Abstract
Both schizophrenia (SCZ) and autism spectrum disorders (ASD) are neuropsychiatric disorders with overlapping genetic etiology. Protocadherin 15 (PCDH15), which encodes a member of the cadherin super family that contributes to neural development and function, has been cited as a risk gene for neuropsychiatric disorders. Recently, rare variants of large effect have been paid attention to understand the etiopathology of these complex disorders. Thus, we evaluated the impacts of rare, single-nucleotide variants (SNVs) in PCDH15 on SCZ or ASD. First, we conducted coding exon-targeted resequencing of PCDH15 with next-generation sequencing technology in 562 Japanese patients (370 SCZ and 192 ASD) and detected 16 heterozygous SNVs. We then performed association analyses on 2,096 cases (1,714 SCZ and 382 ASD) and 1,917 controls with six novel variants of these 16 SNVs. Of these six variants, four (p.R219K, p.T281A, p.D642N, c.3010-1G>C) were ultra-rare variants (minor allele frequency < 0.0005) that may increase disease susceptibility. Finally, no statistically significant association between any of these rare, heterozygous PCDH15 point variants and SCZ or ASD was found. Our results suggest that a larger sample size of resequencing subjects is necessary to detect associations between rare PCDH15 variants and neuropsychiatric disorders.
Introduction
Schizophrenia (SCZ) and autism spectrum disorders (ASD) are neurodevelopmental in origin. While SCZ and ASD are regarded as separate clinical entities, etiological, clinical, and genetic overlap between them have been discovered [1,2]. Genetic factors make substantial contributions to the etiology of both conditions; heritability is estimated to be a minimum of 80% for each [3]. Thousands of trait- and disease-associated common genetic variants confer increased risk of developing either condition [4,5,6], however, they may explain less than half of the total variation in risk of SCZ [7,8] and ASD [9]. Recent studies suggest that rare copy-number variants (CNVs) and rare single-nucleotide variants (SNVs) may explain additional disease risk or trait variability [10,11,12,13,14,15]. A significant excess of rare, disruptive SNVs has been detected in those genes that have previously been implicated as candidate risk genes for SCZ and/or ASD [16,17,18]. Thus, deep sequencing of candidate genes might be a good way for elucidating the pathogenesis of these neuropsychiatric disorders [19].
PCDH15 is a member of the largest group in the cadherin superfamily that involved in generating neural diversity for neuronal differentiation and synapse formation [20]. PCDH15 is primarily recognized as a gene that forms tip-link filaments in sensory hair cells and associated with Usher syndrome type 1F (OMIM 602083) [21]. Notably, more than 20% of these patients exhibit neuropsychiatric symptoms [22]. A GWAS identified PCDH15 as relevant to neurocognitive processes [23]. In mice, PCDH15 is expressed throughout the brain and central nervous system (CNS) during embryogenesis [24], and influences serotonin transporter expression in the adult CNS [25]. Rare, exonic CNVs in PCDH15 were recently identified in ASD [26] and bipolar disorder (BD) patients [27,28]. These findings strongly suggest that PCDH15 is a promising candidate risk gene for neuropsychiatric disorders because several neuropsychiatric disorders including SCZ, ASD, and BD share genetic risk factors [3,4,5,6,13,27,29,30]. To our knowledge, however, no published study has focused on rare PCDH15 variants in cases of neuropsychiatric disorders.
Our hypothesis was that rare PCDH15 variants might confer susceptibility to neuropsychiatric pathogenesis. To increase statistical power and detect shared risk, we combined SCZ and ASD samples in a study cohort [6,31,32]. First, we performed targeted-region sequencing of PCDH15 coding exons in 562 Japanese patients; we then conducted single-variant association analysis in an independent case-control set comprising 4,013 samples to identify putative variants with large effect.
Materials and Methods
Study samples
Two independent Japanese sample groups were used in this study. For the targeted-resequencing discovery cohort, 370 SCZ (mean age ± SD, 49.7 ± 14.8 years; 53.0% male) and 192 ASD (mean age ± SD = 16.3 ± 8.4 years; 77.6% male) individuals participated. For genetic association analysis, the case control sample set comprised 1,714 SCZ (46.3 ± 15.1 years; 51.2% male), 382 ASD (19.6 ± 10.7 years; 77.8% male), and 1,917 control subjects (44.7 ± 14.7 years; 55.3% male). All subjects were unrelated, living on the mainland of Japan, and self-identified as Japanese. All patients fulfilled the criteria listed in Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-5) for SCZ or ASD. Healthy control subjects were selected from the general population and had no history of mental disorders based on questionnaire responses from the subjects themselves during the sample inclusion step. The study was explained to all participants and/or their parents both verbally and in writing. Written informed consent was obtained from the participants and from the parents of the patients under 20 years old. All procedures performed in this study involving human participants were approved by the Ethics Committee of the Nagoya University Graduate School of Medicine and conducted in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Sample preparation
DNA was extracted from peripheral blood or saliva from each SCZ, ASD, and control participant. For DNA extraction, we used the QIAamp DNA Blood Kit or Tissue Kit (Qiagen Ltd. Hilden, Germany). The quantity of extracted DNA was estimated using the Qubit® dsDNA BR Assay Kit (Life Technologies, Carlsbad, CA, USA) on a Qubit® 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s recommended protocol.
Library preparation and resequencing
The next-generation sequencing technology on the Ion Torrent PGM™ was used to resequence the PCDH15 coding regions (Ensembl Transcript ID: ENST00000320301; 1995 amino acids) via the protocols described in the Ion AmpliSeq™ Library Preparation User Guide (Thermo Fisher Scientific, Rev.5; MAN0006735), Ion PGM™ Template OT2 200 Kit (Thermo Fisher Scientific, Rev. 5; MAN0007220), and Ion PGM™ Sequencing 200 Kit (Thermo Fisher Scientific, Rev. 3; MAN0007273). After target-specific PCR amplification, amplicons were purified and pooled. Libraries were then prepared to obtain 200-bp PCR fragments flanked by adaptor and barcode sequences; these sequences allowed sequencing and sample identification respectively. The concentration of each library was determined with the Ion Library TaqMan Quantitation Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). Amplified libraries were subjected to emulsion PCR and subsequent enrichment for template-positive Ion Sphere™ particles (ISPs) with the Ion OneTouch™ system (Life Technologies, Carlsbad, CA, USA). ISPs were enriched and sequenced in a 200-bp configuration run using 318 chips (Life Technologies, Carlsbad, CA, USA).
Data analysis
Sequence reads were run through a data analysis pipeline on the Ion Torrent platform-specific pipeline software, Torrent Suite™ version 4.4 (Life Technologies, Carlsbad, CA, USA) to generate sequence reads filtered according to the pipeline software quality-controls and to remove poor signal reads. Reads assembling and variant identification were performed with the Ingenuity Variant Analysis software™ (http://www.ingenuity.com/variants) from Ingenuity Systems using Fastq files containing sequence reads and the Ion Ampliseq Designer BED file software to map the amplicons with default parameters, (call quality >20 and read depth >10). Candidate variants were defined as exonic or splice-site variants with allele frequencies of ≤1% in the following six public exome databases: dbSNP Build 139 (http://www.ncbi.nlm.nih.gov/projects/SNP/), the 1000 Genomes Project (http://www.1000genomes.org), NHLBI ESP exomes (http://evs.gs.washington.edu/EVS/), the Human Genetic Variation Database (http://www.genome.med.kyoto-u.ac.jp/SnpDB/), the Exome Aggregation Consortium (http://exac.broadinstitute.org) and the Genebook (http://atgu.mgh.harvard.edu/~spurcell/genebook) [17, 27]. To identify deleterious effects caused by amino acid substitution, Sorting Intolerant From Tolerant (SIFT) [33] and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) [34] were used for in silico prediction of functional consequences. Additional clinical variant annotations were obtained from NCBI ClinVar (last accessed July 2015; http://www.ncbi.nlm.nih.gov/clinvar/) [35]. To analyze the potential effect of detected variants on putative splicing regulatory elements as exonic splicing enhancer and exonic splicing silencer, we used Splicing-based Analysis of Variants (SPANR) (http://tools.genes.toronto.edu) [36]. Evolutionary conservation was assessed with Evola ver. 7.5 (http://www.h-invitational.jp/evola/search.html) [37]. De novo analysis was performed when DNA samples from parents were available.
Sanger sequencing with the ABI 3130xl Genetic Analyzer (Life Technologies, Carlsbad, CA, USA), and standard methods were used to confirm each candidate variant. Sequence analysis software version 6.0 (Applied Biosystems, Foster City, CA, USA) was used to analyze all sequence data. Primer sequences for validating each variant are available in S1 Table.
Genetic association analysis
An ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA) and TaqMan assays with custom probes were used to genotype putative deleterious variants. Custom probe sequences are available in S2 Table. Each 384-microtiter plate contained two non-template controls and two samples with the variant. The reactions and data analysis were performed using Genotyping Master Mix and Sequence Detection Systems, respectively, according to the standard protocols (Applied Biosystems, Foster City, CA, USA). Differences in genotype distribution between cases and controls were tested with one-sided, Fisher’s exact tests.
We computed the effective sample size and statistical power using a web browser program, Genetic Power Calculator developed by Purcell et al. (http://pngu.mgh.harvard.edu/~purcell/gpc/) [38].
Results
Variation screening of all PCDH15 coding exons
Nucleotide sequence data reported have been deposited in the DNA Data Bank of Japan (DDBJ) databases (http://www.ddbj.nig.ac.jp) under the accession number DRA004490.
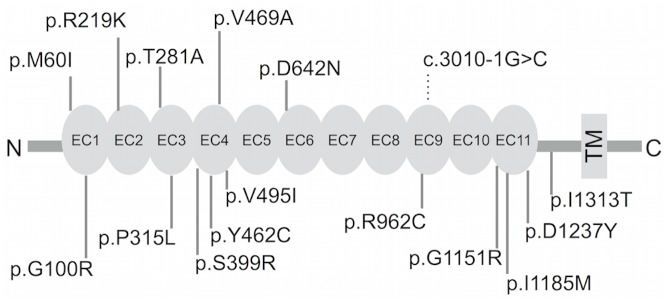
We sequenced PCDH15 exon and exon-intron boundary in genomic DNA isolated from Japanese patient sample (n = 562). Of 17 SNVs and three indels detected by the Ion Torrent PGM™, one SNV and three indels were not validated by Sanger sequencing. In total, we evaluated one splice-site variant and 15 missense variants (Table 1). Analyzing the frequency of rare SNVs in ASD and SCZ individuals, 8.9% of ASD (17/192) and 5.6% of SCZ (17/370) were identified as carriers, pointing to a higher frequency of rare SNVs in ASD (p = 0.037). Nonsense and frameshift variants were not found. Each variant detected was heterozygous. Each of the 15 missense SNVs was located in the coding region of the extracellular domain (Fig 1).
Table 1. Rare PCDH15 SNVs identified in this study.
| Chr. | Position (GRCh38) | Ref | Val | Amino Acid changes | Case | Gender | Inheritance status | SIFT | Polyphen-2 | dbSNP | 1000 Genomes | HGVDa | ClinVar | ExACb |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 53840365 | A | G | p.I1313T | 1 ASD | M | Maternal | Damaging | Possibly Damaging | rs147250420 | _ | 0/3/1 | _ | 2/121412 |
| 10 | 53866650 | C | A | p.D1237Y | 1 SCZ | F | Damaging | Probably Damaging | rs371278220 | _ | 0/1/299 | _ | C>T 3/120638 | |
| 10 | 53866804 | T | C | p.I1185M | 1 ASD | F | Paternal | Damaging | Probably Damaging | _ | _ | 0/3/796 | _ | 12/121316 |
| 1 SCZ | M | Paternal | ||||||||||||
| 10 | 53903293 | C | T | p.G1151R | 4ASD | F | Paternal | Damaging | Probably Damaging | rs149478475 | 0.0028 | 0/21/1084 | Likely benign | 153/121170 |
| M | Maternal | |||||||||||||
| M | Maternal | |||||||||||||
| M | Maternal | |||||||||||||
| 3 SCZ | 1F | |||||||||||||
| 2M | ||||||||||||||
| 10 | 53959845 | C | G | c.3010-1G>C | 1 ASD | M | Maternal | _ | _ | _ | _ | _ | _ | _ |
| 10 | 53961877 | G | A | p.R962C | 1 ASD | M | Paternal | Tolerated | Possibly Damaging | rs201816080 | 0.0014 | 0/7/1130 | Likely benign | 109/121132 |
| 2 SCZ | F | |||||||||||||
| M | ||||||||||||||
| 10 | 54090057 | C | T | p.D642N | 1 SCZ | F | Damaging | Probably Damaging | _ | _ | _ | _ | _ | |
| 10 | 54183551 | C | T | p.V495I | 1 SCZ | M | Tolerated | Benign | rs187727835 | 0.0004 | 0/1/428 | _ | 1/121400 | |
| 10 | 54185168 | A | G | p.V469A | 1 ASD | M | Maternal | Damaging | Possibly Damaging | _ | _ | _ | _ | 3/121336 |
| 10 | 54185189 | T | C | p.Y462C | 1 ASD | M | Maternal | Damaging | Possibly Damaging | rs201284699 | 0.0004 | _ | _ | 11/121346 |
| 10 | 54195793 | T | G | p.S399R | 2 SCZ | 2M | Tolerated | Benign | rs199786639 | 0.0002 | 0/4/763 | Uncertain significance | 31/121404 | |
| 10 | 54236864 | G | A | p.P315L | 3 ASD | M | Maternal | Damaging | Probably Damaging | rs138299477 | 0.0004 | 0/13/1096 | _ | 8/121380 |
| M | Paternal | |||||||||||||
| M | ||||||||||||||
| 5 SCZ | 3F | |||||||||||||
| 2M | ||||||||||||||
| 10 | 54317306 | T | C | p.T281A | 1 ASD | M | Maternal | Tolerated | Possibly Damaging | _ | _ | _ | _ | 1/121250 |
| 10 | 54329645 | C | T | p.R219K | 1 ASD | M | Maternal | Tolerated | Possibly Damaging | _ | _ | _ | _ | 1/121090 |
| 10 | 54378892 | C | T | p.G100R | 1 ASD | M | Tolerated | Probably Damaging | rs140716525 | 0.0002 | 0/3/431 | _ | 16/120962 | |
| 1 SCZ | M | |||||||||||||
| 10 | 54378920 | C | T | p.M60I | 1 ASD | F | Paternal | Tolerated | Benign | _ | _ | 0/1/367 | _ | _ |
Note: Amino acid position is based on NCBI reference sequence NP_149045. Chr, chromosome; Ref, reference; Val, variant; M, male; F, female; ASD, autism spectrum disorders; SCZ, schizophrenia; dbSNP, dbSNP build 139 (http://www.ncbi.nlm.nih.gov/projects/SNP/); 1000 Genomes, the 1000 Genomes Project (http://www.1000genomes.org); HGVD, the Human Genetic Variation Database (http://www.genome.med.kyoto-u.ac.jp/SnpDB/); ClinVar, NCBI ClinVar (last accessed July 2015; http://www.ncbi.nlm.nih.gov/clinvar/); ExAC, Exome Aggregation Consortium (http://exac.broadinstitute.org). Rare nonsynonymous SNVs in the Genebook (http://atgu.mgh.harvard.edu/~spurcell/genebook) were not detected in our study.
a homozygous for a minor allele / heterozygote / homozygous for a major allele
b minor allele count / total allele count
Fig 1. Location for each variant of interest.
PCDH15 protein structure is based on NCBI Reference Sequence NP_149045. Each variant was located in the extracellular domain. EC: extracellular cadherin repeat, TM: Transmembrane.
Of the 15 missense variants, 12 were predicted to be damaging with in silico prediction tools (Table 1). No variants were located in conserved sequences of the cadherin-specific motifs (XEX, DXD, DYE, XDX, and DXNDN) required for calcium binding and rigidification of the extracellular cadherin (EC) domains [39]. Based on the in silico predictions, the 15 missense variants were not expected to affect splicing, but one splice-site variant (c.3010-1G>C) was (result not shown).
We were able to determine inheritance status for 16 cases. Among these 16 cases, 10 involved mother-to-son variant transmission, three involved father-to-son transmission, and three others involved father-to-daughter transmission (Table 1). An affected brother shared the p.T281A variant (S1 Fig). An unaffected brother shared the p.G1151R variant, but an unaffected sister of the same patient did not. No de novo variants were found in these 16 cases.
We regarded four missense variants (p.R219K, p.T281A, p.V469A, p.D642N) and the splice-site variant (c.3010-1G>C) as novel ones because they were predicted to be damaging or to affect splicing based on in silico predictions and because each was not registered in any of the public databases described in the Materials and Methods. p.M60I was included in the association analysis because it was previously detected in a Japanese boy with developmental delay and hearing loss [40], although it was neither classified as damaging in the in silico analysis nor absent from the Human Genetic Variation Database. Each of these six SNVs was located in a genomic region that is highly conserved among eight vertebrate species (Table 2). Brief information and results of segregation analysis are in S1 Fig.
Table 2. Multiple alignments of amino acid sequences for PCDH15 eight vertebrate homologs.
| Variant | Reference | p.M60I | p.R219K | p.T281A | p.V469A | p.D642N |
|---|---|---|---|---|---|---|
| Human | NP_149045 | LVDNMLIKG | VLRKRLNYE | CRPLTYQAA | LLQPVDREE | LQATDREGD |
| Chimpanzee | XP_507798.3 | LVDNMLIKG | VLRKRLNYE | CRPLTYQAA | LLQPVDREE | LQATDREGD |
| Orangutan | ENSPPYT00000002926 | _ | VLRKRLNYE | CRPLTYQAA | LLQPVDREE | LQATDREGD |
| Macaque | XP_001098443.1 | LVDNMLIKG | VLRKRLNYE | CRPLTYQAA | LLQPVDREE | LQATDREGD |
| Mouse | ENSMUST00000105426 | LVDNMLIKG | VLRKRLNYE | CRPLTYQAA | LLQPVDREE | LQATDREGD |
| Rat | XP_001080000.1 | LVDNMLIKG | VLRKRLNYE | CRPLTYQAA | LLQPVDREE | LQATDREGD |
| Chicken | ABC79282.1 | LVDNMLIKG | VLRERLNYE | CRPLTYQAS | LLQPVDREA | LQAFDREGD |
| Zebrafish | AAW50924.1 | LVENMQING | VLRERLNYE | CKPLTYRAS | LLRPVDHEE | IQATDREKD |
Genetic association analysis
For our sample set of cases (n = 2,096) and controls (n = 1,917), we computed a statistical power of >80% using the following parameters: disease prevalence of 0.01, observed rare-allele frequency of 0.0021, odds ratio for dominant effect of ≥ 3.59, and type I error rate of 0.0083 (using a Bonferroni correction by a factor of 6, based on the 6 SNVs investigated). An overview and each phenotype of genetic association analysis of the six novel variants are presented in Table 3 and S3 Table. Of the six, four novel SNVs (p.R219K, p.T281A, p.D642N, c.3010-1G>C) remained as singleton observations after genotyping of all cases and controls. We found no statistically significant association for any of the six rare heterozygous point variants in PCDH15 with case-control analysis. Post hoc calculation of statistical power based on a minor allele frequency of 0.00048 (p.M60I, Table 3) revealed that good power accrues for odds ratio ≥ 8.96 or with an increase of the sample size to nearly 20,000 individuals (cases + controls).
Table 3. Association analysis of novel rare SNVs.
| Case | Control | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Exona | Ref | Val | Position (GRCh38) | Variant | Genotype countb | Minor allele frequency | P valuec | Genotype countb | Minor allele frequency |
| 5' side of 23 | C | G | 10:53959845 | c.3010-1G>C | 0/0/2085 | 0 | 1 | 0/0/1909 | 0 |
| 16 | C | T | 10:54090057 | p.D642N | 0/0/2087 | 0 | 1 | 0/0/1905 | 0 |
| 12 | A | G | 10:54185168 | p.V469A | 0/1/2091 | 0.00024 | 0.52 | 0/0/1908 | 0 |
| 8 | T | C | 10:54317306 | p.T281A | 0/0/2091 | 0 | 1 | 0/0/1911 | 0 |
| 7 | C | T | 10:54329645 | p.R219K | 0/0/2086 | 0 | 1 | 0/0/1915 | 0 |
| 4 | C | T | 10:54378920 | p.M60I | 0/2/2090 | 0.00048 | 0.19 | 0/5/1906 | 0.0013 |
Note: Ref, reference; Val, variant
a Based on ENST00000320301;
b homozygous for a minor allele / heterozygote / homozygous for a major allele;
C P values were calculated by one-tailed Fisher’s exact test
Discussion
To our knowledge, this is the first study to investigate the contribution of rare PCDH15 variants to neuropsychiatric disorders and susceptibility to these disorders. We conducted targeted resequencing of coding exons in PCDH15 for 562 Japanese patients and detected 16 heterozygous SNVs as condition-related candidate genes. More rare SNVs were detected from samples of ASD than those of SCZ. Of these 16 SNVs, five SNVs (p.R219K, p.T281A, p.V469A, p.D642N, c.3010-1G>C) were selected because they were both predicted to be protein-damaging by in silico analysis and not registered in public databases or found with a very low frequency in ExAC. p.M60I was selected because it previously implicated in developmental delay [40]. An independent association analysis was then performed with a cohort comprising 2,096 cases and 1,917 controls. Our a priori calculation indicated our sample size was appropriately powered to determine statistical significance of SNVs. To assume the odds ratio for dominant effect of rare SNVs would be more than 3.59 seems reasonable according to previous studies that reported odds ratios from 1.88 [41] to 7.1 [32]. Of these six SNVs, four variants (p.R219K, p.T281A, p.D642N, c.3010-1G>C) were not detected in our case control samples, in public databases or found with a very low frequency in ExAC. Although a number of similar studies have identified statistical associations between rare SNVs and SCZ [32,41,42,43], we found no statistically significant association between any of these rare heterozygous PCDH15 SNVs and either neuropsychiatric disorder.
We find it interesting that all protein-coding SNVs observed in the resequencing cohort were located within the PCDH15 extracellular domain (Fig 1), which may interact with other proteins. Of the 15 protein-damaging SNVs, 12 predicted by in silico analysis might change the biological functions of PCDH15. PCDH15 plays an essential role in maintenance of normal retinal and cochlear function [39]. Atypical processing of peripheral sensory inputs plays a crucial role in both SCZ and ASD pathology [44,45,46]. Taken together, deleterious protein changes will induce sensory processing differences contribute to SCZ and ASD symptoms. Notably, splicing misregulation has been implicated in neuropsychiatric disorders [36,47]; c.3010-1G>C also might be a promising candidate for a causal variant in these disease etiopathologies.
In this study, all inheritance statuses were either from apparently unaffected parents or of unknown origin, suggesting variable penetrance (Table 1; S1 Fig). Each candidate variant of maternal origin was transmitted to an affected son; this finding is similar to previous findings [48]. While de novo variants have been the focus of research on SCZ and ASD pathogenesis, inherited variants also contribute substantially to these complex diseases [49]. In addition, evolutionary theory predicts that deleterious alleles are likely to be especially rare because of purifying selection [50,51]. Recent large-scale genetic studies report that ultra-rare, private, and inherited-truncating variants in conserved genes are highly enriched in patient populations, especially in genes that closely involved in neurodevelopment [8,17,48,52,53]. The inherited ultra-rare variants (p.R219K, p.T281A, p.D642N, c.3010-1G>C) within highly conserved regions (Table 2) could increase susceptibility to development of a neuropsychiatric disorder.
There are several explanations for our inability to find statistical evidence for a causative role in SCZ and/or ASD for any of these rare PCDH15 SNVs. First, due to extremely low minor-allele frequencies (< 0.0005) as revealed by the association analysis or to odds ratios lower than expected, we could neither confirm nor dismiss the significance of rare PCDH15 variants in either neuropsychiatric disorder. Post hoc calculations revealed that a larger, higher-powered sample should be sought to reveal relationships between neuropsychiatric disorders and PCDH15 variants. Secondly, we focused on the shared genetic risk to increase the statistical power in this study. Considering that the burden of rare PCDH15 was statistically greater in ASD cases than in SCZ, further research may be needed to provide similarities and differences between SCZ and ASD. Thirdly, we focused on the ENST00000320301 transcript, but PCDH15, like many other neuronal proteins, is structurally diversified through the differential inclusion and exclusion of exons. We did not cover the promoter, untranslated regions, or intronic regions of PCDH15, which contain potentially disease-relevant regions. Fourthly, the lack of DNA from a sufficient number of patient family members prevented us from monitoring variant segregation. Finally, although 81% of the SNVs (13/16) identified in this study were predicted to be protein-disrupting or splicing-altering based on in silico analysis, the exact molecular mechanisms and networks affected by PCDH15 variants in SCZ and ASD remain unclear. Useful model systems that can address these questions will be needed to assess the impact of the SNVs discovered here.
Conclusions
We explored the role of rare PCDH15 SNVs in Japanese SCZ and ASD patients. We found four ultra-rare variants (p.R219K, p.T281A, p.D642N, c.3010-1G>C) that may increase disease susceptibility. No statistically significant association between any rare, heterozygous point PCDH15 variant and neuropsychiatric disorders was detected. A much larger sample size is needed to elucidate the relevance of rare PCDH15 variants to neuropsychiatric disorders.
Supporting Information
The genotypes of the tested individuals are indicated on the lower-side. All comorbidities were diagnosed by experienced psychiatrists according to Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-5) criteria. Note: 1Autism Spectrum Disorder; 2Interectual Disability; 3Attention-Deficit/Hyperactivity Disorder; 4Tic Disorder; 5Epilepsy; 6Schizophrenia.
(PDF)
(PDF)
Note: A TaqMan probe consists with a FAM or VIC dye label on the 5' end, and nonfluorescent quencher (NFQ) on the 3' end.
(PDF)
Note: Ref, reference; Val, variant. a Based on ENST00000320301; b homozygous for a minor allele / heterozygote / homozygous for a major allele; C P values were calculated by one-tailed Fisher’s exact test.
(PDF)
Acknowledgments
We are grateful to all of the patients, their families, and control individuals who contributed to this study.
Data Availability
Nucleotide sequence data reported have been deposited in the DNA Data Bank of Japan (DDBJ) databases (http://www.ddbj.nig.ac.jp) under the accession number DRA004490.
Funding Statement
The current research was supported by research grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; the Ministry of Health, Labour, and Welfare of Japan; “Integrated research on neuropsychiatric disorders” carried out under the Strategic Research Program for Brain Sciences from the Japan Agency for Medical Research and Development (AMED); the Brain Mapping by Integrated Neurotechnologies for Disease Studies (Brain/MINDS) project of AMED; Grant-in-Aid for Scientific Research on Innovative Areas “Glial assembly: a new regulatory machinery of brain function and disorders”; and Innovative Areas “Comprehensive Brain Science Network.”
References
- 1.Rapoport J, Chavez A, Greenstein D, Addington A, Gogtay N. Autism spectrum disorders and childhood-onset schizophrenia: clinical and biological contributions to a relation revisited. J Am Acad Child Adolesc Psychiatry. 2009;48(1):10–8. 10.1097/CHI.0b013e31818b1c63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13(8):537–51. 10.1038/nrg3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullivan PF, Magnusson C, Reichenberg A, Boman M, Dalman C, Davidson M, et al. Family history of schizophrenia and bipolar disorder as risk factors for autism. Arch Gen Psychiatry. 2012;69(11):1099–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craddock N, Owen MJ. The Kraepelinian dichotomy—going, going… but still not gone. Br J Psychiatry. 2010;196(2):92–5. 10.1192/bjp.bp.109.073429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cross-Disorder Group of the Psychiatric Genomics C, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45(9): 984–94 10.1038/ng.2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cross-Disorder Group of the Psychiatric Genomics C, et al. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. The Lancet. 2013;381(9875):1371–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.International Schizophrenia C, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–52. 10.1038/nature08185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mowry BJ, Gratten J. The emerging spectrum of allelic variation in schizophrenia: current evidence and strategies for the identification and functional characterization of common and rare variants. Mol Psychiatry. 2013;18:38–52. 10.1038/mp.2012.34 [DOI] [PubMed] [Google Scholar]
- 9.Stein JL, Parikshak NN, Geschwind DH. Rare inherited variation in autism: beginning to see the forest and a few trees. Neuron. 2013;77(2):209–11. 10.1016/j.neuron.2013.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007;80(4):727–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141(2):210–7. 10.1016/j.cell.2010.03.032 [DOI] [PubMed] [Google Scholar]
- 12.Moens LN, De Rijk P, Reumers J, Van den Bossche MJ, Glassee W, De Zutter S, et al. Sequencing of DISC1 pathway genes reveals increased burden of rare missense variants in schizophrenia patients from a northern Swedish population. PLoS One. 2011;6(8):e23450 10.1371/journal.pone.0023450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148(6):1223–41. 10.1016/j.cell.2012.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515(7526):216–21. 10.1038/nature13908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuk O, Schaffner SF, Samocha K, Do R, Hechter E, Kathiresan S, et al. Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci U S A. 2014;111(4):E455–64. 10.1073/pnas.1322563111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515(7526):209–15. 10.1038/nature13772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506(7487):185–90. 10.1038/nature12975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–7. 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S, Abecasis GR, Boehnke M, Lin X. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95;5–23. 10.1016/j.ajhg.2014.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirano S, Takeichi M. Cadherins in brain morphogenesis and wiring. Physiol Rev. 2012;92(2):597–634. 10.1152/physrev.00014.2011 [DOI] [PubMed] [Google Scholar]
- 21.Ahmed ZM, Riazuddin S, Bernstein SL, Ahmed Z, Khan S, Griffith AJ, et al. Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am J Hum Genet. 2001;69(1):25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dammeyer J. Children with Usher syndrome: mental and behavioral disorders. Behav Brain Funct. 2012;8:16 10.1186/1744-9081-8-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oikkonen J, Huang Y, Onkamo P, Ukkola-Vuoti L, Raijas P, Karma K, et al. A genome-wide linkage and association study of musical aptitude identifies loci containing genes related to inner ear development and neurocognitive functions. Mol Psychiatry. 2014. 10.1038/mp.2014.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murcia CL, Woychik RP. Expression of Pcdh15 in the inner ear, nervous system and various epithelia of the developing embryo. Mechanisms of Development. 2001;105:163–6. [DOI] [PubMed] [Google Scholar]
- 25.Ye R, Carneiro AM, Han Q, Airey D, Sanders-Bush E, Zhang B, et al. Quantitative trait loci mapping and gene network analysis implicate protocadherin-15 as a determinant of brain serotonin transporter expression. Genes Brain Behav. 2014;13(3):261–75. 10.1111/gbb.12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sorte HS, Gjevik E, Sponheim E, Eikid KL, Rodningen OK. Copy number variation findings among 50 children and adolescents with autism spectrum disorder. Psychiatric Genetics. 2013;23:61–9. 10.1097/YPG.0b013e32835d718b [DOI] [PubMed] [Google Scholar]
- 27.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506(7487):179–84. 10.1038/nature12929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noor A, Lionel AC, Cohen-Woods S, Moghimi N, Rucker J, Fennell A, et al. Copy number variant study of bipolar disorder in Canadian and UK populations implicates synaptic genes. Am J Med Genet B Neuropsychiatr Genet. 2014;165B(4):303–13. 10.1002/ajmg.b.32232 [DOI] [PubMed] [Google Scholar]
- 29.Pettersson E, Larsson H, Lichtenstein P. Common psychiatric disorders share the same genetic origin: a multivariate sibling study of the Swedish population. Mol Psychiatry. 2015. 10.1038/mp.2015.116 [DOI] [PubMed] [Google Scholar]
- 30.Selten JP, Lundberg M, Rai D, Magnusson C. Risks for nonaffective psychotic disorder and bipolar disorder in young people with autism spectrum disorder: a population-based study. JAMA Psychiatry. 2015;72(5):483–9. 10.1001/jamapsychiatry.2014.3059 [DOI] [PubMed] [Google Scholar]
- 31.Hommer RE, Swedo SE. Schizophrenia and autism-related disorders. Schizophr Bull. 2015;41(2):313–4. 10.1093/schbul/sbu188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura H, Tsuboi D, Wang C, Kushima I, Koide T, Ikeda M, et al. Identification of rare, single-nucleotide mutations in NDE1 and their contributions to schizophrenia susceptibility. Schizophr Bull. 2015;41(3):744–53. 10.1093/schbul/sbu147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–81. 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- 34.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42(Database issue):D980–5. 10.1093/nar/gkt1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiong HY, Alipanahi B, Lee LJ, Bretschneider H, Merico D, Yuen RK, et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347(6218):1254806 10.1126/science.1254806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuya A, Sakate R, Kawahara Y, Koyanagi KO, Sato Y, Fujii Y, et al. Evola: Ortholog database of all human genes in H-InvDB with manual curation of phylogenetic trees. Nucleic Acids Res. 2008;36(Database issue):D787–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purcell S C S, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. [DOI] [PubMed] [Google Scholar]
- 39.Sotomayor M, Weihofen WA, Gaudet R, Corey DP. Structure of a force-conveying cadherin bond essential for inner-ear mechanotransduction. Nature. 2012;492(7427):128–32. 10.1038/nature11590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakashima M, Takano K, Osaka H, Aida N, Tsurusaki Y, Miyake N, et al. Causative novel PNKP mutations and concomitant PCDH15 mutations in a patient with microcephaly with early-onset seizures and developmental delay syndrome and hearing loss. J Hum Genet. 2014;59(8):471–4. 10.1038/jhg.2014.51 [DOI] [PubMed] [Google Scholar]
- 41.Hu X, Zhang B, Liu W, Paciga S, He W, Lanz TA, et al. A survey of rare coding variants in candidate genes in schizophrenia by deep sequencing. Mol Psychiatry. 2014;19(8):857–8. 10.1038/mp.2013.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song W, Li W, Feng J, Heston LL, Scaringe WA, Sommer SS. Identification of high risk DISC1 structural variants with a 2% attributable risk for schizophrenia. Biochem Biophys Res Commun. 2008;367(3):700–6. 10.1016/j.bbrc.2007.12.117 [DOI] [PubMed] [Google Scholar]
- 43.Kushima I, Nakamura Y, Aleksic B, Ikeda M, Ito Y, Shiino T, et al. Resequencing and association analysis of the KALRN and EPHB1 genes and their contribution to schizophrenia susceptibility. Schizophr Bull. 2012;38(3):552–60. 10.1093/schbul/sbq118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shergill S, Samson G, Bays PM, Frith CD, Wolpert DM. Evidence for Sensory Prediction Deficits in Schizophrenia. Am J psychiatry. 2005;162:2394–86. [DOI] [PubMed] [Google Scholar]
- 45.Girard MPP, Brigitte MA, Bonnabau H, Malauzat D. Experimental Pain Hypersensitivity in Schizophrenic Patients. The Clinical Journal of Pain. 2011;27(3):790–5. [DOI] [PubMed] [Google Scholar]
- 46.Brandwein AB, Foxe JJ, Butler JS, Frey HP, Bates JC, Shulman LH, et al. Neurophysiological indices of atypical auditory processing and multisensory integration are associated with symptom severity in autism. J Autism Dev Disord. 2015;45(1):230–44. 10.1007/s10803-014-2212-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Irimia M, Weatheritt RJ, Ellis JD, Parikshak NN, Gonatopoulos-Pournatzis T, Babor M, et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 2014;159(7):1511–23. 10.1016/j.cell.2014.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47(6):582–8. 10.1038/ng.3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoischen A, Krumm N, Eichler EE. Prioritization of neurodevelopmental disease genes by discovery of new mutations. Nat Neurosci. 2014;17(6):764–72. 10.1038/nn.3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2011;13(2):135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Genomes Project C, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bourgeron T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat Rev Neurosci. 2015;16(9):551–63. 10.1038/nrn3992 [DOI] [PubMed] [Google Scholar]
- 53.Kenny EM, Cormican P, Furlong S, Heron E, Kenny G, Fahey C, et al. Excess of rare novel loss-of-function variants in synaptic genes in schizophrenia and autism spectrum disorders. Mol Psychiatry. 2014;19(8):872–9. 10.1038/mp.2013.127 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The genotypes of the tested individuals are indicated on the lower-side. All comorbidities were diagnosed by experienced psychiatrists according to Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-5) criteria. Note: 1Autism Spectrum Disorder; 2Interectual Disability; 3Attention-Deficit/Hyperactivity Disorder; 4Tic Disorder; 5Epilepsy; 6Schizophrenia.
(PDF)
(PDF)
Note: A TaqMan probe consists with a FAM or VIC dye label on the 5' end, and nonfluorescent quencher (NFQ) on the 3' end.
(PDF)
Note: Ref, reference; Val, variant. a Based on ENST00000320301; b homozygous for a minor allele / heterozygote / homozygous for a major allele; C P values were calculated by one-tailed Fisher’s exact test.
(PDF)
Data Availability Statement
Nucleotide sequence data reported have been deposited in the DNA Data Bank of Japan (DDBJ) databases (http://www.ddbj.nig.ac.jp) under the accession number DRA004490.