Abstract
Since its inception more than 30 years ago, the baculovirus expression vector system (BEVS) has been used prolifically to produce heterologous proteins for research and development. In the cell, a cornerstone of biological activity are multiprotein complexes, catalyzing essential functions. BEVS has been uniquely successful to unlock such complex assemblies for high-resolution structural and functional analysis. Synthetic biology approaches have been implemented to optimize multigene assembly methods, accelerating upstream processes. Specialized baculoviral genomes are being created with functions tailored to enhance production of particular target protein classes. Here we comment on current and emerging developments in the field and their potential to accelerate protein complex research.
Keywords: automation, baculovirus, DNA recombineering, insect cell culture, multiprotein complex, protein modification, recombinant expression, structural biology, synthetic biology
Introduction
In eukaryotes, protein complexes composed of many subunits carry out most essential physiological processes, and are intensely researched today. Some complexes are highly abundant in cells, and can be purified with relative ease from endogenous source material in the quality and quantity required for high resolution analysis of their structure and mechanism. Proteasomes, ribosomes, globins and RNA polymerases are impressive examples. However, this appears to the exception rather that the rule. Our insight into the overwhelming complexity of cellular biology increases rapidly, and it appears that the majority of the protein complexes which function in our body are characterized by low abundance and, moreover, by compositional heterogeneity, which thoroughly complicates or even rules out their extraction from tissue.
Recombinant production affords a convenient means to resolve this bottleneck. Consequently, recombinant protein production has become a dominant force for producing protein specimens today. A number of expression systems exist, utilizing prokaryotic or eukaryotic host organisms, or also cell-free systems, each with their own merit.1,2 Among these, heterologous expression in mammalian cells or insect cells, respectively, have emerged as methods of choice for eukaryotic protein specimens, exemplified by a rapidly growing number of entries in the Protein Data Bank (PDB) using these expression hosts. Mammalian expression has been determinant for the production of large, multi-domain, sometimes highly glycosylated eukaryotic cell surface receptors and their supramolecular assemblies.3 For eukaryotic multiprotein complexes with many subunits, the baculovirus expression vector system (BEVS) has turned out to be particularly powerful, unlocking the structure and mechanism of many important complex assemblies that had remained inaccessible to detailed analysis beforehand.4,5 Today, baculovirus is prolifically used to accelerate a wide range of applications in the life sciences (Fig. 1).
Figure 1.
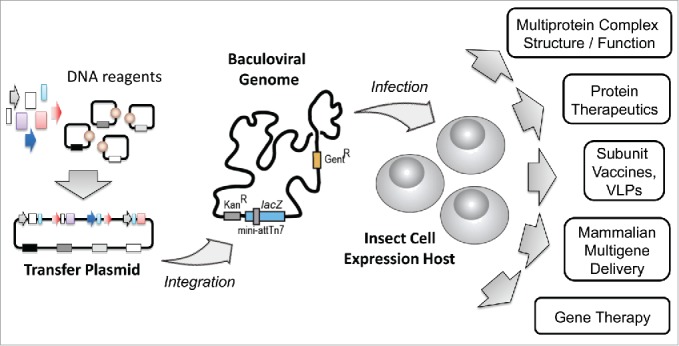
Baculovirus Expression Vector System BEVS. The BEVS can be conceptualized on 3 levels. DNA elements comprising genes of interest, promoters, terminators, transcriptional enhancers and others are combined with plasmid backbone modules into (multi)gene expression cassettes on transfer plasmids (left). Transfer plasmids are then integrated into a baculoviral genome by using transposition or recombination methods to yield a composite baculoviral genome containing all heterologous DNA elements (center). Recombinant baculovirus containing the composite genome is then used to infect insect cell cultures to produce materials for a multitude of down-stream applications (right). All three levels, transfer plasmid assembly, the baculoviral genome and the expression host cell, can be engineered by exploiting powerful synthetic biology tools, for optimizing and maximizing system performance.
Baculovirus Expression: The Origins
The first report illustrating the enormous potential of a recombinant baculovirus was published in 1983. Max Summers, Gale Smith and colleagues used Autographa californica multiple nucleopolyhedrovirus (AcMNPV) to express recombinant human INF-β.6 They have substituted the gene of a viral protein, polyhedrin (polh), with the human gene encoding for INF-β. Since this hallmark study, the procedures were step-by-step improved to render this expression system user friendly, involving engineering of recombinant viruses and development of time and cost-effective protocols for protein production in insect cell cultures.
The original procedure to generate recombinant baculoviruses was based on homologous recombination into the polh locus, in insect cells, between a viral genome and a transfer plasmid harboring the gene(s) of interest. This was not overly efficient, and required inspection of infected cells by eye to identify the polh deficient phenotype which would lack granular shapes that were birefringent. This approach was much improved when suitable restriction enzymes were identified to cleave the baculovirus within the polh locus, thus linearizing the baculoviral genome and eliminating non recombined genomes.7 Now, recircularization of the viral genome, and thus live and infectious baculovirus generation depended on the success of heterologous gene integration, which still occurred by homologous recombination in the insect cell which thereby invited they own destruction, after releasing recombinant baculovirions to infect further cells in the culture. This was already a much improved procedure, and in the hands of experienced users could be applied relatively straight-forwardly in the research laboratory.
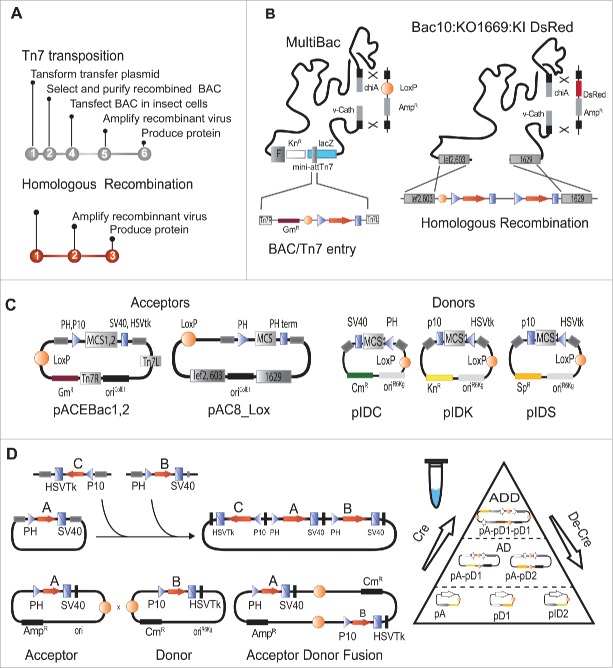
A breakthrough which propelled baculovirus expression and rendered it genuinely accessible to many laboratories was the construction of an artificial chromosome (BAC) made up of the baculoviral genome supplemented by DNA elements that enabled stable propagation of this BAC at very low copy number (1-2) in E.coli cells. This approach profoundly simplified the generation of recombinant baculovirus, removing laborious and time consuming interventions to clonally isolate producer viruses by plaque-purification (Fig. 2A). The first and to date most popular approach for inserting foreign genes into the BAC relies on Tn7-mediated transposition.8 This BAC/Tn7 entry system is used by several popular systems including the classical Bac-to-Bac™ system from Invitrogen / Life sciences, and also by the more recent MultiBac system which in recent years became the lead technology for BEVS-mediated production of multiprotein complexes for many applications in academia and industry.4,9-11 In the BAC/Tn7 approach, prior to the production of the recombinant virus, the gene of interest is cloned into a transfer plasmid harboring the expression cassette which is flanked by short DNA sequences representing the right and left arms of the Tn7 transposon. The sequenced transfer plasmid is then transformed into custom E. coli DH10 cells which contain the BAC and in addition a helper plasmid that supplies the Tn7 transposase needed to carry out the transposition into the Tn7 attachment site that is present on the BAC. The resulting recombinant BAC DNA (bacmid) is extracted and can be verified easily by a PCR reaction with appropriate primers. The recombinant bacmid DNA is then used to transfect insect cells using a lipidic tranfection reagent in small cell cultures. Infected cells then realize the viral genome, giving rise to live baculovirions that can then be used to infect larger cell cultures to produce and purify the protein specimen of interest (Figs. 1, 2).
Figure 2.
Baculovirus multigene expression tools. (A) Flowcharts for production of recombinant viruses and protein expression protein production using either Tn7-mediated transposition (top) or homologous recombination (bottom) are shown. (B) Construction of recombinant baculovirus genomes are shown in a schamtic representation. Transposition of an expression cassette embedded between the Tn7L and Tn7R DNA sequence into the Tn7 attachment site of the baculoviral genome is illustrated on the left (from Ref. 8). Homologous recombination combining a linearized baculoviral DNA genome and a transfer plasmid for inserting the gene(s) of interest is shown on the right (from Ref. 13). Most currently used viral genomes contain deletions of the genes encoding for chiA and V-cath, a chitinase and protease, respectively, shown to be detrimental for protein production and stability. This locus can be used to insert foreign genes including fluorescent makers such as YFP or the DsRed protein to monitor virus performance and protein production. A LoxP site-specific recombination site, inserted at this position, allows to generate composite viral genomes by in vivo Cre-LoxP fusion in E. coli with a customized transfer plasmids containing a conditional replication origin.9 (C) Multigene assembly DNA tools used in tandem recombineering (TR) are shown. So-called Acceptor and Donor plasmids contain multiple cloning sites (MCS) to insert genes of interest by conventional or samless cloning methods of choice. Genes are placed typically under the control of late baculoviral promoters (PH or p10) and eukaryotic polyadenylation signals (PH ter, SV40 or HSVtk). Acceptors and Donors possess multiplications modules (gray rectangles) located on both sides of the expression cassette as well a loxP site (circle filled in orange). (D) For multi-gene construct generation using multiplication modules, individual expression cassettes are excised by digestion with a pair of endonucleases and inserted via compatible restriction sites into the multiplication module of a progenitor plasmid. Following ligation, the restriction sites used for integration are eliminated and multiplication can be repeated iteratively using the intact multiplication module in the inserted cassette.9 Plasmids thus charged with several genes of interest can be fused by Cre mediated recombination via the LoxP sites. Acceptors have a regular origin of replication (ori ColE1), whereas donors have a conditional origin derived from R6Kγ phage (ori R6Kγ), facilitating multigene assembly.21,30,39
Numerous variations to the BAC approach have been implemented. A BAC was constructed carrying a disabling mutation in an essential gene (ORF1692) which impairs virus replication. This detrimental mutation is repaired when the mutant BAC is co-transfected with a transfer plasmid containing the expression cassette and a wild type copy of the mutated gene. Homologous recombination between the viral DNA and the transfer plasmid restores replication, eliminates the bacterial replicon in the polyhedrin locus, and integrates the gene of interest (Figs. 2A, B). As replication of not recombined viral genomes is ruled out here, clonal selection procedures are not required. This effectively generates a time-saving one-step procedure.12-14 Ready-to-use linearized DNA Baculovirus DNAs are available commercially (for example OET's FlashBac) but can also be prepared with reasonable cost and effort from appropriate bacterial strains.13,15 This approach lends itself to automation and thus to high-throughput applications when a large number of constructs are to be screened in expression tests. Automation of the BAC/Tn7 approach is also possible, but far more laborious as many more steps to prepare bacmid DNA have to be considered. Recently, a set of transfer plasmids, called OmniBac, combines the options to use both the Tn7/BAC system and classical heterologous recombination approach using the same entry reagents, thus allowing exploitation of the advantages of either system without having to reclone the heterologous genes of interest.16,17
Protein expression has drawn vast benefit from advances in synthetic biology and recombineering technologies that allow targeted modification of the baculovirus genome.10,16,18 Most platforms for virus production now rely on engineered viral genomes in which genes considered to be detrimental for protein production or stability have been deleted. Among the first genes that were removed following the original polh deletion are v-cath and chiA, that encode a cathepsin protease and a chitinase, respectively, which are involved in the liquefaction of infected larvae.9,19 This was followed up by elimination of further genes (p10, p26, p73).20 The loci for v-cath and chiA are juxtaposed in the viral genome, and this region has been utilized to insert genes encoding maker proteins such as yellow fluorescent protein (YFP) or sea anemone red fluorescent protein (dsRed), as well as chaperones or post-translational modification enzymes by site specific recombination, thus generating novel functionalized genomes (Fig. 2B). Additions can include kinases, phosphates as well as enzymes that allow to synthetize complex carbohydrates found on many secreted proteins of mammalian origin.10,21,22
Synthetic biology is emerging as a profound game-changer in the life sciences and synthetic biology approaches are opening up entirely new avenues that may transform recombinant protein production in the future. We are only beginning to scratch at the surface of what is possible. The design and synthesis of the entire one megabase Mycoplasma mycoides bacterial genome has been achieved.23 We now have all tools in hand to entirely rewrite the 130 kilobases baculovirus genome and create synthetic versions comprising only DNA elements essential for laboratory applications. Synthetic baculoviral genomes with improved genetic stability and customized for optimal protein production of specific classes of proteins at high levels, will benefit for applications ranging from drug discovery to industrial protein therapeutics production.16
Strategies for Protein Complex Production
Expression levels and solubility of recombinant proteins expressed by BEVS can range from a few micrograms to hundreds of milligrams per liter, and cannot be predicted. Typically, constructs need to be modified by truncation, mutation or deletion of low complexity regions. The use of adapted affinity tags is often the only means to obtain pure protein.24,25 Numerous pipelines for expression screening and protein production at medium to large scale (250 ml-6L) have been established.15,26,27
Preparation of multiprotein complexes in particular for structural biology applications has its own challenges and specifics. Sometimes, functional subunits of a given complex can be produced in isolation. Then, protein complexes may be assembled in vitro from these individually expressed subunits. Reconstitution methods are particularly useful when the complex is short-lived and thus cannot be purified intact. One or several components may not be proteinaceous, such as in protein-DNA or protein-RNA complexes or enzymatic assemblies with co-enzymes and prosthetic factors, or when a small-molecular ligand is required to stabilize a complex.28 Here, components are expressed individually, and revised expression experiments can be easily accommodated to modify the composition of the complex by replacing one or more subunits by mutant versions for example to define protein functional domains, optimize solubility or to reduce conformational heterogeneity by eliminating disordered regions that often interfere with high-resolution structural studies.
In many cases, however, in vitro reconstitution is not applicable as individual subunits of a complex often cannot be expressed and manipulated in absence of their protein partners. Then, co-expression is a vital prerequisite.29 In BEVS, this can be achieved by co-infection with multiple viruses, each expressing a single polypeptide, or, alternatively, by infection with a single virus driving the expression of all foreign genes required. If a large number of proteins need to be co-expressed, a combination of both approaches can be applied.4,30 This strategy has been particularly successfully for the reconstitution, structural and functional analysis of large multi-protein complexes including large human general transcription factors TFIID 31,32 and TFIIH33,34 and 2 essential multiprotein components of the gene regulation machinery.
Co-expression of many gene products from several viruses each encoding for a single or limited number of heterologous proteins exploits the capacity of insect cells to be simultaneously infected by multiple viruses. This approach affords a certain flexibility to adjust protein expression ratios by controlling the multiplicity of infection (MOI) of the individual viruses which are administered, and has been used successfully for mapping protein-protein interactions, and for providing additional factors for post-translational modifications and chaperones for enhancing proper folding. Co-infection can work reasonably well for small-scale experiments when a small number (2-3) baculoviruses are added, but scale-up is inefficient and logistically demanding, in particular for the production of complex assemblies that would require co-infection of a large number of viruses, and viral titers for all of these would need to be carefully gauged. The uptake of the individual viruses by the cells in an infected culture will follow a Poisson distribution, resulting in subpopulations of cells infected by subsets of viruses.35 Furthermore, it was shown that excessive viral load can inhibit protein expression.36 Using a single viral genome that contains all heterologous genes circumvents these issues, thus ensuring co-expression of all heterologous genes in every cell in an infected culture. Several early baculovirus transfer plasmids were designed for accepting 2-4 genes (pFastBac™ Dual from Invitrogen / Life Sciences, and pAcUW51, pAcAB, pAcAB4.37,38
A theoretically unlimited number of heterologous genes can be co-expressed by the more recent MultiBac system, which exploits a method called tandem-recombineering (TR) based on seamless gene integration by sequence and ligation independent cloning techniques into an array of small transfer plasmid modules which are then conjoined by site-specific recombination using the Cre-Lox fusion reaction.39 Multigene expression cassettes are then inserted into the MultiBac baculoviral genome by Tn7 transposition into a Tn7 attachment site (in the polh locus) and by site-specific recombination into a LoxP site on the viral backbone (in the v-cath-chiA locus) (Fig. 2B).9,21 A series of transfer plasmids were developed to facilitate multigene cassette assembly (Figs. 2C, D). A large and growing set of tools now exist to assemble DNA elements into large functional multigene circuits, including homing endonuclease based fragment assembly, one-step isothermal in vitro recombination (Gibson cloning), or restriction free (Rf) cloning to name a few39,40,41 all of which can be used for multigene expression with the MultiBac system.
In case of complexes with many different subunits or containing large proteins, the size of the insert and clustering of identical promoter and terminator sequences may lead to genetic instability of the recombinant beculoviral genome, resulting in deletions notably of the inserted foreign DNA. This can be avoided by rigorously adhering to virus amplification protocols designed to minimize such damage.21 Alternatively, heterologous genes can be spread over multiple insertion sites in the baculoviral genome by iterative homologous recombination steps carried out on a BAC in E.coli cells.42,43 Applying these strategies can significantly increase the number of genes that can be co-expressed while preserving the stability of the viral genome.
Stoichiometry of Complex Subunits
Assembly and activity of cellular multiprotein complexes critically depends on correct stoichiometry of the subunits present in the complex. In the cell, elaborate mechanisms involving chaperones, assembly factors and transport between cellular compartments exist that assist and regulate complex assembly. A recombinant expression experiment typically aims at substantial overexpression in a heterologous host cell, in which corresponding mechanisms may or may not exist, but will likely fail to cope with very high level expression of the foreign proteins. This challenge often needs to be addressed during expression and during purification by tight control of the proper stoichiometry of the individual subunits that make up the complex.
Both for complexes reconstituted in vitro and for complexes assembled in situ within the expression host cell, the desired physiological complex will need to be separated from excess subunits and partially assembled sub-complexes, that would otherwise give rise to heterogenous sample preparations impeding down-stream applications, notably structural and mechanistic analyses that require ultra-pure sample. Purification schemes typically combine orthogonal techniques in a sequential manner that has to be customized with care to yield physiological assemblies that are homogenous in composition.44 On the other hand, it may be essential to tightly control the amounts of subunits already when they are expressed for successful complex assembly, prior to extraction. This sometimes imposing challenge was recently addressed in an elegant fashion by adapting a strategy used by certain viruses in nature to realize their proteome. For example Coronavirus expresses its gene product repertoire from 2 large open reading frames that give rise to polyproteins that are co-translationally processed by highly specific proteases, also encoded in the ORFs, into the individual protein components. A recombinant approach which recapitulates this strategy was implemented in the MultiBac system recently, exploiting the action of tobacco etch virus (TEV) protease to process a synthetic polyprotein that was inserted into the MultiBac baculoviral genome by means of a customized transfer plasmid.11,45 This approach, ComplexLink, was successfully applied to produce, for the first time, influenza polymerase that could be crystallized unlocking the atomic structure of this complex which has remained elusive for decades.46
Baculovirus Expression: Stay tuned!
Baculovirus-mediated production of heterologous proteins in insect cells has made come a long way since its inception in the Summers lab many years ago.47,48 Proteins for a wide range of applications, in basic and applied research, from structural biology to vaccinology have benefitted substantially from this powerful and versatile system. Many proteins and protein complexes were produced, often for the first time, by BEVS, in the quality and quantity to enable high resolution structural ad mechanistic analysis, advancing fundamental insight and accelerating drug discovery. Much has been done over the years to develop improved reagents and robust protocols, which already allow efficient application of the system also by non-specialist users, and BEVS is firmly established a work-horse for eukaryotic protein production in many laboratories, in academia and industry. Improved viral backbones with customized functionalities for particular protein classes and applications are being developed, compellingly undersocirng the enormous potential of this system at the forefront of life science research.
Conceptually, BEVS can be understood as a system operating at 3 distinct levels, all of which are amenable to optimization by engineering (Fig. 1). The first level is the transfer reagents which should enable rapid and flexible assembly of multicomponent gene expression circuits, ideally in a highly parallelized, automated manner suited in a robotics enviornment. Promoters, terminators, gene boundaries and affinity tags should be freely exchangable at this level, with minimal investment for the scientists carrying out the experiment. Methods from synthetic biology are essential for achieving this objective, and are being successfully implemented in BEVS, substantially increasing throughput. Moreover, gene synthesis has become thoroughly affordable, effectively multiplying the options to design and lead to success multiparameter expression experiments.
The baculoviral genome constitutes the second level in this concept. The benefit of modifying the baluloviral genome was already evident very early on, when the polh gene was removed from the viral backbone. Before, recombinant baculovirus had to be identifed by a specialist user observing the loss of occluded virus phenotype in infected cells, which occurred with very low efficiency. This improved considerably when the polh locus was disrupted, and dramatically when the baculoviral genome was linearized by endonuclease cleavage in the polh locus. The polh deletion was soon followed by further knock-outs of genes non-essential in laboratory culture, which exhibited a negative effect recombinant protein production, notably v-cath and chiA, and others. Today, with the advent of powerful technologies to assemble large DNAs, the complete de novo synthesis of customized baculoviral genomes, devoid of unnecessary or detrimental elements becomes a genuine possibility. These synthetic baculoviral genomes containing multiple helper functions such as chaperones and modifiers and characterized by improved genomic stability, can be tailored to specific applications, for optimized, maximum level recombinant expression of target proteins.
Genome engineering does not need to be restricted to the baculovirus. Much effort is being devoted currently in the community to the improvement of the characteristics of bacterial host organisms, with major ongoing synthetic biology genome rewiring efforts aimed at generating new and improved chassis for a wide variety of purposes.49 The number of available cell types for BEVS pale to insignificance when compared to the very large number of E.coli strains and engineered variants that are at disposal for recombinant expression in this prokaryotic host. Undoubtedly, it is much more demanding to engineer a eukaryotic cell. The advent of powerful genome engineering tools, notably CRISPR/Cas has revolutionized eukaryotic genome engineering with transformative potential.50 We anticipate that these technologies will be profusely exploited in the near future to extend optimization to the host cell genome, to create specialized cell strains with defined functionalities. Coupled to proteomics and metabolic flux analyses during heterologous expression, genome engineering could be likewise applied to eliminate negative factors in the host cell genome, by targeted deletions ideally in high-throughput. Moreover, helper functionalities such as chaperones and modifiers, but also selected components of a protein complex under investigation or even complete complexes could then be outsourced conveniently from the baculovirus to the host cell genome. This could open entirely new possibilities for drug discovery, greatly accelerating research and progress. For instance, in the pharmaceutical industry, hundreds of kinases are produced, often with baculoviral systems, and many remain problematic and rely on accessory factors. Multiprotein membrane protein targets are increasingly in focus, and are basically not accessible with available methods. Moreover, a major focus in the industrial biotechnology community is on assembling and testing synthetic metabolic circuits and signaling cascades, for which the here described technologies may be uniquely useful. We expect that engineering on all levels will ever improve and optimize this versatile and powerful expression system, enabling researchers to address the most complex question in the biology of health and disease states in the years to come.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank all members of our laboratories for helpful discussion.
Funding
This work was funded by the CNRS, the INSERM, the Université de Strasbourg (UdS), the Alsace Region and the French Infrastructure for Integrated Structural Biology (FRISBI), Instruct, part of the European Strategy Forum on Research Infrastructures (ESFRI) and supported by national member subscriptions. AP acknowledges support from the Agence Nationale de la Recherche (ANR), the Institut National du Cancer (INCA), the Fondation pour la Recherche sur le Cancer (ARC), the Fondation pour la Recherche Médicale (FRM) and La Ligue contre le Cancer. IB acknowledges support from ANR, ARC and the European Commission (EC) Framework Program (FP) 7 project ComplexINC (279039).
References
- 1.Almo SC, Garforth SJ, Hillerich BS, Love JD, Seidel RD, Burley SK. Protein production from the structural genomics perspective: achievements and future needs. Curr Opin Struct Biol 2013; 23(3):335; PMID:23642905; http://dx.doi.org/ 10.1016/j.sbi.2013.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger I, Mayr LM. Protein production for structural biology: new solutions to new challenges. Curr Opin Struct Biol 2013; 23(3):317; PMID:23735677; http://dx.doi.org/ 10.1016/j.sbi.2013.05.004 [DOI] [PubMed] [Google Scholar]
- 3.Aricescu AR, Owens RJ. Expression of recombinant glycoproteins in mammalian cells: towards an integrative approach to structural biology. Curr Opin Struc Biol 2013; 23(3):345; http://dx.doi.org/ 10.1016/j.sbi.2013.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieniossek C, TImasaki , Takagi Y, Berger I. MultiBac: expanding the research toolbox for multiprotein complexes. Trends Biochem Sci 2012; 37(2):49; PMID:22154230; http://dx.doi.org/ 10.1016/j.tibs.2011.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barford D, Takagi Y, Schultz P, Berger I. Baculovirus expression: tackling the complexity challenge. Curr Opin Struct Biol 2013; 23(3):357; PMID:23628287; http://dx.doi.org/ 10.1016/j.sbi.2013.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith GE, Summers MD, Fraser MJ. Production of human beta interferon in insect cells infected with a baculovirus expression vector. Mol Cell Biol 1983; 3(12):2156; PMID:6318086; http://dx.doi.org/ 10.1128/MCB.3.12.2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitts PA, Ayres MD, Possee RD. Linearization of baculovirus DNA enhances the recovery of recombinant virus expression vectors. Nucleic Acids Res 1990; 18(19):5667; PMID:2216760; http://dx.doi.org/ 10.1093/nar/18.19.5667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luckow VA, Lee SC, Barry GF, Olins PO. Efficient Generation of Infectious Recombinant Baculoviruses by Site-Specific Transposon-Mediated Insertion of Foreign Genes into a Baculovirus Genome Propagated in Escherichia-Coli. J Virol 1993; 67(8):4566; PMID:8392598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berger I, Fitzgerald DJ, Richmond TJ. Baculovirus expression system for heterologous multiprotein complexes. Nat Biotechnol 2004; 22(12):1583; PMID:15568020; http://dx.doi.org/ 10.1038/nbt1036 [DOI] [PubMed] [Google Scholar]
- 10.Palmberger D, Wilson IB, Berger I, Grabherr R, Rendic D. SweetBac: a new approach for the production of mammalianised glycoproteins in insect cells. PLoS One 2012; 7(4): e34226; PMID:22485160; http://dx.doi.org/ 10.1371/journal.pone.0034226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crepin T, Swale C, Monod A, Garzoni F, Chaillet M, Berger I. Polyproteins in structural biology. Curr Opin Struct Biol 2015; 32:139; PMID:25996897; http://dx.doi.org/ 10.1016/j.sbi.2015.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Je YH, Chang JH, Choi J, Roh YJ, Jin BR, Reilly O’, Kang SK. A defective viral genome maintained in Escherichia coli for the generation of baculovirus expression vectors. Biotechnol Lett 2001; 23:575; http://dx.doi.org/ 10.1023/A:1010301404445 [DOI] [Google Scholar]
- 13.Zhao Y, Chapman DA, Jones IM. Improving baculovirus recombination. Nucleic Acids Res 2003; 31(2): E6; PMID:12527795; http://dx.doi.org/ 10.1093/nar/gng006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Possee RD, Hitchman RB, Richards KS, Mann SG, Siaterli E, Nixon CP, Irving H, Assenberg R, Alderton D, Owens RJ, King LA. Generation of baculovirus vectors for the high-throughput production of proteins in insect cells. Biotechnol Bioeng 2008; 101(6):1115; PMID:18781697; http://dx.doi.org/ 10.1002/bit.22002 [DOI] [PubMed] [Google Scholar]
- 15.Osz-Papai J, Radu L, Abdulrahman W, Kolb-Cheynel I, Troffer-Charlier N, Birck C, Poterszman A. Insect cells-baculovirus system for the production of difficult to express proteins. Methods Mol Biol 2015; 1258:181; PMID:25447865; http://dx.doi.org/ 10.1007/978-1-4939-2205-5_10 [DOI] [PubMed] [Google Scholar]
- 16.Vijayachandran LS, Thimiri Govinda Raj DB, Edelweiss E, Gupta K, Maier J, Gordeliy V, Fitzgerald DJ, Berger I. Gene gymnastics: Synthetic biology for baculovirus expression vector system engineering. Bioengineered 2013; 4(5):279; PMID:23328086; http://dx.doi.org/ 10.4161/bioe.22966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raj DB, Vijayachandran LS, Berger I. OmniBac: universal multigene transfer plasmids for baculovirus expression vector systems. Methods Mol Biol 2014; 1091:123; PMID:24203327; http://dx.doi.org/ 10.1007/978-1-62703-691-7_7 [DOI] [PubMed] [Google Scholar]
- 18.Roy P, Noad R. Use of bacterial artificial chromosomes in baculovirus research and recombinant protein expression: current trends and future perspectives. ISRN Microbiol 2012; 2012:628797; PMID:23762754; http://dx.doi.org/ 10.5402/2012/628797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaba SA, Salcedo AM, Wafula PO, Vlak JM, van Oers MM. Development of a chitinase and v-cathepsin negative bacmid for improved integrity of secreted recombinant proteins. J Virol Methods 2004; 122(1):113; PMID:15488628; http://dx.doi.org/ 10.1016/j.jviromet.2004.07.006 [DOI] [PubMed] [Google Scholar]
- 20.Hitchman RB, Possee RD, Crombie AT, Chambers A, Ho K, Siaterli E, Lissina O, Sternard H, Novy R, Loomis K, Bird LE, Owens RJ, King LA. Genetic modification of a baculovirus vector for increased expression in insect cells. Cell Biol Toxicol 2010; 26(1):57; PMID:19655260; http://dx.doi.org/ 10.1007/s10565-009-9133-y [DOI] [PubMed] [Google Scholar]
- 21.Fitzgerald DJ, Berger P, Schaffitzel C, Yamada K, Richmond TJ, Berger I. Protein complex expression by using multigene baculoviral vectors. Nat Methods 2006; 3(12):1021; PMID:17117155; http://dx.doi.org/ 10.1038/nmeth983 [DOI] [PubMed] [Google Scholar]
- 22.Geisler C, Mabashi-Asazuma H, Jarvis DL. An Overview and History of Glyco-Engineering in Insect Expression Systems. Methods Mol Biol 2015; 1321:131; PMID:26082220; http://dx.doi.org/ 10.1007/978-1-4939-2760-9_10 [DOI] [PubMed] [Google Scholar]
- 23.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, et al.. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 2010; 329(5987):52; PMID:20488990; http://dx.doi.org/ 10.1126/science.1190719 [DOI] [PubMed] [Google Scholar]
- 24.Abdulrahman W, Uhring M, Kolb-Cheynel I, Garnier JM, Moras D, Rochel N, Busso D, Poterszman A. A set of baculovirus transfer vectors for screening of affinity tags and parallel expression strategies. Anal Biochem 2009; 385(2):383; PMID:19061853; http://dx.doi.org/ 10.1016/j.ab.2008.10.044 [DOI] [PubMed] [Google Scholar]
- 25.Hu NJ, Rada H, Rahman N, Nettleship JE, Bird L, Iwata S, Drew D, Cameron AD, Owens RJ. GFP-based expression screening of membrane proteins in insect cells using the baculovirus system. Methods Mol Biol 2015; 1261:197; PMID:25502201; http://dx.doi.org/ 10.1007/978-1-4939-2230-7_11 [DOI] [PubMed] [Google Scholar]
- 26.Unger T, Peleg Y. Recombinant protein expression in the baculovirus-infected insect cell system. Methods Mol Biol 2012; 800:187; PMID:21964790; http://dx.doi.org/ 10.1007/978-1-61779-349-3_13 [DOI] [PubMed] [Google Scholar]
- 27.Mahajan P, Strain-Damerell C, Gileadi O, Burgess-Brown NA. Medium-throughput production of recombinant human proteins: protein production in insect cells. Methods Mol Biol 2014; 1091:95; PMID:24203326; http://dx.doi.org/ 10.1007/978-1-62703-691-7_6 [DOI] [PubMed] [Google Scholar]
- 28.Perrakis A, Musacchio A, Cusack S, Petosa C. Investigating a macromolecular complex: the toolkit of methods. J Struct Biol 2011; 175(2):106; PMID:21620973; http://dx.doi.org/ 10.1016/j.jsb.2011.05.014 [DOI] [PubMed] [Google Scholar]
- 29.Sokolenko S, George S, Wagner A, Tuladhar A, Andrich JM, Aucoin MG. Co-expression vs. co-infection using baculovirus expression vectors in insect cell culture: Benefits and drawbacks. Biotechnol Adv 2012; 30(3):766; PMID:22297133; http://dx.doi.org/ 10.1016/j.biotechadv.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdulrahman W, Radu L, Garzoni F, Kolesnikova O, Gupta K, Osz-Papai J, Berger I, Poterszman A. The production of multiprotein complexes in insect cells using the baculovirus expression system. Methods Mol Biol 2015; 1261:91; PMID:25502195; http://dx.doi.org/ 10.1007/978-1-4939-2230-7_5 [DOI] [PubMed] [Google Scholar]
- 31.Bieniossek C, Papai G, Schaffitzel C, Garzoni F, Chaillet M, Scheer E, Papadopoulos P, Tora L, Schultz P, Berger I. The architecture of human general transcription factor TFIID core complex. Nature 2013; 493(7434):699; PMID:23292512 [DOI] [PubMed] [Google Scholar]
- 32.Trowitzsch S, Viola C, Scheer E, Conic S, Chavant V, Fournier M, Papai G, Ebong IO, Schaffitzel C, Zou J, et al.. Cytoplasmic TAF2-TAF8-TAF10 complex provides evidence for nuclear holo-TFIID assembly from preformed submodules. Nat Commun 2015; 6:6011; PMID:25586196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdulrahman W, Iltis I, Radu L, Braun C, Maglott-Roth A, Giraudon C, Egly JM, Poterszman A. ARCH domain of XPD, an anchoring platform for CAK that conditions TFIIH DNA repair and transcription activities. Proc Natl Acad Sci U S A 2013; 110(8):E633; PMID:23382212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuper J, Braun C, Elias A, Michels G, Sauer F, Schmitt DR, Poterszman A, Egly JM, Kisker C. In TFIIH, XPD helicase is exclusively devoted to DNA repair. PLoS Biol 2014; 12(9):e1001954; PMID:25268380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vijayachandran LS, Viola C, Garzoni F, Trowitzsch S, Bieniossek C, Chaillet M, Schaffitzel C, Busso D, Romier C, Poterszman A, et al.. Robots, pipelines, polyproteins: enabling multiprotein expression in prokaryotic and eukaryotic cells. J Struct Biol 2011; 175(2):198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mena JA, Ramirez OT, Palomares LA. Population kinetics during simultaneous infection of insect cells with two different recombinant baculoviruses for the production of rotavirus-like particles. BMC Biotechnol 2007; 7:39; PMID:17610729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weyer U, Possee RD. A baculovirus dual expression vector derived from the Autographa californica nuclear polyhedrosis virus polyhedrin and p10 promoters: co-expression of two influenza virus genes in insect cells. J Gen Virol 1991; 72 (Pt 12):2967; PMID:1765769 [DOI] [PubMed] [Google Scholar]
- 38.Belyaev AS, Roy P. Development of baculovirus triple and quadruple expression vectors: co-expression of three or four bluetongue virus proteins and the synthesis of bluetongue virus-like particles in insect cells. Nucleic Acids Res 1993; 21(5):1219; PMID:8385313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haffke M, Viola C, Nie Y, Berger I. Tandem recombineering by SLIC cloning and Cre-LoxP fusion to generate multigene expression constructs for protein complex research. Methods Mol Biol 2013; 1073:131; PMID:23996444 [DOI] [PubMed] [Google Scholar]
- 40.Gibson DG. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol 2011; 498:349; PMID:21601685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peleg Y, Unger T. Application of the Restriction-Free (RF) cloning for multicomponents assembly. Methods Mol Biol 2014; 1116:73; PMID:24395358 [DOI] [PubMed] [Google Scholar]
- 42.Noad RJ, Stewart M, Boyce M, Celma CC, Willison KR, Roy P. Multigene expression of protein complexes by iterative modification of genomic Bacmid DNA. BMC Mol Biol 2009; 10:87; PMID:19725957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanai Y, Athmaram TN, Stewart M, Roy P. Multiple large foreign protein expression by a single recombinant baculovirus: a system for production of multivalent vaccines. Protein Expr Purif 2013; 91(1):77; PMID:23872366 [DOI] [PubMed] [Google Scholar]
- 44.Frey S, Gorlich D. Purification of protein complexes of defined subunit stoichiometry using a set of orthogonal, tag-cleaving proteases. J Chromatogr A 2014; 1337:106; PMID:24636567 [DOI] [PubMed] [Google Scholar]
- 45.Nie Y, Bellon-Echeverria I, Trowitzsch S, Bieniossek C, Berger I. Multiprotein complex production in insect cells by using polyproteins. Methods Mol Biol 2014; 1091:131; PMID:24203328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reich S, Guilligay D, Pflug A, Malet H, Berger I, Crepin T, Hart D, Lunardi T, Nanao M, Ruigrok RW, Cusack S. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 2014; 516(7531):361; PMID:25409151; http://dx.doi.org/ 10.1038/nature14009 [DOI] [PubMed] [Google Scholar]
- 47.Summers MD. Milestones leading to the genetic engineering of baculoviruses as exrpession vectors systems and viral pesticides. Advances in Virus Resarch 2006; 68(4):3; http://dx.doi.org/ 10.1016/S0065-3527(06)68001-9 [DOI] [PubMed] [Google Scholar]
- 48.van Oers MM, Pijlman GP, Vlak JM. Thirty years of baculovirus-insect cell protein expression: from dark horse to mainstream technology. J Gen Virol 2015; 96(Pt 1):6; PMID:25246703; http://dx.doi.org/ 10.1099/vir.0.067108-0 [DOI] [PubMed] [Google Scholar]
- 49.Juhas M. On the road to synthetic life: the minimal cell and genome-scale engineering. Crit Rev Biotechnol 2015; 1:1-8; PMID:25578717; http://dx.doi.org/ 10.3109/07388551.2014.989423 [DOI] [PubMed] [Google Scholar]
- 50.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014; 346(6213):1258096; PMID:25430774; http://dx.doi.org/ 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]