Direct binding by CCA1 protein in the morning defines a strong transcriptionally repressed state of PRR5, a mechanism used for temporal regulation of a set of evening-expressed genes, including PRR5.
Abstract
The circadian clock is a biological timekeeping system that provides organisms with the ability to adapt to day-night cycles. Timing of the expression of four members of the Arabidopsis thaliana PSEUDO-RESPONSE REGULATOR (PRR) family is crucial for proper clock function, and transcriptional control of PRRs remains incompletely defined. Here, we demonstrate that direct regulation of PRR5 by CIRCADIAN CLOCK-ASSOCIATED1 (CCA1) determines the repression state of PRR5 in the morning. Chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) analyses indicated that CCA1 associates with three separate regions upstream of PRR5. CCA1 and its homolog LATE ELONGATED HYPOCOTYL (LHY) suppressed PRR5 promoter activity in a transient assay. The regions bound by CCA1 in the PRR5 promoter gave rhythmic patterns with troughs in the morning, when CCA1 and LHY are at high levels. Furthermore, ChIP-seq revealed that CCA1 associates with at least 449 loci with 863 adjacent genes. Importantly, this gene set contains genes that are repressed but upregulated in cca1 lhy double mutants in the morning. This study shows that direct binding by CCA1 in the morning provides strong repression of PRR5, and repression by CCA1 also temporally regulates an evening-expressed gene set that includes PRR5.
INTRODUCTION
The circadian clock is a timekeeping system that provides organisms with a mechanism to adapt to 24-h day-night cycles. The clock activates biological processes at specific times during the daily cycle through synchronous expression of genes involved in related biological processes, such as preparing for colder temperatures in the evening or anticipating infection by pathogens at dawn (Fowler et al., 2005; Wang et al., 2011). Recent studies indicate that there is broad conservation among similar genetic networks in which clock-associated transcription factors (TFs) directly regulate clock output, despite differences in the actual clock-associated proteins (Smith et al., 2010; Abruzzi et al., 2011; Huang et al., 2012; Koike et al., 2012; Nakamichi et al., 2012; Liu et al., 2013; Markson et al., 2013).
In Arabidopsis thaliana, transcription and translation feedback loops among clock-associated genes are required to maintain circadian clock functionality (Nakamichi, 2011; Pokhilko et al., 2012; Carré and Veflingstad, 2013; Chow and Kay, 2013; Hsu and Harmer, 2014; McClung, 2014). Two single Myb-related TFs, CIRCADIAN CLOCK-ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), directly repress expression of TIMING OF CAB EXPRESSION1 (TOC1), EARLY FLOWERING4 (ELF4), and LUXARRHYTHMO (LUX), all of which are expressed during the evening hours (Alabadí et al., 2001; Hazen et al., 2005; Perales and Más, 2007; Li et al., 2011). CCA1 and LHY repress such target genes through the corepressor DE-ETIOLATED1 (DET1) (Lau et al., 2011). CCA1 and LHY are required for activating expression of PSEUDO-RESPONSE REGULATOR9 (PRR9) and PRR7 (Farré et al., 2005), but the molecular details of this activation remain unknown. As part of one feedback loop, CCA1 and LHY are directly repressed by morning-expressed PRR9, midday-expressed PRR7, afternoon-expressed PRR5, and evening-expressed TOC1, thus forming a continuous set of repression events that extends from noon until about midnight (Nakamichi et al., 2010; Gendron et al., 2012; Huang et al., 2012; Wang et al., 2013). Three other clock-associated proteins, NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED GENE1 (LNK1), LNK2, and REVEILLE8 (RVE8), form complexes that activate PRR5 expression in the afternoon (Rawat et al., 2011; Hsu et al., 2013; Rugnone et al., 2013; Xie et al., 2014). Induction of PRR5 by RVE8 occurs in the afternoon, but not in the morning, suggesting that repression supercedes LNK-RVE8 activation of PRR5 transcription in the morning. The exact mechanism for this repression is not known (Hsu et al., 2013).
TFs involved in clock feedback loops also directly regulate clock output pathways by controlling key TFs for each biological process. Chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) analyses combined with transcriptomics experiments indicate that clock-associated PRR family proteins directly repress key TFs involved in photoperiodic flowering, hypocotyl elongation, and cold stress responses (Huang et al., 2012; Nakamichi et al., 2012; Liu et al., 2013; Liu et al., 2016). Genome-wide gene expression analyses using a chemically induced gene expression system revealed the potential targets of RVE8 and TOC1 (Gendron et al., 2012; Hsu et al., 2013). Though it was suggested that ELF3, ELF4, and LUX form an evening complex (EC) that directly regulates TFs involved in hypocotyl elongation (Nusinow et al., 2011; Herrero et al., 2012), and that CCA1 directly regulates TFs involved in cold or oxidative stress responses and flowering time regulation (Dong et al., 2011; Lai et al., 2012; Seaton et al., 2015), a more thorough understanding of genome-wide gene regulation mediated by ECs and CCA1 has been limited by a lack of data that could be supplied by genomic approaches like ChIP-seq or genome-wide gene expression profiling.
In this study, we performed an in silico survey of the upstream region of PRR5 and found evening elements (EEs) that could potentially be bound by RVEs, CCA1, and LHY. ChIP-seq indicated that CCA1 associates with three separate upstream regions of PRR5 in vivo. Time-course ChIP followed by quantitative PCR (ChIP-qPCR), gene expression analysis in cca1 lhy mutants, and promoter-reporter analyses showed that PRR5 is repressed by CCA1. Furthermore, ChIP-seq coupled with a genome-wide expression profile indicated that there are 113 potential target genes of CCA1. This gene set contains genes that are known to be repressed in the morning, suggesting that CCA1 associates with and mostly suppresses them in the morning, which results in evening-phased gene expression.
RESULTS
CCA1 Associates with PRR5 Upstream Regions in Vivo
To understand the circadian transcriptional regulation of PRR5, we surveyed cis-acting elements that could be involved in circadian expression upstream of PRR5 (5′ region to the PRR5 coding sequence) because the region between −1416 and the PRR5 start codon (where +1 indicates the adenine of the start codon) controls the rhythmic transcriptional activation characteristic of circadian clock cycles (Ueoka-Nakanishi et al., 2012). A number of potential regulatory cis-acting elements, including the LUX binding site (GATA/TGC) (Helfer et al., 2011), TCP binding site (GGNCCCAC) (Pruneda-Paz et al., 2009), protein box (ATGGGCC) (Michael et al., 2008), and morning element (AACCACGAAAA) (Harmer and Kay, 2005) were absent, whereas a G-box (CACGTG) (Schindler et al., 1992), four EEs (AAAATATCT) (Harmer et al., 2000), and a CCA1 binding site (CBS; AACAATCT or AAAAATCT) (Wang et al., 1997) were found in the region between −1416 and the start codon of PRR5 (Figure 1A). Genomic sequences that coimmunoprecipitate with PRRs in vivo are enriched for G-boxes (Huang et al., 2012; Nakamichi et al., 2012; Liu et al., 2013; Liu et al., 2016), and the regulatory elements EE and CBS are directly recognized by RVEs/CCA1/LHY in vitro and by REV8 in vivo (Wang et al., 1997; Alabadí et al., 2001; Rawat et al., 2011), but previous work had left it unclear whether or not CCA1 and LHY associate with PRR5 upstream in vivo.
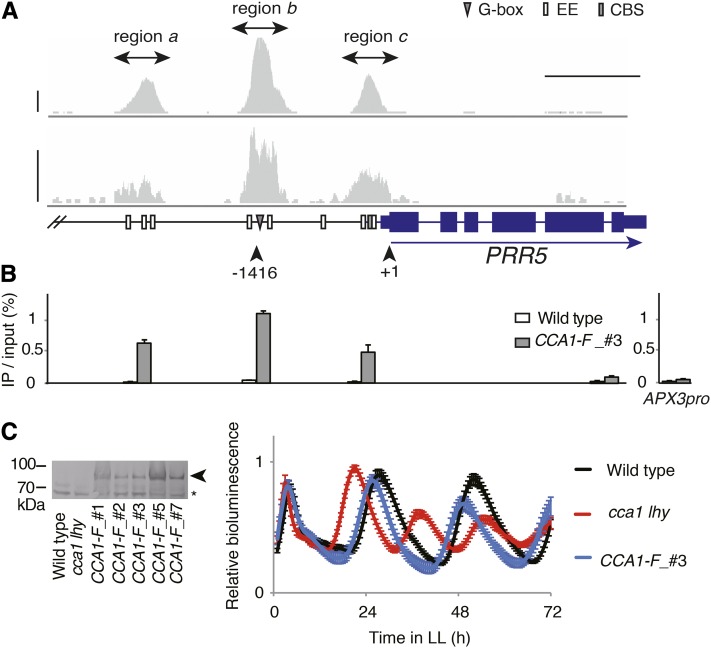
Figure 1.
CCA1 Associates with PRR5 Upstream.
(A) Position of ChIP peak reads determined by ChIP-seq assays in CCA1-F_#3 plants and potential cis-elements upstream of PRR5. Upper and lower panels were determined by IonPGM and GAII sequencing, respectively. Horizontal bar indicates 1 kb, and vertical bars indicate 50 reads.
(B) ChIP-qPCR assays around PRR5. White and gray bars show percentages of amplicons in ChIP DNA relative to input DNA from wild-type and CCA1-F_#3 plants, respectively. APX3pro indicates the amplicon located upstream of APX3. Error bars are the sd of three technical replicates. Similar results were obtained from independent experiments.
(C) CCA1-FLAG protein expressed under CCA1 promoter in cca1 lhy mutant (left, CCA1-F). The arrow and asterisk show CCA1-FLAG fusion protein and nonspecific bands, respectively. Circadian rhythm determined by bioluminescence of luciferase under the control of CCA1 (CCA1pro:LUC) in CCA1-F_#3 plants (right). Error bars indicate sd. Periods of the wild type, cca1 lhy, and CCA1-F_#3 were 25.4 ± 0.28 (n = 16), 17.5 ± 0.14 (n = 10), and 23.7 ± 0.40 h (n = 11), respectively. Note that CCA1-F_#1, 2, 5, and 7 also complemented the short period of cca1 lhy.
To determine whether CCA1 associates with the upstream region of PRR5 in vivo, we performed ChIP-seq using transgenic plants expressing CCA1-FLAG under the control of the native promoter in a cca1 lhy double mutant background (CCA1pro:CCA1-FLAG/cca1 lhy, namely, CCA1-F). We used cca1 lhy double mutants as parental plants because cca1 single mutants occasionally display subtle confounding phenotypes (Mizoguchi et al., 2002; Niwa et al., 2007), which might cause difficulties in interpreting the biological functionality of exogenous CCA1-FLAG protein. We found CCA1-FLAG protein in five independent transgenic lines (Figure 1C, left panel #1, 2, 3, 5, and 7). Real-time luminescence imaging demonstrated that introduction of CCA1-FLAG resulted in at least partial complementation of cca1 lhy, thus confirming the biological functionality of CCA1-FLAG (Figure 1C). However, CCA1 did not form heterodimers with LHY in the transgenic plants. CCA1-F_#3 was chosen as the representative plant line for subsequent ChIP-seq analyses (Figure 1C). Plants were grown under 12-h-light/12-h-dark conditions (LD) and harvested at Zeitgeber time 3 (ZT3; 3 h after lights were turned on), the time when CCA1 is normally expressed (Wang and Tobin, 1998). DNA in the anti-FLAG antibody-immunoprecipitated fraction from CCA1-F plants (ChIP DNA) was used to make a DNA library for deep sequencing with the Ion Personal Genetics Machine system (IonPGM). Sequence reads were mapped to the Arabidopsis genome TAIR10, and a mapping profile around the PRR5 locus was visualized using the Integrative Genomics Viewer (Figure 1A). ChIP DNAs from other biological samples were used to make an additional DNA library for deep sequencing by Illumina GA II (Figure 1A). CCA1-FLAG associates with three distinct regions upstream of PRR5 (−2754 to −2457 [region a], −1547 to −1171 [region b], and −379 to −84 [region c]; Figure 1A). Regions a, b, and c contain 3, 2, and 2 EEs, respectively. An EE is also found at −716, but that particular motif sequence was not bound by CCA1-FLAG. ChIP-qPCR analysis further supported the conclusion that CCA1-FLAG associates with the upstream regions of PRR5, but less with the coding sequence of PRR5 or upstream regions of ASCORBATE PEROXIDASE3 (APX3), whose expression is not under clock control (Figure 1B). Thus, two independent ChIP-seq experiments indicate that CCA1-FLAG associates with PRR5 upstream in vivo but does not associate with all of the EEs located upstream of PRR5 (Figure 1A).
CCA1 and LHY Repress PRR5 Transcription
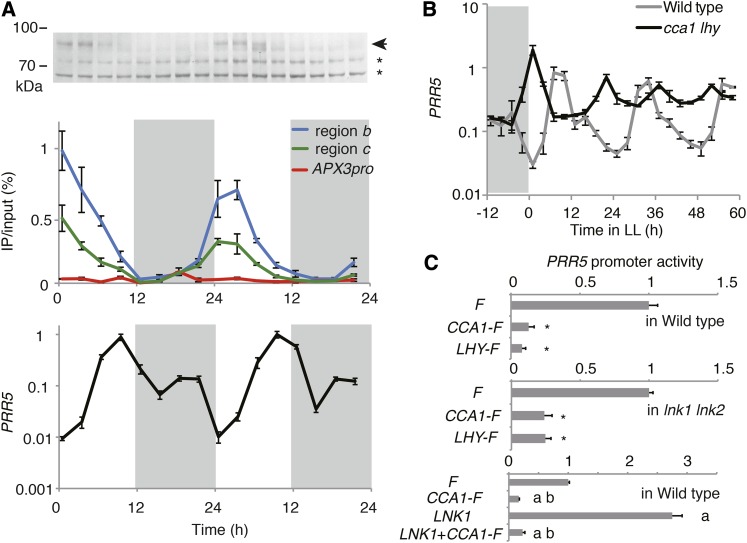
Since CCA1 protein expression is mostly limited in the morning (Wang and Tobin, 1998), we investigated the amount of association between CCA1 and the upstream region of PRR5 in CCA1-F_#3 plants grown under LD (Figure 2A). CCA1-FLAG protein accumulated from ZT0 to ZT6, and the protein associated with the upstream region of PRR5 during the same time frame (Figure 2A, top and middle panels), indicating that the timing of CCA1 expression is mostly responsible for limiting the association between CCA1 and PRR5. To understand the effect of CCA1 on PRR5 expression in vivo, PRR5 expression was measured from the same biological samples used for ChIP-qPCR (Figure 2A, bottom). PRR5 was suppressed from ZT0 and ZT3 and induced from ZT6 to a maximum at ZT9, followed by a decrease from ZT12. The temporal pattern of association between CCA1 and PRR5 as well as the timing of expression of PRR5 suggested either that CCA1 binding represses PRR5 transcription from ZT0 until ZT3, or CCA1 binding triggers activation of PRR5 transcription at ZT6. PRR5 expression in cca1 lhy double mutants was measured to determine which of these hypotheses was appropriate. Plants were initially grown under LD conditions and then transferred to constant light conditions (LL). Expression of PRR5 reached its maximum at ZT9 under LD, and subjective afternoon under LL, in wild-type plants (Figure 2B), whereas PRR5 peaks were temporally advanced under both LD and LL conditions in cca1 lhy plants. We also found that trough levels of PRR5 were elevated in the cca1 lhy background compared with the wild type under both LD and LL, suggesting that CCA1 and LHY downregulate PRR5 in the morning.
Figure 2.
CCA1 and LHY Repress PRR5 Expression.
(A) Expression of CCA1-FLAG protein (top) by immunoblot, association between CCA1-FLAG and PRR5 upstream (middle), and expression of PRR5 (bottom) in CCA1-F_#3 plants harvested simultaneously. CCA1-FLAG was determined by probing with anti-FLAG antibody. Arrow and asterisk are CCA1-FLAG and nonspecific bands, respectively (top). The ChIP-qPCR for APX3pro was performed as negative control on the CCA1-FLAG-bound locus (middle). Regions b and c are upstream of PRR5 as in Figure 1B. Expression of PRR5 was normalized to IPP2 expression, and its maxim value was set to 1 (bottom). White and gray areas indicate light (day) and dark (night) periods.
(B) PRR5 expression in cca1 lhy under LD and LL conditions. Gray area indicates the period of darkness.
(C) Effect of CCA1-FLAG (CCA1-F) and LHY-FLAG (LHY-F) on PRR5 promoter activity in wild-type protoplasts (top), lnk1 lnk2 double mutants protoplasts (middle), and with overexpression of LNK1 (bottom).
Error bars indicate the sd of three technical replicates in (A) and three biological replicates in (B) and (C). Asterisks show a significant difference in PRR5 activity compared with coexpression with FLAG only (F) (Student’s t test; P < 0.05). The “a” and “b” indicate statistical differences (P < 0.05) determined by one-way ANOVA compared with F and LNK1, respectively.
We next examined the extent of CCA1 and LHY regulation on PRR5 transcription activity in Arabidopsis mesophyll protoplasts. The circadian clock is maintained in these cells, as indicated by luciferase activity under the control of the PRR5 promoter (PRR5pro:LUC) or CCA1 promoter (CCA1pro:LUC), as reported previously (Kim and Somers, 2010) (Supplemental Figure 1). Then, reporter plasmid PRR5pro:LUC and effector plasmids harboring CCA1-FLAG, LHY-FLAG, or a negative control plasmid containing FLAG only, all under the control of the CaMV 35S promoter (35Spro:CCA1-F, LHY-F, or F) were cotransfected into mesophyll protoplasts. Cotransfection with 35Spro:CCA1-F or 35Spro:LHY-F resulted in lower PRR5pro:LUC activity than the control plasmid, suggesting that CCA1 and LHY independently suppress PRR5 transcription (Figure 2C, top).
To determine whether CCA1 and LHY affect PRR5 transcription indirectly via transcriptional activators of PRR5, LNK1, and LNK2 (Rugnone et al., 2013; Xie et al., 2014), 35Spro:CCA1-F or LHY-F was cotransfected into protoplasts prepared from lnk1 lnk2 double mutant plants (lnk1-4 lnk2-1). These plants have long circadian periods and elongated hypocotyls, resembling phenotypes reported in previous studies (Rugnone et al., 2013; Xie et al., 2014), due to a lack of full-length LNK1 and LNK2 transcripts (Supplemental Figure 2). Both 35Spro:CCA1-F and LHY-F reduced PRR5pro:LUC activity in the lnk1 lnk2 background (Figure 2C, middle). To further examine the relationship between CCA1 and LNK1 in determining PRR5 transcriptional activity, we cotransfected the PRR5pro:LUC reporter, 35Spro:CCA1-F, and 35Spro:LNK1 into wild-type mesophyll protoplasts and measured PRR5 promoter activity. Introduction of 35Spro:LNK1 resulted in activation of PRR5 promoter activity at twice the levels of that of the FLAG control (Figure 2C, bottom), which was consistent with previous studies (Rugnone et al., 2013; Xie et al., 2014). Cointroduction of 35Spro:CCA1-F and 35Spro:LNK1 suppressed PRR5 promoter activity to a similar level as the 35Spro:CCA1-F construct by itself (Figure 2C, bottom), implying that CCA1 is epistatic to LNK1 in PRR5 transcriptional regulation.
Association of CCA1 with the Promoter Region of PRR5 Relates to Circadian Expression
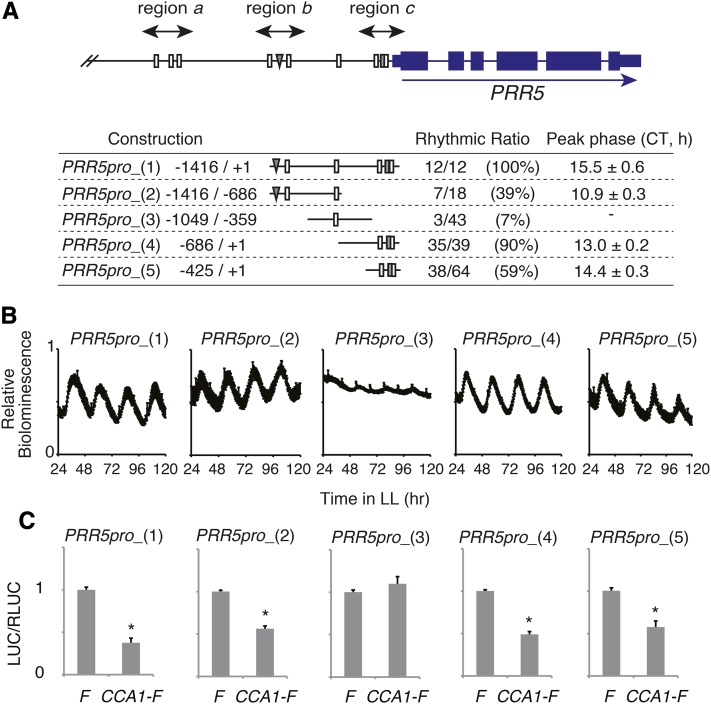
The region between −1416 and the start codon of PRR5 confers circadian transcriptional activation (Ueoka-Nakanishi et al., 2012). To further examine the transcriptional regulation of this region, we introduced a series of truncations of the PRR5 upstream region fused to a luciferase reporter into Arabidopsis (Figure 3). Constructions from −1416 to the start codon [PRR5pro_(1)], −1416 to −686 [PRR5pro_(2)], −686 to the start codon [PRR5pro_(4)], and −425 to the start codon [PRR5pro_(5)] gave robust rhythmic luciferase activities with peaks in the evening (CT10.9 to 15.5) and troughs in the morning (Figures 3A and 3B). All of these constructs contain regions b or c (Figure 1A). Although the region from −1049 to −359 contains an EE [PRR5pro_(3)], this construct conferred constant luciferase expression. In transient assays, introduction of CCA1-F resulted in suppression of promoter activity under the control of PRR5pro_(1), PRR5pro_(2), PRR5pro_(4), and PRR5pro_(5) but did not affect promoter activity of PRR5pro_(3) (Figure 3C). Regions b and c, both of which contain EEs that are bound by CCA1 in vivo, thus may act as cis-regulatory regions for evening-phase activation and morning-phase repression of PRR5.
Figure 3.
Regions b and c Upstream of PRR5 Give Robust Evening-Phase Rhythmic Expression.
(A) Bioluminescence pattern of luciferase fusion constructs. PRR5pro_(3) was arrhythmic.
(B) Real-time bioluminescence of PRR5pro:LUC transgenic plants under LL. The error bars indicate sd of biological replicates (n > 12).
(C) Effect of CCA1-FLAG (CCA1-F) on PRR5pro:LUC reporters. The error bars represent the se of three biological replicates. Asterisks indicate a significant change in LUC/RLUC activity compared with coexpression with FLAG (F) (Student’s t test; P < 0.05).
CCA1-Bound Genes on a Genomic Scale
Although CCA1 binds to EE and CBS motifs in vitro, our ChIP-seq analyses combined with promoter analyses indicate that CCA1 does not associate with every EE motif located in the PRR5 promoter region in vivo (Figures 1A and 3). To identify in vivo CCA1 binding regions on a genomic scale and to examine whether they are involved in circadian gene expression, we identified genomic loci bound by CCA1 according to a false discovery rate (FDR) q-value < 10−20 in two ChIP-seq data sets (Supplemental Figure 3 and Supplemental Data Sets 1A to 1C) and examined expression patterns of genes adjacent to these loci.
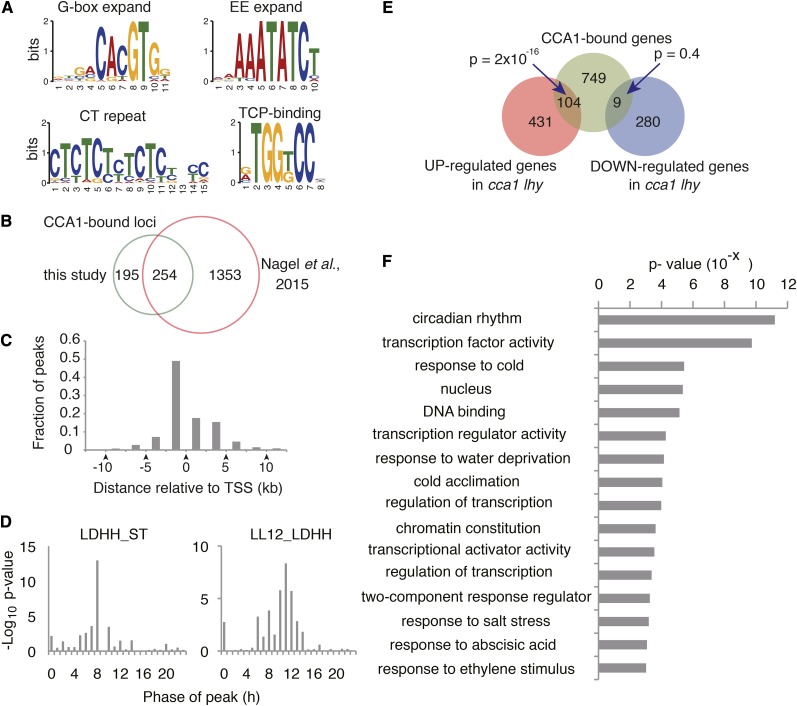
As indicated by two independent ChIP-seq studies, 449 loci were bound by CCA1, with 863 adjacent genes (Supplemental Data Sets 1C and 1D). A Multiple Motif Elicitation tool (Machanick and Bailey, 2011) found that 449 CCA1-bound loci were significantly enriched for G-box expanded, EE expanded, CT repeated, and TCP binding sequences (Figure 4A). EE expanded sequences contain typical EE (AAAATATCT), but not CBS (AACAATCT or AAAAATCT) motifs. CCA1 was originally identified as a transcription factor that binds to a CBS in the promoter region of a gene encoding chlorophyll a/b binding protein 1 (CAB1, At1g29930) in vitro (Wang et al., 1997). In our ChIP-seq analyses, CCA1 associates with the promoter regions of CAB1 and CAB2 (At1g29920), but binding values (q-value = 10−48.55 and 10−23.53 by PGM and GAII, respectively) were not as high as for other sites. For example, region b of the region upstream of PRR5 upstream has q-values of 10−121.04 by PGM and 10−105.63 by GAII (Supplemental Data Set 1). CCA1 binds less efficiently to upstream regions of the other CAB genes containing CBS (i.e., At1g29910, At2g34420, At2g34430, At3g54890, and At5g01530, each with q-values > 10−20). These results suggest that CCA1 binds to CBS in vitro, but prefers to bind to EE than to CBS in vivo. CCA1 also associates with EE-like sequences with higher binding affinity than with CBS-like sequences in vivo, as demonstrated recently by a ChIP-seq study with CCA1-GFP-expressing Wassilewskija accession plants (Nagel et al., 2015). To further understand CCA1 binding on a genomic scale, we compared our ChIP-seq data with that generated by Nagel et al. and found that 254 of 449-CCA1 binding loci were found in common with Nagel et al. (Figure 4B), suggesting that these two independent ChIP-seq studies are complementary and confirmatory despite the different genetic backgrounds used.
Figure 4.
CCA1-Bound Genes on a Genomic Scale.
(A) Enriched sequences in CCA1-bound loci.
(B) Concordance between CCA1-bound loci in this study and 1607 CCA1-bound loci under LD by Nagel et al. (2015).
(C) Positions of ChIP peaks relative to TSS in CCA1-bound genes.
(D) Phase enrichment of expression peaks of 863 CCA1-bound genes, calculated by Phaser under LDHH_ST and LL12_LDHH conditions.
(E) Overlap between CCA1-bound genes and genes whose expression differed from cca1 lhy double mutants. Fisher’s exact test indicates significant overlap between CCA1-bound genes and upregulated genes in cca1 lhy.
(F) eGO analysis for CCA1 target genes.
There are 517 CCA1 binding sites located within the upstream regions of transcriptional start sites (TSSs), which excludes the 5′ untranslated region (UTR) (Figure 4C). Moreover, 191 of the 517 CCA1 binding sites are within 500 bp upstream of the TSS. A total of 346 CCA1 binding sites are found in the downstream regions of TSSs, including the 5′ UTR, coding sequence, and 3′ UTR (Figure 4C). These data imply that CCA1 preferentially associates within regulatory sequences, most often located upstream of TSSs.
An analysis of the expression patterns of the 863 CCA1-bound genes was performed using a public microarray database with the Web-based tool Phaser (Mockler et al., 2007) (Figure 4D). The set of 863 genes was enriched for evening-phased genes but also contained morning-phased genes. Together, these 863 genes make up the set of CCA1-bound genes in vivo, a group that potentially contains CCA1 target genes, but may contain several false-positive targets due to the inherent technical limitations of ChIP, deep sequencing, and mapping procedures.
To discover genes directly targeted by CCA1 in another way, genes whose expression was altered in the cca1 lhy compared with the wild type were explored. There were 535 upregulated and 289 downregulated genes in cca1 lhy plants compared with the wild type (FDR q < 0.01) at ZT1, at a time when native CCA1 and LHY proteins are expressed (Supplemental Data Set 2). This set of genes contains direct targets of CCA1, but also includes genes that are indirect targets of CCA1. Overlap between upregulated genes in cca1 lhy and CCA1-bound genes was statistically significant (Fisher’s exact test P = 2 * 10−16, 104 genes), whereas overlap between downregulated genes and CCA1-bound genes was not (P = 0.4, nine genes), suggesting that CCA1 and LHY downregulate CCA1-bound genes (Figure 4E; Supplemental Data Set 3). The 113 genes whose expression was altered in cca1 lhy mutants and bound by CCA1 have been annotated as potential CCA1 target genes (Figure 4E; Supplemental Data Set 3).
The expression patterns of the 113 CCA1-potential targets were also examined using Phaser. The genes whose expression peaked between time 10 and 14 under LL conditions were significantly enriched among the 113 genes (Supplemental Figure 4). Evening-phased genes were also significantly enriched in the 535 upregulated genes of cca1 lhy double mutants. On the other hand, genes expressed in the late nighttime and early morning were enriched among the downregulated genes of cca1 lhy (Supplemental Figure 4). Taken together, these results indicate that direct regulation of CCA1 is crucial for diurnal and circadian expression patterns with troughs in the morning.
To explore the biological functions of each of the 113 CCA1-potential target genes, significantly enriched Gene Ontology (eGO) analysis was performed. “Circadian rhythm” was the most enriched category in the gene set (P < 10−11) (Figure 4F). “Transcription factor activity,” “response to cold,” “nucleus,” and “DNA binding” were also enriched (P < 10−5). There was enrichment of TF-related terms when eGO analysis was performed for the 863 CCA1 binding genes (Supplemental Figure 5). Because eGO terms indicate a relationship to TF, we further focused on the TF set of genes (Table 1). CCA1-potential target TFs include five AP2 genes (DEHYDRATION-RESPONSIVE ELEMENT BINDINAG 2A [DREB2A], DREB2B, DREB2C, DREB2H, and DECREASE WAX BIOSYNTHESIS [DEWAX]), three PRRs (TOC1, PRR7, and PRR5), three MYB (RVE7, myeloblastosis family transcription factor like-2 [MYBL2], and At1g17460), two B-BOX DOMAIN PROTEIN (BBX8 and BBX13), two GARP (LUX and BROTHER OF LUX ARRHYTHMO [BOA; also known as NOX]), two bHLH (ABSCISIC ACID-RESPONSIVE KINASE SUBSTRATE2 [AKS2] and ACTIVATION-TAGGED BRI1 SUPRESSOR1-INTERACTING FACTOR1 [AIF1]), a bZIP ([ABA INSENSITIVE5 [ABI5]), a TCP (CCA1 HIKING EXPEDITION [CHE]), C2H2 (BALDIBIS [BIB]), ARF6 (a DNA binding auxin response factor), a NAC (NAC019), a HSF (HSFC1), a CCAAT TF (NF-YB2), GAI, RGA, and SCR (GRAS) (SCR-LIKE13 [SCL13]). Three PRRs, LUX, BOA, and CHE, are all involved in circadian clock control (Pruneda-Paz et al., 2009; Dai et al., 2011). DREB2A and DREB2B are drought stress response proteins (Liu et al., 1998), DREB2C, AKS2, and ABI5 are involved in ABA signaling (Finkelstein and Lynch, 2000; Lee et al., 2010; Takahashi et al., 2013), DREB2C is also in heat stress response (Lim et al., 2007), DEWAX is involved in wax biosynthesis regulation (Go et al., 2014), ARF6 is required for flower maturation (Nagpal et al., 2005), AIF1 and MYBL2 are brassinosteroid signaling proteins (Wang et al., 2009; Ye et al., 2012), MYBL2 is important for anthocyanin biosynthesis (Matsui et al., 2008), BIB is involved in root development (Long et al., 2015), SNF-YB2 helps control flowering time (Cai et al., 2007), SCL13 is a part of red light signaling (Torres-Galea et al., 2006), RVE7 regulates cotyledon opening and flowering time (Kuno et al., 2003), NAC019 is a water stress regulator (Tran et al., 2004), and HSFC1 responds to heat stress (Guan et al., 2014). These data imply that CCA1 influences diverse physiological processes partly through regulating this set of TFs.
Table 1. CCA1 Potential Direct Target Genes Encoding TFs.
| Gene Name | Physiological Processes | Reference |
|---|---|---|
| DREB2A (At5g05410) | Drought stress response | Liu et al. (1998) |
| DREB2B (At3g11020) | ||
| DREB2C (At2g40340) | Heat stress response | Lim et al. (2007) |
| ABA signaling | Lee et al. (2010) | |
| DEWAX (At5g61590) | Wax synthesis regulation | Go et al. (2014) |
| TOC1 (At5g61380) | Circadian clock | Gendron et al. (2012) |
| PRR7 (At5g02810) | Nakamichi et al. (2010) | |
| PRR5 (At5g24470) | Hazen et al. (2005) | |
| LUX (At3g46640) | Dai et al. (2011) | |
| BOA (At5g59570) | Pruneda-Paz et al. (2009) | |
| CHE (At5g08330) | ||
| AKS2 (At1g05805) | ABA signaling | Takahashi et al. (2013) |
| ABI5 (At2g36270) | Finkelstein and Lynch (2000) | |
| ARF6 (At1g30330) | Flower maturation | Nagpal et al. (2005) |
| AIF1 (At3g05800) | BR signaling | Wang et al. (2009) |
| MYBL2 (At1g71030) | BR signaling | Ye et al. (2012) |
| Anthocyanin biosynthesis | Matsui et al. (2008) | |
| BIB (At3g45260) | Root development | Long et al. (2015) |
| SCL13 (At4g17230) | Red light signaling | Torres-Galea et al. (2006) |
| RVE7 (At1g18330) | Cotyledon opening | Kuno et al. (2003) |
| Flowering time | ||
| NAC019 (At1g52890) | Water stress response | Tran et al. (2004) |
| HSFC1 (At3g24520) | Heat stress response | Guan et al. (2014) |
| NF-YB2 (At5g47640) | Flowering time | Cai et al. (2007) |
| DREB2H (At2g40350) | Unknown | |
| BBX8 (At5g48250) | ||
| BBX13 (At1g28050) | ||
| TRF-like 3 (At1g17460) |
BR, brassinosteroid; ABA, abscisic acid.
CCA1 Represses Several Clock-Associated Genes
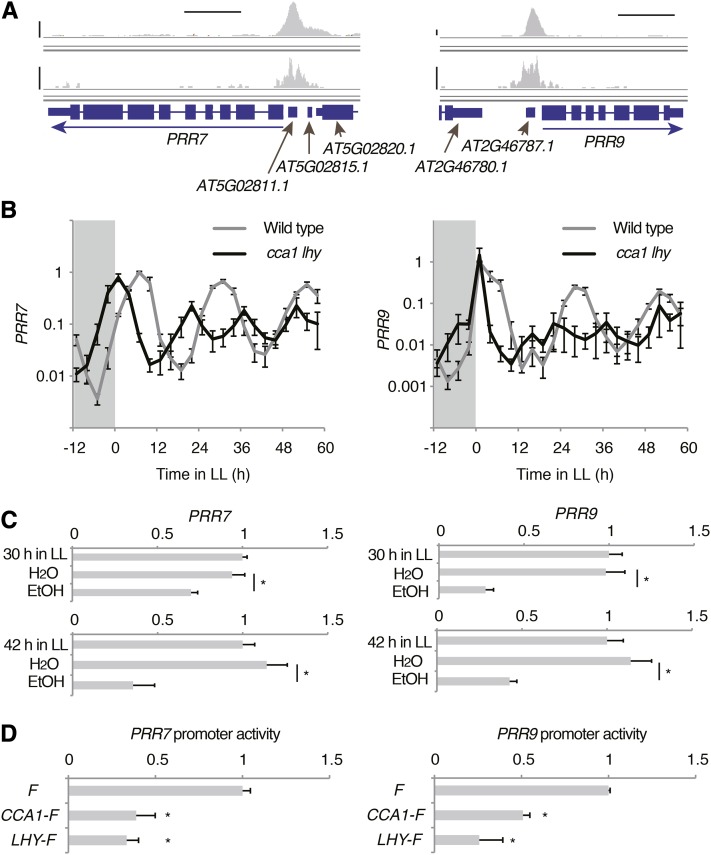
Among the 113 potential CCA1 target genes, we found several clock-associated genes: PRR7, PRR5, TOC1, ELF4, GI, LUX, BOA/NOX, and CHE (Figure 5A; Supplemental Figure 6 and Supplemental Data Set 3). TOC1, ELF4, GI, LUX, BOA, and CHE are repressed by CCA1 and LHY (Alabadí et al., 2001; Hazen et al., 2005; Mizoguchi et al., 2005; Pruneda-Paz et al., 2009; Dai et al., 2011; Li et al., 2011), and expression of these genes was significantly elevated in cca1 lhy plants at ZT 1 (Supplemental Figure 7). PRR7 may be activated by CCA1 and LHY (Farré et al., 2005). We also found that PRR9 was bound by CCA1 (Figure 5A), though PRR9 expression in cca1 lhy at ZT1 was comparable to the wild type (Supplemental Figure 7). To understand the effects of CCA1 and LHY activity on PRR7 and PRR9, expression was measured in cca1 lhy plants under LD and LL conditions (Figure 5B). PRR7 expression was upregulated around dawn, but downregulated from noon to evening relative to the wild type under LD (Figure 5B). PRR7 peak levels were lower and trough levels were higher in cca1 lhy mutants than in wild-type plants under LL conditions. PRR9 expression was highly suppressed in cca1 lhy plants, except at 1 h after lights were turned on (Figure 5B). To further understand the effect of CCA1 on PRR7 and PRR9 expression, CCA1 was transiently induced under the control of an ethanol-inducible promoter system (Alc:CCA1; Knowles et al., 2008). Alc:CCA1 seedlings were initially grown under LD and transferred to LL. Seedlings were then treated with ethanol for 20 min at the following subjective noon (i.e., 30 h after lights on), when native CCA1 is less expressed, and PRR9 and PRR7 expression was analyzed 2 h after ethanol treatment (Figure 5C). Ethanol treatment resulted in higher CCA1 expression (Supplemental Figure 8) and lower PRR9 and PRR7 expression compared with the control treatment (Figure 5C). Expression of the CCA1 targets PRR5 and TOC1 was also downregulated by ethanol treatment at subjective noon (Supplemental Figure 8). We found that expression of PRR9, PRR7, PRR5, and TOC1 was significantly decreased with ethanol treatment at subjective midnight (i.e., 42 h after lights on) (Figure 5C; Supplemental Figure 8). These results indicate that CCA1 does not directly activate PRR9 and PRR7, but it potentially suppresses them. To understand the effect of CCA1 and LHY on PRR7 and PRR9 transcription, we performed transient expression assays using PRR9pro:LUC or PRR7pro:LUC reporters and 35Spro:CCA1-F and LHY-F effectors (Figure 5D). Expression of CCA1-F and LHY-F did not result in any activation of PRR7 or PRR9 promoter activity. Rather, CCA1-F and LHY-F significantly decreased PRR7 and PRR9 promoter activities (Figure 5D), but the degrees of CCA1-F- or LHY-F-dependent suppression of these promoter activities were relatively small compared with PRR5 promoter activity (Figure 2C). These results suggest that CCA1 and LHY do not activate PRR7 and PRR9 transcription, but rather that CCA1 and LHY directly suppress PRR7 and PRR9.
Figure 5.
CCA1 and LHY Do Not Activate PRR9 and PRR7 Transcription.
(A) Visualization of ChIP-seq data around PRR7 and PRR9. Upper and lower panels were determined by IonPGM and GAII sequencer, respectively. Horizontal bar indicates 1 kb, and vertical bars indicate 50 reads.
(B) PRR7 and PRR9 mRNA expression in cca1 lhy under LD and LL.
(C) Effect of transient induction of CCA1 on PRR7 and PRR9 expression. CCA1 was transiently expressed by 20 min exposure to ethanol vapor at subjective noon (30 h in LL) or midnight (42 h in LL). Gene expression before treatment (labeled as 30 h in LL or 42 h in LL), control treatment (water), and ethanol treatment was analyzed.
(D) Effect of CCA1-FLAG (CCA1-F) and LHY-FLAG (LHY-F) on PRR7 and PRR9 promoter activity. Error bars indicate the se of three biological replicates in (B) and (D) and four biological replicates in (C). Asterisks indicate a significant difference (Student’s t test; P < 0.05) in (C) and compared with coexpression with FLAG (F) (Student’s t test; P < 0.05) in (D).
CCA1 and PRR5 Interact to Shape Target Gene Expression Phases
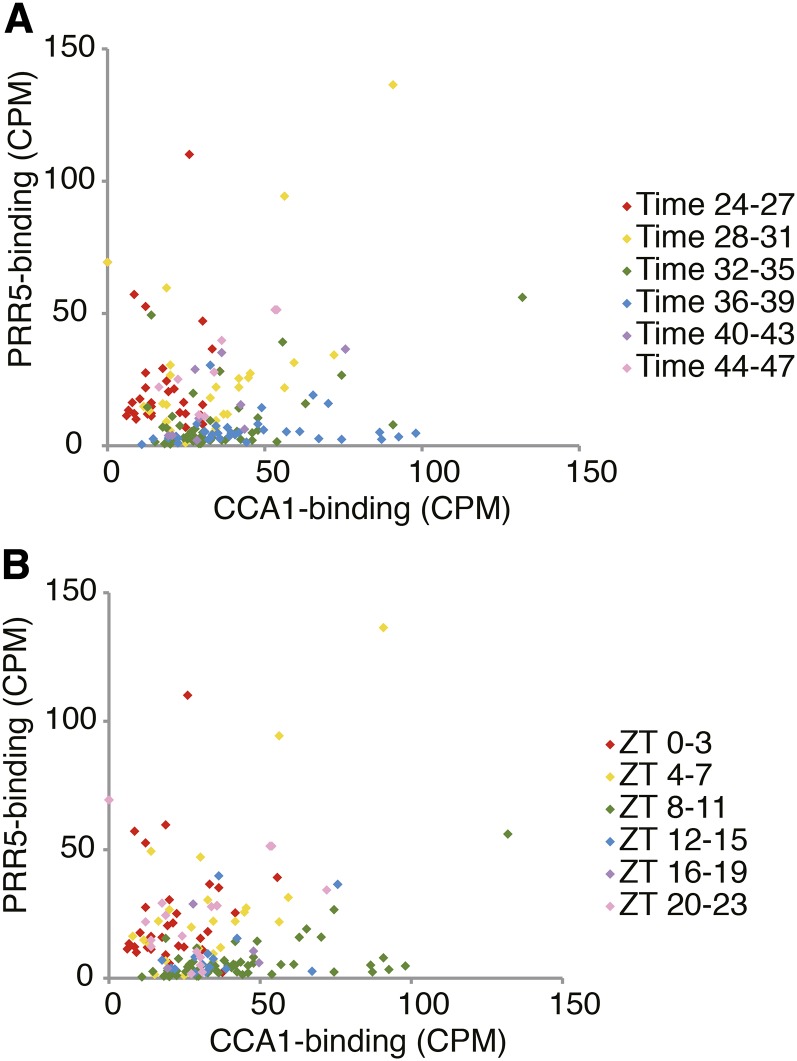
ChIP-seq studies suggest that the target genes of PRR7, PRR5, and TOC1 are mostly expressed from dawn until early morning (Huang et al., 2012; Nakamichi et al., 2012; Liu et al., 2013). In order to examine whether CCA1 and PRR5 interact within the circadian clock transcriptional network, we compared potential CCA1 targets (Supplemental Data Set 3) and PRR5 targets (Nakamichi et al., 2012) (Figure 6). The expression peaks of genes preferentially bound by PRR5 occurred between dawn and early morning (pink and red dots in Figure 6). Genes preferentially bound by CCA1 were expressed around evening (green and blue dots in Figure 6). Notably, genes bound by both CCA1 and PRR5 tend to be expressed toward the middle of the day (yellow dots in Figure 6). These patterns suggest that binding by different classes of TFs, namely, CCA1 and PRR5, shape and refine temporal gene expression in the clock transcriptional network.
Figure 6.
CCA1 and PRR5 Shape Expression Timing of Potential Targets.
Sequence reads in counts per million in each ChIP peak responsible for CCA1 or PRR5 target genes were determined and plotted on x and y axes, respectively. Expression peaks of these genes are colored with red (for early morning), yellow (around noon), green (afternoon), blue (evening), purple (before midnight), and pink (dawn). Gene expression data for (A) and (B) are LL12_LDHH and LDHH_ST, respectively.
DISCUSSION
CCA1 Represses PRR5 in the Arabidopsis Circadian Clock
The Arabidopsis circadian clock operates through a system of transcriptional feedback loops composed of clock-associated transcriptional factors, including the PRRs. Expression timing of the PRR family (PRR9, PRR7, PRR5, and TOC1) differs throughout the day (Matsushika et al., 2000), suggesting that there are different molecular mechanisms underlying regulation of PRR genes in the clock genetic circuit. Based on earlier data, TOC1 and PRR5 were proposed as nighttime repressors of PRR5 (Huang et al., 2012; Nakamichi et al., 2012). More recent studies suggest that the RVE8-LNK complex activates PRR5 transcription (Rugnone et al., 2013; Xie et al., 2014), but RVE8- and LNK-dependent PRR5 activation is highly attenuated in the morning (Hsu et al., 2013). In this study, we found that CCA1 associates with the PRR5 promoter in the morning, CCA1 and LHY suppress PRR5 transcription, and morning repression of PRR5 is attenuated in cca1 lhy double mutant plants (Figure 2). In addition, CCA1 and LHY suppress PRR5 promoter activity even in lnk1 lnk2 double mutants and in cells cotransfected with LNK1 (Figure 2C). These data suggest that CCA1 and LHY are strong repressors of PRR5 in the morning, as well as candidates for the molecular masking of RVE8- and LNK-dependent transcriptional activation of PRR5 (Hsu et al., 2013).
PRR9 and PRR7 are regulated by environmental cues such as light and temperature changes, which may continually reset the phase of the clock to coincide with external growth conditions and the plant’s internal energy state (Ito et al., 2005; Haydon et al., 2013; Kolmos et al., 2014; Mizuno et al., 2014). PRR9 and PRR7 are expressed in the morning, and these genes are directly repressed by components of the EC (Dixon et al., 2011; Helfer et al., 2011; Mizuno et al., 2014). In addition, given that PRR9 and PRR7 transcript levels are lower in the cca1 lhy double mutant and that CCA1 binds to the PRR9 and PRR7 upstream regions in vitro, CCA1 and LHY were assumed to be implicated in activation of PRR9 and PRR7 expression (Farré et al., 2005). In this study, we found that CCA1 associates with the upstream regions of PRR9 and PRR7, and peak levels of PRR9 and PRR7 under LL were lower in cca1 lhy plants than in the wild type (Figure 5B). However, transient induction of CCA1 by the ethanol-inducible system resulted in downregulation of PRR9 and PRR7 at subjective noon or midnight (Figure 5C), and CCA1 and LHY suppressed PRR9 and PRR7 promoters in transient assays (Figure 5D). In addition, transient induction of CCA1 and LHY alone did not result in any activation of PRR9 and PRR7 (Knowles et al., 2008), and induction of LHY alone resulted in lower expression of these PRRs (Adams et al., 2015). These studies suggest that CCA1 and LHY act as transcriptional repressors of PRR9 and PRR7.
Although transcription of PRR9 and PRR7 is suppressed by CCA1 and LHY (Figure 5D), PRR9 mRNA contents are downregulated in cca1 lhy, and continuous induction of LHY increases these PRR mRNAs, indicating that CCA1 and LHY indirectly activate PRR9 and PRR7 in a clock genetic network system (Figure 5B) (Farré et al., 2005; Adams et al., 2015). It is possible to imagine that CCA1 and LHY activate PRR9 and PRR7 expression through direct repression of PRR5, TOC1, LUX, and ELF4 (Figure 1; Supplemental Figure 7), all of which encode transcriptional repressors of PRR9 and PRR7 (Dixon et al., 2011; Helfer et al., 2011; Huang et al., 2012; Nakamichi et al., 2012). A similar molecular model was developed recently (Fogelmark and Troein, 2014). In addition, ELF3, which encodes a component of the EC, is epistatic to CCA1 and LHY for PRR9 and PRR7 expression (Dixon et al., 2011).
Molecular Mechanisms of CCA1-Dependent Transcriptional Control
In the circadian clock system, PRR9, PRR7, and PRR5 proteins repress transcription of target genes in concert with TOPLESS-related proteins as corepressors, along with histone deacetylase (Wang et al., 2013). EAR (ethylene-responsive element binding factor-associated amphiphilic repression)-like motifs within the three PRR proteins are sufficient to bind to TOPLESS-related proteins, which subsequently recruit histone deacetylases to suppress target gene transcription (Nakamichi et al., 2010; Wang et al., 2013). However, there are no known archetypal repression or activation domains in CCA1 or LHY, implying that these proteins are not active transcriptional repressors or activators, though CCA1 represses TOC1 by accelerating histone deacetylation around the TOC1 promoter (Perales and Más, 2007). This work demonstrates that CCA1 and LHY also repress transcription of PRR5, PRR7, and PRR9 (Figures 2 and 5). CCA1 and LHY apparently regulate gene transcription by interacting with other classes of regulators like DET1 as a corepressor for TOC1 transcription (Lau et al., 2011). This type of property may give CCA1 and LHY molecular plasticity (i.e., as a weak or strong repressor or activator of transcription) with regulatory control in a context-dependent manner. This ability to fine-tune a number of coordinated processes may explain the results of a recent article describing CCA1 suppression and/or activation of an unexpectedly high number of genes, including noncycling genes (Nagel et al., 2015).
Binding Preference of CCA1 on a Genomic Scale in Vivo
Although CCA1 and LHY form heterodimers (Lu et al., 2009), expression of CCA1-FLAG mostly complemented the short-period phenotype of cca1 lhy (Figure 1C), implying that CCA1-LHY heterodimers are not necessary for normal clock functioning. Because the CCA1-F transgenic line for ChIP-seq was generated in a cca1 lhy genetic background, our ChIP-seq data exclude loci bound by CCA1-LHY heterodimers. The absence of CCA1-LHY heterodimers could explain at least part of the difference between the set of loci identified in our ChIP-seq data and the set identified by Nagel et al. (2015). The set from Nagel et al. likely contains loci bound by CCA1-LHY heterodimers, as well as CCA1 homodimers, which would account for the many loci that were not found in our ChIP-seq study (Figure 4B). It may thus be possible to determine which of the loci are dependent on CCA1-LHY heterodimers versus CCA1 homodimers for expression by comparing the two data sets. However, a caveat would be that these two ChIP-seq studies were performed using different procedures, which may influence CCA1 binding on a genomic scale.
It was previously demonstrated that CCA1 binds to EE or CBS in vitro (Wang et al., 1997; Farré et al., 2005). However, our ChIP-seq analyses showed that CCA1 does not associate with all of the EE or CBS, indicating that there are differences in CCA1 binding preferences in vivo (Figures 1A and 4A). This is likely due to sequence-dependent contexts within the broader regulatory region or to the requirement for competitive binding elements that might be present in vivo but not in vitro. Preferential binding at EE sites by CCA1 was also detected in a ChIP-seq study using CCA1-GFP-expressing plants in a wild-type Wassilewskija accession background (Nagel et al., 2015).
Association of CCA1 with the EEs of PRR5 is efficient at regions a, b, and c, but less so with the EE between regions b and c (Figure 1). In addition, regions containing EE bound by CCA1 were sufficient to confer a rhythmic pattern, but EE not bound by CCA1 were not (Figure 3). These data indicate that binding of the trans-factor CCA1 to EE cis-elements is crucial for forming or maintaining a rhythmic pattern. EEs located upstream of TOC1 and GI are crucial for maintaining a rhythmic pattern with peaks in the evening (Harmer et al., 2000; Berns et al., 2014). Our ChIP-seq analyses indicated that there are high-resolution CCA1 binding profiles at TOC1 and GI, as well as at ELF4, LUX, and BOX, all of which are implicated in circadian clock functions (Supplemental Figure 6). Upstream regions of TOC1 and GI, which were previously shown to be sufficient for rhythmic patterning (Harmer et al., 2000; Berns et al., 2014), were also bound by CCA1 in vivo (Supplemental Figure 6). There are also CCA1 binding site preferences around these clock-associated genes (Supplemental Figure 6). The distribution and locations of these CCA1 preference sites on a genomic scale can be assessed using ChIP-seq studies. A thorough understanding of which loci are regulated by clock-associated transcription factors can provide a better understanding of global regulation by clock-associated transcription factors and, thus, the overall mechanism of circadian rhythms in plants. The expression of many CCA1-bound genes was not significantly different between in the cca1 lhy double mutant and the wild type at ZT1 (Figure 4C, 749 genes). To explain this apparent paradox, at least two possibilities should be considered. First, although we performed two different types of ChIP-seq analysis, using IonPGM and Illumina GA II pipelines to minimize the inherent limitations associated with ChIP, deep sequencing, and mapping procedures, there may still be false positives among targeted loci. Second, CCA1 binding may be only one of a number of regulatory constraints on expression of target genes. In fact, G-box and TCP binding sites were also enriched in DNA coimmunoprecipitated with CCA1, suggesting that CCA1 interacts with additional transcription factors that recognize these elements (Figure 4A). Nagel et al. (2015) also reported that DNA sequences containing G-box, protein box, and E-box, presumably bound by other classes of TFs, are enriched in the ChIP fraction of CCA1-GFP. Because our RNA-seq methodology used whole seedlings, neither tissue-specific nor age-specific gene expression could be determined. Different plant tissues have their own specific diurnal transcriptomes (Endo et al., 2014); thus, tissue-specific CCA1 targets may be assessed in future studies by examining the individual tissue-specific transcriptomes of cca1 lhy mutants.
The Circadian Clock Transcriptional Network Involves Other Transcription Factors, and Additional Layers of Regulation
This study demonstrated that CCA1 and PRR5, which are different types of clock-associated proteins expressed at different times of day (Wang and Tobin, 1998; Nakamichi et al., 2010), share some target loci (Figure 6). Our data suggest that combinatorial binding of these proteins may cause shifts in the timing of gene expression of many of the target genes (Figure 6). Given that other classes of clock-associated transcription factors (e.g., LUX, RVEs, and LNKs) are expressed at various times of day, further analyses of these transcription factors, and their interactions with CCA1 and with the PRRs, may reveal mechanisms underlying a variety of gene expression timing events on a genomic scale.
The Arabidopsis circadian clock coordinates genome-wide gene expression within the plant’s daily cycle. Comparative gene expression profiles indicate that evening-phase genes are expressed under many diurnal and circadian conditions (Michael et al., 2008). ChIP-seq revealed that CCA1 directly represses a set of evening-phase genes, including genes involved in circadian clock, ABA signaling, brassinosteroid signaling, wax biosynthesis, drought stress response, flowering time regulation, flower maturation, anthocyanin biosynthesis, and root development (Supplemental Data Set 3), implying that CCA1 regulates these processes via its direct targets. The number of potential CCA1 targets and CCA1-bound genes (113 and 863 genes, respectively) are fewer than the previously estimated 1000 to 2000 evening-phase genes (Michael et al., 2008), indicating that other transcriptional regulators and posttranscriptional control elements may involve evening-phase transcript accumulation. Control of cyclic genes by different regulatory layers (e.g., histone modification, transcription, splicing, and mRNA degradation) seem to be crucial for the accurate expression of output pathways in mammals and fungi (Sanchez et al., 2010; Koike et al., 2012; Hurley et al., 2014), each of which has a different type of central clock mechanism than plants.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana Col-0 accession plants were used as the wild type. CCA1pro:LUC plant construction was reported previously (Nakamichi et al., 2005). cca1 lhy CCA1pro:LUC (cca1-1 lhy-11 CCA1pro:LUC) was reported previously (Mizoguchi et al., 2002; Yamashino et al., 2008). T-DNA insertion lines lnk1-1 (SALK_024353), lnk1-4 (SALK_142366), lnk2-1 (GABI_484F07), and lnk2-2 (SALK_116103) were obtained from the ABRC and GABI Kat T-DNA insertion collections. PCR amplification (primers are listed in Supplemental Table 1) followed by reverse transcription suggested that these four lines were knockout mutants (Supplemental Figure 2). lnk1 lnk2 double mutants were generated by crossing lnk1-4 and lnk2-1. Alc:CCA1 (CS67790) (Knowles et al., 2008) was obtained from the ABRC. To obtain CCA1pro:CCA1-FLAG/cca1 lhy CCA1pro:LUC plants (CCA1-F), a region containing the CCA1 promoter and coding sequences from 863 bp upstream of the inferred start codon through the coding region was amplified with KOD-Plus-Neo DNA polymerase (Toyobo) from the genomic DNA of Col-0 using primers listed in Supplemental Table 1. To obtain genomic DNA, 200 mg of Col-0 seedlings was flash-frozen in liquid nitrogen and crushed with zirconia beads (ZB-50; Tomy) in a Tissue Lyser II (Qiagen). The resulting powdered sample was suspended in 100 μL of TE buffer and the supernatant was processed with NucleoSpin Gel and PCR cleanup (Takara). Amplified DNA was cloned into pENTR/D-TOPO (Life Technologies), generating entry plasmid pENTR/D-CCA1pro:CCA1. The DNA sequence of pENTR/D-CCA1pro:CCA1 was validated by Sanger sequencing. The pENTR/D-CCA1pro:CCA1 plasmid was incubated with Gateway LR clonase (Life Technologies) and modified pBA-PF5 binary vector (Kiba et al., 2007), generating a C-terminal 3*FLAG fusion construct (pBA-CCA1pro:CCA1-FLAG). pBA-CCA1pro:CCA1-FLAG was then transferred into cca1 lhy CCA1pro:LUC plants by an Agrobacterium tumefaciens-mediated method (Bechtold et al., 1993). Ten independent T1 transformants were selected on MS media (Murashige and Skoog, 1962) containing 10 mg L−1 of bialaphos sodium salt (022-15413; Wako), and exogenous CCA1-FLAG protein in T2 plants was detected by immunoblotting using anti-FLAG antibody (F3165; Sigma-Aldrich). Although At5g24470.1 can serve as one gene model for PRR5, our RNA-seq analysis showed that the first 220 bp of At5g24470.1 are not transcribed. In addition, we previously overexpressed PRR5 (the first candidate start codon in the transcript is 328 to 330 bp downstream of the 5′ end of At5g24470.1), which resulted in altered phenotypes related to circadian function (Sato et al., 2002; Nakamichi et al., 2012). Based on these observations, we used the chromosomal sequence of PRR5, rather than At5g24470.1. To generate PRR5pro_(1):LUC transgenic plants, the PRR5 promoter (PRR5pro; 1413 bp upstream of the inferred start codon) was amplified by PCR from Col-0 genomic DNA and cloned into the HindIII and NcoI sites of a modified pSP-luc+ vector (originally from Promega) using an In-Fusion HD cloning kit (Takara). The resulting PRR5pro_(1):LUC region was then cloned into the HindIII and SacI sites of binary vector pABH (Nakamichi et al., 2004). Truncated PRR5pro:LUC sequences were also cloned into pABH. pABH vectors were transformed into Col-0 by Agrobacterium as above. Plants were grown on MS containing 2% sucrose and 0.3% gellan gum under 12 h white light by bulb (70 µmol s−1 m−2)/12-h-dark conditions.
Bioluminescence-Based Circadian Rhythm of Transgenic Plants
Bioluminescence-based circadian rhythms of T3 CCA1-F plants were analyzed by autoluminescence (CL96; Churitsu) as described previously (Figure 1) (Onai and Ishiura, 2005). Briefly, Arabidopsis seedlings were grown on MS plates without sucrose under 12 h light/12 h dark for 4 d, and each seedling was transferred into a well of a 96-well plate. Twenty microliters of MS liquid medium containing 2% sucrose and 250 µM d-Luciferin potassium salt (Wako) was added to each well. The 96-well plate was further incubated under 12-h-light/12-h-dark conditions for 1 d and transferred to LL conditions in a CL96 platform at ZT0 of the next day. Period length and amplitude values were automatically calculated using the CL96 platform with embedded software. Amplitudes lower than 0.1 were annotated as noncyclic. Maximum values of individual luciferase activities were normalized to unity. T1 plants resistant to 20 mg L−1 hygromycin were analyzed by CL96 (Figure 3).
Immunoblotting
Two hundred milligrams of whole CCA1-F plants were flash-frozen with liquid nitrogen at ZT1 and crushed with zirconia beads in a Tissue Lyser II. The resulting powdered sample was suspended in 200 μL of 2× SM buffer, incubated at 95°C for 5 min, and centrifuged for 5 min at 14,000 rpm. Supernatants were loaded onto a Super Sep Ace 10 to 20% gradient gel (Wako) and blotted onto Hybond-P (GE Healthcare). Anti-FLAG antibody (F3165; Sigma-Aldrich) and goat anti-mouse IgG conjugated with alkaline phosphatase (170-6520; Bio-Rad) were used as primary and secondary antibodies to detect FLAG-fusion protein.
ChIP-Seq Analysis
ChIP-seq was performed as described previously with Illumina GA II (Nakamichi et al., 2012). A 5-g sample of CCA1-F_#3 plants flash-frozen at ZT3 was used for ChIP-seq analysis. DNA library construction was performed with a ChIP-Seq Sample Prep Kit (Illumina) as described previously (Nakamichi et al., 2012). To prepare a ChIP DNA library for IonPGM, ChIP was performed on a 5-g ZT3 sample of CCA1-F_#3 plants. ChIP DNA was used to generate a DNA library with an Ion Plus Fragment library kit (4471252; Life Technologies). The resulting DNA library was then analyzed by IonPGM with an Ion PGM Template OT2 200 kit (4480974; Life Technologies), Ion PGM Sequencing 200 Kit version 2 (4482006; Life Technologies), and Ion 318 Chip Kit version 2 (4484354; Life Technologies).
ChIP-Seq Data Analysis
Base-calling of sequence reads obtained by Illumina GA II was performed with GA II pipeline software. ChIP DNA sequence reads in the FASTQ format were compared against the reference genome TAIR10 and mapped to the Arabidopsis genome using Bowtie software (Langmead et al., 2009), with the parameter ‘–t –p 8 –n 3 –m 1 –a –best –strata –sam’. The resulting sequence alignment/map (SAM) file was converted into a binary alignment/map (BAM) format file by Samtools 0.1.18 (Li et al., 2009). Base-calling of sequence reads obtained by IonPGM was performed with IonPGM pipeline software. Mapping of these sequence reads was performed by Torrent Suite Software (Life Technologies) using default parameters. Significant ChIP DNA peaks (FDR q < 10−20) were annotated as CCA1-FLAG binding loci using Model-based Analysis of ChIP-Seq (MACS2) software (Zhang et al., 2008), with the genome size parameter dm (1.8 * 108). Forward- and reverse-peak distributions were validated by MACS2 and drawn by R (http://www.r-project.org/). Peaks from forward and reverse strands were within 200 bp, indicating that DNA fragment size in the ChIP library was acceptable. BAM and indexed BAM files were used for visualization of mapping patterns using the Integrative Genomics Viewer 1.5.64. The nearest gene for each peak was found by ChIPpeakAnno (Zhu et al., 2010), and the other side of each peak was searched manually. Sequences of ChIP peaks (Supplemental Data Set 1) were analyzed with the MEME-ChIP open Web tool (http://meme-suite.org) (Machanick and Bailey, 2011) to identify enriched DNA sequences in ChIP-DNA (Figure 4A). The data set of 1607 CCA1-bound loci under LD described by Nagel et al. (2015) (Supplemental Table 2 in their article) was compared with 449 CCA1-bound loci (Figure 4B; Supplemental Data Set 1). ChIP-seq data have been deposited with NCBI under accession number GSE67903.
ChIP-qPCR Analysis
ChIP-qPCR analyses were performed as described previously (Nakamichi et al., 2010) using an Eco Real-Time PCR system (Illumina). Primers for ChIP-qPCR are listed in Supplemental Table 1.
RT-qPCR Analysis
Two hundred milligrams of tissue was frozen with liquid nitrogen and crushed with zirconia beads in a Tissue Lyser II. Powdered samples were then used for RNA isolation with Illustra RNAspin (25-0500-72; GE Healthcare). RT-qPCR was performed as described previously (Nakamichi et al., 2010) using an Eco Real-Time PCR system. Gene expression was normalized against IPP2, and maximal values were set to 1. Primers for RT-qPCR are listed in Supplemental Table 1. Estimation of period length of lnk1 lnk2 (Supplemental Figure 2) was done by R with a script as previously reported (Yoshida et al., 2009).
Transient Expression Assays
pSP-PRR5pro_(1):LUC was used as a reporter plasmid to measure PRR5 promoter activity. To generate a reporter plasmid harboring luciferase under the control of the PRR7 promoter, an 898-bp fragment upstream of the inferred start codon of PRR7 was amplified and cloned into HindIII and NcoI sites in a modified pSP-luc+ vector. To generate a reporter plasmid harboring luciferase under the control of the PRR9 promoter, a 760-bp fragment upstream of the inferred initiation codon of PRR9 was amplified and cloned into the HindIII and NcoI sites of a modified pSP-luc+ vector by In-fusion reaction. To make an effector plasmid harboring CCA1 under the control of the 35S CaMV promoter (35Spro:CCA1-FLAG), CCA1 cDNA without the stop codon was amplified and cloned into pBS-FLAG (in which the 35S CaMV promoter, 3-FLAG, and NOS terminator were assembled 5′ to 3′ in pBluescript) between its XbaI and BamHI sites. Similarly, LHY cDNA was amplified and cloned into pBS-FLAG between BamHI and NcoI, generating 35Spro:LHY-FLAG. LNK1, including its stop codon, was cloned into pENTR/D-TOPO (pENTR/D-LNK1). pENTR/D-LNK1 was incubated with Gateway LR clonase and modified pBS-GW-FLAG vector (Nakamichi et al., 2012), generating 35Spro:LNK1. Transient expression assays using mesophyll protoplasts were performed as previously described (Yoo et al., 2007), with minor modifications. Arabidopsis plants grown under 16-h-light/8-h-dark conditions for 18 to 21 d were used for preparing protoplasts. Expression buffer (0.5× MS, 2 mM MES, pH 5.7, and 0.4 M mannitol) was used instead of WI. After incubation of protoplasts in expression buffer for 20 h under 16-h-light/8-h-dark conditions, luminescence was measured with the Dual-Luciferase Reporter Assay System (Promega) in an EnSpire Multimode Plate Reader (Perkin-Elmer). Usually, dual-luciferase assays were performed between ZT6 and ZT8. Data were obtained from at least three biological replicates. To analyze circadian rhythms in isolated mesophyll protoplasts, protoplasts were transfected with CCA1pro:LUC (Nakamichi et al., 2010) or PRR5pro:LUC plasmids at a concentration of 2 µg for each transfection and incubated in a 96-well plate in the dark for 12 h. After incubation, protoplasts were transferred to constant light and bioluminescence was measured by CL96. Primers for cloning are listed in Supplemental Table 1.
Measurement of Hypocotyl Lengths
Wild-type and lnk1 lnk2 plants were sown on MS plates containing 2% sucrose. After 2 d of incubation in the dark at 4°C, plates were incubated under 12 h white light (70 to 80 µmol m−2 s−1)/12 h dark for 6 d. Hypocotyl lengths were measured with Image J (http://imagej.nih.gov/ij).
RNA-Seq Analysis
Wild-type and cca1 lhy plants were grown on MS plates containing 2% sucrose under LD for 2 weeks after germination. Three biological replicates of whole plants were harvested at ZT1, and RNA was extracted for RNA-seq with Illustra RNAspin (25-0500-72; GE Healthcare). Two micrograms of total RNA was used to generate an RNA-seq library using TruSeq RNA sample Preparation Kits v2 (Illumina). Libraries were then sequenced by Illumina GA II. Base-calling of sequence reads was performed using Illumina GA II pipeline software. The reads were mapped to Arabidopsis TAIR10 (http://www.arabidopsis.org/) by Bowtie (Langmead et al., 2009), and the number of reads mapped to the reference was counted. Reads were then normalized to counts per million. Genes whose expression in cca1 lhy was different from those in the wild type (FDR q < 0.01) were annotated as “upregulated genes in cca1 lhy” (Supplemental Data Set 2A) or “downregulated genes in cca1 lhy” (Supplemental Data Set 2B) by EdgeR (Robinson et al., 2010). Overlaps between CCA1-bound genes and upregulated or downregulated genes in cca1 lhy were analyzed by Fisher’s exact one-tailed test in R (http://www.r-project.org/). RNA-seq data were deposited in DDBJ (http://www.ddbj.nig.ac.jp/) under the BioProject ID PRJDB3468 and accession ID DRA003474.
eGO Analysis
eGO analysis was done as previously described (Tsukagoshi et al., 2010).
Determination of Gene Expression Peaks
Phaser was used to examine the expression peaks of gene sets (113 potential CCA1 targets, 863 CCA1 binding genes, 535 upregulated genes in cca1 lhy, and 289 downregulated genes in cca1 lhy) using LL12_LDLL or LDHH_ST (http://mocklerlab.org/tools) (Figure 4; Supplemental Figure 4). The correction cutoff value was set to 0.7 in Phaser. To examine the relationship between expression peaks and CCA1- or PRR5-bound loci, sequence reads for each peak of the locus were set to counts per million, and the peak phase of the target locus was determined by Phaser (Figure 6).
Ethanol Induction in Alc:CCA1 Plants
Seeds were plated on MS containing 2% sucrose and kept in the dark at 4°C for 2 d. Seedlings were then grown under LD for 12 d and transferred to LL. Filter papers soaked in 1% ethanol in water (v/v) or water only were adhered to the underside of the plate lid for 20 min at 30 or 42 h after lights were turned on. Plants were harvested 2 h after ethanol treatment.
Accession Numbers
Sequence data for the genes described in this article have been deposited in The Arabidopsis Information Resource under the following accession numbers: APX3 (At4g35000), BOA/NOX (At5g59570), CCA1 (At2g46830), CHE (At5g08330), ELF3 (At2g25930), ELF4 (At2g40080), GI (At1g22770), IPP2 (At3g02780), LHY (At1g01060), LNK1 (At5g64170), LNK2 (At3g54500), LUX (At3g46640), PRR9 (At2g46790), PRR7 (At5g02810), PRR5 (At5g24470), and TOC1 (At5g61380). Accession numbers of CCA1 potential targets are listed in Table 1 and in Supplemental Data Set 3.
Supplemental Data
Supplemental Figure 1. PRR5 and CCA1 promoter activities in Arabidopsis mesophyll protoplasts.
Supplemental Figure 2. The lnk1 lnk2 mutants used in this study.
Supplemental Figure 3. Overlap between CCA1 binding loci determined by IonPGM and Illumina GAII.
Supplemental Figure 4. Peak-phase enrichment in the sets of 113 CCA1-potential target genes, 535 upregulated genes in cca1 lhy, and 289 downregulated genes in cca1 lhy.
Supplemental Figure 5. eGO analysis for CCA1-bound genes.
Supplemental Figure 6. CCA1 binding profiles around clock-associated genes.
Supplemental Figure 7. Expression of clock-associated genes in cca1 lhy at ZT1.
Supplemental Figure 8. Expression of CCA1, TOC1, and PRR5 upon ethanol in Alc:CCA1.
Supplemental Table 1. Primers used in this study.
Supplemental Data Set 1. CCA1-bound loci determined by IonPGM and Illumina GA II, and CCA1-bound genes.
Supplemental Data Set 2. Genes up- and downregulated in cca1 lhy mutants compared with the wild type at ZT1.
Supplemental Data Set 3. Potential CCA1 target genes.
Supplementary Material
Acknowledgments
We thank the ABRC and GABI Kat for sending seeds; T. Mizuno, T. Yamashino, and T. Mizoguchi for sharing plants; T. Kiba for kindly gifting pBS-35S:FLAG and modified pBA-PF5 vectors; Y. Takahashi, Y. Mizutani, and H. Sugimoto for technical suggestions on conducting transient assays; E. Ando for creating a macro in Excel for microarray data analysis; K. Onai for advice about CL96 setup; and Y. Toda for critical reading of the manuscript. We also thank each member of the T. Kinoshita laboratory for discussions about this research. This work was supported by Japan Science and Technology Agency Precursory Research for Embryonic Science and Technology Grant 20109, Ministry of Education, Culture, Sports, Science, and Technology Grant-in-Aid for Young Scientist 26870267, and The Naito Foundation to N.N.
AUTHOR CONTRIBUTIONS
M.K. and N.N. conceived the research. M.K., S.T., and N.N. generated vectors and transgenic plants. M.K. and S.T. performed ChIP, qPCR, RT-qPCR, and immunoblotting analysis. S.T. generated DNA libraries for deep sequencing and performed deep DNA sequencing with IonPGM. T.S. performed deep DNA sequencing with Illumina GA II. T.S., T.H., T.K., and N.N. performed data analysis of deep sequences. M.K. performed real-time luciferase imaging. K.T. performed dual-luciferase reporter assay using mesophyll protoplast. N.N. wrote the article.
Glossary
- TF
transcription factor
- EC
evening complex
- ChIP-seq
chromatin immunoprecipitation followed by deep sequencing
- EE
evening element
- ChIP-qPCR
ChIP followed by quantitative PCR
- CBS
CCA1 binding site
- LD
12-h-light/12-h-dark
- IonPGM
Ion Personal Genetics Machine
- LL
constant light
- FDR
false discovery rate
- UTR
untranslated region
- TSS
transcriptional start site
- eGO
enriched Gene Ontology
Footnotes
Articles can be viewed online without a subscription.
References
- Abruzzi K.C., Rodriguez J., Menet J.S., Desrochers J., Zadina A., Luo W., Tkachev S., Rosbash M. (2011). Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Dev. 25: 2374–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams S., Manfield I., Stockley P., Carré I.A. (2015). Revised morning loops of the Arabidopsis circadian clock based on analyses of direct regulatory interactions. PLoS One 10: e0143943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadí D., Oyama T., Yanovsky M.J., Harmon F.G., Más P., Kay S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883. [DOI] [PubMed] [Google Scholar]
- Bechtold N., Ellis J., Pelletier G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316: 1194–1199. [Google Scholar]
- Berns M.C., Nordström K., Cremer F., Tóth R., Hartke M., Simon S., Klasen J.R., Bürstel I., Coupland G. (2014). Evening expression of arabidopsis GIGANTEA is controlled by combinatorial interactions among evolutionarily conserved regulatory motifs. Plant Cell 26: 3999–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Ballif J., Endo S., Davis E., Liang M., Chen D., DeWald D., Kreps J., Zhu T., Wu Y. (2007). A putative CCAAT-binding transcription factor is a regulator of flowering timing in Arabidopsis. Plant Physiol. 145: 98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carré I., Veflingstad S.R. (2013). Emerging design principles in the Arabidopsis circadian clock. Semin. Cell Dev. Biol. 24: 393–398. [DOI] [PubMed] [Google Scholar]
- Chow B.Y., Kay S.A. (2013). Global approaches for telling time: omics and the Arabidopsis circadian clock. Semin. Cell Dev. Biol. 24: 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S., Wei X., Pei L., Thompson R.L., Liu Y., Heard J.E., Ruff T.G., Beachy R.N. (2011). BROTHER OF LUX ARRHYTHMO is a component of the Arabidopsis circadian clock. Plant Cell 23: 961–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon L.E., Knox K., Kozma-Bognar L., Southern M.M., Pokhilko A., Millar A.J. (2011). Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr. Biol. 21: 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M.A., Farré E.M., Thomashow M.F. (2011). Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 7241–7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M., Shimizu H., Nohales M.A., Araki T., Kay S.A. (2014). Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 515: 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré E.M., Harmer S.L., Harmon F.G., Yanovsky M.J., Kay S.A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15: 47–54. [DOI] [PubMed] [Google Scholar]
- Finkelstein R.R., Lynch T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogelmark K., Troein C. (2014). Rethinking transcriptional activation in the Arabidopsis circadian clock. PLOS Comput. Biol. 10: e1003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S.G., Cook D., Thomashow M.F. (2005). Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 137: 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron J.M., Pruneda-Paz J.L., Doherty C.J., Gross A.M., Kang S.E., Kay S.A. (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 109: 3167–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go Y.S., Kim H., Kim H.J., Suh M.C. (2014). Arabidopsis cuticular wax biosynthesis is negatively regulated by the DEWAX gene encoding an AP2/ERF-type transcription factor. Plant Cell 26: 1666–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Q., Yue X., Zeng H., Zhu J. (2014). The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell 26: 438–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer S.L., Hogenesch J.B., Straume M., Chang H.S., Han B., Zhu T., Wang X., Kreps J.A., Kay S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113. [DOI] [PubMed] [Google Scholar]
- Harmer S.L., Kay S.A. (2005). Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell 17: 1926–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon M.J., Mielczarek O., Robertson F.C., Hubbard K.E., Webb A.A. (2013). Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 502: 689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen S.P., Schultz T.F., Pruneda-Paz J.L., Borevitz J.O., Ecker J.R., Kay S.A. (2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 102: 10387–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer A., Nusinow D.A., Chow B.Y., Gehrke A.R., Bulyk M.L., Kay S.A. (2011). LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 21: 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E., et al. (2012). EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.Y., Devisetty U.K., Harmer S.L. (2013). Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife 2: e00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.Y., Harmer S.L. (2014). Wheels within wheels: the plant circadian system. Trends Plant Sci. 19: 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Pérez-García P., Pokhilko A., Millar A.J., Antoshechkin I., Riechmann J.L., Mas P. (2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79. [DOI] [PubMed] [Google Scholar]
- Hurley J.M., et al. (2014). Analysis of clock-regulated genes in Neurospora reveals widespread posttranscriptional control of metabolic potential. Proc. Natl. Acad. Sci. USA 111: 16995–17002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Nakamichi N., Matsushika A., Fujimori T., Yamashino T., Mizuno T. (2005). Molecular dissection of the promoter of the light-induced and circadian-controlled APRR9 gene encoding a clock-associated component of Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 69: 382–390. [DOI] [PubMed] [Google Scholar]
- Kiba T., Henriques R., Sakakibara H., Chua N.H. (2007). Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19: 2516–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Somers D.E. (2010). Rapid assessment of gene function in the circadian clock using artificial microRNA in Arabidopsis mesophyll protoplasts. Plant Physiol. 154: 611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles S.M., Lu S.X., Tobin E.M. (2008). Testing time: can ethanol-induced pulses of proposed oscillator components phase shift rhythms in Arabidopsis? J. Biol. Rhythms 23: 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N., Yoo S.H., Huang H.C., Kumar V., Lee C., Kim T.K., Takahashi J.S. (2012). Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338: 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmos E., Chow B.Y., Pruneda-Paz J.L., Kay S.A. (2014). HsfB2b-mediated repression of PRR7 directs abiotic stress responses of the circadian clock. Proc. Natl. Acad. Sci. USA 111: 16172–16177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno N., Møller S.G., Shinomura T., Xu X., Chua N.H., Furuya M. (2003). The novel MYB protein EARLY-PHYTOCHROME-RESPONSIVE1 is a component of a slave circadian oscillator in Arabidopsis. Plant Cell 15: 2476–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A.G., Doherty C.J., Mueller-Roeber B., Kay S.A., Schippers J.H., Dijkwel P.P. (2012). CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl. Acad. Sci. USA 109: 17129–17134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O.S., Huang X., Charron J.B., Lee J.H., Li G., Deng X.W. (2011). Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Mol. Cell 43: 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.J., Kang J.Y., Park H.J., Kim M.D., Bae M.S., Choi H.I., Kim S.Y. (2010). DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol. 153: 716–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., et al. (2011). Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat. Cell Biol. 13: 616–622. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C.J., Hwang J.E., Chen H., Hong J.K., Yang K.A., Choi M.S., Lee K.O., Chung W.S., Lee S.Y., Lim C.O. (2007). Over-expression of the Arabidopsis DRE/CRT-binding transcription factor DREB2C enhances thermotolerance. Biochem. Biophys. Res. Commun. 362: 431–436. [DOI] [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Carlsson J., Takeuchi T., Newton L., Farré E.M. (2013). Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. 76: 101–114. [DOI] [PubMed] [Google Scholar]
- Liu T.L., Newton L., Liu M.J., Shiu S.H., Farré E.M. (2016). A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol. 170: 528–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y., Smet W., Cruz-Ramírez A., Castelijns B., de Jonge W., Mähönen A.P., Bouchet B.P., Perez G.S., Akhmanova A., Scheres B., Blilou I. (2015). Arabidopsis BIRD zinc finger proteins jointly stabilize tissue boundaries by confining the cell fate regulator SHORT-ROOT and contributing to fate specification. Plant Cell 27: 1185–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.X., Knowles S.M., Andronis C., Ong M.S., Tobin E.M. (2009). CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 150: 834–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P., Bailey T.L. (2011). MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markson J.S., Piechura J.R., Puszynska A.M., O’Shea E.K. (2013). Circadian control of global gene expression by the cyanobacterial master regulator RpaA. Cell 155: 1396–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K., Umemura Y., Ohme-Takagi M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 55: 954–967. [DOI] [PubMed] [Google Scholar]
- Matsushika A., Makino S., Kojima M., Mizuno T. (2000). Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol. 41: 1002–1012. [DOI] [PubMed] [Google Scholar]
- McClung C.R. (2014). Wheels within wheels: new transcriptional feedback loops in the Arabidopsis circadian clock. F1000Prime Rep. 6: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T.P., et al. (2008). Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T., Wheatley K., Hanzawa Y., Wright L., Mizoguchi M., Song H.R., Carré I.A., Coupland G. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2: 629–641. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T., Wright L., Fujiwara S., Cremer F., Lee K., Onouchi H., Mouradov A., Fowler S., Kamada H., Putterill J., Coupland G. (2005). Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17: 2255–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Nomoto Y., Oka H., Kitayama M., Takeuchi A., Tsubouchi M., Yamashino T. (2014). Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana. Plant Cell Physiol. 55: 958–976. [DOI] [PubMed] [Google Scholar]
- Mockler T.C., Michael T.P., Priest H.D., Shen R., Sullivan C.M., Givan S.A., McEntee C., Kay S.A., Chory J. (2007). The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb. Symp. Quant. Biol. 72: 353–363. [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio-assays with tobacco tissue culture. Physiol. Plant. 15: 473–497. [Google Scholar]
- Nagel D.H., Doherty C.J., Pruneda-Paz J.L., Schmitz R.J., Ecker J.R., Kay S.A. (2015). Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. USA 112: E4802–E4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P., Ellis C.M., Weber H., Ploense S.E., Barkawi L.S., Guilfoyle T.J., Hagen G., Alonso J.M., Cohen J.D., Farmer E.E., Ecker J.R., Reed J.W. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132: 4107–4118. [DOI] [PubMed] [Google Scholar]
- Nakamichi N. (2011). Molecular mechanisms underlying the Arabidopsis circadian clock. Plant Cell Physiol. 52: 1709–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Ito S., Oyama T., Yamashino T., Kondo T., Mizuno T. (2004). Characterization of plant circadian rhythms by employing Arabidopsis cultured cells with bioluminescence reporters. Plant Cell Physiol. 45: 57–67. [DOI] [PubMed] [Google Scholar]
- Nakamichi N., Kiba T., Henriques R., Mizuno T., Chua N.H., Sakakibara H. (2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Kiba T., Kamioka M., Suzuki T., Yamashino T., Higashiyama T., Sakakibara H., Mizuno T. (2012). Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. USA 109: 17123–17128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Kita M., Ito S., Yamashino T., Mizuno T. (2005). PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 46: 686–698. [DOI] [PubMed] [Google Scholar]
- Niwa Y., Ito S., Nakamichi N., Mizoguchi T., Niinuma K., Yamashino T., Mizuno T. (2007). Genetic linkages of the circadian clock-associated genes, TOC1, CCA1 and LHY, in the photoperiodic control of flowering time in Arabidopsis thaliana. Plant Cell Physiol. 48: 925–937. [DOI] [PubMed] [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farré E.M., Kay S.A. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai K., Ishiura M. (2005). PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 10: 963–972. [DOI] [PubMed] [Google Scholar]
- Perales M., Más P. (2007). A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 19: 2111–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A., Fernández A.P., Edwards K.D., Southern M.M., Halliday K.J., Millar A.J. (2012). The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 8: 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Breton G., Para A., Kay S.A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat R., Takahashi N., Hsu P.Y., Jones M.A., Schwartz J., Salemi M.R., Phinney B.S., Harmer S.L. (2011). REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 7: e1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugnone M.L., Faigón Soverna A., Sanchez S.E., Schlaen R.G., Hernando C.E., Seymour D.K., Mancini E., Chernomoretz A., Weigel D., Más P., Yanovsky M.J. (2013). LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc. Natl. Acad. Sci. USA 110: 12120–12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez S.E., et al. (2010). A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468: 112–116. [DOI] [PubMed] [Google Scholar]
- Sato E., Nakamichi N., Yamashino T., Mizuno T. (2002). Aberrant expression of the Arabidopsis circadian-regulated APRR5 gene belonging to the APRR1/TOC1 quintet results in early flowering and hypersensitiveness to light in early photomorphogenesis. Plant Cell Physiol. 43: 1374–1385. [DOI] [PubMed] [Google Scholar]
- Schindler U., Terzaghi W., Beckmann H., Kadesch T., Cashmore A.R. (1992). DNA binding site preferences and transcriptional activation properties of the Arabidopsis transcription factor GBF1. EMBO J. 11: 1275–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaton D.D., Smith R.W., Song Y.H., MacGregor D.R., Stewart K., Steel G., Foreman J., Penfield S., Imaizumi T., Millar A.J., Halliday K.J. (2015). Linked circadian outputs control elongation growth and flowering in response to photoperiod and temperature. Mol. Syst. Biol. 11: 776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.M., et al. (2010). Transcription factors in light and circadian clock signaling networks revealed by genomewide mapping of direct targets for neurospora white collar complex. Eukaryot. Cell 9: 1549–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Ebisu Y., Kinoshita T., Doi M., Okuma E., Murata Y., Shimazaki K. (2013). bHLH transcription factors that facilitate K⁺ uptake during stomatal opening are repressed by abscisic acid through phosphorylation. Sci. Signal. 6: ra48. [DOI] [PubMed] [Google Scholar]
- Torres-Galea P., Huang L.F., Chua N.H., Bolle C. (2006). The GRAS protein SCL13 is a positive regulator of phytochrome-dependent red light signaling, but can also modulate phytochrome A responses. Mol. Genet. Genomics 276: 13–30. [DOI] [PubMed] [Google Scholar]
- Tran L.S., Nakashima K., Sakuma Y., Simpson S.D., Fujita Y., Maruyama K., Fujita M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H., Busch W., Benfey P.N. (2010). Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616. [DOI] [PubMed] [Google Scholar]
- Ueoka-Nakanishi H., Yamashino T., Ishida K., Kamioka M., Nakamichi N., Mizuno T. (2012). Molecular mechanisms of circadian rhythm in Lotus japonicus and Arabidopsis thaliana are sufficiently compatible to regulate heterologous core clock genes robustly. Biosci. Biotechnol. Biochem. 76: 2332–2334. [DOI] [PubMed] [Google Scholar]
- Wang H., Zhu Y., Fujioka S., Asami T., Li J., Li J. (2009). Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 21: 3781–3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Kim J., Somers D.E. (2013). Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. USA 110: 761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Barnaby J.Y., Tada Y., Li H., Tör M., Caldelari D., Lee D.U., Fu X.D., Dong X. (2011). Timing of plant immune responses by a central circadian regulator. Nature 470: 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Kenigsbuch D., Sun L., Harel E., Ong M.S., Tobin E.M. (1997). A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9: 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Tobin E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217. [DOI] [PubMed] [Google Scholar]
- Xie Q., et al. (2014). LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell 26: 2843–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashino T., Ito S., Niwa Y., Kunihiro A., Nakamichi N., Mizuno T. (2008). Involvement of Arabidopsis clock-associated pseudo-response regulators in diurnal oscillations of gene expression in the presence of environmental time cues. Plant Cell Physiol. 49: 1839–1850. [DOI] [PubMed] [Google Scholar]
- Ye H., Li L., Guo H., Yin Y. (2012). MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 20142–20147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yoshida T., Murayama Y., Ito H., Kageyama H., Kondo T. (2009). Nonparametric entrainment of the in vitro circadian phosphorylation rhythm of cyanobacterial KaiC by temperature cycle. Proc. Natl. Acad. Sci. USA 106: 1648–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L.J., Gazin C., Lawson N.D., Pagès H., Lin S.M., Lapointe D.S., Green M.R. (2010). ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.