ABSTRACT
Amniotic fluid stem cells (AFSC) represent an attractive potential cell source for fetal and pediatric cell-based therapies. However, upgrading them to pluripotency confers refractoriness toward senescence, higher proliferation rate and unlimited differentiation potential. AFSC were observed to rapidly and efficiently reacquire pluripotency which together with their easy recovery makes them an attractive cell source for reprogramming. The reprogramming process as well as the resulting iPSC epigenome could potentially benefit from the unspecialized nature of AFSC. iPSC derived from AFSC also have potential in disease modeling, such as Down syndrome or β-thalassemia. Previous experiments involving AFSC reprogramming have largely relied on integrative vector transgene delivery and undefined serum-containing, feeder-dependent culture. Here, we describe non-integrative oriP/EBNA-1 episomal plasmid-based reprogramming of AFSC into iPSC and culture in fully chemically defined xeno-free conditions represented by vitronectin coating and E8 medium, a system that we found uniquely suited for this purpose. The derived AF-iPSC lines uniformly expressed a set of pluripotency markers Oct3/4, Nanog, Sox2, SSEA-1, SSEA-4, TRA-1-60, TRA-1-81 in a pattern typical for human primed PSC. Additionally, the cells formed teratomas, and were deemed pluripotent by PluriTest, a global expression microarray-based in-silico pluripotency assay. However, we found that the PluriTest scores were borderline, indicating a unique pluripotent signature in the defined condition. In the light of potential future clinical translation of iPSC technology, non-integrating reprogramming and chemically defined culture are more acceptable.
KEYWORDS: amniotic fluid stem cells, episomal reprogramming, E8, induced pluripotent stem cells, PluriTest, vitronectin, xeno-free culture
Introduction
In humans, dermal fibroblasts represent a common cell source for generation of induced pluripotent stem cells (iPSC). However, the requirement for skin biopsies and the need to expand fibroblast cells for several passages in vitro before reprogramming renders these cells an inconvenient source for generating patient-specific stem cells.1 Amniotic fluid stem cells (AFSC), on the contrary, can be easily and rapidly isolated from second trimester amniocentesis samples, representing source cells for reprogramming into autologous iPSC that can be performed before birth and used in future therapies. AFSC, representing fetal mesenchymal stem cells, have been shown to be broadly multipotent, bordering on pluripotency,2 with a high proliferation potential. These characteristics make them highly amenable for reprogramming. AFSC themselves are being explored in light of their potential to be used in tissue engineering-based therapies directly.3-5 However, proliferation and differentiation capacity of mesenchymal stem cells dwindles with prolonged culture6 and aberrant DNA methylation pattern at specific CpG sites were observed in late-passage mesenchymal stromal cells.7 Epigenetic instability was observed in the form of loss of parental allele-specific imprinting of the genes encoding insulin-like growth factor 2 (IGF2), H19, small nuclear ribonucleoprotein polypeptide N gene (SNRPN), and mesoderm-specific transcript (MEST), eliciting unwanted activity of these alleles in AFSC beyond 8 passages.8 Loss of imprinting is implicated in a large variety of human tumors.9
iPSC tend to retain methylation signatures associated with tissues that the source cells for reprogramming are isolated from and these signatures render the differentiation of iPSC biased toward their tissue of origin.10 Considering that the AFSC are isolated early in the fetal development and that their phenotype is mesenchymal but partially poised on the verge of pluripotency,2,11 their level of commitment is low and thus conceivably allows their epigenetic landscape to be more open to remodeling. Therefore, iPSC derived from AFSC have the potential to address the differentiation bias of iPSC derived from more differentiated cells as differentiation stage of cells has been shown to have a strong impact on the efficiency and kinetics of reprogramming.12 Upgrading AFSC to full pluripotency is an attractive option that has the potential to provide iPSC that can undergo dozens of passages, be expanded in very high numbers, possibly in scalable suspension bioreactors,13 and are capable of differentiating into any cell type of the body while maintaining genetic stability for over 25 passages and more than 3 months in culture in serum-free conditions.14 Indeed, AFSC were found to be more rapidly and efficiently reprogrammed into iPSC compared to adult cells.15,16 Transcriptome analysis revealed that the expression of key senescence-associated genes is down-regulated upon the induction of pluripotency in primary AFSC.17 In addition to potential regenerative medicine applications, AFSC possessing trisomy 21 mutation can be used to derive iPSC to serve in modeling of the Down syndrome as impaired neurogenesis has recently been observed using these iPSC.18 AFSC from β-thalassemia patients were found to serve as a rapid and efficient cell source for reprogramming into iPSC.19 However, in these studies, virus-based integrating reprogramming approach or integrating approach with subsequent excision and feeder-based iPSC culture were used. Means of reprogramming somatic cells into iPSC that entail either permanent or transient incorporation of vectors carrying reprogramming factors into the cell genome bears the risk of insertional mutagenesis.
To address this issue a system of 3 non-integrating episomal oriP/EBNA1 plasmid vectors involving reprogramming factors OCT4, SOX2, NANOG, c-Myc, KLF4, coexpression linkers IRES2 and SV40 large T gene (SV40LT, to offset the toxicity of c-Myc) was developed.20 The reprogramming plasmids are gradually lost from proliferating cells producing iPSC free of transgenes potentially suitable for therapies. Low DNA sequence variation was indeed found in iPSC derived using non-integrating plasmid vectors.20 This combination of plasmids was used to derive iPSC in fully chemically defined medium E8 with recombinant truncated human vitronectin used as a culture vessel attachment factor, forming a xeno-free culture system ready for the clinics.14
To overcome the limitations of serum-supplemented culture systems and integrative reprogramming approaches, we sought to identify the optimal route of derivation of high-quality iPSC from AFSC in fully chemically defined conditions using non-integrating episomal plasmids. This approach addresses the inconsistencies arising from undefined nature of animal serum and eliminates xeno-components. We derived iPSC from AFSC collected from 3 patients using episomal and 1 using retroviral reprogramming. Episomal iPSC were thoroughly characterized applying the most stringent methods. We also characterized the partially reprogrammed cells arising from incomplete pluripotency reacquisition which has a potential benefit for cancer research.
Results
Characterization of amniotic fluid stem cells
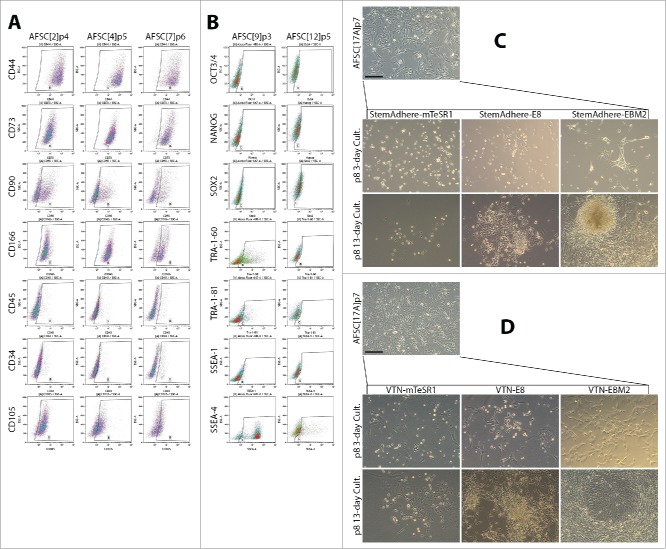
Amniotic fluid stem cells have previously been characterized as predominantly mesenchymal. Their ability to adhere to tissue culture plastic makes them the dominant cell type in primary cultures, despite amniotic fluid containing heterogeneous populations. In our laboratory setting, 100% of amniotic fluid samples from patients (n = 24) yielded primary cultures highly proliferative for at least 8 passages. The source amniotic fluid mixed with FCS-supplemented EBM-2 medium with growth factors (bFGF, EGF, IGF) provided an ideal environment for adaptation of the cells to adherent culture. The process took 5 days and another 5 days were needed for expansion before the first passaging. Expression of MSC markers in AFSC was measured by flow cytometry and revealed, with some variability, expression of CD44, CD73, CD90, CD166 and CD105 antigens, while hematopoietic stem cell markers CD34 and CD45 were mostly negative (Fig. 1A, Table 1). Expression of ESC markers Oct3/4 and Nanog was low, while dim Sox2 signal was observed in over 30% of the cells. Interestingly, TRA-1-60, TRA-1-81 and SSEA-4 antigen expression was variable and reached 40%, 14% and 87% respectively, in AFSC9, while AFSC12 exhibited poor evidence of the expression of these markers (Fig. 1B, Table 1). Albeit inconsistent with pluripotency, these characteristics could contribute to amenability of AFSC toward reprogramming.
Figure 1.
Characterization of AFSC and optimization of the culture condition following episomal reprogramming. (A) Expression of MSC markers in AFSC measured by flow cytometry. The expression pattern is similar to a typical MSC pattern, with some variability in CD90 and CD166. CD34 and CD45 expression were low. (B) Flow cytometric analysis of ESC marker expression in AFSC. No Oct3/4 expression was observed, Nanog expression was negligible, dim Sox2 signal was seen in over 30% of the cells. TRA antigen expression was observed in AFSC9 but not AFSC12. SSEA-1 expression was negligible, a bright SSEA-4 signal was found to be high in AFSC9 but much lower in AFSC12. The pattern of ESC marker expression is not consistent with pluripotency, (C, D). Morphological transformation of AFSC in response to episomal plasmid reprogramming, secondary passaging on day 9 and subsequent culturing on StemAdhere-coated plates in 2 different pluripotency-supporting media – mTeSR-1, E8. AFSC growth medium was used as a control (C); and on VTN-coated plates in the same media (D). VTN in combination with the E8 medium supported emerging colonies of epithelioid cells while StemAdhere and mTeSR-1 medium were non-permissive. Scale bars = 200 µm.
Table 1.
Expression of MSC and ESC markers in AFSC measured by flow cytometry shown as percent positive cells. Corresponding isotype controls IgG1, IgG3, IgG2a, IgM precede actual markers and were matched across lines tested. AFSC showed predominantly a mesenchymal stem cell phenotype and ESC marker expression pattern inconsistent with pluripotency. However, expression of TRA-1-60, TRA-1-81 and SSEA-4 was detected in AFSC9 p = passage number.
| MSC markes [%] |
ESC markers [%] |
||||||
|---|---|---|---|---|---|---|---|
| AFSC[2] | AFSC[4] | AFSC[7] | AFSC[9] | AFSC[12] | |||
| p4 | p5 | p6 | p3 | p5 | |||
| IgG1κ | 3.87 | 3.92 | 4.05 | IgG1κ | 2.05 | 2.04 | |
| CD44 | 99.92 | 99.95 | 98.17 | Oct3/4 | 1.80 | 1.72 | |
| CD73 | 99.98 | 100.00 | 96.07 | IgG1κ | 2.08 | 2.04 | |
| CD90 | 54.23 | 40.43 | 31.31 | Nanog | 5.71 | 6.01 | |
| CD166 | 99.80 | 99.98 | 62.82 | IgG1κ | 2.06 | 2.14 | |
| CD45 | 5.13 | 26.46 | 5.09 | Sox2 | 32.14 | 36.12 | |
| IgG1κ | 3.32 | 3.45 | 3.70 | IgMκ | 2.09 | 2.00 | |
| CD34 | 18.49 | 20.17 | 9.06 | TRA-1-60 | 40.29 | 1.35 | |
| IgG2a | 3.07 | 3.24 | 6.04 | IgMκ | 2.04 | 2.02 | |
| CD105 | 92.45 | 57.50 | 35.31 | TRA-1-81 | 14.49 | 1.85 | |
| Cy2 2nd | 2.23 | 0.85 | 2.64 | IgG1κ | 2.17 | 2.12 | |
| SSEA-1 | 6.00 | 2.54 | |||||
| IgG3κ | 2.19 | 2.14 | |||||
| SSEA-4 | 86.84 | 28.98 | |||||
Optimizing culture conditions for AFSC reprogramming
For reprogramming purposes, we chose the Neon® Transfection System as an efficient tool of plasmid delivery into the AFSC. The optimal set of transfection parameters recommended by the manufacturer for mesenchymal stem cells is 990 V, 40 ms and 1 pulse. We lowered the voltage to 950 V to help preserve the viability of the cells. Applying these parameters, we transfected AFSC17A with 3 reprogramming episomal plasmids and plated the cells into EBM-2 supplemented with 5% FCS and growth factors (bFGF, EGF, IGF), a medium supporting growth and proliferation of AFSC. 9 days later, colonies of visibly smaller cells emerged, morphologically resembling epithelial cells indicating their mesenchymal to epithelial transition. In these initial cultures, 9 colonies that emerged from 200.000 transfected cells were observed when 2 µg of M2L, 3 µg of ET2K and 3 µg of EN2K plasmids per 1 million cells was used. Increasing the concentrations of all plasmids to 5 µg did not result in higher numbers of MET cell colonies as only 1 colony with the above-described morphology was observed, likely due to reprogramming factor imbalance. On day 9, these initial cultures underwent a secondary split, preventing overgrowth and enabling testing of multiple culture conditions suitable for reprogramming. We passaged the cells into combinations of 2 different recombinant extracellular matrix attachment factors – StemAdhere (Fig. 1C) and VTN (Fig. 1D) – and 2 different pluripotent stem cell-specific media – E8, a serum-free and chemically defined pluripotent stem cell supporting medium; mTeSR-1, a rich, serum-free and defined pluripotent stem cell supporting medium containing 1% of highly purified BSA. EBM-2 supplemented with 5% FCS and growth factors (bFGF, EGF, IGF), a medium supporting growth and proliferation of AFSC was used as a control condition. mTeSR-1 medium and StemAdhere coating selected for cells that had already undergone the MET and thus have acquired the epithelial morphology while cells with typical spindle-shaped AFSC morphology were not present. The epithelioid cells that survived and attached but did not proliferate as 10 days later, the number of adherent epithelial-like cells was unchanged. The combination of StemAdhere coating and mTeSR-1 medium seemed to be highly selective toward epithelioid cells and did not support the source AFSC, therefore, this condition did not support reprogramming. The cells survived, however, their proliferation stalled and the acquisition of morphology typical for fully reprogrammed cells was not observed. A similar effect was observed for cells grown on StemAdhere coating in E8 medium in that only epithelioid cell attached and survived. This condition did not support survival and reprogramming of the source AFSC. AFSC attached and survived on StemAdhere-coated culture vessels only in the control AFSC serum-containing growth medium.
VTN was much more supportive for AFSC growth and proliferation and therefore, had a greater potential to support reprogramming considering that the growth of source somatic cells is necessary in its initial stages. VTN in combination of mTeSR-1 was not sufficient to support robust cell growth of both AFSC and partially reprogrammed cells as it still seemed to be highly pluripotent stem cell-specific. VTN in combination with the E8 medium provided an optimal support to epithelioid cells. This condition also supported survival of AFSC and, for a limited time, their proliferation. This transient growth support of AFSC in this condition is important for the reprogramming process. The epithelioid colonies were mechanically picked and transferred onto VTN-coated vessels and cultured in E8 medium. The cells did not grow in compact colonies but stayed in form of partially reprogrammed cells, some colonies failed but some proliferated and their numbers expanded, allowing for their characterization. Epithelioid clones mechanically picked from VTN-E8 cultures transferred on VTN-coated vessels and cultured in mTeSR-1 medium survived only briefly and ultimately failed, which was consistent with the inability of mTeSR-1 to support epithelioid cell growth.
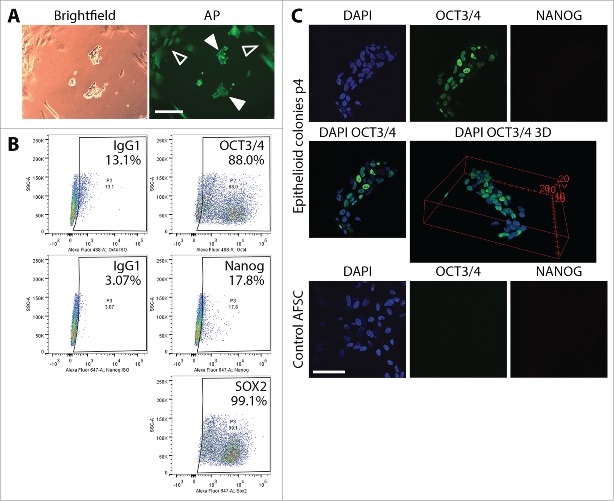
To characterize proliferating epithelioid partially reprogrammed cells, we performed alkaline phosphatase staining using the AP Live Stain, flow cytometry analysis of the expression of ESC markers Oct4, Sox2 and Nanog, as well as confocal microscopy immunofluorescence analysis of the expression of Oct3/4 and Nanog. AP Live Stain permits identification of reprogrammed colonies and potentially marking them for further expansion. AP Live Stain of epithelioid cells cultured for 18 days following reprogramming plasmid transfection, showed that background fluorescence associated with the source AFSC was relatively high. Epithelioid cells were seen on Figure 2A (filled arrows). However, discreetly bright AP signal beyond source AFSC background signal (hollow arrows) was not observed in these cells, which was consistent with incomplete reprogramming of the cells. A flow cytometric analysis of ESC marker expression in the transformed epithelioid cells cultured for 14 days following mechanical picking (30 day post-transfection) on VTN-coated plates in E8 medium showed expression of Oct3/4 in approximately 75%, dim Nanog signal in 15% and Sox2 expression in around 96% of the population (Fig. 2B). TRA antigen expression was not observed. Confocal microscopy analysis corroborated the flow cytometry data. Overlay image of Oct3/4 and DAPI fluorescent signals as well as 3D reconstruction (Fig. 2C) of the stained colony of the epithelioid cells revealed heterogeneous expression of Oct3/4 in the colony. AFSC at passage 4 were used as a control, revealing neither Oct3/4 nor Nanog expression.
Figure 2.
Characterization of partially reprogrammed epithelioid cells resulting from episomal reprogramming of AFSC. (A) Alkaline phosphatase staining of colonies of partially reprogrammed cells before mechanical picking and expansion. Discreetly bright AP signal beyond background fluorescence (hollow arrows) was not observed, confirming incomplete reprogramming (filled arrows). (B) Flow cytometric analysis of Oct4, Nanog and Sox2 in mechanically picked epithelioid colonies cultured for 14 days (30 days post-transfection) on VTN-coated plates in E8 medium. The profile corroborated incomplete reprogramming. Scale bars = 200µm. (C) Immunocytochemical staining and confocal microscopy visualization of ESC markers Oct4 and Nanog in epithelioid cell colonies expanded for 4 passages post-transfection. Oct4 expression was clearly heterogeneous among the individual cells in the colonies ranging from cells where Oct4 is completely absent to Oct4 being represented by a strong fluorescent signal. Nanog expression was not observed. As a control, we stained unreprogrammed AFSC for the same markers. These cells exhibited no Oct4 and Nanog expression at all. Scale bar = 50 µm.
VTN/E8 support retroviral reprogramming of AFSC to pluripotency
In order to test the ability of the chemically defined combination of VTN coating and E8 medium to support the process of full reprogramming of AFSC into iPSC, we performed reprogramming of AFSC12 at passage 4 by means of retroviral transduction introducing Oct4, Sox2, Klf-4, c-myc (“Yamanaka factors”). The host cells were cultured on VTN-coated vessels in AFSC growth medium for 6 days following transduction. On day 6, the medium was switched to E8 when epithelioid cells emerged. Ultimately, the cells acquired typical pluripotent stem cell morphology – high nucleus-to-cytoplasm ratio, growth in compact flat colonies with well-defined, phase-bright edges (Fig. 3D). Thus, VTN/E8 was readily permissive for the retroviral reprogramming of AFSC. Flow cytometry analysis confirmed typical expression of ESC markers in the reprogrammed cells at passage 10, showing a pattern similar to one of control episomally derived iPSC line IISH1i-BM1 (Fig. 3A, Table 2). Leaving c-myc out of the reprogramming cocktail led to no signs of reprogramming.
Figure 3.
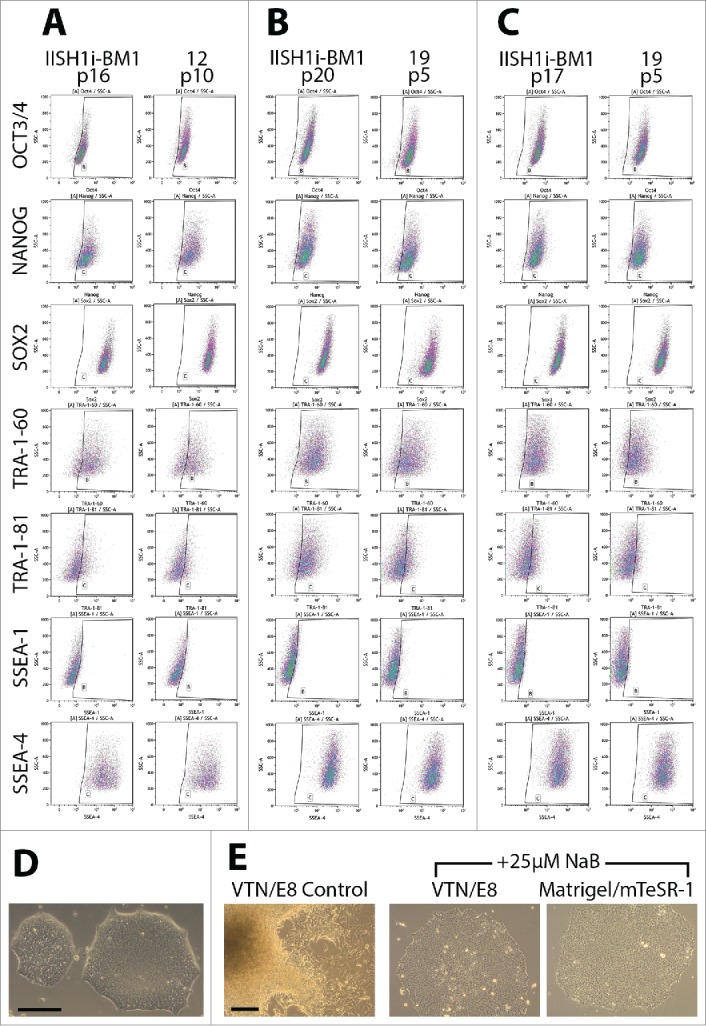
Analysis of ESC marker expression in fully reprogrammed AFSC and facilitation of full pluripotency acquisition by butyrate treatment in AFSC reprogrammed using episomal plasmids. (A) Flow cytometric analysis of ESC marker expression in fully reprogrammed AF-iPSC[12] (retroviral reprogramming) at passage 10 cultured on VTN-coated plates in E8 medium. The expression profile was analyzed alongside an episomally derived IISH1i-BM1 control iPSC line. (B) Flow cytometric analysis of ESC marker expression in AFSC reprogrammed using episomal plasmids in E8 medium on VTN. All 3 lines of AF-iPSC (19, 20A, D1) showed ESC marker expression profile comparable to that of the control line. (C) Flow cytometric analysis of the expression of ESC-specific markers in iPSC grown on Matrigel-coated plates in mTeSR-1 medium. As observed for VTN/E8-grown lines, all 3 lines of AF-iPSC showed ESC marker expression profile comparable to that of the control line. Matrigel and mTeSR-1 medium did not support the episomal reprogramming process and only fully reprogrammed AF-iPSC could be expanded in this condition. (D) Morphology of the colonies of retrovirally reprogrammed AF-iPSC[12], consistent with pluripotency. (E) Differences in morphology of partially reprogrammed AFSC cultured with no histone deacetylase inhibitor versus ultimately fully reprogrammed cells in conditions supplemented with 25 µM sodium butyrate – E8 medium on VTN and mTeSR-1 medium on Matrigel. Partially reprogrammed epithelioid cells were small and formed compact dome-shaped colonies, with a phase-bright edge. Fully reprogrammed cells were bigger with high nucleus-to-cytoplasm ratio and formed flat colonies with a clearly defined phase-bright edge representing the typical pluripotent stem cell colony morphology. Sodium butyrate enabled progression of partially reprogrammed cells to full pluripotency, otherwise the episomal reprogramming efficiency was prohibitively low. Scale bars = 200 µm.
Table 2.
Expression of ESC markers measured by flow cytometry and shown as percent positive cells in early-passage fully reprogrammed retroviral AF-iPSC (19, 20A, D1), episomal AF-iPSC grown in VTN/E8 and episomal AF-iPSC grown in Matrigel/mTeSR-1 conditions. IISH1i-BM1 control iPSC line was included. All lines show ESC marker expression pattern consistent with pluripotency.
| Retroviral [%] |
Episomal VTN/E8 [%] |
Episomal Matrigel/mTeSR-1 [%] |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IISH1i BM1 | 12 | IISH1i BM1 | 19 | 20A | D1 | IISH1i BM1 | 19 | 20A | D1 | |
| p16 | p10 | p20 | p5 | p5 | p6 | p17 | p5 | p5 | p4 | |
| IgG1κ | 2.02 | 2.06 | 2.33 | 2.00 | 2.06 | 2.07 | 2.07 | 2.04 | 2.11 | 2.08 |
| Oct3/4 | 92.12 | 95.65 | 99.60 | 96.90 | 98.22 | 97.23 | 99.47 | 99.06 | 96.59 | 97.56 |
| IgG1κ | 2.04 | 2.01 | 2.10 | 1.98 | 2.01 | 2.07 | 2.05 | 2.05 | 2.10 | 2.01 |
| Nanog | 90.24 | 93.87 | 96.35 | 88.21 | 93.64 | 88.01 | 98.97 | 95.16 | 95.06 | 88.11 |
| Sox2 | 100.00 | 100.00 | 99.99 | 99.97 | 99.93 | 99.92 | 99.95 | 99.98 | 99.84 | 99.73 |
| IgMκ | 3.08 | 3.23 | 3.17 | 3.03 | 3.08 | 3.11 | 3.09 | 3.13 | 3.12 | 3.04 |
| TRA-1-60 | 79.41 | 65.58 | 96.76 | 81.31 | 76.85 | 94.84 | 92.99 | 78.50 | 72.21 | 89.74 |
| IgMκ | 3.04 | 3.31 | 3.25 | 2.94 | 2.99 | 3.05 | 3.13 | 3.03 | 3.09 | 3.12 |
| TRA-1-81 | 27.79 | 22.67 | 76.66 | 27.84 | 29.20 | 69.77 | 29.48 | 14.06 | 8.95 | 27.18 |
| IgG1κ | 3.22 | 3.38 | 3.04 | 3.09 | 3.05 | 3.02 | 3.11 | 3.04 | 2.98 | 3.04 |
| SSEA-1 | 5.90 | 6.34 | 7.49 | 6.39 | 9.93 | 11.62 | 4.11 | 4.72 | 5.56 | 4.78 |
| IgG3κ | 3.30 | 3.40 | 3.02 | 3.18 | 3.06 | 2.97 | 3.00 | 3.07 | 2.94 | 3.00 |
| SSEA-4 | 94.34 | 99.67 | 99.96 | 99.90 | 99.86 | 99.90 | 99.88 | 99.86 | 98.81 | 99.53 |
VTN/E8 support episomal reprogramming of AFSC to pluripotency
Considering only VTN coating together with the E8 medium presented a permissive condition for reprogramming, we revisited non-integrative reprogramming in this condition using episomal plasmids. Transfecting AFSC lines derived from 3 patients with the reprogramming plasmids led to formation of epithelioid cell colonies. They were first visible 5-6 days following transfection. Secondary split was performed to prevent overgrowth of the source AFSC. At the time of the secondary split, on average 9.7 ± 1.7 of MET colonies had emerged per 100.000 cells. Supplementing transfected AFSC with 25 µM of sodium butyrate (a histone deacetylase inhibitor) at the time of the secondary split elicited progression further in the direction of pluripotency reacquisition. 12 days following secondary split, colonies of cells with typical ESC/iPSC morphology were ready for mechanical picking and expansion. The picked colonies were transferred and expanded equally well on VTN-coated vessels in E8 medium (3 clones each patient) and Matrigel-coated vessels in mTeSR-1 medium (3 clones each patient). Without sodium butyrate, the MET cells never progressed toward full pluripotency (Fig. 3E). The expression of ESC markers at an early passage by flow cytometry (including Oct3/4, Nanog, Sox2, TRA-1-60, TRA-1-81, SSEA-1 and SSEA-4) in 3 lines of episomally reprogrammed AFSC (AF-iPSC) – 19, 20A, and D1 – grown in VTN-coated vessels in E8 medium was consistent with the control IISH1i-BM1 iPSC line (Fig. 3B, Table 2). The same pattern was observed for the expression of ESC markers in AF-iPSC at an early passage grown on Matrigel in mTeSR-1 medium (Fig. 3C, Table 2).
Mature iPSC derived from AFSC display typical pluripotency characteristics
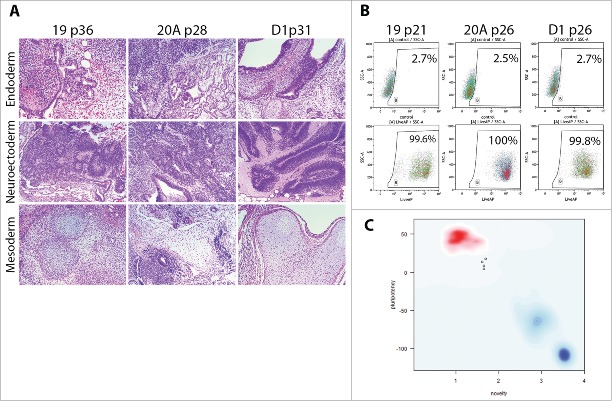
In their mature state at passages over 20, AF-iPSC display typical characteristics of pluripotent stem cells. Flow cytometric analysis of ESC markers Oct3/4A, Nanog, Sox2, TRA-1-60, TRA-1-81 and SSEA-4 revealed their expression in the absolute majority of cells of all 3 iPSC lines (19, 20A, and D1) approaching 100%. SSEA-1 was expressed in negligible portions of the cell populations analyzed. The ESC marker expression pattern was identical to the WA25 control ESC line, however, minor differences in Nanog and TRA antigen expression levels were observed (Fig. 4, Table 3). Subsequent confocal microscopy-based immunocytochemical analysis confirmed the presence and localization of the individual ESC markers in all AF-iPSC lines in parallel (Fig. 5). All AF-iPSC lines formed teratomas upon injection of the cells subcutaneously into immunocompromised scid-beige mice within 6 to 9 weeks. The cells were injected as clumps to prevent dissociation-induced cell death. H&E staining and subsequent histological analysis confirmed presence of tissues representative of all 3 germ layers, demonstrating pluripotency of the injected cells (line 19 shown on Fig. 6A). Alkaline phosphatase activity was assayed using AP Live Stain in all lines in parallel. Flow cytometry revealed alkaline phosphatase to be active in the majority of the cells (Fig. 6B). We subjected global transcriptome data derived from AF-iPSC lines and WA25 to an alternative measure of pluripotency known as PluriTest. All lines were deemed pluripotent by the test, however, they were flagged as “further evaluate” and their pluripotency and novelty scores intersected at points clustered off the red spot on the pluripotency/novelty plot. This spot represents the cluster area of validated PSC lines grown in feeder layer-based culture conditions and used for PluriTest training. These data indicate a unique transcriptional profile of PSC lines cultured on VTN-coated vessels in E8 medium (Fig. 6C, Table 4).
Figure 4.
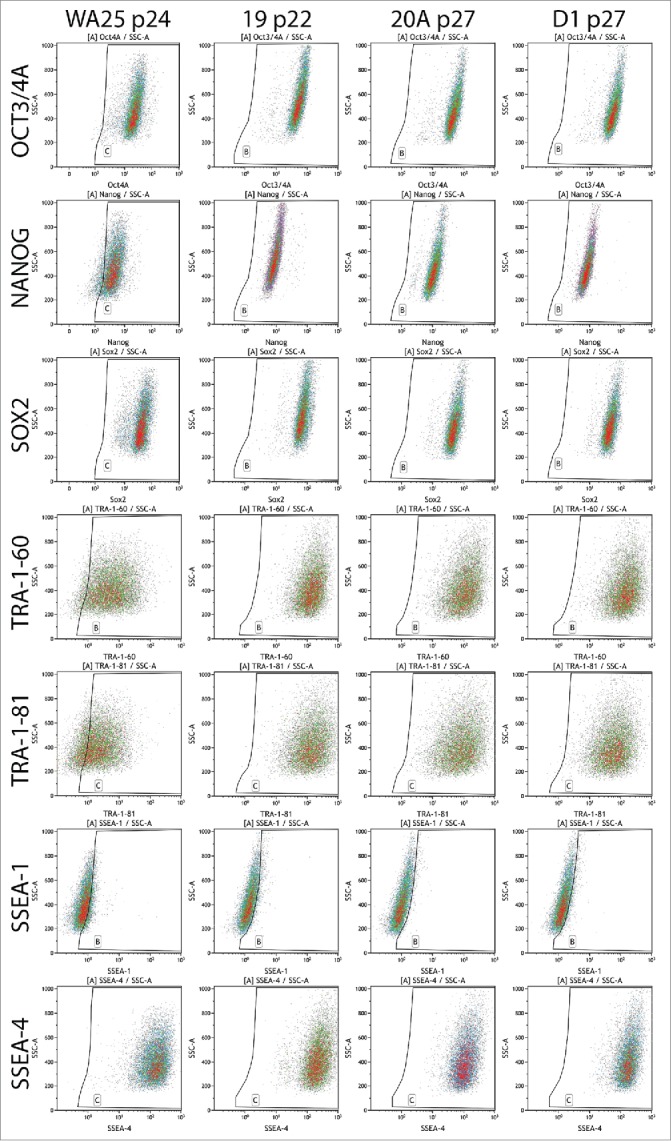
Flow cytometric analysis of the expression of ESC markers in AF-iPSC in their mature state grown in VTN/E8. All 3 lines of AF-iPSC (19, 20A, D1) showed typical ESC marker expression profile, comparable to that of a control ESC line WA25 also derived and cultured in VTN/E8. The WA25 line showed slightly lower intensities of Nanog and TRA antigen expression and it was more prone to spontaneous differentiation in culture.
Table 3.
Flow cytometric analysis of the expression of ESC markers in AF-iPSC (19, 20A, D1) in their mature state grown in VTN/E8 Control ESC line WA25 was included. All lines showed typical ESC marker expression profile, consistent with pluripotency.
| [%] |
|||||
|---|---|---|---|---|---|
| WA25 | 19 | 20A | D1 | ||
| p24 | p22 | p27 | p27 | ||
| IgG1κ | 1.96 | 1.51 | 1.51 | 1.48 | |
| Oct3/4A | 99.67 | 99.99 | 99.99 | 99.98 | |
| Nanog | 89.72 | 99.98 | 100.00 | 100.00 | |
| Sox2 | 99.98 | 99.96 | 99.95 | 100.00 | |
| IgMκ | 2.67 | 2.81 | 2.44 | 2.56 | |
| TRA-1-60 | 92.88 | 99.99 | 99.98 | 99.91 | |
| IgMκ | 2.68 | 2.81 | 2.41 | 2.45 | |
| TRA-1-81 | 74.52 | 99.98 | 99.97 | 99.80 | |
| IgG1κ | 2.05 | 2.31 | 2.01 | 2.30 | |
| SSEA-1 | 14.00 | 6.15 | 2.30 | 5.74 | |
| IgG3κ | 2.13 | 2.13 | 2.04 | 2.08 | |
| SSEA-4 | 99.96 | 100.00 | 100.00 | 100.00 | |
Figure 5.
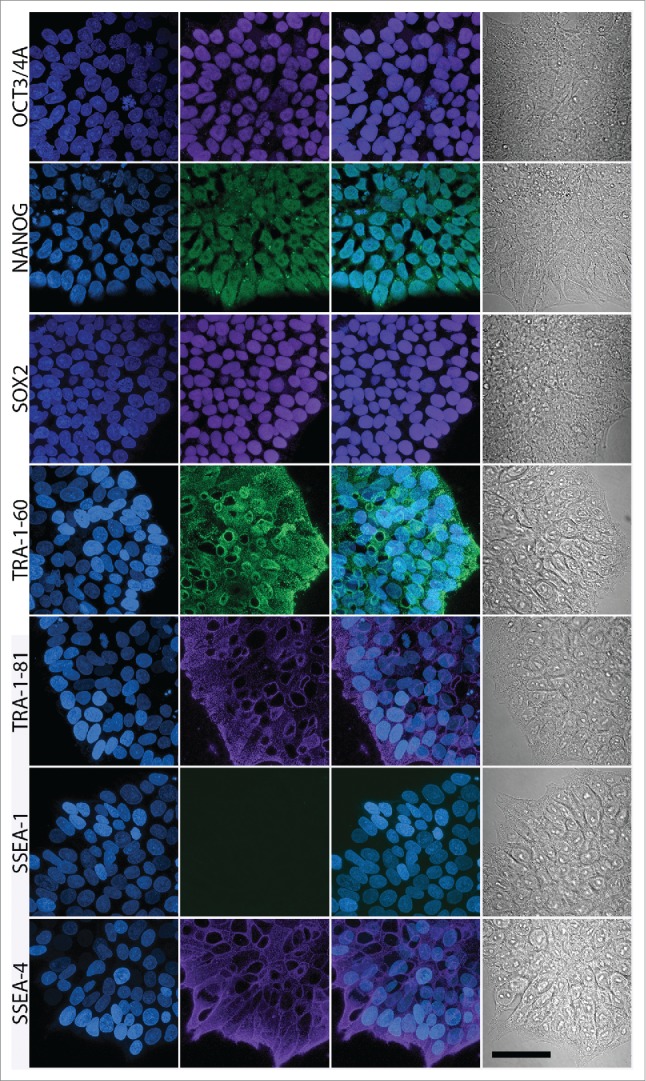
Confocal microscopy-based immunocytochemical analysis of the expression of ESC markers in AF-iPSC in their mature state grown in VTN/E8. Images of colonies of D1 line are depicted on the figure. Oct3/4A, Nanog and Sox2 displayed clear signal localized in the nucleus while TRA-1-60, TRA-1-81 and SSEA-4 displayed surface localization. SSEA-1 was negative. The projections were reconstructed from the fluorescence signal spanning all of the individual scanned slices using the Maximum Intensity Projection algorithm. The first column contains DAPI images, second column contains marker fluorescence, third column contains DAPI and marker overlay images and the fourth column contains a single-slice leftover transmitted light-based representation of the same colonies. Scale bar = 50 µm.
Figure 6.
Demonstration of pluripotency in AF-iPSC by 3 methods – in-vivo xenografts, alkaline phosphatase activity and transcriptomics (A). Histological analysis (H&E staining) of teratomas formed by means of in-vivo differentiation of AF-iPSC (19, 20A, D1). Cell clumps were injected subcutaneously into scid-beige mice and allowed to proliferate and differentiate for 6 to 9 weeks. The resulting teratomas showed presence of various tissues representative of all 3 germ layers – endoderm, neuroectoderm, mesoderm – demonstrating pluripotency of the injected cells. (B). Alkaline phosphatase activity in AF-iPSC measured by flow cytometry using AP Live Stain. The majority of the cells displayed bright fluorescence, supporting their pluripotency. (C). PluriTest - global gene expression microarray data-based in-silico evaluation of pluripotency. AF-iPSC and WA25 (control ESC line) were tested. Y axis – pluripotency score, X axis – novelty score. Red spot represents cluster area of validated PSC lines grown in standard feeder layer- and KSR-based culture condition. Blue spot represents the cluster area of differentiated cells. Faint blue spot represents the cluster area of partially pluripotent stem cells. The pluripotency and novelty score both indicated pluripotency of the lines tested, however, the plot revealed a deviation from the red cluster area indicating a unique expression signature possibly attributable to the defined culture condition.
Table 4.
PluriTest - global gene expression microarray data-based in-silico evaluation of pluripotency. “pluri-raw” – pluripotency score, “novelty” – novelty score, RMSD – root mean square deviation. AF-iPSC lines (19, 20A, D1) and WA25 control ESC line were deemed pluripotent by the test, however, pluripotency scores were relatively lower and novelty scores relatively higher compared to values typical for PSC grown in undefined feeder-based culture conditions.
| pluri-raw | pluri logit-p | novelty | novelty logit-p | RMSD | |
|---|---|---|---|---|---|
| 19 p19 | 13.74 | 1 | 1.62 | 0.08 | 0.47 |
| 20A p23 | 17.63 | 1 | 1.7 | 0.16 | 0.48 |
| D1 p23 | 8.35 | 1 | 1.66 | 0.11 | 0.47 |
| WA25 p21 | 4.57 | 0.99 | 1.66 | 0.11 | 0.46 |
Discussion
AFSC are an attractive cell source for reprogramming due to their unspecialized nature and the fact that they can be easily isolated and cultured before birth.2-5 In order for the clinical translation of in-vitro cell-based therapies to proceed, chemically defined media free of animal components for growth and proliferation of the cells need to be developed. Chemically defined E8 medium free of animal components for growth and proliferation of pluripotent stem cells has been developed. The medium also supports growth and proliferation of fibroblasts.14 Reprogramming process has been shown to be more efficient when chemically defined media were used.14,24
Expression of MSC markers indicated a mesenchymal phenotype of AFSC but with some unique properties as variability and deviation in CD90, CD166 and CD105 was observed, while markers of hematopoietic stem cells CD34 and CD45 were low. ESC markers Oct3/4, Nanog, Sox2, TRA and SSEA antigens were not expressed in a pluripotent pattern, however, weak Sox2 expression, variable TRA antigen and SSEA-4 expression was observed. Along with a high proliferative potential, these features could render AFSC highly amenable toward reprogramming. To this end, we sought to reprogram AFSC using episomal plasmids, an approach that would enable derivation of transgene-free iPSC. We used a combination of oriP/EBNA1-based episomal vectors described previously.20 Generally, in the absence of drug selection, oriP/EBNA-1 episomes are lost at around 5% per cell generation due to defects in plasmid synthesis and partitioning and thus, cells devoid of plasmids can easily be isolated.20,25 The transient expression of pluripotency genes forces the expression of the endogenous pluripotency genes.26
Reprogramming plasmids introduced into the source AFSC gave rise to epithelioid cell colonies. Acquisition of epithelial cell properties, reduced size and compact cell-cell interaction, through MET is one of the earliest events in reprogramming of somatic cells to iPSCs.26-30 Another 4 days of culture allowed for optimal number and size of the epithelioid colonies while further culture would lead to overgrowth of unreprogrammed AFSC. Secondary passaging allowed for testing of reprogramming supporting capacity of 2 different ECM matrix culture vessel coatings – VTN and StemAdhere; and 2 different media – E8, mTeSR-1 (rich, BSA-containing medium). AFSC growth medium was included as a control. Culture conditions involving StemAdhere coating and mTeSR-1 provided poor or no support to the source AFSC, disabling the roughly 3-week long reprogramming process. On the other hand, VTN in combination with E8 medium showed excellent ability to support reprogramming because the survival of the AFSC in this condition was limited but not eliminated, tipping the balance in favor of cells in the process of pluripotency acquisition.
Partially reprogrammed epithelioid cells continued to slowly proliferate in VTN/E8. Alkaline phosphatase was not active in these cells. Flow cytometry analysis revealed activation of Oct3/4 and Sox2 while Nanog and TRA antigens were expressed at low levels or absent. Confocal microscopy analysis corroborated the flow cytometry data. Oct3/4 expression was heterogeneous. Activation of ESC markers is not complete at intermediate stages of reprogramming.31-33 Some data indicate that activation of Nanog is a rate-limiting step in reprogramming and tends to be activated later in the process.34 TRA antigens have been implicated to represent markers capable of distinguishing between partially and fully reprogrammed cells.24 The morphology as well as the expression pattern of ESC markers in transformed AFSC demonstrated that the progression toward pluripotency was arrested at the partially reprogrammed stage. Notably, incomplete reprogramming has recently been implicated in driving of dysplasia leading to undifferentiated tumor development in various tissues in mice by means of altered epigenetic regulation.35 Therefore, the partially reprogrammed cells described here as a product of episomal reprogramming could provide a window of opportunity in studying epigenetic dysregulation leading to cancerous transformation. VTN/E8 seems to reproducibly provide ideal conditions for their derivation and expansion in the absence of butyrate. Parallels between tumorigenesis and reprogramming/dedifferentiation have indeed been the subject of speculation for some time. An alternative view of cancer based on these parallels may be of clinical benefit.36
After identifying that VTN coating and E8 medium were optimal for partially reprogrammed MET cell growth, we confirmed that the combination was indeed readily permissible for reprogramming of AFSC into cells with fully reprogrammed morphological and molecular characteristics. This was initially achieved using a much more efficient retroviral approach. However, the progression of partially reprogrammed cells to full pluripotency had to be facilitated in order to make the episomal plasmid-based reprogramming possible. Drastic chromatin reorganizations of the somatic state into the ESC-like state were implicated in the reprogramming process numerous times. Histone acetylation was shown to be enriched in ESC compared to differentiated cells.37-39 Butyrate improves reprogramming by reducing the frequency of partially reprogrammed cells.14,32,40,41 Treatment of the transfected AFSC starting at the time of the secondary split with 25 µM of sodium butyrate enabled progression to full pluripotency, otherwise, the reprogramming efficiency was prohibitively low. The pluripotent colonies possessed typical morphological features of primed pluripotent stem cells – high nucleus-to-cytoplasm ratio, prominent nuclei and nucleoli, flat colonies with phase-bright edges – and were expanded equally well on VTN-coated vessels in E8 medium as well as Matrigel-coated vessels in mTeSR-1 medium. At an early passage, flow cytometric analysis of the expression of ESC markers revealed similar profiles across AF-iPSC lines and the control episomally derived IISH1i-BM1 iPSC line. The expression profile observations were made for both culture system combinations – VTN/E8 and Matrigel/mTeSR-1.
We cultured the derived lines of iPSC for almost 40 passages. In their mature state at passages over 20, the cells exhibited typical characteristics of pluripotent stem cells. Flow cytometry and confocal microscopy confirmed typical expression pattern of ESC markers, identical to WA25 ESC line derived and cultured in VTN/E8 condition, representing an ideal control. However, WA25 exhibited a relatively higher frequency of spontaneous differentiation in culture, consistent across multiple passages, which reflected Nanog and TRA antigen fluorescence measured at slightly lower values. Alkaline phosphatase was active in all 3 iPSC lines. The data are indicative of homogeneous and high-quality cultures. Furthermore, the antibody we used to profile Oct3/4 expression was specific to its pluripotency-related splice variant – Oct3/4A – active in ESC and embryos beyond the 8-cell stage as opposed to Oct3/4B that is not related to pluripotency in these stages of embryonic development as well as lines of embryonic stem cells. Antibodies that bind the Oct3/4B can potentially give false-positive signals.42 Fluorescent intensity corresponding to Nanog expression was approximately 1 log decade dimmer compared to Oct3/4A and Sox2. Confocal microscopy analyses showed similar differences in the fluorescent signal and the images were consistent with those published in a recent paper showing Nanog expression in conventional human pluripotent stem cells. Bright Nanog fluorescence was then shown to be a feature of pluripotent stem cells that the authors converted into cells with naïve properties.43
When injected subcutaneously into immunocompromised scid-beige mice, the iPSC proliferated and differentiated into teratomas consisting of tissues representative of all 3 germ layers, demonstrating their pluripotency. However, teratoma formation assay has been a target of criticism in the recent years due to the fact that it is time-consuming, expensive and more importantly, it lacks standardization. This assay would be highly impractical for validation of pluripotency in patient-specific iPSC lines derived for the purposes of potential therapies. Furthermore, partially reprogrammed cells have also been shown to generate teratomas.24,44 The need for a fast, reliable, sensitive and inexpensive pluripotency validation tool can be addressed by deployment of the so-called “PluriTest.”45 This bioinformatic test processes global expression microarray profiles acquired on the Illumina platform (HT-12 BeadChip) and calculates the degree of pluripotency against a matrix of profiles of well-established human pluripotent stem cell lines. The test returns values for 2 different classifiers – pluripotency and novelty scores – developed based on 2 differently constructed metagene models. The pluripotency score is a measure as to what extent a query transcriptional profile contains a pluripotent signature; and the novelty score is a measure of how much of the signal from the query can be explained by the normal PSC lines contained in the matrix. The higher the pluripotency score and the lower the novelty score, the more the gene expression pattern resembles that of pluripotent stem cells in the matrix. The test can easily discern partially and fully reprogrammed cells with high resolution. The AF-iPSC lines were deemed pluripotent by PluriTest. Owing to its high sensitivity, slight variations in pluripotent signature can be picked up by the test so it is well-suited for selection of clones with the most faithfully reprogrammed configuration. AF-iPSC lines and the control ESC line WA25 localized slightly off the red spot that represents the cluster area of validated PSC lines which were cultured on conventional MEF layers in KO-SR-supplemented medium. WA25 is an ESC line that has been derived in VTN/E8 conditions and not simply adapted to it later. The culture condition differences are therefore the most likely reason for our observation of the deviation. To this date, few reports exist on PluriTest validation of PSC lines grown in xeno-free, chemically defined conditions. A recent study described episomal derivation of iPSC from novel urine cells as well as adipose tissue-derived stromal cells under xeno-free conditions.46 Among other tests, the authors utilized PluriTest to determine the degree of pluripotency in the novel iPSC lines, along with the control H9 human embryonic stem cell line, also adapted to the xeno-free conditions. The individual lines clustered in an area similar to that of AF-iPSC described here, including the H9 control line (that exhibited the highest pluripotency score). Albeit the cells presented in this study as well as our study were deemed pluripotent, it cannot be ruled out that for the purposes of pluripotency degree calculation in PSC lines cultured in chemically defined conditions, PluriTest might need to be re-evaluated to account for slight deviations from profiles of PSC cultured in conventional conditions. However, this would mean rebuilding the entire PSC matrix of PluriTest. Also, it cannot be ruled out that chemically defined conditions that are often much more simple than the conventional counterparts are not permissive to maintenance of the highest grade of pluripotency in PSC that is only discernible using the high resolution of tests like PluriTest. The latter would signify the need for optimization of chemically defined culture conditions.
iPSC derived from AFSC are potentially an optimal cell source for perinatal cell-based therapies but are also ideal for modeling diseases like Down syndrome. Impaired neurogenesis has been observed using iPSC derived from AFSC possessing trisomy 21.18 AFSC from β-thalassemia patients were found to serve as a rapid and efficient cell source for reprogramming into iPSC.19 Additionally, their potential to provide a universal cell source for allogeneic cardiac cell therapies has been explored.47 However, in these studies, virus-based integrating reprogramming approach or integrating approach with subsequent excision was used as well as feeder-based culture system. More recently, transgene-free iPSC were derived from AFSC employing a Sendai virus-based vector and were demonstrated to efficiently differentiate into neural progenitors capable of engrafting into the brain parenchyma of rats.48 However, the iPSC were derived and cultured in undefined ESC-conditioned medium on MEF layers though they could be transferred onto feeder-free surfaces. Our work complements this study in that it provides a method for derivation and culture of iPSC from AFSC in a chemically defined medium. We also analyzed the transcriptome of these cells using a global expression microarray system and PluriTest capable of scrutinizing fine details of the reprogramming outcome. Finally, we demonstrated the ability of these cells to differentiate into tissues representative of all 3 germ layers in a teratoma formation assay. While this manuscript was in preparation, a study described reprograming several adult and fetal cell types, including amniotic fluid stem cells, using oriP/EBNA-1 episomal plasmids.49 The calculated reprogramming efficiencies and the effect of additional reprogramming modulators miR302/367 and Mbd3 are of practical value. The iPSC were derived using the E8 medium, however, Geltrex was used as the culture matrix which, together with Matrigel, are undefined mixtures of extracellular matrix proteins derived from mouse Engelbreth-Holm-Swarm sarcoma. Additionally, teratoma formation and PluriTest scores were not reported in this study.
Conclusion
Reprogramming of somatic cells using episomal plasmids carries a compelling promise of a technology potentially capable of producing induced pluripotent stem cells free of integrated transgene sequences. AFSC represent a cell source that is easy to isolate and reprogram. iPSC derived from AFSC could potentially be utilized in a range of pediatric interventions, as they could be derived before birth, especially with recent advances in the reprogramming process. AFSC appear to have unique culture or growth factor requirements. The narrow range of optimal source AFSC growth and proliferation conditions limits the choice of conditions for reprogramming, therefore, identification of VTN/E8 as permissive is valuable. This condition is also uniquely suited for expansion of partially reprogrammed epithelioid cells that may be of interest in cancer research. An in-vitro model unaffected by inconsistencies of serum-supplemented culture and uncompromised by potential genetic instability resulting from transgene integration, would be desirable for both regenerative medicine applications and disease modeling.
Materials and methods
Human amniotic fluid stem cell culture
Human amniotic fluid stem cells (AFSC) were cultured in EBM-2 basal medium (CC-3156, Lonza, http://www.lonza.com/products-services/bio-research/primary-cells/human-cells-and-media/endothelial-cells-and-media/endothelial-cell-growth-media-kits.aspx) supplemented with 20% (intial stages of AFSC line derivation) or 5% (initial stages of reprogramming) fetal calf serum (S1520, BioWest, http://www.biowest.net/products/serum/fetal-bovine-serum/8311-fetal-bovine-serum-fbs-usa.html), with selected recombinant growth factors – basic fibroblast growth factor 50 ng/ml (GF003, Millipore, http://www.emdmillipore.com/US/en/product/Fibroblast-Growth-Factor-basic-Protein%2C-Human-recombinant,MM_NF-GF003), epidermal growth factor 25 ng/ml (GF144, Millipore, http://www.emdmillipore.com/US/en/product/Epidermal-Growth-Factor-Protein%2C-Human-recombinant,MM_NF-GF144) and insulin-like growth factor 20 ng/ml (GF138, Millipore, http://www.emdmillipore.com/US/en/product/Insulin-like-Growth-Factor-I-Protein%2C-Recombinant-human,MM_NF-GF138). AFSC cell lines were obtained by mixing 2.5 ml of amniotic fluid with 3.5ml of culture medium, plating onto a T25 tissue culture flask and incubating at 37°C and 5% CO2. The first medium change was performed 4–5 days after seeding procedure to allow cell adherence and clonal proliferation. On approximately day 10, the adherent cell cultures were harvested using StemPro® Accutase® (A1110501, Thermo Fisher Scientific, https://www.thermofisher.com/order/catalog/product/A1110501?ICID=search-a1110501) and replated onto larger culture flasks and expanded up to passage 4 to 6.
Retroviral reprogramming of AFSC
Viral particle assembly was carried out in Plat-E cells transfected with individual pMXs vectors carrying GFP or Oct4, Sox2, Klf4, c-Myc, and secreted into the DMEM medium (11960-044, Thermo Fisher Scientific, https://www.thermofisher.com/order/catalog/product/11960044?ICID=search-11960044) supplemented with 5% FCS and GlutaMAX (35050-061, Thermo Fisher Scientific, https://www.thermofisher.com/order/catalog/product/35050061?ICID=search-35050061) overnight following transfection. Media were collected, 0.22 µm filtered and used for AFSC transduction immediately. 3 days prior to transduction, 5000 cells/cm2 were seeded in a 6-well plate coated with recombinant human vitronectin – Vitronectin XF™ (07180, StemCell Technologies, http://www.stemcell.com/en/Products/All-Products/Vitronectin-XF.aspx) – in EBM-2 + 5% FCS and the growth factors bFGF 20 ng/ml, EGF 25 ng/ml and IGF 20 ng/ml. On the day of transduction, AFSC culture medium was replaced by a mixture of virus-containing media of equal volumes with 2 μg/mL polybrene (H9268, Sigma-Aldrich, http://www.sigmaaldrich.com/catalog/search?term+h9268&interface+All&N+0&mode+match%20partialmax&lang+en®ion+US&focus+product) and supplemented with bFGF, IGF and EGF. The source AFSC were exposed either to viruses carrying all 4 reprogramming genes (Oct4, Sox 2, Klf-4, c-myc) or 3 (c-myc excluded). Transduction media were discarded after an overnight incubation and for the following 5 days cells were cultured in EBM-2 + 5% FBS and growth factors (bFGF, EGF, IGF) with medium changes every other day. On day 5, the medium was replaced with E8 medium.
Episomal reprogramming of AFSC
AFSC cells were reprogrammed by means of transfection with non-integrating episomal/EBNA plasmids as previously published.20 Plasmids were purchased in form of E. coli bacterial stab cultures and were as follows: pEP4 E02S EN2K (Oct4+Sox2, Nanog+Klf4; 20925 Addgene, http://www.addgene.org/20925/), pEP4 E02S ET2K (Oct4+Sox2, SV40LT+Klf4; 20927 Addgene, http://www.addgene.org/20927/), pCEP4-M2L (c-Myc+LIN28; 20926 Addgene, http://www.addgene.org/20926/). Stab cultures were streaked on Luria agar plates containing 100µg/ml of Ampicillin. The plates were incubated for 8-12 hours at 37°C until individual bacterial colonies appeared. The colonies were then transferred into stirred, air exchange-allowing, bacterial culture bottles in Terrific Broth medium to expand the bacterial cells in suspension. Suspension cultures were maintained at 37°C for another 8-12 hours and while still in log-phase of growth, bacteria were harvested by centrifugation at 6000 g for 15 min.
Plasmid DNA was isolated from bacterial pellets using a commercial kit (12381, Qiagen, https://www.qiagen.com/us/search.aspx?q=12381#&&pg=1) following the manufacturer's instruction. In principle, the bacterial cells were left to undergo alkaline lysis. DNA was then bound to a proprietary resin in a column. The resin with the bound DNA was then washed and subsequently eluted. DNA was precipitated using isopropanol, resuspended in TE buffer and the yield measured using the NanoDrop 2000 device (Thermo Fisher Scientific, http://www.nanodrop.com/Productnd2000overview.aspx). All three plasmids were transfected into the source cells using the Neon Transfection System (Thermo Fisher Scientific, https://www.thermofisher.com/order/catalog/product/MPK5000?ICID=search-product) according to the manufacturer's instruction. The plasmid amounts per 1 million transfected cells were 3.0 µg pEP4 EO2S ET2K, 3.0 µg pEP4 EO2S EN2K, 2.0 µg pCEP4-M2L (combination #19).20 The transfection parameters were 950 V, 40 ms and 1 pulse. Following transfection, the cells were plated into 6-well plates and kept in the AFSC medium for the first 5-9 days. Then, secondary split was performed and the cells were plated directly into the iPSC medium. The medium was changed daily.
iPSC culture and freezing
For cell expansion, colonies of fully reprogrammed cells with pluripotent stem cell-like morphology were first mechanically picked from the original plate and then passaged using EDTA (15575-020, Thermo Fisher Scientific, https://www.thermofisher.com/order/catalog/product/15575020?ICID=search-15575020) diluted in PBS to 0.5mM.22 Medium was removed, wells washed with EDTA and incubated with 1mL of EDTA per 10 cm2 at RT for 5 minutes. EDTA was aspirated afterwards. Partially dissociated colonies were mechanically released using the E8 medium and plated into freshly coated wells. The iPSC culture media were as follows: E8 medium (05940, StemCell Technologies, http://www.stemcell.com/en/Products/All-Products/TeSR-E8.aspx), mTeSR-1 medium (05850, StemCell Technologies, http://www.stemcell.com/en/Products/All-Products/mTeSR1.aspx). iPSC culture surfaces used were as follows: StemAdhere™ Defined Matrix (07170, StemCell Technologies) and VTN. Control induced pluripotent stem cell line IISH1i-BM123 and an embryonic stem cell line WA25 used in this study were obtained from WiCell Research Institute (http://www.wicell.org/home/stem-cell-lines/order-stem-cell-lines/obtain-stem-cell-lines.cmsx).
For the purpose of cryopreservation, pellets of EDTA-harvested cell clumps were resuspended in CryoStor™CS10 (07930 StemCell Technologies, http://www.stemcell.com/en/Products/All-Products/CryoStorCS10.aspx), transferred into cryovials and placed into freezing containers providing 1°C/min cooling at −80°C. Following overnight incubation, the cryovials were transferred to liquid nitrogen storage.
Alkaline phosphatase staining
Partially reprogrammed epithelioid cells were subjected to alkaline phosphatase staining using Alkaline Phosphatase Live Stain kit (AP Live Stain, A14353, Thermo Fisher Scientific, https://www.thermofisher.com/order/catalog/product/A14353?ICID=search-a14353) according to the manufacturer's instruction for the purpose of identifying reprogrammed cells. Medium was discarded from the adherent cultures, changed for DMEM/F-12 containing diluted alkaline phosphatase substrate and incubated for 30min. Then the colonies were washed twice with DMEM/F-12 and inspected under an EVOS FL fluorescent microscope (Thermo Fisher Scientific, https://www.thermofisher.com/order/catalog/product/AMF4300?ICID=search-product). In mature iPSC, alkaline phosphatase activity was measured using flow cytometry. Substrate diluted at the ratio of 1:2000 in the cell suspension was incubated for 30min, washed twice with DMEM/F-12 and analyzed using FACSCanto II flow cytometer using 20mW 488nm excitation-502LP-530/30BP for detection (BD Biosciences, http://www.bdbiosciences.com/us/instruments/research/cell-analyzers/bd-facscanto-ii/m/744810).
Flow cytometry
AFSC were harvested using StemPro Accutase for 7 min, iPSC were harvested using EDTA for 15 min or StemPro Accutase for 10 min. For the purpose of nuclear transcription factor staining, the cells were washed with PBS, fixed in 2% formaldehyde diluted in PBS (28908, Thermo Fisher Scientific, https://www.thermofisher.com/order/catalog/product/28908?ICID=search-product) for 20 min at RT and permeabilized by BD Perm Buffer III (558050, BD Biosciences, http://www.bdbiosciences.com/us/applications/research/t-cell-immunology/th-1-cells/intracellular-markers/cell-signalling-and-transcription-factors/buffers/perm-buffer-iii/p/558050) for 30 min on ice. Incubation with directly labeled mouse anti-human monoclonal antibodies (listed below) was carried out at 4°C for 45 min, followed by 3 washes. For the purpose of surface antigen staining, the cells were washed with PBS + 2% FCS, incubated with the directly labeled mouse anti-human monoclonal antibodies listed below for 20 min at RT and subsequently washed. The cell suspension was analyzed using a FACSCanto II flow cytometer. AlexaFluor 488 was detected with 20mW 488 nm excitation-502LP-530/30BP. AlexaFluor 647 was detected with 17mW 633nm excitation-685LP-660/20BP. The resulting data analysis was performed using FlowJo 10.0.7 (TreeStar, http://www.flowjo.com) and Kaluza 1.2 and 1.3 (Beckman Coulter, https://www.beckmancoulter.com/wsrportal/page/itemDetails?itemNumber=B16407#2/10//0/25/1/0/asc/2/B16407///0/1//0/).
Confocal microscopy
For the purpose of immunocytochemical labeling of AFSC and iPSC, the cells were grown in E8 medium on VTN-coated 24-well glass-bottom dishes (P24G-1.5-13-F, MatTek- http://www.glass-bottom-dishes.com/pages/dish-selection-1.html). For surface antigen staining, the wells were washed with PBS + 2% FCS. The cells were incubated with the directly labeled mouse anti-human monoclonal antibodies listed below for 20 min at RT and then washed with PBS + 2% FCS. For nuclear transcription factor staining, the wells were washed with PBS, fixed with 2% formaldehyde for 20 min at RT and permeabilized using BD Perm Buffer III. Incubation with directly labeled mouse anti-human monoclonal antibodies listed below was carried out at 4°C for 45 min. Three washing steps were performed with PBS + 2% FCS, each incubated for 10 min. DAPI (D1306, Thermo Fisher Scientific, https://www.thermofisher.com/order/catalog/product/D1306?icid=fr-dapi-1) was added into the washing solution (800nM solution) and incubated for 5 min during one of the washing steps. Colonies were imaged using either a Leica TSC SP2 confocal microscope (http://www.leica-microsystems.com/products/confocal-microscopes/details/product/leica-tcs-sp8/) or Nikon Ti-E with A1r-SI and N-STORM confocal microscope (http://www.nikoninstruments.com/Products/Light-Microscope-Systems/Super-Resolution/N-STORM-Super-Resolution).
List of antibodies used in flow cytometry and immunohistochemistry experiments
Antibodies were purchased from the following sources: BD Biosciences (http://www.bdbiosciences.com/us/reagents/c/reagents), BioLegend http://www.biolegend.com/productstab), Immunotools (http://www.immunotools.de/html/english.html) and Jackson Immunoresearch (https://www.jacksonimmuno.com/catalog/22).
Intracellular staining: mouse anti-human monoclonal AlexaFluor 488-conjugated anti-Oct3/4 (560253, BD) or AlexaFluor 488-conjugated anti-Oct3/4A (561628, BD), AlexaFluor 647-conjugated anti-Oct3/4A (562252, BD), AlexaFluor 488-conjugated anti-Nanog (560791, BD) AlexaFluor 647-conjugated anti-Nanog (561300, BD), AlexaFluor 647-conjugated anti-Sox2 (561593, BD). Surface staining: mouse anti-human monoclonal AlexaFluor 488-conjugated anti-TRA-1-60 (330614, BioLegend), AlexaFluor 647-conjugated anti-TRA-1-81 (330706, BioLegend), AlexaFluor 488-conjugated anti-SSEA-1 (560120, BD), AlexaFluor 647-conjugated anti-SSEA-4 (560308, BD). Isotype controls for these markers were matched in concentrations: AlexaFluor 488-conjugated mouse IgG1κ (557782, BD), AlexaFluor 647-conjugated mouse IgG1κ (557783, BD), AlexaFluor 488-conjugated mouse IgMκ (401617, BioLegend), AlexaFluor 647-conjugated mouse IgMκ (401618, BioLegend), AlexaFluor 488-conjugated mouse IgG1κ (400129, BioLegend), AlexaFluor 647-conjugated mouse IgG3κ (401321, BioLegend).
Antibodies used in flow cytometric analysis of AFSC for the purpose of mesenchymal stem cell marker expression analysis were: mouse anti-human monoclonal IgG1κ isotype control (400102, BioLegend), CD44 (sc-59758, Santa Cruz), CD73 (344002, BioLegend), CD90 (328102, BioLegend), CD166 (343902, BioLegend), FITC-conjugated IgG1κ isotype control (21275513S, ImmunoTools), FITC-conjugated CD34 (21270341, ImmunoTools) and goat anti-mouse Cy2-conjugated secondary antibody (115-225-003, Jackson ImmunoResearch).
Teratoma formation assay
All animal experiments were approved by the IACUC of the University of South Alabama (protocol number 277067-4) and carried out at the vivarium of the Department of Comparative Medicine at the College of Medicine (Assurance Number A3288-01; AAALAC accredited). iPSC colonies were harvested from the 6-well culture plates by EDTA as described above and resuspended in 1:1 mixture of E8 medium and Matrigel. Suspension of cell clumps containing between 500.000 to 1 million of cells were injected subcutaneously into both flanks of immunocompromised scid-beige mice (Taconic Biosciences, http://www.taconic.com/mouse-model/scid-beige), 3 animals per line. 6 to 9 weeks post-injection, the teratomas were excised, fixed in formalin and processed for standard H&E histological analysis. The presence of tissues representative of all 3 germ layers was verified by a pathologist.
PluriTest transcriptional profiling
RNA was extracted from iPSC cultures using RNeasy Plus Mini kit (74134, Qiagen, https://www.qiagen.com/us/shop/sample-technologies/rna-sample-technologies/total-rna/rneasy-plus-micro-and-mini-kits). Whole genome expression analysis was performed by the HudsonAlpha Institute for Biotechnology Genomic Services Laboratory (http://gsl.hudsonalpha.org/index) using the Human HT-12 v4 Expression BeadChip microarray system (Illumina, http://www.illumina.com/products/humanht_12_expression_beadchip_kits_v4.html). For pluripotency level analysis, the raw idat files were uploaded to PluriTest through a web-based interface at www.pluritest.org.
Abbreviations
- AAALAC
Association of Assessment and Accreditation of Laboratory Animal Care
- AF-iPSC
induced pluripotent stem cells derived from amniotic fluid stem cells
- AFSC
amniotic fluid stem cells
- AP
alkaline phosphatase
- bFGF
basic fibroblast growth factor
- BSA
bovine serum albumin
- DAPI -
4′,6-diamidino-2-phenylindole
- E8
pluripotent stem cell-specific growth medium published by Chen et al. (2011)
- EDTA
ethylenediaminetetraacetic acid
- ESC
embryonic stem cells
- FCS
fetal calf serum
- IACUC
Institutional Animal Care and Use Committee
- IGF
insulin-like growth factor
- iPSC
induced pluripotent stem cells
- IRES2
internal ribosome entry site
- KO-SR
KnockOut™ Serum Replacer;
- MET
mesenchymal-to-epithelial transition
- MSC
mesenchymal stem cells
- oriP/EBNA-1
plasmid origin of viral replication/Epstein-Barr nuclear antigen-1
- PSC
pluripotent stem cells
- RT
room temperature
- SV40LT
SV40 large T antigen
- TE -
Tris/EDTA
- TGFβ
transforming growth factor β
- VTN
vitronectin (Vitronectin XF™)
Disclosure of potential conflicts of interest
The authors have no potential conflicts of interest to disclose.
Acknowledgments
We would like to thank Dr. Nicole Gross and Dr. Josef Achermann for procuring amniotic fluid samples and sharing their experience; Ursula Steckholzer at the SCRM; I-Hsuan Wang and the rest of Dr. Urs Greber's lab, at the Institute of Molecular Life Sciences of the University of Zurich, for technical assistance. Finally, we would like to thank Dr. Joel Andrews for help as well as sharing his valuable imaging expertise.
Funding
This work was supported by the Fonds Medizinische Forschung at the University of Zurich, Forschungskredit of the University of Zurich, The SCIEX NMSCh 10.216 and 12.176 Fellowships, The Swiss Society of Cardiology, The Swiss National Science Foundation [320030-122273] and [310030-143·992], The 7th Framework Programme, Life Valve, European Commission [242008], Alfred und Anneliese Sutter-Stöttner Stiftung, Olga Mayenfisch Foundation, EMDO Foundation, Start-up Grant 2012 of the University Hospital Zurich, The APRC of Slovakia - Grant LPP-0119-09, internal funding of the Mitchell Cancer Institute. Jaroslav Slamecka was a SCIEX NMSCh Fellow in 2012 [10.216] and 2013 [12.176] at the University Hospital Zurich.
References
- 1.Loh Y-H, Agarwal S, Park I-H, Urbach A, Huo H, Heffner GC, Kim K, Miller JD, Ng K, Daley GQ. Generation of induced pluripotent stem cells from human blood. Blood 2009; 113:5476-9; PMID:19299331; http://dx.doi.org/ 10.1182/blood-2009-02-204800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Coppi P, Bartsch G Jr, Siddiqui MM, Xu T, Santos CC, Perin L, Mostoslavsky G, Serre AC, Snyder EY, Yoo JJ, et al.. Isolation of amniotic stem cell lines with potential for therapy. Nat Biotechnol 2007; 25:100-6; PMID:17206138; http://dx.doi.org/ 10.1038/nbt1274 [DOI] [PubMed] [Google Scholar]
- 3.Schmidt D, Achermann J, Odermatt B, Breymann C, Mol A, Genoni M, Zund G, Hoerstrup SP. Prenatally fabricated autologous human living heart valves based on amniotic fluid derived progenitor cells as single cell source. Circulation 2007; 116:I64-70; PMID:17846327; http://dx.doi.org/ 10.1161/CIRCULATIONAHA.107.184051 [DOI] [PubMed] [Google Scholar]
- 4.Schmidt D, Achermann J, Odermatt B, Genoni M, Zund G, Hoerstrup SP. Cryopreserved amniotic fluid-derived cells: a lifelong autologous fetal stem cell source for heart valve tissue engineering. J Heart Valve Dis 2008; 17:446-55; PMID:18751475 [PubMed] [Google Scholar]
- 5.Weber B, Emmert MY, Behr L, Schoenauer R, Brokopp C, Drögemüller C, Modregger P, Stampanoni M, Vats D, Rudin M, et al.. Prenatally engineered autologous amniotic fluid stem cell-based heart valves in the fetal circulation. Biomaterials 2012; 33:4031-43; PMID:22421386; http://dx.doi.org/ 10.1016/j.biomaterials.2011.11.087 [DOI] [PubMed] [Google Scholar]
- 6.Zaim M, Karaman S, Cetin G, Isik S. Donor age and long-term culture affect differentiation and proliferation of human bone marrow mesenchymal stem cells. Ann Hematol 2012; 91:1175-86; PMID:22395436; http://dx.doi.org/ 10.1007/s00277-012-1438-x [DOI] [PubMed] [Google Scholar]
- 7.Bork S, Pfister S, Witt H, Horn P, Korn B, Ho AD, Wagner W. DNA methylation pattern changes upon long-term culture and aging of human mesenchymal stromal cells. Aging Cell 2010; 9:54-63; PMID:19895632; http://dx.doi.org/ 10.1111/j.1474-9726.2009.00535.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phermthai T, Suksompong S, Tirawanchai N, Issaragrisil S, Julavijitphong S, Wichitwiengrat S, Silpsorn D, Pokathikorn P. Epigenetic Analysis and Suitability of Amniotic Fluid Stem Cells for Research and Therapeutic Purposes. Stem Cells Dev 2013; 22:1319-28; PMID:23249260; http://dx.doi.org/ 10.1089/scd.2012.0371 [DOI] [PubMed] [Google Scholar]
- 9.Jelinic P, Shaw P. Loss of imprinting and cancer. J Pathol 2007; 211:261-68; PMID:17177177; http://dx.doi.org/ 10.1002/path.2116 [DOI] [PubMed] [Google Scholar]
- 10.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, et al.. Epigenetic memory in induced pluripotent stem cells. Nature 2010; 467:285-90; PMID:20644535; http://dx.doi.org/ 10.1038/nature09342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moschidou D, Mukherjee S, Blundell MP, Drews K, Jones GN, Abdulrazzak H, Nowakowska B, Phoolchund A, Lay K, Ramasamy TS, et. al. Valproic acid confers functional pluripotency to human amniotic fluid stem cells in a transgene-free approach. Mol Ther 2012; 20:1953-67; PMID:22760542; http://dx.doi.org/ 10.1038/mt.2012.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eminli S, Foudi A, Stadtfeld M, Maherali N, Ahfeldt T, Mostoslavsky G, Hock H, Hochedlinger K. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nat Genet 2009. ; 41:968-76; PMID:19668214; http://dx.doi.org/ 10.1038/ng.428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zweigerdt R, Olmer R, Singh H, Haverich A, Martin U. Scalable expansion of human pluripotent stem cells in suspension culture. Nat Prot 2011; 6:689-700; http://dx.doi.org/ 10.1038/nprot.2011.318 [DOI] [PubMed] [Google Scholar]
- 14.Chen G, Gulbrason DR, Hou Z, Bolin J, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, et al.. Chemically defined conditions for human iPSC derivation and culture. Nat Methods 2011; 8:424-29; PMID:21478862; http://dx.doi.org/ 10.1038/nmeth.1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li C, Zhou J, Shi G, Ma Y, Yang Y, Gu J, Yu H, Jin S, Wei Z, Chen F, Jin Y. Pluripotency can be rapidly and efficiently induced in human amniotic fluid-derived cells. Hum Mol Genet 2009; 18:4340-49; PMID:19679563; http://dx.doi.org/ 10.1093/hmg/ddp386 [DOI] [PubMed] [Google Scholar]
- 16.Galende E, Karakikes I, Edelmann L, Desnick RJ, Kerenyi T, Khoueiry G, Lafferty J, McGinn JT, Brodman M, Fuster V, et al.. Amniotic fluid cells are more efficiently reprogrammed to pluripotency than adult cells. Cell Reprogramm 2010; 12:117-25; http://dx.doi.org/ 10.1089/cell.2009.0077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolfrum K, Wang Y, Prigione A, Sperling K, Lehrach H, Adjaye J. The LARGE principle of cellular reprogramming: lost, acquired and retained gene expression in foreskin and amniotic fluid-derived human iPS cells. PLoS One 2010; 5:e13703; PMID:21060825; http://dx.doi.org/ 10.1371/journal.pone.0013703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu HE, Yang YC, Chen SM, Su HL, Huang PC, Tsai MS, Wang TH, Tseng CP, Hwang SW. Modeling neurogenesis impairment in Down syndrome with induced pluripotent stem cells from Trisomy 21 amniotic fluid cells. Exp Cell Res 2013; 319:498-505; PMID:23041301; http://dx.doi.org/ 10.1016/j.yexcr.2012.09.017 [DOI] [PubMed] [Google Scholar]
- 19.Fan Y, Luo Y, Chen X, Li Q, Sun X. Generation of human β-thalassemia induced pluripotent stem cells from amniotic fluid cells using a single excisable lentiviral stem cell cassette. J Reprod Dev 2012; 58:404-9; PMID:22498813; http://dx.doi.org/ 10.1262/jrd.2011-046 [DOI] [PubMed] [Google Scholar]
- 20.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science 2009; 324:797-801; PMID:19325077; http://dx.doi.org/ 10.1126/science.1172482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng L, Hansen NF, Zhao L, Du Y, Zou C, Donovan FX, Chou BK, Zhou G, Li S, Dowey SN, et al.. Low incidence of DNA sequence variation in human induced pluripotent stem cells generated by nonintegrating plasmid expression. Cell Stem Cell 2012; 10:337-44; PMID:22385660; http://dx.doi.org/ 10.1016/j.stem.2012.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beers J, Gulbranson DR, George N, Siniscalchi LI, Jones J, Thomson JA, Chen G. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat Protoc 2012; 7:2029-40; PMID:23099485; http://dx.doi.org/ 10.1038/nprot.2012.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu K, Yu J, Suknuntha K, Tian S, Montgomery K, Choi KD, Stewart R, Thomson JA, Slukvin II. Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood 2011; 117:e109-19; PMID:21296996; http://dx.doi.org/ 10.1182/blood-2010-07-298331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan EM, Chan EM, Ratanasirintrawoot S, Park I-H, Manos PD, Loh Y-H, Huo H, Miller JD, Hartung O, Rho J, et al.. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol 2009; 27:1033-37; PMID:19826408; http://dx.doi.org/ 10.1038/nbt.1580 [DOI] [PubMed] [Google Scholar]
- 25.Nanbo A, Sugden A, Sugden B. The coupling of synthesis and partitioning of EBV's plasmid replicon is revealed in live cells. EMBO J 2007; 26:4252-62; PMID:17853891; http://dx.doi.org/ 10.1038/sj.emboj.7601853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang G, Zhang Y. Embryonic stem cell and induced pluripotent stem cell: an epigenetic perspective. Cell Res 2013; 23:49-69; PMID:23247625; http://dx.doi.org/ 10.1038/cr.2012.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang G, He J, Zhang Y. Kdm2b promotes induced pluripotent stem cell generation by facilitating gene activation early in reprogramming. Nat Cell Biol 2012; 14:457-66; PMID:22522173; http://dx.doi.org/ 10.1038/ncb2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, et al.. Chromatin-modifying enzymes as modulators of reprogramming. Nature 2012; 483:598-602; PMID:22388813; http://dx.doi.org/ 10.1038/nature10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al.. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 2010; 7:51-63; PMID:20621050; http://dx.doi.org/ 10.1016/j.stem.2010.04.014 [DOI] [PubMed] [Google Scholar]
- 30.Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 2010; 7:64-77; PMID:20621051; http://dx.doi.org/ 10.1016/j.stem.2010.04.015 [DOI] [PubMed] [Google Scholar]
- 31.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature 2008; 454:49-55; PMID:18509334; http://dx.doi.org/ 10.1038/nature07056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang G, Taranova O, Xia K, Zhang Y. Butyrate promotes induced pluripotent stem cell generation. J Biol Chem 2010; 285:25516-21; PMID:20554530; http://dx.doi.org/ 10.1074/jbc.M110.142059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 2012; 150:1209-22; PMID:22980981; http://dx.doi.org/ 10.1016/j.cell.2012.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Theunissen TW, Jaenisch R. Molecular control of induced pluripotency. Cell Stem Cell 2014; 14:720-34; PMID:24905163; http://dx.doi.org/ 10.1016/j.stem.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohnishi K, Semi K, Yamamoto T, Shimizu M, Tanaka A, Mitsunaga K, Okita K, Osafune K, Arioka Y, Maeda T, et al.. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell 2014; 156:663-77; PMID:24529372; http://dx.doi.org/ 10.1016/j.cell.2014.01.005 [DOI] [PubMed] [Google Scholar]
- 36.Goding CR, Pei D, Lu X. Cancer: pathological nuclear reprogramming. Nature Reviews Cancer 2014; 14:568-73; PMID:25030952; http://dx.doi.org/ 10.1038/nrc3781 [DOI] [PubMed] [Google Scholar]
- 37.Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, et al.. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2008; 2:437-47; PMID:18462694; http://dx.doi.org/ 10.1016/j.stem.2008.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krejci J, Uhlirova R, Galiova G, Kozubek S, Smigova J, Bartova E. Genome-wide reduction in H3K9 acetylation during human embryonic stem cell differentiation. J Cell Physiol 2009; 219:677-87; PMID:19202556; http://dx.doi.org/ 10.1002/jcp.21714 [DOI] [PubMed] [Google Scholar]
- 39.Li X, Li L, Pandey R, Byun JS, Gardner K, Qin Z, Dou Y. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell 2012; 11:163-78; PMID:22862943; http://dx.doi.org/ 10.1016/j.stem.2012.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mali P, Chou BK, Yen J, Ye Z, Zou J, Dowey S, Brodsky RA, Ohm JE, Yu W, Baylin SB, et al.. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells 2010; 28:713-20; PMID:20201064; http://dx.doi.org/ 10.1002/stem.402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huangfu D,R Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small molecule compounds. Nat Biotechnol 2008; 26:795-97; PMID:18568017; http://dx.doi.org/ 10.1038/nbt1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martí M, Mulero L, Pardo C, Morera C, Carrió M, Laricchia-Robbio L, Esteban CR, Belmonte JCI. Characterization of pluripotent stem cells. Nat Prot 2013; 8:223-53; PMID:Can't; http://dx.doi.org/ 10.1038/nprot.2012.154 [DOI] [PubMed] [Google Scholar]
- 43.Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W, et al.. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 2014; 158:1254-69; PMID:25215486; http://dx.doi.org/ 10.1016/j.cell.2014.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Müller FJ, Goldmann J, Löser P, Loring JF. A call to standardize teratoma assays used to define human pluripotent cell lines. Cell Stem Cell 2010; 6:412-14; PMID:20452314; http://dx.doi.org/ 10.1016/j.stem.2010.04.009 [DOI] [PubMed] [Google Scholar]
- 45.Müller FJ, Schuldt BM, Williams R, Mason D, Altun G, Papapetrou EP, Danner S, Goldmann JE, Herbst A, Schmidt NO, et al.. A bioinformatic assay for pluripotency in human cells. Nat Methods 2013; 8:315-17; http://dx.doi.org/ 10.1038/nmeth.1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee KI, Kim HT, Hwang DY. Footprint- and xeno-free human iPSCs derived from urine cells using extracellular matrix-based culture conditions. Biomaterials 2014; 35:8330-38; PMID:24994040; http://dx.doi.org/ 10.1016/j.biomaterials.2014.05.059 [DOI] [PubMed] [Google Scholar]
- 47.Parolini O, Soncini M, Evangelista M, Schmidt D. Amniotic membrane and amniotic fluid-derived cells: potential tools for regenerative medicine? Regen Med 2009; 4:275-91; PMID:19317646; http://dx.doi.org/ 10.2217/17460751.4.2.275 [DOI] [PubMed] [Google Scholar]
- 48.Jiang G, Di Bernardo J, Maiden MM, Villa-Diaz LG, Mabrouk OS, Krebsbach PH, O'Shea KS, Kunisaki SM. Human transgene-free amniotic fluid-derived induced pluripotent stem cells for autologous cell therapy. Stem Cells Dev 2014; 23:2613-25; PMID:25014361; http://dx.doi.org/ 10.1089/scd.2014.0110 [DOI] [PubMed] [Google Scholar]
- 49.Drozd AM, Walczak MP, Piaskowski S, Stoczynska-Fidelus E, Rieske P, Grzela DP. Generation of human iPSCs from cells of fibroblastic and epithelial origin by means of the oriP/EBNA-1 episomal reprogramming system. Stem Cell Res Ther 2015; 6:122; PMID:26088261; http://dx.doi.org/ 10.1186/s13287-015-0112-3 [DOI] [PMC free article] [PubMed] [Google Scholar]