the progesterone (P) as a substrate for the following mutagenesis studies to create mutants with the altered regiospecificity of 3-oxo-D4-steroids hydroxylation from the 15 to the 11-position (Table 1).65 Despite a low homology model of CYP106A2, its predictions about the haem charge neutralization, catalysis and redox–partner interaction correlated well with the features of other cytochrome P450 structures. Supported by the results of molecular docking, the broader regioselectivity and the increase of 11α-, 9α- and 6α-hydroxylation activities up to fourfold for the mutants A395L and G397P were found in comparison with the wild type of the enzyme.65
The computational method was used to introduce amidase activity and estimate its reaction barriers in Candida antarctica lipase B (CalB) mutants (Table 1).68 The qualitative activity of 15 out of 22 mutants was correctly predicted to obtain the enzymes with 0.5-3-fold wild type activity. The point mutations are selected based on different design principles: introduction of structural rearrangements in the active site to change the binding site properties of the active site (residues P38, G39, G41, T42, T103); introduction of space to accommodate the substrate (W104, L278, A282, I285, V286); introduction of dipolar interactions between the enzyme and the substrate (A132, A141, I189); reduction of polarity in the active site (D223). For the analysis, the reaction barrier was defined by the difference between the highest energy point on the reaction profile and the energy corresponding to the enzyme substrate complex. Six residues G39, T103, W104, A141, I189, L278 was assumed to contribute strongest to increased the activity and the preferred residues for their substitutions were defined at each position. Remarkably, the single mutants at the position 104 were predicted to have low activity and to distinguish by very high activity in the combination with other mutation at the position 189.68
The same effect was observed in the double mutant of the recombinant hybrid mannan-binding holothurian lectin CmAP/MBL-AJ (Table 1).44 It has been experimentally confirmed that the double mutation A137N/F159K has synergy effect on the lectin-binding activity of CmAP/MBL-AJ enhancing its activity by 25 ± 5%, 40 ± 2% and 28 ± 3% in comparison with the wild type lectin and the single mutants A137N and F159K, respectively. In silico mutagenesis calculations and molecular docking with the model oligosaccharide have revealed that the double mutation A137N/F159K lead to the significant rearrangement of the amino acid residues around the lectin binding site, where the residue Leu in the mutant position F159K becomes able to react with the ligand forming the additional 2 ionic and one hydrogen bonds (Fig. 2)
Figure 2.
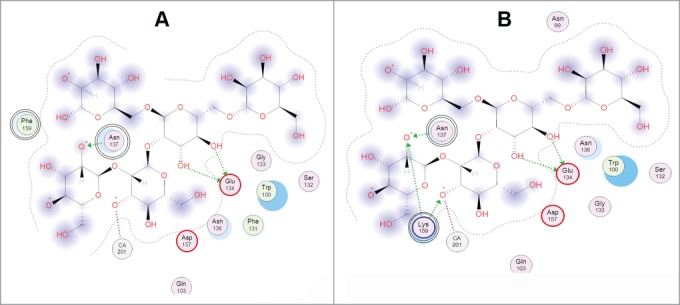
2D-diagram of the model oligosaccharide binding with the single mutant A137N of the Far Eastern holothurian mannan-binding lectin MBL-AJ and its contacts with the oligosaccharide (left). 2D-diagram of the model oligosaccharide binding with the MBL-AJ double mutant F159K and its contacts with the oligosaccharide (right). The residue substitution positions 137 and 159 were indicated by double circles.
The double mutation A137N/F159K in the lectin MBL-AJ resulted to synergistic positive effect increasing its binding activity by 25 ± 5%, 40 ± 2% and 28 ± 3% in comparison with the wild type MBL-AJ, and single A137N and F159K mutants, respectively, due to rearrangement of the amino acid residues around the lectin binding site leading to hydrogen bonding of Lys159 with OH-groups of the ligand mannose residues.44
Generally, site saturation mutagenesis is very useful tool for enhancing drastically specific activity in recombinant proteins.66,69 By combining V546C variants with other amino acid replacements in the Trametes multicolor pyranose 2-oxidase (P2Ox) for carbohydrate biotransformations in food applications or as the anodic bio-component in biofuel cells, it was found that the mutant V546C/E542K had 4.4- and 17-fold increased kcat for 1,4-benzoquinone (BQ) compared to the wild-type enzyme when D-glucose and D-galactose, respectively, and were the saturating substrates. While the V546C/T169G mutant showed approximately 50-fold higher kcat for BQ with D-galactose in excess (Table 1).66
The highest activity in the cyclodextrin glycosyltransferase (CGTase) from Paenibacillus macerans for the synthesis of vitamin C derivatives was achieved by the site saturation mutagenesis of Tyr 195, Tyr 260 and Glu 265 resulted in the triple mutant Y260R/Q265K/Y195S finding (Table 1).69 The triple mutation enhanced maltodextrin specificity by 60% in comparison with the wild type enzyme, altering the hydrogen bonding interactions between the side chain of the residues at these positions and the substrate sugars.69
The significance of the amide side chain in Asp 175 of the endo-β-N-acetylglucosaminidase from Mucor hiemalis (Endo-M) for promoting oxazoline transglycosylation in the second step of the catalysis was confirmed, when the N175Q mutant was found to possess dramatically enhanced glycosynthase-like activity with sugar oxazoline in comparison with the N175A and a transglycosidase-like activity with nature N-glycan as well (Table 1).67
Changing Specificity
The interaction between the substrate and the enzyme is so complex that only few researches on modifying the substrate specificity of enzymes have been reported.71-75
Three mutants (I387A, I387C and I387S) of aminopeptidase from Bacillus subtilis (BSAP) with a broad substrate specificity toward p-nitroanilides (pNAs) derivatives of Leu, Arg and Lys hydrolyzed Phe-pNA, which was undetectable in wild-type BSAP (Table 1).74 In docking simulation, bestatin was docked into the predicted structure of BSAP as the substrate analog. The result showed that there were several residues in direct contact with the side chain of the substrate. It has been suggested that the change in the environment of the substrate-binding region by this mutation leads to a subtle orientation shift of the bound substrate. This shift changed the distance between the substrate and the catalytic residues, reducing the side chain of the residue. The Phe residue with the large side chains does not suit the substrate-binding pocket due to its large side chain. Therefore, the replacement of Ile 387 by a small residue can expand the substrate-binding pocket of BSAP. However, the I387A mutation weakened the hydrophobic interaction in substrate binding region, which led to the poorer thermostability.74
The entirely new functions have been found to arise even from the single mutations altering protein dynamics. The S68P mutation converted the guanylate kinase enzyme (GKenz), which catalyzes phosphotransfer from ATP to GMP, into a protein-binding domain (GKdom) found in membrane associate guanylate kinases (MAGUK) that function in mitotic spindle orientation and cell adhesion (Table 1).71 A dramatic functional change suggested that the MAGUK family proteins with a defining protein-binding feature have diverged from the enzymes GKenz, which is broadly distributed throughout evolution. With respect to the all known GKenz sequences, Ser is conserved at this position. Although the Pro mutation abrogated catalytic activity in GKenz as revealed by crystalization, it retained the ability to bind ATP and altered the protein's conformational response to ligand binding.71
The mutations introduced into neprilysin (NEP), a transmembrane zinc metallopeptidase that degrades a wide range of peptides, have fundamentally altered the cleavage site preference of the enzyme (Table 1).75 Cleavage of amyloid β 1–40 (Ab1–40) at Phe20-Ala21 by double mutant G399V/G714K has not been previously observed in NEP that was largely driven by a decrease in KM, which may result from the substrate being able to adopt an altered binding conformation at the active site. The alteration was in the shape and size of the pocket containing the active site compared with the wild-type enzyme. The new variant of NEP preferable to increase its activity 20-fold at the desired substrate Ab1–40, and reduced activity at peptides where the potential for unwanted side effects exist, can be efficient in the degradation of amyloid β in vivo as a therapeutic for the treatment of Alzheimer's disease.75
Overall, the narrowing of substrate specificity is more derivable result in the changing of protein specificity. The flavin-dependent tryptamine-specific mutant Y455W of the halogenase RebH was engineered to install chlorine preferentially onto tryptamine rather than the native substrate tryptophan (Table 1).72 The mutant gene was transformed into the alkaloid-producing plant Madagascar periwinkle (Catharanthus roseus) for de novo production of the halogenated alkaloid 12-chloro-19,20-dihydroakuammicine. The resulting tissue cultures did not accumulate 7-chlorotryptophan due to the changing of the enzyme RebH substrate specificity from tryptophan to tryptamine.72
The disadvantage of the glucose 1-dehydrogenase IV (BmGlcDH-IV) used for a clinical assay to examine blood glucose levels is its broad substrate specificity. The G259A variant of BmGlcDH-IV exhibited the narrowest substrate specificity toward only D-glucose, while retaining comparable catalytic activity and thermostability to the wild-type enzyme (Table 1).73 Its activity with D-xylose, D-mannose and D-galactose was not detected at pH 8.0. To understand the structural basis for the broadened substrate specificity in the mutant, crystal structures of G259A was determined. The electron density for the C-terminal residues 259–261 (chain A) and 260–261 (chain B) was completely absent, suggesting that these residues at the C-terminal region are protruded into the solvent region, and that the conformation of the C-terminal region is disordered. The residues of a-helix in the structure of the G259A mutant had relatively high B-factor values (60 A°) and moved slightly away from the active site, probably due to the lack of electrostatic interactions between Arg 257 and Asp 208. The side chain amino group of Lys 199 derived from the α-helix formed hydrogen bonds with both O4 of D-glucose and the C-terminal carboxyl, and also moved away from the active site. The observation implies that the mobility of the C-terminal region and the α-helix might be related to sugar-binding selectivity of the enzyme.73
It is evident that the effects of point mutations relate to electronic structure and protein dynamics.76,77 Changes in geometrical parameters introduced by a mutation are usually limited to the local mutational site. However, this local structural modification could affect the global protein dynamics through correlated motions of particular amino acid residues even far from the mutation site.47,76 Transition state stabilization is a result of conformational modifications and reorganization around the active site. As for the electrostatic effect created by the polar protein environment and electrostatic media for optimum catalysis, the effect of global polarization in the electronic structure was found to be a small catalytic element during the process, and the proteins structures are generally robust against external electrostatic perturbations.76 The protein structures have a certain flexibility, which allows them to slightly modulate their conformations to maximize the transition state stabilization in response to the steric perturbations induced by mutations. Therefore, the enzyme most be effectively designed to stabilize the transition state of the reactive substrate, including algorithms based on evolutionary principles.76,77
Hybrid Proteins and Multifunctionality
Hybrid protein can be defined as a chimeric multifunctional polypeptide originated from the modified DNA sequence combining 2 or more unrelated parent sequences. The synthesis of hybrid proteins by domain fusion or the ligation of chemically or biologically synthesized peptides improves apparently a functional unit utility and is generally used for biological probing approach such as biosensors and bioassays, molecular imaging, targeted cancer or anti-inflammatory therapy, regulated drug delivery.41-44,78-81 Hybrid proteins may possess 2 or more functional activities realized by different modes of action indicating that the molecules of hybrid act as several distinct pharmacophores. Many pharmaceutically or industrially relevant enzymatic processes can also utilize hybrid enzymes with multiple domains.82-85 There are terminal-tagging schemes of a hybrid protein production, when one functional domain is tagged at the N- or C-terminal ends of other functional domain through a covalently linked short amino acid sequence (linker or spacer).86 Linker can be rigid (Pro-rich) to prohibit undesired interactions or steric effects between the discrete domains, and flexible (Gly-rich) to connect domains without interfering with the function of each domain in a single protein. It improves the recombinant protein's properties such as foldability, solubility and functionality.44,82,86 The decision of whose domain should follow downstream of C-terminal end depends on the many factors, particularly the possibility of quick folding during the recombinant production or the involvement of the N- or C-terminal ends in the active site formation or oligomerization.
Thus the Bacillus phytase, when expressed in E. coli, was found in inclusion bodies, whereas its endoglucanase was found in the active soluble form (Table 1).82 A chimeric gene construct coding for these fused enzymes allowed their production in soluble form. The hybrid enzyme exhibited both endoglucanase and phytase activities across broad pH (4.0–8.0) and temperature (25–75 C) ranges, and it might have served as a potential feed additive for enhanced nutrition uptake in monogastric animals.82
An improvement in endoglucanase (Cel5A) and endoxylanase (XylT) catalytic activities was observed in the bifunctional enzymes generated by the fusion of 2 genes encoding these heat-active enzymes (Table 1).84 The N-end to C-end fused xylanase Cel5A–XylT and glucanase XylT–Cel5A were active on both β-glucan and beechwood xylan. The specific activity of the hybrid enzymes toward xylan was significantly raised when compared to the nature xylanase XylT. The fusion constructs were found to be active from 40 to 100 °C for endoglucanase and from 40 to 90 °C for endoxylanase. However, the temperature optimums were lowered from 90 to 80 °C for the endoglucanase and from 80 to 70 C for the endoxylanase, underlining the relationship between the catalytic domain residues surroundings and their function. XylT in the hybrid XylT–Cel5A was less stable at higher temperatures compared to Cel5A–XylT.84
For the improvement of the glucanohydrolases utility in the prevention of dental biofilm formation, the mutanase (α-1,3-glucanase) of Paenibacillus humicus and dextranase (α-1,6-glucanase) of Streptococcus mutans were fused, synthesized in E.coli and partially purified to hydrolyze water-insoluble glucans (WIGs) produced by cariogenic pathogens on the tooth surfaces (Table 1).85 The chimeric glucanase reduced the formation of the total amount of WIGs in a dose-dependent manner, and the significant WIGs reduction in the adherent fraction were observed. Moreover, the chimeric glucanase was able to decompose biofilm, being 4.1 times more effective toward glucan inhibition of biofilm formation than a mixture of dextranase and mutanase.85
In contrast to the end-to-end fusion, there are only a few studies showing the use of insertional fusion to created multifunctional hybrid proteins.43,83 For-example, the Bacillus subtilis chimeric bifunctional enzyme laccase/β-1,3–1,4-glucanase that catalyzes the hydrolysis of both plant cell wall β-glucans and oxidation of aromatic compounds was created by an insertion fusion of the bglS and cotA genes (Table 1).83 The hydrolytic activity of the chimera was 20% higher against natural milled sugarcane bagasse as compared with equimolar mixtures of the separate parental enzymes. Molecular dynamics simulations indicated approximation of the 2 catalytic domains in the chimeric enzyme, and the formation of an inter-domain interface may underlie the improved catalytic function. Contacts were observed in the chimeric laccase loops Gly319–Gly320, Asn354–Ala359 and Leu634–Ser638 between side chain atoms of one domain and main chain polar groups of other domain, and main chain interactions between the 2 domains. It was also observed that the conformation of the loops in the laccase domain altered because of the formation of the CotA–BglS domain interface, increasing in the channel diameter for the water product exit.83
A set of methods was developed for subcloning, expression, purification and application of chimeric proteins containing a protein domains of interest such as cytotoxic and imaging agents fused to C-terminal moiety derived from the antigen recognition domains (Fc region) of immunoglobulins in the studies of protein-ligand interactions.87-90 However, the C-terminal end of luciferases is a part of the active site and thus tagging an epitope to it invades the optical intensities for the broad use in bioassays. The problem of the limited functionality of the hybrid epitope was solved by creating a copepod luciferase Aluc30 of Renilla reniformis, embedding epitopes in the middle of the N-terminal region with a high optical intensity (Table 1).43 Molecular modeling with specific softwares the molecular structure of ALuc30 was optimized by a molecular mechanics (MM) calculation based on a Polak–Ribiere algorithm template-based modeling (TBM) is the only reliable method for high-resolution structure prediction. The supersecondary structure revealed EF hand-like motifs between helices 5 and 6, and helices 7 and 8. Interestingly, the deepest end of the cave provided a large hole for iodide, exerting the excellent substrate specificity to coelenterazine derivative (CTZi) reported as the poorest substrate for the existing marine luciferases and photoproteins. This drastic contrast was explainable by a unique size selection effect of the Aluc30 pocket to the residues of CTZ.43
The targeting Fc-based hybrid proteins have been recently shown to be advantaged by organic or inorganic nanoparticles. For example, nanoconstructs consisting of targeting biopolymer molecules and inorganic photoluminescent nanocrystals were suggested to serve as promising carriers for the targeted delivery and detection deep in the living tissue of a wide variety of cytotoxic and imaging agents, which determines their particular potential for personalized optical diagnosis of malignant tumors (Table 1).80
There are non-Ig scaffolds for the engineering of the hybrid proteins, which constitute a distinct family of proteins with functions in ligand binding. Lectins with the databased or experimentally prescribed target specificities can be easily generated. Such molecules with a narrow specificity, which are able to bind selectively to carbohydrates, have also a key importance in the development of research related with mechanism of cancerogenesis or inflammation at the molecular level as well as for designing drugs targeted to a relevant molecule.44,78,79,81 To develop a less complex chimeric fusion protein then the native human lectin MBL with similar ligand recognition and enhanced effector functions, the MBL carbohydrate recognition domain and L-ficolin (L-FCN) collagenous domain were fused and applied to reduce more significantly infection by wild type Ebola virus (Table 1).79 It has been proposed that alterations in the quaternary structure of the hybrid lectin L-FCN/MBL76 resulted in greater flexibility in the collagenous region, enhancing cooperativity between the carbohydrate recognition domains and their cognate ligands, complement activation, and calreticulin binding dynamics. L-FCN/MBL chimeric proteins should be considered as potential novel therapeutics.
The human lipocalins derivations named anticalins with picomolar affinities have been developed for 3 classes of ligands and already reached the clinical trial stage.81 Due to their very small size and simple composition of a single polypeptide chain, which also facilitates the construction of bifunctional fusion proteins, anticalins promise benefits as a next class of biopharmaceuticals.
A method for producing a highly carbohydrate-specific bifunctional hybrid holothurian lectin MBL-AJ, whose expression and purification were monitored by the alkaline phosphatase CmAP activity, has been developed for improving the enzyme-linked MBL-AJ-based assay (ELLA) for cervical cancer diagnosis (Table 1).44 The hybrid protein CmAP/MBL-AJ was produced in E. coli cells as a fully soluble and bifunctional homodimer, whose one subunit consisted of the mature proteins of the highly active alkaline phosphatase CmAP (55 kDa) and holothurian lectin MBL-AJ (17 kDa) connected through a flexible peptide linker (G4S)3 (Fig. 3). The molecular modeling has shown that CmAP/MBL-AJ dimer consists of the 2-subunit lectin part associated with the 2 molecules of alkaline phosphatase functioning independently from each other.44 An attempt to fuse CmAP and MBL-AJ through a rigid Pro-reach linker led to decrease significantly in alkaline phosphatase activity in the hybrid protein CmAP/MBL-AJ (unpublished data). Probably, the psychrophilic nature of the movable marine alkaline phosphatase requires more degree of freedom for the fulfillment of its function.47
Figure 3.
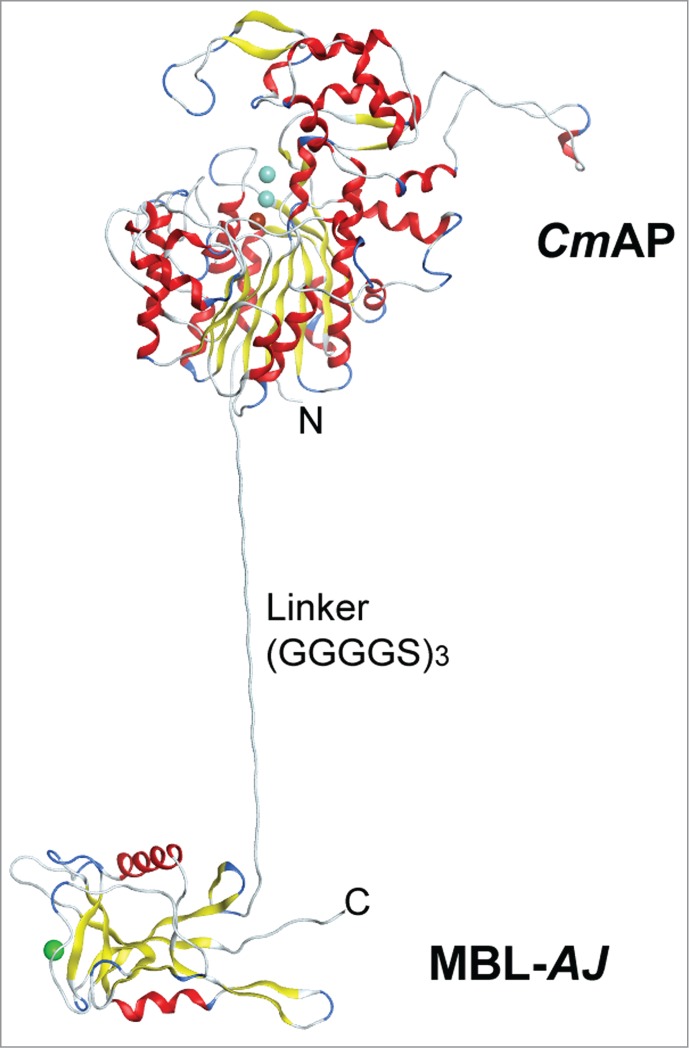
The structural model of the Far Eastern holothurian mannan-binding hybrid lectin MBL-AJ monomer with the alkaline phosphatase CmAP (CmAP/MBL-AJ). C-terminal end of the CmAP subunit is linked via flexible linker (G4S)3 with N-terminal end of the lectin MBL-AJ subunit that allow to function domains independently from each other.44 Three spheres in CmAP module indicate 2 ions Zn2+ and one ion Mg2+; one sphere in MBL-AJ indicates ion Ca2+. α-Helixes and β-strands are shown as ribbons. Unordered structures are shown as threads.
There are hybrid enzymes possesing not only protein-protein interaction activity, but also other specific binding activity. TAL nucleases (TALNs) produced by fusion of the restriction enzyme FokI endonuclease domain with the highly specific DNA-binding domains (transcription activator-like (TAL) effectors) were created for targeted genome editing.91 When expressed in yeast, TALNs promoted DNA homologous recombination of a LacZ gene containing paired AvrXa7 or asymmetric AvrXa7/PthXo1 target sequences. The results demonstrate the feasibility of creating a tool box of novel TALNs with potential for targeted genome modification in organisms lacking facile mechanisms for targeted gene knockout and homologous recombination.
The hybrid proteins consisted of immunogenic sequences such as multiepitope fusion antigens and/or toxoids have been designed and tested as potential vaccines or inflammation diagnosis tools.92,93 The bioinformatics analysis of immunogenic sequences of the tumor-associated antigens MUC1 and HER2 allowed combining it in the hybrid protein for using as the breast cancer antigens in ELISA for detection of antibodies against MUC1 or HER2 in human serum (Table 1).92 The optimized chimeric gene composed of the structural subunits of colonization factor antigens (CFA), labile toxin subunit B and the binding subunit of heat-labile and heat-stable toxoid, was designed to provide broad-spectrum protection against 7 enterotoxigenic E. coli (ETEC) strains, the most common cause of bacterial diarrhea, inducing anti-CFA and antitoxin immunity (Table 1).93
The above will show that alterations in protein's thermoactivity, stability, pH-dependence and foldability are generally special cases for the rational improvement of the valuable protein function. Beneficial mutations that lead to increased thermostability or decreased thermoactivity, improved pH-optimum or solubility can be involved in many different mechanisms determined by the laws of physics and chemistry including solvent interactions, structural support, and electrostatic balance.59,94-100 Consequently, the search for variants with improved function is best treated as a combinatorial optimization problem, in which a number of parameters must be optimised simultaneously to achieve a successful outcome imitating the nature's manner.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
Funding
This work was supported by the Russian Foundation for Basic Research (15-04-08654), the Program “Far East” (15-I-5-020) and the Scientific Foundation of the Far Eastern Federal University (14-08-06-10_u).
References
- 1.Stalz H, Roth U, Schleuder D, Macht M, Haebel S, Strupat K, Peter-Katalinic J, Hanisch FG. The Geodia cydonium galectin exhibits prototype and chimera-type characteristics and a unique sequence polymorphism within its carbohydrate recognition domain. Glycobiology 2006; 16:402-14; PMID:16449348 [DOI] [PubMed] [Google Scholar]
- 2.Ruan K-H, Cervantes V, So SP. Engineering of a novel hybrid enzyme: an anti-inflammatory drug target with triple catalytic activities directly converting arachidonic acid into the inflammatory prostaglandin E2. Prot Engin Design Select 2009; 22:733-40; http:/dx.doi.org/ 10.1093/protein/gzp058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruel L, Sulzenbacher G, Tison MC, Pujol A, Nicoletti C, Perrier J, Galinier A, Ropartz D, Fons M, Pompeo F, et al.. α-Galactosidase/sucrose kinase (AgaSK), a novel bifunctional enzyme from the human microbiome coupling galactosidase and kinase activities. J Biol Chem 2011; 286:40814-23; PMID:21931163; http://dx.doi.org/ 10.1074/jbc.M111.286039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meceron R, Foucault M, Haser R, Mattes R, Watzlawick H, Gouet P. Crystal structure of liganded rat peroxisomal multifunctional enzyme type 1: a flexible molecule with two interconnected active sites. J Biol Chem 2012; 28:39642-52; http://dx.doi.org/ 10.1074/jbc.M112.394114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aydin C, Mukherjee S, Hanson AM, Frick DN, Schiffer CA. The interdomain interface in bifunctional enzyme protein 3/4A (NS3/4A) regulates protease and helicase activities Prot Sci 2013; 22:1786-98; PMID:24123290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasaragod P, Venkatesan R, Kiema TR, Hiltunen JK, Wierenga RK. Crystal structure of liganded rat peroxisomal multifunctional enzyme type 1: a flexible molecule with two interconnected active sites. J Biol Chem 2010; 285:24089-98; PMID:20463028; http://dx.doi.org/ 10.1074/jbc.M110.117606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jia B, Cheong GW, Zhang S. Multifunctional enzymes in archaea: promiscuity and moonlight. Extremophiles 2013; 17:193-203; PMID:23283522; http://dx.doi.org/ 10.1007/s00792-012-0509-1 [DOI] [PubMed] [Google Scholar]
- 8.Sankhala RS, Lokareddy RK, Cingolani G. Structure of Human PIR1, an Atypical Dual-Specificity Phosphatase. Biochemistry 2014; 53:862-871; PMID:24447265; http://dx.doi.org/ 10.1021/bi401240x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu X. Functional divergence in protein (family) sequence evolution. Genetica 2003; 118:133-41; PMID:12868604; http://dx.doi.org/ 10.1023/A:1024197424306 [DOI] [PubMed] [Google Scholar]
- 10.Philippe H, Casane D, Gribaldo S, Lopez P, Meunier J. Heterotachy and functional shift in protein evolution. IUBMB Life 2003; 55:257-65 [DOI] [PubMed] [Google Scholar]
- 11.Siltberg-Liberles J, Grahnen JA, Liberles DA. The evolution of protein structures and structural ensembles under functional constraint. Genes (Basel) 2011; 2:748-62; PMID:24710290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harms MJ, Thornton JW. Evolutionary biochemistry: revealing the historical and physical causes of protein properties. Nat Rev Genet 2013; 14:559-71; PMID:23864121; http://dx.doi.org/ 10.1038/nrg3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liberles DA, Teichmann SA, Bahar I, Bastolla U, Bloom J, Bornberg-Bauer E, Colwell LJ, de Koning APJ, Dokholyan NV, Echave J, et al.. The interface of protein structure, protein biophysics, and molecular evolution. Prot Sci 2012; 21:769-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynch M. Evolutionary diversification of the multimeric states of proteins. Proc Natl Acad Sci U S A 2013; 110:E2821-28; PMID:23836639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang TT, del Valle Marcos ML, Hwang JK, Echave J. A mechanistic stress model of protein evolution accounts for site-specific evolutionary rates and their relationship with packing density and flexibility. BMC Evol Biol 2014; 14:78; PMID:24716445; http://dx.doi.org/ 10.1186/1471-2148-14-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcos ML, Echave J. Too packed to change: side-chain packing and site-specific substitution rates in protein evolution. PeerJ 2015; 3:e911; PMID:25922797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulenburg C, Hilvert D. Protein conformational disorder and enzyme catalysis. Top Curr Chem 2013; 337:41-67; PMID:23536241; http://dx.doi.org/ 10.1007/128_2012_411 [DOI] [PubMed] [Google Scholar]
- 18.van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT. Classification of intrinsically disordered regions and proteins. Chem Rev 2014; 114:6589-631; PMID:24773235; http://dx.doi.org/ 10.1021/cr400525m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z, Huang Y. Advantages of proteins being disordered. Prot Sci 2014; 23:539-50; http://dx.doi.org/ 10.1002/pro.2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warnecke T, Weber CC, Hurst LD. Why there is more to protein evolution than protein function: splicing, nucleosomes and dual-coding sequence. Biochem Soc Trans 2009; 37:756-61; PMID:19614589; http://dx.doi.org/ 10.1042/BST0370756 [DOI] [PubMed] [Google Scholar]
- 21.Nishida H, Kondo S, Nojiri H, Noma K, Oshima K. Evolutionary mechanisms of microbial genomes. Int J Evol Biol 2011; 2011:319479; PMID:21716727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaparro-Riggers JF, Polizzi KM, Bommarius AS. Better library design: data-driven protein engineering. Biotechnol J 2007; 2:180-191; PMID:17183506; http://dx.doi.org/ 10.1002/biot.200600170 [DOI] [PubMed] [Google Scholar]
- 23.Basit N, Wechsler H. Prediction of enzyme mutant activity using computational mutagenesis and incremental transduction. Adv Bioinformatics 2011; 2011:958129; PMID:22007208; http://dx.doi.org/ 10.1155/2011/958129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vyas VK, Ukawala RD, Ghate M, Chintha C. Homology modeling a fast tool for drug discovery: current perspectives. Indian J Pharm Sci 2012; 74:1-17; PMID:23204616; http://dx.doi.org/ 10.4103/0250-474X.102537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor WR, Hamilton RS, Sadowski MI. Prediction of contacts from correlated sequence substitutions. Curr Opin Struct Biol 2013; 23:473-79; PMID:23680395; http://dx.doi.org/ 10.1016/j.sbi.2013.04.001 [DOI] [PubMed] [Google Scholar]
- 26.Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, et al.. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013; 29:845-54; PMID:23407358; http://dx.doi.org/ 10.1093/bioinformatics/btt055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rose PW, Prli A, Bi C, Bluhm WF, Christie CH, Dutta S, Green RK, Goodsell DS, Westbrook JD, Woo J, et al.. The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nuc Acids Res 2014; 43:D345-56; http://dx.doi.org/ 10.1093/nar/gku1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baker M. Protein engineering: navigating between chance and reason. Nat methods 2011; 8:623-26; PMID:21799494; http://dx.doi.org/ 10.1038/nmeth.1654 [DOI] [PubMed] [Google Scholar]
- 29.Koga N, Tatsumi-Koga R, Liu G, Xiao R, Acton TB, Montelione GT, Baker D. Principles for designing ideal protein structures. Nature 2012; 8: 491(7423):222-7; PMID:23135467; http://dx.doi.org/ 10.1038/nature11600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang WH, Otting G, Jackson CJ. Protein engineering with unnatural amino acids. Curr Opin Struct Biol 2013; 23:581-87; PMID:23835227; http://dx.doi.org/ 10.1016/j.sbi.2013.06.009 [DOI] [PubMed] [Google Scholar]
- 31.Yadahalli S, Hemanth VV, Rao G, Gosavi S. Modeling non-native interactions in designed proteins. Isr J Chem 2014; 54:1230-40; http://dx.doi.org/ 10.1002/ijch.201400035 [DOI] [Google Scholar]
- 32.Matthaei JF, DiMaio F, Richards JJ, Pozzo LD, Baker D, Baneyx F. Designing two-dimensional protein arrays through fusion of multimers and computational interface redesign. Nano Lett 2015; PMID:25986921 [DOI] [PubMed] [Google Scholar]
- 33.Dimitrov DS. Therapeutic proteins. Methods Mol Biol 2012; 899:1-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang P. Nanoscale engineering for smart biocatalysts with fine-tuned properties and functionalities. Top Catal 2012; 55(16-18):1107-13; PMID:2275251922752519 [Google Scholar]
- 35.Kruger B, Liang C, Prell F, Fieselmann A, Moya A, Schuster S, Volker U, Dandekar T. Metabolic adaptation and protein complexes in prokaryotes. Metabolites 2012; 2:940-58; PMID:24957769; http://dx.doi.org/ 10.3390/metabo2040940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carrera J, Estrela R, Luo J, Rai N, Tsoukalas A, Tagkopoulos I. An integrative, multi-scale, genome-wide model reveals the phenotypic landscape of Escherichia coli. Mol Syst Biol 2014; 10:735; PMID:24987114; http://dx.doi.org/ 10.15252/msb.20145108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winkler U, Saftel W, Stabenau H. A new type of a multifunctional - oxidation enzyme in Euglena. Plant Physiol 2003; 131:753-62; PMID:12586899; http://dx.doi.org/ 10.1104/pp.013151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nam H, Lewis NE, Lerman JA, Lee DH, Chang RL, Kim D, Palsson BO. Network context and selection in the evolution to enzyme specificity. Science 2012; 337:1101-04; PMID:22936779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohrer C, RajBhandary UL. Protein Engineering Nucleic Acids and Molecular Biology, edited by Gross HJ, 2009. Springer-Verlag Berlin Heidelberg [Google Scholar]
- 40.Turanli-Yildiz B, Alkim C, Cakar ZP. Protein Engineering Methods and Applications. Protein Engineering, edited by Kaumaya P, 2012. InTech, http://www.intechopen.com/books/protein-engineering/protein-engineering-methods-and-applications [Google Scholar]
- 41.Meunier B. Hybrid molecules with a dual mode of action: dream or reality? Acc Chem Res 2008; 41:69-77; PMID:17665872; http://dx.doi.org/ 10.1021/ar7000843 [DOI] [PubMed] [Google Scholar]
- 42.Palmer AE, Qin Y, Park JG, McCombs JE. Design and application of genetically encoded biosensors. Trends Biotechnol 2011; 29:144-52; PMID:21251723; http://dx.doi.org/ 10.1016/j.tibtech.2010.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SB, Izumi H. Functional artificial luciferases as an optical readout for bioassays Sung Bae Kim, Hiroshi Izumi. Biochem Biophys Res Comm 2014; 448:418-23; PMID:24802399; http://dx.doi.org/ 10.1016/j.bbrc.2014.04.128 [DOI] [PubMed] [Google Scholar]
- 44.Balabanova L, Golotin V, Kovalchuk S, Bulgakov A, Likhatskaya G, Son O, Rasskazov V. A novel bifunctional hybrid with marine bacterium alkaline phosphatase and Far Eastern holothurian mannan-binding lectin activities. PLoS One 2014; 9(11):e112729; http://dx.doi.org/ 10.1371/journal.pone.0112729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McLachlan MJ, Sullivan RP, Zhao H. Directed enzyme evolution and high-throughput screening Biocatalysis for the pharmaceutical industry: discovery, development, and manufacturing, edited by Tao J, Lin GQ, Liese A, 2009. John Wiley & Sons Asia (Pte) Ltd [Google Scholar]
- 46.Amin SR, Erdin S, Ward RM, Lua RC, Lichtarge O. Prediction and experimental validation of enzyme substrate specificity in protein structures. Proc Natl Acad Sci U S A 2013; 110:E4195-202; PMID:24145433; http://dx.doi.org/ 10.1073/pnas.1305162110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Golotin V, Balabanova L, Likhatskaya G, Rasskazov V. Recombinant production and characterization of a highly active alkaline phosphatase from marine bacterium Cobetia marina. Mar Biotechnol 2015; 17:130-43 [DOI] [PubMed] [Google Scholar]
- 48.Takehiro N, Takeshi S, Tomoaki M, Tetsuya Y. Directed evolution of proteins through in vitro protein synthesis in liposomes. J Nucleic Acids 2012; 2012:923214; http://dx.doi.org/ 10.1155/2012/923214; PMID:22957209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yong KJ, Scott DJ. Rapid directed evolution of stabilized proteins with cellular high-throughput encapsulation solubilization and screening (CHESS). Biotechnol Bioeng 2015; 112:438-46; PMID:25220691; http://dx.doi.org/ 10.1002/bit.25451 [DOI] [PubMed] [Google Scholar]
- 50.Raymond A, Lovell S, Lorimer D, Walchli J, Mixon M, Wallace E, Thompkins K, Archer K, Burgin A, Stewart L. Combined protein construct and synthetic gene engineering for heterologous protein expression and crystallization using Gene Composer. BMC Biotechnol 2009; 9:37; PMID:19383143; http://dx.doi.org/ 10.1186/1472-6750-9-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Makino T, Skretas G, Georgiou G. Strain engineering for improved expression of recombinant proteins in bacteria. Microb Cell Fact 2011; 10:32.; PMID:21569582; http://dx.doi.org/ 10.1186/1475-2859-10-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schaepe S, Levisauskas D, Simutis R, Lübbert A. Data-based optimization of protein production processes. Biotechnol Lett 2014; 36:929-35; PMID:24557076; http://dx.doi.org/ 10.1007/s10529-013-1448-3 [DOI] [PubMed] [Google Scholar]
- 53.Gomes S, Leonor IB, Mano JF, Reis RL, Kaplan DL. Natural and genetically engineered proteins for tissue engineering. Prog Polym Sci 2012; 37:1-17; PMID:22058578; http://dx.doi.org/ 10.1016/j.progpolymsci.2011.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parrilli E, Papa R, Tutino ML, Sannia G, Engineering of a psychrophilic bacterium for the bioremediation of aromatic compounds. Bioeng Bugs 2010; 1:213-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mukherjee AK, Rai SK, Bordoloi NK. Biodegradation of waste chicken-feathers by an alkaline b-keratinase (Mukartinase) purified from a mutant Brevibacillus sp. strain AS-S10-II. Int Biodeterior Biodegradation 2011; 65:1229-37; http://dx.doi.org/ 10.1016/j.ibiod.2011.09.007 [DOI] [Google Scholar]
- 56.Al-Zahrani NHM. Improving ability of Pseudomonas putida strain to biosynthesis of L-tryptophan. Life Sci J 2014; 11:476-81 [Google Scholar]
- 57.Sadhu S, Ghosh PK, Aditya G, Maiti TK. Optimization and strain improvement by mutation for enhanced cellulase production by Bacillus sp. (MTCC10046) isolated from cow dung. J King Saud Univ Sci 2014; 26:323-32; http://dx.doi.org/ 10.1016/j.jksus.2014.06.001 [DOI] [Google Scholar]
- 58.Blagodatski A, Katanaev VL. Technologies of directed protein evolution in vivo. Cell Mol Life Sci 2011; 68:1207-14; PMID:21190058; http://dx.doi.org/ 10.1007/s00018-010-0610-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Çelik E, Çalık P. Production of recombinant proteins by yeast cells. Biotechnol Adv 2012; 30(5):1108-18 [DOI] [PubMed] [Google Scholar]
- 60.Nielsen J. Production of biopharmaceutical proteins by yeast. Bioengineered 2013; 4:207-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gopal GJ, Kumar A. Strategies for the production of recombinant protein in Escherichia coli. Protein J 2013; 32:419-25; PMID:23897421; http://dx.doi.org/ 10.1007/s10930-013-9502-5 [DOI] [PubMed] [Google Scholar]
- 62.Weinstock MT, Jacobsen MT, Kay MS. Synthesis and folding of a mirror-image enzyme reveals ambidextrous chaperone activity. Proc Natl Acad Sci U S A 2014; 111:11679-84; PMID:25071217; http://dx.doi.org/ 10.1073/pnas.1410900111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jakob F, Lehmann C, Martinez R, Schwaneberg V. Increasing protein production by directed vector backbone evolution. AMB Express 2013; 3:39; PMID:23890095; http://dx.doi.org/ 10.1186/2191-0855-3-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rahman RNZRA, Zakaria II, Salleh AB, Basri M. Enzymatic properties and mutational studies of chalcone synthase from Physcomitrella patens. Int J Mol Sci 2012; 13:9673-91; PMID:22949824; http://dx.doi.org/ 10.3390/ijms13089673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lisurek M, Simgen B, Antes I, Bernhardt R. Theoretical and experimental evaluation of a CYP106A2 low homology model and production of mutants with changed activity and selectivity of hydroxylation. ChemBioChem 2008; 9:1439-49; PMID:18481342; http://dx.doi.org/ 10.1002/cbic.200700670 [DOI] [PubMed] [Google Scholar]
- 66.Spadiut O, Pisanelli I, Maischberger T, Peterbauer C, Gorton L, Chaiyen P, Haltrich D. Engineering of pyranose 2-oxidase: improvement for biofuel cell and food applications through semi-rational protein design. J Biotechnol 2009; 139:250-57; PMID:19095017; http://dx.doi.org/ 10.1016/j.jbiotec.2008.11.004 [DOI] [PubMed] [Google Scholar]
- 67.Umekawa M, Li C, Higashiyama T, Huang W, Ashida H, Yamamoto K, Wang L-X. Efficient glycosynthase mutant derived from Mucor hiemalis endo-β-N- acetylglucosaminidase capable of transferring oligosaccharide from both sugar Oxazoline and natural N-glycan. J Biol Chem 2010; 285:511-21; PMID:19880511; http://dx.doi.org/ 10.1074/jbc.M109.059832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hediger MR, De Vico L, Rannes JB, Jäckel C, Besenmatter W, Svendsen A, Jensen JH. In silico screening of 393 mutants facilitates enzyme engineering of amidase activity in CalB. PeerJ 2013; 1:e145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Han R, Liu L, Shin H-D, Chen RR, Li J, Du G, Chend J. Systems engineering of tyrosine 195, tyrosine 260, and glutamine 265 in cyclodextrin glycosyltransferase from Paenibacillus macerans to enhance maltodextrin specificity for 2-O-D-glucopyranosyl-L-ascorbic acid synthesis. App Environ Microbiol 2013; 79:672-77; http://dx.doi.org/ 10.1128/AEM.02883-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dong Q, Wang J, Yang H, Wei C, Yu J, Zhang Y, Huang Y, Zhang XE, Wei H. Construction of a chimeric lysin Ply187N-V12C with extended lytic activity against staphylococci and streptococci. Microb Biotechnol 2014; 8:210-20; PMID:25219798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnston CA, Whitney DS, Volkman BF, Doe CQ, Prehoda KE. Conversion of the enzyme guanylate kinase into a mitotic-spindle orienting protein by a single mutation that inhibits GMP-induced closing. Proc Natl Acad Sci U S A 2011; 108:E973-8; PMID:21990344; http://dx.doi.org/ 10.1073/pnas.1104365108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Glenn WS, Nims E, O'Connor SE. Reengineering a tryptophan halogenase to preferentially chlorinate a direct alkaloid precursor. J Am Chem Soc 2011; 133:19346-9; PMID:22050348; http://dx.doi.org/ 10.1021/ja2089348 [DOI] [PubMed] [Google Scholar]
- 73.Nishioka T, Yasutake Y, Nishiya Y, Tamura T. Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose1-dehydrogenase IV. FEBS J 2012; 279:3264-75; PMID:22804868; http://dx.doi.org/ 10.1111/j.1742-4658.2012.08713.x [DOI] [PubMed] [Google Scholar]
- 74.Gao X, Cui W, Ning Ding, Zhongmei Liu, Yaping Tian, Zhemin Zhou. Structure-based approach to alter the substrate specificity of Bacillus subtilis aminopeptidase. Prion 2013; 7:328-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Webster CI, Burrell M, Olsson L-L, Fowler SB, Digby S, Sandercock A, Snijder A, Tebbe J, Haupts U, Grudzinska J, et al.. Engineering neprilysin activity and specificity to create a novel therapeutic for Alzheimer's disease. PLoS One 2014; 9:e104001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ishida T. Effects of point mutation on enzymatic activity: correlation between protein electronic structure and motion in chorismate mutase reaction. J Am Chem Soc 2010; 132:7104-18; PMID:20426479; http://dx.doi.org/ 10.1021/ja100744h [DOI] [PubMed] [Google Scholar]
- 77.Currin A, Swainston N, Day PJ, Kell DB. Synthetic biology for the directed evolution of protein biocatalysts: navigating sequence space intelligently. Chem Soc Rev 2015; 44:1172-239; PMID:25503938; http://dx.doi.org/ 10.1039/C4CS00351A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Melo FHM, Butera D, Medeiros RS, Andrade LNS, Nonogaki S, Soares FA, Alvarez RA, da Silva AMM, Chammas R. Biological applications of a chimeric probe for the assessment of galectin-3 ligands. J Histochem Cytochem 2007; 55:1015-26; PMID:17565118; http://dx.doi.org/ 10.1369/jhc.7A7174.2007 [DOI] [PubMed] [Google Scholar]
- 79.Michelow SC, Dong M, Mungall BA, Yantosca LM, Lear C, Ji X, Karpel M, Rootes CL, Brudner M, Houen G, et al.. A novel L-ficolin/mannose-binding lectin chimeric molecule with enhanced activity against Ebola virus. J Biol Chem 2010; 285:24729-39; PMID:20516066; http://dx.doi.org/ 10.1074/jbc.M110.106260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grebenik EA, Generalova AN, Nechaev AV, Khaydukov EV, Mironova KE, Stremovskiy OA, Lebedenko EN, Zvyagin AV, Deyev SM. Specific visualization of tumor cells using upconversion nanophosphors. Acta Naturae 2014; 6:48-53; PMID:25558394 [PMC free article] [PubMed] [Google Scholar]
- 81.Richter A, Eggenstein E, Skerra A. Anticalins: Exploiting a non-Ig scaffold with hypervariable loops for the engineering of binding proteins. FEBS Lett 2014; 588:213-18; PMID:24239535; http://dx.doi.org/ 10.1016/j.febslet.2013.11.006 [DOI] [PubMed] [Google Scholar]
- 82.Reddy VA, Venu K, Rao DE, Rao KV, Reddy VD. Chimeric gene construct coding for bi-functional enzyme endowed with endoglucanase and phytase activities. Arch Microbiol 2009; 191:171-5; PMID:18987844; http://dx.doi.org/ 10.1007/s00203-008-0437-8 [DOI] [PubMed] [Google Scholar]
- 83.Furtado GP, Ribeiro LF, Lourenzoni MR, Ward RJ. A designed bifunctional laccase/b-1,3–1,4-glucanase enzyme shows synergistic sugar release from milled sugarcane bagasse. Protein Eng Des Sel 2013; 26:15-23; PMID:23012443 [DOI] [PubMed] [Google Scholar]
- 84.Rizk M, Elleuche S, Antranikian G. Generating bifunctional fusion enzymes composed of heat-active endoglucanase (Cel5A) and endoxylanase (XylT). Biotechnol Lett 2015; 37:139-45; PMID:25214221; http://dx.doi.org/ 10.1007/s10529-014-1654-7 [DOI] [PubMed] [Google Scholar]
- 85.Otsuka R, Imai S, Murata T, Nomura Y, Okamoto M, Tsumori H, Kakuta E, Hanada N, Momoi Y. Application of chimeric glucanase comprising mutanase and dextranase for prevention of dental biofilm formation. Microbiol Immunol 2015; 59:28-36; PMID:25411090; http://dx.doi.org/ 10.1111/1348-0421.12214 [DOI] [PubMed] [Google Scholar]
- 86.Reddy Chichili VP, Kumar V, Sivaraman J. Linkers in the structural biology of protein–protein interactions. Prot Sci 2013; 22:153-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Deyev SM, Lebedenko EN. Modern technologies for creating synthetic antibodies for clinical application. Acta naturae 2009; 1:32-50; PMID:22649585 [PMC free article] [PubMed] [Google Scholar]
- 88.Cannon JP, O'Driscoll M, Litman GW. Construction, expression, and purification of chimeric protein reagents based on immunoglobulin fc regions. Methods Mol Biol 2011; 48:51-67; http://dx.doi.org/ 10.1007/978-1-61779-139-0_4 [DOI] [PubMed] [Google Scholar]
- 89.Banta S, Dooley K, Shur O. Replacing antibodies: engineering new binding proteins. Annu Rev Biomed Eng 2013; 15:93-113; PMID:23642248; http://dx.doi.org/ 10.1146/annurev-bioeng-071812-152412 [DOI] [PubMed] [Google Scholar]
- 90.Alewine C, Hassan R, Pastan I. Advances in anticancer immunotoxin therapy. The Oncologist 2015; 20:176-85; PMID:25561510; http://dx.doi.org/ 10.1634/theoncologist.2014-0358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nuc Acids Res 2011; 39:359-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gheybi E, Amani J, Salmanian AH, Mashayekhi F, Khodi S. Designing a recombinant chimeric construct contain MUC1 and HER2 extracellular domain for prediagnostic breast cancer. Tumour Biol 2014; 35:11489-97; PMID:25128064; http://dx.doi.org/ 10.1007/s13277-014-2483-y [DOI] [PubMed] [Google Scholar]
- 93.Ruan X, Sack DA, Zhang W. Genetic fusions of a CFA/I/II/IV MEFA (Multiepitope Fusion Antigen) and a toxoid fusion of heat-stable toxin (STa) and heat-labile toxin (LT) of enterotoxigenic Escherichia coli (ETEC) retain broad anti-CFA and antitoxin antigenicity. PLoS One 2015; 10(3):e0121623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ugwu SO, Apte SP. The effect of buffers on protein conformational stability. Pharmaceutical Technology 2004; 86-113 [Google Scholar]
- 95.Zarrine-Afsar A, Zhang Z, Schweiker KL, Makhatadze GI, Davidson AR, Chan HS. Kinetic consequences of native state optimization of surface-exposed electrostatic interactions in the Fyn SH3 domain proteins. Adv Exp Med Biol 2012; 80:858-70 [DOI] [PubMed] [Google Scholar]
- 96.Griffin MD, Gerrard JA. The relationship between oligomeric state and protein function. Adv Exp Med Biol 2012; 747:74-90; PMID:22949112; http://dx.doi.org/ 10.1007/978-1-4614-3229-6_5 [DOI] [PubMed] [Google Scholar]
- 97.Karan R, Capes MD, Sarma SD. Function and biotechnology of extremophilic enzymes in low water activity. Aquat Biosyst 2012, 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Struvay C, Feller G. Optimization to low temperature activity in psychrophilic enzymes. Int J Mol Sci 2012; 13:11643-65; PMID:23109875; http://dx.doi.org/ 10.3390/ijms130911643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Morelli A, Haugner J, Seelig B. Thermostable artificial enzyme isolated by in vitro selection. PLoS One 2014; 9(11):e112028; http://dx.doi.org/ 10.1371/journal.pone.0113256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yu H, Huang H. Engineering proteins for thermostability through rigidifying flexible sites. Biotechnol Adv 2014; 32:308-15; PMID:24211474; http://dx.doi.org/ 10.1016/j.biotechadv.2013.10.012 [DOI] [PubMed] [Google Scholar]


