The biosynthesis pathway to the C25 linear precursor of defensive sesterterpenoids, which highly accumulate in glandular trichomes of Leucosceptrum canum, was revealed through transcriptomic analysis.
Abstract
Plant sesterterpenoids, an important class of terpenoids, are widely distributed in various plants, including food crops. However, little is known about their biosynthesis. Here, we cloned and functionally characterized a plant geranylfarnesyl diphosphate synthase (Lc-GFDPS), the enzyme producing the C25 prenyl diphosphate precursor to all sesterterpenoids, from the glandular trichomes of the woody plant Leucosceptrum canum. GFDPS catalyzed the formation of GFDP after expression in Escherichia coli. Overexpressing GFDPS in Arabidopsis thaliana also gave an extract catalyzing GFDP formation. GFDPS was strongly expressed in glandular trichomes, and its transcript profile was completely in accordance with the sesterterpenoid accumulation pattern. GFDPS is localized to the plastids, and inhibitor studies indicated its use of isoprenyl diphosphate substrates supplied by the 2-C-methyl-d-erythritol 4-phosphate pathway. Application of a jasmonate defense hormone induced GFDPS transcript and sesterterpenoid accumulation, while reducing feeding and growth of the generalist insect Spodoptera exigua, suggesting that these C25 terpenoids play a defensive role. Phylogenetic analysis suggested that GFDPS probably evolved from plant geranylgeranyl diphosphate synthase under the influence of positive selection. The isolation of GFDPS provides a model for investigating sesterterpenoid formation in other species and a tool for manipulating the formation of this group in plants and other organisms.
INTRODUCTION
Plants biosynthesize a large number of highly diversified secondary metabolites that play crucial roles in adaptation to biotic and abiotic stresses (Dixon, 2001; Nakabayashi and Saito, 2015). Terpenoids constitute the largest class of plant secondary metabolites, with more than 30,000 different compounds having been reported (Broun and Somerville, 2001; Trapp and Croteau, 2001), many of which play important protective roles against biotic stresses, such as herbivores and pathogens (Gershenzon and Dudareva, 2007). Terpenoids with C25 carbon frameworks derived from five isoprene units are known as sesterterpenoids. The distribution of sesterterpenoids in plants is widespread, including lichens, diatoms, ferns, monocotyledons, and dicotyledons (Supplemental Data Set 1). In vascular plants, sesterterpenoids are a characteristic feature of the genus Salvia (Lamiaceae) (Ebrahimi et al., 2014), with 57 sesterterpenoids isolated from 10 different Salvia species (Rustaiyan et al., 1982; Rustaiyan and Sadjadi, 1987; Rustaiyan and Koussari, 1988; Gonzalez et al., 1989; Moghaddam et al., 1995, 1998, 2010; Topcu et al., 1996a, 1996b; Ulubelen et al., 1996; Cioffi et al., 2008; Dal Piaz et al., 2009, 2010; Ebrahimi et al., 2014; Moridi Farimani and Mazarei, 2014) and were reported to be present in two important food crops, wheat (Triticum aestivum) (Akihisa et al., 1999) and potato (Solanum tuberosum) (Toyoda et al., 1969). Plant sesterterpenoids have been shown to exhibit a broad range of biological activities, serving as insect antifeedant (Luo et al., 2010, 2011, 2013a, 2013b; Li et al., 2013) and antifungal (Luo et al., 2010; Li et al., 2013) agents, as cytostatic agents against human lung cancer cell (Rowland et al., 2001), as inhibitors of tubulin tyrosine ligase (Dal Piaz et al., 2009, 2010), as prolylendopeptidase inhibitors (Choudhary et al., 2004a), and as enhancers of interleukin-2 gene expression (Kawahara et al., 1999). However, our knowledge of sesterterpenoid biosynthesis and evolution in plants is rather limited, and genes and enzymes involved in plant sesterterpenoid formation have only very recently been reported from species of the Brassicaceae (Nagel et al., 2015; Wang et al., 2015). It would be interesting to learn if these genes arose from genes forming larger, better known groups of terpenoids, such as those of C20 diterpenoids, by duplication, divergence, and neofunctionalization (Pichersky and Lewinsohn, 2011; Kliebenstein and Osbourn, 2012), or if they were introduced from other sources, such as microorganisms, through horizontal gene transfer (Copley and Dhillon, 2002; Richards et al., 2006; Emiliani et al., 2009).
Sesterterpenoids have been regarded to be biosynthesized from the intermediate geranylfarnesyl diphosphate (GFDP) (Wang et al., 2013) by GFDP synthase (GFDPS), a member of the isoprenyl diphosphate synthase (IDS) family. To date, GFDPSs have been identified in a few prokaryotes. A GFDPS was originally isolated from the archaeon Natronobacterium pharaonis (Tachibana, 1994). A GFDPS gene with high similarity to archaeal geranylgeranyl diphosphate synthase (GGDPS) was subsequently cloned from the archaeon Aeropyrum pernix (Tachibana et al., 2000). Recently, another GFDPS gene was identified from the archaeon Methanosarcina mazei, which was predicted to have arisen from long-chain IDSs (Ogawa et al., 2010). Other research on the biosynthesis of sesterterpenoids has mainly focused on in vivo feeding experiments with isotopically labeled precursors. For instance, mangicols, a class of sesterterpenoids with anti-inflammatory activity isolated from the marine fungus Fusarium heterosporum, have been demonstrated to use GFDP as their biosynthetic precursor through feeding of labeled sodium acetate (Renner et al., 2000). However, little is known about the biosynthesis of plant sesterterpenoids, except for an analysis of the formation of the linear C25 precursor GFDP in the Brassicaceae (Nagel et al., 2015; Wang et al., 2015).
Recently, we found that two Himalayan-Chinese species of the Lamiaceae, Leucosceptrum canum and Colquhounia coccinea var mollis, harbored two unique classes of defensive sesterterpenoids, namely, leucosceptroids and colquhounoids, respectively, which are closely related due to their common 5/6/5 core structure but are distinguished from each other based on differences in stereochemistry at C-6 and C-7 and later modifications (Luo et al., 2010; Li et al., 2013). Subsequent investigation led to the isolation and identification of over 100 defensive sesterterpenoids from the leaves and flowers of the above two plants (Luo et al., 2011, 2013a, 2013b, 2014). In addition, Choudhary and coworkers reported the isolation of several sesterterpenoids from L. canum of Nepalese origin, but their skeletons are different from those discovered in the Chinese plant (Choudhary et al., 2004a, 2004b, 2007). Curiously, diterpenoids rather than sesterterpenoids were found in Colquhounia seguinii, a sister species of C. coccinea var mollis (Li et al., 2014), and no sesterterpenoids were detected in other congeneric species, including Colquhounia compta and Colquhounia vestita, suggesting that the biosynthesis of sesterterpenoids in some Lamiaceae lineages may be of recent evolutionary origin.
To learn more about the molecular and biochemical bases of sesterterpenoid formation in plants and its evolution, we investigated the IDSs of L. canum, a large woody member of the Lamiaceae up to 10 m high with flowers containing colored nectar. Here, we report the cloning, functional characterization, and subcellular localization of the plant GFDP synthase gene Lc-GFDPS. Our results shed light on the ecological role of sesterterpenoids in this species and indicate that this GFDPS probably arose from an ancestral GGDPS through gene duplication and neofunctionalization driven by positive selection.
RESULTS
Sesterterpenoids Are Highly Enriched in L. canum Glandular Trichomes
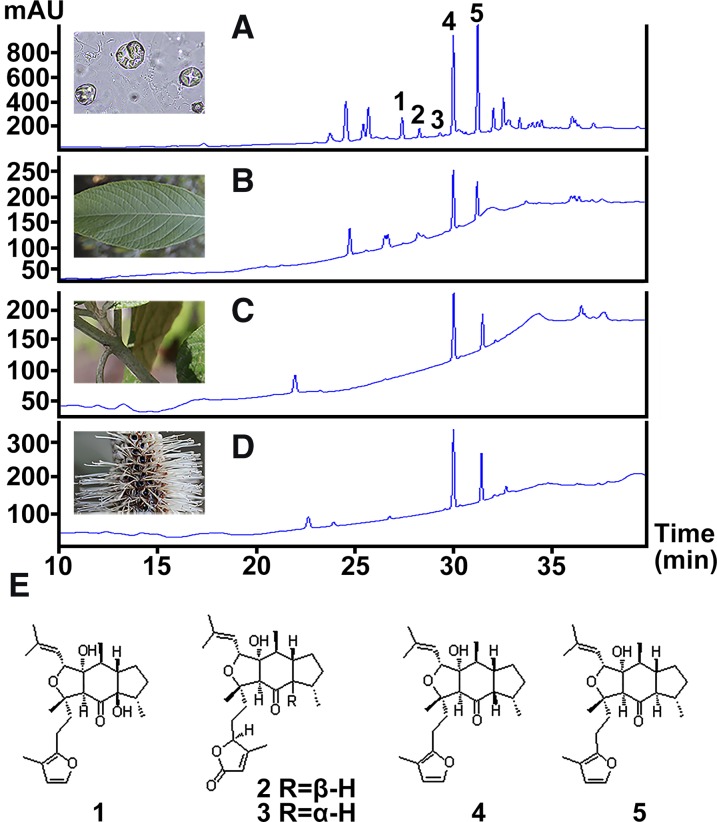
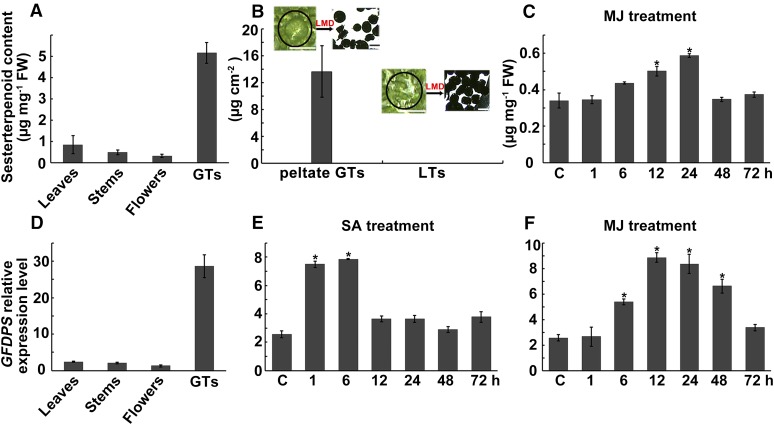
Our previous study of L. canum identified two sesterterpenoids possessing a unique C25 skeleton, the leucosceptroids A and B, and found these to be present in glandular trichomes (GTs) on surfaces of the aerial parts of the plant (Luo et al., 2010). This suggested that the biosynthesis of these sesterterpenoids was probably also accomplished in GTs, since terpenoids accumulating in these structures in other plant species have been previously reported to be formed in situ (McCaskill et al., 1992; Wagner et al., 2004; Wang et al., 2008; Xie et al., 2008; Schilmiller et al., 2009). To investigate the relative distribution of sesterterpenoids in GTs compared with various whole organs of L. canum, HPLC analysis was performed on extracts of GTs, whole leaves, young stems, and flowers. In addition to previously reported leucosceptroids A (1, tR = 27.3 min) and B (5, tR = 31.8 min) (Luo et al., 2010), a number of other peaks were also detected (Figure 1). These were not observed previously, possibly due to sampling having been conducted at a different time of year. To identify these unknown peaks, GT exudates were collected by wiping the fresh leaves with an acetone-soaked cotton swab, and the major components were separated by column chromatography followed by semipreparative HPLC purification. Through comparison of their 1H NMR and 13C NMR spectra with those of the sesterterpenoids previously isolated from L. canum, three additional peaks with retention times of 28.0, 29.0, and 30.9 min were identified as leucosceptroids I (2) and J (3) (Luo et al., 2013b), and 11-βH-leucosceptroid B (4) (Huang et al., 2013), respectively. Considering that compounds 4 and 5 were major peaks in all organs, the combined amount of these two compounds was quantified by HPLC using purified compounds as external standards. The content of sesterterpenoids in GTs (5.2 μg mg−1 fresh weight) was 8- to 15-fold higher than the average contents in whole leaves, young stems, and flowers (all samples still contained GTs) (Figure 2A). Additionally, peltate (shield-shaped) GTs and other neighboring leaf tissues (LTs) that did not contain peltate GTs were carefully selected under the microscope and dissected using laser microdissection (LMD). Approximately 200 peltate GTs or LTs with a total area of 1.25 to 1.87 mm2 were collected, respectively. The contents of compounds 4 and 5 were then quantified via ultraperformance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS). The combined amount of compounds 4 and 5 in peltate GTs was 13.63 ± 3.84 μg cm−2, but neither compound was detectable in LTs (Figure 2B), indicating that sesterterpenoids are highly enriched in GTs compared with other tissues. Thus, the GTs of L. canum provide an excellent system to study the biosynthetic pathway of plant sesterterpenoids.
Figure 1.
Sesterterpenoids in Different Organs of L. canum.
(A) to (D) HPLC analysis of sesterterpenoids in GTs (A), whole leaves (B), young stems (C), and flowers (D).
(E) The structures of five major sesterterpenoids, 1 to 5, corresponding to the peaks in (A).
Figure 2.
Sesterterpenoid Content and IDS3 (=GFDPS) Transcript Level.
(A) to (C) Sesterterpenoid contents in different organs of L. canum (A), in peltate GTs and neighboring LTs (B), and in leaves after MJ treatment (C). The insets illustrate how tissue was collected for GT and LT samples via laser microdissection.
(D) to (F) IDS3 expression level (relative to that of Actin) in different organs of L. canum (D) and in leaves after SA (E) and MJ (F) treatments.
Values represent the mean ± sd of biological replicates. Asterisks indicate significant differences from control plants (P < 0.05 by significant one-way ANOVA tests; n = 3 to 5). FW, fresh weight. Bar = 50 μm.
GFDP Is Found in L. canum Glandular Trichomes
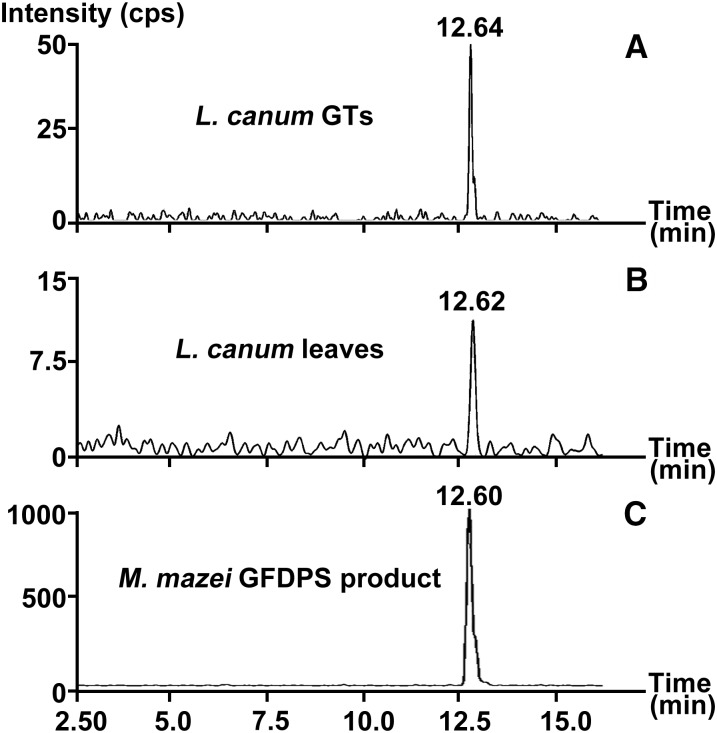
Sesterterpenoids have been postulated to be biosynthesized from the C25 prenyl diphosphate precursor GFDP in several organisms (Renner et al., 2000; Matsuda et al., 2015; Qin et al., 2015; Ye et al., 2015). We therefore looked for GFDP in L. canum. Using UPLC-MS/MS, GFDP was detected in GTs and whole leaves (containing GTs) of L. canum (Figure 3). When the relative level of GFDP was determined with geranylgeranyl diphosphate (GGDP) as an external standard, the accumulation of GFDP in GTs was 0.011 ± 0.003 μg mg−1 fresh weight, which was ∼3 times higher than that in whole leaves (0.004 ± 0.001 μg mg−1 fresh weight). Given this enrichment, which parallels that of the sesterterpenoids, GFDP is likely the biosynthetic precursor of sesterterpenoids in L. canum.
Figure 3.
Analysis of GFDP in GTs and Whole Leaves of L. canum.
Ion chromatograms with MRM transition of m/z 517 > 79 of L. canum GT extracts (A), L. canum leaf extracts (B), and M. mazei GFDPS assay products (C).
Cloning, Expression and Characterization of a GFDP Synthase
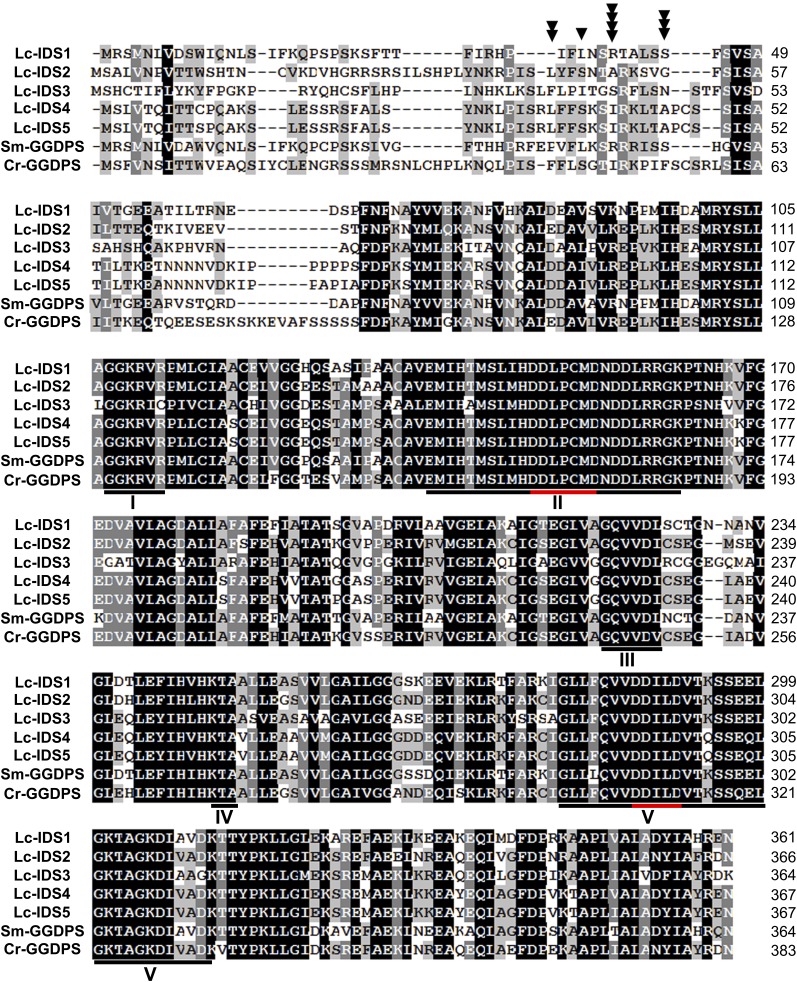
To investigate the plant sesterterpenoid biosynthetic pathway, high-throughput sequencing of mRNA isolated from L. canum GTs was undertaken (Supplemental Figures 1 and 2). The mRNA sequences were searched with known IDS sequences, which suggested 15 unigenes as candidate GFDPSs (Supplemental Data Set 2). To obtain full-length sequences, 5′ and 3′ RACE was performed, which yielded 10 different candidates designated as IDS1–10 (Table 1). In particular, IDS1–5 showed high similarity to other plant GGDPS sequences. Alignment of the deduced amino acid sequences with the corresponding sequences from functionally characterized plant GGDPSs revealed the presence of five conserved regions in IDS1–5, including two conserved aspartate-rich motifs (DDLPCMD and DDILD) (Figure 4). The amino acid sequence of IDS6 shares 94% identity with farnesyl diphosphate synthase (FDPS) from peppermint (Mentha × piperita), while Lc-IDS7 is 87% identical to solanesyl diphosphate synthase from rubber tree (Hevea brasiliensis). Unlike Lc-IDS1–7, Lc-IDS8–10 lack either one or two typical aspartate-rich motifs and show high similarity to the geranyl diphosphate synthase (GDPS) small subunit from Salvia miltiorrhiza, with identities ranging from 61 to 88%. The properties of these Lc-IDSs, coupled with the sequence homology of archaeal GFDPSs (Tachibana et al., 2000; Ogawa et al., 2010) to archaeal GGDP sequences, led us to suspect that L. canum GFDPS might be similar to plant GGDPS. Thus, the five GGDPS-like IDSs were targeted as candidate GFDPSs in L. canum.
Table 1. Bioinformatic Analysis of 10 Candidate GFDP Synthases from the Glandular Trichomes of L. canum.
| Name | ORF Length (bp) | Peptide Length (aa) | Estimated Molecular Mass (kD) | Estimated pI | Functional Annotation (BLASTP in Database Nr) | Identity (%) | Predicted Subcellular Localization (Using Plant-mPLoc) |
|---|---|---|---|---|---|---|---|
| IDS1 | 1086 | 361 | 39.0 | 5.63 | GGDPS from S. miltiorrhiza | 85 | Chloroplast, plastid |
| IDS2 | 1101 | 366 | 39.9 | 5.82 | GDPS.LSU from Catharanthus roseus | 75 | Chloroplast, plastid |
| IDS3 | 1095 | 364 | 39.5 | 6.95 | GGDPS from Croton sublyratus | 63 | Chloroplast |
| IDS4 | 1104 | 367 | 39.8 | 5.76 | GDPS.LSU from C. roseus | 70 | Chloroplast, plastid |
| IDS5 | 1104 | 367 | 39.8 | 5.76 | GDPS.LSU from C. roseus | 71 | Chloroplast, plastid |
| IDS6 | 1050 | 349 | 40.0 | 5.64 | FDPS from M. piperita | 94 | Mitochondrion |
| IDS7 | 1266 | 421 | 46.4 | 5.33 | Putative SDPS from S. miltiorrhiza | 87 | Chloroplast |
| IDS8 | 975 | 324 | 35.4 | 5.21 | GDPS.SSU II from S. miltiorrhiza | 88 | Chloroplast |
| IDS9 | 915 | 304 | 32.6 | 6.21 | GDPS.SSU I from S. miltiorrhiza | 61 | Chloroplast |
| IDS10 | 993 | 330 | 36.1 | 6.03 | GDPS.SSU II from S. miltiorrhiza | 70 | Chloroplast |
Figure 4.
Alignment of Deduced Amino Acid Sequences of Lc-IDS1-5 with Known Plant GGDPS.
Sm-GGDPS, S. miltiorrhiza GGDPS (accession number ACJ66778); Cr-GGDPS, Catharanthus roseus GGDPS (accession number AEI53622). Identical amino acids are highlighted in black. Identical and similar residues in at least four out of six sequences are highlighted in gray. Five conserved regions (I to V) are indicated by black lines, in which the conserved Asp-rich motifs are highlighted by red lines. The truncation site for IDS1 is indicated by a triangle, IDS2 by two triangles, IDS3 by three triangles, and IDS4 and IDS5 by four triangles.
The transcript profiles of five GGDPS-like candidates were analyzed using qRT-PCR with the actin gene as an internal control. As shown in Figure 2D, IDS3 was expressed predominantly in GTs where sesterterpenoids accumulated, while IDS1 was abundantly expressed in all organs examined, IDS2 was expressed in leaves and flowers, and IDS4 and IDS5 were mainly expressed in flowers (Supplemental Figure 3). These results suggest that IDS3 is the most likely candidate GFDPS gene in L. canum. The full-length cDNA of IDS3 is 1382 bp in length with an open reading frame (ORF) of 1095 bp encoding a protein of 364 amino acids with a calculated molecular mass of 39.5 kD.
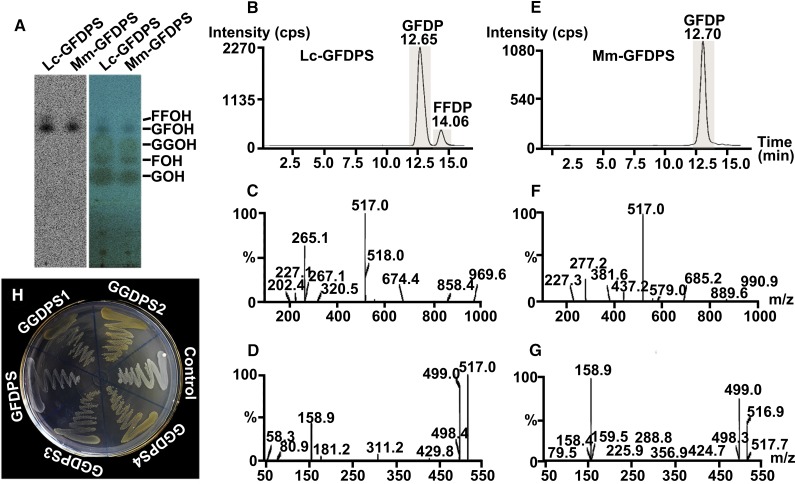
For functional characterization of GFDPS, the protein encoded by IDS3 was expressed in Escherichia coli after truncation of the N-terminal predicted signal sequence and was subsequently purified with Ni-NTA agarose columns (Supplemental Figure 4). When incubated with isopentenyl diphosphate (IDP) and an allylic substrate, i.e., dimethylallyl diphosphate (DMADP), geranyl diphosphate (GDP), farnesyl diphosphate (FDP), or GGDP, the purified recombinant IDS3 protein catalyzed the formation of GFDP, the linear precursor of sesterterpenoids, as its major product, as well as a minor C30 prenyl diphosphate by-product (tentatively identified as farnesylfarnesyl diphosphate). The identity of GFDP was established through comparison of its Rf value on radio-thin-layer chromatography (radio-TLC) with that of the product of a positive control, M. mazei GFDPS (Ogawa et al., 2010) (Figure 5A). The product was further confirmed through comparison of its retention time, MS, and MS/MS spectra with those of the product of M. mazei GFDPS (Figures 5B to 5G), using UPLC-MS/MS in multiple reaction monitoring (MRM) mode with an MRM transition of m/z 517 > 79. Based on its major product, IDS3 was therefore renamed as L. canum GFDP synthase (GFDPS). In addition, the other four GGDPS-like IDSs (1, 2, 4, and 5) were also expressed in a similar way, and their in vitro enzyme activities were identified by radio-TLC. All purified recombinant proteins catalyzed the formation of GGDP as their major product, with GFDP as a minor by-product (Supplemental Figure 5). Based on their product profiles, IDSs 1, 2, 4, and 5 were renamed as L. canum GGDP synthases 1–4 (GGDPS1–4), respectively.
Figure 5.
Enzyme Activity Assay of GFDPS.
(A) Product identification by radio-TLC: 14C image (left) and merged image of 14C image and iodine coloration (right) of hydrolyzed products of assays of Lc-GFDPS and M. mazei GFDPS performed with [1-14C]IDP and GGDP and separated by TLC. Mm-GFDPS, M. mazei GFDPS; GOH, geraniol; FOH, farnesol; and GGOH, geranylgeraniol, indicate the authentic standards; GFOH, geranylfarnesol, indicates the major enzyme assay product; FFOH, farnesylfarnesol, indicates the minor enzyme assay product.
(B) and (E) Ion chromatograms with MRM transition of Lc-GFDPS and M. mazei GFDPS assay products from assays conducted with IDP and GGDP. GFDP indicates the major enzyme assay product; FFDP, farnesylfarnesyl diphosphate, indicates the minor enzyme assay product.
(C) and (D) MS and MS/MS spectra of the peaks with retention time of 12.65 min from assays with Lc-GFDPS (B).
(F) and (G) MS and MS/MS spectra of the peaks with retention time of 12.70 min from assays with M. mazei GFDPS (E).
(H) Complementation assay of GFDPS and GGDPSs in E. coli with carotenoid gene cluster from E. uredovora lacking GGDPS.
The purified recombinant GFDPS was not active when Mn2+ was used as cofactor, and increased Mg2+ concentration (to 20 mM) did not significantly improve the enzyme activity. However, the GFDPS activity was dramatically enhanced by adding the detergent Tween 20 to the enzyme assay system (Supplemental Table 1). Under these conditions, the product profile of recombinant GFDPS was examined using different ratios of isoprenyl substrates (1:1, 3:1, or 5:1 of IDP with DMADP, GDP, FDP, or GGDP), and it was found that GFDP was always the predominant product (Supplemental Table 2). Kinetic analysis using Lineweaver-Burk plots revealed that the recombinant GFDPS preferred allylic diphosphates as substrates in the order of GGDP = FDP > GDP > DMADP (Table 2). The kinetic constants for GGDP and IDP were in the same range as those for other IDSs (Schmidt and Gershenzon, 2007; Nagel et al., 2015).
Table 2. Kinetic Parameters for the Purified Recombinant GFDPS.
| Varying Substrate | Fixed Substrate | Km (μM) | Kcat × 10−6 (s−1) | Kcat/Km (s−1 M−1) |
|---|---|---|---|---|
| DMADP (0–100 μM) | IDP (100 μM) | 34.89 | 16.90 | 0.48 |
| GDP (0–50 μM) | IDP (100 μM) | 6.28 | 6.16 | 0.95 |
| FDP (0–50 μM) | IDP (100 μM) | 4.10 | 7.19 | 2.11 |
| GGDP (0–25 μM) | IDP (100 μM) | 1.65 | 5.17 | 2.96 |
| IDP (0–50 μM) | GGDP (100 μM) | 3.04 | 8.20 | 2.95 |
All results are presented as the averages of duplicate experiments.
Considering that GFDPS had high sequence similarity to the above-mentioned four GGDPSs, with identity ranging from 61 to 70%, but no GGDPS activity was detected in in vitro enzymatic assays, we further investigated this protein. A complementation assay was performed by cotransforming GFDPS and pACCAR25ΔcrtE, which carried the carotenoid biosynthesis gene cluster except for GGDPS from Erwinia uredovora (Kainou et al., 1999). GGDPS1–4 were cotransformed into E. coli strain BL21 (DE3), with pACCAR25ΔcrtE as a positive control. The colonies harboring GGDPS1–4 in the background of pACCAR25ΔcrtE produced the expected yellow color pigments, indicating their ability to produce GGDP that was subsequently converted to carotenoids. However, no yellow-colored colonies were observed in the transformants containing GFDPS and pACCAR25ΔcrtE (Figure 5H). These results indicate that GFDPS lacks GGDPS activity.
Arabidopsis thaliana Plants Overexpressing GFDPS Have GFDP Synthase Activity
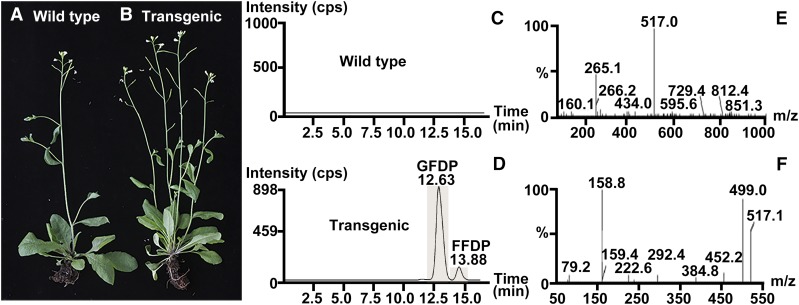
To determine whether GFDPS possesses GFDP synthase activity in vivo as found for the heterologously expressed protein in vitro, we generated transgenic Arabidopsis plants overexpressing Lc-GFDPS driven by a double CaMV 35S promoter. A set of 58 independently transformed lines were analyzed by RT-PCR and DNA sequencing, and the GFDPS transcript level determined by qRT-PCR was comparable to the reference gene actin (0.99 ± 0.03 of the actin gene) in transgenic lines. To measure GFDPS activity in transgenic Arabidopsis, enzyme assays were performed with crude protein extracts of leaves. We found that the GFDPS-overexpressing lines catalyzed the formation of GFDP when incubated with the substrates IDP and any of several allylic cosubstrates (DMADP, GDP, FDP, or GGDP), while the wild-type Arabidopsis did not (Figures 6C to 6F), confirming the enzymatic activity of GFDPS in planta. In addition, after the initiation of flowering, we found that several stalks bolted from the rosettes of the transgenic Arabidopsis plants (Figure 6B) in comparison to the typical single stalk that bolted from the rosette of wild-type plants (Figure 6A), an observation that warrants further in-depth investigation.
Figure 6.
Overexpression of GFDPS in Arabidopsis and Enzyme Assay with Crude Protein Extracts.
(A) and (B) Morphology of wild-type control and GFDPS-overexpressing (transgenic) Arabidopsis plants.
(C) and (D) Ion chromatograms with MRM transition of the enzyme assay products of crude protein extracts from wild-type control and GFDPS-overexpressing (transgenic) Arabidopsis plants assayed with IDP and GGDP. GFDP indicates the major enzyme assay product; FFDP, farnesylfarnesyl diphosphate, indicates the minor enzyme assay product.
(E) and (F) MS and MS/MS spectra of the peak with retention time of 12.63 min in (D).
L. canum GFDP Synthase Is Targeted to Chloroplast and Uses Substrates Derived from the MEP Pathway
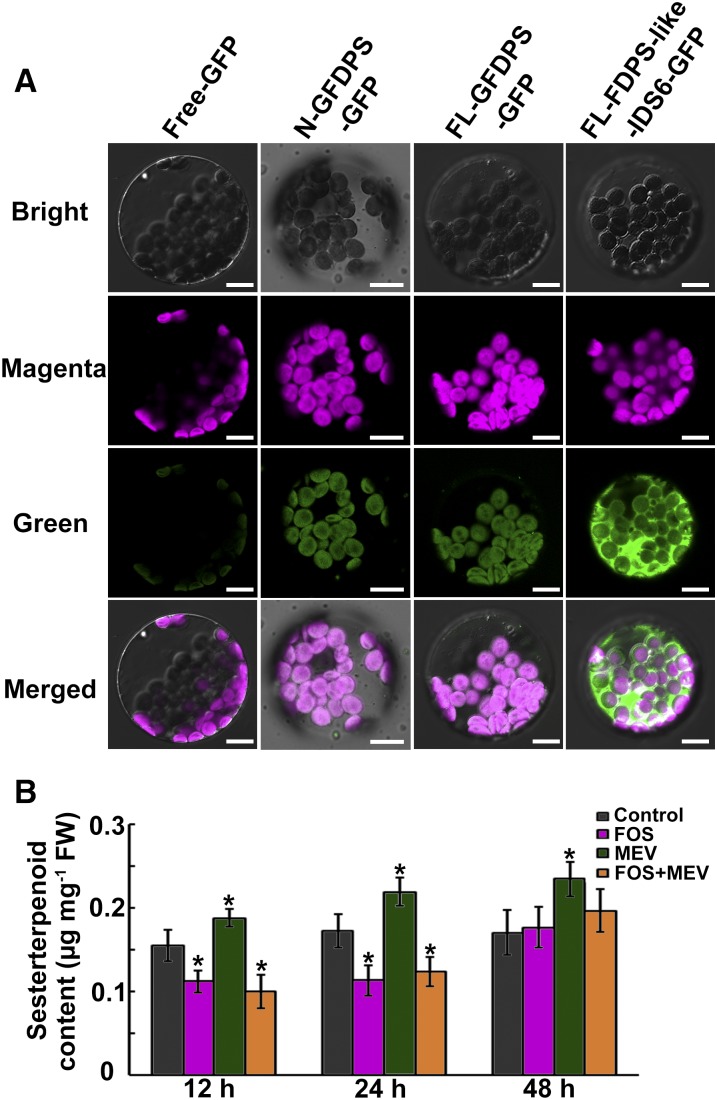
Although GFDPS was predicted to be localized to chloroplasts by Plant-Ploc (http://www.csbio.sjtu.edu.cn/bioinf/plant/) and LocTree3 (https://www.rostlab.org/services/loctree2/) software, the predicted score for chloroplast localization was only 0.207 (possible range: 0 to 1.000; the higher the score, the more accurate the prediction) with Target P software (http://www.cbs.dtu.dk/services/TargetP/). To test the subcellular predictions experimentally, recombinant plasmids containing the DNA constructs encoding the first 51 amino acids of GFDPS or the full-length cDNA of GFDPS in fusion with the GFP gene were introduced into Arabidopsis leaf protoplasts. Meanwhile, the GFP signals for GGDPS1–4, which were predicted to be chloroplast localized by Target P (with high scores), and the above-mentioned FDPS-like IDS6, which was predicted to be cytosol-localized, were also tested for comparison (Figure 7A; Supplemental Figure 6). All GFP signals for GFDPS and GGDPSs were found exclusively in the chloroplasts (Figure 7A; Supplemental Figure 6), while the GFP signals for FDPS-like IDS6 produced a typical cytosolic localization pattern (Figure 7A), indicating that GFDPS is targeted to the chloroplast.
Figure 7.
Subcellular Localization of GFDPS and Sesterterpenoid Content in L. canum after Inhibitor Treatments.
(A) GFP fusion proteins in Arabidopsis leaf mesophyll protoplasts visualized by laser confocal microscopy. Chloroplasts are represented by magenta chlorophyll autofluorescence. Green fluorescence indicates GFP signal. Free-GFP, N-GFDPS-GFP, FL-GFDPS-GFP, and FL-FDPS-like-IDS6-GFP indicate Arabidopsis leaf protoplasts harboring the plasmids containing the empty vector fused to GFP, the N-terminal predicted transit peptide of GFDPS fused to GFP, the full-length cDNA of GFDPS fused to GFP, and the full-length cDNA of FDPS-like IDS6 fused to GFP, respectively. Bar = 10 μm.
(B) Sesterterpenoid contents in L. canum leaves after FOS, MEV, and FOS+MEV treatments. Values represent the mean ± sd. Asterisks indicate significant differences from control plants (P < 0.05 by significant one-way ANOVA tests; n = 9). FW, fresh weight.
Since GFDPS is localized to plastids, it is likely that sesterterpenoids in L. canum are biosynthesized via the plastidial 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway. However, analysis of the L. canum GT transcriptome revealed sequences for all steps of the mevalonic acid (MVA) pathway as well as nearly all steps of the MEP pathway (Supplemental Data Set 2). To investigate this question further, mevinolin (MEV), a specific inhibitor of 3-hydroxy-3-methylglutaryl CoA reductase in the MVA pathway (Lichtenthaler, 2000), and fosmidomycin (FOS), a specific inhibitor of 1-deoxy-d-xylulose 5-phosphate reductoisomerase in the MEP pathway (Kuzuyama et al., 1998; Lichtenthaler, 2000), were used for an in vivo inhibition study. The accumulation of MVA-derived β-sitosterol and MEP-derived phytol were measured to determine the efficiency of inhibition. The decrease in levels of β-sitosterol with MEV and phytol with FOS (55.18% ± 3.21% and 79.35% ± 6.43% of controls, respectively, at 24 h after treatment; Supplemental Figure 7) suggested that the inhibitors were penetrating the plant tissue and having the expected effect. HPLC analysis indicated that compound 5 was present most prominently in each L. canum plantlet; therefore, it was used as a marker sesterterpenoid. When the plantlets were treated with MEV, the content of compound 5 increased significantly (134.21% ± 7.09% of controls at 24 h after treatment, one-way ANOVA: F = 13.861, P < 0.01) (Figure 7B). However, FOS treatment resulted in a dramatic decrease in compound 5 content (63.26% ± 4.78% of controls at 24 h after treatment, one-way ANOVA: F = 8.254, P < 0.05) (Figure 7B). When the plantlets were subjected to treatment with both inhibitors simultaneously, there was also a sharp drop in compound 5 content (68.72% ± 3.90% of controls at 24 h after treatment, one-way ANOVA: F = 10.599, P < 0.05) as observed for FOS treatment, suggesting that the plastidial MEP pathway is the primary supplier of isoprenyl diphosphate precursors for sesterterpenoid biosynthesis (Figure 8).
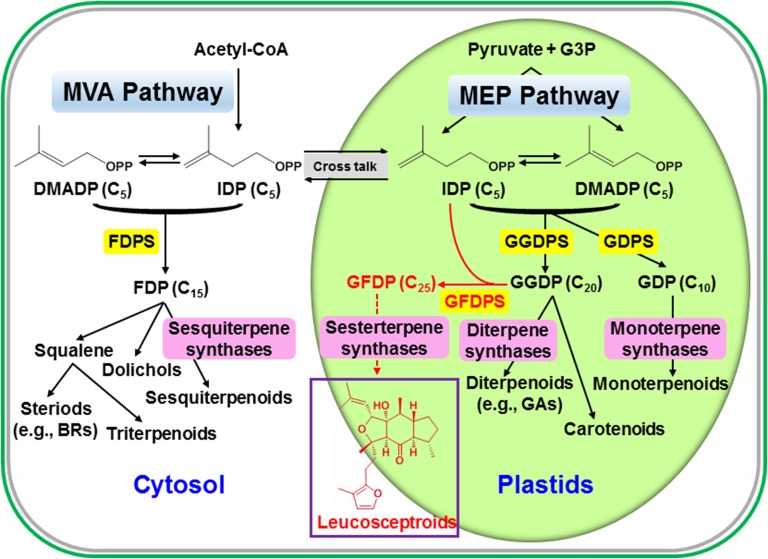
Figure 8.
Outline of the Basic Pathways of Terpenoid Metabolism, Including the Formation of Sesterterpenoids.
The biosynthesis of the C25 precursor GFDP to leucosceptroid sesterterpenoids is catalyzed by GFDPS that uses substrates GGDP and IDP derived from the plastidial MEP pathway (shown in red).
L. canum GFDPS Transcript, Sesterterpenoids Formation and Defense Function Are Stimulated by Jasmonates
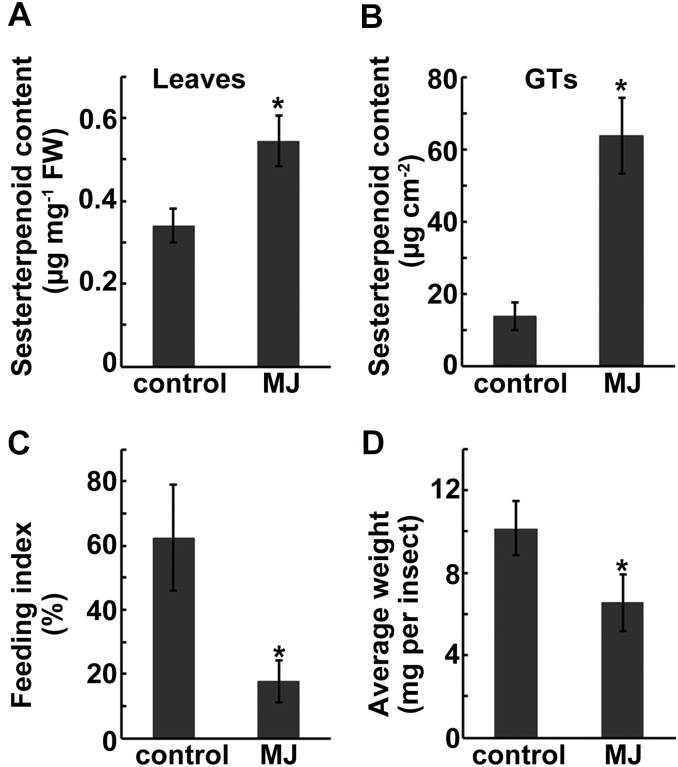
Since our previous work had indicated that the sesterterpenoids of L. canum function as defense compounds against insect herbivores and pathogens (Luo et al., 2010, 2011, 2013a, 2013b, 2014), we were also interested in investigating whether GFDPS expression and sesterterpenoid accumulation could be induced by the defense hormones methyl jasmonate (MJ) and salicylic acid (SA). The qRT-PCR results show that treatment of the plant with 100 μM MJ or 100 μM SA significantly stimulated the expression of GFDPS in the leaves of L. canum saplings (Figures 2E and 2F). The highest levels of GFDPS transcript appeared at ∼12 and 24 h after treatment with MJ (one-way ANOVA: F = 11.337, P < 0.05). In accordance with the GFDPS transcript level, the accumulation of two major sesterterpenoids (compounds 4 and 5) in whole leaves increased after MJ treatment, reaching their maximum levels after 24 h (one-way ANOVA: F = 5.917, P < 0.05) (Figure 2C). The level of these two compounds in GTs of MJ-sprayed L. canum (12 to 24 h after treatment), as determined by laser microdissection coupled with UPLC-MS/MS, was 63.78 ± 10.52 μg cm−2, which was 4.7-fold higher than that of untreated GTs (Figure 9B). In neighboring leaf tissue, compounds 4 and 5 were still under the detection limit, regardless of whether the plants were treated with the phytohormones. In contrast to MJ, no obvious changes in sesterterpenoid content were observed after SA treatment (one-way ANOVA: F = 0.146, not significant). These results suggest that MJ signaling might be involved in regulating GFDPS gene expression and thereby the accumulation of its product and thus shed light on the molecular regulation of sesterterpenoid accumulation in L. canum.
Figure 9.
Sesterterpenoid Contents of the Whole Leaves and GTs of L. canum after MJ Treatment and Increased Resistance of Leaves to Herbivores.
(A) and (B) Sesterterpenoid contents in whole leaves at 12 h after MJ treatment and in GTs at 12 to 24 h after MJ treatment.
(C) Feeding index of leaves at 12 h after MJ treatment.
(D) Average weight of beet armyworm larvae after 5 d of feeding on artificial diet containing MJ-treated or untreated control leaves.
Values represent the mean ± sd. Asterisks indicate significant differences from control plants (significant one-way ANOVA tests: (A) P < 0.05, n = 5; (B) P < 0.05, n = 3; (C) P < 0.05, n = 5; (D) P < 0.01, n = 50.
To determine whether the increased sesterterpenoid formation after MJ treatment (Figure 9A) could contribute to increased plant defense, a generalist insect herbivore, beet armyworm (Spodoptera exigua), which has been found to feed on L. canum leaves in the laboratory, was used. Leaves collected at 12 h after L. canum treatment with MJ showed a potent antifeedant effect against S. exigua, with a feeding index significantly less than that of the unsprayed control (17.7% ± 0.65% versus 62.5% ± 0.17%, one-way ANOVA: F = 19.06, P < 0.05) (Figure 9C). The growth inhibitory effect of MJ-treated L. canum against S. exigua was tested by feeding the insect with an artificial diet containing MJ-treated leaves or control. After 5 d, the growth of S. exigua on MJ-sprayed foliage was significantly inhibited compared with the unsprayed control (6.55 ± 1.30 versus 10.16 ± 1.70 mg per insect; one-way ANOVA: F = 46.769, P < 0.01) (Figure 9D). These results indicate that the MJ-induced increase in sesterterpenoid levels in L. canum could contribute to enhanced defense against insect pests.
Positive Selection Drives Neofunctionalization of GFDP Synthase
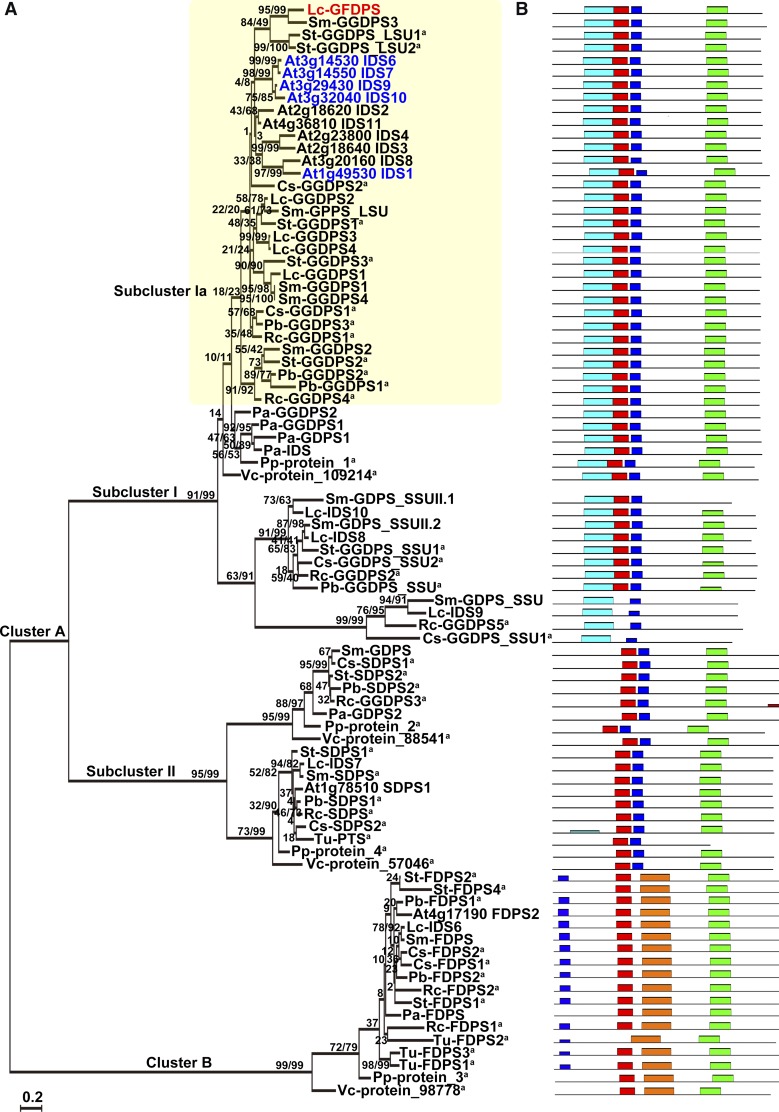
To gain insight into the evolution of GFDPS, IDS genes from 10 representative plants (one green alga, one moss, one conifer, one monocot, and six dicots) were compared. The number of predicted IDS genes ranged from 4 to 15, with Volvox carteri possessing the fewest copies and vascular plants containing the most. A phylogenetic tree of IDSs was then constructed with the maximum likelihood method. As shown in Figure 10A, two main clusters, A and B, were identified, with cluster A containing two subclusters, I and II. The conserved motifs of the encoded proteins were then predicted using MEME software (http://meme.nbcr.net/meme/) (Figure 10B; Supplemental Table 3) (Bailey et al., 2009). The topological structure of the phylogenetic tree was congruent with the biochemical function rather than with the taxonomic origin of the IDSs, as genes for all the plant species tested were found in each of the three clusters, as noted for previously constructed IDS phylogenic trees (Tholl et al., 2004; Schmidt and Gershenzon, 2008; Hsieh et al., 2011). Therefore, it can be postulated that the differentiation of IDSs into these three categories occurred before the differentiation of the plants in this phylogeny. The evolution of any increase in IDS gene number in vascular plants is thus largely due to gene duplication after the divergence of their biochemical functions. However, some new functions have arisen, such as the evolution of GFDPS activity.
Figure 10.
Phylogenetic Analysis and Conserved Motifs of Plant IDSs.
(A) Phylogenetic analysis of Lc-GFDPS and other Lc-IDSs along with their homologous sequences in 11 representative plants (plant species abbreviations are listed in Supplemental Data Set 3). Numbers above branches show bootstrap support values inferred from maximum likelihood and distance analyses, respectively. GDPS.LSU, GDPS large subunit; GDPS.SSU, GDPS small subunit; PTS, prenyltransferases; SDPS, solanesyl diphosphate synthases. aPredicted protein.
(B) The conserved motifs of each IDS corresponding to the phylogenetic tree (conserved motifs represented by different colored bars are listed in Supplemental Table 4).
Lc-GFDPS clearly clustered in subcluster Ia, which is dominated by GGDPS sequences, albeit not all have been biochemically confirmed. Since Lc-GFDPS contains identical conserved motifs to plant GGDPSs (Figure 10B), the new function of Lc-GFDPS was probably due to alteration of individual amino acids. To explore the evolution of Lc-GFDPS, the codon substitution patterns were analyzed with a maximum likelihood (PAML) approach implementing a branch-site model (Yang, 1998) by setting Lc-GFDPS in the foreground and other homologs in subcluster Ia as the background. Significant positive selection was detected in Lc-GFDPS (P < 0.001) (Table 3), suggesting that positive selection was a driving force for the functional diversification of plant GFDPS.
Table 3. Molecular Evolutionary Analysis of Lc-GFDPS.
| Branch-Site Model | Parameters | LnL | Pdf=1 | Positive Selected Sitesa | ||||
|---|---|---|---|---|---|---|---|---|
| Null | Site class | ω0 | ω1 | ω2a | ω2b | −13624.6953 | ||
| Proportion | 0.78662 | 0.04379 | 0.16064 | 0.00894 | ||||
| Background | 0.06289 | 1.00000 | 0.06289 | 1.00000 | ||||
| Foreground | 0.06289 | 1.00000 | 1.00000 | 1.00000 | ||||
| Alternative | Site class | ω0 | ω1 | ω2a | ω2b | −13618.1028 | P < 0.001 | 108 L (0.981*) |
| Proportion | 0.88027 | 0.04712 | 0.06892 | 0.00369 | 137 A (1.000**) | |||
| Background | 0.06257 | 1.00000 | 0.06257 | 1.00000 | 202 K (0.955*) | |||
| Foreground | 0.06257 | 1.00000 | 999.00000 | 999.00000 | 321 L (0.968*) | |||
Posterior probabilities calculated by Bayes Empirical Bayes analysis. LnL, Log likelihoods.
The amino acids and their positions refer to those of GFDPS.
DISCUSSION
GFDP Synthase Provides the Precursor for Sesterterpenoid Formation in the Glandular Trichomes of L. canum
Sesterterpenoids have been reported from widespread sources (vascular plants, lichens, terrestrial fungi, insects, and various marine organisms especially sponges) and have attracted extensive interest due to their novel and complex structures as well as various biological activities (Liu et al., 2007; Wang et al., 2013; Zhang and Liu, 2015). However, their biosynthetic pathways have not been well investigated to date, with only a few related genes and enzymes reported from microorganisms (Tachibana, 1994; Tachibana et al., 2000; Ogawa et al., 2010; Chiba et al., 2013; Matsuda et al., 2015; Qin et al., 2015; Ye et al., 2015). In this study, we cloned and functionally identified a GFDP synthase gene from GTs of L. canum (a woody member of the Lamiaceae) through high-throughput sequencing of GT mRNA in combination with RACE technology. The GFDP synthase activity was first demonstrated by an in vitro enzyme assay of the recombinant protein expressed heterologously in E. coli and then confirmed by overexpression of this gene in Arabidopsis and analysis of GFDPS activity in crude extracts. Additionally, the transcript profile of GFDPS was completely in accordance with the sesterterpenoid accumulation pattern in L. canum, further indicating that Lc-GFDPS might be involved in sesterterpenoid biosynthesis.
The deduced protein sequence of Lc-GFDPS contains two highly conserved aspartate-rich motifs, DDLPCMD and DDILD, and shows high similarity to plant GGDPSs, especially GGDPSs from L. canum, with identity ranging from 61 to 70%. Lc-GFDPS shares 52 to 62% identity with Arabidopsis GFDPSs and GFDPSs from other Brassicaceae plants (Nagel et al., 2015; Wang et al., 2015), but only 33 and 34% identity with M. mazei GFDPS and A. pernix GFDPS, respectively. In Arabidopsis, GFDPS activity is associated with a change in the 5th amino acid in front of the first aspartate-rich motif from Met in GGDPS to Ala or Ser in GFDPS. However, Lc-GFDPS does not have an Ala or Ser residue at this position, but rather a Met, suggesting that the molecular basis of GFDPS formation in L. canum might be different from that in the Brassicaceae.
GFDPS catalyzed the formation of GFDP as its major product and a C30 prenyl diphosphate (likely farnesylfarnesyl diphosphate) as a minor by-product from the condensation of IDP with any of several allylic cosubstrates after expression in E. coli. Interestingly, no GGDPS activity was detected in either in vitro or in vivo enzymatic assays. The GFDPS enzymes reported from Brassicaceae plants and archaea all produce GGDP or GDP as minor products, in addition to GFDP (Tachibana et al., 2000; Ogawa et al., 2010; Nagel et al., 2015; Wang et al., 2015). Lc-GFDPS preferred GGDP or FDP as its allylic cosubstrate and showed much less activity with GDP and DMADP, which is similar to the substrate specificity of Arabidopsis IDS9 and M. mazei GFDPS but distinct from that of Arabidopsis IDS7 and A. pernix GFDPS (Tachibana et al., 2000; Ogawa et al., 2010; Nagel et al., 2015; Wang et al., 2015). These results point to differences in the GFDPS of various organisms and are consistent with independent recruitment of this enzyme in different plant lineages.
The discovery of GFDP synthase and its encoding gene in L. canum provides strong support for the formation of sesterterpenoids via GFDP, which is consistent with most previous suggestions about the pathway of sesterterpenoid biosynthesis in archaea, fungi, and marine sponges (Renner et al., 2000; Ogawa et al., 2010; Qin et al., 2015; Ye et al., 2015). However, the sesterterpenoids isolated from the plant Aletris farinosa were proposed to be derived from an oxidogeranylfarnesol precursor via mechanistic studies (Challinor et al., 2015). Moreover, other sesterterpenoids were reported to be generated by several classic triterpenoid synthases (Sato et al., 2013; Shinozaki et al., 2013; Ueda et al., 2015), indicating that alternate routes to sesterterpenoid formation might exist in different organisms, making this an interesting topic for further investigation.
Leucosceptroid Sesterterpenoids Are Biosynthesized in L. canum GTs and Function in Defense
Plant GTs are well known for their capacity to biosynthesize various secondary metabolites and thus have been regarded as so-called biochemical factories (Tissier, 2012). In particular, a wide variety of terpenoids have been reported to form and accumulate in GTs (Lange and Turner, 2013). In our previous work, we reported that the GTs of L. canum could accumulate leucosceptroid sesterterpenoids with a unique tetracyclic skeleton (Luo et al., 2010). Subsequent study using LMD coupled with UPLC-MS/MS disclosed another new class of sesterterpenoids, the colquhounoids, from the peltate GTs of C. coccinea var mollis (Li et al., 2013). In this study, three additional sesterterpenoids, leucosceptroids I, J, and 11-βH-leucosceptroid B, were identified from L. canum GTs, besides the already known leucosceptroids A and B. The content of these sesterterpenoids in isolated GTs was dramatically higher than their average contents in whole leaves, young stems, and flowers (all samples still contained GTs). Further investigation using LMD coupled with UPLC-MS/MS confirmed that sesterterpenoids were highly accumulated in peltate GTs but were undetectable in nonglandular leaf tissues, suggesting that leucosceptroid sesterterpenoids are specific secondary metabolites of the peltate GTs of L. canum. These findings, coupled with the observation that GFDPS is much more abundantly expressed in GTs, with the expression level significantly higher than in other organs, indicate that leucosceptroid sesterterpenoids are not only accumulated but are also biosynthesized in L. canum GTs.
Plant GT secondary metabolites (especially terpenoids) have been frequently reported to function in chemical defense against both biotic and abiotic stresses (Wagner et al., 2004; Li et al., 2014). Our previous studies have demonstrated that two major GT-specific sesterterpenoids (compounds 1 and 5) of L. canum showed antifeedant activities against the larvae of two generalist insect herbivores, the beet armyworm and cotton bollworm (Helicoverpa armigera), and their antifeedant activities were positively correlated with compound concentration (Luo et al., 2010). This study provides further evidence of the protective role of these trichome-stored substances at the molecular level by showing that the defensive hormone MJ could induce both GFDPS expression and sesterterpenoid accumulation while also enhancing the antifeedant and growth inhibitory effects of L. canum against the larvae of beet armyworm.
GFDP Synthase Is Localized to the Plastids of GT Cells and Uses Substrates Derived from MEP Pathway
Given the diversity of terpenoid pathway organization in different groups of organisms, sesterterpenoids can be expected to be biosynthesized via different routes. For example, in species of the pathogenic fungus Bipolaris, a group of phytotoxic sesterterpenoids known as ophiobolins are produced from GFDP via the MVA pathway (Canonica et al., 1967; Nozoe et al., 1967; Kawaguchi et al., 1973). In the diatom Rhizosolenia setigera, the Δ7(20)-haslene sesterterpenoids are generated through the MVA pathway, but the structurally similar Δ6(17)-haslenes are instead biosynthesized via the MEP pathway in the diatom Haslea ostrearia (Massé et al., 2004). In vascular plants, the MEP and MVA pathways both operate, but in different cellular compartments. Generally, the MEP pathway, localized in the plastids, provides substrates to GDPS and GGDPS, two enzymes that are also localized in plastids (Hemmerlin et al., 2012). On the other hand, the cytosolic-localized MVA pathway furnishes substrate for FDPS, which is also present in the cytosol (Hemmerlin et al., 2012). In L. canum, the presence of N-terminal signal peptides on the GFDPS suggested a plastidial location for GFDPS, which was confirmed by GFP-fusion experiments showing that GFDPS was confined to the chloroplasts of Arabidopsis leaf-mesophyll protoplasts.
Given its plastidial localization, GFDPS likely derives its substrates from the MEP pathway. This conclusion was substantiated by in vivo inhibitor studies with MEV and FOS, which have been frequently used to study the operation of the MVA and MEP pathways in plant terpenoid biosynthesis (Laule et al., 2003). The decrease in sesterterpenoid content upon treatment of the plant with FOS or FOS+MEV together, coupled with the significant increase in sesterterpenoid content upon MEV treatment, indicated that sesterterpenoid biosynthesis in L. canum uses isoprenyl diphosphate substrates derived from the plastidial MEP pathway (illustrated in Figure 8). This conclusion is consistent with the biochemical properties of GFDPS, which was found to use GGDP as its preferred allylic cosubstrate; GGDP is also formed in plastids (Hemmerlin et al., 2012). The increase in sesterterpenoid accumulation upon MEV treatment has a precedent in previous studies that have shown how blocking one terpenoid biosynthetic pathway stimulates increases in terpenoid levels by the other pathway (Laule et al., 2003; Ramak et al., 2013; Mansouri and Salari, 2014). Despite the formation of L. canum sesterterpenoids from MEP pathway substrates, the GT transcriptome of this species contains sequences for nearly all steps of both the MEP and MVA pathways. Hence, in addition to sesterterpenoids, other types of terpenoids (e.g., sesquiterpenoids and triterpenoids) might be formed in these structures utilizing substrate supplied by the MVA pathway.
GFDP Synthase Evolved from a GGDP Synthase via Positive Selection
Our comprehensive phylogenetic analysis demonstrated that Lc-GFDPS clustered with plant GGDPSs and that this clade expanded mainly via gene duplication. Plant GGDPSs are involved in the production of a large group of primary isoprenoid compounds, including the chlorophylls, tocopherols, gibberellins, plastoquinones, carotenoids, and carotenoid breakdown products (e.g., the hormones abscisic acid and strigolactones), which play crucial roles in plant growth and development (Tholl and Lee, 2011; Zi et al., 2014). In addition, GGDPSs also produce GGDP for the diterpenoids of secondary metabolism. Given the designation of sesterterpenoids as secondary metabolites, GFDPS is also an enzyme of secondary metabolism, and the origin of GFDPS from GGDPS provides another example of the origin of secondary metabolism from primary metabolic pathways (Qi et al., 2006; Weng et al., 2012; Kliebenstein, 2013), which has been well illustrated for other genes of terpenoid metabolism (Trapp and Croteau, 2001; Matsuba et al., 2013). The plastidial localization of Lc-GFDPS, its use of isoprenyl diphosphate substrates derived from the MEP pathway, and the comparable kinetic parameters (Km) of Lc-GFDPS with Lc-GGDPS (Supplemental Table 4) and other plant GGDPSs (Schmidt and Gershenzon, 2007) are all consistent with the evolution of GFDPS from GGDPS. Considering that plants have the capacity to biosynthesize new secondary metabolites to adapt to environmental stresses (Kliebenstein and Osbourn, 2012) and the finding that the novel leucosceptroid and colquhounoid sesterterpenoids play crucial roles in plant defense against herbivorous insects and pathogens, we hypothesize that herbivore or pathogen pressure has led to the evolution of sesterterpenoid formation.
To date, sesterterpenoids have been isolated from at least 40 plant species belonging to 25 genera and 19 families, with certain genera of the Lamiaceae (Salvia, Leucosceptrum, and Colquhounia) being particularly rich in these compounds (Supplemental Data Set 1). More sesterterpenoid-producing plants will undoubtedly be discovered upon screening with modern analytical techniques. The scattered distribution pattern of sesterterpenoids in plants may arise from repeated origins of the GFDPS gene in separate lineages. Alternatively, GFDPS may be more basal in the plant kingdom, but its presence may be unrecognized because it appears to be just another member of the GGDPS family, a group of genes found in all species investigated to date (Dewick, 2002; Tholl and Lee, 2011). Very recently, GFDPSs similar to GGDPSs were reported from Arabidopsis and other Brassicaceae plants (Nagel et al., 2015; Wang et al., 2015), although no sesterterpenoids were detected, supporting the proposition that GFDPSs are basal in the plant kingdom. In such a scenario, sesterterpenoids might be very widespread but missed because of low concentrations or their presence only under specific growth conditions or in particular organs. Comprehensive analysis of plant extracts and characterization of IDS genes will provide more information about the distribution and evolution of sesterterpenoids.
METHODS
Plant Material and Treatments
Adult trees of Leucosceptrum canum were grown in the Botanical Garden of Kunming Institute of Botany, Chinese Academy of Sciences. L. canum saplings, germinated from the seeds collected from adult L. canum, were propagated and grown in a greenhouse under four highly efficient full-spectrum fluorescent lamps (KDT5 type, 28 W, λ ≥ 0.98) at 23°C with a 16-h-light/8-h-dark cycle.
For quantification of sesterterpenoids in different organs, specific organs of L. canum were harvested and ground into a powder in liquid nitrogen using a mortar and pestle. One hundred milligrams fresh weight of each sample was extracted with 1 mL methanol in an ultrasonic bath for 30 min. The extract was centrifuged at 13,400g for 10 min, and the clear supernatant of each sample was transferred into the sample vial for HPLC with diode array detector (HPLC-DAD; Agilent) analysis as described below. Each organ was analyzed with five independent biological replicates.
For FOS and MEV treatments, 30-d-old L. canum saplings grown in Murashige and Skoog liquid medium were used. FOS sodium salt (Sigma-Aldrich) was prepared in distilled water, while MEV (Sigma-Aldrich) was converted to the water soluble salt as described (Kita et al., 1980; Laule et al., 2003). L. canum saplings were transferred to Murashige and Skoog liquid medium supplemented with 100 μM FOS, 10 μM MEV, both inhibitors (100 μM FOS plus 10 μM MEV), or no additions as a control. The leaves were harvested at 12, 24, and 48 h after inhibitor addition and ground into powder in liquid nitrogen. One hundred milligrams fresh weight of each sample was extracted with 1 mL methanol in an ultrasonic bath for 30 min and analyzed for sesterterpenoid and phytol content using HPLC-DAD as described below. Another 100 mg of each sample was extracted with 5 mL of 20% KOH in ethanol-water (3:1, v/v) at 60°C for 1.5 h and analyzed for β-sitosterol content using gas chromatography-mass spectrometry (Agilent) as previously described (Opitz et al., 2014). Every measurement was repeated with nine independent biological replicates.
For MJ and SA treatments, 60-d-old L. canum saplings were removed from the greenhouse to avoid contamination of control plants. A quantity of 150 mL of 100 μM MJ or SA (Sigma-Aldrich) in 0.01% (aq) ethanol was sprayed onto four L. canum saplings, and control plants were sprayed with pure 0.01% ethanol solution. The leaves were harvested at 1, 6, 12, 24, 48, and 72 h after spraying and ground into powder in liquid nitrogen. Total RNA was extracted for qRT-PCR, and 100 mg fresh weight of each sample was extracted with 1 mL methanol in an ultrasonic bath for 30 min for sesterterpenoid analysis with HPLC-DAD as described below. Every measurement was repeated with five independent biological replicates.
Collection of GTs from L. canum
For collection of GTs for mRNA sequencing, qRT-PCR, and GFDP and sesterterpenoid quantification, L. canum leaves from adult trees were detached and immediately surface-frozen above a mortar filled with liquid nitrogen. Using a paintbrush briefly frozen with liquid nitrogen, leaves were brushed along the main veins to detach GTs, which were then combined in the mortar filled with liquid nitrogen. After removing any hairs of the paintbrush that had become detached, the collected GT sample was quickly checked for its purity under a microscope and then ground to a fine powder for subsequent RNA extraction or chemical analysis. Approximately 100 mg GTs was extracted with 1 mL methanol for detection and quantification of sesterterpenoids, while another 500 mg GTs was extracted with 5 mL of methanol/water (7:3, v/v) for GFDP analysis as previously described (Nagel et al., 2014) in an ultrasonic bath for 30 min. Each experiment was repeated with three to five independent biological replicates.
For further quantification of sesterterpenoids in laser-microdissected peltate GTs and LTs, a Leica LMD7000 system (Leica) was used to precisely collect peltate GTs and the neighboring nonglandular LT, respectively, as previously described (Li et al., 2013, 2014). The collected GTs and LTs were extracted with 200 μL methanol in an ultrasonic bath for 30 min and analyzed using UPLC-MS/MS (Waters) as described below. The experiment was repeated with three independent biological replicates.
Sesterterpenoid and GFDP Analyses Using HPLC-DAD and UPLC-MS/MS
To isolate the compounds in GTs for structural identification, L. canum GT exudates were collected by wiping the fresh leaves with acetone-soaked cotton swabs and then the extracts were evaporated to dryness under reduced pressure. The crude extracts were chromatographed on an MCI gel column with gradient elution of methanol-water (1:1, 7:3, 9:1, and 1:0, v/v) to afford four fractions. The third fraction (9:1 eluent) was combined and dried in vacuo and then suspended in 1 mL methanol. After centrifugation at 13,400g for 10 min, the supernatant was subjected to semipreparative HPLC (Agilent) to yield sesterterpenoids 1 to 5. Compounds 1 and 5 were identified through TLC comparison with standards isolated during our previous investigations (Luo et al., 2010). 1H and 13C spectra of 2 to 4 were measured on a Bruker Avance III 600 spectrometer with TMS as an internal standard.
For quantification of sesterterpenoids in different organs of L. canum, and in leaves after various treatments (MJ, SA, FOS, MEV, and FOS+MEV), HPLC-DAD was employed using an Agilent 1200 system; 20 μL of each sample was injected onto a Thermo-Scientific BDS Hypersil C18 column (5 μm, 4.6 × 250 mm) at a flow rate of 1 mL/min. The mobile phase composed of solvent A (water) and solvent B (methanol) was used (0 to 30.00 min, linear gradient of 5 to 95% of B; 30.01 to 40.00 min, isocratic 95% of B), and the peaks were detected at 208 nm. Calibration curves for the standard compounds 4 and 5 were prepared. Triplicate injections were performed at five concentrations (50, 100, 200, 500, and 1000 μg mL−1). The linearity of each standard curve was made by plotting the peak area versus concentration. The equations and correlation coefficients obtained from the linearity studies were y = 0.1638x − 12.571 (R2 = 0.9994) and y = 0.0599x − 8.0106 (R2 = 0.9990) for 4 and 5, respectively.
For quantification of sesterterpenoids in laser-microdissected GTs and LTs with UPLC-MS/MS, samples were analyzed on a Waters Xevo TQ-S spectrometer equipped with a turbo ion spray (electrospray) source and a triple quadrupole ion path (Waters). The LC conditions were optimized based on the separation patterns of compounds 4 and 5 (column, Zorbax SB-C18, 5 μm, 4.6 × 150 mm; mobile phase, acetonitrile in water, gradient 5 to 95% in 45 min; detection, 210 nm; injection volume, 20 μL; column temperature, 30°C; flow rate, 1 mL/min). Positive ionization was applied with the following parameters: source temperature 250°C, ion source gas (nebulizer gas) 7.0 bar (400 L/h of nitrogen), cone voltage 30 V (250 L/h of nitrogen), capillary voltage 1.5 kV. Calibration curves for the standard compounds 4 and 5 were prepared. Triplicate injections were performed at five concentrations (0.1, 0.5, 1, 5, and 10 μg mL−1), and each standard curve constructed by plotting the peak area versus concentration and fitting a line to the data. The equations and correlation coefficients were y = 252206x + 3488.93 (R2 = 0.999871) and y = 248070x − 1206.02 (R2 = 0.999957) for 4 and 5, respectively.
For detection and quantification of GFDP in the enzyme assay, GTs, and whole leaves, the same UPLC-MS/MS instrument as described above was used. An Acquity BEH-C18 UPLC column (1.7 μm, 2.1 × 50 mm; Waters) was employed, and the mass spectrometer was set to the MRM mode with MRM transitions of 517 > 79. The mobile phase consisting of 5 mM ammonium bicarbonate in water as solvent A and acetonitrile as solvent B was used, with a flow rate at 0.2 mL min−1. Separation was achieved using a gradient elution: isocratic 0% B (0 min), linear to 10% B (0 to 2 min), linear to 64% B (2 to 12 min), and linear to 100% B (12 to 14 min). The injection volume for samples and standards was 5 μL. The negative electrospray ionization mode was used with the following parameters: mass range 2 to 2048 D, capillary voltage 1.8 kV, cone voltage 30 V, source temperature 150°C, desolvation temperature 350°C, cone gas flow 150 L/h, and MS/MS mode collision energy 20.00 eV. A calibration curve for commercial GGDP was prepared as an external standard. Triplicate injections were performed at five concentrations (0.1, 0.5, 1, 5, and 10 μg mL−1), and the standard curve was constructed by plotting the peak area versus concentration and fitting a line. The equation and correlation coefficient obtained was y = 0.1433x + 12.097 (R2 = 0.9996).
L. canum GT RNA Extraction, Transcriptomic Sequencing, Data Processing, de Novo Assembly, and Unigene Functional Annotation
Total RNA from L. canum GTs was extracted using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. mRNA purified using Sera-mag magnetic oligo(dT) beads (Ambion) was sequenced using the Illumina HiSeq 2000 platform at the Beijing Genomics Institute in Shenzhen, China. Transcriptome de novo assembly was performed with a short-read assembly program (Trinity). Unigenes were then aligned by BLASTx (e-value < 10−5) to Nr (NCBI nonredundant protein) protein databases. To assign putative gene function, unigenes were aligned to protein databases, Nr, KEGG (Kyoto Encyclopedia of Genes and Genomes), SwissProt, COG (Clusters of Orthologous Groups of proteins), and GO (Cluster of Orthologous Groups of proteins) by BLASTx with e-value < 10−5, and the retrieved proteins with the highest sequence similarity with the given unigenes were used for protein functional annotation.
Isolation, Expression, Purification, and Enzyme Assay of GFDPS from L. canum GTs
The full-length sequences of IDSs were cloned according to the manufacturer’s instructions for the SMART RACE cDNA amplification kit (Clontech). The truncated ORF sequence of the GFDPS lacking 51 amino acids at the N terminus was cloned into the cold-inducible expression vector pCold TF (Takara), which contained a six histidine residue tag at the N terminus. After confirming the correct orientation of the insert and the presence of an uninterrupted reading frame by sequencing five to eight independent clones, the positive clones were transferred into the Escherichia coli strain Rosetta (DE3) (Novagen).
Bacterial cultures expressing GFDPS protein were grown to an OD of 0.6, and transformants were induced with 0.5 mM IPTG overnight at 16°C. The cell pellets were harvested and resuspended in extraction buffer containing 100 mM MOPSO (pH 7.5), 500 mM NaCl, 10 mM imidazole, and 5 mM DTT and sonicated. The recombinant proteins were individually purified over Ni-NTA agarose columns (Qiagen) following the manufacturer’s instructions and were finally eluted with 250 mM imidazole in the assay buffer. Both crude lysate and the purified protein were then checked by SDS-PAGE. Protein concentration was determined using the Bradford method with BSA as standard.
For determination of the in vitro activity of GFDPS by radio-TLC, an enzymatic activity assay was performed in a similar way to other plant IDS assays described previously (Wang and Dixon, 2009) in a final volume of 50 μL containing 20 mM MOPSO (pH 7.5), 10 mM MgCl2, 10% (v/v) glycerol, 2 mM DTT, 0.5 μL [1-14C] IDP (final concentration 20 μM), 0.25 μg unlabeled IDP (final concentration 17 μM), 0.5 μg of each allylic cosubstrate DMADP (final concentration 34 μM), GDP (final concentration 27 μM), FDP (final concentration 23 μM), or GGDP (final concentration 22 μM) (Sigma-Aldrich), and 10 μg enzyme extract. The assay mixture was incubated for 6 h at 30°C. To stop the assay reaction and hydrolyze all diphosphate esters, 250 μL of 0.2 M Tris solution (pH 9.0) containing 1 unit potato apyrase (Sigma-Aldrich) and 1 unit alkaline phosphatase from calf intestine (Takara) were added to each assay, which was immediately overlaid with 300 μL hexane. After incubation overnight at 30°C, the assay mixture was extracted with hexane by vigorous mixing for 15 s, and standards, geraniol, farnesol, and geranylgeraniol (Sigma-Aldrich), were added into the hexane extracts. Subsequently, the extracts were evaporated under nitrogen and analyzed by radio-TLC using benzene/ethyl acetate (9:1, v/v) as the developing system. The distribution of radioactivity was detected using a Typhoon Trio variable mode imager (GE Healthcare Biosciences). Authentic geraniol, farnesol, and geranylgeraniol were visualized in iodine vapor. The enzyme assays with each allylic substrate were repeated with at least five independent experimental replicates.
For determination of the in vitro activity of GFDPS using UPLC-MS/MS, the enzymatic activity assay was performed in a total volume of 200 μL containing 5% Tween 20 (v/v), 2.5 μg IDP (final concentration 42.5 μM), 2.5 μg of each allylic cosubstrate DMADP (final concentration 42.5 μM), GDP (final concentration 33.75 μM), FDP (final concentration 28.75 μM), or GGDP (final concentration 27.5 μM), and 200 μg protein per assay. The assay mixture was incubated for 1 h at 30°C and then stopped by freezing in liquid nitrogen. After centrifugation at 13,400g for 10 min, the water-soluble layer was transferred into a sample vial for UPLC-MS/MS analysis of GFDP as described above. The enzyme assays with each allylic substrate were repeated with at least three independent experimental replicates and four technical replicates. For determination of the product profile, three different ratios of isoprenyl substrates (100, 300, or 500 μM IDP, respectively) with 100 μM of each allylic cosubstrate [DMADP, GDP, FDP (E,E-configuration), or GGDP] were used. Each experiment was repeated with three independent replicates. For determination of the apparent kinetic constants of allylic substrates by UPLC-MS/MS, six different concentrations of the substrate DMADP (0, 20, 40, 60, 80, and 100 μM), GDP (0, 10, 20, 30, 40, and 50 μM), FDP (0, 10, 20, 30, 40, and 50 μM), and GGDP (0, 5, 10, 15, 20, and 25 μM) were used in the presence of a fixed concentration of 100 μM IDP. For determination of apparent kinetic constants of IDP, six different concentrations of IDP (0, 10, 20, 30, 40, and 50 μM) and a fixed 50 μM concentration of GGDP were used. The kinetics reactions were incubated at 30°C for 20 min. The reactions were stopped by immediately freezing the sample in liquid nitrogen, and the relative concentration of GFDP in each sample was analyzed with UPLC-MS/MS using commercial GGDP as an external standard. Km and Kcat values were calculated using hyperbolic regression analysis methods with Hyper32 software (version 1.0.0; http://homepage.ntlworld.com/john.easterby/hyper32.html). Each experiment was performed in duplicate.
The kinetics analyses of purified recombinant GGDPSs were performed in a final volume of 50 μL containing 20 mM MOPSO (pH 7.5), 10 mM MgCl2, 10% (v/v) glycerol, 2 mM DTT, a fixed 30 μM IDP (7 μM [1-14C]IDP combined with 23 μM unlabeled IDP), and an allylic cosubstrate DMADP (0, 2, 5, 10, 20, and 50 μM), GDP (0, 2, 5, 10, 20, and 50 μM), FDP (0, 2, 5, 10, 20, and 50 μM), or GGDP (0, 2, 5, 10, 20, and 50 μM). The assay reaction was incubated for 30 min at 30°C. To stop the assay reaction and hydrolyze the diphosphate esters, 5 μL of 3 n HCl was added to each assay and then each mixture was immediately overlaid with 300 μL hexane. After incubation for 20 min at 30°C, the assay mixture was extracted twice with hexane (600 μL in total) by vigorous mixing for 15 s. Then, 500 μL of supernatant hexane was used for total radioactivity determination by liquid scintillation counting. Km and Kcat values were calculated using hyperbolic regression analysis methods with Hyper32 software (version 1.0.0; http://homepage.ntlworld.com/john.easterby/hyper32.html). Each experiment was repeated with three technical replicates.
For the complementation assay of IDSs from L. canum, the plasmid (pACCAR25ΔcrtE) carrying all carotenoid cluster genes from Erwinia uredovora except for GGDPS (Kainou et al., 1999) was used. Plasmids pCold TF, pCold TF/Lc-GFDPS, and pCold TF/Lc-GGDPS1–4 were cotransformed with pACCAR25ΔcrtE into BL21 (DE3) Star cells (Invitrogen). The transformants were selected on Luria-Bertani agar medium containing 34 μg mL−1 chloramphenicol and 100 μg mL−1 ampicillin.
Generation of Transgenic Arabidopsis Plants and Crude Protein Extraction
Full-length GFDPS cDNA was cloned into the modified pCAMBIA1300 binary vector containing a double CaMV 35S promoter. The plasmid was sequenced and introduced into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis thaliana Col-0 using the floral dip method (Clough and Bent, 1998). The Arabidopsis Col-0 plants were grown in a greenhouse under four highly efficient full-spectrum fluorescent lamps (KDT5 type, 28 W, λ ≥ 0.98) at 23°C with a 16-h-light/8-h-dark cycle. The seeds of T0 transgenic lines were initially selected on 25 μg mL−1 hygromycin B (Sigma-Aldrich). T1 and T2 lines, confirmed by DNA sequencing and qRT-PCR, were used for the enzyme and qRT-PCR assays.
Plant leaves were ground with a mortar and pestle under liquid nitrogen to a fine powder. Cooled extraction buffer containing 250 mM MOPSO (pH 6.8), 5 mM ascorbic acid, 5 mM sodium bisulfite, 5 mM DTT, 10 mM MgCl2, 1 mM EDTA, 10% (v/v) glycerol, 1% (w/v) polyvinylpyrrolidone (Mr = 10,000), and 0.1% (v/v) Tween 20 was added to frozen tissue in a ratio of 1:5 (tissue [g]/buffer [mL]) (Nagel et al., 2012). The extract was shaken at 4°C, 1400 rpm for 30 min. After centrifugation at 20,000g at 4°C for 10 min, the supernatant was passed through a 4-mL Amicon Ultra-4 centrifugal filter with a 3-kD molecular mass cutoff (Millipore) to exchange the buffer to 20 mM MOPSO (pH 7.5), 10 mM MgCl2, 10% (v/v) glycerol, and 2 mM DTT according to the manufacturer’s protocol. Protein concentration was determined using the Bradford method with BSA as standard. The enzyme assay was then performed as described above and repeated with three independent experimental replicates.
qRT-PCR Analysis
qRT-PCR was performed with the actin gene as an internal standard using the ΔΔCT method (Livak and Schmittgen, 2001) with UltraSYBR mixture reagent (with ROX) (CWBiotech) on an Applied Biosystems 7500 instrument (Life Technologies) according to the manufacturer’s instructions. Every measurement was repeated with five independent biological replicates, each of which was represented by three technical replicates.
GFDP Synthase Subcellular Localization
The DNA sequence encoding the N-terminal predicted transit peptide (the first 51 amino acids of GFDPS), full-length L. canum GFDPS cDNA, full-length cDNA of GGDPS1–4 cDNA, and FDPS-like IDS6 were separately amplified from cDNA of L. canum leaves and cloned into the pJIT163-hGFP vector which contains a C-terminal eGFP tag under the control of the CaMV 35S promoter (Xu et al., 2013). The plasmids were confirmed by DNA sequencing and then introduced into Arabidopsis leaf protoplasts, which were prepared as previously described (Yoo et al., 2007). An empty vector fused to GFP was introduced into Arabidopsis leaf protoplasts as a control. After incubation for 12 h in the dark at 28°, the GFP signals were observed and imaged under a confocal laser scanning microscope (Olympus). A 488-nm excitation laser and a 520-nm emission filter were used for detecting GFP signals; a 633-nm excitation laser and a 650-nm emission filter were used for detecting red chlorophyll autofluorescence.
Antifeedant and Growth Inhibition Assays
Beet armyworms (Spodoptera exigua) were purchased from the Pilot-Scale Base of Bio-Pesticides, Institute of Zoology, Chinese Academy of Sciences. The larvae were reared on an artificial diet in the laboratory under controlled photoperiod (light:dark, 16:8 h) and temperature (25°C ± 2°C). For the antifeedant assay of L. canum leaves, a dual-choice bioassay was performed as previously described (Luo et al., 2010). Leaves of L. canum were collected at 12 h after the plant was sprayed with MJ or 0.01% (aq) ethanol (control) and then cut into 1.1-cm-diameter discs using a cork borer. Two MJ-treated leaf discs and two controls were set in alternating position in the same Petri dish (90 mm in diameter), with moistened filter paper at the bottom. Two 3rd instar larvae were starved for 4 to 5 h prior to each assay and then placed at center of the Petri dish. After feeding for 15 h, areas of leaf discs consumed were measured. The feeding index was calculated according to the formula T/C ×100, where C and T represent the beginning leaf area and leaf area after consumption, respectively. The experiment was repeated with five replicates.
For the growth inhibitor assay, leaves of L. canum were collected at 12 h after the plant was sprayed with MJ or 0.01% (aq) ethanol (control), ground into powder in liquid nitrogen using a mortar and pestle, and then mixed with artificial diet in a ratio of 1:4 (w/w). Fifty first instar beet armyworm larvae with beginning mass of ∼1.5 mg per insect were selected to test insect growth inhibition by the leaf-containing diet. After 5 d of feeding, the weights of insects were determined and compared.
Phylogenetic Analysis
The amino acid sequences (Supplemental Data Set 3) were obtained from the NCBI database, aligned using the ClustalX 2.0 program (Larkin et al., 2007), and then manually adjusted to optimize the alignments (Supplemental File 1). The phylogenetic tree was constructed with MEGA6 software (Tamura et al., 2013) based on the maximum likelihood method with LG+G model and neighbor-joining method. One thousand bootstrap replicates were performed in each analysis to obtain the confidence support.
The molecular evolution of Lc-GFDPS was analyzed using the codeml program in the PAML 4.8 package. The branch-site model was used for testing the significance of positive selection. The positive sites with high posterior probabilities (>0.95) were obtained through Bayes Empirical Bayes analysis (Yang, 1998).
Statistical Analysis
ANOVA followed by a Duncan’s test was used (SPSS 19.0), where differences between means were assessed and significance was determined at α = 0.05. Correlation significance was assessed using an F test (SPSS 19.0) with α = 0.05 and α = 0.01.
Oligonucleotide Primers
The complete list of oligonucleotide primers used in this study is available in Supplemental Data Set 4.
Accession Numbers
Files containing the raw read sequences are available from the National Center for Biotechnology Information (NCBI) Short Read Archive under accession number SAMN04376921. Sequences data from this article can be found in the GenBank database under the following accession numbers: L. canum GFDPS, KT312959; GGDPS1, KT312957; GGDPS2, KT312958; GGDPS3, KT312960; GGDPS4, KT312961; IDS6, KT324653; IDS7, KT324654; IDS8, KT324655; IDS9, KT324656; and IDS10, KT324657.
Supplemental Data
Supplemental Figure 1. GO Classification of Unigenes of the Glandular Trichome Transcriptome of Leucosceptrum canum.
Supplemental Figure 2. KEGG Pathways Significantly Enriched for Unigenes of the Glandular Trichome Transcriptome of L. canum.
Supplemental Figure 3. Transcript Level Analysis of IDS1, IDS2, IDS4, and IDS5 in Different Organs of L. canum.
Supplemental Figure 4. SDS-PAGE Analysis of Recombinant Lc-IDS1-5 Proteins Expressed in E. coli.
Supplemental Figure 5. In Vitro Enzyme Activity Assay of GGDPSs.
Supplemental Figure 6. Subcellular Localization of GGDPSs.
Supplemental Figure 7. The Contents of β-Sitosterol and Phytol in the Leaves of L. canum after MEV and FOS Treatments.
Supplemental Table 1. Enzyme Activity of Purified Recombinant GFDPS Protein Enhanced by Tween 20.
Supplemental Table 2. Product Profiles of Purified Recombinant GFDPS Protein.
Supplemental Table 3. Conserved Motifs Represented by Different Colored Bars.
Supplemental Table 4. Kinetic Parameters for the Purified Recombinant GGDPSs.
Supplemental Data Set 1. Sesterterpenoids Identified from Plants.
Supplemental Data Set 2. The Unigenes of Early Enzymes in Terpenoid Biosynthesis in the Glandular Trichome Transcriptome of L. canum.
Supplemental Data Set 3. List of Protein Sequences Used for Sequence Comparison and Phylogeny.
Supplemental Data Set 4. The Oligonucleotide Primers Used in This Study (5′ to 3′).
Supplemental File 1. Alignments Used to Produce the Phylogenetic Tree.
Supplementary Material
Acknowledgments
We thank Hisashi Hemmi (Nagoya University, Aichi, Japan) for kindly providing the plasmid containing Methanosarcina mazei GFDPS, Makoto Kawamukai (Shimane University, Matsue, Japan) for kindly providing pACCAR25ΔcrtE plasmid, Raimund Nagel (Max Planck Institute for Chemical Ecology, Jena, Germany), Guo Wei (Institute of Genetics and Developmental Biology, Beijing, China), and Qing-Wen Chen (Institute of Genetics and Developmental Biology, Beijing, China) for help with enzyme assay, Jian-Hong Yang (Kunming Institute of Botany, Kunming, China) for UPLC-MS/MS analysis, and Ai-Zhong Liu (Kunming Institute of Botany, Kunming, China), Yu Song (Xishuangbanna Tropical Botanical Garden, Xishuangbanna, China), and Ting Sun (Kunming Institute of Botany, Kunming, China) for help with GFDPS evolutionary analysis. This research was financially supported by the National Science Fund for Distinguished Young Scholars (31525005), the NSFC-Yunnan Joint Fund (U1202263), the National Basic Research Program of China (973 Program) on Biological Control of Key Crop Pathogenic Nematodes (2013CB127505), the National Natural Science Foundation of China (31100222 and 31470395), the “Hundred Talents Program” of the Chinese Academy of Sciences (awarded to S.-H. Li), the State Key Laboratory of Phytochemistry and Plant Resources in West China (P2015-ZZ12), and the Max Planck Society.
AUTHOR CONTRIBUTIONS
Y.L., S.-H. Li, J.G., B.S., and G.-D.W. designed the research. Y.L., S.-H. Luo, A.S., M.G., C.K., M.-J.Y., S.-X.J., and C.-H.L. carried out the experimental work. G.-D.W. contributed new reagents/analytical tools. Y.L. and G.-L.S analyzed the data. Y.L., S.-H. Li, J.G., and B.S. wrote the article.
Glossary
- GFDP
geranylfarnesyl diphosphate
- GGDPS
geranylgeranyl diphosphate synthase
- GT
glandular trichome
- LT
leaf tissue
- LMD
laser microdissection
- UPLC-MS/MS
ultraperformance liquid chromatography-tandem mass spectrometry
- GGDP
geranylgeranyl diphosphate
- FDPS
farnesyl diphosphate synthase
- GDPS
geranyl diphosphate synthase
- ORF
open reading frame
- IDP
isopentenyl diphosphate
- DMADP
dimethylallyl diphosphate
- GDP
geranyl diphosphate
- FDP
farnesyl diphosphate
- radio-TLC
radio-thin-layer chromatography
- MRM
multiple reaction monitoring
- MEP
2-C-methyl-d-erythritol 4-phosphate
- MVA
mevalonic acid
- MEV
mevinolin
- FOS
fosmidomycin
- MJ
methyl jasmonate
- SA
salicylic acid
- HPLC-DAD
HPLC with diode array detector
References
- Akihisa T., Koike K., Kimura Y., Sashida N., Matsumoto T., Ukiya M., Nikaido T. (1999). Acyclic and incompletely cyclized triterpene alcohols in the seed oils of theaceae and gramineae. Lipids 34: 1151–1157. [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P., Somerville C. (2001). Progress in plant metabolic engineering. Proc. Natl. Acad. Sci. USA 98: 8925–8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canonica L., Fiecchi A., Kienle M.G., Ranzi B.M., Scala A., Salvator T., Pella E. (1967). The biosynthesis of ophiobolins. Tetrahedron Lett. 35: 3371–3376. [Google Scholar]
- Challinor V.L., Johnston R.C., Bernhardt P.V., Lehmann R.P., Krenske E.H., De Voss J.J. (2015). Biosynthetic insights provided by unusual sesterterpenes from the medicinal herb Aletris farinosa. Chem. Sci. (Camb.) 6: 5740–5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba R., Minami A., Gomi K., Oikawa H. (2013). Identification of ophiobolin F synthase by a genome mining approach: a sesterterpene synthase from Aspergillus clavatus. Org. Lett. 15: 594–597. [DOI] [PubMed] [Google Scholar]
- Choudhary M.I., Ranjit R., Atta-ur-Rahman A.U., Shrestha T.M., Yasin A., Parvez M. (2004a). Leucosceptrine--a novel sesterterpene with prolylendopeptidase inhibitory activity from Leucosceptrum canum. J. Org. Chem. 69: 2906–2909. [DOI] [PubMed] [Google Scholar]
- Choudhary M.I., Ranjit R., Atta-ur-Rahman, Hussain S., Devkota K.P., Shrestha T.M., Parvez M. (2004b). Novel sesterterpenes from Leucosceptrum canum of nepalese origin. Org. Lett. 6: 4139–4142. [DOI] [PubMed] [Google Scholar]
- Choudhary M.I., Ranjit R., Atta-ur-Rahman, Devkota K.P., Shrestha T.M. (2007). Two new leucosesterterpenes from Leucosceptrum canum. Z. Naturforsch. C 62: 587–592. [Google Scholar]
- Cioffi G., Bader A., Malafronte A., Dal Piaz F., De Tommasi N. (2008). Secondary metabolites from the aerial parts of Salvia palaestina Bentham. Phytochemistry 69: 1005–1012. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Copley S.D., Dhillon J.K. (2002). Lateral gene transfer and parallel evolution in the history of glutathione biosynthesis genes. Genome Biol. 3: h0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Piaz F., Imparato S., Lepore L., Bader A., De Tommasi N. (2010). A fast and efficient LC-MS/MS method for detection, identification and quantitative analysis of bioactive sesterterpenes in Salvia dominica crude extracts. J. Pharm. Biomed. Anal. 51: 70–77. [DOI] [PubMed] [Google Scholar]
- Dal Piaz F., Vassallo A., Lepore L., Tosco A., Bader A., De Tommasi N. (2009). Sesterterpenes as tubulin tyrosine ligase inhibitors. First insight of structure-activity relationships and discovery of new lead. J. Med. Chem. 52: 3814–3828. [DOI] [PubMed] [Google Scholar]
- Dewick P.M. (2002). The biosynthesis of C5-C25 terpenoid compounds. Nat. Prod. Rep. 19: 181–222. [DOI] [PubMed] [Google Scholar]
- Dixon R.A. (2001). Natural products and plant disease resistance. Nature 411: 843–847. [DOI] [PubMed] [Google Scholar]
- Ebrahimi S.N., Farimani M.M., Mirzania F., Soltanipoor M.A., De Mieri M., Hamburger M. (2014). Manoyloxide sesterterpenoids from Salvia mirzayanii. J. Nat. Prod. 77: 848–854. [DOI] [PubMed] [Google Scholar]
- Emiliani G., Fondi M., Fani R., Gribaldo S. (2009). A horizontal gene transfer at the origin of phenylpropanoid metabolism: a key adaptation of plants to land. Biol. Direct 4: 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenzon J., Dudareva N. (2007). The function of terpene natural products in the natural world. Nat. Chem. Biol. 3: 408–414. [DOI] [PubMed] [Google Scholar]
- Gonzalez M.S., Sansegundo J.M., Grande M.C., Medarde M., Bellido I.S. (1989). Sesterterpene lactones from Salvia aethiopis. Salviaethiopisolide and 13-epi-salviaethiopisolide. Tetrahedron 45: 3575–3582. [Google Scholar]
- Hemmerlin A., Harwood J.L., Bach T.J. (2012). A raison d’être for two distinct pathways in the early steps of plant isoprenoid biosynthesis? Prog. Lipid Res. 51: 95–148. [DOI] [PubMed] [Google Scholar]
- Hsieh F.L., Chang T.H., Ko T.P., Wang A.H. (2011). Structure and mechanism of an Arabidopsis medium/long-chain-length prenyl pyrophosphate synthase. Plant Physiol. 155: 1079–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Song L., Xu J., Zhu G., Liu B. (2013). Asymmetric total synthesis of leucosceptroid B. Angew. Chem. Int. Ed. Engl. 52: 952–955. [DOI] [PubMed] [Google Scholar]
- Kainou T., Kawamura K., Tanaka K., Matsuda H., Kawamukai M. (1999). Identification of the GGPS1 genes encoding geranylgeranyl diphosphate synthases from mouse and human. Biochim. Biophys. Acta 1437: 333–340. [DOI] [PubMed] [Google Scholar]
- Kawaguchi A., Nozoe S., Okuda S. (1973). Subcellular distribution of sesterterpene- and sterol-biosynthesizing activities in Cochliobolus heterostrophus. Biochim. Biophys. Acta 296: 615–623. [PubMed] [Google Scholar]
- Kawahara N., Nozawa M., Kurata A., Hakamatsuka T., Sekita S., Satake M. (1999). A novel sesterterpenoid, nitiol, as a potent enhancer of IL-2 gene expression in a human T cell line, from the Peruvian folk medicine “Hercumpuri” (Gentianella nitida). Chem. Pharm. Bull. (Tokyo) 47: 1344–1345. [DOI] [PubMed] [Google Scholar]
- Kita T., Brown M.S., Goldstein J.L. (1980). Feedback regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in livers of mice treated with mevinolin, a competitive inhibitor of the reductase. J. Clin. Invest. 66: 1094–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D.J. (2013). Making new molecules--evolution of structures for novel metabolites in plants. Curr. Opin. Plant Biol. 16: 112–117. [DOI] [PubMed] [Google Scholar]
- Kliebenstein D.J., Osbourn A. (2012). Making new molecules - evolution of pathways for novel metabolites in plants. Curr. Opin. Plant Biol. 15: 415–423. [DOI] [PubMed] [Google Scholar]
- Kuzuyama T., Shimizu T., Takahashi S., Seto H. (1998). Fosmidomycin, a specific inhibitor of 1-deoxy-D-xylulose 5-phosphate reductoisomerase in the nonmevalonate pathway for terpenoid biosynthesis. Tetrahedron Lett. 39: 7913–7916. [Google Scholar]
- Lange B.M., Turner G.W. (2013). Terpenoid biosynthesis in trichomes--current status and future opportunities. Plant Biotechnol. J. 11: 2–22. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Laule O., Fürholz A., Chang H.S., Zhu T., Wang X., Heifetz P.B., Gruissem W., Lange M. (2003). Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100: 6866–6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.H., Jing S.X., Luo S.H., Shi W., Hua J., Liu Y., Li X.N., Schneider B., Gershenzon J., Li S.H. (2013). Peltate glandular trichomes of Colquhounia coccinea var. mollis harbor a new class of defensive sesterterpenoids. Org. Lett. 15: 1694–1697. [DOI] [PubMed] [Google Scholar]
- Li C.H., Liu Y., Hua J., Luo S.H., Li S.H. (2014). Peltate glandular trichomes of Colquhounia seguinii harbor new defensive clerodane diterpenoids. J. Integr. Plant Biol. 56: 928–940. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H.K. (2000). Non-mevalonate isoprenoid biosynthesis: enzymes, genes and inhibitors. Biochem. Soc. Trans. 28: 785–789. [PubMed] [Google Scholar]
- Liu Y., Wang L., Jung J.H., Zhang S. (2007). Sesterterpenoids. Nat. Prod. Rep. 24: 1401–1429. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Luo S.H., Hua J., Li C.H., Liu Y., Li X.N., Zhao X., Li S.H. (2013a). Unusual antifeedant spiro-sesterterpenoid from the flowers of Leucosceptrum canum. Tetrahedron Lett. 54: 235–237. [Google Scholar]
- Luo S.H., Hua J., Niu X.M., Liu Y., Li C.H., Zhou Y.Y., Jing S.X., Zhao X., Li S.H. (2013b). Defense sesterterpenoid lactones from Leucosceptrum canum. Phytochemistry 86: 29–35. [DOI] [PubMed] [Google Scholar]
- Luo S.H., Hugelshofer C.L., Hua J., Jing S.X., Li C.H., Liu Y., Li X.N., Zhao X., Magauer T., Li S.H. (2014). Unraveling the metabolic pathway in Leucosceptrum canum by isolation of new defensive leucosceptroid degradation products and biomimetic model synthesis. Org. Lett. 16: 6416–6419. [DOI] [PubMed] [Google Scholar]
- Luo S.H., Luo Q., Niu X.M., Xie M.J., Zhao X., Schneider B., Gershenzon J., Li S.H. (2010). Glandular trichomes of Leucosceptrum canum harbor defensive sesterterpenoids. Angew. Chem. Int. Ed. Engl. 49: 4471–4475. [DOI] [PubMed] [Google Scholar]
- Luo S.H., Weng L.H., Xie M.J., Li X.N., Hua J., Zhao X., Li S.H. (2011). Defensive sesterterpenoids with unusual antipodal cyclopentenones from the leaves of Leucosceptrum canum. Org. Lett. 13: 1864–1867. [DOI] [PubMed] [Google Scholar]
- Mansouri H., Salari F. (2014). Influence of mevinolin on chloroplast terpenoids in Cannabis sativa. Physiol. Mol. Biol. Plants 20: 273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé G., Belt S.T., Rowland S.J., Rohmer M. (2004). Isoprenoid biosynthesis in the diatoms Rhizosolenia setigera (Brightwell) and Haslea ostrearia (Simonsen). Proc. Natl. Acad. Sci. USA 101: 4413–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuba Y., et al. (2013). Evolution of a complex locus for terpene biosynthesis in Solanum. Plant Cell 25: 2022–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y., Mitsuhashi T., Quan Z., Abe I. (2015). Molecular basis for stellatic acid biosynthesis: A genome mining approach for discovery of sesterterpene synthases. Org. Lett. 17: 4644–4647. [DOI] [PubMed] [Google Scholar]
- McCaskill D., Gershenzon J., Croteau R. (1992). Morphology and monoterpene biosynthetic capabilities of secretory cell clusters isolated from glandular trichomes of peppermint (Mentha piperita L.). Planta 187: 445–454. [DOI] [PubMed] [Google Scholar]
- Moghaddam F.M., Amiri R., Alam M., Hossain M.B., van der Helm D. (1998). Structure and absolute stereochemistry of salvimirzacolide, a new sesterterpene from Salvia mirzayanii. J. Nat. Prod. 61: 279–281. [DOI] [PubMed] [Google Scholar]
- Moghaddam F.M., Farimani M.M., Seirafi M., Taheri S., Khavasi H.R., Sendker J., Proksch P., Wray V., Edrada R. (2010). Sesterterpenoids and other constituents of Salvia sahendica. J. Nat. Prod. 73: 1601–1605. [DOI] [PubMed] [Google Scholar]
- Moghaddam F.M., Zaynizadeh B., Ruedi P. (1995). Salvileucolide methylester, a sesterterpene from Salvia sahendica. Phytochemistry 39: 715–716. [Google Scholar]
- Moridi Farimani M., Mazarei Z. (2014). Sesterterpenoids and other constituents from Salvia lachnocalyx Hedge. Fitoterapia 98: 234–240. [DOI] [PubMed] [Google Scholar]
- Nagel R., Berasategui A., Paetz C., Gershenzon J., Schmidt A. (2014). Overexpression of an isoprenyl diphosphate synthase in spruce leads to unexpected terpene diversion products that function in plant defense. Plant Physiol. 164: 555–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel R., Bernholz C., Vranová E., Košuth J., Bergau N., Ludwig S., Wessjohann L., Gershenzon J., Tissier A., Schmidt A. (2015). Arabidopsis thaliana isoprenyl diphosphate synthases produce the C25 intermediate geranylfarnesyl diphosphate. Plant J. 84: 847–859. [DOI] [PubMed] [Google Scholar]
- Nagel R., Gershenzon J., Schmidt A. (2012). Nonradioactive assay for detecting isoprenyl diphosphate synthase activity in crude plant extracts using liquid chromatography coupled with tandem mass spectrometry. Anal. Biochem. 422: 33–38. [DOI] [PubMed] [Google Scholar]
- Nakabayashi R., Saito K. (2015). Integrated metabolomics for abiotic stress responses in plants. Curr. Opin. Plant Biol. 24: 10–16. [DOI] [PubMed] [Google Scholar]
- Nozoe S., Morisaki M., Tsuda K., Okuda S. (1967). Biogenesis of ophiobolins. Origin of oxygen atoms in ophiobolins. Tetrahedron Lett. 8: 3365–3368. [Google Scholar]
- Ogawa T., Yoshimura T., Hemmi H. (2010). Geranylfarnesyl diphosphate synthase from Methanosarcina mazei: Different role, different evolution. Biochem. Biophys. Res. Commun. 393: 16–20. [DOI] [PubMed] [Google Scholar]
- Opitz S., Nes W.D., Gershenzon J. (2014). Both methylerythritol phosphate and mevalonate pathways contribute to biosynthesis of each of the major isoprenoid classes in young cotton seedlings. Phytochemistry 98: 110–119. [DOI] [PubMed] [Google Scholar]
- Pichersky E., Lewinsohn E. (2011). Convergent evolution in plant specialized metabolism. Annu. Rev. Plant Biol. 62: 549–566. [DOI] [PubMed] [Google Scholar]
- Qi X., Bakht S., Qin B., Leggett M., Hemmings A., Mellon F., Eagles J., Werck-Reichhart D., Schaller H., Lesot A., Melton R., Osbourn A. (2006). A different function for a member of an ancient and highly conserved cytochrome P450 family: from essential sterols to plant defense. Proc. Natl. Acad. Sci. USA 103: 18848–18853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin B., Matsuda Y., Mori T., Okada M., Quan Z., Mitsuhashi T., Wakimoto T., Abe I. (2015). An unusual chimeric diterpene synthase from Emericella variecolor and its functional conversion into a sesterterpene synthase by domain swapping. Angew. Chem. Int. Ed. Engl. 55: 1658–1661. [DOI] [PubMed] [Google Scholar]
- Ramak P., Kazempour Osaloo S., Ebrahimzadeh H., Sharifi M., Behmanesh M. (2013). Inhibition of the mevalonate pathway enhances carvacrol biosynthesis and DXR gene expression in shoot cultures of Satureja khuzistanica Jamzad. J. Plant Physiol. 170: 1187–1193. [DOI] [PubMed] [Google Scholar]
- Renner M.K., Jensen P.R., Fenical W. (2000). Mangicols: structures and biosynthesis of A new class of sesterterpene polyols from a marine fungus of the genus Fusarium. J. Org. Chem. 65: 4843–4852. [DOI] [PubMed] [Google Scholar]
- Richards T.A., Dacks J.B., Campbell S.A., Blanchard J.L., Foster P.G., McLeod R., Roberts C.W. (2006). Evolutionary origins of the eukaryotic shikimate pathway: gene fusions, horizontal gene transfer, and endosymbiotic replacements. Eukaryot. Cell 5: 1517–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland S.J., Belt S.T., Wraige E.J., Massé G., Roussakis C., Robert J.M. (2001). Effects of temperature on polyunsaturation in cytostatic lipids of Haslea ostrearia. Phytochemistry 56: 597–602. [DOI] [PubMed] [Google Scholar]
- Rustaiyan A., Koussari S. (1988). Further sesterterpenes from Salvia hypoleuca. Phytochemistry 27: 1767–1769. [Google Scholar]
- Rustaiyan A., Niknejad A., Nazarians L., Jakupovic J., Bohlmann F. (1982). Sesterterpenes from Salvia hypoleuca. Phytochemistry 21: 1812–1813. [Google Scholar]
- Rustaiyan A., Sadjadi A. (1987). Salvisyriacolide, a sesterterpene from Salvia syriaca. Phytochemistry 26: 3078–3079. [Google Scholar]
- Sato T., Yamaga H., Kashima S., Murata Y., Shinada T., Nakano C., Hoshino T. (2013). Identification of novel sesterterpene/triterpene synthase from Bacillus clausii. ChemBioChem 14: 822–825. [DOI] [PubMed] [Google Scholar]
- Schilmiller A.L., Schauvinhold I., Larson M., Xu R., Charbonneau A.L., Schmidt A., Wilkerson C., Last R.L., Pichersky E. (2009). Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc. Natl. Acad. Sci. USA 106: 10865–10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Gershenzon J. (2007). Cloning and characterization of isoprenyl diphosphate synthases with farnesyl diphosphate and geranylgeranyl diphosphate synthase activity from Norway spruce (Picea abies) and their relation to induced oleoresin formation. Phytochemistry 68: 2649–2659. [DOI] [PubMed] [Google Scholar]
- Schmidt A., Gershenzon J. (2008). Cloning and characterization of two different types of geranyl diphosphate synthases from Norway spruce (Picea abies). Phytochemistry 69: 49–57. [DOI] [PubMed] [Google Scholar]
- Shinozaki J., Shibuya M., Ebizuka Y., Masuda K. (2013). Cyclization of all-E- and 2Z-geranylfarnesols by a bacterial triterpene synthase: insight into sesterterpene biosynthesis in Aleuritopteris ferns. Biosci. Biotechnol. Biochem. 77: 2278–2282. [DOI] [PubMed] [Google Scholar]
- Tachibana A. (1994). A novel prenyltransferase, farnesylgeranyl diphosphate synthase, from the haloalkaliphilic archaeon, Natronobacterium pharaonis. FEBS Lett. 341: 291–294. [DOI] [PubMed] [Google Scholar]
- Tachibana A., Yano Y., Otani S., Nomura N., Sako Y., Taniguchi M. (2000). Novel prenyltransferase gene encoding farnesylgeranyl diphosphate synthase from a hyperthermophilic archaeon, Aeropyrum pernix. Molecularevolution with alteration in product specificity. Eur. J. Biochem. 267: 321–328. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D., Kish C.M., Orlova I., Sherman D., Gershenzon J., Pichersky E., Dudareva N. (2004). Formation of monoterpenes in Antirrhinum majus and Clarkia breweri flowers involves heterodimeric geranyl diphosphate synthases. Plant Cell 16: 977–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D., and Lee S. (2011). Terpene specialized metabolism in Arabidopsis thaliana. The Arabidopsis Book 9: e0143, doi/10.1199/tab.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A. (2012). Glandular trichomes: what comes after expressed sequence tags? Plant J. 70: 51–68. [DOI] [PubMed] [Google Scholar]
- Topcu G., Ulubelen A., Tam T.C.M., Che C.T. (1996a). Sesterterpenes and other constituents of Salvia yosgadensis. Phytochemistry 42: 1089–1092. [Google Scholar]
- Topcu G., Ulubelen A., Tam T.C.M., Che C.T. (1996b). Norditerpenes and norsesterterpenes from Salvia yosgadensis. J. Nat. Prod. 59: 113–116. [Google Scholar]
- Toyoda M., Asahina M., Fukawa H., Shimizu T. (1969). Isolation of new acyclic C 25-isoprenyl alcohol from potato leaves. Tetrahedron Lett. 55: 4879–4882. [Google Scholar]
- Trapp S.C., Croteau R.B. (2001). Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics 158: 811–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda D., Yamaga H., Murakami M., Totsuka Y., Shinada T., Sato T. (2015). Biosynthesis of sesterterpenes, head-to-tail triterpenes, and sesquarterpenes in Bacillus clausii: Identification of multifunctional enzymes and analysis of isoprenoid metabolites. ChemBioChem 16: 1371–1377. [DOI] [PubMed] [Google Scholar]
- Ulubelen A., Topcu G., Sonmez U., Eris C., Ozgen U. (1996). Norsesterterpenes and diterpenes from the aerial parts of Salvia limbata. Phytochemistry 43: 431–434. [Google Scholar]
- Wagner G.J., Wang E., Shepherd R.W. (2004). New approaches for studying and exploiting an old protuberance, the plant trichome. Ann. Bot. (Lond.) 93: 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Chen Q., Fan D., Li J., Wang G., Zhang P. (2015). Structural analyses of short-chain prenyltransferases identify an evolutionarily conserved GFPPS clade in Brassicaceae plants. Mol. Plant 9: 195–204. [DOI] [PubMed] [Google Scholar]
- Wang G., Dixon R.A. (2009). Heterodimeric geranyl(geranyl)diphosphate synthase from hop (Humulus lupulus) and the evolution of monoterpene biosynthesis. Proc. Natl. Acad. Sci. USA 106: 9914–9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Tian L., Aziz N., Broun P., Dai X., He J., King A., Zhao P.X., Dixon R.A. (2008). Terpene biosynthesis in glandular trichomes of hop. Plant Physiol. 148: 1254–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Yang B., Lin X.P., Zhou X.F., Liu Y. (2013). Sesterterpenoids. Nat. Prod. Rep. 30: 455–473. [DOI] [PubMed] [Google Scholar]
- Weng J.K., Philippe R.N., Noel J.P. (2012). The rise of chemodiversity in plants. Science 336: 1667–1670. [DOI] [PubMed] [Google Scholar]
- Xie Z., Kapteyn J., Gang D.R. (2008). A systems biology investigation of the MEP/terpenoid and shikimate/phenylpropanoid pathways points to multiple levels of metabolic control in sweet basil glandular trichomes. Plant J. 54: 349–361. [DOI] [PubMed] [Google Scholar]
- Xu H., Zhang F., Liu B., Huhman D.V., Sumner L.W., Dixon R.A., Wang G. (2013). Characterization of the formation of branched short-chain fatty acid:CoAs for bitter acid biosynthesis in hop glandular trichomes. Mol. Plant 6: 1301–1317. [DOI] [PubMed] [Google Scholar]
- Yang Z. (1998). Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15: 568–573. [DOI] [PubMed] [Google Scholar]
- Ye Y., Minami A., Mandi A., Liu C., Taniguchi T., Kuzuyama T., Monde K., Gomi K., Oikawa H. (2015). Genome mining for sesterterpenes using bifunctional terpene synthases reveals a unified intermediate of di/sesterterpenes. J. Am. Chem. Soc. 137: 11846–11853. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang C., Liu Y. (2015). Targeting cancer with sesterterpenoids: the new potential antitumor drugs. J. Nat. Med. 69: 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi J., Mafu S., Peters R.J. (2014). To gibberellins and beyond! Surveying the evolution of (di)terpenoid metabolism. Annu. Rev. Plant Biol. 65: 259–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.