Abstract
The intricate shaping of the facial skeleton is essential for function of the vertebrate jaw and middle ear. While much has been learned about the signaling pathways and transcription factors that control facial patterning, the downstream cellular mechanisms dictating skeletal shapes have remained unclear. Here we present genetic evidence in zebrafish that three major signaling pathways − Jagged-Notch, Endothelin1 (Edn1), and Bmp − regulate the pattern of facial cartilage and bone formation by controlling the timing of cartilage differentiation along the dorsoventral axis of the pharyngeal arches. A genomic analysis of purified facial skeletal precursors in mutant and overexpression embryos revealed a core set of differentiation genes that were commonly repressed by Jagged-Notch and induced by Edn1. Further analysis of the pre-cartilage condensation gene barx1, as well as in vivo imaging of cartilage differentiation, revealed that cartilage forms first in regions of high Edn1 and low Jagged-Notch activity. Consistent with a role of Jagged-Notch signaling in restricting cartilage differentiation, loss of Notch pathway components resulted in expanded barx1 expression in the dorsal arches, with mutation of barx1 rescuing some aspects of dorsal skeletal patterning in jag1b mutants. We also identified prrx1a and prrx1b as negative Edn1 and positive Bmp targets that function in parallel to Jagged-Notch signaling to restrict the formation of dorsal barx1+ pre-cartilage condensations. Simultaneous loss of jag1b and prrx1a/b better rescued lower facial defects of edn1 mutants than loss of either pathway alone, showing that combined overactivation of Jagged-Notch and Bmp/Prrx1 pathways contribute to the absence of cartilage differentiation in the edn1 mutant lower face. These findings support a model in which Notch-mediated restriction of cartilage differentiation, particularly in the second pharyngeal arch, helps to establish a distinct skeletal pattern in the upper face.
Author Summary
The exquisite functions of the vertebrate face require the precise formation of its underlying bones. Remarkably, many of the genes required to shape the facial skeleton are the same from fish to man. In this study, we use the powerful zebrafish system to understand how the skeletal components of the face acquire different shapes during development. To do so, we analyze a series of mutants that disrupt patterning of the facial skeleton, and then assess how the genes affected in these mutants control cell fate in skeletal progenitor cells. From these genetic studies, we found that several pathways converge to control when and where progenitor cells commit to a cartilage fate, thus controlling the size and shape of cartilage templates for the later-arising bones. Our work thus reveals how regulating the timing of when progenitor cells make skeleton helps to shape the bones of the zebrafish face. As mutations in many of the genes studied are implicated in human craniofacial defects, differences in the timing of progenitor cell differentiation may also explain the wonderful diversity of human faces.
Introduction
Morphogenesis of the facial skeleton in zebrafish is tightly linked with the early differentiation of pharyngeal arch neural crest-derived cells (NCCs) into cartilage. Shortly after migration into the pharyngeal arches, NCCs form a series of pre-cartilage condensations that prefigure the distinct shapes of the later cartilage-replacement bones. As near-isometric growth of these cartilages during the later larval period largely preserves these initial shapes [1], early patterning, not later growth, is the major determinant of facial skeletal shaping. Identifying the local signals that sculpt and arrange early condensations in specific regions of the developing arches is therefore critical to understanding how the facial skeletal bauplan is established.
Genetic studies in a wide range of vertebrates has revealed that patterning of arch NCCs along the dorsoventral axis is an important early step in regionalization of the face, with ventral (distal) cells generating the lower jaw and hyoid bone, maxillary cells forming the upper jaw, and more posteriorly located dorsal (proximal) cells making the lateral upper face. These dorsoventral domains are established in large part by interactions between dorsal Jagged-Notch, ventral/intermediate Endothelin1 (Edn1), and ventral Bmp signaling. Mutation of Edn1 signaling components and key downstream targets (e.g. Dlx5/6) in mice and zebrafish results in homeotic transformations and/or losses of skeletal elements derived from the ventral and intermediate domains of the arches, such that the lower jaw adopts an ectopic upper jaw morphology [2–12]. Downregulation of Bmp signaling results in a similar loss of ventral arch-derived structures in mice and zebrafish [13–16], whereas loss of the Notch ligand jag1b in zebrafish conversely affects bones and cartilages of the upper/dorsal face, particularly those from the second arch and the dorsal-posterior region of the first arch [17]. These pathways are actively antagonistic: Edn1 and Bmp signaling prevent jag1b expression in ventral/intermediate cells, Notch signaling blocks the expression of Edn1 target genes dorsally (e.g. dlx3b/5a/6a, msxe, nkx3.2) [17], and Jagged-Notch and Edn1 signaling limit Bmp activity to the most ventral arches in part through upregulation of the Bmp antagonist Gremlin2 in the intermediate domain [13, 14]. The end result of these interactions is the establishment of a distinct dorsal domain (excluding the anterior/maxillary region of the first arch, which is not patterned by Notch [17]) and the subdivision of an initial ventral arch region into distinct ventral/lower and intermediate regions [14]. How this dorsoventral patterning is translated into region-specific cartilage shapes has, however, remained unresolved.
Previous microarray studies of dissected arches in mice lacking the key Edn1 target genes Dlx5/6 [18] or overexpressing Bmp4 [16] revealed a number of misregulated ventral- and dorsal-specific genes. However, an overarching logic by which the Edn1 and Bmp pathways impart region-specific skeletal shapes remained elusive, with the role of Notch signaling in this process even less clear. In the present study, we perform genome-wide expression analyses of purified arch NCCs to correlate how gene expression patterns change over time in wild-type zebrafish with how gene expression is affected by reduction or elevation of Edn1 or Jagged-Notch signaling. In so doing, we find a prominent role for Jagged-Notch signaling in repressing, and Edn1 in activating, the expression of a set of genes that are strongly induced as arch progenitors mature and begin to acquire cartilage fates, implying that Notch and Edn1 signaling exert opposite effects on cartilage differentiation within the arches.
Two such downstream effectors identified in our genomic analysis are the pre-cartilage condensation marker barx1 (inhibited by Notch, activated by Edn1) and the early progenitor markers prrx1a and prrx1b (inhibited by Edn1). In mouse and chick, the homologs of prrx1a and prrx1b (Prrx1/PRRX1, previously called Prx1 or mHox) are expressed in uncondensed preskeletogenic mesenchyme [19–22], whereas Barx1/BARX1 is found in cells of nascent pre-cartilage condensations that have not or are just beginning to upregulate Sox9 [23–25]. Studies using a Prrx1 proximal promoter to drive lacZ expression [22, 26] or Cre recombinase [27] revealed that the cells that make up the limb skeleton and associated connective tissues all pass through a Prrx1+ state at some point during their differentiation program. Though no similarly definitive lineage-tracing studies exist for Barx1, corollary evidence suggests that most populations of Barx1+ cells mature into Sox9+ chondrocytes [25, 28–31]. In mammals, PRRX1 is required to repress cartilage differentiation in certain parts of the face: Prrx1 mouse mutants develop a large ectopic cartilage in place of the dermal squamosal bone on the side of the skull (derived from the dorsal first arch) as well as an aberrant sigmoidal process off of a shortened Meckel’s cartilage; these mutants also show chondrification of the stylohyoid ligament between the styloid process and Reichert’s cartilage (second arch), among numerous other craniofacial and limb skeletal defects [21, 22]. By contrast, impaired cartilage development is observed in barx1 mutant zebrafish, particularly in the ventral/lower face [32]. In edn1 mutant zebrafish and mice mutant for the Edn1 receptor (Ednra2), defects in ventral and intermediate facial structures are preceded by a loss and shift of Barx1/barx1 expression, particularly in the second arch [10, 33]. These studies indicate that although cartilage differentiation does not strictly require Barx1, chondrogenesis in the ventral and intermediate arches is most sensitive to its loss. Here we demonstrate that early arch patterning pathways compete to drive (Edn1) or restrict (Jagged-Notch, Bmp) the commitment of NCCs to chondrogenic differentiation, in part through antagonistic regulation of barx1 and prrx1a/b. These region-specific differences in the timing and extent of cartilage formation thus establish the template for the later formation of uniquely shaped bones in the upper and lower face.
Results
Widespread antagonistic control of arch gene expression by Edn1 and Notch signaling
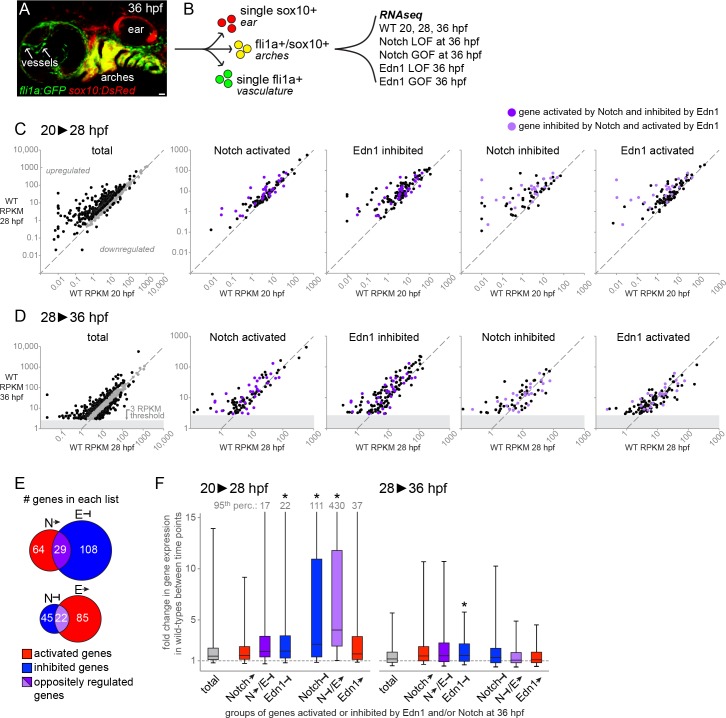
In an unbiased approach towards understanding facial skeletal patterning, we first performed a global gene expression analysis of pharyngeal arch NCCs at three time-points in wild-type embryos. To purify arch NCCs, we conducted fluorescence-activated cell sorting (FACS) on dissociated cells doubly positive for sox10:DsRed and fli1a:EGFP transgenes (and single-positive and double-negative cells for comparison) (Fig 1A and 1B). sox10:DsRed labels all NCCs and the ear, and fli1a:EGFP labels arch NCCs, blood vessels, and macrophages. These transgenes uniquely intersect within the arch NCC population, allowing us to selectively enrich for these cells shortly after NCC migration into the arches (20 hours post-fertilization, hpf) and during the initiation of pre-cartilage condensation formation (28 and 36 hpf). cDNA libraries were then constructed for each cell population and subjected to next-generation sequencing. To remove genes with low expression in the arches, we excluded genes with RPKM values ≤ 3 in the wild-type 36 hpf sample. However, a number of genes with known expression in the erthyroid lineage (e.g. hemoglobin genes hbae1/3, hbbe1/3), macrophages (e.g. mfap4 [34]), and the ear (e.g. mvp [35] and oc90 [36]) were found in this filtered list, suggesting some degree of contamination of the GFP/DsRed double-positive population by single-positive fli1a:EGFP or sox10:DsRed cells. We therefore further filtered for genes with expression ratios of 1.5-fold or higher in the double-positive cells relative to both single-positive populations. This left 536, 668, and 741 arch-enriched genes in the 20, 28, and 36 hpf samples, respectively, with the latter group comprising the “total” arch gene list in Fig 1 (also see S1 Table).
Fig 1. Jagged-Notch signaling represses genes strongly induced during pharyngeal arch differentiation.
(A) 36 hpf fli1a:EGFP; sox10:DsRed embryo showing colocalization of GFP and DsRed in the arches (yellow), with DsRed-only cells (red) in the ear and GFP-only cells (green) in the vasculature (and macrophages). Scale bar = 20 μm. (B) Double-positive embryos were dissociated and subjected to FACS to isolate GFP/DsRed+ arch cells (yellow) for RNA sequencing. Wild-type embryos were profiled at three stages, and Notch and Edn1 loss- and gain-of-function (LOF, GOF) models at 36 hpf only. (C-D) Scatterplots depicting changes in expression (RPKM) among the genes on each list from 20 to 28 hpf (C) and 28 to 36 hpf (D). Genes to the left of the diagonal were upregulated, while genes to the right were downregulated. For the ‘total’ arch NCC-enriched gene charts, black points indicate genes with fold-changes ≥ 1.5, and grey points indicate genes with fold changes ≤ 1.5. In the other charts, black points correspond to genes uniquely present on a given list, whereas purple points indicate genes oppositely regulated by Notch and Edn1 (dark purple: Notch-activated/Edn1-inhibited; light purple: Notch-inhibited/Edn1-activated). The grey blocks in D reflect the filtering threshold (RPKM ≥ 3 at 36 hpf in wild types) used to generate the total list of arch NCC-enriched genes. (E) Venn diagrams display the numbers of genes on each list, with genes shared between the Notch-activated/Edn1-inhibited or Notch-inhibited/Edn1-activated lists indicated in dark and light purple, respectively. (F) Summary of gene expression changes (RPKM) in wild types between 20 and 28 hpf (left chart) and 28 and 36 hpf (right chart), for genes classified as activated (red) or inhibited (blue) by Notch or Edn1 signaling. The data are shown as the median fold-change value for all genes on a given list (black central line), with flanking second and third quartiles as the lower and upper boxes, respectively, and the 5th and 95th percentiles indicated by the whiskers. For the lists in which the 95th percentile value exceeded the upper boundary of the chart, the value is indicated at the top of the upper whisker. Compared with the total list of arch NCC-enriched genes, genes inhibited by Notch signaling and those that were both inhibited by Notch and activated by Edn1 (light purple) showed a significantly larger increase in expression from 20 to 28 hpf (p < 0.001). Genes inhibited by Edn1 also increased slightly but significantly from 20–28 and 28–36 hpf relative to the total list. The gray dashed line indicates a fold-change value of 1 (no change in expression) between the two time points.
In order to understand how Edn1 and Notch signaling control the expression of these arch NCC-enriched genes, we next performed FACS purification and next-generation cDNA sequencing of GFP/DsRed double-positive cells from 36 hpf embryos with gain or loss of each signaling pathway. Specifically, we compared fold-change differences between edn1 mutants and stage-matched controls, jag1b mutants and wild-type siblings, and hsp70I:Gal4; UAS:Edn1 or hsp70I:Gal4; UAS:NICD (Notch1 intracellular domain) versus hsp70I:Gal4 controls (subjected to a 20–24 hpf heat-shock to overactivate Edn1 or Notch signaling) (see Methods). The top 20 genes up- and down-regulated in the Edn1 and Notch mutant and overexpression datasets (prior to filtering for arch NCC-enriched genes) are presented in S2 Table. Known targets of Notch (e.g. jag1b, hey2 and her2/4/15 genes) and Edn1 (e.g. dlx3b/4a/4b/6a and Evf1/2) are highly represented in these lists. All subsequent analyses were performed using the filtered list of 741 genes with arch-enriched expression in the 36 hpf wild-type sample. To identify those genes most strongly regulated by the Edn1 or Notch pathway, we divided the fold-change of the overexpression (OE) sample by the fold-change of the corresponding mutant (mut) sample. Genes considered ‘activated’ by the Edn1 or Notch pathways had an OE-fold-change/mut-fold-change ratio ≥ 1.5 as well as an OE-RPKM/control-RPKM ratio ≥ 1. Genes considered ‘inhibited’ by Edn1 or Notch had an OE-fold-change/mut-fold-change ratio ≤ 0.667 and a mutant-RPKM/control-RPKM ratio ≥ 1. Lastly, we performed one further refinement for the Notch lists by analyzing genes affected by treatment of embryos with the γ-secretase inhibitor dibenzazepine (DBZ), which blocks processing of the Notch receptor into its active intracellular form [37], starting at 24 hpf. After FACS-purification and next-generation sequencing of double-positive cells from 36 hpf embryos, we calculated the fold-change between DBZ-treated and control samples. Eleven of the top 20 genes downregulated in DBZ-treated embryos belong to the Her/Hes/Hey family of Notch targets [38, 39] (7 of which were shared with the jag1b mutant list) (S2 Table), showing that DBZ is primarily affecting Notch signaling in this experiment. However, as γ-secretase inhibitors such as DBZ are also known to affect other signaling pathways [40], we only used the DBZ dataset to further refine the lists generated from the jag1b and NICD analyses. Specifically, we excluded genes from the ‘Notch activated’ list that were not also elevated in NICD versus DBZ (fold-change ratio ≥ 1.25) and from the ‘Notch inhibited’ list those not also decreased in NICD versus DBZ (fold-change ratio ≤ 0.8). These filtered gene lists (Fig 1E, S3–S6 Tables) were then used for the global analyses described below.
Consistent with our previous data that the Edn1 and Jagged-Notch signaling pathways antagonize one another during facial development [17], we observed a disproportionately high number of genes oppositely regulated by these pathways. Of the 67 ‘Notch-inhibited’ genes and 107 ‘Edn1-activated’ genes, 22 were in common (Fig 1E). Conversely, 29 genes were in common between the 93 ‘Notch-activated’ and 137 ‘Edn1-inhibited’ genes (Fig 1E). These commonly regulated genes include many known positive Edn1 targets (e.g. dlx3b, dlx4a, dlx4b, epha4b, Evf1/2, msxe, and notch2) [5, 17, 18, 33, 41, 42] and negative Edn1 targets (e.g. jag1b and pou3f3a/b) [17, 18]. Smaller groups of genes were co-regulated by Notch and Edn1 in the same direction (positive, n = 9; negative, n = 6; S3–S6 Tables), as we previously observed for the BMP antagonist grem2 [14].
Jagged-Notch signaling represses genes strongly induced at the onset of facial skeletogenesis
We next examined whether genes activated or inhibited by Notch or Edn1 presented any common temporal signatures during arch development in wild-type embryos. To do so, we first determined the fold changes in wild-type RPKM values for the 741 total arch genes from 20 to 28 hpf and from 28 to 36 hpf (Fig 1C, 1D and 1F; S1 Table). Total arch genes increased by a median of 1.46-fold between 20 and 28 hpf, and 1.18-fold between 28 and 36 hpf. In contrast, we found that the subset of genes that we had annotated as ‘Notch inhibited’ increased 2.62-fold from 20 to 28 hpf in wild types, with many of these upregulated more than 10-fold (p < 0.001). These strongly upregulated genes presented a range of expression levels at 20 hpf, showing that the stronger upregulation of ‘Notch inhibited’ genes is likely not an artifact of these having very low initial expression levels. ‘Edn1 inhibited’ genes also displayed a modest but significantly higher upregulation than total arch genes (median 1.95-fold increase; p < 0.001), though no significant differences were observed for ‘Notch activated’ genes (median = 1.51). Although the ‘Edn1 activated’ genes were not more highly upregulated than total arch genes (median = 1.68), the subset of ‘Edn1 activated’ genes in common with ‘Notch inhibited’ genes were the most strongly induced (median 4.01 fold increase; p < 0.001). Of these 22 common genes, 8 were induced more than 10-fold between 20 and 28 hpf, out of only 48 total >10-fold-upregulated arch genes. At later stages (28–36 hpf; Fig 1D and 1F), only ‘Edn1 inhibited’ genes (median 1.52-fold increase) showed a small but significant difference (p < 0.001) relative to all arch genes (median 1.18-fold increase). In summary, these data show that genes commonly inhibited by Notch and activated by Edn1 are some of the most highly induced during early arch differentiation, consistent with a global role for Notch repression in limiting the differentiation of NCCs in the dorsal arches.
Accelerated condensation and cartilage formation in the lower face
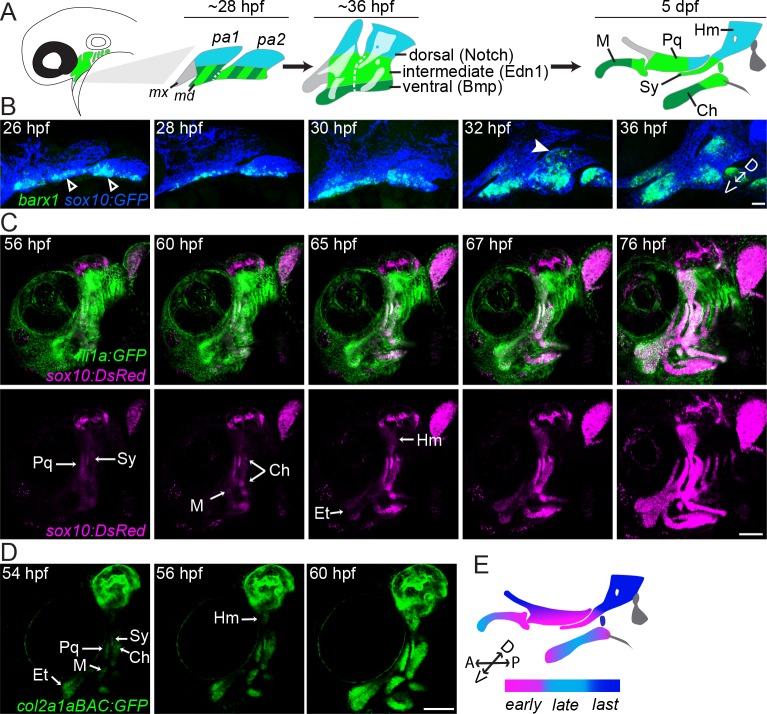
Given the genome-wide role of dorsal Jagged-Notch signaling in repressing strongly induced genes during arch maturation, we investigated whether this might reflect a delay in cartilage differentiation in the dorsal domain versus the rest of the arches. From our genomic analysis, we observed that barx1, which marks early pre-cartilage condensations [23, 29], was negatively regulated by Notch signaling (~5-fold lower in NICD versus jag1b mutant; S3 Table) and 12.5-fold upregulated between 20 and 28 hpf in wild-type NCCs (S1 Table). By examining a time-course of barx1 expression (also see [29, 32]), we find barx1 to be confined to the intermediate/ventral portions of the first and second arches at 26–28 hpf, with maxillary first arch and dorsal second arch expression not initiating until 30–32 hpf (Fig 2B). To determine whether this delay reflects later cartilage differentiation in the dorsal second arch, we made time-lapse recordings of fish expressing sox10:DsRed (which shows biphasic expression–first in all NCCs and later in differentiating chondrocytes) along with the arch NCC transgene fli1a:EGFP or the chondrocyte transgene col2a1aBAC:GFP (Fig 2C and 2D and S1 and S2 Movies). In both cases, chondrocyte differentiation was first evident in the palatoquadrate cartilage (Pq, primarily an intermediate first arch element with a small amount of dorsal contribution at its posterior end), the symplectic cartilage (Sy, intermediate second arch), at either end of the ceratohyal cartilage (Ch, ventral-intermediate second arch), and the proximal portion of Meckel’s cartilage (M, ventral-intermediate first arch). Chondrocyte transgene expression then spread into the center of the Ch and more ventral portions of the M cartilage. The last elements to differentiate were the hyomandibular cartilage (Hm, dorsal second arch) and the pterygoid process cartilage (Ptp, maxillary) (schematized in Fig 2A and 2E). We also observed that sox9a, an early marker of pre-chondrocytes [43–46] that is positively regulated by Edn1 signaling (S6 Table), was expressed only in ventral-intermediate arch NCCs at 36 hpf, with expression spreading to dorsal arch NCCs by 48 hpf (Fig 3A and 3K). Our findings point to cartilage differentiation occurring first in discrete zones, primarily within the intermediate arches, then spreading to other ventral regions and lastly to dorsal regions, consistent with previous studies based on Alcian Blue staining of sulfated proteoglycans typical of cartilage [47].
Fig 2. Accelerated cartilage differentiation in ventral-intermediate arch NCCs.
(A) Schematic of pharyngeal arch patterning in zebrafish. At early patterning stages (~28 hpf), the first two pharyngeal arches (pa1, pa2) are divided into distinct dorsal (blue) and ventral/intermediate (green stripe) domains, with the latter resolving into intermediate (light green) and ventral (dark green) domains by 36 hpf. Notch activity governs the dorsal domain, Edn1 the intermediate domain, and Bmp signaling the ventral domain. The anterior maxillary domain (grey) is not significantly influenced by any of these pathways. The facial cartilages of the larval skeleton (5 dpf) are color-coded based on their arch origins. Hm, hyomandibula; Pq, palatoquadrate; M, Meckel’s; Sy, symplectic; Ch, ceratohyal. (B) barx1 (green) is upregulated ventrally (≤ 26 hpf, white open arrowhead) well before dorsal second arch expression can be detected (~32 hpf, white arrowhead). NCCs express the sox10:GFP transgene (blue). Shown are maximum intensity projections of confocal z-stacks of single-color in situs co-stained with a GFP antibody. The orientation of the dorsal (D)-ventral (V) axis is indicated. (C) Stills from a time-lapse movie (see S1 Movie) show the emergence of facial cartilages (sox10:DsRed+, magenta) from fli1a:EGFP+ ectomesenchyme (green). sox10:DsRed+ chondrocytes appear in a stereotyped sequence within the facial cartilages, with cells of the intermediate Sy and Pq cartilages detectable first at 56 hpf, followed by the ventral M and Ch cartilages at 60 hpf and the dorsal Hm at 65 hpf. (D) The same sequence of cartilage differentiation is seen slightly earlier in stills from a time-lapse movie of col2a1aBAC:GFP fish (see S2 Movie). The time-lapses in B and C were performed with a 20x objective using 0.5x digital magnification. Et, ethmoid cartilage. (E) Color-coded schematic of the sequence of chondrocyte differentiation in the facial skeleton. The orientations of the D-V and anterior (A)-posterior (P) axes are indicated. Scale bar in B = 20 μm; scale bars in C, D = 100 μm.
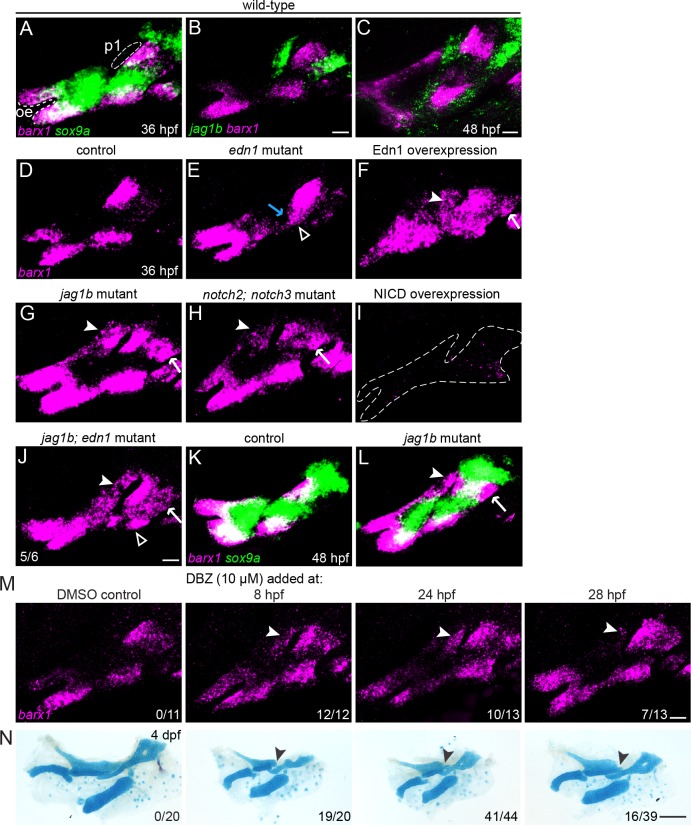
Fig 3. Regulation of barx1+ condensations by Edn1 and Notch.
(A) At 36 hpf, the intermediate sox9a domain (green) only partially overlaps with zones of barx1 expression (magenta) at the ventral and dorsal poles of each arch. The oral ectoderm (oe) and first pharyngeal pouch (p1) are shown for reference. (B, C) jag1b (green) and barx1 (magenta) are anti-correlated in dorsal NCCs at 36 and 48 hpf. (D-J) barx1 expression at 36 hpf in the first and second arches of wild-type controls, mutants, and overexpression embryos. Open arrowheads show the loss of ventral barx1 in edn1 mutants (E) and its restoration in 5/6 jag1b; edn1 mutants (J). The blue arrow in E indicates weak upregulation of barx1 in the intermediate domain of edn1 mutants. Upregulation of barx1 in the dorsal first arch (white arrowhead) and dorsal second arch (white arrow) is seen in Edn1-overexpressing embryos (F), jag1b mutants (G), notch2; notch3 mutants (H), and jag1b; edn1 mutants (J). Dotted lines in (I) show the arches of NICD-overexpression embryos in which barx1 is nearly absent. (K, L) Ectopic barx1 persists in jag1b mutants at least until 48 hpf, but no ectopic expression of sox9a is observed. (M, N) Representative barx1 expression patterns and skeletal preparations in embryos treated with the Notch inhibitor DBZ (10 μM) starting at the indicated time points. Earlier exposure to the DBZ inhibitor correlated with stronger ectopic barx1 expression (M) and more severe and penetrant Notch-type skeletal phenotypes (N). Fractions indicate the number of embryos in each treatment that exhibited unambiguous ectopic barx1 expression in the dorsal first arch (arrowheads in M) or showed posterior Pq malformations (arrowheads in N. DBZ treatment also caused systemic effects, including spinal curvature and cardiac edema, which reduced bone mineralization and led to a general reduction in the size of the craniofacial skeleton. Scale bars in B, C, J, L, M = 20 μm; scale bar in N = 100 μm.
Jagged-Notch inhibits and Edn1 promotes barx1+ condensations
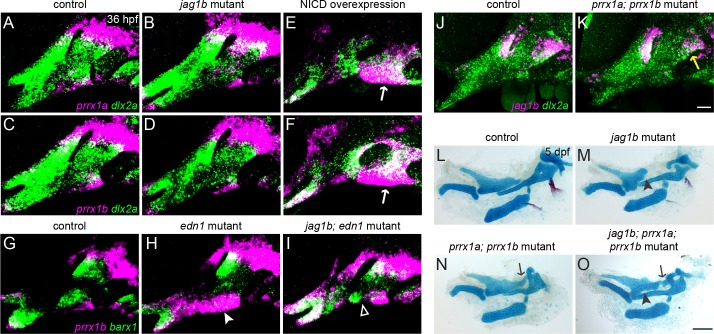
The earlier chondrogenic differentiation in intermediate/ventral arch cells relative to dorsal cells led us to hypothesize that antagonism between dorsal Jagged-Notch and ventral Edn1 signaling may serve to establish barx1+ condensations earlier and/or more extensively in the lower face. As reported previously [10], we find that barx1 expression is lost from the ventral second but not first arch of edn1 mutants at 36 hpf (Fig 3E). Conversely, elevation of Edn1 signaling (via 20–24 hpf heat-shock induction of hsp70I:Gal4; UAS:Edn1 fish) resulted in an expansion of barx1 expression throughout the arches (Fig 3F). In contrast, we find Jagged-Notch signaling to be required to restrict dorsal barx1 expression, consistent with our RNAseq data (S3 Table) and the mutually exclusive expression of barx1 and jag1b at 36 and 48 hpf (Fig 3B and 3C). In jag1b mutants, barx1 expression expands into the dorsal-posterior regions of both the first and second arch (Fig 3G; similar to Edn1 overexpression (Fig 3F)), domains that correlate precisely with jag1b expression at this stage (Fig 3B). Conversely, jag1b expression is unaltered in barx1 mutants (S1 Fig), indicating that Jagged-Notch signaling functions largely upstream of barx1 and not vice versa. This ectopic dorsal barx1 expression was also observed in notch2; notch3 double mutants (Fig 3H), which display similar facial cartilage defects to jag1b mutants (consistent with notch2 and notch3, but not notch1a or notch1b, being expressed in arch NCCs; S2 Fig). Reciprocally, forced activation of Notch signaling in heat-shock-treated hsp70I:Gal4; UAS:NICD fish eliminated nearly all barx1 expression in the arches (Fig 3I). Finally, we find that the positive effect of Edn1 on ventral second arch barx1 expression can be explained at least in part by the previously reported role of Edn1 in blocking jag1b expression [17], as mutation of jag1b partially restored ventral second arch barx1 expression in edn1 mutants (Fig 3J).
We next examined whether the ectopic dorsal expression of barx1 persisted in jag1b mutants, as well as the consequences of this for cartilage differentiation. At 36 hpf, sox9a expression marks the nascent cartilages in the ventral-intermediate arches that are the first to differentiate, with barx1 expression in a partially overlapping set of cells nearer to the poles of each arch (Fig 3A). By 48 hpf, sox9a expression has spread into the nascent dorsal cartilages yet remains only minimally overlapping with barx1 (Fig 3K, also see S3 Fig). These results are consistent with previous literature showing that Barx1 is expressed in nascent pre-cartilage condensations that have not or are just beginning to upregulate Sox9 [23–25]. In jag1b mutants at 48 hpf, we observe an expansion of barx1 but not sox9a expression in the dorsal first and second arch (Fig 3L), suggesting that a subset of dorsal arch NCCs may be trapped in an early barx1+ condensation state in the absence of Jagged-Notch signaling. This failure of ectopic dorsal barx1+ cells to transition to a more mature sox9a+ state may help explain why the dorsal cartilages of jag1b mutants are truncated rather than expanded (Fig 4B).
Fig 4. Loss of barx1 rescues some features of the jag1b skeletal phenotype.
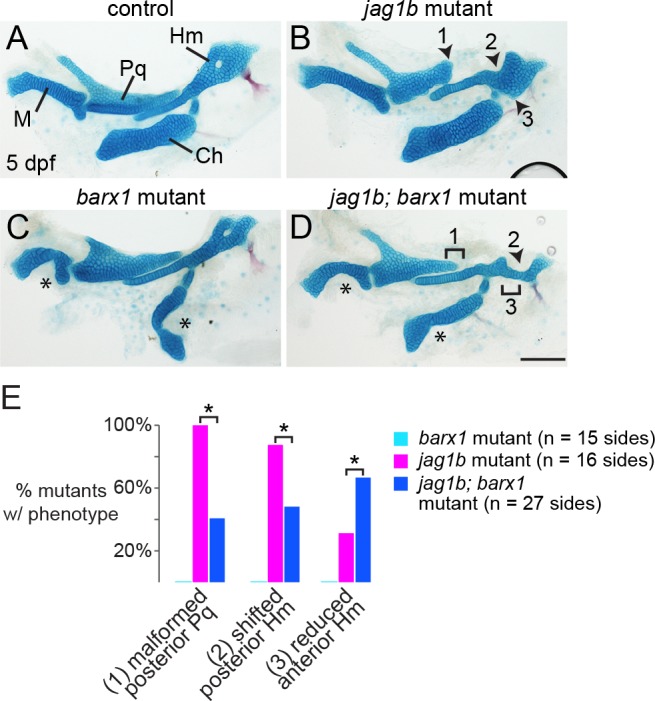
(A, B) Alcian staining of dissected jag1b mutant larval facial cartilages derived from the first and second arches show malformation of the posterior Pq (1), reduction of the anterior Hm (2), and a shift of the posterior Hm (3) such that it abuts the ventral Ch cartilage. (C) In barx1 mutants, the ventral M and Ch cartilages are reduced (asterisks). (D) In jag1b; barx1 mutants, the Pq truncation (1) and posterior Hm shift (3) are variably rescued, yet loss of the anterior Hm (2) becomes more penetrant. Scale bar = 100 μm. (E) Proportions of mutant larvae exhibiting the indicated phenotypes. None of these skeletal defects were observed in controls or barx1 mutants. For each phenotype, differences between the genotype groups were significant by Chi-Square test at p < 0.0001 (asterisks).
Because ectopic dorsal barx1 expression correlated with dorsal cartilage defects in jag1b and notch2; notch3 mutants, we next investigated whether this reflected a common early requirement for Jagged-Notch signaling for both processes. To temporally inhibit Notch signaling, we treated embryos at different stages with 10 μM DBZ, and evaluated the effects on barx1 expression and skeletal patterning. Although DBZ may also affect other signaling pathways [40], our RNAseq analysis showed that the majority of the most strongly downregulated genes were canonical Notch targets (S2 Table). This analysis focused on the first arch phenotypes, which have proved the most penetrant and consistent across all of our Notch loss-of-function models. Compared with DMSO-treated controls, treatment of embryos with DBZ starting at 8 hpf resulted in a highly penetrant expansion of barx1 expression into the posterior dorsal first arch (12/12), as well as dorsal cartilage defects similar to jag1b mutants (19/20 with Pq malformations; Fig 3M and 3N). DBZ treatment starting at 24 hpf resulted in a weaker and less penetrant barx1 expansion (10/13 embryos with ectopic first arch barx1) and milder dorsal cartilage defects (41/44 with moderate Pq malformations). In contrast, treatment at 28 hpf only mildly affected barx1 expression in 7/13 embryos, with only 16/39 embryos displaying weak dorsal cartilage malformations (Fig 3M and 3N). Treatments initiated at 32 hpf did not affect barx1 expression or skeletal patterning. Inhibition of Notch signaling at these stages also had other effects on embryo development, including cardiac edema, which likely contributed to the general reductions in cartilage size. In summary, we observe a tight correlation between barx1 expression changes and subsequent malformations of dorsal cartilages in Notch-deficient embryos, with the requirement for Notch inhibition by approximately 24 hpf being consistent with the predicted global effects of Notch in repressing arch gene induction between 20–28 hpf (Fig 1F).
Loss of barx1 partially rescues the jag1b mutant skeletal phenotype
We next investigated the extent to which the ectopic dorsal expression of barx1 in Notch pathway mutants contributes to the dorsal cartilage malformations. In particular, jag1b mutants display several characteristic changes in cartilages of the upper face, including truncation of the posterior end of Pq (i.e. the portion deriving from dorsal first arch NCCs; Fig 2A) and a variable reduction of the anterior part of Hm (Fig 4A and 4B). jag1b mutants also display a highly penetrant posterior shift of Hm such that it sits closer to the ventral Ch cartilage [17]. In barx1 mutants, the dorsal cartilages are largely unaffected, with there instead being conspicuous reductions of the ventral M and Ch cartilages (Fig 4C) [32]. In jag1b; barx1 mutants, we observed an incompletely penetrant rescue of posterior Pq (truncation in 11/27 double mutants versus 16/16 jag1b mutants, p < 0.0001) and the position of Hm (posterior shift in 13/27 double mutants versus 14/16 jag1b mutants, p < 0.0001) (Fig 4D and 4E). However, ventral M and Ch defects were not restored, and the anterior Hm was more prominently diminished (loss in 18/27 double mutants versus 5/16 jag1b mutants, p < 0.0001). Of note, the two regions of skeletal rescue (posterior Pq and posterior Hm) correlate precisely with the earlier ectopic expression of barx1 in dorsal-posterior first and second arch domains of jag1b mutants (Fig 3G), suggesting that the ectopic barx1 may account in part for these phenotypes. On the other hand, the incompletely penetrant rescue of these elements, in addition to exacerbated phenotypes in other regions (e.g. anterior Hm), indicates the presence of other causative changes in jag1b mutants beyond barx1 misexpression.
Fate maps reveal Notch dependency for dorsal cartilage growth
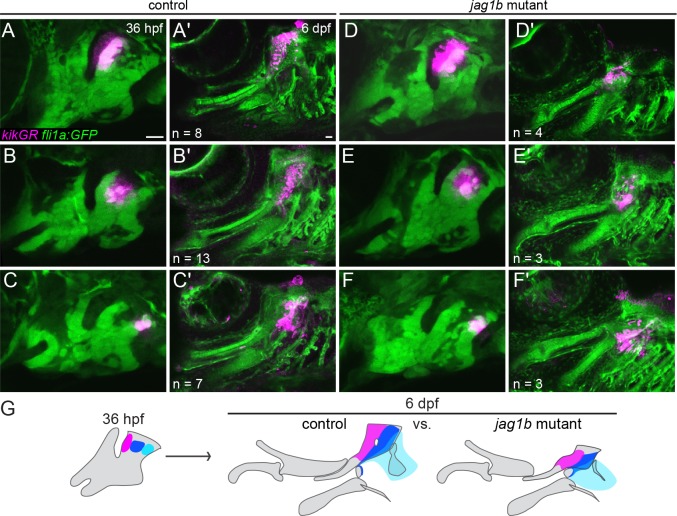
The finding that posterior-dorsal cells ectopically express barx1 but fail to turn on sox9a in jag1b mutants, as well as the fact that cartilage defects were only modestly rescued in jag1b; barx1 double mutants, suggest that Notch signaling has additional roles in dorsal cartilage development. In order to better understand the reductions of dorsal cartilage in jag1b mutants, we used photoconversion of the kikGR protein to follow the fate of dorsal second arch NCCs in wild types versus mutants (Fig 5). When wild-type cells were converted at 36 hpf and then re-imaged at 6 days post fertilization (dpf), we found that anterior dorsal second arch NCCs contributed to the anterior portion of the Hm cartilage, central dorsal second arch NCCs to the posterior portion of Hm, and posterior dorsal second arch NCCs to a small amount of Hm and the opercle bone to which it attaches (Fig 5A–5C). In jag1b mutants, all three domains contributed to similar portions of the malformed Hm cartilage as in wild types (Fig 5D–5F), indicating no major shift in the fate map of skeletal precursors in mutants. However, whereas cells from all three regions spread along the dorsoventral axis in wild types, cells from comparable domains in jag1b mutants gave rise to much smaller domains of cartilage (Fig 5G). These results suggest that Jagged-Notch signaling is also required for the expansion of the dorsal second arch NCCs that generate cartilage in the upper face.
Fig 5. Reduced growth of Hm cartilage in jag1b mutants.
(A-F) kikGR RNA was injected into control fli1a:EGFP or jag1b; fli1a:EGFP embryos, and kikGR protein was photoconverted in small groups of GFP+ arch NCCs using a UV laser at 36 hpf. The same larvae were then reimaged at 6 dpf to assess contributions of kikGR-converted cells (magenta) to cartilage. In both controls (A-C) and jag1b mutants (D-F), photoconverted cells from the anterior, center, and posterior regions of the dorsal second arch contributed to the anterior Hm (A,D), posterior Hm (B,E) and posterior edge of the Hm and opercle bone (C,F). Relative to controls, labeled NCCs from jag1b mutants contributed to qualitatively smaller domains of cartilage by 6 dpf. Reproducible differences were seen in each mutant or control examined, with n numbers listed in each panel. (G) Summary of these fate maps showing contribution to anterior Hm (red), posterior Hm (dark blue), and opercle bone (light blue). Scale bars = 20 μm.
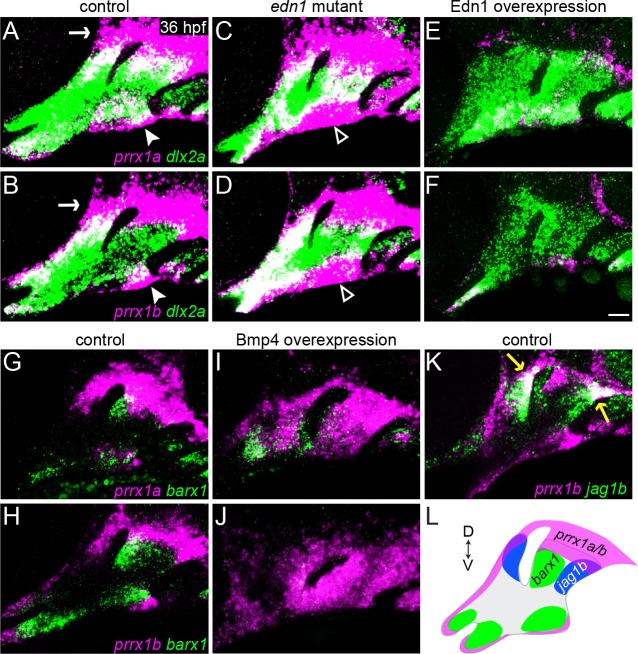
Edn1 inhibits and Bmp signaling promotes arch expression of prrx1a and prrx1b
While loss of Jagged-Notch signaling can rescue barx1 expression and ventral cartilage development in edn1 mutants (Fig 3J; [17]), the partial and largely second-arch nature of this rescue implies the presence of other important pathways downstream of Edn1. Our expression analysis of sorted arch NCCs identified two genes implicated in early skeletogenic mesenchyme identity, prrx1a and prrx1b, which, like jag1b, were upregulated in edn1 mutants and downregulated in Edn1-overexpressing embryos (S5 Table). Loss of the homologous Prrx1 gene in mice results in ectopic dorsal facial cartilage [21, 48], implying that Prrx1 genes may also restrict cartilage formation in the upper face. We thus reasoned that Edn1-mediated repression of prrx1a and prrx1b could help to explain the observed acceleration of cartilage differentiation in the intermediate domain (Fig 2B–2E). Consistently, we observed that expression of prrx1a and prrx1b was largely excluded from NCCs in the intermediate domain, instead being confined to the dorsal-most and ventral-most poles of the first two arches in 36 hpf wild types (Fig 6A and 6B). However, as predicted by our RNAseq analysis, prrx1a and prrx1b expression was upregulated along the ventral border and expanded into the intermediate arches of edn1 mutants, and lost in Edn1-overexpressing embryos (Fig 6C–6F), in accord with the elevated ventral Prrx1 expression observed in Dlx5/6 mutant mice [6]. Conversely, overactivation of Bmp4 signaling (via 20–24 hpf heat-shock induction of hsp70I:Gal4; UAS:Bmp4 fish) upregulated prrx1a and prrx1b expression throughout the arches (Fig 6G–6J), in accord with previous findings that Bmp signaling promotes genes associated with progenitor status and self-renewal in arch NCCs [16]. Positive regulation by the ventral Bmp signal, combined with negative regulation by intermediate Edn1, could help explain the restriction of prrx1a/b expression to the ventral pole of the arches.
Fig 6. prrx1a and prrx1b are repressed by Edn1 and activated by Bmp4 signaling.
(A-F) Two-color fluorescent in situs of 36 hpf wild-type embryos show that, relative to all arch NCCs (dlx2a, green), prrx1a and prrx1b (magenta) are expressed in dorsal arch NCCs and mesenchyme surrounding the ear (white arrow), as well as in a more limited ventral arch domain (white arrowhead). prrx1a and prrx1b are upregulated in ventral arch NCCs (white open arrowhead) of edn1 mutants and nearly lost upon overexpression of Edn1 in hsp70I:Gal4; UAS:Edn1 embryos subjected to a 20–24 hpf heat-shock treatment. (G, H) prrx1a/b and barx1 are expressed complementarily in the arches of wild types. (I, J) Overexpression of Bmp4 in hsp70I:Gal4; UAS:Bmp4 embryos heat-shocked from 20–24 hpf resulted in broad upregulation of prrx1a/b throughout the arches, with barx1 restricted to domains showing lower prrx1a/b expression. (K) prrx1b overlaps only slightly with jag1b expression at the dorsal-posterior tips of the first and second arches (yellow arrows). (L) Schematic depicting the expression patterns of prrx1a/b (magenta), barx1 (green), and jag1b (blue). Scale bar = 20 μm.
We next examined whether the Bmp4 induction of prrx1a/b is mediated by Hand2, a strong Bmp target that is specifically expressed in NCCs at the ventral border of the arches in both mice and fish [5, 49], domains that closely overlap with ventral prrx1a/b expression. While Hand2/hand2 expression requires positive input from both the Edn1/Dlx and Bmp pathways [5, 6, 13, 49, 50], overexpression of Bmp4 –but not Edn1 –induces its widespread ectopic expression [13, 14], similar to the patterns observed here for prrx1a/b (Fig 6G–6J). However, consistent with previous results in mice [51], prrx1a and prrx1b were expressed largely normally in hand2 mutants, with a limited expansion of prrx1a in the ventral domain (S4 Fig). Thus, Bmp signaling appears to positively regulate prrx1a/b expression largely independently of Hand2 function.
Prrx1 genes are required to repress barx1 expression and cartilage formation in the dorsal arches
In order to interrogate Prrx1 function in zebrafish, we used TALENs to generate prrx1ael558 and prrx1bel491 mutant alleles resulting in early truncation of the Prrx1a and Prrx1b proteins upstream of the conserved DNA-binding homeobox domains (S5 Fig). Whereas prrx1a and prrx1b single mutants did not show craniofacial defects during larval stages (consistent with their identical expression patterns), double homozygous mutants exhibited highly penetrant abnormalities affecting dorsal skeletal elements of the first two arches (Fig 7A–7F). Identical dorsal skeletal phenotypes were seen in double mutants carrying prrx1ab1246 and prrx1bb1247 alleles independently generated by CRISPR-mediated mutagenesis (S5 Fig). In the first arch of double mutant larvae, ectopic cartilage develops along the dorsomedial surface of the Pq cartilage in place of the dermal entopterygoid bone. This extra cartilage is occasionally fused with the trabecular cartilages of the neurocranium. In approximately 40% of double mutant embryos, Pq also extended dorsal-posteriorly to fuse with the otic capsule. In the second arch, the top of the Hm cartilage is malformed, with two highly penetrant cartilaginous fusions to the anterior and middle parts of the otic capsule. The foramen of the Hm, a channel for the VIIth cranial nerve and the anterior lateral line nerve [52], is absent, and the opercle bone is reduced. Despite the expression of prrx1a/b at both the dorsal and ventral poles of the arches, double mutants had no detectable defects in ventral cartilages.
Fig 7. Combined loss of prrx1a and prrx1b results in ectopic dorsal cartilage.
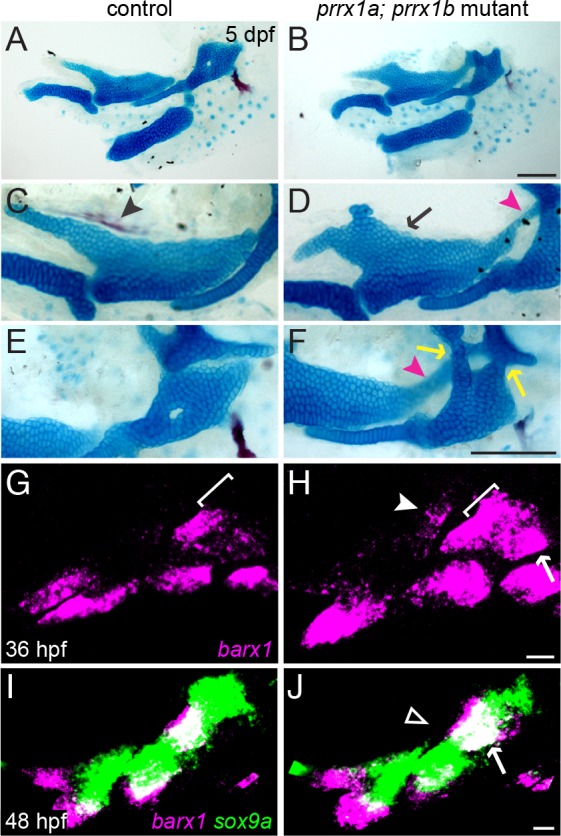
(A-F) prrx1a; prrx1b mutants develop ectopic cartilage, both from the dorsal-medial surface of Pq (black arrow) and connecting Pq to the otic cartilage (magenta arrowheads), as well as fusions of Hm to the otic cartilage (yellow arrows). The entopterygoid dermal bone (black arrowhead) that normally forms along the dorsal-medial surface of the Pq is also lost. (G, H) In prrx1a; prrx1b mutants at 36 hpf, barx1 (magenta) is ectopically upregulated in the dorsal first arch (white arrowhead), along the dorsal border of the second arch (white bracket, compare with G), and in the posterior dorsal second arch (white arrow). (I, J) By 48 hpf in prrx1a; prrx1b mutants, ectopic barx1 expression is no longer evident in the posterior first arch (open white arrowhead), and the second arch barx1+ domain is slightly larger than the sibling control (white arrow). sox9a expression (green) is largely normal at this stage in prrx1a; prrx1b mutants. Scale bars in B and F = 100 μm; scale bars in H and J = 20 μm.
Consistent with the ectopic dorsal cartilage, we also found that double mutants displayed ectopic barx1 expression at earlier stages (36 hpf) in dorsal arch regions that generate the parts of Pq, Hm, and otic cartilages affected in mutants (Fig 7G and 7H). This upregulation of barx1 in double mutants is consistent with the near mutually exclusive expression of prrx1a/b and barx1 in 36 hpf wild-type embryos (Fig 6G, 6H and 6L). In contrast to jag1b mutants (Fig 3L), these ectopic barx1 expression domains largely disappeared by 48 hpf (Fig 7I and 7J), perhaps accounting for the ectopic formation of cartilage in Prrx1 but not Notch pathway mutants. Interestingly, despite hand2 being expressed in a similar domain to prrx1a/b in the ventral arches, barx1 has been reported to be lost in hand2 mutants [32], opposite to the barx1 expansion we observe in prrx1a/b mutants. However, hand2 expression was unaffected in prrx1a; prrx1b mutants (S4 Fig), similar to previous observations in Prrx1-/- mice [53], suggesting that Prrx1 and Hand2 act antagonistically and independently to regulate barx1 expression and chondrogenesis in the ventral second arch.
Prrx1a/b and Jagged-Notch signaling function largely independently in the dorsal arches
Despite both prrx1a/b and jag1b expression being mutually exclusive to barx1, we found only limited overlap between these genes (Fig 6K and 6L). We therefore hypothesized that these pathways function independently to limit barx1 expression and cartilage formation in distinct domains of the arches. Consistently, we observed no defect in prrx1a or prrx1b expression in 36 hpf jag1b mutants, although forced activation of Notch signaling expanded prrx1a and prrx1b expression ventrally and decreased it dorsally (Fig 8A–8F). Loss of jag1b also partially rescued the ventral expansion of prrx1b observed in edn1 mutants, especially in the second arch (Fig 8G–8I). These findings indicate that, although jag1b is not required for prrx1a/b expression, high levels of Notch signaling (either artificially or by loss of Edn1) can induce prrx1b expression ventrally. Reciprocally, a subset of prrx1a; prrx1b mutants showed a modest reduction of jag1b expression limited to the dorsal posterior second arch (Fig 8J and 8K). To further clarify the genetic interaction between these genes, we analyzed jag1b; prrx1a; prrx1b triple mutants (Fig 8L–8O). In 9/13 triple mutant sides examined, we observed the ectopic posterior extension and fusion of the Pq cartilage to the ear (and not the Pq truncations seen in Notch pathway mutants), indicating that prrx1a/b are largely epistatic to jag1b with respect to the ectopic Pq phenotype. However, in addition to this extra cartilage, skeletons of the triple mutants (but not prrx1a; prrx1b double mutants) showed irregular gaps within the body of Pq, reminiscent of abnormalities seen in Notch pathway mutants. These observations support the triple mutant phenotype being largely additive, in line with Prrx1a/b and Jagged-Notch signaling having distinct roles in regulating condensation and cartilage formation in the upper face.
Fig 8. Partially overlapping functions of Prrx1a/b and Jagged-Notch signaling in dorsal cartilage development.
(A-F) The expression of prrx1a and prrx1b (magenta) is largely normal in jag1b mutants (B, D) but is upregulated in ventral arch NCCs (white arrows) and reduced in dorsal NCCs upon forced Notch activation in hsp70I:Gal4; UAS:NICD embryos subjected to a 20–24 hpf heat-shock treatment. dlx2a expression (green) marks all arch NCCs. (G-I) edn1 mutants display a loss of ventral barx1 expression (green) and gain of prrx1b (magenta) (white arrowhead) (H). In jag1b; edn1 mutants, there is partial recovery of ventral barx1 expression in the second arch (white open arrowhead), which corresponds to regions where the ectopic expression of prrx1b is restored to control levels. (J, K) In 7/10 prrx1a; prrx1b mutants, jag1b expression is partially reduced in the dorsal second arch (yellow arrow). (L-O) Dissections of facial cartilage and bone derived from the first two arches show additive phenotypes in jag1b; prrx1a; prrx1b triple mutants. Similar to prrx1a; prrx1b double mutants, triple mutants display ectopic cartilage connecting Pq to the otic cartilage (black arrows). However, similar to jag1b single mutants, jag1b; prrx1a; prrx1b triple mutants also display irregularities in the main body of Pq (black arrowheads). Scale bar in K = 20 μm; scale bar in O = 100 μm.
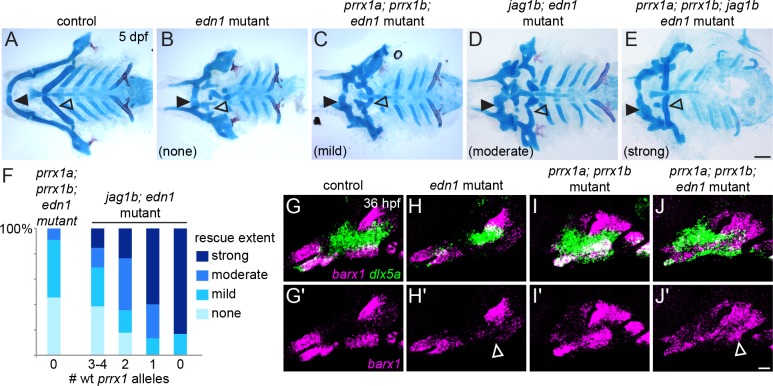
Simultaneous loss of Prrx1 genes and Jagged-Notch signaling further improves ventral skeletal development in edn1 mutants
Because ventral prrx1a/b expression increases in the ventral arches of edn1 mutants (Fig 6C and 6D), we speculated that increased repression of cartilage differentiation by Prrx1 proteins might contribute to the ventral skeletal losses seen in edn1 mutants. Indeed, we found that homozygous loss of both prrx1a and prrx1b resulted in a modest rescue of ventral cartilage formation in edn1 mutants, particularly in the second arch (Fig 9B and 9C), as well as rescue of ventral barx1 in the second arch and dlx5a expression in both the first and second arch (Fig 9G–9J). The partial rescue of ventral cartilage in prrx1a; prrx1b; edn1 mutants, as well as the earlier recovery of barx1 and dlx5a expression, are qualitatively similar to the phenotypes seen in jag1b; edn1 mutants (Fig 9D and [17]). By contrast, there was no rescue of ventral hand2 expression, consistent with our finding that Prrx1a/b do not regulate hand2 (S4 Fig). As Hand2 normally restricts Dlx expression into the ventral-most arches [54, 55], the lack of hand2 recovery in the triple mutants may explain the ectopic ventral expansion of dlx5a in the prrx1a; prrx1b; edn1 mutants.
Fig 9. Simultaneous loss of prrx1a/b and jag1b further improves ventral cartilages in edn1 mutants.
(A-E) Ventral views of dissected facial skeletons. edn1 mutants have much reduced ventral cartilage in the first (black closed arrowhead) and second (black open arrowhead) arches. Whereas jag1b; edn1 and prrx1a; prrx1b; edn1 mutants show some restoration of ventral cartilage, primarily in the second arch, quadruple prrx1a; prrx1b; jag1b; edn1 mutants show a prominent rescue of second arch Ch cartilage (black open arrowhead) and increased length of first arch M cartilage (black closed arrowhead). (F) Quantification of skeletal rescue in prrx1a; prrx1b; edn1 mutants (left column) and jag1b; edn1 mutants with decreasing numbers of wild-type prrx1a/b alleles (e.g. 4 wild-type alleles = pure jag1b; edn1 mutants; 0 wild-type alleles = quadruple mutant). Examples of no, mild, moderate, or strong rescue of the ceratohyal cartilage are shown in (B-E). Numbers in each genotype, from left to right: 11, 13, 17, 15, 6. (G-J) edn1 mutants show a loss of ventral barx1 expression (magenta, white open arrowhead) and a partial reduction of dlx5a (green), which are partially restored in prrx1a; prrx1b; edn1 triple mutants. The magenta channel is shown by itself in (G'-J'). Scale bar in E = 100 μm; scale bar in J' = 20 μm.
In jag1b; edn1 mutants, the partial recovery of ventral barx1 expression correlated with zones where prrx1b expression was reduced to control levels (Fig 8I). We therefore asked whether the remaining areas of elevated Prrx1 expression in jag1b; edn1 mutants might account for the incomplete rescue. Consistently, we found that progressive reduction of prrx1a/b gene dosage in jag1b; edn1 mutants resulted in a progressively better rescue of ventral cartilages, with 5/6 quadruple homozygous prrx1a; prrx1b; jag1b; edn1 mutants showing a prominent rescue of the ventral second arch-derived Ch cartilage and improved elongation of the first arch-derived M cartilage (Fig 9E and 9F). However, even in these quadruple mutants, the ‘rescued’ ventral cartilages are still smaller than in wild types, and the dorsal skeletal phenotypes associated with jag1b and prrx1a; prrx1b mutants are still present. These findings reveal important parallel contributions of ectopic Prrx1 and Jagged-Notch activity to the ventral craniofacial defects of edn1 mutants, yet indicate that Edn1 has additional functions beyond inhibiting Prrx1 and Notch activity.
Discussion
Parallel roles of Jagged-Notch and Prrx1 genes in restraining cartilage differentiation in the pharyngeal arches
RNAseq analyses of facial NCCs confirmed our previous findings that Notch acts oppositely to Edn1 during pharyngeal arch development [17]. At a mechanistic level, this global analysis revealed that a major function of Notch signaling is to repress the expression of some of the most strongly upregulated genes in early arch development. These include the homologs of a number of genes implicated in mesenchymal condensation, chondrogenesis, and general skeletogenesis in mammals: e.g. barx1 [23, 31, 32], ctgfb [56], col6a1 and col6a6 [57, 58], and tbx22 [59, 60]. As we only profiled global gene expression patterns in mutants and overexpression embryos at 36 hpf, we cannot conclude whether Notch represses these highly-induced genes only at this later stage, or whether it also restrains their initial upregulation.
The concept of Notch limiting differentiation is becoming a common theme in many developmental and regeneration contexts. For example, sustained Notch signaling in preskeletogenic mesenchyme in vivo or mesenchymal progenitors in vitro severely abrogates cartilage formation, with cells inappropriately maintained in a precursor state [61–64]. Likewise, Notch signaling promotes regeneration of the caudal fin of zebrafish by maintaining the blastema in a proliferative, undifferentiated state [65, 66]. Though Notch can also promote differentiation in other contexts (e.g. stimulating maturation and hypertrophy in committed chondrocytes [reviewed by [67]]), our findings are consistent with the large body of literature describing roles for Notch in resisting differentiation of progenitor cell populations, in this case specifically in the dorsal arches.
Our genomic analysis also identified two Prrx1 homologs (prrx1a and prrx1b) as negative targets of Edn1 that function in parallel to Jagged-Notch signaling to restrain cartilage differentiation, yet these pathways appear to do so in different ways (Fig 10). jag1b and prrx1a/b are expressed in largely non-overlapping domains and are generally not required for the other’s expression. The skeletal phenotypes of Notch and Prrx1 mutants also differ in critical ways. Mutants in both pathways develop ectopic barx1+ condensations and malformed cartilages in the dorsal arches, but only prrx1a; prrx1b mutants form ectopic dorsal cartilage. One potential explanation is that ectopic barx1 expression persists in dorsal NCCs at later stages in jag1b but not prrx1a; prrx1b mutants. Perhaps, Jagged-Notch signaling is also required for ectopic barx1+ cells to progress to a sox9a+ chondrogenic state. Further, our fate-mapping studies show that dorsal arch NCCs expand less in jag1b mutants compared to wild types, which could be due to persistent barx1 expression restricting the proliferation of chondrogenic cells. However, loss of barx1 improved only a subset of skeletal defects in jag1b mutants, suggesting that skeletal changes in Notch-deficient embryos result from more than just ectopic barx1 expression. In prrx1a; prrx1b mutants, the release of dorsal cells from a transient barx1+ state may instead allow these cells to reach a critical threshold for making ectopic cartilage (as proposed for the Prrx1 mouse mutant [68]).
Fig 10. Model of genetic interactions regulating the timing of chondrogenesis in the zebrafish pharyngeal arches.
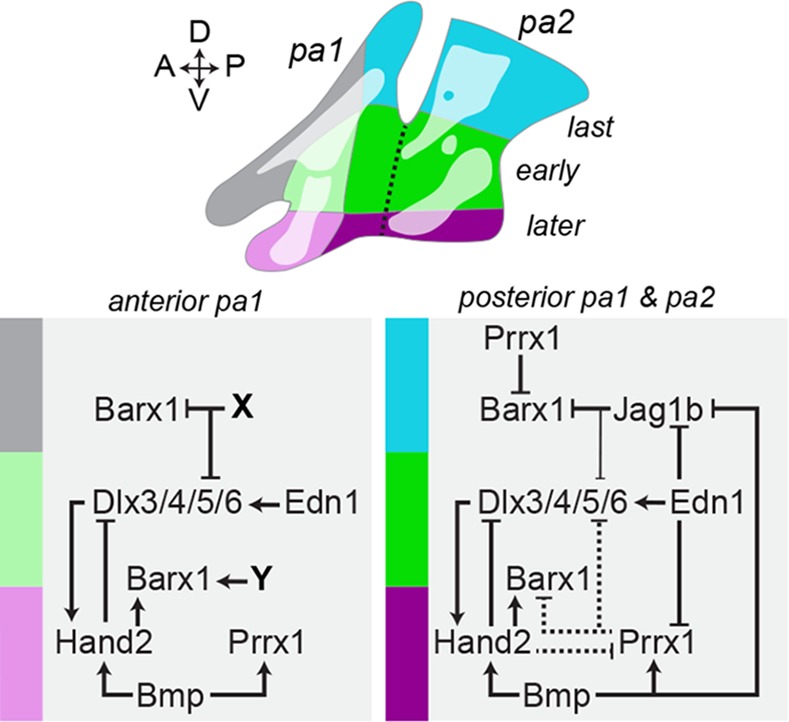
Schematic depicting the approximate origins of the facial cartilages in the early (green), later (purple) and latest (blue and grey) zones of chondrogenesis. In posterior pharyngeal arch 1 (pa1) and pharyngeal arch 2 (pa2), antagonism between Jag1b-Notch and Edn1/Bmp signaling restrains cartilage formation dorsally. Edn1 signaling, possibly acting through Dlx5/6 genes, suppresses dorsal jag1b [17] and ventral prrx1a/b expression in the intermediate domain (bright green), which allows the earliest initiation of barx1 expression and chondrogenic differentiation. In the ventral domain (purple), Bmp signaling helps to repress intermediate/ventral jag1b expression [13], while persistent expression of prrx1a/b–potentially driven by Bmp signaling in a Hand2-independent manner–may help to explain the later onset of chondrogenesis. Hand2 promotes barx1 [32] while limiting prrx1a (but not prrx1b) and ventral Dlx expansion [54]. In the dorsal domain (blue), stronger and/or longer repression of barx1 by combined Prrx1 and Jagged-Notch signaling results in the latest chondrogenic differentiation. Jag1b weakly represses the dorsal expansion of Dlx genes [17]. Prrx1a/b and Jag1b also feed back onto Edn1 signaling to help restrict dlx5a expression to the intermediate domain. In anterior pa1, a similar sequence of intermediate to ventral to dorsal/maxillary chondrogenesis is observed, yet Jag1b-Notch and Prrx1 seem to play a less important role, potentially due to different factors (X, Y) restricting barx1 in the maxillary domain (grey) and promoting barx1 in the mandibular domain.
The finding that prrx1a; prrx1b double mutants presented skeletal defects only in dorsal elements was somewhat unexpected, given the expression of prrx1a and prrx1b in both dorsal and ventral arch regions. While homozygous or dominant-negative mutations in PRRX1 have been associated with loss of the lower jaw in humans [69–73], Prrx1 mutant mice are similar to zebrafish mutants in displaying ectopic dorsal cartilage, but dissimilar in showing minor abnormalities of the lower jaw [21]; Prrx1; Prrx2 double mutants display much more pronounced jaw reductions [22, 48, 53]. As zebrafish lack a Prrx2 homolog [74, 75], the lack of lower jaw defects in prrx1a; prrx1b double mutants could reflect redundancy with other pathways, or, alternatively, the evolution of different requirements for Prrx1 genes between fish and mammals. At a molecular level, the ectopic barx1 expression we observe in the dorsal arches of prrx1a; prrx1b mutant fish is reminiscent of the medial expansion of Barx1 seen in the ventral first arch of Prrx1; Prrx2 mutant mice [53].
While our data implicate Prrx1 genes and Jagged-Notch as two important negative targets of Edn1 in the ventral arches of zebrafish, the fact that edn1 mutant phenotypes are only partially rescued by the combined loss of Prrx1a/b and Jag1b suggests other yet to be identified key targets of Edn1. Indeed, our RNAseq analysis revealed two different classes of genes activated by Edn1: (1) those that are highly upregulated during early arch development and also inhibited by Notch (including many well-known Edn1 targets such as dlx3b/4a/4b/6a, hand2, epha4b, Evf1/2, and msxe) and (2) those that are Notch-independent and only modestly upregulated during early arch development. Given that jag1b itself is negatively regulated by Edn1 signaling [17], many of the genes on the first list may in fact be Notch targets that are only indirectly stimulated by Edn1. Functional interrogation of these two classes of target genes should help uncover additional functions of Edn1 in arch development.
Our findings in zebrafish also support a greater role for the Jagged-Notch and Prrx1 pathways in patterning the second arch and posterior half of the first arch compared with the anterior portion of the first arch, which generates the bulk of the lower and upper jaw skeleton (Fig 10). For example, jag1b is expressed in only a limited posterior dorsal domain of the first arch and not in the maxillary or mandibular prominences [17], and first arch-derived skeletal structures are less affected than second arch-derived structures in jag1b and prrx1a; prrx1b mutants. barx1 expression is also primarily lost in ventral NCCs of the second but not first arch in Edn1 pathway mutants [10, 33], consistent with the more pronounced upregulation of jag1b and prrx1a/b in this domain. This second arch bias is also reflected by greater rescue of second versus first arch ventral cartilages upon loss of Prrx1 and Notch signaling in edn1 mutants. Given the very different arrangements of cartilage and bone in the second versus first arch, it is not surprising that programs that restrict cartilage formation have distinct roles in each arch. In the future, it will be interesting to explore how Hoxa2 and Hoxb2, which confer second arch identity [76–79], impact the Notch- and Prrx1-based cartilage restriction programs we have identified.
We also note that other major signaling pathways, such as Bmp, Fgf, Tgfβ, Shh, and Wnt, also influence the spatiotemporal patterns of differentiation within the arches–including control of prrx1a/b and barx1 expression [13, 14, 25, 29, 80–86]. For example, our work indicates that Bmp signaling likely helps to establish prrx1a/b expression at the ventral poles of the zebrafish arches. Whether Bmp signaling regulates Prrx1 genes in other vertebrates remains unclear, as previous studies did not detect changes in Prrx1 expression in conditional Bmp4 deletion mice [15] or chicken mandibular explants exposed to exogenous Bmp ligands and antagonists [80]. Future work will need to integrate these other key patterning programs into the model to more fully explain how the timing and extent of chondrogenesis is precisely controlled in the developing face.
Shaping the facial skeleton through temporal control of chondrogenic differentiation
Heterochrony in skeletal differentiation is an important mechanism contributing to the evolution of morphological differences between species [reviewed by [87], also see [88, 89] and references therein]. This concept of variation in developmental timing of homologous structures between species has been proposed to explain, for example, differences in beak length and morphology, as well as the shape of Meckel’s cartilage between quail and duck [90–92]. Our work supports the idea that differential developmental timing can also be a critical driving force for varying skeletal structure within an individual. In the arches of zebrafish, chondrocyte differentiation invariably occurs first in intermediate/ventral before dorsal regions [47]. We have found that these events are presaged by an earlier initiation of barx1 and sox9a expression in intermediate/ventral relative to dorsal arch NCCs, with Jagged-Notch and Prrx1a/b circumscribing the size of the later-forming dorsal condensations.
In a previously proposed ‘hinge-and-caps’ model [93, 94], arch polarity is established by differential signaling in the intermediate regions of the arches (i.e., ‘hinges’) versus the dorsal and ventral poles of the arches (i.e., ‘caps’). Our work provides potential cellular correlates to these hinges and caps in zebrafish, particularly in the second arch and posterior portion of the first arch [95]. We propose that the poles of the arches, or caps, represent progenitor domains, consistent with their expression of the mesenchyme progenitor marker Prrx1 in many species [22, 80, 96]. In contrast, the intermediate arches, or hinges, reflect the sites of initial chondrogenesis, as evidenced by their earlier expression of sox9a. Whereas this model predicts that Prrx1 expression at both the ventral and dorsal poles would restrict chondrogenesis relative to the intermediate hinges, dorsal-specific Jagged-Notch signaling would further restrict chondrogenesis in dorsal relative to ventral regions. This model would explain our observations that cartilages generally form first in the intermediate regions, then spread next to the ventral pole, and lastly to the dorsal pole due to combined repressive effects of Prrx1 and Jagged-Notch. However, the timing of cartilage differentiation is clearly more complex. For example, our time-lapse imaging revealed that the Ch cartilage first undergoes chondrogenesis at its tips and then later in its center, potentially correlating with expression of the Bmp target gene msxe in a subset of ventral second arch Ch precursors [14]. Hence, layering of additional signaling pathways, such as Bmp, may further refine the timing of cartilage differentiation within the arches.
Given the expression of Prrx1 homologs at the ventral and dorsal poles of the arches from sharks through mammals [96], and conserved expression of Jag1 in the dorsal arches of mice [97, 98], it appears likely that a similarly regulated intermediate—ventral—dorsal gradient of chondrogenesis may be conserved across vertebrates. For example, in human embryos, Meckel’s cartilage (ventral) differentiates before those elements that form in more proximal/dorsal positions (i.e. the malleus, sphenoid, and styloid process) [99]. On the other hand, differences in the timing and extent of cartilage differentiation might account for the striking differences in facial form between species. In larval zebrafish, the majority of the bony visceral skeleton arises through cartilage templates in the first two arches. In contrast, much of the mammalian facial skeleton forms through direct ossification, with exceptions including Meckel’s cartilage in the lower jaw and the ossicles of the middle ear. These differences might be reflected in the fact that loss of the pre-cartilage marker Barx1/barx1 has more profound effects on the facial skeleton of zebrafish than mice [32, 100, 101], and, reciprocally, that loss of Prrx1 genes impacts lower jaw development in mammals but not fish [21, 22, 48]. It will therefore be interesting to examine whether differences in the requirements and/or regulation of Prrx1 and Barx1 genes underlie differences in the extent and timing of chondrogenesis between species.
An unanswered question is how heterochrony in cartilage differentiation might translate to the distinct shapes of skeletal elements along the dorsoventral axis. One possibility is that dorsoventral differences in the timing at which progenitors commit to a cartilage fate influences the duration and types of signals they encounter from the surrounding endoderm and ectoderm. For example, Jagged-Notch signaling in the dorsal posterior second arch would protect progenitors from early chondrogenesis, thus allowing these cells to receive later osteogenic cues that direct them to form the large, fan-shaped opercle bone. Such an interpretation is consistent with the reciprocal expansion of barx1+ pre-cartilage condensations and loss of opercle bone in jag1b mutants, and the formation of an ectopic opercle bone upon forced expression of JAG1 in ventral regions [17]. Another possibility is that the timing of condensation formation and subsequent chondrogenesis influences the degree of proliferative expansion of elements in different arch domains [1]. In conclusion, our study revisits heterochrony, a fundamental concept of evolutionary biology, from a developmental perspective, showing that the timing and extent of cartilage differentiation within specific arch regions contributes to the diversity of skeletal shapes within the skull.
Materials and Methods
Ethics statement
All zebrafish (Danio rerio) were maintained and handled in strict accordance with good animal practices as defined by the relevant national and local animal welfare bodies. Zebrafish embryos were anesthetized for time-lapse imaging or prior to fixation by adding tricaine to their water. All animal experiments performed in this study were approved by the Institutional Animal Care and Use Committee of the University of Southern California (No. 10885, 20193).
Zebrafish lines
Zebrafish (Danio rerio) embryos were reared at 28.5°C and staged as previously described [102]. The following transgenic lines were maintained as heterozygotes: Tg(fli1a:EGFP)y1 [103], Tg(sox10:DsRed-Express)el10 [104], Tg(col2a1aBAC:GFP) [105], Tg(hsp70I:Gal4)kca4/+ and Tg(UAS:myc-Notch1a-intra)kca3 (hereafter UAS:NICD) [106], Tg(UAS:Edn1;α-crystallin:Cerulean)el249 and Tg(UAS:Bmp4;cmlc2:GFP)el49 (hereafter UAS:Edn1 and UAS:Bmp4, respectively) [14]. The hsp70I:Gal4 and UAS:NICD lines do not contain selectable markers and were genotyped using primers for Gal4 (F: 5′-CTCCCAAAACCAAAAGGTCTCC-3′; R: 5′-TGAAGCCAATCTATCTGTGACGG-3′) and UAS:NICD (F: 5’-CATCGCGTCTCAGCCTCAC-3’; R: 5’-CGGAATCGTTTATTGGTGTCG-3’). For the UAS:Edn1 line, in cases where it was not possible to ascertain α-crystallin:Cerulean expression in living animals, individuals carrying the transgene were identified by genotyping for the lens marker (F: 5’-TGGTGCAGATGAACTTCAGG-3’ and R: 5’- GCATGCAGACAGCAGCAATA-3’). Gal4 expression was induced in hsp70I:Gal4; UAS:NICD, hsp70I:Gal4; UAS:Edn1, and hsp70I:Gal4; UAS:Bmp4 embryos by heat-shocking from 20–24 hpf in a 40°C incubator. The sucker/edn1tf216 [5], jag1bb1105 [17], barx1fh331 [32], notch3fh332 [107], and Df(Chr1)hand2s6 [108, 109] mutant lines were described previously and genotyped by PCR using GoTaq (Promega, Madison, WI) with published primer sequences followed by digestion with the appropriate restriction enzymes.
Three new mutant lines (notch2el515, prrx1ael558, prrx1bel491) were generated for this study via TALEN-mediated mutagenesis. The notch2el515 allele was generated with the same TALEN pair used for the previously reported notch2el517 allele [110]. Exon 2 (of 4) of prrx1a was targeted with TALENs that recognize the following sequences: Left: 5’-CGTTGAGCTGCTCGTCTGGA-3’; Right: 5’-TGTTTCGCCTCTGTTTACGC-3’, and exon 1 (of 5) of prrx1b was targeted with TALENs that recognize the following sequences: Left: 5’-TGGCGAAACGGGCAGGACTA-3’; Right: 5’-TGTATCACTGCCACTCGTTA-3’. TALEN constructs were produced using a PCR-based platform [111]. The TALEN plasmids were linearized by StuI digestion (New England Biolabs, Ipswich, MA), and RNAs were synthesized with the mMessage mMachine T7 Ultra kit (Ambion/Life Technologies, Carlsbad, CA, USA). TALEN RNAs (100 ng/μl) were injected into 1‐cell-stage embryos. Germline founders were identified among the injected individuals by screening outcrossed progeny by PCR followed by restriction digestion. The primers used to identify mutations in each gene are listed in S7 Table. Stable mutant alleles predicted to result in immediate stop codons or frameshifts followed by stop codons were identified by sequencing PCR products in the F1 generation. The notch2el515 allele consists of a 2-bp deletion and a single nucleotide polymorphism (SNP) that destroy a ClaI site in the target region and result in an immediate stop after aa 208 (of 2471), within the extracellular EGF-like domains. The prrx1ael558 allele is an 8-bp deletion that destroys a BseRI site and produces a frameshift after aa 90 (of 245; upstream of the homeodomain at aa 101–155), causing the addition of one incorrect amino acid followed by a stop codon. The prrx1bel491 allele is a 2-bp insertion that abolishes a HinfI site and causes a frameshift after aa 68 (of 245; upstream of the homeodomain at aa 87–165), resulting in the addition of four incorrect amino acids followed by a stop codon. Two additional alleles, prrx1ab1246 and prrx1bb1247, were independently generated via CRISPR-mediated mutagenesis. CRISPR gRNA templates were produced via PCR following a published protocol [112], and gRNAs were synthesized with the MEGAScript T7 transcription kit (Ambion) and column-purified with the mirVana miRNA isolation kit (Ambion). Cas9 RNA was transcribed from pT3TS-nCas9n with the T3 mMessage kit (Ambion) and purified with an RNeasy Mini Kit (Qiagen, Hilden, Germany) [112]. gRNAs (25 ng/μl) plus Cas9 RNA (50 ng/μl) were injected into 1‐cell-stage embryos, and stable lines were identified by sequencing as described above. The prrx1ab1246 allele is an 11-bp deletion that causes a frameshift after aa 62, which results in the incorporation of 28 additional amino acids followed by a stop codon. The prrx1bb1247 allele consists of an 8-bp deletion that causes a frameshift after aa 24 and the inclusion of 29 incorrect amino acids before termination. All animal experiments performed in this study were approved by the Institutional Animal Care and Use Committee of the University of Southern California.
Preparation of FACS-sorted cell populations for RNA sequencing
For RNA sequencing experiments, fli1a:EGFP fish were crossed to the sox10:DsRed line, and doubly transgenic fli1a:EGFP; sox10:DsRed fish were further crossed to the edn1, jag1b, hsp70I:Gal4, UAS:Edn1, and UAS:NICD lines. Each of these lines were then separately incrossed to generate embryos for FACS sorting. Wild-type fli1a:EGFP; sox10:DsRed (20, 28, and 36 hpf) embryos were sorted for co-expression of GFP and DsRed expression under a fluorescent dissecting stereomicroscope (Leica M165 FC, Wetzlar, Germany) prior to dissociation. Single-positive and double-negative embryos were also saved as controls for FACS. Mutant edn1; fli1a:EGFP; sox10:DsRed embryos were selected under the fluorescent microscope at approximately 34 hpf based on the reduced distance between the bottom of the first pharyngeal pouch and the ventral border of the arches. To identify jag1b mutants and doubly-transgenic hsp70I:Gal4; UAS:Edn1 or hsp70I:Gal4; UAS:NICD individuals, we genotyped cell lysates of tail biopsies collected from anesthetized individual 24-hpf fli1a:EGFP; sox10:DsRed double-positive embryos. To induce Edn1 or NICD overexpression in the hsp70I:Gal4; UAS:Edn1 and hsp70I:Gal4; UAS:NICD lines, embryos were heat-shocked from 20–24 hpf in an incubator set at 40°C. As another means of inhibiting Notch signaling, fli1a:EGFP; sox10:DsRed embryos were treated with dibenzazepine (DBZ; Tocris, Bristol, UK; final concentration of 10 μM in embryo medium) from 24–36 hpf. The number of embryos used for each sort and the number of cells obtained are presented in S9 Table.
To facilitate FACS analyses at the 36 hpf time point, embryos were moved at 27 hpf to an incubator set at 22°C to delay their development such that they reached an approximation of the 36 hpf stage the following morning. fli1a:EGFP; sox10:DsRed double-positive embryos were dissociated following [113], with minor modifications. Briefly, 30–40 dechorionated embryos were incubated in fresh Ringer’s solution for 5–10 minutes and agitated by pipetting to remove the yolk. The deyolked embryos were then mixed with a protease solution containing 0.25% trypsin (Life Technologies), 1 mM EDTA, and 2 mg/ml Collagenase P (Roche Life Science, Indianapolis, IN) in PBS and incubated at 28.5°C for 15 min, pipetting up and down every 5 min to aid the dissociation. The reaction was stopped by the addition of a 6x stop solution consisting of 6 mM CaCl2 and 30% fetal bovine serum (FBS) in PBS. The cells were pelleted via centrifugation at 2000 rpm for 5 min at 4°C, resuspended in suspension medium (1% FBS, 0.8 mM CaCl2, 50 U/ml penicillin, and 0.05 mg/ml streptomycin (Sigma-Aldrich, St. Louis, MO) in phenol red-free Leibovitz’s L15 medium (Life Technologies)), pelleted again as above, and then resuspended in 500 μl suspension medium and placed on ice. Cells were sorted by FACS for GFP and DsRed expression on a MoFlo Astrios instrument (Beckman-Coulter, Brea, CA, USA). GFP/DsRed double-positive, double-negative, and single-positive populations were collected directly into RLT lysis buffer (Qiagen). Total RNA was immediately extracted using the RNeasy Micro kit (Qiagen) following the manufacturer’s protocol and quantified on a NanoDrop 2000 spectrophotometer (NanoDrop Products, Wilmington, DE, USA).
cDNA library preparation and RNA sequencing
The quality and quantity of extracted RNA were assessed on a Bioanalyzer Pico RNA chip (Agilent, Santa Clara, CA). cDNA was then made from the extracted RNA using the SMARTer V3 kit (Clontech, Mountain View, CA), according to the manufacturer’s instructions. The number of amplification cycles for cDNA synthesis was estimated based on input amounts of RNA. The size and amount of the resulting cDNA were then confirmed by Bioanalyzer. Sonication was performed on a S2 ultrasonicator (Covaris, Woburn, MA) according to Clontech’s recommended conditions. DNA libraries were constructed using the Kapa Hyper prep kit (Kapa Biosystems, Wilmington, MA) and NextFlex adapters (Bioo Scientific, Austin, TX). Libraries were visualized by Bioanalyzer analysis and quantified by qPCR (Kapa library quantification kit). Sequencing was performed on Illumina HiSeq 2000 (50-bp paired end reads) and NextSeq 500 (75-bp paired end reads) machines (Illumina, San Diego, CA). DNA libraries were constructed and sequencing was performed at the Norris Cancer Center Molecular Genomics Next Gen Sequencing Core at USC.
Sequencing data analysis
Raw sequencing data in Fastq format was imported into the Partek Flow interface for alignment and quantification. Pre-alignment QC showed that the reads from all samples had generally high quality, with the average Phred quality score for each sample being above 30. Reads were then trimmed from both ends based on Phred quality score with a minimum end quality level of 20 and a minimum acceptable read length of 25. The TopHat 2 algorithm was used to align the trimmed reads to the zebrafish GRCz10 genome assembly (Ensembl_v80). Aligned reads were then quantified using the Partek E/M algorithm with default parameters to yield the RPKM values. RNAseq files have been deposited in NCBI’s Gene Expression Omnibus and are accessible through the GEO Series accession number GSE72985. Filtered gene lists were derived in MS Excel as described in the Results section. Six genes on the list of arch NCC-enriched genes had passed the ≥ 3 RPKM threshold at 36 hpf but had RPKM values of 0 in the 20 hpf sample, leading to a division error that would have precluded their inclusion in the temporal expression analysis; we thus set the 20 hpf RPKM value for these genes to 0.01 based on the lowest positive RPKM values in the dataset.
Skeletal analysis and in situ hybridization
Alcian Blue and Alizarin Red staining to detect cartilage and bone, respectively, was performed on 4–6 dpf larvae as previously described [114]. Two-color fluorescent in situ hybridizations were carried out as previously reported [17]. Published probes used in this study include dlx2a [115], dlx5a [10], notch2, jag1b [17], and sox9a [44]. Partial cDNAs for barx1, notch1a, notch1b, notch3, prrx1a, and prrx1b were cloned into the pCR-Blunt II-TOPO vector (Life Technologies) and sequence-verified prior to plasmid linearization and in vitro transcription with Sp6 or T7 polymerase (Roche) (S8 Table).
Notch inhibitor treatments
To determine when Notch signaling affects skeletal patterning, we treated embryos with the γ-secretase inhibitor DBZ. DBZ dissolved in dimethyl sulfoxide (DMSO; 10 mM stock) was added to embryo medium to a final concentration of 10 μM. Embryos (n = 30–50 per treatment) were incubated in this solution starting at 8, 24, or 28 hpf until fixation at 36 (8 hpf group) or 42 hpf (other groups) for in situs or at 4 dpf for Alcian and Alizarin staining. In the groups used for skeletal staining, the DBZ solution was refreshed at 48 hpf and thoroughly washed out at 56 hpf. Clutch-mate controls were exposed to the same concentration of DMSO. Embryos were dechorionated at 24 hpf to improve drug accessibility.
Fate maps
Fate-mapping was performed with the green-to-red photoconvertible kikGR protein [116]. kikGR RNA was injected into embryos from a jag1b; fli1a:EGFP cross at the one-cell stage. At 36 hpf, embryos were anesthetized in Tricaine (Sigma-Aldrich) and mounted in 0.2% agarose for confocal imaging. Small groups of cells in the dorsal arches were selected using the region of interest tool in the Zeiss LSM software and exposed to the UV 405 laser until the red photoconverted protein became apparent (typically ~10 seconds using 50% laser power). The same animals were reimaged at 6 dpf to determine the destination of the photoconverted cells and then genotyped for the jag1bb1105 mutation.
Imaging
Confocal images of in situ hybridizations (~30 μM z-stacks) were captured on a Zeiss LSM5 microscope using ZEN software. Time-lapse imaging of doubly transgenic fli1a:EGFP; sox10:DsRed and col2a1aBAC:GFP; sox10:DsRed larvae followed [117], with ~130 μM of z-stacks collected every 10 or 12 minutes starting at 48 hpf. Skeletal preparations were photographed using a Leica DM2500 microscope. Image levels were adjusted in Adobe Photoshop CS6, with care taken to apply identical adjustments to images from the same data set and to avoid removing information from the image.
Data analysis
To analyze changes in gene expression between 20 and 28 hpf and 28 and 36 hpf for the total arch and Edn1- and Notch-regulated gene lists, we first calculated the median and quartile values for each list. The full lists were then collectively compared first by a Kruskal-Wallis test and then pairwise by Mann-Whitney U tests, with the Bonferroni correction applied to an α-value of 0.05 to account for multiple comparisons. Chi-square was used to compare the proportions of embryos showing different skeletal phenotypes in jag1b, barx1, and jag1b; barx1 mutants, with p < 0.05 considered significant. JMP 7.0 software (SAS) was used for statistical analysis. Numbers for each experiment are presented in S10 Table.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
jag1b expression (magenta) in posterior-dorsal cells of the first and second arches is indistinguishable between controls (A) and barx1 mutants (B) at 36 hpf. dlx2a (green) marks all arch NCCs. Maximum intensity projections of confocal z-stacks. Scale bar = 20 μm.
(TIF)
(A-D) Expression of Notch receptors in the pharyngeal arches. At 36 hpf, notch1a (A, magenta) and notch1b (B, magenta) are expressed in the ectodermal cleft adjacent to the first pharyngeal pouch but are undetectable in dlx2a+ NCCs (green). By contrast, notch2 (C) is strongly expressed in intermediate/dorsal NCCs (white arrow), and notch3 is expressed in dorsal second arch NCCs (white arrow), arch core mesoderm (dlx2a-negative, orange arrow), and the first ectodermal cleft (white arrowhead). (E-I) Mutation of notch2 (F) or notch3 (G) alone does not affect skeletal patterning, though a subset of notch2-/-; notch3+/- mutants (H) show some jag1b-like dorsal skeletal defects. Combined loss of both genes (I) results in a severe phenotype (two examples shown in I, I') consisting of a significant reduction in the size of the Hm and a shift towards a more posterior position (black open arrowhead), variable fusions of the second arch joint (black arrow), and abnormalities in the posterior Pq (black arrowhead). The overall size reduction and failure of bone mineralization are likely non-specific consequences of cardiac edema. Scale bar in D = 20 μm; scale bar in I = 100 μm.
(TIF)
Individual confocal sections of a barx1 (magenta) and sox9a (green) in situ hybridization in a wild-type embryo at 48 hpf, showing that the two markers are largely mutually exclusive, with the exception of the ventral tip of the forming Meckel’s and ceratohyal cartilages (white arrowheads in z12). Scale bars in all panels = 20 μm.
(TIF)
In hand2 mutants, expression of prrx1a (A, B; magenta) is slightly upregulated (arrowhead) in ventral cells, and prrx1b (C, D; magenta) is unaltered. (E, F) hand2 expression (magenta) is unaffected in prrx1a; prrx1b mutants. dlx2a (green) marks all arch NCCs. (G-J) hand2 expression (magenta) is not rescued in prrx1a; prrx1b; edn1 triple mutants (J) compared with edn1 single mutants (I). Dashed lines indicate approximate arch boundaries. Scale bar = 20 μm.
(TIF)
(A, B) Independently derived prrx1ab1246; prrx1bb1247 double mutants (B, B') phenocopy prrx1ael558; prrx1bel491 mutants (Fig 7B). Note the expanded cartilage in the palatoquadrate (black arrowhead) and the fusions between the hyomandibula and otic cartilage (black arrow). Scale bar = 100 μm. (C) Schematic of prrx1a/b TALEN (el558, el491) and CRISPR (b1246, b1247) alleles. In all cases, the mutant allele produces a frameshift (black) and premature stop codon upstream of the homeobox (Hx) and OAR domains (O).
(TIF)
Time-lapse movie of a fli1a:EGFP; sox10:DsRed embryo showing the invariant sequence of emergence of differentiating chondrocytes (sox10:DsRed+, magenta) from arch ectomesenchyme (fli1a:EGFP+, green) first in intermediate and then ventral elements and finally in the dorsal hyomandibula. Imaging of the 125-μM z-stack was initiated at 48 hpf and continued for 24 hpf. Stills from this movie are presented in Fig 2B.
(MP4)
Time-lapse movie of a col2a1aBAC:GFP; sox10:DsRed embryo showing that, similar to sox10:DsRed (magenta), a GFP transgene (green) driven by regulatory information in the col2a1a genomic locus is also upregulated in intermediate/ventral cartilages prior to the dorsal hyomandibula, though GFP expression can be detected several hours before DsRed becomes apparent, likely due to transgene effects. Imaging of the 150-μM z-stack was initiated at 48 hpf and continued for 24 hpf. Stills from this movie are presented in Fig 2C.
(MP4)
Acknowledgments
We thank Megan Matsutani and Jennifer DeKoeyer Crump for fish care, Lora Barsky at the USC Stem Cell Flow Cytometry Core Facility for assisting with the FACS analysis, and Charles Nicolet at the Norris Cancer Center Molecular Genomics Next Gen Sequencing Core at USC for performing the RNA sequencing. We also thank USC’s Norris Medical Library Bioinformatics Service for assisting with sequencing data analysis.
Data Availability
RNAseq files have been deposited in NCBI’s Gene Expression Omnibus and are accessible through the GEO Series accession number GSE72985. All other relevant data are contained within the paper and the Supporting Information files.
Funding Statement
This work was supported by the National Institute of Dental and Craniofacial Research (http://www.nidcr.nih.gov/) (R01 DE018405 to JGC and K99/R00 DE024190 to JTN), as well as by the March of Dimes (http://www.marchofdimes.org) (JGC), the National Institute on Deafness and Other Communication Disorders (http://www.nidcd.nih.gov/) (5T32DC009975 fellowship to LB) and the A.P. Giannini Foundation (http://apgianninifoundation.org/) (postdoctoral fellowship to LB). The bioinformatics software and computing resources used in the analysis were funded by the USC Office of Research and the Norris Medical Library. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kimmel CB, Miller CT, Kruze G, Ullmann B, BreMiller RA, Larison KD, et al. The shaping of pharyngeal cartilages during early development of the zebrafish. Dev Biol. 1998;203(2):245–63. Epub 1998/11/11. 10.1006/dbio.1998.9016 . [DOI] [PubMed] [Google Scholar]
- 2.Kurihara Y, Kurihara H, Suzuki H, Kodama T, Maemura K, Nagai R, et al. Elevated blood pressure and craniofacial abnormalities in mice deficient in endothelin-1. Nature. 1994;368(6473):703–10. Epub 1994/04/21. 10.1038/368703a0 . [DOI] [PubMed] [Google Scholar]
- 3.Clouthier DE, Hosoda K, Richardson JA, Williams SC, Yanagisawa H, Kuwaki T, et al. Cranial and cardiac neural crest defects in endothelin-A receptor-deficient mice. Development. 1998;125(5):813–24. Epub 1998/05/09. . [DOI] [PubMed] [Google Scholar]
- 4.Yanagisawa H, Yanagisawa M, Kapur RP, Richardson JA, Williams SC, Clouthier DE, et al. Dual genetic pathways of endothelin-mediated intercellular signaling revealed by targeted disruption of endothelin converting enzyme-1 gene. Development. 1998;125(5):825–36. Epub 1998/05/09. . [DOI] [PubMed] [Google Scholar]
- 5.Miller CT, Schilling TF, Lee K, Parker J, Kimmel CB. sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development. 2000;127(17):3815–28. Epub 2000/08/10. . [DOI] [PubMed] [Google Scholar]
- 6.Depew MJ, Lufkin T, Rubenstein JL. Specification of jaw subdivisions by Dlx genes. Science. 2002;298(5592):381–5. Epub 2002/08/24. 10.1126/science.1075703 . [DOI] [PubMed] [Google Scholar]
- 7.Beverdam A, Merlo GR, Paleari L, Mantero S, Genova F, Barbieri O, et al. Jaw transformation with gain of symmetry after Dlx5/Dlx6 inactivation: mirror of the past? Genesis. 2002;34(4):221–7. Epub 2002/11/16. 10.1002/gene.10156 . [DOI] [PubMed] [Google Scholar]
- 8.Ruest LB, Xiang X, Lim KC, Levi G, Clouthier DE. Endothelin-A receptor-dependent and -independent signaling pathways in establishing mandibular identity. Development. 2004;131(18):4413–23. Epub 2004/08/13. 10.1242/dev.01291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozeki H, Kurihara Y, Tonami K, Watatani S, Kurihara H. Endothelin-1 regulates the dorsoventral branchial arch patterning in mice. Mech Dev. 2004;121(4):387–95. Epub 2004/04/28. 10.1016/j.mod.2004.02.002 . [DOI] [PubMed] [Google Scholar]
- 10.Walker MB, Miller CT, Coffin Talbot J, Stock DW, Kimmel CB. Zebrafish furin mutants reveal intricacies in regulating Endothelin1 signaling in craniofacial patterning. Dev Biol. 2006;295(1):194–205. Epub 2006/05/09. 10.1016/j.ydbio.2006.03.028 . [DOI] [PubMed] [Google Scholar]
- 11.Walker MB, Miller CT, Swartz ME, Eberhart JK, Kimmel CB. phospholipase C, beta 3 is required for Endothelin1 regulation of pharyngeal arch patterning in zebrafish. Dev Biol. 2007;304(1):194–207. Epub 2007/01/24. 10.1016/j.ydbio.2006.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato T, Kurihara Y, Asai R, Kawamura Y, Tonami K, Uchijima Y, et al. An endothelin-1 switch specifies maxillomandibular identity. Proc Natl Acad Sci U S A. 2008;105(48):18806–11. Epub 2008/11/20. 10.1073/pnas.0807345105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexander C, Zuniga E, Blitz IL, Wada N, Le Pabic P, Javidan Y, et al. Combinatorial roles for BMPs and Endothelin 1 in patterning the dorsal-ventral axis of the craniofacial skeleton. Development. 2011;138(23):5135–46. Epub 2011/10/28. 10.1242/dev.067801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuniga E, Rippen M, Alexander C, Schilling TF, Crump JG. Gremlin 2 regulates distinct roles of BMP and Endothelin 1 signaling in dorsoventral patterning of the facial skeleton. Development. 2011;138(23):5147–56. Epub 2011/10/28. 10.1242/dev.067785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu W, Selever J, Murali D, Sun X, Brugger SM, Ma L, et al. Threshold-specific requirements for Bmp4 in mandibular development. Dev Biol. 2005;283(2):282–93. Epub 2005/06/07. 10.1016/j.ydbio.2005.04.019 . [DOI] [PubMed] [Google Scholar]
- 16.Bonilla-Claudio M, Wang J, Bai Y, Klysik E, Selever J, Martin JF. Bmp signaling regulates a dose-dependent transcriptional program to control facial skeletal development. Development. 2012;139(4):709–19. 10.1242/dev.073197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuniga E, Stellabotte F, Crump JG. Jagged-Notch signaling ensures dorsal skeletal identity in the vertebrate face. Development. 2010;137(11):1843–52. Epub 2010/05/01. 10.1242/dev.049056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeong J, Li X, McEvilly RJ, Rosenfeld MG, Lufkin T, Rubenstein JL. Dlx genes pattern mammalian jaw primordium by regulating both lower jaw-specific and upper jaw-specific genetic programs. Development. 2008;135(17):2905–16. Epub 2008/08/14. 10.1242/dev.019778 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cserjesi P, Lilly B, Bryson L, Wang Y, Sassoon DA, Olson EN. MHox: a mesodermally restricted homeodomain protein that binds an essential site in the muscle creatine kinase enhancer. Development. 1992;115(4):1087–101. . [DOI] [PubMed] [Google Scholar]
- 20.Kuratani S, Martin JF, Wawersik S, Lilly B, Eichele G, Olson EN. The expression pattern of the chick homeobox gene gMHox suggests a role in patterning of the limbs and face and in compartmentalization of somites. Dev Biol. 1994;161(2):357–69. Epub 1994/02/01. 10.1006/dbio.1994.1037 . [DOI] [PubMed] [Google Scholar]
- 21.Martin JF, Bradley A, Olson EN. The paired-like homeo box gene MHox is required for early events of skeletogenesis in multiple lineages. Genes Dev. 1995;9(10):1237–49. Epub 1995/05/15. . [DOI] [PubMed] [Google Scholar]
- 22.Lu MF, Cheng HT, Kern MJ, Potter SS, Tran B, Diekwisch TG, et al. prx-1 functions cooperatively with another paired-related homeobox gene, prx-2, to maintain cell fates within the craniofacial mesenchyme. Development. 1999;126(3):495–504. Epub 1999/01/07. . [DOI] [PubMed] [Google Scholar]
- 23.Tissier-Seta JP, Mucchielli ML, Mark M, Mattei MG, Goridis C, Brunet JF. Barx1, a new mouse homeodomain transcription factor expressed in cranio-facial ectomesenchyme and the stomach. Mech Dev. 1995;51(1):3–15. Epub 1995/05/01. . [DOI] [PubMed] [Google Scholar]
- 24.Thomas BL, Tucker AS, Qui M, Ferguson CA, Hardcastle Z, Rubenstein JL, et al. Role of Dlx-1 and Dlx-2 genes in patterning of the murine dentition. Development. 1997;124(23):4811–8. . [DOI] [PubMed] [Google Scholar]
- 25.Barlow AJ, Bogardi JP, Ladher R, Francis-West PH. Expression of chick Barx-1 and its differential regulation by FGF-8 and BMP signaling in the maxillary primordia. Dev Dyn. 1999;214(4):291–302. Epub 1999/04/23. . [DOI] [PubMed] [Google Scholar]
- 26.Martin JF, Olson EN. Identification of a prx1 limb enhancer. Genesis. 2000;26(4):225–9. . [PubMed] [Google Scholar]
- 27.Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33(2):77–80. Epub 2002/07/12. 10.1002/gene.10092 . [DOI] [PubMed] [Google Scholar]
- 28.Lorda-Diez CI, Montero JA, Diaz-Mendoza MJ, Garcia-Porrero JA, Hurle JM. Defining the earliest transcriptional steps of chondrogenic progenitor specification during the formation of the digits in the embryonic limb. PLoS One. 2011;6(9):e24546 10.1371/journal.pone.0024546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sperber SM, Dawid IB. barx1 is necessary for ectomesenchyme proliferation and osteochondroprogenitor condensation in the zebrafish pharyngeal arches. Dev Biol. 2008;321(1):101–10. Epub 2008/07/02. 10.1016/j.ydbio.2008.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meulemans D, Bronner-Fraser M. Insights from amphioxus into the evolution of vertebrate cartilage. PLoS One. 2007;2(8):e787 10.1371/journal.pone.0000787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meech R, Edelman DB, Jones FS, Makarenkova HP. The homeobox transcription factor Barx2 regulates chondrogenesis during limb development. Development. 2005;132(9):2135–46. 10.1242/dev.01811 . [DOI] [PubMed] [Google Scholar]
- 32.Nichols JT, Pan L, Moens CB, Kimmel CB. barx1 represses joints and promotes cartilage in the craniofacial skeleton. Development. 2013;140(13):2765–75. Epub 2013/05/24. 10.1242/dev.090639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clouthier DE, Williams SC, Yanagisawa H, Wieduwilt M, Richardson JA, Yanagisawa M. Signaling pathways crucial for craniofacial development revealed by endothelin-A receptor-deficient mice. Dev Biol. 2000;217(1):10–24. Epub 2000/01/08. 10.1006/dbio.1999.9527 . [DOI] [PubMed] [Google Scholar]
- 34.Zakrzewska A, Cui C, Stockhammer OW, Benard EL, Spaink HP, Meijer AH. Macrophage-specific gene functions in Spi1-directed innate immunity. Blood. 2010;116(3):e1–11. 10.1182/blood-2010-01-262873 . [DOI] [PubMed] [Google Scholar]
- 35.Thisse B, Pflumio S, Fürthauer M, Loppin B, Heyer V, Degrave A, et al. Expression of the zebrafish genome during embryogenesis. ZFIN Direct Data Submission (http://zfinorg). 2001. [Google Scholar]
- 36.Petko JA, Millimaki BB, Canfield VA, Riley BB, Levenson R. Otoc1: a novel otoconin-90 ortholog required for otolith mineralization in zebrafish. Dev Neurobiol. 2008;68(2):209–22. Epub 2007/11/15. 10.1002/dneu.20587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ichida JK, Tcw J, Williams LA, Carter AC, Shi Y, Moura MT, et al. Notch inhibition allows oncogene-independent generation of iPS cells. Nat Chem Biol. 2014;10(8):632–9. 10.1038/nchembio.1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis RL, Turner DL. Vertebrate hairy and Enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene. 2001;20(58):8342–57. 10.1038/sj.onc.1205094 . [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi T, Kageyama R. Expression dynamics and functions of Hes factors in development and diseases. Curr Top Dev Biol. 2014;110:263–83. . [DOI] [PubMed] [Google Scholar]
- 40.Golde TE, Koo EH, Felsenstein KM, Osborne BA, Miele L. gamma-Secretase inhibitors and modulators. Biochim Biophys Acta. 2013;1828(12):2898–907. 10.1016/j.bbamem.2013.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson GW, Mahon KA. Differential and overlapping expression domains of Dlx-2 and Dlx-3 suggest distinct roles for Distal-less homeobox genes in craniofacial development. Mech Dev. 1994;48(3):199–215. . [DOI] [PubMed] [Google Scholar]
- 42.Qiu M, Bulfone A, Ghattas I, Meneses JJ, Christensen L, Sharpe PT, et al. Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev Biol. 1997;185(2):165–84. Epub 1997/05/15. 10.1006/dbio.1997.8556 . [DOI] [PubMed] [Google Scholar]
- 43.Wright E, Hargrave MR, Christiansen J, Cooper L, Kun J, Evans T, et al. The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nat Genet. 1995;9(1):15–20. 10.1038/ng0195-15 . [DOI] [PubMed] [Google Scholar]
- 44.Yan YL, Miller CT, Nissen RM, Singer A, Liu D, Kirn A, et al. A zebrafish sox9 gene required for cartilage morphogenesis. Development. 2002;129(21):5065–79. . [DOI] [PubMed] [Google Scholar]
- 45.Eames BF, Sharpe PT, Helms JA. Hierarchy revealed in the specification of three skeletal fates by Sox9 and Runx2. Dev Biol. 2004;274(1):188–200. 10.1016/j.ydbio.2004.07.006 . [DOI] [PubMed] [Google Scholar]
- 46.Yan YL, Willoughby J, Liu D, Crump JG, Wilson C, Miller CT, et al. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. 2005;132(5):1069–83. Epub 2005/02/04. 10.1242/dev.01674 . [DOI] [PubMed] [Google Scholar]
- 47.Schilling TF, Kimmel CB. Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development. 1997;124(15):2945–60. Epub 1997/08/01. . [DOI] [PubMed] [Google Scholar]
- 48.ten Berge D, Brouwer A, Korving J, Martin JF, Meijlink F. Prx1 and Prx2 in skeletogenesis: roles in the craniofacial region, inner ear and limbs. Development. 1998;125(19):3831–42. Epub 1998/09/08. . [DOI] [PubMed] [Google Scholar]
- 49.Thomas T, Kurihara H, Yamagishi H, Kurihara Y, Yazaki Y, Olson EN, et al. A signaling cascade involving endothelin-1, dHAND and msx1 regulates development of neural-crest-derived branchial arch mesenchyme. Development. 1998;125(16):3005–14. . [DOI] [PubMed] [Google Scholar]
- 50.Charite J, McFadden DG, Merlo G, Levi G, Clouthier DE, Yanagisawa M, et al. Role of Dlx6 in regulation of an endothelin-1-dependent, dHAND branchial arch enhancer. Genes Dev. 2001;15(22):3039–49. 10.1101/gad.931701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barbosa AC, Funato N, Chapman S, McKee MD, Richardson JA, Olson EN, et al. Hand transcription factors cooperatively regulate development of the distal midline mesenchyme. Dev Biol. 2007;310(1):154–68. 10.1016/j.ydbio.2007.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cubbage CC, Mabee PM. Development of the cranium and paired fins in the zebrafish Danio rerio (Ostariophysi, cyprinidae). J Morphol. 1996;229(2):121–60. . [DOI] [PubMed] [Google Scholar]
- 53.Balic A, Adams D, Mina M. Prx1 and Prx2 cooperatively regulate the morphogenesis of the medial region of the mandibular process. Dev Dyn. 2009;238(10):2599–613. Epub 2009/09/25. 10.1002/dvdy.22092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Talbot JC, Johnson SL, Kimmel CB. hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development. 2010;137(15):2507–17. Epub 2010/06/25. 10.1242/dev.049700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barron F, Woods C, Kuhn K, Bishop J, Howard MJ, Clouthier DE. Downregulation of Dlx5 and Dlx6 expression by Hand2 is essential for initiation of tongue morphogenesis. Development. 2011;138(11):2249–59. 10.1242/dev.056929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arnott JA, Lambi AG, Mundy C, Hendesi H, Pixley RA, Owen TA, et al. The role of connective tissue growth factor (CTGF/CCN2) in skeletogenesis. Crit Rev Eukaryot Gene Expr. 2011;21(1):43–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alexopoulos LG, Youn I, Bonaldo P, Guilak F. Developmental and osteoarthritic changes in Col6a1-knockout mice: biomechanics of type VI collagen in the cartilage pericellular matrix. Arthritis Rheum. 2009;60(3):771–9. 10.1002/art.24293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Christensen SE, Coles JM, Zelenski NA, Furman BD, Leddy HA, Zauscher S, et al. Altered trabecular bone structure and delayed cartilage degeneration in the knees of collagen VI null mice. PLoS One. 2012;7(3):e33397 10.1371/journal.pone.0033397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pauws E, Hoshino A, Bentley L, Prajapati S, Keller C, Hammond P, et al. Tbx22null mice have a submucous cleft palate due to reduced palatal bone formation and also display ankyloglossia and choanal atresia phenotypes. Hum Mol Genet. 2009;18(21):4171–9. 10.1093/hmg/ddp368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Braybrook C, Doudney K, Marcano AC, Arnason A, Bjornsson A, Patton MA, et al. The T-box transcription factor gene TBX22 is mutated in X-linked cleft palate and ankyloglossia. Nat Genet. 2001;29(2):179–83. 10.1038/ng730 . [DOI] [PubMed] [Google Scholar]
- 61.Kohn A, Dong Y, Mirando AJ, Jesse AM, Honjo T, Zuscik MJ, et al. Cartilage-specific RBPjkappa-dependent and -independent Notch signals regulate cartilage and bone development. Development. 2012;139(6):1198–212. Epub 2012/02/23. 10.1242/dev.070649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dong Y, Jesse AM, Kohn A, Gunnell LM, Honjo T, Zuscik MJ, et al. RBPjkappa-dependent Notch signaling regulates mesenchymal progenitor cell proliferation and differentiation during skeletal development. Development. 2010;137(9):1461–71. Epub 2010/03/26. 10.1242/dev.042911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oldershaw RA, Tew SR, Russell AM, Meade K, Hawkins R, McKay TR, et al. Notch signaling through Jagged-1 is necessary to initiate chondrogenesis in human bone marrow stromal cells but must be switched off to complete chondrogenesis. Stem Cells. 2008;26(3):666–74. Epub 2008/01/15. 10.1634/stemcells.2007-0806 . [DOI] [PubMed] [Google Scholar]
- 64.Mead TJ, Yutzey KE. Notch pathway regulation of chondrocyte differentiation and proliferation during appendicular and axial skeleton development. Proc Natl Acad Sci U S A. 2009;106(34):14420–5. 10.1073/pnas.0902306106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Munch J, Gonzalez-Rajal A, de la Pompa JL. Notch regulates blastema proliferation and prevents differentiation during adult zebrafish fin regeneration. Development. 2013;140(7):1402–11. 10.1242/dev.087346 . [DOI] [PubMed] [Google Scholar]
- 66.Grotek B, Wehner D, Weidinger G. Notch signaling coordinates cellular proliferation with differentiation during zebrafish fin regeneration. Development. 2013;140(7):1412–23. 10.1242/dev.087452 . [DOI] [PubMed] [Google Scholar]
- 67.Chen S, Lee BH, Bae Y. Notch signaling in skeletal stem cells. Calcif Tissue Int. 2014;94(1):68–77. 10.1007/s00223-013-9773-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith KK, Schneider RA. Have gene knockouts caused evolutionary reversals in the mammalian first arch? Bioessays. 1998;20(3):245–55. Epub 1998/06/19. . [DOI] [PubMed] [Google Scholar]
- 69.Sergi C, Kamnasaran D. PRRX1 is mutated in a fetus with agnathia-otocephaly. Clin Genet. 2011;79(3):293–5. Epub 2011/02/08. 10.1111/j.1399-0004.2010.01531.x . [DOI] [PubMed] [Google Scholar]
- 70.Celik T, Simsek PO, Sozen T, Ozyuncu O, Utine GE, Talim B, et al. PRRX1 is mutated in an otocephalic newborn infant conceived by consanguineous parents. Clin Genet. 2012;81(3):294–7. Epub 2012/01/04. 10.1111/j.1399-0004.2011.01730.x . [DOI] [PubMed] [Google Scholar]
- 71.Herman S, Delio M, Morrow B, Samanich J. Agnathia-otocephaly complex: a case report and examination of the OTX2 and PRRX1 genes. Gene. 2012;494(1):124–9. Epub 2011/12/27. 10.1016/j.gene.2011.11.033 . [DOI] [PubMed] [Google Scholar]
- 72.Donnelly M, Todd E, Wheeler M, Winn VD, Kamnasaran D. Prenatal diagnosis and identification of heterozygous frameshift mutation in PRRX1 in an infant with agnathia-otocephaly. Prenat Diagn. 2012;32(9):903–5. Epub 2012/06/08. 10.1002/pd.3910 . [DOI] [PubMed] [Google Scholar]
- 73.Dasouki M, Andrews B, Parimi P, Kamnasaran D. Recurrent agnathia-otocephaly caused by DNA replication slippage in PRRX1. Am J Med Genet A. 2013;161(4):803–8. Epub 2013/02/28. 10.1002/ajmg.a.35879 . [DOI] [PubMed] [Google Scholar]
- 74.Hernandez-Vega A, Minguillon C. The Prx1 limb enhancers: targeted gene expression in developing zebrafish pectoral fins. Dev Dyn. 2011;240(8):1977–88. Epub 2011/06/16. 10.1002/dvdy.22678 . [DOI] [PubMed] [Google Scholar]
- 75.Braasch I, Guiguen Y, Loker R, Letaw JH, Ferrara A, Bobe J, et al. Connectivity of vertebrate genomes: Paired-related homeobox (Prrx) genes in spotted gar, basal teleosts, and tetrapods. Comparative biochemistry and physiology Toxicology & pharmacology. 2014;163:24–36. 10.1016/j.cbpc.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hunter MP, Prince VE. Zebrafish hox paralogue group 2 genes function redundantly as selector genes to pattern the second pharyngeal arch. Dev Biol. 2002;247(2):367–89. Epub 2002/06/28. . [DOI] [PubMed] [Google Scholar]
- 77.Gendron-Maguire M, Mallo M, Zhang M, Gridley T. Hoxa-2 mutant mice exhibit homeotic transformation of skeletal elements derived from cranial neural crest. Cell. 1993;75(7):1317–31. Epub 1993/12/31. . [DOI] [PubMed] [Google Scholar]
- 78.Rijli FM, Mark M, Lakkaraju S, Dierich A, Dolle P, Chambon P. A homeotic transformation is generated in the rostral branchial region of the head by disruption of Hoxa-2, which acts as a selector gene. Cell. 1993;75(7):1333–49. . [DOI] [PubMed] [Google Scholar]
- 79.O'Gorman S. Second branchial arch lineages of the middle ear of wild-type and Hoxa2 mutant mice. Dev Dyn. 2005;234(1):124–31. Epub 2005/04/30. 10.1002/dvdy.20402 . [DOI] [PubMed] [Google Scholar]
- 80.Doufexi AE, Mina M. Signaling pathways regulating the expression of Prx1 and Prx2 in the chick mandibular mesenchyme. Dev Dyn. 2008;237(11):3115–27. Epub 2008/10/23. 10.1002/dvdy.21762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alexander C, Piloto S, Le Pabic P, Schilling TF. Wnt signaling interacts with bmp and edn1 to regulate dorsal-ventral patterning and growth of the craniofacial skeleton. PLoS Genet. 2014;10(7):e1004479 10.1371/journal.pgen.1004479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tucker AS, Yamada G, Grigoriou M, Pachnis V, Sharpe PT. Fgf-8 determines rostral-caudal polarity in the first branchial arch. Development. 1999;126(1):51–61. Epub 1998/12/03. . [DOI] [PubMed] [Google Scholar]
- 83.David NB, Saint-Etienne L, Tsang M, Schilling TF, Rosa FM. Requirement for endoderm and FGF3 in ventral head skeleton formation. Development. 2002;129(19):4457–68. Epub 2002/09/12. . [DOI] [PubMed] [Google Scholar]
- 84.Crump JG, Maves L, Lawson ND, Weinstein BM, Kimmel CB. An essential role for Fgfs in endodermal pouch formation influences later craniofacial skeletal patterning. Development. 2004;131(22):5703–16. Epub 2004/10/29. 10.1242/dev.01444 . [DOI] [PubMed] [Google Scholar]
- 85.Larbuisson A, Dalcq J, Martial JA, Muller M. Fgf receptors Fgfr1a and Fgfr2 control the function of pharyngeal endoderm in late cranial cartilage development. Differentiation. 2013;86(4–5):192–206. Epub 2013/11/02. 10.1016/j.diff.2013.07.006 . [DOI] [PubMed] [Google Scholar]
- 86.Wurdak H, Ittner LM, Lang KS, Leveen P, Suter U, Fischer JA, et al. Inactivation of TGFbeta signaling in neural crest stem cells leads to multiple defects reminiscent of DiGeorge syndrome. Genes Dev. 2005;19(5):530–5. 10.1101/gad.317405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smith KK. Time's arrow: heterochrony and the evolution of development. Int J Dev Biol. 2003;47(7–8):613–21. . [PubMed] [Google Scholar]
- 88.Mitgutsch C, Wimmer C, Sanchez-Villagra MR, Hahnloser R, Schneider RA. Timing of ossification in duck, quail, and zebra finch: intraspecific variation, heterochronies, and life history evolution. Zoolog Sci. 2011;28(7):491–500. 10.2108/zsj.28.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koyabu D, Werneburg I, Morimoto N, Zollikofer CP, Forasiepi AM, Endo H, et al. Mammalian skull heterochrony reveals modular evolution and a link between cranial development and brain size. Nat Commun. 2014;5:3625 10.1038/ncomms4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eames BF, Schneider RA. The genesis of cartilage size and shape during development and evolution. Development. 2008;135(23):3947–58. 10.1242/dev.023309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schneider RA, Helms JA. The cellular and molecular origins of beak morphology. Science. 2003;299(5606):565–8. 10.1126/science.1077827 . [DOI] [PubMed] [Google Scholar]
- 92.Tucker AS, Lumsden A. Neural crest cells provide species-specific patterning information in the developing branchial skeleton. Evol Dev. 2004;6(1):32–40. . [DOI] [PubMed] [Google Scholar]
- 93.Depew MJ, Compagnucci C. Tweaking the hinge and caps: testing a model of the organization of jaws. J Exp Zool B Mol Dev Evol. 2008;310(4):315–35. 10.1002/jez.b.21205 . [DOI] [PubMed] [Google Scholar]
- 94.Depew MJ, Simpson CA, Morasso M, Rubenstein JL. Reassessing the Dlx code: the genetic regulation of branchial arch skeletal pattern and development. J Anat. 2005;207(5):501–61. Epub 2005/11/30. 10.1111/j.1469-7580.2005.00487.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Medeiros DM, Crump JG. New perspectives on pharyngeal dorsoventral patterning in development and evolution of the vertebrate jaw. Dev Biol. 2012;371(2):121–35. Epub 2012/09/11. 10.1016/j.ydbio.2012.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Compagnucci C, Debiais-Thibaud M, Coolen M, Fish J, Griffin JN, Bertocchini F, et al. Pattern and polarity in the development and evolution of the gnathostome jaw: both conservation and heterotopy in the branchial arches of the shark, Scyliorhinus canicula. Dev Biol. 2013;377(2):428–48. Epub 2013/03/12. 10.1016/j.ydbio.2013.02.022 . [DOI] [PubMed] [Google Scholar]
- 97.Mitsiadis TA, Henrique D, Thesleff I, Lendahl U. Mouse Serrate-1 (Jagged-1): expression in the developing tooth is regulated by epithelial-mesenchymal interactions and fibroblast growth factor-4. Development. 1997;124(8):1473–83. . [DOI] [PubMed] [Google Scholar]
- 98.Kiernan AE, Xu J, Gridley T. The Notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genet. 2006;2(1):e4 Epub 2006/01/18. 10.1371/journal.pgen.0020004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tanaka O. Time of the appearance of cartilage centers in human embryos with special reference to its individual difference. Okajimas Folia Anat Jpn. 1976;53(4):173–98. . [DOI] [PubMed] [Google Scholar]
- 100.Kim BM, Miletich I, Mao J, McMahon AP, Sharpe PA, Shivdasani RA. Independent functions and mechanisms for homeobox gene Barx1 in patterning mouse stomach and spleen. Development. 2007;134(20):3603–13. 10.1242/dev.009308 . [DOI] [PubMed] [Google Scholar]
- 101.Miletich I, Yu WY, Zhang R, Yang K, Caixeta de Andrade S, Pereira SF, et al. Developmental stalling and organ-autonomous regulation of morphogenesis. Proc Natl Acad Sci U S A. 2011;108(48):19270–5. 10.1073/pnas.1112801108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. 10.1002/aja.1002030302 . [DOI] [PubMed] [Google Scholar]
- 103.Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248(2):307–18. Epub 2002/08/09. . [DOI] [PubMed] [Google Scholar]
- 104.Das A, Crump JG. Bmps and id2a act upstream of Twist1 to restrict ectomesenchyme potential of the cranial neural crest. PLoS Genet. 2012;8(5):e1002710 Epub 2012/05/17. 10.1371/journal.pgen.1002710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Askary A, Mork L, Paul S, He X, Izuhara AK, Gopalakrishnan S, et al. Iroquois proteins promote skeletal joint formation by maintaining chondrocytes in an immature state. Dev Cell. 2015;35(3):358–65. 10.1016/j.devcel.2015.10.004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Scheer N, Campos-Ortega JA. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech Dev. 1999;80(2):153–8. Epub 1999/03/12. . [DOI] [PubMed] [Google Scholar]
- 107.Alunni A, Krecsmarik M, Bosco A, Galant S, Pan L, Moens CB, et al. Notch3 signaling gates cell cycle entry and limits neural stem cell amplification in the adult pallium. Development. 2013;140(16):3335–47. 10.1242/dev.095018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yelon D, Ticho B, Halpern ME, Ruvinsky I, Ho RK, Silver LM, et al. The bHLH transcription factor hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development. 2000;127(12):2573–82. . [DOI] [PubMed] [Google Scholar]
- 109.Sasaki MM, Nichols JT, Kimmel CB. edn1 and hand2 interact in early regulation of pharyngeal arch outgrowth during zebrafish development. PLoS One. 2013;8(6):e67522 Epub 2013/07/05. 10.1371/journal.pone.0067522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim AD, Melick CH, Clements WK, Stachura DL, Distel M, Panakova D, et al. Discrete Notch signaling requirements in the specification of hematopoietic stem cells. Embo J. 2014;33(20):2363–73. 10.15252/embj.201488784 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012;7(1):171–92. Epub 2012/01/10. 10.1038/nprot.2011.431 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A. 2013;110(34):13904–9. 10.1073/pnas.1308335110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Covassin L, Amigo JD, Suzuki K, Teplyuk V, Straubhaar J, Lawson ND. Global analysis of hematopoietic and vascular endothelial gene expression by tissue specific microarray profiling in zebrafish. Dev Biol. 2006;299(2):551–62. 10.1016/j.ydbio.2006.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Walker MB, Kimmel CB. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech Histochem. 2007;82(1):23–8. Epub 2007/05/19. 10.1080/10520290701333558 . [DOI] [PubMed] [Google Scholar]
- 115.Akimenko MA, Ekker M, Wegner J, Lin W, Westerfield M. Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J Neurosci. 1994;14(6):3475–86. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tsutsui H, Karasawa S, Shimizu H, Nukina N, Miyawaki A. Semi-rational engineering of a coral fluorescent protein into an efficient highlighter. EMBO Rep. 2005;6(3):233–8. 10.1038/sj.embor.7400361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Crump JG, Swartz ME, Kimmel CB. An integrin-dependent role of pouch endoderm in hyoid cartilage development. PLoS Biol. 2004;2(9):E244 Epub 2004/07/23. 10.1371/journal.pbio.0020244 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
jag1b expression (magenta) in posterior-dorsal cells of the first and second arches is indistinguishable between controls (A) and barx1 mutants (B) at 36 hpf. dlx2a (green) marks all arch NCCs. Maximum intensity projections of confocal z-stacks. Scale bar = 20 μm.
(TIF)
(A-D) Expression of Notch receptors in the pharyngeal arches. At 36 hpf, notch1a (A, magenta) and notch1b (B, magenta) are expressed in the ectodermal cleft adjacent to the first pharyngeal pouch but are undetectable in dlx2a+ NCCs (green). By contrast, notch2 (C) is strongly expressed in intermediate/dorsal NCCs (white arrow), and notch3 is expressed in dorsal second arch NCCs (white arrow), arch core mesoderm (dlx2a-negative, orange arrow), and the first ectodermal cleft (white arrowhead). (E-I) Mutation of notch2 (F) or notch3 (G) alone does not affect skeletal patterning, though a subset of notch2-/-; notch3+/- mutants (H) show some jag1b-like dorsal skeletal defects. Combined loss of both genes (I) results in a severe phenotype (two examples shown in I, I') consisting of a significant reduction in the size of the Hm and a shift towards a more posterior position (black open arrowhead), variable fusions of the second arch joint (black arrow), and abnormalities in the posterior Pq (black arrowhead). The overall size reduction and failure of bone mineralization are likely non-specific consequences of cardiac edema. Scale bar in D = 20 μm; scale bar in I = 100 μm.
(TIF)
Individual confocal sections of a barx1 (magenta) and sox9a (green) in situ hybridization in a wild-type embryo at 48 hpf, showing that the two markers are largely mutually exclusive, with the exception of the ventral tip of the forming Meckel’s and ceratohyal cartilages (white arrowheads in z12). Scale bars in all panels = 20 μm.
(TIF)
In hand2 mutants, expression of prrx1a (A, B; magenta) is slightly upregulated (arrowhead) in ventral cells, and prrx1b (C, D; magenta) is unaltered. (E, F) hand2 expression (magenta) is unaffected in prrx1a; prrx1b mutants. dlx2a (green) marks all arch NCCs. (G-J) hand2 expression (magenta) is not rescued in prrx1a; prrx1b; edn1 triple mutants (J) compared with edn1 single mutants (I). Dashed lines indicate approximate arch boundaries. Scale bar = 20 μm.
(TIF)
(A, B) Independently derived prrx1ab1246; prrx1bb1247 double mutants (B, B') phenocopy prrx1ael558; prrx1bel491 mutants (Fig 7B). Note the expanded cartilage in the palatoquadrate (black arrowhead) and the fusions between the hyomandibula and otic cartilage (black arrow). Scale bar = 100 μm. (C) Schematic of prrx1a/b TALEN (el558, el491) and CRISPR (b1246, b1247) alleles. In all cases, the mutant allele produces a frameshift (black) and premature stop codon upstream of the homeobox (Hx) and OAR domains (O).
(TIF)
Time-lapse movie of a fli1a:EGFP; sox10:DsRed embryo showing the invariant sequence of emergence of differentiating chondrocytes (sox10:DsRed+, magenta) from arch ectomesenchyme (fli1a:EGFP+, green) first in intermediate and then ventral elements and finally in the dorsal hyomandibula. Imaging of the 125-μM z-stack was initiated at 48 hpf and continued for 24 hpf. Stills from this movie are presented in Fig 2B.
(MP4)
Time-lapse movie of a col2a1aBAC:GFP; sox10:DsRed embryo showing that, similar to sox10:DsRed (magenta), a GFP transgene (green) driven by regulatory information in the col2a1a genomic locus is also upregulated in intermediate/ventral cartilages prior to the dorsal hyomandibula, though GFP expression can be detected several hours before DsRed becomes apparent, likely due to transgene effects. Imaging of the 150-μM z-stack was initiated at 48 hpf and continued for 24 hpf. Stills from this movie are presented in Fig 2C.
(MP4)
Data Availability Statement
RNAseq files have been deposited in NCBI’s Gene Expression Omnibus and are accessible through the GEO Series accession number GSE72985. All other relevant data are contained within the paper and the Supporting Information files.