Abstract
Geraniol is a valuable acyclic monoterpene alcohol and has many applications in the perfume industries, pharmacy and others. It has been hypothesized that phosphatases can convert geranyl diphosphate (GPP) into geraniol. However, whether and which phosphatases can transform GPP to geraniol has remained unanswered up till now. In this paper, the catalysis abilities of 4 different types of phosphatases were studied with GPP as substrate in vitro. They are bifunctional diacylglycerol diphosphate phosphatase (DPP1) and lipid phosphate phosphatase (LPP1) from Saccharomyces cerevisiae, ADP-ribose pyrophosphatase (NudF) and alkaline phosphatase (PhoA) from Escherichia coli. The results show that just PhoA from E. coli can convert GPP into geraniol. Moreover, in order to confirm the ability of PhoA in vivo, the heterologous mevalonate pathway and geranyl diphosphate synthase gene from Abies grandis were co-overexpressed in E. coli with PhoA gene and 5.3 ± 0.2 mg/l geraniol was produced from glucose in flask-culture. Finally, we also evaluated the fed-batch fermentation of this engineered E. coli and a maximum concentration of 99.3 mg/l geraniol was produced while the conversion efficiency of glucose to geranoid (gram to gram) was 0.51%. Our results offer a new option for geraniol biosynthesis and promote the industrial bio-production of geraniol.
Keywords: Biosynthesis, engineered Escherichia coli, geraniol, PhoA, Phosphatase
Introduction
The monoterpene geraniol, which is emitted from flowers, has an important role in flavor and fragrance industries due to its pleasant rose-like odor.1 Geraniol also exhibits huge potential in pharmacy and agrochemistry.2,3 Fractional distillation of plant essential oils is the major method for geraniol manufacture, but high cost and other limitations, such as weather dependence and plant diseases, limited the supplies of geraniol.4 Converting renewable resources into monoterpene products by engineered microorganisms was interesting technology and developed quickly recently year, which have the advantages of fast growth, no need for land during their growth and sustainable development.5,6
Geraniol is likely to be synthesized from geranyl diphosphate (GPP), the universal precursor of all monoterpenes, which can be produced from both the methylerythritol 4-phosphate pathway and the mevalonate (MVA) pathway in plants.7,8 Although many microorganisms carry out the methylerythritol 4-phosphate pathway or MVA pathway to supply the intermediates dimethylallyl pyrophosphate and isopentenyl pyrophosphate, they are unable to produce the monoterpenes because of the absence of monoterpene synthase. In recent year, Iijima et al.9 first purified and characterized geraniol synthase (GES) from peltate glands of Ocimum basilicum as a member of the terpene synthase family and proved that GES can catalyze geraniol formation from GPP by the addition of a hydroxyl group to a carbocation intermediate. Then, the generation of geraniol by GES has been demonstrated in engineered microbial and a recent study proved that geraniol could be generated at a level of 0.185 mg l−1 by simply over-expressing GES in E. coli.10 However, the structure of geraniol, whose carbon skeleton is identical to that of its precursor GPP, hypothetically allows for an alternative mechanism of simply breaking the phosphoester bond by a phosphatase to generate geraniol.9 Moreover, geraniol formation via endogenous dephosphorylation of GPP has been observed in engineered E. coli and yeast.11,12 But, whether and which phosphatases can transform GPP to geraniol has remained unanswered up till now. The catalysis ability of phosphatases toward geraniol formation was investigated in this study, and it will show a new option for geraniol biosynthesis.
Results and Discussion
Phosphatases activity on geranyl diphosphate (GPP)
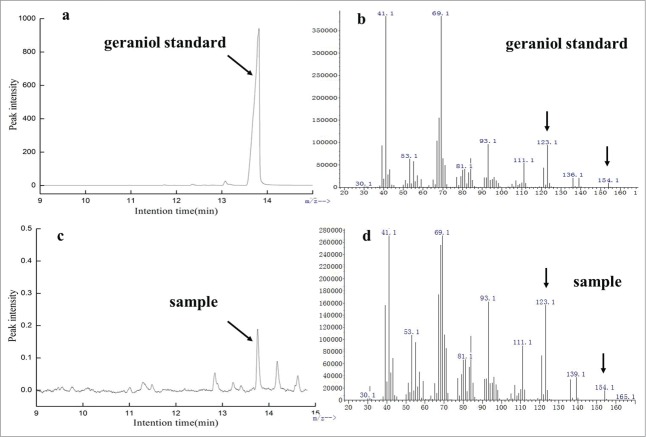
With the aim to study the catalysis ability of phosphatases to form geraniol from GPP, 4 different types of phosphatases genes were studied. These four phosphatases were bifunctional diacylglycerol diphosphate phosphatase (DPP1) from Saccharomyces cerevisiae, lipid phosphate phosphatase (LPP1) from Saccharomyces cerevisiae, ADP-ribose pyrophosphatase (NudF) from E. coli and alkaline phosphatase (PhoA) from E. coli. The crude protein samples taken from cells were analyzed by SDS-PAGE. The theoretical molecular mass of DPP1, LPP1, NudF and PhoA were known as 32 kDa, 30 kDa, 23 kDa and 52 kDa, which were determined from their amino acids.14 The results of SDS-PAGE show that all phosphatases were expressed and matched their theoretical molecular mass respectively (Fig. 1). Then, the crude proteins were used to detect the phosphatase activity toward GPP in vitro. The GC-MS result of reaction shows that only PhoA can act on geranyl diphosphate to form geraniol (Fig. 2), while geraniol was not detected in catalysis reactions by the crude proteins of DPP1, LPP1 and NudF. The control also didn't detect the generation of geraniol. Each reaction does 3 parallels.
Figure 1.
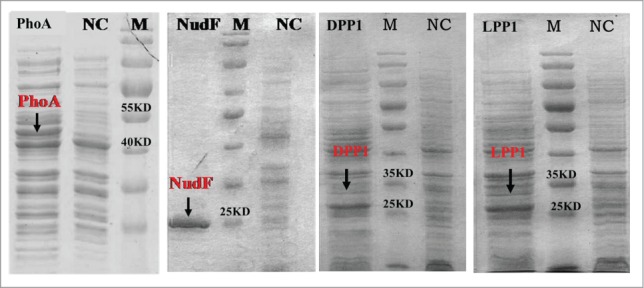
SDS-PAGE analysis of phosphatases protein expressed in E. coli. Lane M, unstained protein molecular weight marker; NC, crude protein extracts from E. coli BL21(DE3); PhoA, crude protein extracts from strain LWG2 4 h after induction; NudF, crude protein extracts from strain LWG5 4 h after induction; DPP1, crude protein extracts from strain LWG3 6 h after induction; LPP1, crude protein extracts from strain LWG2 6 h after induction.
Figure 2.
GC-MS analysis of the product of reaction catalyzed by phosphatases with geranyl diphosphate as substrate. By comparing with the authoritative geraniol (a, b), the capacity of geraniol formation was verified in the reaction catalysis by alkaline phosphatase PhoA from E. coli (c, d). (a, c) total ion chromatogram (TIC); (b, d) mass spectrum. Geraniol was not detected either in control or reactions catalyzed by bifunctional diacylglycerol diphosphate phosphatase (DPP1), lipid phosphate phosphatase (LPP1) and ADP-ribose pyrophosphatase (NudF) respectively. The experiment was performed in triplicate.
This result shows the different catalysis ability of phosphatase. DPP1 and LPP1 were proved to produce geranylgeraniol by removing the phosphatate of geranylgeranyl diphosphate.13 However, they cannot use GPP as substrate to form geraniol in this study, which is consistent with the result no significant increase in geraniol formation by overexpressed DPP1 in vivo, as proved by Oswald.11 NudF can remove the pyrophosphate from IPP and DMAPP to form prenol and isoprenol by introducing NudF into the strain that employed the whole MVA pathway, but not useful in this reaction.14 Alkaline phosphatase (PhoA) is a nonspecific phosphomonoesterase which can remove the inorganic phosphate to form corresponding alcohols.15 The PhoA activity was studied and proved with other phosphorylated substrates, such as 4-nitrophenyl phosphate and p-nitrophenyl phosphate, but this paper first proves the catalysis ability of PhoA with GPP as substrate in vitro, and our finding opens the door to biosynthesis of geraniol with alkaline phosphatase.
Utilization of PhoA in the bioproduction of geraniol from glucose in E. coli
In order to biosynthesize geraniol from glucose by phosphatase, sufficient GPP should be obtained in E. coli. Considering the limited supply of IPP and DMAPP from the native methylerythritol 4-phosphate pathway, the heterologous MVA pathway and gene GPPS2 from Abies grandis were co-overexpressed in E. coli, which have been confirmed can provide sufficient GPP in our pervious study (Fig. 3). To simplify the task of engineering a 7-gene pathway, they were separated into 2 operons and cloned into 2 compatible plasmids. The gene phoA was introduced into plasmid pYJM26, which made up of genes GPPS2 and upper portion of MVA pathway with pACYCDuet-1 vector. The resulting plasmid was named as pLWG9. The other plasmid named pYJM14 contains bottom portion of MVA pathway. The plasmids pLWG9 and pYJM14 were transformed into E. coli BL21 (DE3) strain simultaneously, resulting in strain LWG9. Growing in 100 ml shake-flask, about 5.3 ± 0.2 mg/l geraniol was produced from glucose when LWG9 was grown in 100 ml shake flask for 48h at pH7, 180 rpm and 30°C. As control, strain LWG10 was constructed by transforming plasmids pYJM26 and pYJM14 into E. coli BL21 (DE3). Geraniol production was not detected during 48h of culture by strain LWG10, though geraniol formation via endogenous dephosphorylation of GPP has been observed in E. coli with supplementation of mevalonate, which bearing both GPPS mutated form E. coli and bottom portion of MVA pathway.12 The failure of geraniol production by strain LWG10 might be ascribed to that the supply of GPP by whole MVA pathway from glucose was lower than by the bottom portion of MVA pathway from mevalonate and the low native PhoA catalytic activity limited the conversion of GPP to geraniol in E. coli.
Figure 3.
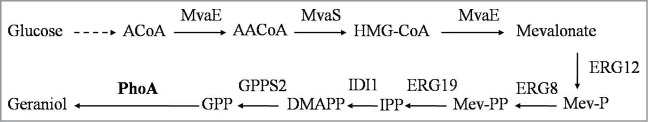
Production of genraniol from glucose with PhoA via the MVA pathways used in this study. The enzymes involved in this pathway: MvaE, Enterococcus faecalis acetyl-CoA acetyltransferase /HMG- CoA reductase; MvaS, Enterococcus faecalis HMG-CoA synthase; ERG12, Saccharomyces cerevisiae mevalonate kinase; ERG8, Saccharomyces cerevisiae phosphomevalonate kinase; ERG19, Saccharomyces cerevisiae mevalonate pyrophosphate decarboxylase; IDI1, Saccharomyces cerevisiae IPP isomerase; GPPS2, Abies grandis geranyl diphosphate synthase; PhoA, Escherichia coli alkaline phosphatase. Pathway intermediates: A-CoA, acetyl-CoA; AA-CoA, acetoacetyl-CoA; HMG-CoA, hydroxymethylglutaryl-CoA; Mev-P, mevalonate 5-phosphate; Mev-PP, mevalonate pyrophosphate. IPP,isopentenyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate; GPP, geranyl diphosphate.
Fed-batch fermentation was carried out using the engineered E. coli strain LWG9. As is shown in Figure 4, geraniol production increased rapidly from 0 h to 10 h after induction and reached 98.7 mg/l at 14 h of culture with an average productivity of 16.7 mg g cdw−1 l−1. Then, geraniol production keeps relatively stable till 32h of culture. Considering that geraniol could be dehydrogenated and transformed into other geranoids in E. coli by YjgB, geranoids were analyzed by GC–MS.12 Geranial, the oxidative product of geraniol, was detected after 6h of culture, while nerol, the cis-isomer of geraniol, was detected after 24h of culture in this study. Geraniol, geranial and nerol formations were sequentially observed during culture, which is consistent with the geranoid formation mechanism that geranial and nerol are formed via geraniol as proposed by Iijima et al.16 and Zhou et al.12,16 Moreover, considering the mechanism of phosphatase, it is hard to form nerol and geranial directly from GPP, which supported above geranoid formation mechanism from another aspect. At 28 h of culture, the total geranoid production was increased to 151.9 mg/l while the geraniol production reached 99.3 mg/l and the geraniol dehydrogenation rate in strain LWG9 was 1.88 mg l−1 h−1. The conversion efficiency of glucose to geranoid (gram to gram) just reached 0.51%.
Figure 4.
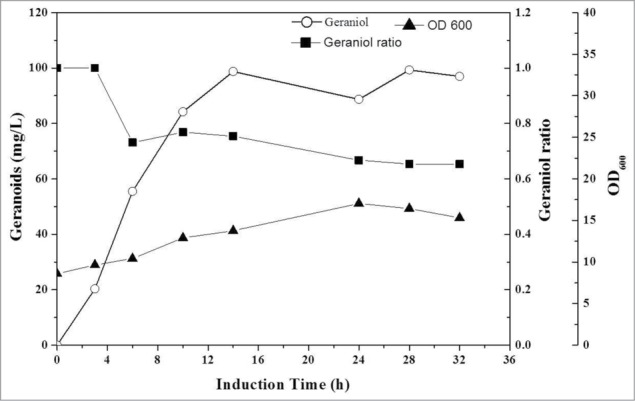
The fermentation course of geraniol (□) production by LWG9. Induction was carried out when OD600 (▴) reached 20 using 1mM IPTG. Nerol and geranial were observed during fermentation with geraniol biosynthesis and (▪) representing the ratio of geraniol to these geranoids.
Although the ability of PhoA to form geraniol was proved in this study, the geraniol production and productivity need to be improved in the future. The main reason for the poor status of strain LWG9 may lie in the low activity of wild type of PhoA. Meanwhile, overexpression of many heterologous genes may be another reason. To resolve the above-mentioned problems, many possible improvements can be used to enhance geraniol production. One approach is to mutate and screen the high activity PhoA for geraniol production. The previous studies about PhoA including the structural and mutant studies will provide a large amount of data for PhoA optimizing. Another approach is engineering of the host including: employing a chromosome integration technique to decrease the cell growth burden on the host that results from overexpression of heterologous genes.
Conclusions
In conclusion, the catalysis ability of phosphatases with GPP as substrate to produce geraniol was first studies. Four different types of phosphatases, DPP1, LPP1, NudF and PhoA were screened and only PhoA from E. coli can form geraniol from GPP in vitro. Moreover, a new pathway with PhoA for geraniol bioproduction from glucose was established in engineered E. coli and 5.3 mg/l geraniol was biosynthesized under shake-flask culture. Finally, we also evaluated the fed-batch fermentation of geraniol and a maximum concentration of 99.3 mg/l was reached. This study provided a new sustainable production strategy for geraniol in E.coli.
Materials and Methods
Medium and culture conditions
The Luria Broth (LB) medium (10 g/L tryptone, 10 g/L NaCl, and 5 g/L yeast extract) was used for gene cloning and shake-flask fermentation. For geraniol production, recombinant strains were cultured in shake-flask or fed-batch fermentation with the medium (pH 7) containing glucose 20 g/l, K2HPO4 9.8 g/l, beef extract 5 g/l, ferric ammonium citrate0.3 g/l, citric acid monohydrate 2.1 g/l, MgSO4 0.06 g/l and 1 ml trace element solution which includes (NH4)6 Mo7O24·4H2O 0.37 g/l, ZnSO4·7 H2O 0.29 g/l, H3BO4 2.47 g/l, CuSO4·5H2O 0.25 g/l, and MnCl2·4H2O 1.58 g/l. Appropriate antibiotics were added to the culture medium according to selectable marker gene of each plasmid listed in Table 1 at the following concentration: ampicillin (Amp, 100 mg/ml), kanamycin (Kan, 50 mg/ml), and chloramphenicol (Cm, 34 mg/ml).
Table 1.
Strains and plasmids used in this stud
| Name | Relevant characteristics | References |
|---|---|---|
| Strains | ||
| BL21(DE3) | F−ompT hsdSB (rB− mB− ) gal dcm rne131 λ(DE3) | Invitrogen |
| LWG2 | E.coli BL21(DE3)/ pYY11 | This study |
| LWG3 | E.coli BL21(DE3)/ pYY12 | This study |
| LWG4 | E.coli BL21(DE3)/ pYY13 | This study |
| LWG5 | E.coli BL21(DE3)/ pYY16 | This study |
| LWG9 | E.coli BL21(DE3)/ pLWG9, pYJM14 | This study |
| LWG10 | E.coli BL21(DE3)/ p YJM26, pYJM14 | This study |
| Plasmids | ||
| pACYCDuet-1 | P15A (pACYC184), Cmr | Novagen |
| pLWG9 | pACYCDuet-1 carrying mvaE and mvaS from Enterococcus faecalis, GPPS2 from Abiesgrandis, ‘phoA from Escherichia coli, Cmr | This study |
| pYY11 | pCOLADuet-1 carrying ‘phoA from Escherichia coli, Kanr | 14 |
| pYY12 | pCOLADuet-1 carrying ‘DPP1 from Saccharomyces cerevisiae, Kanr | 14 |
| pYY13 | pCOLADuet-1 carrying ‘LPP1 from Saccharomyces cerevisiae, Kanr | 14 |
| pYY16 | pCOLADuet-1 carrying nudF from Escherichia coli, Kanr | 14 |
| pYJM26 | pACYCDuet-1 carrying mvaE and mvaS from Enterococcus faecalis, GPPS2 from Abiesgrandis, Cmr | 17 |
| pYJM14 | pTrcHis2B carrying ERG12, ERG8, ERG19 and IDI1 from Saccharomyces cerevisiae,Ampr | 17 |
Strains and plasmids
All strains and plasmids used in this study are listed in Table 1. Four different phosphatase genes, alkaline phosphatase (PhoA) gene from E. coli, ADP-ribose pyrophosphatase (NudF) gene, bifunctional diacylglycerol diphosphate phosphatase (DPP1) gene and lipid phosphate phosphatase (LPP1) gene were cloned into pCOLADuet-1 creating pYY11, pYY12, pYY13 and pYY16 respectively in our previous study,14 and LWG2, LWG3, LWG4, LWG5 were formed by transferred these plasmid into E. coli BL21 (DE3) separately.
For geraniol production from glucose, plasmids of the whole pathway for geraniol synthesis were constructed. Plasmid pYJM26 and pYJM14, which harboring the heterologous mevalonate pathway and geranyl diphosphate synthase gene from Abies grandis, were constructed in our lab's early study.17 The phoA gene was PCR-amplified from plasmid DNA of pYY11 with the primer sets phoA-rbs-F (GGAAGATCTAGGAGGTAAAAAATATGCGGACACCAGAAATGCCTGTTC) and phoA-R (CACCTCGAGTTATTTCAGCCCCAGAGCGGC). The PCR product was digested with BglII and XhoI respectively and then ligated into the corresponding sites of pYJM26 cut with the same restriction enzymes, creating pLWG9. The pLWG9 and pYJM14 were transformed into the BL21 (DE3) competent cell to form LWG9.
Enzyme extraction and assay
The four strains LWG2, LWG3, LWG4 and LWG5 were cultured in LB broth respectively. When the OD600 of the bacterial culture reached 0.6–0.8, the cells were induced by IPTG at a final concentration of 0.1 mM and further incubated at 30°C for 4 h–6 h. Cells were harvested by centrifugation at 6000 g for 5 min, washed with distilled water, and then resuspended in reaction buffer of each enzyme. All the extraction procedures were carried out at 4°C. The cells were sonicated on ice for 10 min(3 s pulse on, 3 s pulse off, 40 W, Sonics VCX130, China). The corresponding supernatants were then analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and prepared for activity assays.
The phosphatase activity toward geranyl diphosphate (GPP) was assayed as the formation of geraniol. The activity of alkaline phosphatase (PhoA) was measured as described by Kojima et al.18 with the following modification: the incubation mixture contained 1 M Tris-Cl, pH 8.0, 10 mM MgSO4, 50 μM ZnSO4, 5 μM GPP and the enzyme extract (about 0.5 mg protein), incubated for 30 min at 37°C. The activity of bifunctional diacylglycerol diphosphate phosphatase (DPP1) and lipid phosphate phosphatase (LPP1) were assayed using a modification of existing methods.13,19 The assay mixture (100 μl) was comprised of citrate buffer (120 mM, pH 4.3), 5 mM MgCl2, the enzyme extract (about 0.5 mg protein), and 5 μM GPP and was incubated for 30 min at 37°C. The activity of ADP-ribose pyrophosphatase (NudF) was measured as described by Dunn et al.20 with the following modification: the incubation mixture contained 50 mM Tris-Cl (pH 8.0), 2 mM MgCl2, 5 μM GPP and the enzyme extract (about 0.5 mg protein) and was incubated for 15 min at 37°C. The products of the reactions were analyzed by GC-MS. Every reaction has 3 parallels and cell extract with empty pCOLADuet-1 was used as control.
Shake-flask cultures
A single colony of LWG9 was grown up in LB broth overnight at 37°C. The culture was used to inoculate the same medium (1:100 dilution) and grown at 37°C until the culture reached an OD600 of 0.6–0.8. IPTG was added to a final concentration of 0.1 mM, and the culture was further incubated at 30°C for 48 h at 180 rpm. The samples were added with the same volume of ethyl acetate, vortexed briefly, and centrifuged to separate the phases and the organic phase was analyzed by GC-MS. The strain LWG10 was used as control. The experiment was performed in triplicate.
Fed-batch fermentation
The strain LWG9 grew up overnight at 37°C in 100 ml of fermentation medium. The overnight culture was used to inoculate a 5L fermentor (BIOSTAT Bplus MO 5 L, Sartorius,Germany) containing 2L fermentation medium. The temperature was maintained at 30°C, the pH was maintained at 7.0 via automated addition of ammonia. The stirring speed was first set at 400 rpm and then linked to the dissolved oxygen (DO) concentration to maintain a 20% saturation of DO. The expression of plasmid-born exogenous genes for geraniol production was initiated at an OD600 about 20 by adding IPTG at a final concentration of 1 mM. During the course of fermentation, the residual glucose was measured using a glucose analyzer (SBA-40D, China) and was maintained below 1 g/l by adding 70% glucose solution during the course of fermentation. To harvest geranoid from the culture broth during fermentation, 2-phase culture was carried out by adding 200 ml isopropyl myristate at 4 h after induction. The samples were collected on time and the organic phase was separated by centrifugation at 13000 rpm for 10 min, then added with 10 volume of ethyl acetate and analyzed by GC-MS.
Geraniol characterization by GC-MS
Putative geraniol products were identified by GC-MS. A HP-INNOWAX capillary column (30 m×0.25 mm; 0.25 μm film thickness; Agilent Technologies) was used. The separation conditions were an initial column temperature of 50°C for 1 min, an increase of 10°C/min to 250°C, where it was held for 5 min. Peak identification was based on a relative retention time and total ion mass spectral comparison with an external standard (Sigma-Aldrich, USA). The peak areas were converted into geraniol or nerol concentrations in comparison with standard curves plotted with set of known concentrations of standards.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was financially supported by Qingdao Institute of Bioenergy and Bioprocess Technology Director Innovation Foundation for Young Scientists, National High Technology Research and Development Program of China (863 Program, No. 2013AA050703–2), National Natural Science Foundation (21202179) and Taishan Scholars Climbing Program of Shandong(No.tspd20150210).
References
- 1.Chen W, Viljoen AM. Geraniol – a review of a commercially important fragrance material. S Afr J Bot 2010; 76:643-51; http://dx.doi.org/ 10.1016/j.sajb.2010.05.008 [DOI] [Google Scholar]
- 2.Carnesecchi S, Bras-Gonçalves R, Bradaia A, Zeisel M, Gossé F, Poupon M-F, Raul F. Geraniol, a component of plant essential oils, modulates DNA synthesis and potentiates 5-fluorouracil efficacy on human colon tumor xenografts. Cancer Lett 2004; 215:53-9; PMID:15374632; http://dx.doi.org/ 10.1016/j.canlet.2004.06.019 [DOI] [PubMed] [Google Scholar]
- 3.Polo MP, Crespo R, de Bravo MG. Geraniol and simvastatin show a synergistic effect on a human hepatocarcinoma cell line. Cell Biochem Funct 2011; 29:452-8; PMID:21735455; http://dx.doi.org/ 10.1002/cbf.1772 [DOI] [PubMed] [Google Scholar]
- 4.Chang MC, Keasling JD. Production of isoprenoid pharmaceuticals by engineered microbes. Nat Chem Biol 2006; 2:674-81; PMID:17108985; http://dx.doi.org/ 10.1038/nchembio836 [DOI] [PubMed] [Google Scholar]
- 5.Clomburg JM, Gonzalez R. Biofuel production in Escherichia coli: the role of metabolic engineering and synthetic biology. Appl Microbiol Biotechnol 2010; 86:419-34; PMID:20143230; http://dx.doi.org/ 10.1007/s00253-010-2446-1 [DOI] [PubMed] [Google Scholar]
- 6.Leonard E, Lim KH, Saw PN, Koffas MA. Engineering central metabolic pathways for high-level flavonoid production in Escherichia coli. Appl Environ Microbiol 2007; 73:3877-86; PMID:17468269; http://dx.doi.org/ 10.1128/AEM.00200-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rohmer M. The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat Product Rep 1999; 16:565-74; PMID:10584331; http://dx.doi.org/ 10.1039/a709175c [DOI] [PubMed] [Google Scholar]
- 8.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature 1990; 343:425-30; PMID:1967820; http://dx.doi.org/ 10.1038/343425a0 [DOI] [PubMed] [Google Scholar]
- 9.Iijima Y, Gang DR, Fridman E, Lewinsohn E, Pichersky E. Characterization of geraniol synthase from the peltate glands of sweet basil. Plant Physiol 2004; 134:370-9; PMID:14657409; http://dx.doi.org/ 10.1104/pp.103.032946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer MJ, Meyer S, Claudel P, Perrin M, Ginglinger JF, Gertz C, Masson JE, Werck-Reinhardt D, Hugueney P, Karst F. Specificity of Ocimum basilicum geraniol synthase modified by its expression in different heterologous systems. J Biotechnol 2013; 163:24-9; PMID:23108028; http://dx.doi.org/ 10.1016/j.jbiotec.2012.10.012 [DOI] [PubMed] [Google Scholar]
- 11.Oswald M, Fischer M, Dirninger N, Karst F. Monoterpenoid biosynthesis in Saccharomyces cerevisiae. FEMS Yeast Res 2007; 7:413-21; PMID:17096665; http://dx.doi.org/ 10.1111/j.1567-1364.2006.00172.x [DOI] [PubMed] [Google Scholar]
- 12.Zhou J, Wang C, Yoon S-H, Jang H-J, Choi E-S, Kim S-W. Engineering Escherichia coli for selective geraniol production with minimized endogenous dehydrogenation. J Biotechnol 2014; 169:42-50; PMID:24269531; http://dx.doi.org/ 10.1016/j.jbiotec.2013.11.009 [DOI] [PubMed] [Google Scholar]
- 13.Tokuhiro K, Muramatsu M, Ohto C, Kawaguchi T, Obata S, Muramoto N, Hirai M, Takahashi H, Kondo A, Sakuradani E, et al.. Overproduction of geranylgeraniol by metabolically engineered Saccharomyces cerevisiae. Appl Environ Microbiol 2009; 75:5536-43; PMID:19592534; http://dx.doi.org/ 10.1128/AEM.00277-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Y, Liu Q, Li L, Qin W, Yang J, Zhang H, Jiang X, Cheng T, Liu W, Xu X, et al.. Metabolic engineering of Escherichia coli for high-specificity production of isoprenol and prenol as next generation of biofuels. Biotechnol Biofuels 2013; 6:57; PMID:23618128; http://dx.doi.org/ 10.1186/1754-6834-6-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim EE, Wyckoff HW. Reaction mechanism of alkaline phosphatase based on crystal structures. Two-metal ion catalysis. J Mol Biol 1991; 218:449-64; PMID:2010919; http://dx.doi.org/ 10.1016/0022-2836(91)90724-K [DOI] [PubMed] [Google Scholar]
- 16.Iijima Y, Wang G, Fridman E, Pichersky E. Analysis of the enzymatic formation of citral in the glands of sweet basil. Arch Biochem Biophys 2006; 448:141-9; PMID:16150417; http://dx.doi.org/ 10.1016/j.abb.2005.07.026 [DOI] [PubMed] [Google Scholar]
- 17.Yang J, Nie Q, Ren M, Feng H, Jiang X, Zheng Y, Liu M, Zhang H, Xian M. Metabolic engineering of Escherichia coli for the biosynthesis of alpha-pinene. Biotechnol Biofuels 2013; 6:60; PMID:23631625; http://dx.doi.org/ 10.1186/1754-6834-6-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kojima M, Ayabe K, Ueda H. Importance of terminal residues on circularly permutated Escherichia coli alkaline phosphatase with high specific activity. J Biosci Bioeng 2005; 100:197-202; PMID:16198264; http://dx.doi.org/ 10.1263/jbb.100.197 [DOI] [PubMed] [Google Scholar]
- 19.Bansal VS, Vaidya S. Characterization of 2 Distinct Allyl Pyrophosphatase Activities from Rat-Liver Microsomes. Arch Biochem Biophys 1994; 315:393-9; PMID:7986083; http://dx.doi.org/ 10.1006/abbi.1994.1516 [DOI] [PubMed] [Google Scholar]
- 20.Dunn CA, O'Handley SF, Frick DN, Bessman MJ. Studies on the ADP-ribose pyrophosphatase subfamily of the Nudix hydrolases and tentative identification of trgB, a gene associated with tellurite resistance. J Biol Chem 1999; 274:32318-24; PMID:10542272; http://dx.doi.org/ 10.1074/jbc.274.45.32318 [DOI] [PubMed] [Google Scholar]