Abstract
The regulated loading of the Mcm2-7 DNA helicase into pre-replicative complexes (pre-RCs) at multiple replication origins ensures precise once per cell cycle replication in eukaryotic cells. Origin Recognition Complex (ORC), Cdc6 and Cdt1 load Mcm2-7 into a double hexamer bound around duplex DNA in an ATP-dependent reaction, but the molecular mechanism of this origin ‘licensing’ is still poorly understood. Here we show that both Mcm2-7 hexamers are recruited to origins by an essential, conserved C-terminal domain of Mcm3 which interacts with and stimulates the ATPase activity of ORC•Cdc6. ATP hydrolysis can promote Mcm2-7 loading, but can also promote Mcm2-7 release if components are missing or if ORC has been inactivated by cyclin-dependent kinase phosphorylation. Our work provides new insights into how origins are licensed and reveals a novel ATPase-dependent mechanism contributing to precise once per cell cycle replication.
Stable genome inheritance requires that replication origins initiate efficiently during S phase, and that re-initiation from these origins is subsequently prevented. This is accomplished by first licensing origins during G1 phase with a pre-RC containing an inactive double hexamer of the Mcm2-7 helicase, and then activating the helicase during S phase, when pre-RCs can no longer be assembled1-9.
Mcm3 recruits Cdt1•Mcm2-7 to ORC•Cdc6
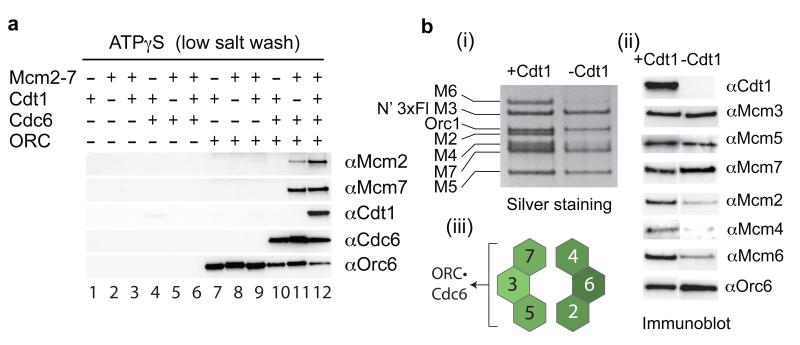
We have previously described a system in which pre-RCs can be assembled on DNA with purified proteins7. When ATP is present, ORC and Cdc6 load Cdt1•Mcm2-7 heptamers into a salt-resistant Mcm2-7 double hexamer with concomitant release of Cdc6 and Cdt1; in ATPγS all pre-RC components are recruited in low salt but are removed by a high salt wash (HSW) (Supp. Fig. 1a). To investigate the individual roles of Cdt1 and Mcm2-7, we purified each separately (Supp. Fig. 1b-d) and showed each protein was functional for loading when re-assembled into a Cdt1•Mcm2-7 complex (Supp. Fig. 2). Consistent with our previous results7, none of the pre-RC components bound to DNA under recruitment conditions (ATPγS, low salt) in the absence of ORC (Fig. 1a, lanes 1-6). In the absence of Cdc6 (lanes 7-9), ORC bound DNA, but could not recruit Cdt1, Mcm2-7 or Cdt1•Mcm2-7. Cdc6 was recruited to DNA in an ORC-dependent manner (compare lanes 4 and 10) consistent with the formation of an essential ORC•Cdc6 complex10. Cdt1 was not recruited in the presence of ORC•Cdc6 (lane 10), but we found significant ORC- and Cdc6-dependent recruitment of Mcm2-7 subunits in the absence of Cdt1 (compare lane 11 to lanes 6 and 9). Cdt1 was only recruited when Mcm2-7 was present along with ORC and Cdc6 (lane 12). From this we conclude that Mcm2-7 can be recruited to ORC•Cdc6 without Cdt1 and that Cdt1 recruitment requires Mcm2-7.
Figure 1. Mcm3 is necessary and sufficient for Mcm2-7 recruitment.
a) Protein requirements for Mcm2-7 recruitment. ORC, Cdc6, Cdt1 and Mcm2-7 were purified as described in Supplementary Methods and in Supp. Fig. 1. After incubation of the indicated proteins with DNA beads in the presence of ATPγS, beads were isolated and washed with low salt. DNA was uncoupled with irradiation as described in Supplementary Methods and bound proteins were analysed by immunoblot with the indicated antibodies. b) Recruitment of Mcm2-7 by ORC and Cdc6 was performed as in Fig. 1a. Reactions contained Mcm2-7 either with (+) or without (−) Cdt1. Bound proteins were visualised by silver staining (i) or immunoblot (ii). Panel (iii) summarises the result in Fig. 1b (i and ii).
The absence of Cdt1 did not affect Mcm7 recruitment, but did reduce the amount of Mcm2 recruited (Fig. 1a, compare lanes 11 and 12). To explore this further, we tested for the presence of each of the six Mcm2-7 subunits after Mcm2-7 recruitment in ATPγS in the presence or absence of Cdt1 using silver staining (Fig. 1b i) and immunoblotting (Fig. 1b ii). We found that Mcm3, 5 and 7 were recruited to ORC•Cdc6 similarly in the presence and absence of Cdt1 (Fig. 1b); the recruitment of Mcm2, 4 and 6, however, was significantly reduced without Cdt1. This suggests that Mcm3, 5 and 7, which are immediate neighbours in the Mcm2-7 ring, are recruited to ORC•Cdc6 directly without Cdt1, but Cdt1 plays some role in recruiting Mcm2, 4 and 6 (Fig. 1B iii).
The extreme C-terminus of Mcm3 recruits Cdt1•Mcm2-7
We took several approaches to address which Mcm2-7 subunits were involved in the Cdt1-independent recruitment. We tested each of the six Mcm2-7 subunits individually and found that only Mcm3 could be recruited in a Cdc6-dependent manner without the other subunits (Supp. Fig. 3a,b,f). We next assembled Mcm2-7 complexes containing all subunit or lacking either Mcm3 or Mcm4 (Supp. Fig. 3c,d). Mcm5 and 7 were recruited to ORC•Cdc6 in the absence of Cdt1 by the complete complex (Supp. Fig. 3f, lanes 1 and 5) and by Δ4 (lanes 3 and 7), but not by Δ3 (lanes 2 and 6). Taken together, these results indicate that Mcm3 is critical for recruiting Mcm5 and 7 to ORC•Cdc6 in the absence of Cdt1.
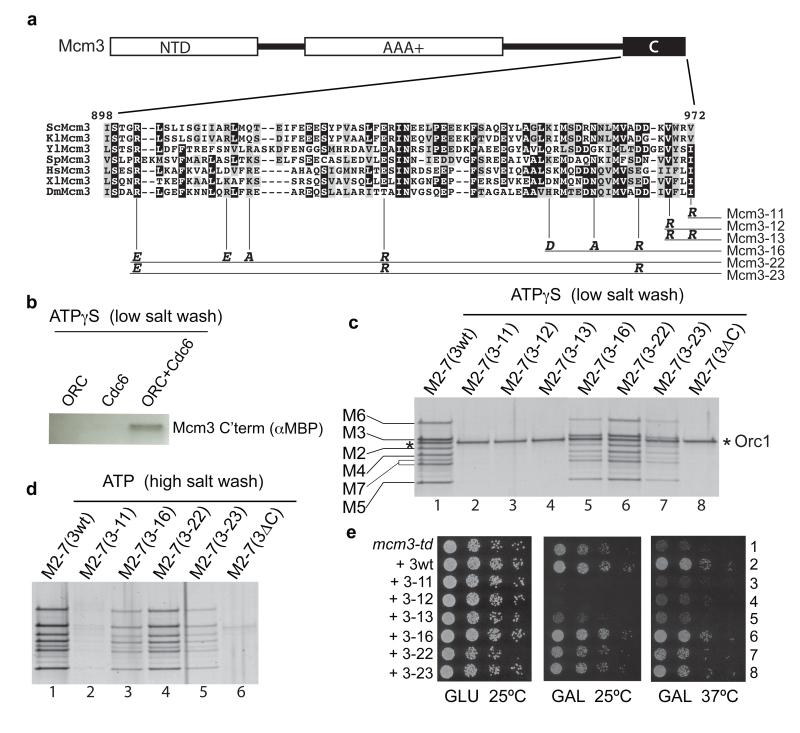
Mcm3 comprises an N-terminal domain (NTD) and a AAA+ domain, which are both found in all Mcm2-7 subunits11,12, and an extended C-terminal tail of unknown function containing a conserved domain at the extreme C-terminus (‘C’ in Fig. 2a) which is not found in other Mcm2-7 subunits (Fig. 2a). A version of Mcm3 containing a 3X FLAG tag at its C-terminus could be assembled into a stable heptameric Cdt1•Mcm2-7 complex, but was unable to recruit or load Mcm2-7 (Supp. Fig. 4). Moreover, a small fragment containing the C-terminal 194 amino acids residues of Mcm3 was recruited in an ORC- and Cdc6-dependent manner (Fig. 2b). These results pointed to the C-terminus as being crucial for Mcm3 recruitment.
Figure 2. The C-terminus of Mcm3 is required for Mcm2-7 recruitment.
a) The domain architecture of Mcm3 and alignment of the Mcm3 C-terminus. The alignment includes Mcm3 from a variety of eukaryotic species (Sc, Saccharomyces cerevisiae; Kl, Kluyveromyces lactis; Yl, Yarrowia lipolytica; Sp, Schizosaccharomyces pombe; Hs, Homo sapiens; Xl, Xenopus laevis; Dm, Drosophila melanogaster). Residue numbers above alignment correspond to S. cerevisiae Mcm3. The position of various mutants is shown by vertical lines, and the mutant amino acid residue is shown at the bottom of the line. Allele names are on the right. The numbering of alleles begins with 11 to prevent confusion with any existing mcm3 mutant alleles. b) Recruitment assay performed with a C-terminal MBP tagged Mcm3 fragment (last 194 aa) and with the indicated proteins. c) Cdt1•Mcm2-7 complexes containing wild type and mutant Mcm3 were tested for recruitment or d) for loading, proteins were analysed by silver staining. e) Wild type and mutant Mcm3 alleles were expressed from a galactose-inducible promoter in strains containing the mcm3-td degron mutant. A dilution series was tested for growth on plates containing either 2% D-glucose or 2% D-galactose at either 25°C or 37°C as indicated.
Based on the conservation of the Mcm3 C-terminus (Fig. 2a), and the fact that a tag at the C-terminus interfered with its function, we generated a series of C domain amino acid substitution mutants in full-length Mcm3 (Fig. 2a). We assembled the mutant proteins into Cdt1•Mcm2-7 complexes (Supp. Fig. 5) and tested their ability to recruit and load Mcm2-7. The single mutants Mcm3-11 and Mcm3-12, and the Mcm3-13 double mutant were completely defective in recruitment of Cdt1•Mcm2-7 to ORC•Cdc6 (Fig. 2c, lanes 2-4) as was Mcm3 lacking its entire C-terminal domain (3ΔC, Fig. 2c, lane 8). In these experiments, Orc1 serves as a useful loading control. These mutant proteins were also not recruited when tested individually in the absence of Cdt1 and the other Mcm2-7 subunits (Supp. Fig. 6). Consistent with their defect in recruitment, Mcm3-11 and Mcm3ΔC were unable to load Mcm2-7 (Fig. 2d, lanes 2 and 6). Mutation of other conserved residues (Mcm3-16, 3-22 and 3-23) generated proteins that were less defective, showing reduced recruitment (Fig. 2c, lanes 5-7 and Supp. Fig. 6) and a commensurate reduction in loading (Fig. 2d, lanes 3-5). Therefore, the C-terminus of Mcm3 is essential for recruitment of all Mcm2-7 subunits. As Mcm2-7 containing Mcm3-11, -12 or -13 is not recruited even when bound to Cdt1, this indicates that Cdt1 cannot recruit Mcm2-7 to ORC•Cdc6 directly. The reduced association of Mcm2,4,6 with ORC•Cdc6 (Fig. 1a,b) therefore likely reflects a role for Cdt1 in retaining or stabilising these subunits after initial recruitment by Mcm3.
We next expressed the mutant Mcm3 proteins in yeast strains harbouring a temperature-sensitive degron mutant of MCM3 (mcm3-td)13. This degron mutant grew well at 25°C, but not at the restrictive temperature, 37°C (Fig. 2e, row 1). The growth defect at 37°C was suppressed by expression of wild-type Mcm3 (3wt, row 2), but not by expression of Mcm3-11, -12 or -13 (rows 3-5) indicating that these proteins are not functional in vivo. Expression of these proteins significantly reduced growth at even at 25°C when the wild-type degron fusion protein was functional (middle panel, GAL 25°C, compare row 1 with rows 3-5), indicating that these mutants act as dominant-negative inhibitors when overexpressed. Some of the other mutants (Mcm3-16, -22, -23) showed mild growth defects at 37°C consistent with their partial defects in Mcm2-7 loading. Supp. Fig. 7 shows that mcm3-11 supported only very slow growth when present as an only copy whilst mcm3-12 and mcm3-13 did not support growth. Taken together, these results indicate that a domain at the extreme C-terminus of Mcm3 is necessary for recruiting Cdt1•Mcm2-7 to ORC•Cdc6, and that this function is essential for viability.
Both Mcm2-7 hexamers require the C-terminus of Mcm3
The results in Figs 1 and 2 show that recruitment of the first Mcm2-7 hexamer occurs by interaction of Mcm3 with ORC•Cdc6. If recruitment of the second hexamer occurred by a different mechanism, one that didn’t involve Mcm3-ORC•Cdc6 interaction but did require recruitment of the first hexamer, then the Mcm3-11 mutant should be able to be recruited as the second hexamer in the presence of wild-type Cdt1•Mcm2-7. To test this, we first generated two different forms of Mcm3 that could be distinguished after SDS-PAGE, one containing an N-terminal fusion to the FLAG tag (N’ 3XFLAG M3), the other containing a larger N-terminal fusion to maltose binding protein (N’ MBP M3). Both proteins supported similar levels of recruitment and loading, and both complemented the mcm3-td mutant at 37°C (Supp. Fig. 8). We engineered the Mcm3-11 mutation into either the MBP fusion (Fig. 3a) or the FLAG fusion (Fig. 3b), generated mutant Cdt1•Mcm2-7 complexes and mixed these complexes in different ratios with Cdt1•Mcm2-7 containing either wild-type FLAG-Mcm3 (Fig. 3a) or wild-type MBP-Mcm3wt (Fig. 3b). These experiments show that the Mcm3-11 mutant was not recruited above background levels in ATP or ATPγS over a wide range of wild type:mutant ratios, showing that the wild-type protein cannot aid recruitment of the Mcm3-11 mutant complex indicating that recruitment of both hexamers requires the Mcm3 C-terminus.
Figure 3. Both Mcm2-7 hexamers must interact with ORC•Cdc6 through Mcm3.
a) Complexes containing wild type 3x FLAG Mcm3 and MBP Mcm3-11 were assembled (IN) and mixed in the indicated ratios, with a fixed amount of the wild type complex (4 pmol) and tested for loading and recruitment as above. b) Loading and recruitment of mixed complexes containing 3x FLAG Mcm3-11 and MBP Mcm3 wt in the indicated ratios.
Mcm3 binding activates the ORC•Cdc6 ATPase
ATP hydrolysis is required for assembly of the Mcm2-7 double hexamer, but what stage of assembly requires ATP hydrolysis is not known. We therefore next tested whether Mcm3 binding affected the ATPase activity of DNA-bound ORC•Cdc6. Neither Mcm3 nor the Mcm3-13 mutant exhibited appreciable ATPase activity by themselves (Fig. 4a), consistent with the fact that, as a AAA+ ATPase, Mcm3 requires an arginine finger from a binding partner (in this case, Mcm5) to promote ATP hydrolysis. ORC, together with Cdc6 (ORC•Cdc6, Fig. 4a), exhibited some ATPase activity in the presence of DNA, which might reflect a basal level of ATPase from DNA-bound ORC•Cdc6 or might be caused by ORC and/or Cdc6 that is not bound to DNA. Addition of Mcm3, but not Mcm3-13, to ORC•Cdc6 resulted in a 3- to 4-fold increase in ATPase activity indicating that the interaction of the C-terminus of Mcm3 stimulates the ATPase of ORC•Cdc6. To rule out any contribution from the Mcm3 ATPase, we examined the effect of the Mcm3 C-terminal fragment lacking the AAA+ domain. Fig. 4b shows that this domain was as effective as full-length Mcm3 in stimulating the ATPase activity of ORC•Cdc6. Reactions in Fig. 4b contained 2.5 pmol each of ORC and Cdc6, yet addition of Mcm3 resulted in approximately 140 pmol of additional ATP hydrolysis (above the level produced by ORC and Cdc6 alone) over 20 min, indicating that Mcm3 binding can induce multiple rounds of ATP binding and hydrolysis by ORC•Cdc6.
Figure 4. Mcm3 binding activates the ORC•Cdc6 ATPase.
The conversion of α32P-ATP to α32P-ADP was monitored as described in Supplementary Methods in reactions containing the indicated proteins (Mcm3 wild type; Mcm3-13 mutant; Mcm3-CT, a polypeptide containing the C-terminal 194 amino acid residues of Mcm3; ORC and Cdc6). Error bars depict standard error of the mean from five reactions.
ATP hydrolysis by ORC•Cdc6 can promote Mcm2-7 release
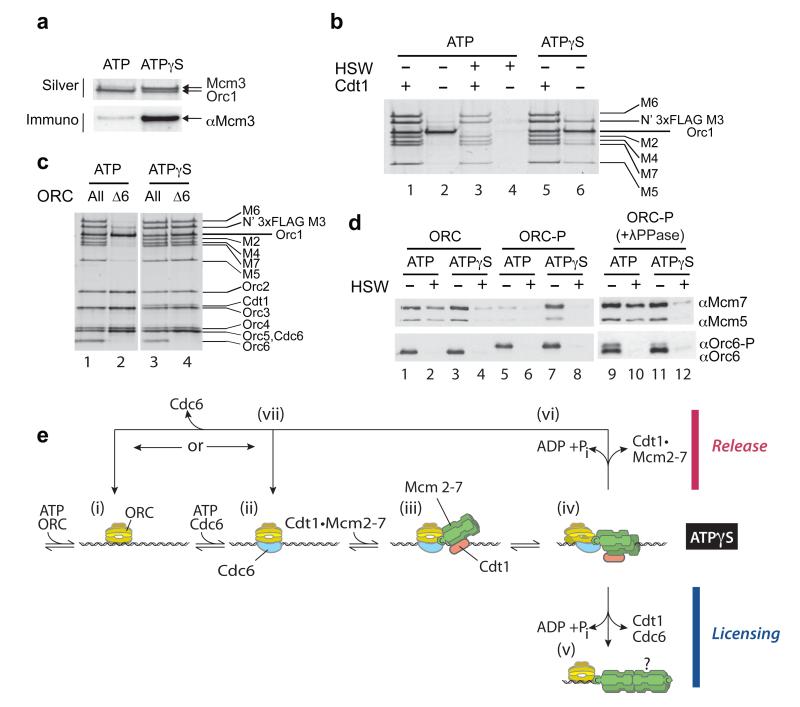
Activation of the ORC•Cdc6 ATPase by the initial recruitment of Mcm3 led us to consider that ATP hydrolysis might play some role early in the Mcm2-7 loading reaction. Supp. Fig. 9a shows that both ORC and Cdc6 are bound to DNA equally well in ATP and ATPγS; however, we found that the stable recruitment of Mcm3 we saw with non-hydrolysable ATPγS (Supp. Fig. 3a,f and Fig. 5a, ATPγS) was lost when hydrolysis was allowed (Fig. 5a, ATP). Similarly, the retention of Mcm3, 5 and 7 seen in ATPγS in the absence of Cdt1 (e.g. Fig. 1) was also lost in the presence of ATP (Fig. 5b, compare lanes 2 and 6). These results indicate that ATP hydrolysis can lead to release of Mcm3 (and associated subunits) when the loading of incomplete Cdt1•Mcm2-7 complexes is attempted.
Figure 5. ATP hydrolysis by ORC•Cdc6 can promote Mcm2-7 release.
a) Mcm3 was tested for recruitment (low salt wash) in reactions containing ORC, Cdc6 and either ATP or ATPγS. Bound proteins were analysed by silver staining (silver) or immunoblotting with anti-Mcm3 antibody (immuno). b) Recruitment and loading assays were performed as above with ORC, Cdc6 and Mcm2-7 either with (+) or without (-) Cdt1 in ATP or ATPγS. Proteins were visualized by silver staining. c) Complexes containing either the complete six subunit ORC complex (All) or a complex lacking Orc6 (Δ6), purified as described in Supplementary Methods, were tested for recruitment (low salt wash) in either ATP or ATPγS. DNA bound proteins were analysed by silver staining. d) Recruitment and loading assays were performed with unphosphorylated ORC (ORC) or CDK-phosphorylated ORC (ORC-P). In lanes 9-12 ORC-P was previously dephosphorylated with lambda phosphatase. e) A revised model for the mechanism of origin licensing which includes an ATPase dependent release step based on experiments in this manuscript. Details of the model are discussed in the text. The “?” in step (v) refers to the assembly of the second hexamer, which is discussed in the text.
This could be because the remaining Mcm2-7 subunits cannot form stable, topologically closed double hexamers around DNA and are, therefore, released during a futile loading reaction with ATP hydrolysis. ORC is not required for Mcm2-7 to remain bound topologically to the DNA after loading (e.g. after high salt wash). So, we next looked at the ability of the full, wild-type Cdt1•Mcm2-7 complex to be recruited and loaded by ORC lacking the Orc6 subunit (ORCΔ6), which can still bind DNA14. ORCΔ6 was able to recruit all six Mcm2-7 subunits in the presence of ATPγS as efficiently as the complete ORC complex (Fig. 5c, lanes 3 and 4). Although the recruited complex was slightly more salt-sensitive than the complex recruited with the complete ORC complex (Supp. Fig. 9b), the only significant difference between Fig. 5c lanes 3 and 4 was the absence of Orc6 in lane 4. Recruitment of Mcm2-7 subunits in the absence of Orc6 still required the C-terminus of Mcm3, because it was completely defective in the Mcm3-11 mutant (Supp. Fig. 9d, lane 4). Strikingly, although ORCΔ6 recruited all six Mcm2-7 subunits in ATPγS, it was unable to stably recruit any of these subunits in the presence of ATP (compare Fig. 5c, lanes 2 and 4). ORCΔ6 and Cdc6 are bound equally well in ATP and ATPγS (Supp. Fig. 9e). Thus, although the entire Mcm2-7 ring is efficiently recruited, ATP hydrolysis promotes release of Cdt1•Mcm2-7 in the absence of Orc6.
To rule out the possibility that the non-physiological absence of Orc6 somehow alters the function of ORC•Cdc6, and to examine the role of ATP-dependent Mcm2-7 release in vivo, we examined a physiologically relevant situation where ORC has been functionally inactivated. Phosphorylation of Orc6, along with Orc2, by cyclin-dependent kinase (CDK) plays a role in preventing re-replication15,16 by blocking the ability of ORC to load Mcm2-717. We phosphorylated purified ORC (ORC-P) with the yeast mitotic CDK, Clb2-Cdc28, and examined its ability to recruit and load Mcm2-7. ORC-P cannot load Mcm2-7 (Fig. 5d, compare lanes 2 and 6), but it can recruit Mcm2-7 subunits as well as the unphosphorylated ORC in the presence of ATPγS (compare lanes 3 and 7). However, ORC-P was unable to accumulate Mcm2-7 subunits in the presence of ATP (compare lanes 1 and 5). Lambda phosphatase treatment of ORC-P restored its ability to recruit and load Mcm2-7 in ATP (Fig. 5d, lanes 9-12). These results show that, as in the cases of Mcm3 alone, Mcm2-7 without Cdt1, and ORCΔ6, the CDK-phosphorylated ORC is competent to recruit Mcm2-7 to ORC•Cdc6, but ATP hydrolysis then promotes Mcm2-7 release.
To provide additional evidence that ATP hydrolysis can promote Mcm2-7 release from ORCΔ6, we have looked at Mcm2-7 recruitment in ATP at reduced temperatures, to slow ATP hydrolysis. In contrast to 30°C (Fig. 5c), the recruitment of Mcm2-7 by ORCΔ6 occurred equally well in ATP and ATPγS at 8°C (Supp. Fig. 10). However, when these reactions were shifted to 30°C after 9 min, recruitment of Mcm2-7 continued in ATPγS but Mcm2-7 was significantly reduced in the presence of ATP, consistent with the idea that elevated temperature promotes ATP hydrolysis and subsequent Mcm2-7 dissociation.
Discussion
Our results support a model for origin licensing wherein Cdt1•Mcm2-7 is first recruited to origins by interaction between the C-terminus of Mcm3 and ORC•Cdc6 (Fig. 5e iii). Cdt1 is not required for this initial recruitment, in contrast to previous experiments in crude extracts17-19. We suggest, by analogy to other AAA+ proteins20, that domain movements in ORC•Cdc6 triggered by Mcm3 binding activate the ATPase of ORC and/or Cdc6 by bringing arginine fingers in contact with ATP binding sites in adjacent subunits (Fig. 5e iv). We further suggest that these domain movements are transmitted to Mcm2-7, leading to destabilisation of Mcm2, 4 and 6 when Cdt1 is missing (Fig. 1a,b). Cdt1 stabilises Mcm2-7 in this new conformation by a mechanism that does not require its interaction with Orc6 (Fig. 5c). Since both hexamers must interact with ORC•Cdc6 (Fig. 3), our results indicate they are either loaded sequentially or there is more than one Mcm3 binding site in ORC•Cdc6 allowing concerted loading.
When all pre-RC components are present and in the correct post-translational modification state, ATP hydrolysis promotes loading of the ring around DNA (Fig. 5e v). However, when criteria for correct licensing are not met, ATP hydrolysis instead is coupled to irreversible dissociation of inappropriate protein assemblies, preventing the accumulation of non-productive complexes (Fig. 5e vi). Cdc6 is bound to ORCΔ6 in both ATP and ATPγS (Supp. Fig. 9e) suggesting either that Cdc6 remains bound to ORC after Cdt1•Mcm2-7 release or that Cdc6 is released, but rebinds quickly (Fig. 5e vii). This ATPase-dependent quality control mechanism contributes to preventing licensing by CDK-phosphorylated ORC outside of G1 phase, and thus, has a role in ensuring once per cell cycle replication. We note that the block to re-replication in metazoans works largely through inhibition and degradation of Cdt121,22, and our results (Fig. 5b) suggest that this will also prevent licensing via this ATPase-dependent mechanism. Although a great deal is known about the multiple pathways contributing to a robust block to re-assembly of pre-RCs after replication in eukaryotes23,24, few quality control mechanisms contributing to efficient pre-RC assembly during G1 phase have been described (see ref 25). The mechanism we have described here may also play a role during G1 phase to ensure assembly of complete, functional pre-RCs.
Full Methods
Generation of Vectors and Yeast Strains
Cdt1•Mcm2-7 overexpressed in yeast (yJF38)
To obtain a yeast strain overexpressing the Cdt1•Mcm2-7 complex, four new vectors were generated using the pRS30_ series27. Making use of the bidirectional inducible GAL1-10 promoter, the six MCM subunits, CDT1 and GAL4 genes were cloned and overexpressed in the strain yJF38. GAL4 is a positive regulator of GAL genes in response to galactose, it was therefore overexpressed together with the MCM2-7 and CDT1 genes to increase protein yield.
GAL4 and MCM2 were amplified using the primer pairs JF1-2 and JF3-4, respectively. GAL4 was cloned into pRS303 (pJF2.1) and MCM2 into pRS306 (pJF5.1) between SmaI and BamHI sites. MCM7 and MCM5 were amplified with JF5-6 and JF7-8 primer pairs and cloned into a SmaI site in pRS304 (pJF4.1) and pRS306 (pJF3.1), respectively. The forward primers inserted a unique AscI site at the 5′ end of the start codon of these genes. MCM3, MCM6, MCM4 and CDT1 were amplified using the following primer pairs: JF9-10, JF11-12, JF13-14 and JF15-16. The PCR products were cloned between SpeI and NotI sites into pJF5.1, 4.1, 3.1 and 2.1, respectively, to yield pJF5.2, 4.2, 3.2 and 2.2. In this case, a unique SgrAI site was inserted at the 5′ end of their start sites. In addition a 3xFLAG tag28 was inserted using a SgrAI site present upstream of MCM3 in pJF5.2 (pJF6.2). Finally, the GAL1-10 promoter was cloned between the AscI and SgrAI sites previously inserted, yielding pJF2, 3, 4 and 6. These vectors were integrated into the strain yJF1 (yJF38).
Cdt1•Mcm2-7 overexpressed in yeast (yJF35, yJF59 and yJF73)
yJF35 and yJF59 are analogous to yJF38 except that the 3xFLAG tag on MCM3 was inserted at its C-terminus in yJF35 and on the C-terminus of CDT1 in yJF59. The vectors integrated into yJF1 were pJF2, 3, 4 and 5. The C-terminal 3xFLAG on MCM3 and CDT1 was inserted afterwards by PCR based tagging using pBP83. pJF2,3,4 and 39 vectors were integrated into the strain yJF1 yielding yJF73.
Mcm2-7 overexpressed in yeast (yJF39)
To obtain a Mcm2-7 complex without Cdt1, a strategy similar to the design of yJF38 was followed, except that pJF2 was not integrated into the final strain and the endogenous CDT1 locus was TAP-tagged29 using pJF21, thus producing yJF39.
ORC overexpressed in yeast (ySD-ORC)
To overexpress ORC in yeast, we modified pJF2, 3 and 5 as follows: (a) In pJF5, MCM2 and MCM3 were replaced with CBP-TEV-ORC1 and ORC2, respectively (pJF19). (b) In pJF2 GAL4 and CDT1 were substituted with ORC3 and ORC4, respectively (pJF17). (c) In pJF3, MCM5 and MCM4 were replaced with ORC5 and ORC6, respectively (pJF18). All ORC genes were codon optimized. Finally, pJF19, 17 and 18 were integrated into yJF1, producing the strain ySD-ORC.
ORC(Δ6) overexpressed in yeast (ySD-ORC(Δ6))
In pJF3, MCM5 was replaced with ORC5 and MCM4 was deleted, producing the vector pJF20. To obtain ORC without Orc6, yJF1 was transformed with pJF17, 19 and pJF20, instead of pJF18. Furthermore, a C- terminal 3xFLAG tag was added to the endogenous copy of ORC6, using pBP83.
MCM3 mutants
MCM3 mutants were cloned into two different vectors: (a) pMAL-C2P to purify recombinant proteins and (b) pRS306 for complementation studies in yeast.
(a) pMAL-C2P was a gift from Satoru Mochida and was derived from pMAL-C2 (NEB) by introducing a PreScission protease site before the EcoRI site in the polylinker region30. MCM3 was amplified from S.cerevisiae genomic DNA using AM51 and AM52 and cloned into pMAL-C2P using XbaI and SalI sites (pAM5). MCM3 contains a KpnI site at position 2690 bp. Mutant fragments of 231 bp in length, between this site and the stop codon were synthesised by GeneArt. The mutated sequences incorporated a NotI, followed by a SalI restriction site at the 3′ end of the stop codon. Using KpnI and SalI sites, the mutant sequences were cloned into pAM5, to yield pJF27 (Mcm3-11), pJF28 (Mcm3-12), pJF29 (Mcm3-13), pJF32 (Mcm3-16), pJF35 (Mcm3-22) and pJF36 (Mcm3-23).
(b) MCM2 in pJF5 was removed, to yield pGC003. MCM3 mutant sequences were excised from pJF27, 28, 29, 32, 35 and 36 vectors using AvrII and NotI sites, and cloned into pGC003 (pJF8, 9, 10, 13, 15 and 16). Mcm2 in pJF6 was removed, to yield pJF37. MBP tag was amplified from pMAL-C2P using the primers JF19 and 20 and cloned into pGC003 using SgrAI unique site, originating pJF38. Finally, these vectors were integrated into a MCM3 degron strain (yKL4313), producing the strains yJF63, 64, 65, 66, 69, 71, 72, 78 and 79.
MCM3 mutants in a Diploid Background
WT C-terminal Mcm3 was amplified from pGC003, C-terminal Mcm3 mutants were amplified from pJF27 (Mcm3-11), pJF28 (Mcm3-12) and pJF29 (Mcm3-13) using primers JF17 and JF18. The PCR products contained the URA3 gene and were used to transform the diploid strain W303 (all amplified products were sequenced prior to transformation). Transformants were selected for on media lacking uracil. These were shown by PCR to be heterozygous for replacement of the C-terminus of Mcm3 with either WT or mutant Mcm3 cassettes as above. Sporulation and tetrad dissection of these transformants was performed to examine their viability.
Fragments of MCM3
A C-terminal fragment of MCM3 (588 bp) was amplified from S.cerevisiae genomic DNA using AM54 and AM52. This PCR product was cloned in pMAL-C2P using XbaI and SalI sites (pAM6).
An N-terminal fragment of MCM3 (2331 bp) was amplified from S.cerevisiae genomic DNA using AM51 and AM53. This PCR product was cloned in pMAL-C2P using XbaI and SalI sites (pAM7).
Individual MCM subunits expressed in bacteria
MCM4 and 5 were amplified by PCR using NC1-2 and NC3-4 primer pairs. These PCR products were cloned between NdeI and XhoI restriction sites into pET22b, to yield pET22b-MCM4 and pET22b-MCM5. MCM6 was excised from pET16b-MCM6 using NdeI and BamHI31 and cloned into pET11a (pET11a-MCM6).
Cdt1 expression in bacteria
CDT1 was amplified from S.cerevisiae genomic DNA using NC5 and NC6 and cloned into pGEX-6p-1 (GE Healthcare), using BamHI and NotI sites (pGEX-6p-1-CDT1).
Cdc6 expression in bacteria
S.cerevisaie CDC6 was amplified from pET15b-CDC6 using the primers AM1 and AM2. The PCR product was cloned between BamHI and XhoI restriction sites in pGEX-6p-1 (GE Healthcare), generating pAM3.
Protein Purifications
Purification of Cdt1•Mcm2-7 from yJF38, yJF35, yJF59 and yJF73
2 L of cells were grown in YP-raffinose at 30°C to a cell density of 4×107 cells/ml and arrested for 3 hours with 100 ng/ml of alpha-factor. Protein expression was induced by adding galactose (2%) and incubating at 30°C for 3-4 hours. Cells were harvested, washed with ice-cold 25 mM Hepes-KOH pH 7.6, 1 M sorbitol, then washed with buffer A (45 mM Hepes-KOH pH 7.6, 0.02% NP-40, 10% glycerol, 5 mM Mg(OAc)2 and 0.1 M K-glutamate). The pellet was resuspended in 0.5 volumes of buffer A/1 mM DTT/protease inhibitors (Roche) and frozen drop-wise in liquid nitrogen. Frozen drops of cells were crushed using a freezer mill (SPEX CertiPrep 6850 Freezer/Mill) with 6 cycles of 2 minutes crushing at rate 15. Frozen cell powder was thawed at room temperature, resuspended in 1 volume of buffer A/1 mM DTT/protease inhibitors, and the concentration of K- glutamate was adjusted to 0.5 M. The suspension was centrifuged for 1 hour at 50000 rpm using a Ti70.1 rotor. The clear phase was recovered, dialyzed twice for one hour against buffer B (25 mM Hepes-KOH pH 7.6, 0.02% NP-40 and 10% glycerol)/0.1 M K glutamate/5 mM Mg(OAc)2, and then centrifuged for 30 minutes at 50000 rpm. Anti-FLAG immunoprecipitation was performed by adding 1 ml packed bead volume of washed anti-FLAG M2 agarose (Sigma) to the supernatant and incubating for 1 hour at 4°C. Beads were recovered, and washed with ten bed resin volumes (BVs) of buffer B/0.1 M K glutamate/1 mM ATP/5 mM Mg(OAc)2. Elution was performed by adding 0.5 mg/ml 3xFLAG peptide. After 20 minutes at 4°C, the flow-through was collected, concentrated using Microcon YM-10, 10000 MWCO (Millipore) and fractionated on a 24 ml Superdex 200 10/300 GL column (GE Healthcare) pre-equilibrated in buffer B/0.1 M K acetate. Peak fractions were pooled and aliquoted.
Purification of Mcm2-7 from yJF39
yJF39 was purified as described for yJF38. The only difference was a pre-incubation step with IgG Sepharose™ 6 Fast Flow beads (GE Healthcare), to remove endogenous Cdt1, before the sample was loaded onto the gel filtration column. 100 μl of IgG packed resin was added to the 3xFLAG peptide-eluted sample and incubated for 30 min at 4°C. The unbound fraction was recovered, concentrated and loaded onto a Superdex 200 10/300 GL column (GE Healthcare), as described for yJF39.
Purification of ORC from ySD-ORC
2 L of cells were grown in YP-raffinose at 30°C to a cell density of 4×107 cells/ml and arrested for 3 hours with 100 ng/ml of alpha-factor. After arrest, protein expression was induced by adding galactose (2%) for 3-4 hours at 30°C. Cells were harvested, washed with ice-cold 25 mM Hepes-KOH pH7.6/1 M sorbitol, then washed with buffer C (25 mM Hepes-KOH pH 7.6, 0.05% NP-40 and 10% glycerol)/0.1 M KCl. The pellet was resuspended in 0.5 volumes of buffer C/0.1 M KCl/1 mM DTT/Protease inhibitors (Roche) and frozen dropwise in liquid nitrogen. Frozen drops of cells were crushed using a freezer mill (SPEX CertiPrep 6850 Freezer/Mill) with 6 cycles of 2 minutes crushing at rate 15. Frozen cell powder was thawed at room temperature, resuspended in 1 volume of buffer C/0.1 M KCl/1 mM DTT/protease inhibitors, and the concentration of KCl was adjusted to 0.5 M. The suspension was centrifuged for one hour at 50000 rpm using a Ti70.1 rotor. The clear phase was recovered, and subjected to calmodulin affinity purification by adding 2 mM CaCl2 and 1.5 ml of packed beads of Calmodulin affinity resin (Stratagene). After 3 hours at 4°C, beads were collected, washed with 10 BVs of buffer C/0.3 M KCl/2 mM CaCl2/2 mM ß-mercaptoethanol. Elution was performed with buffer C/0.3 M KCl/2 mM EGTA/1 mM EDTA/2 mM ß-mercaptoethanol, peak fractions were pooled and subjected to fractionation over a Superdex 200 10/300 GL column (GE Healthcare), pre-equilibrated in buffer C/0.15 mM KCl/2 mM ßmercaptoethanol. Fractions containing ORC were pooled and concentrated over a 1 ml MonoQ™ 5/50 GL column (GE Healthcare) using an elution gradient of 0.15-0.5 M KCl over 10 column volumes (CVs). Peak fractions containing ORC were pooled, dialyzed against buffer C/0.3 M K acetate and stored in aliquots.
Purification of ORC(Δ6) from ySD-ORC(Δ6)
To purify ORC without Orc6, a similar protocol to the purification of ORC was followed, except that FLAG immunoprecipitation of endogenous Orc6 was included, to remove complexes containing Orc6 (Supp. Fig. 9c). This was performed as follows: pooled fractions from the gel filtration were incubated for 30 minutes at 4°C with 1 ml packed bead volume of anti-FLAG M2 agarose (Sigma). Flow through was collected and concentrated over a MonoQ™ 5/50 GL column (GE Healthcare), as described for the purification of ORC.
Proteins expressed in bacteria
All expression plasmids were transformed into BL21 DE3 Codon+ RIL cells (Stratagene) (unless indicated). 3 L of cells (unless indicated) were grown at 37°C to a density of OD600=0.5-0.8. Cells were chilled on ice, and then IPTG was added to 1 mM. Induction was carried out O/N at 18°C.
Purification of MBP-Mcm3
0.5 L of cells expressing MBP-Mcm3 were grown as described above. Cells were harvested, washed once with ice-cold 25 mM Hepes-KOH pH7.6/1M sorbitol, once with buffer D (50 mM Tris-HCl pH 7.5, 0.05% NP-40, 10% glycerol)/1 M NaCl and then the pellet was resuspended in 20 ml of buffer D/1 M NaCl/2 mM ß-mercapethanol/protease inhibitors (Roche). 50 μl of lysozyme (50 mg/ml) was added and the suspension incubated for 20 minutes at 4°C. Cells were kept on ice and sonicated 3 × 30 sec at 15 microns using a sonicator Soniprep 150 (Sanyo). Lysate was centrifuged for 1 hour at 45000 rpm using a Ti45 rotor. The soluble phase was collected and incubated with 2 ml packed amylose bead volume (NEB) at 4°C for 1 hour. Beads were washed with ten BVs of buffer D/0.3 M NaCl/2 mM ß-mercaptoethanol. Elution was performed with buffer D/0.3 M NaCl/2 mM ß-mercaptoethanol/10 mM Maltose. Peak fractions were pooled and 0.5 ml were subjected to fractionation over a Superdex 200 10/300 GL column (GE Healthcare) pre-equilibrated in buffer D/0.3 M NaCl/2 mM ß-mercaptoethanol. The peak fraction, of correct molecular weight, was collected and subjected to PreScission protease cleavage with 100 units of PreScission™ protease (GE healthcare). After 30 minutes at room temperature, 1 volume of buffer D/2 mM ß-mercaptoethanol was added, and the sample was fractionated over a 0.15 ml MonoQ™ PC1.6/5 column (GE Healthcare), using an elution gradient of 0.15-0.7 M NaCl over 50 CVs. Peak fractions containing untagged Mcm3 proteins were kept. The same protocol was used to purify Mcm3 fragments and mutants.
Purification of Mcm2, Mcm7 and Mcm3
To purify these proteins, we followed the protocol described by Davey et al., 2003 (ref 31). The starting cultures were 3 L in volume.
Purification of Mcm5
Cells were grown and expressed as described above. Cells were harvested, washed once with ice-cold 25 mM Hepes-KOH pH 7.6/1 M sorbitol, once with buffer E (50 mM Tris-HCl pH 7.5, 0.05% NP-40, 10% glycerol,1mM EDTA and 1mM DTT)/ 1 M NaCl, and the pellet was resuspended in 40 ml of buffer E/1M NaCl/protease inhibitors (Roche). Cells were sonicated and the lysate centrifuged as described for the purification of MBP-Mcm3. The soluble fraction from centrifugation was treated with 0.3 g/ml Ammonium Sulphate, and stirred for 20 minutes at 4°C. The mixture was centrifuged at 17000 rpm for 20 minutes. The pellet was resuspended in 40 ml of buffer E containing 0.25 g/ml of ammonium sulphate and centrifuged as before. This was repeated with 0.20 mg/ml Ammonium Sulphate. The pellet was resuspended in 30 ml of buffer E and dialyzed against buffer E for one hour at 4°C. After checking that the conductivity of the sample was below that of buffer E/0.1 M NaCl, the sample was applied to a 5 ml FF Q column (GE Healthcare), pre-equilibrated in buffer E/0.1 M NaCl. Elution was done using a gradient of 0.1 M to 0.5 M NaCl over 10 CVs. Peak fractions were pooled and diluted down to a conductivity below that of buffer E/0.1 M NaCl. The sample was then applied to a 5 ml Heparin column (GE healthcare) pre-equilibrated in buffer E/0.1 M NaCl. Flow through was collected and subjected to fractionation over an 8 ml MonoQ™ column (GE Healthcare). Elution was performed using a gradient of 0.1 M to 0.5 M NaCl over 20 CVs. Peak fractions were collected and subjected to fractionation over a Superdex 200 10/300 GL column (GE Healthcare) pre-equilibrated with buffer E/0.3 M NaCl. Peak fractions were pooled, concentrated using a Microcon YM-10, 10000 MWCO (Millipore) and aliquoted.
Purification of Mcm4
3 L of expressing cells were grown and we followed the purification process described by Davey et al., 2003 (ref 31). At the end of the described protocol, we introduced an extra gel filtration step using a Superdex 200 10/300 GL column (GE Healthcare), pre-equilibrated in buffer E/0.3 M NaCl.
Purification of Mcm6
To purify Mcm6, the same protocol as that described for Mcm5 was followed, the difference being that Mcm6 was bound to a 5 ml Heparin column (GE Healthcare). Bound protein was eluted using a gradient of 0.1 to 0.5 M NaCl over 20 CVs. Peak fractions were pooled and the conductivity was reduced to below that of buffer E/0.1 M NaCl. The sample was then applied to an 8 ml MonoQ™ column (GE Healthcare), as described in the purification of Mcm5.
Purification of Cdt1
Expressing cells were grown and induced as described before. Cells were harvested, washed with PBS pH 7.5, and then washed with buffer F (PBS pH 7.5, 10% Glycerol, 1 mM DTT and 0.3 M NaCl). The pellet was resuspended in 40 ml of buffer F/protease inhibitors (Roche) and sonicated 3 × 30 sec using a sonicator Soniprep 150 (Sanyo) at 15 microns. Lysate was centrifuged at 45000 rpm for 60 minutes using a Ti45 rotor. 2 ml of packed bead volume Glutathione Sepharose™ 4Fast Flow (GE Healthcare) were added to the soluble phase and incubated for 2.5 hours at 4°C. The solution was centrifuged at 3000 rpm for 3 minutes, beads were collected, 10 CVs of buffer F were added, and the mixture was incubated for 10 minutes at 4°C. This wash step was repeated with 20 CVs of buffer F, and 10 CVs of buffer E /0.3M NaCl). The pellet was then resuspended in 2 ml of buffer E/0.3M NaCl. 125 units of PreScission Protease (GE Healthcare) were added and the mixture was incubated overnight at 4°C. The flow through was collected and concentrated using a Microcon YM-10, 10000 MWCO (Millipore), then the sample was loaded onto a gel filtration column (Superdex 200 10/300 GL column (GE Healthcare)) pre-equilibrated in buffer B/1 mM EDTA/1 mM DTT/0.3 M NaCl. Peak fractions were pooled, concentrated and aliquoted.
Formation of MCM complexes from individually purified subunits
MCM complexes were formed by combining 10 μg of individually purified subunits and fractionated over a Superdex 200 PC 3.2/30 column (GE Healthcare), pre-equilibrated in buffer B/0.1 M K acetate. High molecular weight fractions containing Cdt1•Mcm2-7 complex were pooled, concentrated and aliquoted. Supp. Fig. 12 shows that this complex can be loaded as efficiently as the complex purified from yeast.
Purification of GST-Cdc6
This protocol is modified from that described by Speck et al., 2005 (ref 10). 1 L of expressing cells were grown at 37°C to O.D600 = 0.6, then induced with 0.5 mM IPTG for 5 hours at 18°C. Cells were harvested at 6000 rpm in an SLA-3000 rotor (Sorvall) for 10 mins. Pellets were resuspended in 50 ml buffer G (50 mM K2HPO4/ KH2PO4 pH7.5, 5 mM MgCl2, 1% Triton X-100 and 1 mM DTT)/ 2 mM ATP/0.15 M KOAc/protease inhibitors (Roche) and 100 μg/ml lysozyme added. The mixture was incubated at 4°C for 30 minutes and sonicated for 2 mins (5 sec off, 5 sec on) at 15 microns. The suspension was centrifuged at 15000 rpm for 15 mins in a SS34 rotor (Sorvall) and the supernatant transferred to 2 ml bed resin glutathione sepharose (GE Healthcare). This was rotated at 4°C for 3 hours. Glutathione beads and bound proteins were recovered and washed with 20 CVs of buffer G/0.15 M KOAc/2 mM ATP. A 50% slurry with buffer G/0.15 M KOAc/2 mM ATP was made and 100 units preScission protease (GE Healthcare) added. The mixture was incubated for 2 hours at 4°C. The flow-through was recovered and the concentration of KOAc diluted to 75 mM with buffer G/2 mM ATP. This was incubated with 2 ml bed resin Hydroxyapatite prewashed in buffer G/0.075 M KOAc/2 mM ATP. The protein-Hydroxyapatite was washed with 5 BVs of buffer G/2 mM ATP and then washed with 5 BVs of buffer G/0.15 M KOAc/15% Glycerol. Cdc6 was eluted with buffer G/0.4 M KOAc/15% glycerol. Peak fractions were pooled and concentrated using a Centricon Plus-20 Centrifugal Filter (Millipore), then aliquoted.
Purification of ORC-P
To purify CDK-phosphorylated ORC (“ORC-P”), ORC was isolated from 50 L of alpha-factor arrested G1 yeast (YDR11) cells via calmodulin pull-down as described8, incubated with TEV protease overnight at 4°C to remove the tag, dialyzed against 25 mM Hepes-KOH pH 7.6 / 0.1 M KCl / 5 mM Mg(OAc)2 / 0.02% NP40 / 10% glycerol / 1 mM DTT, and concentrated to a final volume of 2 ml.
In a parallel preparation, 50 L of YDR12 (MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 cdc15-2 bar1::kanMX pep4::HIS3 ura3::Pgal1,10-CDC6-TAPtcp (URA3)) were grown at 25°C in YP-raffinose to 2×107/ml. Cdc6-TAPTCP expression was induced by addition of 2% galactose for 8 hours at 25°C, upon which the cells arrest with a long-budded phenotype and with replicated DNA in G2/M phase. The cells were harvested by centrifugation, washed twice with cold buffer containing 25 mM Hepes-KOH pH 7.6 / 1 M sorbitol, once with cold buffer containing 45 mM Hepes-KOH pH 7.6 / 0.1 M KCl / 0.02% NP-40 / 10% glycerol, resuspended in ½ volume of packed cell volume of buffer containing 45 mM Hepes-KOH pH 7.6 / 0.1 M KCl / 0.02% NP-40 / 10% glycerol / 2 mM DTT / 2x complete protease inhibitor cocktail (Roche), and the resulting cell suspension frozen dropwise directly in liquid nitrogen. The frozen cell suspension was crushed using a SPEX 6870 Freezer/Mill, the resulting frozen powder thawed on ice, diluted with 1 volume of 45 mM Hepes-KOH pH 7.6 / 0.1 M KCl / 0.02% NP-40 / 10% glycerol / 1 mM DTT, the salt concentration adjusted to 0.3 M KCl, and the resulting lysate clarified by centrifugation in a Beckman 45 Ti rotor at 42,000 rpm for 1 hour. The clarified extract was supplemented with 2 mM CaCl2, and a complex containing Cdc6-TAPTCP·Clb2·Cdc28·Cks1 was isolated from this extract using calmodulin affinity purification in buffer containing 45 mM Hepes-KOH pH 7.6 / 0.3 M KCl / 0.02% NP-40 / 10% glycerol / 1 mM DTT. This partially purified complex was incubated with TEV protease overnight at 4°C to remove the tag from Cdc6, dialyzed against 25 mM Hepes-KOH pH 7.6 / 0.1 M KCl / 5 mM Mg(OAc)2 / 0.02% NP40 / 10% glycerol / 1 mM DTT, and concentrated to a final volume of 2 ml.
The concentrated calmodulin-purified and TEV-protease-digested ORC and Cdc6·Clb2·Cdc28·Cks1 fractions were combined, supplemented with 3 mM ATP, and incubated for 45 min at 30°C. The phosphorylated ORC (ORC-P) resulting from this reaction was re-isolated from the reaction by gel-filtration on a 120 ml Superdex 200 column in a buffer containing 25 mM Hepes-KOH pH 7.6 / 0.1 M KCl / 0.02% NP40 / 10% glycerol / 1 mM DTT, followed by fractionation on a 1 ml MonoQ ion-exchange column as described8.
Loading reaction
In this study, four main differences to the conditions described by Remus et al., 20097 were introduced in the loading reactions. K-glutamate was substituted with K-acetate in the binding and washing buffers. For silver staining purposes we increased the amount of protein used in the assays by four fold compared to ref7. In addition, ARS305 was amplified using an oligonucleotide primer with a photocleavable biotin (Integrated DNA technologies) at one end, as described in Tsakraklides and Bell 201032. We have optimised the photocleavage to minimise DNA damage by irradiating for 10min at 330nM (See Supp. Figure 11). And finally, 2.5 pmol of DNA molecules have been used, instead of the 1 pmol used before.
Loading assays with ORC and ORC-P
Mcm2-7 loading onto immobilized linear 1 kb ARS305-containing DNA using 50 nM purified ORC or ORC-P was performed as described8. To test the effect of ORC-P dephosphorylation by lambda-phosphatase, 20 pmol of purified ORC-P at a concentration of 0.67 mM was supplemented with 20 mM MnCl2, and incubated for 20 minutes at 30°C with either 400 units of l-phosphatase (NEB) or a buffer control prior to addition of the treated ORC-P to the loading reaction.
ATPase assays
Reactions were carried out at 30°C for 20 minutes, buffer contained 25 mM Hepes-KOH, pH 7.6/0.1% NP-40/5 mM Mg(OAc)2/1 mM EDTA/1 mM EGTA/100 mM K-acetate/5% Glycerol/1 mM DTT/100 μM ATP (including 2.5 μCi of [α-32P]ATP). Each reaction contained 2.5 pmol of 1 Kb linear ARS305 DNA. Where indicated, 2.5 pmol of each protein were also included in these reactions. Reactions were stopped by spotting 1 μl of each reaction on PEI-cellulose TLC plates (CamLab). The cellulose membrane was developed in 0.6 M Na2HPO4/NaH2PO4 pH 3.5, and quantified on a Phosphorimager (GE Healthcare).
Antibodies for Western blot analysis
αMcm2 (yN-19, sc-6680, Santa Cruz), αMcm7 (yN-19, sc-6688, Santa Cruz), αMcm4 (yC-19, sc-6685, Santa Cruz), αMcm5 (yC-19, sc-6687, Santa Cruz), αOrc6 (SB49), αCdc6 (98H/5), αCdt133, αFLAG-HRP (Sigma), αPAP-HRP (Sigma), αMBP-HRP (NEB) Antibodies against Mcm6 and Mcm3 were kind gifts from the Labib laboratory34.
Supplementary Material
Acknowledgements
We are grateful to Nicola Cook for help with protein purifications, Anne Early and Lucy Drury for help with strain constructions, Gideon Coster for help with Mcm3 complementation, Satoru Mochida and Boris Pfander for vectors and Karim Labib for antibodies. We also thank members of the Diffley lab for critical reading of the manuscript. This work was funded by Cancer Research UK and grants from the Association for International Cancer Research (10-0270) and the European Research Council (249883 – EUKDNAREP).
References
- 1.Boos D, Frigola J, Diffley JFX. Activation of the replicative DNA helicase: breaking up is hard to do. Curr Opin Cell Biol. 2012 doi: 10.1016/j.ceb.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 2.Tanaka S, Araki H. Regulation of the initiation step of DNA replication by cyclin-dependent kinases. Chromosoma. 2010;119:565–574. doi: 10.1007/s00412-010-0291-8. [DOI] [PubMed] [Google Scholar]
- 3.Mechali M. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat Rev Mol Cell Biol. 2010;11:728–738. doi: 10.1038/nrm2976. [DOI] [PubMed] [Google Scholar]
- 4.Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M. Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem. 2010;79:89–130. doi: 10.1146/annurev.biochem.052308.103205. [DOI] [PubMed] [Google Scholar]
- 5.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 6.Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Remus D, et al. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evrin C, et al. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci U S A. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gambus A, Khoudoli GA, Jones RC, Blow JJ. MCM2-7 form double hexamers at licensed origins in Xenopus egg extract. J Biol Chem. 2011;286:11855–11864. doi: 10.1074/jbc.M110.199521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol. 2005;12:965–971. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tye BK, Sawyer S. The hexameric eukaryotic MCM helicase: building symmetry from nonidentical parts. J Biol Chem. 2000;275:34833–34836. doi: 10.1074/jbc.R000018200. [DOI] [PubMed] [Google Scholar]
- 12.Bochman ML, Schwacha A. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol Mol Biol Rev. 2009;73:652–683. doi: 10.1128/MMBR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Labib K, Tercero JA, Diffley JFX. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- 14.Lee DG, Bell SP. Architecture of the yeast origin recognition complex bound to origins of DNA. Molecular & Cellular Biology. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- 16.Wilmes GM, et al. Interaction of the S-phase cyclin Clb5 with an ‘RXL’ docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes Dev. 2004;18:981–991. doi: 10.1101/gad.1202304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen S, Bell SP. CDK prevents Mcm2-7 helicase loading by inhibiting Cdt1 interaction with Orc6. Genes Dev. 2011;25:363–372. doi: 10.1101/gad.2011511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen S, de Vries MA, Bell SP. Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2-7 loading. Genes Dev. 2007;21:2897–2907. doi: 10.1101/gad.1596807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takara TJ, Bell SP. Multiple Cdt1 molecules act at each origin to load replication-competent Mcm2-7 helicases. EMBO J. 2011;30:4885–4896. doi: 10.1038/emboj.2011.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duderstadt KE, Berger JM. AAA+ ATPases in the initiation of DNA replication. Crit Rev Biochem Mol Biol. 2008;43:163–187. doi: 10.1080/10409230802058296. [DOI] [PubMed] [Google Scholar]
- 21.Fujita M. Cdt1 revisited: complex and tight regulation during the cell cycle and consequences of deregulation in mammalian cells. Cell division. 2006;1:22. doi: 10.1186/1747-1028-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Havens CG, Walter JC. Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes Dev. 2011;25:1568–1582. doi: 10.1101/gad.2068611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diffley JFX. Quality control in the initiation of eukaryotic DNA replication. Phil. Trans. R. Soc. B. 2011;366:3545–3553. doi: 10.1098/rstb.2011.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diffley JFX. The many faces of redundancy in DNA replication control. Cold Spring Harb Symp Quant Biol. 2010;75:135–142. doi: 10.1101/sqb.2010.75.062. [DOI] [PubMed] [Google Scholar]
- 25.Pasion SG, Forsburg SL. Nuclear localization of Schizosaccharomyces pombe Mcm2/Cdc19p requires MCM complex assembly. Mol Biol Cell. 1999;10:4043–4057. doi: 10.1091/mbc.10.12.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Remus D, Beall EL, Botchan MR. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. Embo J. 2004;23:897–907. doi: 10.1038/sj.emboj.7600077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional References
- 27.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gelbart ME, Rechsteiner T, Richmond TJ, Tsukiyama T. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol Cell Biol. 2001;21:2098–2106. doi: 10.1128/MCB.21.6.2098-2106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puig O, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 30.Mochida S, Ikeo S, Gannon J, Hunt T. Regulated activity of PP2A-B55 delta is crucial for controlling entry into and exit from mitosis in Xenopus egg extracts. EMBO J. 2009;28:2777–2785. doi: 10.1038/emboj.2009.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davey MJ, Indiani C, O’Donnell M. Reconstitution of the Mcm2-7p heterohexamer, subunit arrangement, and ATP site architecture. J Biol Chem. 2003;278:4491–4499. doi: 10.1074/jbc.M210511200. [DOI] [PubMed] [Google Scholar]
- 32.Tsakraklides V, Bell SP. Dynamics of pre-replicative complex assembly. J Biol Chem. 2010;285:9437–9443. doi: 10.1074/jbc.M109.072504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka S, Diffley JFX. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2-7 during G1 phase. Nat Cell Biol. 2002;4:198–207. doi: 10.1038/ncb757. [DOI] [PubMed] [Google Scholar]
- 34.Gambus A, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.