ABSTRACT
The Jun Kinase (JNK) signaling pathway responds to diverse stimuli by appropriate and specific cellular responses such as apoptosis, differentiation or proliferation. The mechanisms that mediate this specificity remain largely unknown. The core of this signaling pathway, composed of a JNK protein and a JNK kinase (JNKK), can be activated by various putative JNKK kinases (JNKKK) which are themselves downstream of different adaptor proteins. A proposed hypothesis is that the JNK pathway specific response lies in the combination of a JNKKK and an adaptor protein upstream of the JNKK. We previously showed that the Drosophila homolog of pRb (Rbf1) and a mutant form of Rbf1 (Rbf1D253A) have JNK-dependent pro-apoptotic properties. Rbf1D253A is also able to induce a JNK-dependent abnormal proliferation. Here, we show that Rbf1-induced apoptosis triggers proliferation which depends on the JNK pathway activation. Taking advantage of these phenotypes, we investigated the JNK signaling involved in either Rbf1-induced apoptosis or in proliferation in response to Rbf1-induced apoptosis. We demonstrated that 2 different JNK pathways involving different adaptor proteins and kinases are involved in Rbf1-apoptosis (i.e. Rac1-dTak1-dMekk1-JNK pathway) and in proliferation in response to Rbf1-induced apoptosis (i.e., dTRAF1-Slipper-JNK pathway). Using a transient induction of rbf1, we show that Rbf1-induced apoptosis activates a compensatory proliferation mechanism which also depends on Slipper and dTRAF1. Thus, these 2 proteins seem to be key players of compensatory proliferation in Drosophila.
KEYWORDS: Apoptosis, compensatory proliferation, dTRAF1, JNK, Rbf1, Rac1
Introduction
The retinoblastoma protein (pRb) is inactivated in a wide variety of cancers.1 It plays important roles in many processes implicated in cell fate decisions such as proliferation, apoptosis and differentiation. Consistent with its tumor suppressor role, pRb inhibits cell proliferation.2 Contrarily to this well characterized role in cell cycle regulation, its role in apoptosis remains poorly understood. Indeed, pRb has been found to be either pro- or antiapoptotic.3-8 However, the precise conditions governing pRb properties in apoptosis remain unknown. Interestingly, pRb is stabilized in response to DNA damage.9 Moreover, pRb is required to upregulate pro-apoptotic genes transcription and cell death after a genotoxic stress both in vivo and in vitro.4 To further clarify how the cell status impacts the role of pRb toward apoptosis regulation, Drosophila has been used as a model organism. The overexpression of rbf1 induces apoptosis in proliferative tissues, whereas no cell death is triggered by rbf1 expression in post-mitotic cells.10 Therefore, in Drosophila overexpression of rbf1 can mimic pRb stabilization in response to DNA damage and used to study its pro-apoptotic function
In addition, a form of Rbf1 mutated at a putative conserved caspase cleavage site, Rbf1D253A, retains Rbf1 pro-apoptotic properties, but is also able to increase cell proliferation leading to an overgrowth phenotype.11 This phenotype illustrates the role of Rbf1 in tissue homeostasis and suggests that Rbf1D253A could stimulate a compensatory proliferation mechanism. Activation of the JNK (Jun N-terminal kinase) pathway is required both for Rbf1 and Rbf1D253Ainduced apoptosis and for Rbf1D253A-induced overgrowth phenotype. Thus, fly constitutes a good animal model system to study pro-apoptotic properties of a pRb homolog in vivo in a simple genetic background.
The JNK signaling pathway is conserved among metazoans. It plays an important role in different processes such as tissue morphogenesis, wound healing, immunity, programmed cell death or aging.12,13 It consists of a kinase cascade leading to the phosphorylation and activation of the JNK proteins. The activated JNK translocates into the nucleus where it phosphorylates transcription factors, thus modulating transcriptional activation of target genes. JNK is activated by JNKKs which are themselves activated by a great number of JNKKKs.14 Another level of complexity lies in the mode of activation of the JNKKKs which involves different adaptor proteins: small GTPases of the Ras superfamily, kinases (i.e. : Msn) or scaffold proteins such as Traf1 (TNF receptor associated factor 1). Despite extensive progress in the understanding of the JNK pathway, the mechanisms by which it contributes to pleiotropic effects are poorly defined.
Here, we used Rbf1 and Rbf1D253A-induced phenotypes to tackle the problem of JNK signaling specificity in vivo. We identified the specific adaptor proteins and JNKKKs responsible for the JNK activation leading to apoptosis or proliferation and showed that rbf1-induced apoptosis stimulates a JNK pathway-dependent compensatory proliferation
Results
Both rbf1-induced apoptosis and proliferation in response to apoptosis involve a JNK pathway activation
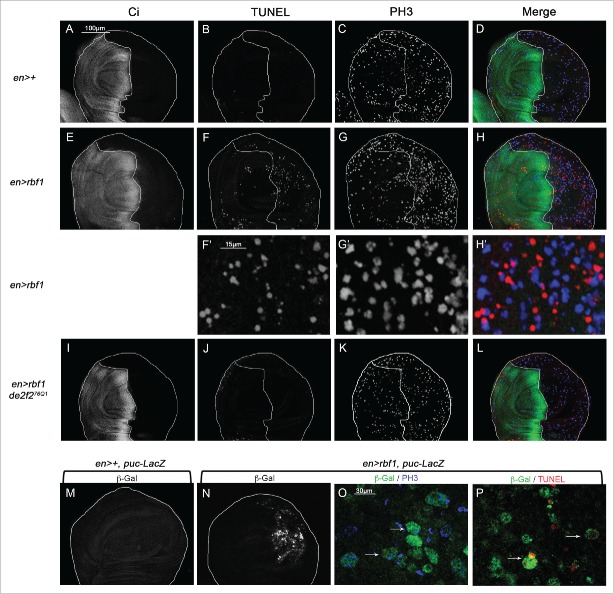
We previously demonstrated that Rbf1D253A, a mutant form of Rbf1, induces both apoptosis and an overgrowth wing phenotype associated with JNK pathway-dependent abnormal non-cell autonomous proliferation. We hypothesized that this mutant form of Rbf1 could deregulate an apoptosisinduced proliferation (AiP) mechanism.10 To test this hypothesis, we first determined whether rbf1-induced apoptosis promotes compensatory proliferation. We performed simultaneously a TUNEL staining to detect apoptotic cells and an anti-phospho-histone H3 (PH3) staining to label mitotic cells in wing imaginal discs of en-Gal4/+ and en-Gal4/+; UAS-rbf1/+ third instar larvae. The en-Gal4 driver allows the expression of rbf1 specifically in the posterior compartment of the wing imaginal disc. We used anti-Ci antibodies to stain the anterior compartment and mark out the posterior compartment (Figure 1A and E). As expected, numerous apoptotic cells were observed in the posterior compartment of en-Gal4/+; UAS-rbf1/+ wing discs, compared with enGal4/+ control wing disc (Figure 1B, F and F'). Interestingly, an increase in PH3 staining was observed in the rbf1-expressing compartment (Figure 1C, G and G'; Figure 1) compared with control. Moreover we observed TUNEL and PH3 labeling in adjacent cells in en-Gal4/+; UAS-rbf1/+ wing imaginal disc (Figure 1H') showing that proliferating cells are close to dying cells. We next tested whether this increase of proliferation is dependent on the presence of apoptotic cells. Since rbf1-induced apoptosis requires the dE2F2 transcription repressor and is inhibited in a heterozygous de2f2 mutant background,15 we used the de2f276Q1 loss of function mutant to inhibit rbf1-induced apoptosis. The inhibition of apoptosis in en-Gal4/de2f276Q1; UAS-rbf1/+ wing imaginal discs was associated with a reduction of PH3 staining compared with en-Gal4/+; UAS-rbf1/+ (Figure 1J, K). A similar result was obtained when apoptosis was decreased in a debcl loss of function context (unpublished data). Thus, the proliferation increase observed in rbf1 overexpressing discs was triggered in response to apoptosis.
Figure 1.
rbf1-induced apoptosis triggers cell proliferation and JNK signaling pathway activation in wing imaginal discs. (A, E, I) anti-Ci staining used to visualize the anterior domain in wing pouch imaginal discs from en-Gal4/+ or en-Gal4/+; UAS-rbf1/+ or en-gal4/de2f276Q1; UAS-rbf1/+ third instar larvae. A line indicates the antero-posterior frontier and the posterior compartment is on the right side. (B, F, J) Visualization of apoptotic cells by TUNEL staining in wing pouch imaginal discs of previously described genotypes. (C, G, K) Visualization of mitotic cells by PH3 staining in wing pouch imaginal discs of previously described genotypes. (F', G' H') High magnification from F, G and H. (D, H, L) Merge of anti-Ci, TUNEL and PH3 staining. (M, N) Visualization of β-Galactosidase in wing pouch imaginal discs from en-Gal4/puc-LacZ or en-Gal4/puc-LacZ; UAS-rbf1/+ third instar larvae. (O) Visualization of β-Galactosidase and PH3 staining in wing pouch imaginal discs from en-Gal4/puc-LacZ; UAS-rbf1/+. (P) Visualization of β-Galactosidase and TUNEL staining in wing pouch imaginal discs from en-Gal4/puc-LacZ; UAS-rbf1/+. Arrows showed double labeled cells.
JNK signaling pathway can be associated with both apoptosis and proliferation processes. To investigate in which cells the JNK pathway is activated consecutively to rbf1 expression, we used the puc-LacZ reporter, which monitors JNK activity.16 rbf1 overexpression caused an activation of the JNK signaling both in apoptotic and in proliferative cells, as PH3 or TUNEL positive cells were detected by the presence of β-Gal staining (Figure 1M-P). Thus the JNK pathway is activated in both apoptotic and proliferating cells.
Altogether, these data indicate that rbf1-induced apoptosis triggers cell proliferation in response to apoptosis and that both events were associated with an activation of the JNK signaling pathway.
The specific adaptor dRac1 and the dTak1 and dMekk1 kinases are required for rbf1-induced apoptosis
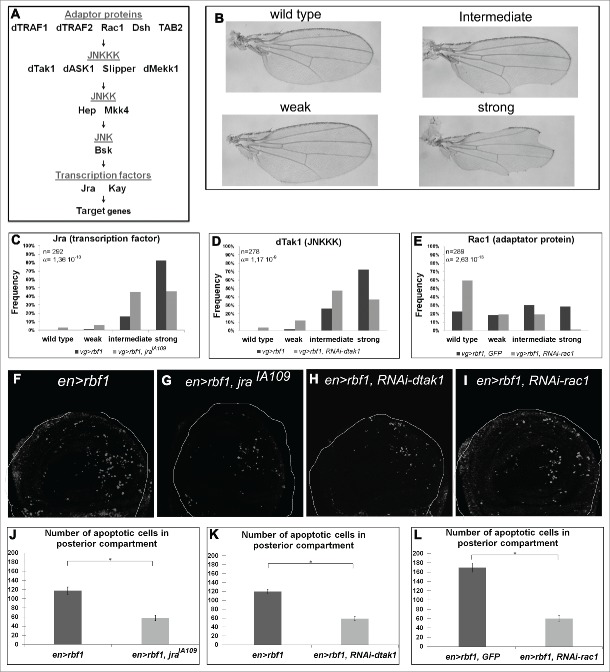
JNK signaling pathway involves a kinase cascade composed of different MAP Kinases (Figure 2A). By co-expressing rbf1 with a bsk-RNAi to disrupt the JNK pathway, we showed that the JNK Bsk was required for Rbf1-induced apoptosis.11 In this study, we tested the involvement of JNK pathway effectors using at least 2 mutant or RNAi lines per effector (Tables S1 and 2). RNAi efficiency has been confirmed previously.17,18 First, to check that the Jun-related antigen (Jra) and the Kayak (Kay) transcription factors were necessary, we performed genetic interaction assays. We took advantage of the fact that overexpression of rbf1, with the vestigial (vg)-Gal4 driver induces notches along the wing margin whose number is correlated with the amount of apoptosis in wing imaginal discs of third instar larvae.10, Thus, we tested whether rbf1-induced adult phenotypes and rbf1-induced apoptosis were altered in a jraIA109 loss of function mutant heterozygous background. Rbf1-induced wing phenotypes were classified into 4 categories according to the number of notches: wild type (no notch), weak, intermediate and strong (Figure 2B). When rbf1 was overexpressed in a jraIA109 heterozygous background, a significant shift toward weaker phenotypes was observed compared with overexpression of rbf1 alone (Figure 2C), showing that jra is necessary for rbf1-induced wing notches phenotype. To confirm that jra was involved in rbf1-induced apoptosis, we performed TUNEL assays on wing imaginal discs of larvae expressing rbf1 under control of en-Gal4 driver (Figure 2F) and quantified the number of apoptotic cells, specifically in the rbf1-overexpressing posterior domain . The number of TUNEL-labeled cells was significantly weaker in en-Gal4/ jraIA109; UASrbf1/+ wing imaginal discs compared with en-Gal4/+; UAS-rbf1/+ discs (Figure 2F-G and J), confirming that the phenotypic distribution shift was correlated with the amount of apoptosis in larvae. We then assessed the involvement of Kay transcription factor, as well as Hep and Mkk4 JNKK in rbf1-induced apoptosis. Fly and larvae co-expressing either RNAi-kay, RNAi-hep or RNAi-mkk4 together with rbf1 were analyzed. The partial depletion of Kay transcription factor or Hep JNKK suppressed both rbf1-induced notches phenotype and apoptosis, whereas RNAi-mkk4 had no effect on these phenotypes (Table 1). The involvement of kay and hep in Rbf1-induced apoptosis was confirmed by using other RNAi or mutant lines (Table S1). Altogether, these results indicate that in addition to Bsk,11 Jra, Kay and Hep have a pivotal role in rbf1-induced apoptosis.
Figure 2.
rbf1-induced apoptosis requires the GTPase Rac1 and the specific kinases dTak1 and dMekk1. (A) Diagram of the JNK signaling pathway. (B) Apoptosis induced by rbf1 in vg-Gal4/+; UAS-rbf1/+ third instar larval wing imaginal discs give rise to notches in adult wings. Adult wing phenotypes are grouped into 4 categories (wild type, weak, intermediate and strong) according to the numbers of notches observed along wing margin (adapted from Milet et al. 2010).10 Comparison of notch wing phenotypes distribution between : (C) vg-Gal4/+; UAS-rbf1/+ and vg-Gal4/ jraIA109; UAS-rbf1/+ flies, Wilcoxon test: n = 292, α=1,36 10−10, Ws=−6,60; (D) vg-Gal4/+; UAS-rbf1/+ and vg-Gal4/ UAS-RNAi dtak1; UAS-rbf1/+ flies, Wilcoxon test: n= 278, α =1,17 10−9, Ws=−6,22; (E) vg-Gal4/UAS-egfp; UAS-rbf1/+ and vg-Gal4/ UAS-RNAi rac1; UAS-rbf1/+ flies, Wilcoxon test: n = 278, α =1,17 10-9, Ws=−6,22. Each experiment presented in C-E was independently performed 3 times. In order to take into account the effect of the genetic background, controls of Figure 2 C and D correspond to a w1118 and y,w[1118];P{attP,y[+],w[3′]} (transformant ID 60100) strains respectively, while control of part E corresponds to the TRIP RNAi stock strain (BL35786). (F-I) Visualization of apoptotic cells by TUNEL staining in wing pouch imaginal discs from en-Gal4/UAS-rbf1 or en-Gal4/ jraIA109; UAS-rbf1/+, or en-Gal4/UAS-RNAi dtak1; UAS-rbf1/+ or en-Gal4/ UAS-RNAi rac1; UAS-rbf1/+ third instar larvae. Posterior compartment is on the right side. (J-L) Comparison of apoptotic cells numbers in the wing pouch imaginal discs between en-Gal4/+; UAS-rbf1/+ and en-Gal4/ jraIA109; UAS-rbf1/+ third instar larvae; between en-Gal4/+; UAS-rbf1/+ and en-Gal4/UAS-RNAi dtak1; UAS-rbf1/+ third instar larvae and between UAS-rbf1/+ or en-Gal4/ UAS- RNAi rac1; UAS-rbf1/+ third instar larvae. Asterisks indicate a statistically significant difference between 2 genotypes (Student's test α < 0.05). For each genotype, quantifications were done for 30 third instar larval wing imaginal discs at least.
Table 1.
Characterization of the JNK signaling pathway effectors required for rbf1-induced apoptosis.
| Adult stage (wing) | Larval stage (wing imaginal disc) | ||
|---|---|---|---|
| Rbf1 | Notch phenotype | Quantification of apoptosis | |
| Transcription factors | jra IA109 | Rescue n = 292, α = 1.36 10–10, Ws = −6.60 | Decrease n = 39, α<10–30 |
| UAS-RNAi-kay | Rescue n = 284, α = 2.12 10–7, Ws = −5.24 | Decrease n = 42, α<10–30 | |
| JNKK | UAS-RNAi-hep | Rescue n = 239, α = 1.36 10–12, Ws = −7.38 | Decrease n= 54, α<10–30 |
| UAS-RNAi-mkk4 | No effect n = 240, α = 0.14, Ws = 1.47 | No effect n = 49, α = 0.47 | |
| JNKKK | UAS-RNAi-dtak1 | Rescue n = 278, α = 1.17 10–9, Ws = −6.22 | Decrease n = 28, α<10–30 |
| UAS-RNAi-dASK1 | No effect n = 232, α = 0.06, Ws = 1.87 | No effect n = 46, α = 0.33 | |
| UAS-RNAi-slipper | No effect n = 194, α = 0.17, Ws = 1.36 | No effect n = 41, α = 0,46 | |
| UAS-RNAi-dmekk1 | Rescue n = 264, α<10 −30, Ws = −12.96 | Decrease n = 53, α<10–30 | |
| Adaptator proteins | UAS-RNAi-dtraf1 | No effect n = 200, α = 0.45, Ws = 0.75 | No effect n = 59, α = 0.44 |
| UAS-RNAi-dtraf2 | No effect n = 217, α = 0.33, Ws = −0.95 | No effect n = 49, α = 0.25 | |
| UAS-RNAi-rac1 | Rescue n = 289, α = 2.63 10–13, Ws = −7.65 | Decrease n = 45, α<10–30 | |
| dsh 1 | No effect n = 165, α = 0.23, Ws = −1.20 | No effect n = 51, α = 0.22 | |
| UAS-RNAi-tab2 | No effect n = 246, α = 0.18, Ws = −1.32 | No effect n = 44, α = 0.67 | |
The JNK signaling pathway can be activated by a wide range of stimuli involving many adaptor proteins and JNKKK (Figure 2A). Thus, we tested whether a specific cascade was necessary for rbf1-induced apoptosis. In order to probe the role of 4 JNKKKs and 5 adaptor proteins in rbf1induced apoptosis, we used several specific RNAi or mutants for each gene to perform genetic interaction tests and TUNEL stainings. The results obtained are summarized in Table 1 and Table S1. The wing phenotypic distribution, TUNEL staining and quantification are shown for dtak1 and rac1 in Figure 2D, H, K and E, I, L respectively. We found a significant rescue of rbf1-induced adult phenotypes and a decrease of rbf1-induced apoptosis for 2 of the JNKKKs tested, dTak1 and dMekk1, and for only one adaptor protein, Rac1 (Table 1). On the contrary, the alteration of the levels of dASK1, Slipper, dTRAF1, dTRAF2, Dsh and TAB2 had no effect on rbf1-induced notches phenotype and apoptosis. The involvement of dtak1, dmekk1 and rac1 in Rbf1-induced apoptosis was confirmed using other RNAi or mutant lines (Supplementary Table 1). All these results indicate that a Rac1/dTak1-dMekk1/Hep/Bsk JNK pathway is necessary for the activation of Jra-Kay transcription factors which induce apoptosis in response to rbf1 overexpression in the wing disc cells.
The specific adaptor dTRAF1 and the kinase Slipper are required for rbf1D253A-induced overgrowth phenotype and rbf1-induced proliferation in response to apoptosis
The overexpression of the rbf1D253A mutant form induces both notches and overgrowth wing phenotype as shown Figure 3A. We previously showed that the kinase Bsk mediates rbf1D253A-induced apoptosis, and is responsible for rbf1D253A-induced overgrowth.11 Thus, we wondered which kinases and adaptor proteins of the JNK pathway were involved in these phenotypes. As previously done for Rbf1, we performed genetic interaction tests, using mutants and RNAi, to disrupt the JNK pathway in vg-Gal4, UAS-rbf1D253A/+ overexpression background. The notches phenotypes were classified and analyzed as described above and overgrowth phenotypes were simultaneously counted. Both rbf1D253A-induced phenotypes were quantified and compared between control and JNK pathway disrupted background. TUNEL staining was performed in parallel on wing imaginal discs of each genotype. The results are presented in Table 2, Table S2 and Figure 3. Interestingly, when rbf1D253A was expressed in jraIA109 mutant heterozygous background or co-expressed with a specific RNAi against kay, hep, dtak1, dmekk1 or rac1, a rescue of the notches phenotype was observed. In all these contexts, quantifications of TUNEL staining in third instar larval wing imaginal discs confirmed that the observed shift of phenotypic distribution was correlated with a decrease in the amount of apoptosis. Therefore, the Rac1-dTak1-dMekk1-JNK pathway necessary for rbf1-induced apoptosis is also involved in rbf1D253A-induced apoptosis. As expected, the observed apoptosis decrease was associated with a suppression of the overgrowth phenotype, which is in agreement with the hypothesis that overgrowth phenotype is triggered by some proliferation in response to rbf1D253A-induced apoptosis. As observed for rbf1-induced apoptosis, the decrease of JNKK Mkk4, JNKKK dASK1 or the adaptor proteins dTRAF2, Dsh and TAB2 had no effect on rbf1D253A-induced apoptosis, notches or overgrowth phenotypes. Surprisingly, the co-expression of rbf1D253A with a RNAi specific to the JNKKK slipper or the adaptor protein dtraf1 significantly enhanced the notches phenotype (Figure 3B and E). Interestingly, these shifts toward stronger phenotypes were not associated with a change in apoptosis level but with a rescue of the overgrowth phenotype (Figure 3C-D and F-G). Thus, the reduction of mRNA levels of slipper or dtraf1 was able to inhibit overgrowth phenotype without inhibiting apoptosis. Slipper and dTRAF1 are the only regulators we found necessary for rbf1D253A-induced overgrowth phenotype but not involved in rbf1D253A-induced apoptosis pathway (Table 2 and Table S2), suggesting that a dTRAF1-Slipper-JNK pathway is necessary for rbf1D253A-induced proliferation in response to apoptosis.
Figure 3.
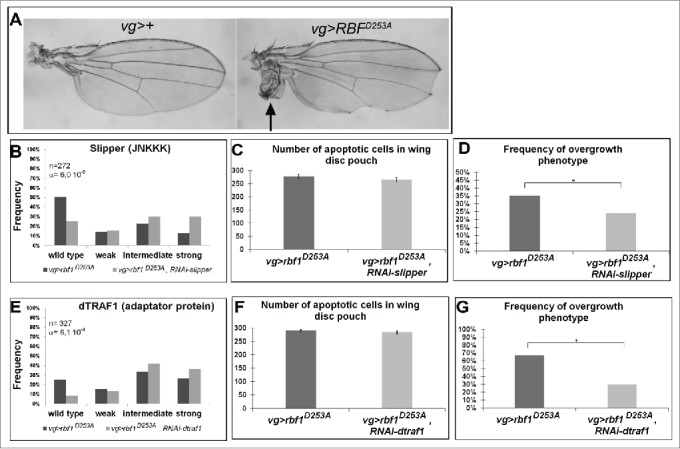
The kinase Slipper and the TNF associated factor dTRAF1 are required for rbf1D253A-induced overgrowth phenotypes but not for rbf1D253A-induced apoptosis. (A) Wild type wing and overgrowth wing phenotype observed in some fly expressing rbf1D253A (adapted from Milet et al. 2014).10 (B, E) Comparison of notch wing phenotypes distribution between vg-Gal4, rbf1D253A/+ and vg-Gal4, rbf1D253A/UAS-RNAi slipper (Wilcoxon test: n = 272, α = 6,0 10-06, Ws = 4,54) and between vg-Gal4, rbf1D253A /and vg-Gal4, rbf1D253A /+; UAS-RNAi dtraf1/+ flies (Wilcoxon test: n = 327, α = 8,1 10-4, Ws = 3,34). (C, F) Comparison of apoptotic cells numbers in the wing pouch imaginal discs between vg-Gal4, rbf1D253A/+ and vg-Gal4, rbf1D253A /UAS-RNAi slipper third instar larvae and between vg-Gal4, rbf1D253A/+ and vg-Gal4, rbf1D253A/+; UAS-RNAi dtraf1/+ third instar larvae. Asterisks indicate a statistically significant difference between 2 genotypes (Student's test α < 0.05). For each genotype, quantifications were done for 30 third instar larval wing imaginal discs at least. (D, G) Frequencies of rbf1D253A-induced ectopic tissue observed in vg-Gal4, rbf1D253A/+ and vg-Gal4, rbf1D253A /UAS-RNAi slipper flies and in vg-Gal4, rbf1D253A/+ and vg-Gal4, rbf1D253A/+; UAS-RNAi dtraf1/+ flies. Asterisks indicate a statistically significant difference between 2 genotypes (Chi2 test α < 0.05). Each experiment presented in B, D, E and G was independently performed 3 times.
Table 2.
Characterization of the JNK signaling pathway effectors required for rbf1D253A-induced apoptosis and overgrowth phenotypes.
| Adult stage (wing) |
Larval stage (wing imaginal disc) |
|||
|---|---|---|---|---|
| Rbf1D253A | Notch phenotype | Overgrowth phenotype | Quantification of apoptosis | |
| Transcription factors | jra IA109 | Rescue n= 417, α = 3.5 10–4, Ws=-3.56 | Rescue n= 511, α = 3.74 10–2 | Decrease n= 52, α<10–30 |
| UAS-RNAi-kay | Rescue n= 417, α = 1.1 10–11, Ws=-7.04 | Rescue n= 417, α = 0.86 10–2 | Decrease n= 45, α<10–30 | |
| JNKK | UAS-RNAi-hep | Rescue n= 348, α = 6.7 10–7, Ws=-5.01 | Rescue n= 384, α<10–30 | Decrease n= 47, α<10–30 |
| UAS-RNAi-mkk4 | No effect n= 207, α = 0.4, Ws=-0.90 | No effect n= 212, α = 0.49 | No effect n= 51, α = 0.53 | |
| JNKKK | UAS-RNAi-dtak1 | Rescue n= 198, α = 1.1 10–8, Ws=-5.82 | Rescue n= 252, α<10–30 | Decrease n= 56, α<10–30 |
| UAS-RNAi-dASK1 | No effect n= 354, α = 8.8 10–2, Ws=-2.61 | No effect n= 370, α = 0.48 | No effect n= 41, α = 0.78 | |
| UAS-RNAi-slipper | Enhancement n= 272, α = 6.0 10–6, Ws = 4.54 | Rescue n= 294, α = 4.33 10–2 | No effect n= 40, α = 0.23 | |
| UAS-RNAi-dmekk1 | Rescue n= 352, α = 2.2 10–6, Ws=-4.76 | Rescue n= 372, α = 1.19 10–2 | Decrease n= 38, α<10–30 | |
| Adaptator proteins | UAS-RNAi-dtraf1 | Enhancement n= 327, α = 8.1 10–4, Ws = 3.34 | Rescue n= 357, α<10–30 | No effect n= 53, α = 0.42 |
| UAS-RNAi-dtraf2 | No effect n= 279, α = 0.5, Ws = 0.64 | No effect n= 336, α = 0.39 | No effect n= 46, α = 0.17 | |
| UAS-RNAi-rac1 | Rescue n= 625, α = 3.4 10–9, Ws=-6.03 | Rescue n= 708, α<10–30 | Decrease n= 49, α<10–30 | |
| dsh 1 | No effect n= 434, α = 4.0 10–3, Ws=-2.87 | No effect n= 476, α = 0.34 | No effect n= 40, α = 0.60 | |
| UAS-RNAi-tab2 | No effect n= 461, α = 1 10–2, Ws=-2.47 | No effect n= 505, α = 0.27 | No effect n= 36, α = 0.91 | |
To test this hypothesis, we investigated whether Slipper and dTraf1 were required for rbf1-apoptosis induced proliferation. We performed PH3 staining in larval wing imaginal discs of en-Gal4/+; UAS-rbf1/+ in control background or in a JNK pathway disrupted background, using the specific mutants or RNAi tested above. The partial depletion of all the effectors necessary for rbf1-induced apoptosis (kay, jra, hep, dtak1, dmekk1 or rac1) induces a decrease in PH3 staining (Table 3 and Table S1), confirming that this proliferation is linked to the apoptosis. Contrarily, co-expression of a RNAi-slipper or a RNAi-dtraf1 with rbf1 significantly reduced proliferation levels (Figure 4) with no effect on apoptosis (Table 1). These results show that a dTRAF1-Slipper-JNK pathway triggers rbf1 AiP.
Table 3.
Characterization of the JNK signaling pathway effectors required for proliferation induced in response to rbf1-induced apoptosis.
| Larval stage (wing imaginal disc) | ||
|---|---|---|
| Rbf1 | Quantification of proliferation | |
| Transcription factors | jra IA109 | Decrease n = 50, α<10–30 |
| UAS-RNAi-kay | Decrease n = 39, α=1.3 10–3 | |
| JNKK | UAS-RNAi-hep | Decrease n = 48, α<10–30 |
| UAS-RNAi-mkk4 | No effect n = 42, α=0.638 | |
| JNKKK | UAS-RNAi-dtak1 | Decrease n = 41, α<10–30 |
| UAS-RNAi-dASK1 | No effect n = 43, α=0.791 | |
| UAS-RNAi-slipper | Decrease n = 47, α<10–30 | |
| UAS-RNAi-dmekk1 | Decrease n = 35, α<10–30 | |
| Adaptator proteins | UAS-RNAi-dtraf1 | Decrease n = 39, α<10–30 |
| UAS-RNAi-dtraf2 | No effect n = 46, α=0.849 | |
| UAS-RNAi-rac1 | Decrease n = 40, α<10–30 | |
| dsh 1 | No effect n = 33, α=0.606 | |
| UAS-RNAi-tab2 | No effect n = 38, α=0.894 | |
Figure 4.
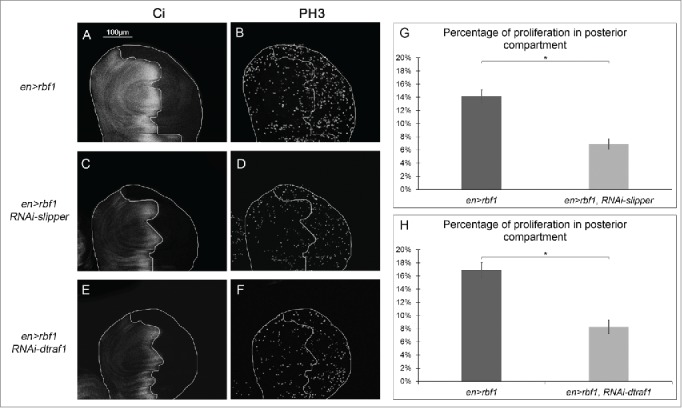
Cell proliferation induced in response to Rbf1-apoptosis requires Slipper and dTRAF1 (A, C, E) anti-Ci staining used to visualize the anterior domain in wing pouch imaginal discs from en-Gal4/+; UAS-rbf1/+ or en-Gal4/UAS-RNAi slipper ; UAS-rbf1/+ or en-Gal4/+; UAS-rbf1/UAS-RNAi dtraf1 third instar larvae. A line indicates the antero-posterior frontier and the posterior compartment is on the right side. (B, D, F) Visualization of mitotic cells by PH3 staining in wing pouch imaginal discs from the previously described genotypes. (G, H) Comparison of proliferation percentage in posterior compartment between wing pouch imaginal discs from en-Gal4/+; UAS-rbf1/+ and en-Gal4/ UAS-RNAi slipper ; UAS-rbf1/+ third instar larvae or between en-Gal4/+; UAS-rbf1/+ and en-Gal4/+; UAS-rbf1/UAS-RNAi dtraf1 third instar larvae. Asterisks indicate a statistically significant difference between 2 genotypes (Student's t-test α < 0.05). Each experiment presented in B, and H was independently performed 3 times.
Therefore, 2 distinct JNK pathways comprising different adaptor proteins and kinases are involved in rbf1-induced apoptosis (i.e., Rac1-dTak1-dMekk1-JNK pathway) and rbf1–induced AiP (i.e. dTRAF1-Slipper-JNK pathway).
dtraf1 overexpression is sufficient to increase the proliferation triggered by Rbf1-induced apoptosis
We suggested that rbf1D253A overgrowth induced-phenotype results from a deregulation of the AiP mechanism.10. We thus wondered whether an overexpression of dtraf1 would be sufficient to enhance rbf1 AiP and thus mimic rbf1D253A-induced overgrowth phenotype. First, to determine whether dtraf1 overexpression could increase the JNK pathway activation triggered by rbf1, we quantified by RT-qPCR the mRNA levels of 2 JNK pathway target genes: puc and mmp1 (Figure 5A). As expected, vg-Gal4/ UAS-dtraf1; UAS-rbf1/+ greatly increased puc and mmp1 expression compared with vg-Gal4/UAS-dtraf1 control. Then, we compared the notches phenotype of vg-Gal4/+; UAS-rbf1/+ with vg-Gal4/UAS-dtraf1; UAS-rbf1/+ flies. We verified that vg-Gal4/ UAS-dtraf1 flies do not exhibit any particular wing phenotype. dtraf1 overexpression triggered a significant shift of rbf1-induced notches phenotype toward weaker phenotypes (Figure 5B), indicating that dtraf1 expression can partially suppress rbf1-induced notches phenotype. We performed TUNEL and PH3 stainings to compare en-Gal4/ UAS-dtraf1 with en-Gal4/+ control wing imaginal discs. dtraf1 overexpression did not modify neither apoptosis nor proliferation levels compared to en-Gal4/+ control (Figure 5 C-H). Interestingly, the co-expression of dtraf1 with rbf1 did not affect rbf1-induced apoptosis (Figure 5H, 5K and 5O), but significantly increased rbf1-induced AiP (Figure 5I; 5L and 5P). Moreover, some rare overgrowth phenotypes were observed in wing of flies co-expressing rbf1 and dtraf1 (Figure 5R). These results show that dtraf1 overexpression is sufficient to enhance rbf1-induced AiP, confirming that overgrowth phenotypes ensue from an excess of proliferation during wing imaginal discs development.
Figure 5.
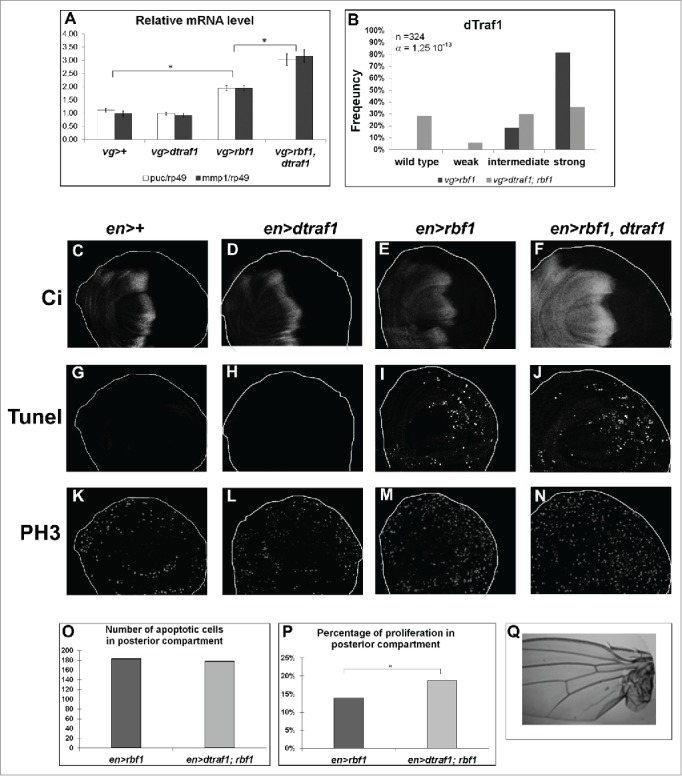
dtraf1 overexpression does not affect Rbf1-induced apoptosis but increases the proliferation and the overgrowth phenotypes induced in response to Rbf1 (A) puc and mmp1 mRNA quantification by RT-qPCR in wing imaginal discs. Data are normalized against rp49 and correspond to the mean of 3 independent experiments. Error bars are the SEM. Asterisks indicate statistical significant difference between 2 genotypes (Student's t-test, α < 0,05). (B) Comparison of notch wing phenotypes distribution between vg-Gal4/+; UAS-rbf1/+ and vg-Gal4/+; UAS-rbf1/UAS-dtraf1 flies. Statistical analysis was performed using Wilcoxon test: n = 324, α = 1,25 10-13, Ws=−7,76 (C, D, E, F) Ci staining used to visualize the anterior domain in wing pouch imaginal discs from en-Gal4/+; +/+ or en-Gal4/ UAS-dtraf1; +/+ or en-Gal4/+; UAS-rbf1/+ or en-Gal4/ UAS-dtraf1; UAS-rbf1/+ third instar larvae. A line indicates the antero-posterior frontier and the posterior compartment is on the rightside. (G, H, I, J) Visualization of apoptotic cells by TUNEL staining in wing pouch imaginal discs from the previously described genotypes. (K, L, M, N) Visualization of mitotic cells by PH3 staining in wing pouch imaginal discs from the previously described genotypes. (0) Comparison of apoptotic cells numbers in posterior compartment of the wing pouch between imaginal discs from en-Gal4/+; UAS-rbf1/+ and en-Gal4/ UAS-dtraf1; UAS-rbf1/+ third instar larvae. Asterisks indicate a statistically significant difference between 2 genotypes (Student's test α < 0.05). For each genotype, quantifications were done for at least 30 third instar larval wing imaginal discs. (P) Comparison of proliferation percentage in posterior compartment of the wing pouch between imaginal discs from en-Gal4/+; UAS-rbf1/+ and en-Gal4/ UAS-dtraf1 ; UAS-rbf1/+ third instar larvae. Asterisks indicate a statistically significant difference between 2 genotypes (Student's test α < 0.05). (R) Overgrowth wing phenotype observed in some fly co-expressing rbf1 and dtraf1. Each experiment presented in B and P was independently performed 3 times.
Rbf1-induced apoptosis activates a compensatory proliferation mechanism that depends on slipper and dtraf1
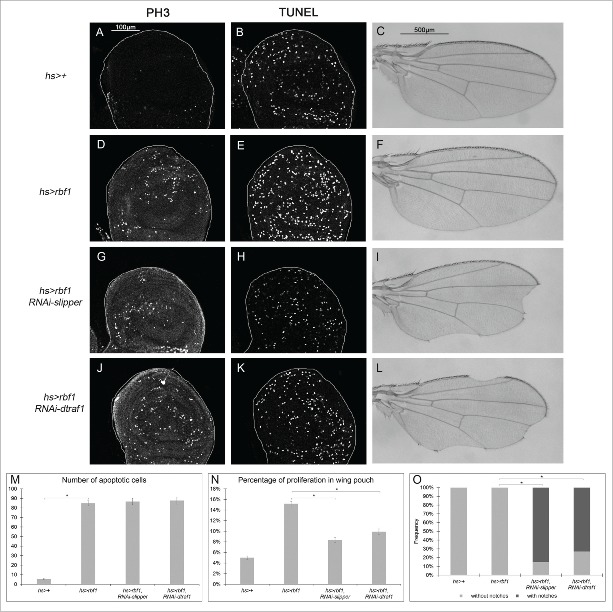
In our assay with vg-Gal4, notches result from a balance between apoptosis and AiP events as suggested by the enhancement of the rbf1D253A-induced notches phenotype by co-expressing slipper or dtraf1 specific RNAi. As vg-Gal4 driver is expressed until the end of pupal development, we thought that apoptosis induced-proliferation may not be sufficient to compensate the loss of tissue induced at the end of development. Thus, we investigated whether a transient induction of rbf1 could activate a compensatory proliferation mechanism able to maintain tissue homeostasis. We used heat shock-Gal4 (hs-Gal4) driver to induce a transient expression of rbf1 and investigated whether this transient induction of rbf1 was able to induce apoptosis and proliferation. Thus, hs-Gal4/+; UAS-rbf1/+ and hs-Gal4/ + control third instar larvae were shocked at 29°C for 10 hours. Their wing imaginal discs were dissected 5 hours after the end of the heat shock and stained for TUNEL or PH3, and the amount of apoptotic or mitotic cells quantified in the wing disc pouch. A significant increase in TUNEL and PH3 staining was observed in hs-Gal4/+; UASrbf1/+ as compared with hs-Gal4/+ control wing discs (Figure 6A-B and D-E), showing that a brief pulse of rbf1 is sufficient to induce apoptosis and AiP in wing imaginal discs. Interestingly, wings of heat shocked hs-Gal4/+; UASrbf1/+ adult flies were normal (Figure 6F and O), indicating that under these conditions the amount of rbf1-induced AiP was sufficient to compensate the cell loss. We then wondered whether the dTRAF1-Slipper-JNK pathway is involved in this compensatory proliferation. Thus, we repeated the same experiment by co-expressing RNAi-slipper or RNAi-dtraf1 with rbf1 using the hsGal4 driver. A significant number of hsGal4/UAS-RNAi-slipper; UAS-rbf1/+ and hsGal4/+; UAS-rbf1/UAS-RNAi-dtraf1 flies presented notched wings (Figure 6I, L, O) compared with hs-Gal4/+; UAS-rbf1/+ flies. We performed TUNEL and PH3 staining on wing imaginal discs of the same genotypes. The reduction of slipper or dtraf1 mRNA levels in wing discs expressing rbf1 did not affect the number of apoptotic cells (Figure 6G, J, M), confirming that Slipper and dTRAF1 are not involved in Rbf1-induced apoptotic cascade. However, a significant decrease of PH3 staining was observed (Figure 6H, K, N). These results highlight that Slipper and dTRAF1 are necessary to trigger the compensatory proliferation in response to Rbf1-induced apoptosis.
Figure 6.
Rbf1-induced apoptosis activates a compensatory proliferation mechanism that depends on slipper and dtraf1 (A, D, G, J) PH3 staining used to visualize the mitotic cells in wing pouch imaginal discs from hs-Gal4/+ or hs-Gal4/+; UAS-rbf1/+ or hs-Gal4/UAS-RNAi slipper; UAS-rbf1/+ or hs-Gal4/+; UAS-rbf1/UAS-RNAi dtraf1 third instar larvae. (B, E, H, K) Visualization of apoptosis cells by TUNEL staining in the wing pouch of imaginal discs from the previously described genotypes. (C, F, I, L) Wing phenotypes observed in some fly from hs-Gal4/+ or hs-Gal4/+; UAS-rbf1/+ or hs-Gal4/UAS-RNAi slipper; UAS-rbf1/+ or hs-Gal4/+; UAS-rbf1/UAS-RNAi dtraf1 genotypes (M) Comparison of apoptotic cells numbers in the wing pouch of imaginal discs from the previously described genotypes. Asterisks indicate a statistically significant difference between 2 genotypes (Student's test α < 0.05). For each genotype, quantifications were done for 30 third instar larval wing imaginal discs at least. (N) Comparison of proliferation percentage in posterior compartment from the genotypes described previously. Asterisks indicate a statistically significant difference between 2 genotypes (Student's test α < 0.05). (0) Frequencies of notches phenotypes observed in genotypes flies described previously. Asterisks indicate a statistically significant difference between 2 genotypes (Chi2 test α < 0.05). Each experiment presented in N and 0 was independently performed 3 times.
Discussion
Compensatory proliferation is one of the main mechanisms allowing the regeneration of a damaged tissue after the induction of massive apoptosis in vivo. Indeed, apoptotic cells are able to promote proliferation of the surrounding cells. This process has been largely studied using “undead cells,” in which apoptosis has been induced but cannot be executed because effector caspases are inhibited by p35. However, there are major differences between undead and genuine apoptotic cells (i.e., not overexpressing p35). Although under p35-expressing conditions JNK activity seems to occur only in the apoptotic cells and not in the neighboring proliferative cells,19 during compensatory mechanisms, the JNK pathway is activated in both cell types.20,21 Moreover, although Dpp and Wg are required in response to undead cells, the regenerative response can occur in absence of these mitogens.22. Because rbf1D253A-induced AiP depends on JNK pathway activation and does not involve Wg,11 the overgrowth phenotypes associated with rbf1D253A expression constitutes a robust model to screen regulators of AiP without using p35.
If the JNK signaling pathway is essential for AiP, the way it is activated and the specific kinases involved remain elusive. Recent results, obtained in the eye imaginal disc, have identified specific components upstream of Bsk involved in the JNK cascade activation in an ey>hid, p35 model of AiP.21 This work permitted the identification of genes acting upstream of JNK including rho1. In our model, we identified a different set of JNK pathway components (Figure 7). Three differences can account for this apparent discrepancy. First, we did not use p35 to inhibit cell death; second, we used another apoptosis inducer (rbf1 versus hid); and third, the nature of the tissue was different (wing vs. eye disc). Indeed, the kinases and their partners present in the cells are probably tissue-specific, which could explain the specificity of the responses. Accordingly, the JNK signaling pathway can lead to a great number of responses including cell death and proliferation depending on the context. This could be partly explained by the large number of possible kinases, adaptors and ligand/receptor combinations. Moreover, these families of proteins have unique and overlapping functions, but the intrinsic properties of the individual family members can confer particular responsiveness to distinct signals. For example, the C-terminal regions of Slpr and dTak1 contribute to their localization and selective integration into the appropriate signaling pathways in a context-dependent manner.23
Figure 7.
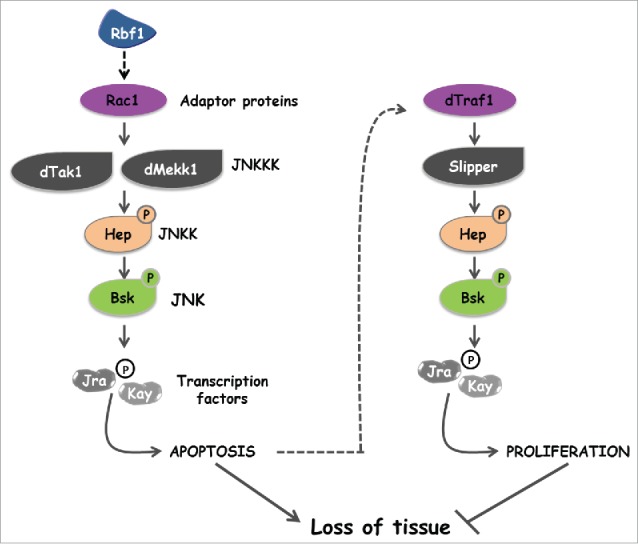
Two specific JNK activation pathways are involved in apoptosis and compensatory proliferation induced in response to Rbf1.
The originality of our approach is to characterize both the effectors of the JNK pathway involved in apoptosis and those involved in AiP in response to the same stimulus. This was facilitated by the duality of the adult phenotypes induced by the rbf1D253A form (notches and overgrowth). In Drosophila, there is a unique JNK, Bsk. Surprisingly, the same kinase and transcription factors Jra and Kay can trigger proliferation or apoptosis. As expected, we have shown that these events do not occur in the same cells. Nevertheless, this highlights that the nature and the combination of the kinases activated upstream of Bsk may lead to somehow different Jra/Kay activation. Alternatively, it is possible that Bsk phosphorylation sites vary depending on the kinases activated upstream in the pathway.
Bsk can be phosphorylated by the JNKK Mkk4 but this event seems restricted to certain particular situations such as the activation of the JNK pathway by the Eiger (the fly TNF) /Wengen complex.24 Mkk4 is not included in our model (Figure 7) suggesting that the activation of the JNK cascade during AiP does not involve Wengen. However, this does not exclude a potential role of Eiger for which the new receptor Grindelwald (Grnd) has recently been characterized.25 As Grnd integrates signals from Eiger and apical polarity determinants to induce JNK-dependent neoplastic growth or apoptosis in a context-dependent manner, it could be involved in our model. Another candidate is Diap1 which has been shown to poly-ubiquitinylate dTRAF1.26 We have previously demonstrated that rbf1-induced apoptosis involves a downregulation of diap1 expression.11 Interestingly, a decrease of Diap1 level can trigger an activation of the JNK pathway.27 Thus, the activation of the JNK signaling cascade involved in Rbf1-induced AiP could result from a decrease in diap1 mRNA.
We have previously shown that the JNK pathway activation induced by rbf1 involved reactive oxygen species (ROS) in dying cells.28 Many data report an activation of the JNK pathway depending on ROS. For example, in mammalian cells, the JNKKK ASK1 has been extensively characterized as a ROS-responsive kinase. Indeed, its activation is regulated by the protein Trx that has a redox-active site and functions as a reactive oxygen species sensor.29,30 As dASK1 is not implicated in the mechanism we describe, we hypothesize that another kinase could be responsible for the activation of JNK pathway.
To date, we do not know how proliferation is induced downstream of the JNK pathway in response to Rbf1induced apoptosis. A JNK-dependent activation of Yorkie, the transcriptional co-activator negatively regulated by Hippo signaling, is essential for regeneration-associated growth.32 Other studies show that a JNK activation could stimulate non-autonomous JAK-STAT signaling for proliferation.33 In the case of an AiP triggered by Dmp53, cell proliferation depends on Notch.34. Thus, each of these pathways may constitute a putative effector of AiP in our model.
In conclusion, our study is the first to identify the JNK effectors involved in either apoptosis or apoptosis-induced proliferation in the same model system. AiP is an important general process for homeostasis in multicellular organisms and its misregulation could be implicated in resistance of tumors to irradiation. Thus, elucidating signaling specificity among JNK pathway in AiP could be of utmost interest in oncology.
Materials and methods
Fly stocks
Flies were raised at 25°C on a standard medium. The UAS-rbf1 and vg-Gal4 strains were generous gifts from J Silber. The en-Gal4 strain was kindly provided by L. Théodore. The hs-Gal4 strain was previously described.35 The following strains were obtained from the Bloomington Stock Center (Bloomington): de2f276Q1 (7436), pucE69 (puc-LacZ, 6762), jraIA109 (3273), UAS-RNAi-rac1 (28985), traf4EP578 (UAS-dtraf1, 17285) and dsh1 (5298). The UAS-RNAi-kay (v27722), UAS-RNAi-mkk4 (v26928), UAS-RNAi-dtak1 (v101357), UAS-RNAi-slipper (v106449), UAS-RNAi-dASK1 (v34892), UAS-RNAi-dmekk1 (v25528), UAS-RNAi-dtraf1 (v21213), UAS-RNAi-dtraf2 (v16125) and UAS-RNAi-tab2 (v37553) strains come from the Vienna Drosophila RNAi Center (VDRC). The UAS-hep-RNAi (4353R2) strain was provided by the National Institute of Genetics stock center (NIG-Fly). In order to use control flies from the same genetic background, the UAS-gfp strain (BL35786) and the host line y,w[1118];P{attP,y[+],w[3′]} (transformant ID 60100) were used as control for TRIP V10 collection and KK VDRC library respectively. We used a w1118 fly stock as the reference for all other strain.
Test of phenotype suppression in the wing
To test the implication of candidate genes in rbf1-induced apoptosis and proliferation, the severity of the notched wing phenotype induced by UAS-rbf1 overexpression led by vg-Gal4 driver was assayed in different genetic backgrounds. For each gene, we verified that the alteration of this gene expression level (overexpression, RNAi or mutant) did not induce any wing phenotype. vg-Gal4>UAS-rbf1 Drosophila females were crossed with males bearing a loss-of function mutation for the different genes or allowing their overexpression. The progenies of all crosses were classified according to the number of notches on the wing margin. Wilcoxon tests were performed as previously described.35 We considered the difference significant when α < 10-3.
To test the implication of candidate genes in rbf1D253A-induced proliferation, the number of flies presenting ectopic tissue in the wing was counted in different genetic backgrounds. Chi2 tests were performed and results were considered to be significant when α < 5%.
Immunostaining and TUNEL staining of imaginal discs
Third instar larvae were dissected in PBS pH 7.6, fixed in PBS-3.7% formaldehyde. Discs were labeled by incubation overnight at 4°C with anti-phospho-histone H3 (rabbit polyclonal antibody, 1/1000, 06-570, Millipore) and anti β-Galactosidase (mouse monoclonal antibody, 1/200, 40-1a, DSHB) in PBT-FCS (PBS, 0,3% Triton, 10% Fetal Calf Serum). Incubation with fluorescently labeled Alexa 488, 568 and 647 secondary antibodies (1/500, Molecular Probes, Thermo Fisher Scientific) was carried in PBT-FCS for 2 h at room temperature. After washing in PBT, TUNEL staining was performed according to manufacturer's instructions (ApopTag Red in situ apoptosis detection kit, Millipore). Discs were mounted in CitifluorTM (Biovalley) and observed with a Leica SPE upright confocal microscope (Leica). Wing discs were finally mounted in CitifluorTM (Biovalley) and observed with a Leica SPE upright confocal microscope. The percentage of proliferation corresponds to the PH3 positive pixel area plotted against the compartment size.
Quantification of TUNEL and PH3 staining was done in the wing pouch (for rbf1D253A or heat shock experiments) or in the posterior compartment (for rbf1 experiments) for at least 30 wing imaginal discs per genotype. Student's t-tests were performed and results were considered to be significant when α<5%.
RNAs extraction and RT-qPCR
Fifty wing imaginal discs per genotype were dissected on ice in serum-free Schneider medium. Total RNAs were extracted from each sample using the RNeasy Mini kit (Qiagen), by following the manufacturer's instructions. RT was performed on each sample using 4.8 mg of RNA incubated with random primer oligonucleotides (Invitrogen) with Recombinant Taq DNA Polymerase (Invitrogen), according to the manufacturer's instructions. Real-time PCR analysis was performed using the ABI Prism 7700 HT apparatus (Applied Biosystems, Life Technologies). Briefly, PCR was performed with the ABsolute blue QPCR SYBR Green ROX mix (Abgene, Thermo Fisher Scientific), using 11 ng of cDNA per RT. Data were normalized against rp49. Three independent RT experiments were performed and the SEM was calculated from these 3 independent samples.
Heat-shock protocol
Crosses were done at 18°C. Early third instar larvae were placed at 29°C during 10h and then returned to 18°C. Larvae were collected 5h after the end of the heat shock for immunostaining or larvae stay at 18°C to allow pupariation and adult wings were observed.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Sébastien Szuplewski, Frédéric Canal, Jessie Colin and Tommaso Villa for their critical readings of the manuscript. We thank Christine Dupuy-Wintz for helpful technical assistance. Confocal microscopy and qPCR experiments were performed on CYMAGES imaging facility and on the genomic platform respectively (University of Versailles/Saint-Quentin-en-Yvelines).
Funding
Amandine Clavier and Adrienne Baillet were supported by the UVSQ and the EPHE.
References
- 1.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell 2002; 2:103-12; PMID:12204530; http://dx.doi.org/ 10.1016/S1535-6108(02)00102-2 [DOI] [PubMed] [Google Scholar]
- 2.Cobrinik D. Pocket proteins and cell cycle control. Oncogene 2005; 24:2796-809; PMID:15838516; http://dx.doi.org/ 10.1038/sj.onc.1208619 [DOI] [PubMed] [Google Scholar]
- 3.Almasan A, Yin Y, Kelly RE, Lee EY, Bradley A, Li W, Bertino JR, Wahl GM. Deficiency of retinoblastoma protein leads to inappropriate S-phase entry, activation of E2F-responsive genes, and apoptosis. Proc Natl Acad Sci U S A 1995; 92:5436-40; PMID:7777526; http://dx.doi.org/ 10.1073/pnas.92.12.5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ianari A, Natale T, Calo E, Ferretti E, Alesse E, Screpanti I, Haigis K, Gulino A, Lees JA. Proapoptotic function of the retinoblastoma tumor suppressor protein. Cancer Cell 2009; 15:184-94; PMID:19249677; http://dx.doi.org/ 10.1016/j.ccr.2009.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hilgendorf KI, Leshchiner ES, Nedelcu S, Maynard MA, Calo E, Ianari A, Walensky LD, Lees JA. The retinoblastoma protein induces apoptosis directly at the mitochondria. Genes Dev 2013; 27:1003-15; PMID:23618872; http://dx.doi.org/ 10.1101/gad.211326.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai KY, Hu Y, Macleod KF, Crowley D, Yamasaki L, Jacks T. Mutation of E2F-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol Cell 1998; 2:293-304; PMID:9774968; http://dx.doi.org/ 10.1016/S1097-2765(00)80274-9 [DOI] [PubMed] [Google Scholar]
- 7.Sharma A, Comstock CE, Knudsen ES, Cao KH, Hess-Wilson JK, Morey LM, Barrera J, Knudsen KE. Retinoblastoma tumor suppressor status is a critical determinant of therapeutic response in prostate cancer cells. Cancer Res 2007; 67:6192-203; PMID:17616676; http://dx.doi.org/ 10.1158/0008-5472.CAN-06-4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huh MS, Parker MH, Scime A, Parks R, Rudnicki MA. Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. J Cell Biol 2004; 166:865-76; PMID:15364961; http://dx.doi.org/ 10.1083/jcb.200403004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palacios G, Talos F, Nemajerova A, Moll UM, Petrenko O. E2F1 plays a direct role in Rb stabilization and p53-independent tumor suppression. Cell Cycle 2008; 7:1776-81; PMID:18583939; http://dx.doi.org/ 10.4161/cc.7.12.6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milet C, Rincheval-Arnold A, Mignotte B, Guenal I. The Drosophila retinoblastoma protein induces apoptosis in proliferating but not in post-mitotic cells. Cell Cycle 2010; 9:97-103; PMID:20016284; http://dx.doi.org/ 10.4161/cc.9.1.10251 [DOI] [PubMed] [Google Scholar]
- 11.Milet C, Rincheval-Arnold A, Morieras A, Clavier A, Garrigue A, Mignotte B, Guenal I. Mutating RBF can enhance its pro-apoptotic activity and uncovers a new role in tissue homeostasis. PLoS One 2014; 9:e102902; PMID:25089524; http://dx.doi.org/ 10.1371/journal.pone.0102902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Igaki T. Correcting developmental errors by apoptosis: lessons from Drosophila JNK signaling. Apoptosis 2009; 14:1021-8; PMID:19466550; http://dx.doi.org/ 10.1007/s10495-009-0361-7 [DOI] [PubMed] [Google Scholar]
- 13.Biteau B, Karpac J, Hwangbo D, Jasper H. Regulation of Drosophila lifespan by JNK signaling. Exp Gerontol 2011; 46:349-54; PMID:21111799; http://dx.doi.org/ 10.1016/j.exger.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhanasekaran DN, Reddy EP. JNK signaling in apoptosis. Oncogene 2008; 27:6245-51; PMID:18931691; http://dx.doi.org/ 10.1038/onc.2008.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clavier A, Baillet A, Rincheval-Arnold A, Coleno-Costes A, Lasbleiz C, Mignotte B, Guenal I. The pro-apoptotic activity of Drosophila Rbf1 involves dE2F2-dependent downregulation of diap1 and buffy mRNA. Cell Death Dis 2014; 5:e1405; PMID:25188515; http://dx.doi.org/ 10.1038/cddis.2014.372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM, Martinez-Arias A. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev 1998; 12:557-70; PMID:9472024; http://dx.doi.org/ 10.1101/gad.12.4.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demay Y, Perochon J, Szuplewski S, Mignotte B, Gaumer S. The PERK pathway independently triggers apoptosis and a Rac1/Slpr/JNK/Dilp8 signaling favoring tissue homeostasis in a chronic ER stress Drosophila model. Cell Death Dis 2014; 5:e1452; PMID:25299777; http://dx.doi.org/ 10.1038/cddis.2014.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchal C, Vinatier G, Sanial M, Plessis A, Pret AM, Limbourg-Bouchon B, Theodore L, Netter S. The HIV-1 Vpu protein induces apoptosis in Drosophila via activation of JNK signaling. PLoS One 2012; 7:e34310; PMID:22479597; http://dx.doi.org/ 10.1371/journal.pone.0034310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryoo HD, Gorenc T, Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev Cell 2004; 7:491-501; PMID:15469838; http://dx.doi.org/ 10.1016/j.devcel.2004.08.019 [DOI] [PubMed] [Google Scholar]
- 20.Bergantinos C, Corominas M, Serras F. Cell death-induced regeneration in wing imaginal discs requires JNK signalling. Development 2010; 137:1169-79; PMID:20215351; http://dx.doi.org/ 10.1242/dev.045559 [DOI] [PubMed] [Google Scholar]
- 21.Fan Y, Wang S, Hernandez J, Yenigun VB, Hertlein G, Fogarty CE, Lindblad JL, Bergmann A. Genetic models of apoptosis-induced proliferation decipher activation of JNK and identify a requirement of EGFR signaling for tissue regenerative responses in Drosophila. PLoS Genet 2014; 10:e1004131; PMID:24497843; http://dx.doi.org/ 10.1371/journal.pgen.1004131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrera SC, Martin R, Morata G. Tissue homeostasis in the wing disc of Drosophila melanogaster: immediate response to massive damage during development. PLoS Genet 2013; 9:e1003446; PMID:23633961; http://dx.doi.org/ 10.1371/journal.pgen.1003446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stronach B, Lennox AL, Garlena RA. Domain specificity of MAP3K family members, MLK and Tak1, for JNK signaling in Drosophila. Genetics 2014; 197:497-513; PMID:24429281; http://dx.doi.org/ 10.1534/genetics.113.160937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geuking P, Narasimamurthy R, Basler K. A genetic screen targeting the tumor necrosis factor/Eiger signaling pathway: identification of Drosophila TAB2 as a functionally conserved component. Genetics 2005; 171:1683-94; PMID:16079232; http://dx.doi.org/ 10.1534/genetics.105.045534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersen DS, Colombani J, Palmerini V, Chakrabandhu K, Boone E, Rothlisberger M, Toggweiler J, Basler K, Mapelli M, Hueber AO, et al.. The Drosophila TNF receptor Grindelwald couples loss of cell polarity and neoplastic growth. Nature 2015; 522(7557):482-6 [DOI] [PubMed] [Google Scholar]
- 26.Kuranaga E, Kanuka H, Igaki T, Sawamoto K, Ichijo H, Okano H, Miura M. Reaper-mediated inhibition of DIAP1-induced DTRAF1 degradation results in activation of JNK in Drosophila. Nat Cell Biol 2002; 4:705-10; PMID:12198495; http://dx.doi.org/ 10.1038/ncb842 [DOI] [PubMed] [Google Scholar]
- 27.Kanda H, Miura M. Regulatory roles of JNK in programmed cell death. J Biochem 2004; 136:1-6; PMID:15269233; http://dx.doi.org/ 10.1093/jb/mvh098 [DOI] [PubMed] [Google Scholar]
- 28.Clavier A, Ruby V, Rincheval-Arnold A, Mignotte B, Guenal I. The Drosophila retinoblastoma protein, Rbf1, induces a Debcl- and Drp1-dependent mitochondrial apoptosis. J Cell Sci 2015; 128:3239-49; PMID:26208635; http://dx.doi.org/ 10.1242/jcs.169896 [DOI] [PubMed] [Google Scholar]
- 29.Son Y, Kim S, Chung HT, Pae HO. Reactive oxygen species in the activation of MAP kinases. Methods Enzymol 2013; 528:27-48; PMID:23849857; http://dx.doi.org/ 10.1016/B978-0-12-405881-1.00002-1 [DOI] [PubMed] [Google Scholar]
- 30.Nakano H, Nakajima A, Sakon-Komazawa S, Piao JH, Xue X, Okumura K. Reactive oxygen species mediate crosstalk between NF-kappaB and JNK. Cell Death Differ 2006; 13:730-7; PMID:16341124; http://dx.doi.org/ 10.1038/sj.cdd.4401830 [DOI] [PubMed] [Google Scholar]
- 31.Zhou S, Yu D, Ning S, Zhang H, Jiang L, He L, Li M, Sun M. Augmented Rac1 Expression and Activity are Associated with Oxidative Stress and Decline of β Cell Function in Obesity. Cell Physiol Biochem 2015; 35:2135-48; PMID:25896148; http://dx.doi.org/ 10.1159/000374019 [DOI] [PubMed] [Google Scholar]
- 32.Sun G, Irvine KD. Ajuba family proteins link JNK to Hippo signaling. Sci Signal 2013; 6:ra81; PMID:24023255; http://dx.doi.org/ 10.1126/scisignal.2004324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu M, Pastor-Pareja JC, Xu T. Interaction between Ras(V12) and scribbled clones induces tumour growth and invasion. Nature 2010; 463:545-8; PMID:20072127; http://dx.doi.org/ 10.1038/nature08702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon R, Aparicio R, Housden BE, Bray S, Busturia A. Drosophila p53 controls Notch expression and balances apoptosis and proliferation. Apoptosis 2014; 19:1430-43; PMID:24858703; http://dx.doi.org/ 10.1007/s10495-014-1000-5 [DOI] [PubMed] [Google Scholar]
- 35.Brun S, Rincheval V, Gaumer S, Mignotte B, Guenal I. reaper and bax initiate two different apoptotic pathways affecting mitochondria and antagonized by bcl-2 in Drosophila. Oncogene 2002; 21:6458-70; PMID:12226749; http://dx.doi.org/ 10.1038/sj.onc.1205839 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.