FRY2/CLP1 interacts with two nonsense-mediated mRNA decay factors, eIF4AIII and UPF3, and is necessary for NMD.
Abstract
Nonsense-mediated decay (NMD) is a posttranscriptional surveillance mechanism in eukaryotes that recognizes and degrades transcripts with premature translation-termination codons. The RNA polymerase II C-terminal domain phosphatase-like protein FIERY2 (FRY2; also known as C-TERMINAL DOMAIN PHOSPHATASE-LIKE1 [CPL1]) plays multiple roles in RNA processing in Arabidopsis thaliana. Here, we found that FRY2/CPL1 interacts with two NMD factors, eIF4AIII and UPF3, and is involved in the dephosphorylation of eIF4AIII. This dephosphorylation retains eIF4AIII in the nucleus and limits its accumulation in the cytoplasm. By analyzing RNA-seq data combined with quantitative RT-PCR validation, we found that a subset of alternatively spliced transcripts and 5′-extended mRNAs with NMD-eliciting features accumulated in the fry2-1 mutant, cycloheximide-treated wild type, and upf3 mutant plants, indicating that FRY2 is essential for the degradation of these NMD transcripts.
INTRODUCTION
The Arabidopsis thaliana FIERY2 (FRY2) locus encodes an RNA polymerase II C-terminal domain phosphatase-like protein (also called C-TERMINAL DOMAIN PHOSPHATASE-LIKE 1, CPL1) that was independently identified in genetic screens for components that regulate the expression of stress-responsive genes (Koiwa et al., 2002; Xiong et al., 2002). The fry2 mutants exhibit increased expression of a stress-inducible reporter as well as certain endogenous stress-responsive genes. The mutants also show increased tolerance to salt stress and to abscisic acid during seed germination but are more sensitive to freezing damage at the seedling stage (Koiwa et al., 2002; Xiong et al., 2002). FRY2 has a phosphatase domain and two double-stranded RNA binding motifs. Previous studies suggested that FRY2 could dephosphorylate a serine residue in the C-terminal heptad repeat domain of RNA polymerase II (Koiwa et al., 2004). In addition, FRY2 can dephosphorylate HYPONASTIC LEAVES1 and is essential for accurate miRNA processing (Manavella et al., 2012). Recently, we found that FRY2 contributes to the regulation of pre-mRNA splicing via interacting with the KH-domain RNA binding protein HOS5 and two splicing factor proteins, RS40 and RS41 (Chen et al., 2013). In addition, FRY2 has been suggested to inhibit transcription by preventing mRNA capping and the transition from transcription initiation to elongation (Jiang et al., 2013). These studies suggest that FRY2 plays important regulatory roles in many aspects of RNA metabolism in Arabidopsis.
Nonsense-mediated decay (NMD) is a posttranscriptional mechanism in eukaryotes that recognizes and degrades transcripts with premature translation-termination codons (PTCs) located at more than 50 bp upstream of the last exon-exon junction (Maquat, 2004; Moore, 2005; Behm-Ansmant and Izaurralde, 2006; Chang et al., 2007; Doma and Parker, 2007; Brogna and Wen, 2009). This process prevents truncated proteins with potentially deleterious impacts from accumulating in cells. In addition, several other kinds of mRNAs have been identified as NMD targets, including mRNAs with 5′ or 3′ transcriptional extensions, mRNA-like noncoding RNAs, and mRNAs with upstream open reading frames (uORFs) (He et al., 2003; Mendell et al., 2004; Rehwinkel et al., 2006; Kurihara et al., 2009; Drechsel et al., 2013). The core NMD machinery comprises three trans-acting factors, called up-frameshift proteins, UPF1, UPF2, and UPF3, all of which are present in yeast, humans, Drosophila melanogaster, Caenorhabditis elegans, and Arabidopsis (Hori and Watanabe, 2005; Arciga-Reyes et al., 2006; Chang et al., 2007; Kerényi et al., 2008; Kim et al., 2009; Shi et al., 2012; Degtiar et al., 2015; Lykke-Andersen and Jensen, 2015). In the NMD pathway, UPF1 is first recruited to mRNAs upon recognition of stop codons by the translation apparatus and then interacts with UPF2 and UPF3 to trigger the decay of PTC-containing mRNAs. UPF1 and UPF2 are known to be phosphoproteins, but the phosphorylation status of UPF3 remains unclear. The phosphorylation and dephosphorylation cycle of UPF1 has been suggested to drive the entire NMD molecular process. In mammals and C. elegans, SMG1, SMG5, SMG6, and SMG7 proteins are responsible for regulating the phosphorylation status of UPF1.
As an important component of the NMD machinery, the exon junction complex (EJC) is deposited onto mRNAs 20 to 25 nucleotides upstream of the exon-exon boundary during pre-mRNA splicing in the nucleus and is involved in NMD by recruiting UPF proteins at the early stage of NMD (Ferraiuolo et al., 2004; Palacios et al., 2004; Shibuya et al., 2004; Singh et al., 2012). The EJC contains more than 20 different proteins, of which the eukaryotic translation initiation factor 4A3 (eIF4AIII), Y14, MAGOH, and MLN51 (also known as BTZ or CASC3) proteins form its core. The DEAD box RNA helicase eIF4AIII serves as an anchor to attach the EJC to its RNA substrate. Its amino acid sequence is highly similar to the amino acid sequences of the translation initiation factors eIF4AI and eIF4AII, two other members of the DEAD box protein family. eIF4AIII is essential for EJC-dependent NMD (Ferraiuolo et al., 2004; Palacios et al., 2004).
In plants, in addition to the UPF1-3 proteins, orthologs of SMG7, SMG1, and the four core EJC components have been experimentally identified (Lloyd and Davies, 2013; Mérai et al., 2013; Nyikó et al., 2013; Shaul, 2015). Some of the regulatory steps of NMD have also been identified. For example, phosphorylation of UPF1 connects NMD complex formation to SMG7-mediated target transcript degradation (Kerényi et al., 2013). SMG7 binds the phosphorylated S/TQ sites of the UPF1 component of the NMD complex and then transports the PTC-containing transcripts into P bodies for degradation (Mérai et al., 2013). In addition, the orthologs of eIF4AIII and the other three core EJC components are essential for NMD in plants (Nyikó et al., 2013).
In this study, we reveal a regulatory role of FRY2 in NMD, whereby FRY2 physically interacts with both the eIF4AIII and UPF3 proteins. FRY2 is associated with the dephosphorylation of eIF4AIII, which limits its cytoplasmic accumulation. Loss of function of FRY2 resulted in the accumulation of NMD targets. Our study provides insights into the molecular mechanism of NMD in eukaryotes.
RESULTS
FRY2 Interacts with eIF4AIII and Is Involved in Its Dephosphorylation
The Arabidopsis fry2-1 mutant was isolated in a genetic screen for components that regulate the expression of the stress-responsive reporter gene RD29A-LUC (the firefly luciferase gene under control of the stress-inducible promoter RD29A). A single nucleotide change (G→A) at the FRY2 locus at the genomic position 11,514,728 of chromosome 4 was identified in the fry2-1 mutant (Supplemental Figure 1A) (Xiong et al., 2002). Because this mutation occurred at the splicing donor recognition site of the eighth exon, it was predicted to disrupt the splicing of the intron in the mutant. This was validated by RT-PCR using intron-flanking primers (Supplemental Figure 1B). This intron retention interrupted the coding sequence and introduced a PTC downstream of the ninth exon, which was predicted to generate a truncated protein that is likely nonfunctional. Interestingly, RNA gel blotting (Supplemental Figure 1C) and quantitative RT-PCR analysis (Supplemental Figure 1D) revealed that the expression level of this PTC-containing FRY2 mRNA was significantly higher in the fry2-1 mutant, being nearly 10-fold upregulated compared with the expression level of FRY2 in the wild-type C24 plants. There are two possible explanations for this increased accumulation of transcripts in the mutant. First, a transcriptional feedback loop could be activated due to the lack of active FRY2 proteins, causing the increased expression of the FRY2 gene. Second, NMD could be deficient in the fry2-1 mutant, resulting in the accumulation of the PTC-containing FRY2 mRNA in the mutant. We compared the expression levels of the FRY2 mRNA between C24 and the mutant following the application of triptolide (TPL), an inhibitor of RNA polymerase I- and II-dependent transcription (Titov et al., 2011). Treatment with TPL is expected to minimize the influence of transcription during investigation into the stability of mRNA. After treatment with TPL for 0, 4, and 8 h, the expression levels of FRY2 mRNA in the wild type and fry2-1 mutant were assessed by RT-qPCR. The FRY2 mRNAs degraded more rapidly in C24 than in fry2-1, whereas the degradation rate of the ACTIN2 reference gene was similar between samples (Supplemental Figures 1E and 1F), suggesting a reduced decay rate of the FRY2 PTC-containing mRNA upon the depletion of the FRY2 protein. Therefore, we suspected that the second possibility better explains the accumulation of the fry2 PTC-containing transcript; thus, here, we explored the possibility that NMD is defective in fry2-1 mutants.
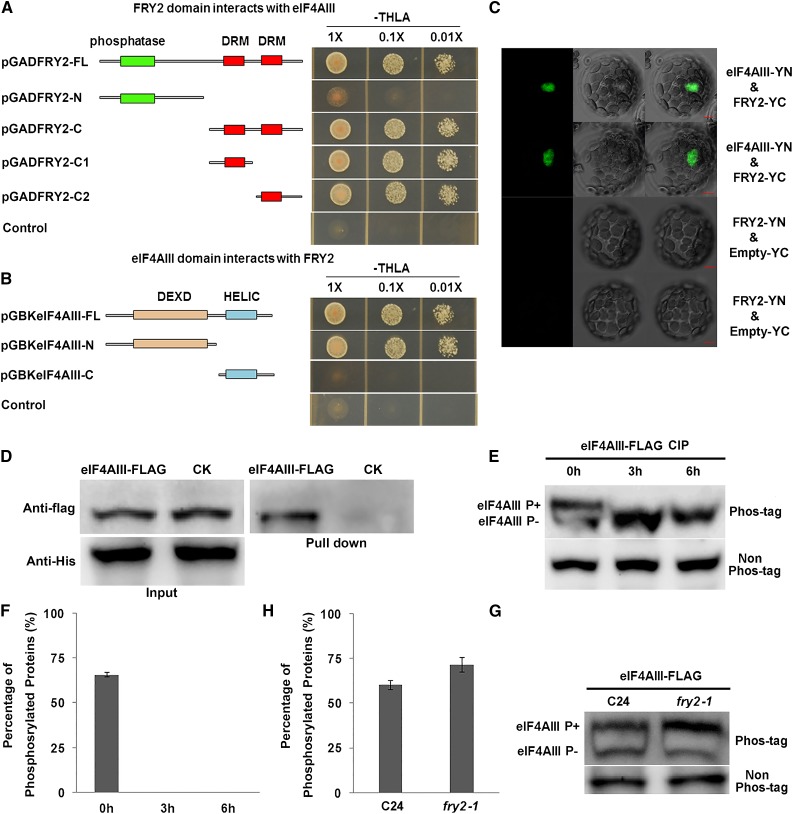
To investigate if FRY2 plays a role in NMD, we first performed yeast two-hybrid screens searching for proteins that interact with FRY2. Interestingly, eIF4AIII was identified as one of the positive clones that strongly interacted with FRY2. eIF4AIII is a key component of the EJC that recruits the NMD factor UPF3 protein at the early stage of NMD and has been shown to be essential for NMD (Le Hir et al., 2001; Palacios et al., 2004; Arciga-Reyes et al., 2006; Chang et al., 2007; Kim et al., 2009; Shi et al., 2012). We found that the interaction occurred between the double-stranded RNA binding motif of FRY2 and the N-terminal DEXD domain of eIF4AIII (Figures 1A and 1B). The interaction between FRY2 and eIF4AIII was further verified in vivo by bimolecular fluorescence complementation (BiFC) assays (Figure 1C) and in vitro by a pull-down experiment (Figure 1D).
Figure 1.
FRY2 Interacts with eIF4AIII and Affects the Phosphorylation Status of eIF4AIII.
(A) and (B) FRY2 interacts with eIF4AIII in yeast. The full-length (FL) or the indicated truncated constructs in pGAD (FRY2) or pGBK (eIF4AIII) were cotransformed into yeast and plated on -tryptophan/-leucine/-histidine/-adenine (-TLHA) media. Empty pGAD vector and eIF4AIII with pGBK were cotransformed into yeast and used as a negative control.
(C) BiFC assays of interaction between FRY2 and eIF4AIII. FRY2 and eIF4AIII were fused to EYFP-N or EYFP-C, respectively. E-YN and E-YC are empty vectors. The combinations of plasmids as indicated were transformed into Arabidopsis protoplasts and the interaction (fluorescence) signals were detected using a confocal microscope. Bars = 5 μm.
(D) In vitro pull-down assay was performed with FLAG-eIF4AIII and His-FRY2. eIF4AIII was pulled down by recombinant His-FRY2 and was detected by anti-FLAG antibody. CK is a FLAG-tagged ribosome release factor 1 (RF1) homolog protein (encoded by the gene AT3G62910) that does not interact with FRY2, which was used as a control.
(E) Phosphorylation level of eIF4AIII. Total proteins were extracted from 12-d-old seedlings expressing FLAG-tagged eIF4AIII and treated with CIP for 0, 3, and 6 h. Proteins were resolved on SDS-PAGE containing Phos-tag (upper panel) or non-Phos-tag (as control), and eIF4AIII was detected by anti-FLAG antibody (α-FLAG). Upper and lower bands were phosphorylated and unphosphorylated eIF4AIII, respectively.
(F) Quantitation of phosphorylation level of eIF4AIII as shown in (D).
(G) Phosphorylation level of eIF4AIII as regulated by FRY2. Proteins were extracted from eIF4AIII-FLAG-expressing C24 and fry2-1 seedlings and separated by SDS-PAGE containing Phos-tag. Non-Phos-tag gel was used as control.
(H) Quantitation for phosphorylation level of eIF4AIII in C24 and fry2-1. Error bars in (F) and (H) indicate the standard deviations that were calculated based on three different experiments.
Because FRY2 is a protein phosphatase, we tested whether there was any change in the phosphorylation status of eIF4AIII in fry2-1 mutants using Phos-tag, a compound that decreases the mobility of phosphorylated proteins in polyacrylamide gels. We generated stable transgenic plants expressing FLAG-tagged eIF4AIII in both wild-type and fry2-1 mutant backgrounds. Crude proteins were extracted from the transgenic plants and were subjected to immunoblot analyses using anti-FLAG antibody. Figures 1E and 1F show that there are two bands of eIF4AIII in protein extracts from the wild type, with the upper band likely corresponding to the phosphorylated form and the lower band the unphosphorylated form. To verify that the two bands differ in their phosphorylation status, total protein extracts from the eIF4AIII-transformed wild-type plants were treated with calf intestinal alkaline phosphatase (CIP) for 3 or 6 h before being subjected to electrophoresis on Phos-tag or non-Phos-tag gels. The amount of protein in the non-Phos-tag control gel was not significantly affected by CIP treatments; however, the amount of the protein in the upper bands of the Phos-tag gel decreased, while that in the lower bands increased, confirming that the upper band is indeed phosphorylated eIF4AIII. Compared with that in the wild type, the abundance of phosphorylated eIF4AIII in the fry2-1 mutant increased significantly and that of unphosphorylated protein decreased (Figures 1G and 1H), suggesting that eIF4AIII is likely an in vivo substrate for the FRY2 phosphatase.
The Phosphorylation Status of eIF4AIII Regulates Its Subcellular Localization
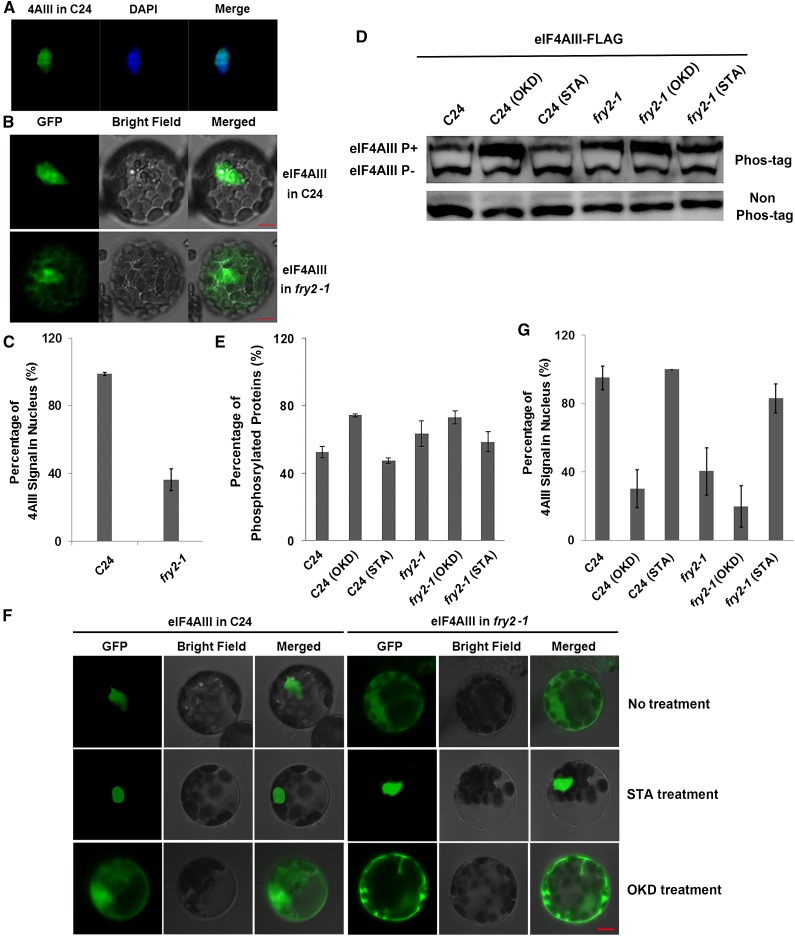
To understand the functional significance of eIF4AIII phosphorylation, we investigated whether its phosphorylation status affects its cellular localization. We made GFP-tagged eIF4AIII cDNA constructs under the control of either the cauliflower mosaic virus 35S promoter or the native promoter and expressed them in leaf protoplasts prepared from the wild-type and fry2-1 mutant seedlings. Regardless of the promoter used, eIF4AIII exclusively localized in the nucleus in the wild-type protoplasts (Figure 2A; Supplemental Figure 2), consistent with a previous report (Koroleva et al., 2009). In fry2-1 mutant protoplasts, however, a significant portion of the protein was found in the cytoplasm, although a portion of the protein was still retained in the nucleus (Figures 2B and 2C). Since the expression level of the native promoter-driven eIF4AIII was very low (Supplemental Figure 2), we decided to use the 35S promoter-driven constructs in all subsequent experiments to facilitate observation.
Figure 2.
Phosphorylation Status of eIF4AIII Determines Its Subcellular Localization.
(A) Nuclear localization of eIF4AIII. GFP-fused eIF4AIII expression construct was transformed into protoplasts of the wild-type C24, and the cell was subjected to 4′,6-diamidino-2-phenylindole (DAPI) staining. Florescence images were taken using a confocal microscope.
(B) Mislocalization of eIF4AIII in fry2-1. GFP-fused eIF4AIII expression construct was transformed into protoplasts of C24 and fry2-1. Images were taken using a confocal microscope. Left panels, GFP signals; middle panels, bright-field images; right panels, merged images. Bars = 5 μm.
(C) Quantitation of the nucleus-versus-cytoplasm partition of the eIF4AIII protein as shown in (B).
(D) Phosphorylation levels of eIF4AIII as affected by STA and OKD treatment. Proteins were extracted from the eIF4AIII-FLAG C24 and eIF4AIII-FLAG fry2-1 seedlings that were treated by STA and OKD, respectively, and then separated by SDS-PAGE containing Phos-tag. Non-Phos-tag gel was used as control. The bands of eIF4AIII were detected by anti-FLAG antibody.
(E) Quantitation of phosphorylation level of eIF4AIII in C24 and fry2-1 as shown in (D). Error bars were calculated based on three different experiments.
(F) Phosphorylation controls the subcellular localization of eIF4AIII. GFP-fused eIF4AIII proteins were expressed in protoplasts of C24 and fry2-1 treated with the protein kinase inhibitor STA and phosphatase inhibitor OKD, respectively.
(G) Quantitation of the nucleus-versus-cytoplasm partition of the eIF4AIII protein as shown in (F). Data in (C) and (G) are means and sd of fluorescence signal intensity from five randomly selected cells.
To further test the notion that excessive phosphorylation of eIF4AIII promotes its cytoplasmic localization, we conducted two sets of experiments to monitor the phosphorylation status and the nucleocytoplasmic partitioning of eIF4AIII. Due to technical difficulties, the phosphorylation assays were conducted with solution-cultured whole seedlings, whereas the nucleocytoplasmic localization assays were performed with isolated protoplasts. First, Phos-tag gel analysis revealed that treatment with the protein kinase inhibitor staurosporine (STA) decreased the phosphorylation of eIF4AIII in the fry2-1 mutant, which made the phosphorylation status of eIF4AIII close to that in the wild type (Figures 2D and 2E). Conversely, the phosphatase inhibitor okadaic acid (OKD) increased eIF4AIII phosphorylation in the wild type, which made its phosphorylation status close to that in the mutant (Figures 2D and 2E). Next, we treated protoplast cells with STA and OKD and checked the localization of eIF4AIII. We observed that treatment with STA had no effect on the localization of eIF4AIII in wild-type cells but completely restored its localization to the nucleus in fry2-1 mutant cells (Figures 2F and 2G). In contrast, treatment with OKD caused the majority of the eIF4AIII protein to shuttle from the nuclei to the cytoplasm in the wild-type cells, similar to what we observed in fry2-1 mutant cells (Figures 2F and 2G). These data consistently indicate that the extent of the phosphorylation of eIF4AIII determines its nucleocytoplasmic partitioning whereby hyperphosphorylation of eIF4AIII increases its cytoplasmic partition and hypophosphorylation decreases its cytoplasmic partition, restricting its localization to the nucleus.
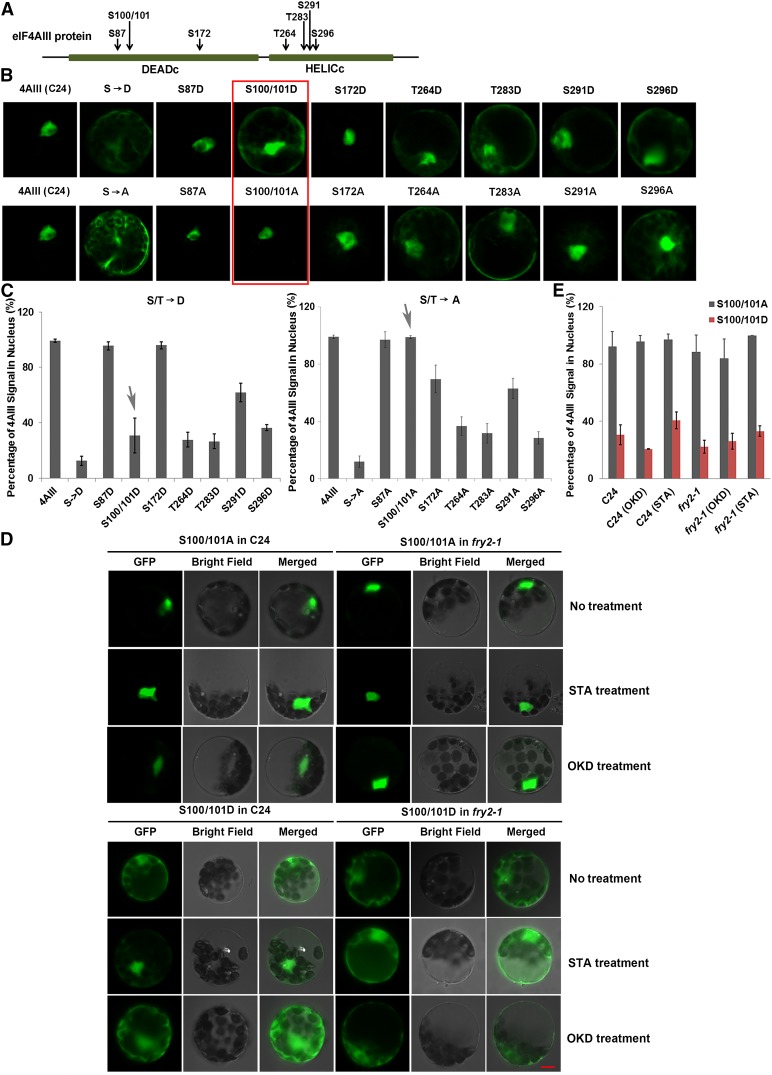
Next, we investigated which residues of eIF4AIII would affect its localization potentially via phosphorylation. We first predicted seven serine/threonine (S/T) residues in the two functional domains as potential phosphorylation sites using the NetPhos 2.0 algorithm (Blom et al., 1999) (Figure 3A). We conducted site-directed mutagenesis to replace all seven serine/threonine residues with aspartate (D) to mimic fully phosphorylated eIF4AIII or with alanine (A) to mimic unphosphorylated eIF4AIII. The mutated eIF4AIII cDNAs with the GFP tag were transformed into the wild-type protoplasts, and we observed the subcellular localization of the translated proteins. The complete phosphomimetics and dephosphomimetics of eIF4AIII (i.e., with all the predicted phosphorylation sites replaced) both diffused throughout the cytoplasm (Figures 3B and 3C), suggesting that mutating all the seven sites might change the structure of the protein, such that both molecules localized to the cytoplasm. Interestingly, by mutating individual serine/threonine residues, we identified two highly conserved residues, Ser-100 and Ser-101, located in the DEAD box domain, as particularly important for localization of eIF4AIII. When both Ser-100 and Ser-101 (S100/101) were mutated to aspartate (S100/101D), the eIF4AIII signal was found in the cytoplasm in addition to the nucleus (Figures 3B and 3C), whereas when they were mutated to alanine (S100/101A), the signal was only found in the nucleus (Figures 3B and 3C). These results indicate that the phosphorylation status of Ser-100 and Ser-101 could modulate the nucleocytoplasmic localization of eIF4AIII, whereas the other residues may be less important in controlling the localization of eIF4AIII since their mutants showed similar localization patterns when mutated either to aspartate or to alanine (Figures 3B and 3C).
Figure 3.
Phosphorylation Status of Two Serine Residues (Ser-100 and Ser-101) of eIF4AIII Is Important for Its Localization.
(A) Diagram of the eIF4AIII structure and positions of the potential phosphorylation residues that were predicted using the NetPhos 2.0 algorithm and were mutagenized to either alanine (A) or aspartic acid (D).
(B) Subcellular localization of eIF4AIII-GFP-fused mutant proteins. S→D and S→A indicate that all seven serine/threonine residues were mutagenized to aspartic acid and alanine to mimic fully phosphorylated and fully unphosphorylated eIF4AIII, respectively. The others were mutagenized at the indicated single serine/threonine residue, and S100/101 (marked by red frame) indicates that two serine residues were simultaneously mutagenized.
(C) Quantitation of the nucleus-versus-cytoplasm partition of the wild and mutant eIF4AIII proteins shown in (B).
(D) GFP-fused eIF4AIII mutant proteins S100/101A and S100/101D were expressed in protoplasts of C24 and fry2-1 treated with STA and OKD, respectively.
(E) Quantitation of the nucleus-versus-cytoplasm partition of the wild type and mutant eIF4AIII proteins as shown in (D). Data in (C) and (E) are means and sd of fluorescence signal intensity from five randomly selected cells. Bars = 5 μm.
To further verify the role of phosphorylation at residues Ser-100 and Ser-101 in controlling the localization of eIF4AIII, we transformed S100/101A and S100/101D eIF4AIII cDNAs into the wild-type and fry2-1 mutant protoplasts and treated the transformed protoplasts with STA or OKD. In the wild-type protoplasts, the S100/101A mutant protein localized in the nucleus, similar to the wild-type eIF4AIII protein. In contrast to the wild-type eIF4AIII protein that was found in the cytoplasm after OKD treatment (Figure 2F), OKD treatment did not alter the nuclear localization of the S100/101A mutant protein in wild-type protoplasts (Figures 3D and 3E). In fry2-1 protoplasts, whereas the wild-type eIF4AIII proteins diffused into the cytoplasm (Figure 2F), the S100/101A mutant protein localized exclusively in the nucleus (Figures 3D and 3E). STA or OKD treatment failed to alter the localization of the mutant protein (Figures 3D and 3E). These results show that the S100/101A eIF4AIII protein localized exclusively in the nucleus and is immune to STA and OKD treatment. Since the S100/101A protein is a dephosphomimetic of eIF4AIII, it could be concluded that the dephosphorylation at Ser-100 and Ser-101 residues may dictate the nuclear localization of eIF4AIII.
The phosphomimetic S100/101D of the eIF4AIII protein diffused into the cytoplasm to various extents. Whereas STA treatment could restore the wild-type eIF4AIII nuclear localization in fry2-1 mutant protoplasts (Figure 2F), the same treatment only slightly promoted the localization of the S100/101D to the nucleus (Figures 3D and 3E). These observations indicate that the phosphorylation status of S100/101 residues largely determines the subcellular localization of the eIF4AIII protein.
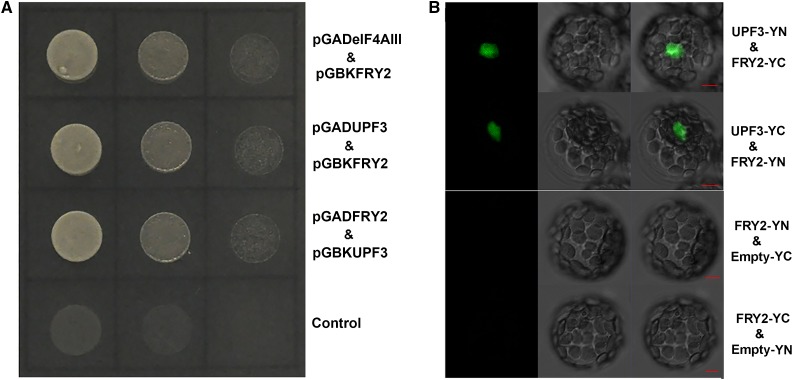
FRY2 Interacts with UPF3
UPF3, as one key NMD factor, is recruited by EJC at the early stage of NMD and becomes a component of EJC (Chang et al., 2007). We tested if there is any link between FRY2 and UPF3. Using yeast two-hybrid and BiFC assays, we observed that FRY2 interacts with UPF3 (Figures 4A and 4B). Phos-tag gel analyses performed with protein extracted from transgenic plants expressing FLAG-tagged UPF3 protein in the wild type or fry2-1 mutant background revealed that there were two bands of UPF3, where the upper and lower bands likely corresponded to the phosphorylated and unphosphorylated forms, respectively. Compared with that in the wild type, the unphosphorylated band of UPF3 in the fry2-1 mutant greatly diminished (Supplemental Figure 3), suggesting that UPF3 could be an in vivo substrate of the FRY2 phosphatase. However, for unknown reasons, CIP treatments of the samples failed to obtain predicted results, but rather both bands were reduced in abundance after the treatment (data not shown), which likely reflects UPF3 proteins in different states of phosphorylation. We also examined the localization of UPF3 in the wild type and fry2-1 mutant. While predominately found in the nucleus, the UPF3 signal was also found in the cytoplasm. However, we did not see a clear difference in the subcellular localization of UPF3 between the wild-type and fry2 mutant protoplasts. Although further investigation into the interaction between FRY2 and UPF3 is necessary, our results suggest that FRY2 may not dramatically affect the localization of UPF3.
Figure 4.
FRY2 Interacts with UPF3.
(A) The full-length (FL) construct of FRY2 or UPF3 in pGAD (FRY2) or pGBK (eIF4AIII) was cotransformed into yeast and plated on -tryptophan/-leucine/-histidine/-adenine (-TLHA) media for interaction validation. Empty pGAD vector and UPF3 with pGBK were cotransformed into yeast and used as a negative control. FRY2 and eIF4AIII were included as a positive control.
(B) BiFC assays of in vivo interaction between FRY2 and UPF3. FRY2 and UPF3 were fused to EYFP-N or EYFP-C, respectively. The combinations of plasmids as indicated were transformed into the protoplasts, and the interaction (fluorescence) signals were detected by confocal microscopy. Bars = 5 μm.
Alternatively Spliced Isoforms with NMD Features Accumulated in the fry2-1 Mutant
The association of FRY2 with both eIF4AIII and UPF3 suggests a possible role of FRY2 in NMD regulation. If FRY2 is necessary for NMD, NMD targets including alternatively spliced isoforms of potential NMD targets would be expected to accumulate in the fry2-1 mutant. Therefore, we analyzed the global change of alternative splicing (AS) upon the deletion of FRY2 by performing RNA-seq of 2-week-old wild-type C24 and fry2-1 mutant seedlings. Based on two libraries (C24 and fry2-1), we generated 52 million reads (101 bp in length), ∼80% of which could be properly aligned to the TAIR10 reference genome sequence (version TAIR10) (Supplemental Figure 4A). Comparison of the mapped reads to the gene model (version TAIR10) revealed that ∼97% of the reads mapped to the exon regions, whereas only ∼2% mapped to intergenic regions (Supplemental Figure 4B). These percentages were consistent with the Arabidopsis gene annotation. By plotting the coverage of reads along each transcript unit, we found a uniform distribution with no obvious 3′/5′ bias (Supplemental Figure 4C), testifying to the high quality of the cDNA libraries. In this analysis, we randomly sampled 40 million properly mapped reads (estimated average ∼57 times coverage on all the expressed transcripts) from each sample to identify or compare AS events. This strategy ensures that the comparison would be performed at the same level.
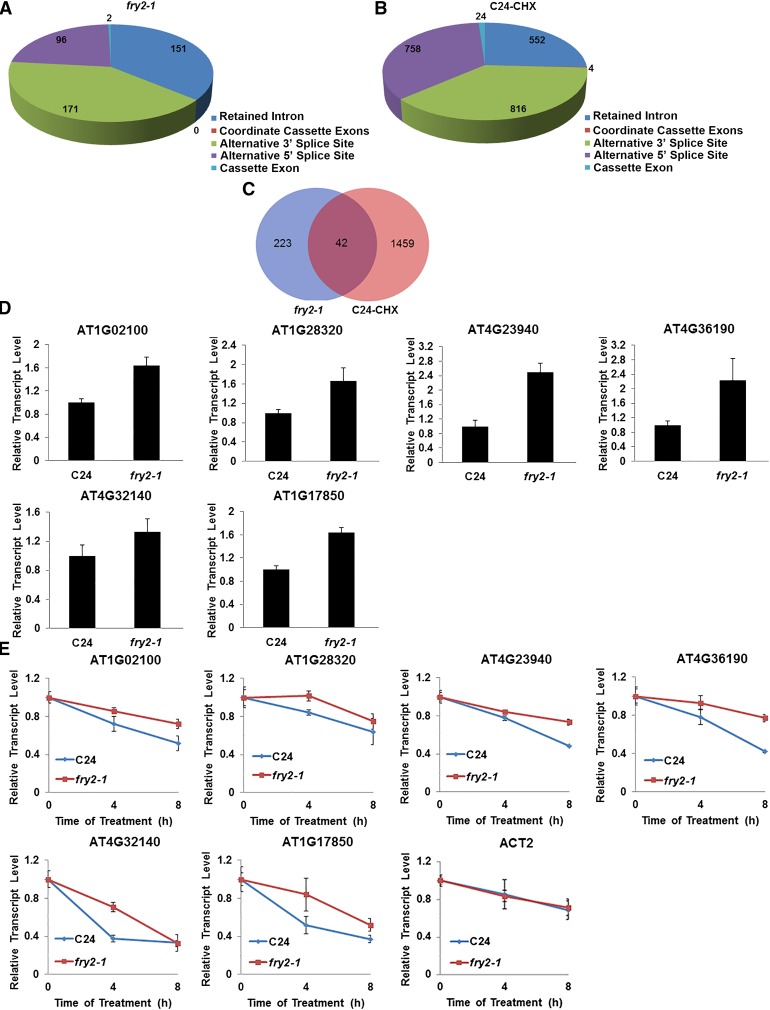
Using a recently published pipeline (see Methods) (Cui et al., 2014), all AS events in each library were identified (Supplemental Figure 4F). Compared with the wild type, 420 AS events including 151 retained introns, 171 alternative 3′ splice sites, 96 alternative 5′ splice sites, and 2 cassette exons were overrepresented in the fry2-1 mutant (P < 0.001, a 4-fold upregulation) (Figure 5A). A previous study has documented that a great portion of introns cannot be removed in the fry2-1 mutant as a result of the role of FRY2 in pre-mRNA splicing via interactions with the splicing-related factors HOS5 and SR40/SR41 (Chen et al., 2013). Therefore, we excluded intron retention from this analysis. Nonetheless, it is unclear whether the accumulation of the AS events, such as alternative 5′ and 3′ splicing site selection, in the fry2-1 mutant is caused by splicing defects or NMD dysfunction in the absence of FRY2. Thus, we compared AS events between the fry2-1 mutant and plants treated with the translation inhibitor cycloheximide (CHX). Because translation is required for NMD (Ishigaki et al., 2001) and treatment with CHX leads to an accumulation of NMD-sensitive transcripts in plants (Kalyna et al., 2012), we treated wild-type C24 plants with CHX and performed RNA-seq on control and treated plants. We achieved high-quality RNA-seq data (Supplemental Figures 4D and 4E), enabling the identification of AS events in the control and treated samples (Supplemental Figure 4G). We found 2154 AS events that accumulated in the treated samples (Figure 5B). Of the 270 alternative 5′ and 3′ splice sites identified in the fry2-1 mutant, 45 were also detected in the CHX-treated C24 (Figure 5C). Through sequence analysis, we found that 75% (34) of these 45 AS variants contain premature stop codons located more than 50 bp upstream of the last exon-exon junction. On the other hand, 59% (133) of AS events that only accumulated in the fry2-1 mutant still contained such PTCs (Supplemental Data Set 1). This result suggested that a great number of these AS variants have the features of NMD targeted transcripts. We validated six selected AS variants with PTCs that were commonly identified in the fry2-1 mutant and CHX-treated C24 using RT-qPCR with specific primers that covered the AS junction region. We found that the levels of these AS transcripts were higher in the fry2-1 mutant than in the wild type (Figure 5D). After treatment with transcriptional inhibitor TPL, the decay rate of these transcripts was slower in the fry2-1 mutant than in the wild type, suggesting a defect in transcript degradation in the mutant (Figure 5E). RT-PCR also showed that the levels of these transcripts are also higher in the CHX-treated plants and the UPF3 mutant, upf3-2, compared with the controls (Supplemental Figure 5). Therefore, these transcripts should be considered NMD targets but not a result of altered AS in the fry2-1 mutant. The FRY2 protein is therefore responsible for regulating the degradation of the AS transcripts with NMD-eliciting features.
Figure 5.
Accumulation of Alternatively Spliced Isoforms in the fry2-1 Mutant.
(A) and (B) Identification and annotation of AS events that were overrepresented in the fry2-1 mutant and the CHX-treated wild type (C24-CHX).
(C) Comparison of the number of AS events in fry2-1 and CHX-treated C24.
(D) The expression level of six representative AS transcripts with premature stop codons in C24 and fry2-1 as measured by RT-qPCR.
(E) These six AS transcripts are more stable in fry2-1. Twelve-day-old seedlings of C24 and fry2-1 were subjected to 200 μM TPL treatment for 0, 4, and 8 h and total RNAs were extracted. RT-qPCR was performed to determine the transcriptional levels. ACT2 was used as a control. Error bars represent the sd (n = 3).
Using the same pipeline (Cui et al., 2014), we performed the same analysis of publicly available RNA-seq data from the other mutant allele of UPF3 (Col-0 background), upf3-1 (Drechsel et al., 2013). Data quality was evaluated (Supplemental Figures 6A to 6C) and AS events in upf3-1 were identified (Supplemental Figure 6D). We found 582 AS events that accumulated in the upf3-1 mutant (Supplemental Figure 6E). Although the background of the upf3-1 mutant is different from that of the fry2-1 mutant, 28 of the alternative 5′ and 3′ splice site selection events found in the fry2-1 mutant were also found in the upf3-1 mutant, 24 of which contain premature stop codons (Supplemental Figure 6F and Supplemental Data Set 1). In total, 56 (21%) out of 270 alternative 5′ and 3′ splice site selection events that accumulated in the fry2-1 mutant also accumulated in the CHX-treated plants or upf3-1 plants. This percentage of overlap is similar to that reported recently where a ∼14% overlap was found between the UPF mutants, upf1 and upf3-1, and CHX-treated samples (Drechsel et al., 2013). Therefore, NMD-associated AS transcripts appear to accumulate in the fry2-1 mutant.
FRY2 Is Necessary for the Degradation of 5′-Extended mRNAs
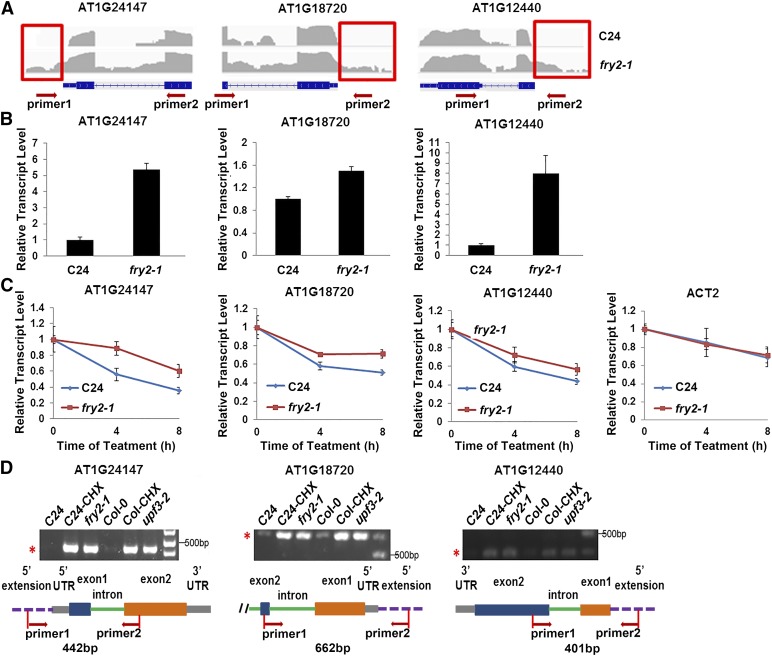
The 5′-extended mRNAs that are transcribed from previously unannotated 5′ flanking sequences or alternative promoters were identified as another group of NMD targets in yeast (Johansson et al., 2007). However, these transcripts had not yet been systematically documented in plants. After assembling the transcripts (see Method), we found that 5′-extended mRNAs accumulated in the fry2-1 mutant. Using Fisher’s exact test on the read counts of transcripts within 500 bp upstream of transcriptional start sites and the corresponding exon read counts between the wild type and mutant, we identified 83 5′-extended mRNAs as abnormal transcripts in the fry2-1 mutant (P < 0.001, a 4-fold upregulation) (Supplemental Data Set 2). In contrast, only 17 5′-extended mRNAs were overrepresented in the wild-type plants. Figure 6A highlights three representative examples visualized by the Integrative Genomics Viewer, which clearly demonstrates that the transcriptional signals were found in the upstream of the transcriptional start sites in the fry2-1 mutant. We further validated 10 selected 5′-extended mRNAs using RT-qPCR with specific primers that covered the 5′-extended region. The levels of these 5′-extended mRNAs were higher in the fry2-1 mutant than in the wild type (Figure 6B; Supplemental Figure 7A). Moreover, we examined the expression levels of these 5′-extended mRNAs in the wild type and fry2-1 mutant following treatment with transcriptional inhibitor TPL over time. We found that seven of these transcripts showed a reduced decay rate in fry2-1 (Figure 6C; Supplemental Figure 7B). In addition, these 5′-extended mRNAs were clearly detected in CHX-treated plants and the upf3-2 mutant but were only nominally or not expressed in the wild-type control plants (Figure 6D; Supplemental Figure 7C). Thus, the accumulation of 5′-extended mRNAs in the fry2-1 mutant may result from defects in NMD due to the depletion of FRY2.
Figure 6.
Accumulation of 5′-Extended mRNAs in the fry2-1 Mutant.
(A) Three representative 5′-extended mRNAs visualized by IGV browser. For the IGV visualization, exon-intron structure of each gene and the 5′-extend region (marked by red frame) are given at the bottom of each panel. The gray peaks indicate RNA-seq read density. For RT-qPCR, the primers (indicated by red arrows) were designed to span the 5′-extended region and the first exon for each gene.
(B) The expression levels of these three 5′-extended mRNAs in C24 and fry2-1 were measured by RT-qPCR.
(C) These 5′-extended mRNAs are more stable in fry2-1. Twelve-day-old seedlings of C24 and fry2-1 were subjected to 200 μM TPL treatment for 0, 4, and 8 h and total RNAs were extracted. RT-qPCR was performed to determine the transcript levels. ACT2 was used as a control. Error bars represent the sd (n = 3).
(D) These 5′-extended mRNAs marked by asterisks in the gel picture were more obviously detected in fry2-1, CHX-treated wild type (C24 or Col-0), and the upf3-2 mutant plants, but not in the control plants. At the bottom, the gene models with boxes, lines, and dashed lines (representing exons, introns, and extended regions) for the 5′-extended mRNAs are shown. The primers were designed to span the 5′-extended region and the second exon for each gene, and the size of PCR product was predicted.
With the fry2-1 mutant, we next studied the features of 5′-extended mRNAs in more detail. First, we found that ∼68% of 5′-extended mRNAs contain a 5′ uORF between 20 to 159 codons in length (Supplemental Data Set 2). This appears to be consistent with the current notion that these RNAs become NMD substrates because alternative transcriptional start sites often lead to at least one transcript with an uORF in the 5′ untranslated region (Miura et al., 2006). Second, using primers that span the 5′-extended region and the second exon, we conducted RT-PCR to examine the splicing of these transcripts. According to the size of the PCR products (Figure 6D), we concluded that the first introns from these 5′-extended mRNAs were normally removed during the pre-mRNA splicing, thereby suggesting that the 5′-extended mRNAs were normally spliced. Third, 5′-extended mRNAs had lower expression levels relative to normal transcripts, with average abundance accounting for ∼5% of the total transcripts (Supplemental Figure 7D). Fourth, the affected genes tended to be downregulated in the fry2-1 mutant (Supplemental Figure 7E), while 500 randomly selected genes as control did not show this pattern (Supplemental Figure 7F), suggesting that the 5′-extended mRNAs were associated with transcriptional repression of the affected genes. This feature is consistent with previous findings in yeast (Thebault et al., 2011).
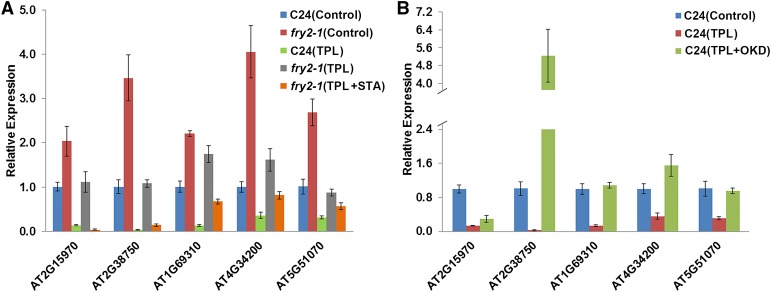
Inhibition of Protein Phosphorylation Rescues NMD Deficiency in the fry2-1 Mutant
In the fry2-1 mutant, some eIF4AIII was excessively phosphorylated and was excluded from the nucleus; meanwhile, there was an accumulation of NMD-targeted transcripts. We posited that efficient NMD might require nuclear localization of eIF4AIII. Because treatment with the protein kinase inhibitor STA restored the nuclear localization of eIF4AIII in the fry2-1 mutant (Figure 2F), we tested whether NMD efficiency would be restored in the fry2-1 mutant after STA treatment. For this, we examined the expression of the NMD-targeted transcripts in protoplast cells of the wild type and the fry2-1 mutant following treatment of STA combined with TPL. Five abundantly accumulated 5′-extended mRNAs in the fry2-1 mutant were selected as NMD targets for this analysis. RT-qPCR measurement of these mRNAs revealed that the transcript levels of these targets in the fry2-1 mutant had decreased toward levels in the wild type after STA treatment (Figure 7A), indicating that the STA successfully rescued NMD deficiency in the fry2-1 mutant. In addition, after treatment with the phosphatase inhibitor OKD, these NMD targets accumulated in the wild-type cells (Figure 7B). This result is consistent with the observation that OKD treatment of the wild-type protoplasts disrupted the phosphorylation and nuclear localization of eIF4AIII, such that treated cells resembled fry2-1 mutants (Figure 2F). These observations suggest a close link between the phosphorylation and localization of eIF4AIII and NMD efficiency.
Figure 7.
Expression of 5′-Extended mRNA in fry2-1 Can Be Rescued by Protein Kinase Inhibitor.
(A) Expression levels of five 5′-extended mRNA in C24 and fry2-1 were examined by RT-qPCR under the control (no treatment), TPL, or TPL and STA treatment.
(B) Expression levels of five 5′-extended mRNAs in C24 were examined by RT-qPCR under the control (no treatment), TPL, or TPL and OKD treatment.
DISCUSSION
Although the EJC core component eIF4AIII has been known to play essential roles in mRNA surveillance and export (Singh et al., 2012), it is unclear how their functions are regulated. We identified FRY2 as a regulator of eIF4AIII, interacting with eIF4AIII and affecting its dephosphorylation. Furthermore, the phosphorylation status of eIF4AIII controls its subcellular localization. We also identified the interaction between FRY2 and UPF3. RNA-seq analysis demonstrated that common NMD-sensitive transcripts accumulated in the fry2-1 mutant, CHX-treated wild-type plants, and upf3 mutants, suggesting that FRY2 is essential for NMD of these transcripts.
The Phosphorylation and Localization of eIF4AIII
In this study, we demonstrated that eIF4AIII is a phosphoprotein and its phosphorylation status regulates its nucleocytoplasmic partition and most likely its function in NMD. According to our Phos-tag gel analyses, ∼50 to 60% of the total eIF4AIII protein was phosphorylated in vivo under our experimental conditions (Figures 1E and 1G). Note that in the wild-type protoplasts, all of the protein appeared to localize in the nucleus; however, in the fry2-1 mutant protoplasts, a large portion of the protein relocated to the cytoplasm. Therefore, it seems that there is no quantitative correlation between the overall phosphorylation status of eIF4AIII (as judged by Phos-tag gel assays) and its localization. In addition to the difference in the experimental systems (e.g., whole seedlings versus protoplasts) in assessing phosphorylation and subcellular localization of the protein, several possible reasons may be responsible for this discrepancy. It is likely that there is a portion of phosphorylated eIF4AIII that remains confined to the nucleus and is not actively involved in nucleocytoplasmic shuttling. Although we noted similar subcellular localizations of the eIF4AIII protein when expressed either under control of its native promoter or the 35S promoter (Figure 2B; Supplemental Figure 2), we cannot rule out the possibility that the higher expression level of eIF4AIII from the 35S promoter causes such nuclear accumulation. More importantly, the bulk of the phosphorylation sites of the eIF4AIII may not be as important in determining its partition between the nucleus and the cytoplasm as some specific sites. When all the potential phosphorylation residues were simultaneously changed either to aspartate (phosphomimetic) or alanine (dephosphomimetic), the mutant proteins localized entirely in the cytoplasm, likely because this disrupted the protein’s structure. By replacing individual residues with phosphomimetic or dephosphomimetic residues, we established that the phosphorylation state of Ser-100 and Ser-101 is of critical importance in determining the nucleocytoplasmic distribution of eIF4AIII (Figure 3B), which was further confirmed using kinase and phosphatase inhibitors (Figure 3D). Nonetheless, it should be noted that additional residues, proteins and/or factors may contribute to the regulation of the intracellular localization of eIF4AIII.
How does phosphorylation control the nucleocytoplasmic shuttling of eIF4AIII? Previous studies have shown that phosphorylation of the Y14 protein, another key component of the EJC, abolished its interaction with other EJC components as well as factors downstream of the EJC (Hsu et al., 2005). Therefore, it is possible that phosphorylation of eIF4AIII similarly disturbs its interaction with other EJC components, causing it to dissociate from the EJC and to diffuse into the cytoplasm, as we observed in the fry2-1 mutant. This hypothesis will be the focus of further studies.
These observations suggest that eIF4AIII may also have a complex cellular behavior that may be regulated by multiple pathways. In addition to its role in NMD, previous studies have shown that eIF4AIII is involved in mRNA export and pre-mRNA splicing. It may have other unknown functions too. Because both eIF4AII (Izaurralde, 2013; Meijer et al., 2013), a closer homolog of eIF4AIII, and FRY2/CPL1 (Manavella et al., 2012; Chen et al., 2015), a regulator of eIF4AIII, are involved in miRNA processing, it is tempting to speculate that eIF4AIII may also have roles in miRNA biogenesis or processing. This hypothesis can be tested in future studies.
FRY2 Is Involved in Plant NMD
There are several lines of evidence to support a regulatory role of FRY2 in NMD. FRY2 interacts with eIF4AIII and UPF3, two important components of NMD. FRY2 also regulates the localization of eIF4AIII via affecting its phosphorylation. RNA-seq analyses revealed that certain alternatively spliced transcripts and 5′-extended transcripts with NMD-eliciting features accumulated in the fry2-1 mutant. The features of these transcripts indicate that their accumulation can be ascribed to a deficiency in NMD rather than a deficiency in splicing. First, most of these transcripts have NMD-eliciting premature stop codons; second, some of these transcripts also accumulated in the CHX-treated plants and the upf3-2 mutant, suggesting that these transcripts are likely NMD targets. Third, these transcripts show a reduced decay rate in the fry2-1 mutant, consistent with their accumulation. These observations indicate that the FRY2 protein is necessary for NMD of certain targets.
FRY2 may regulate NMD via modulating the recruitment, nuclear retention, or function of eIF4AIII or UPF3. Our data has suggested a link between eIF4AIII localization and NMD efficiency. Since loss-of-function mutation in eIF4AIII is likely lethal and we also failed to identify heterozygous insertion lines, we currently are unable to directly test whether the phosphorylation status or localization of eIF4AIII directly regulates NMD.
5′-Extended mRNAs as NMD Targets in Plants
The 5′-extended mRNAs could be transcribed from previously unannotated 5′ flanking sequences or result from usage of alternative promoters/transcription start sites of certain protein-coding genes (Toesca et al., 2011). In the wild-type plants, 5′-extended mRNAs were hardly detectable because they are rapidly degraded by unidentified RNA turnover mechanisms. The 5′-extended mRNAs were first identified as NMD targets in yeast and accumulated in the yeast upf1 mutant (Johansson et al., 2007). Although the biological significance of most of these novel transcripts remains unclear, they likely originate from transcriptional “noise” (Lee et al., 2008). Until now, there have been no reports related to 5′-extended mRNAs in plants. In the fry2-1 mutant, the accumulation of 5′-extended mRNAs allowed us to characterize them in more detail. We found that 5′-extended mRNAs associate with transcriptional repression of the main genes. This finding is consistent with reports in yeast, where certain transcripts associated with upstream transcription can lead to transcriptional repression (Toesca et al., 2011). Therefore, 5′-extended mRNAs may have a universal function in inhibiting the transcription of the affected genes in eukaryotes. Presently, the precise mechanism of 5′-extended mRNA-mediated transcriptional repression is not fully understood. The best-studied example is the 5′-extended RNAs in the Saccharomyces cerevisiae NMD mutants, which can control the repression of a large number of genes located in subtelomeric regions and interfere with binding of the transcriptional activator Zap1p. Whether this transcriptional-interference model is applicable to 5′-extended mRNAs in plants is presently unclear.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) fry2-1 mutant in the C24 background was described previously (Xiong et al., 2002). Seeds of C24 and fry2 were sterilized with 50% bleach in 0.01% Triton X-100 for 5 min and then rinsed five times with sterile water. The sterilized seeds were sown on 0.5× Murashige and Skoog (MS) medium plates supplemented with 3% sucrose. After a 4-d stratification at 4°C, the plates were placed under a 16-h-light and 8-h-dark cycle at 21°C for germination and growth. Twelve days later, the seedlings were harvested for total RNA extraction.
RNA Extraction, Library Construction, and Sequencing
Using TRIzol reagent (Invitrogen), total RNAs were extracted from 12-d-old seedlings of the wild-type C24, the fry2-1 mutant, and CHX-treated C24. For the CHX treatment, 12-d-old seedlings growing on 0.5× MS plates were harvested and incubated by shaking in 0.5× MS solution without (control) or with 20 μM CHX for 5 h before harvesting for RNA extraction. Polyadenylated RNAs were isolated using the Oligotex mRNA Midi Kit (Qiagen). The RNA-seq libraries were constructed using the Illumina Whole Transcriptome Analysis Kit following the standard protocol and sequenced on the HiSeq platform to generate high-quality paired-end reads of 101 nucleotides in length.
Read Alignment, Junction Prediction, and AS Event Annotation
The annotated Arabidopsis gene models were downloaded from TAIR10 (https://www.arabidopsis.org/). TopHat (Trapnell et al., 2009) was used to align the reads against the genome sequences allowing two nucleotide mismatches. TopHat was also used to predict splice junctions that did not permit any mismatches in the anchor region of a spliced alignment. To filter the false positive junctions, the well-studied criteria (i.e., an overhang size by more than 20 bp and at least two reads spanning the junctions) were set as cutoff values (Cui et al., 2014). JuncBASE (Brooks et al., 2011) was used to annotate all AS events, including cassette exons, alternative 5′ splicing sites, alternative 3′ splicing sites, mutually exclusive exons, coordinate cassette exons, alternative first exons, alternative last exons, and intron retention, based on the input genome coordinates of all annotated exons and all confidently identified splice junctions.
The Identification of Differential AS Events
Fisher’s exact tests were also used to identify the differential representation of each AS event. For alternative 5′ splicing sites and 3′ splicing sites and exon skipping events, Fisher’s exact tests were performed on the comparison of the junction-read counts and the corresponding exon-read counts between the control and mutant or treatment. The events with P values < 0.05 were identified as significantly differential events.
Identification of 5′-Extended Transcripts
To identify the 5′-extended transcripts, the transcripts were assembled by the reference annotation-based method that was applied to the Cufflink software (Trapnell et al., 2010). Briefly, the reference transcripts were tiled with sequencing reads that will aid the assembly of novel isoforms. In addition to the two modified parameters (min-isoform-fraction = 0.00001 and pre-mRNA fraction = 0.00001), other parameters were used as default parameters in the assembly. After assembly by Cufflinks, the isoforms with extended 5′ regions (>200 bp) were defined as 5′-extended transcripts. Fisher’s exact tests were used to identify differential representation of each 5′-extended transcript and to compare the counts of the reads in 5′-extended region and the corresponding exon-read counts between C24 and fry2-1. Events with P < 0.01 were considered as significantly different events.
RT-PCR Validation
The selected 5′ extension events were validated by RT-PCR using a set of primers (Supplemental Data Set 3) that were designed based on each event. Total RNAs from the C24 and fry2-1 plants were extracted using Trizol solution (Invitrogen), treated with DNAase I and reverse-transcribed into cDNA (random priming) with SuperScript II reverse transcriptase (Invitrogen).
Quantitative RT-PCR
For the RT reaction, we used 3 μg of total RNAs extracted from the control (water) and 300 mM NaCl-treated C24 and fry2-1 seedlings. RT reactions were conducted with SuperScript III First-Strand Synthesis SuperMix (Invitrogen) in a 20-μL reaction. Random hexamers were used for the first-strand synthesis. The solution was diluted 10 times, and 1 μL of the diluted solution was used as a template in a 10-μL reaction system with 2× SYBR Green SuperMix, ROX (Invitrogen). The quantitative RT-PCRs were performed in triplicate using the ABI 7900HT Fast Real-Time PCR System (Applied Biosystems). Primers for validating the expression of genes with uORF are listed in Supplemental Data Set 3.
Yeast Two-Hybrid Assays
Yeast transformation was performed according to the manufacturer’s instructions (Clontech). To map the interacting domain, truncated forms of FRY2 and eIF4AIII were constructed from both the N and C termini. These truncated mutants were then tested for interaction by cotransforming them into the Y2H gold yeast strain with their corresponding full-length partner (FRY2 in pGAD or eIF4AIII in pGBK) and plating on -tryptophan/-leucine (-TL, for transformation control) and -tryptophan/-leucine/-histidine/-adenine (-TLHA, for selection). Four FRY2 and two eIF4AIII truncated constructs were tested. To test the interaction of FRY2 and eIF4AIII, the corresponding full-length cDNAs were constructed into pGAD or pGBK.
Vector Construction
The cDNA fragments of eIF4AIII, FRY2, and UPF3 were amplified by RT-PCR and cloned into pENTR/D/TOPO (Invitrogen) to generate entry clones. For BiFC, all entry clones were subsequently transferred to the destination vectors nEYFP/pUGW2 and cEYFP/pUGW2 by LR recombination. For protein dephosphorylation assays, the entry clone of eIF4AIII was introduced into pEarleyGate202. For subcellular localization of the fluorescent protein, eIF4AIII was transformed into pEarleyGate 101.
BiFC Analysis
BiFC was performed with transient expression in protoplasts. Arabidopsis protoplasts were isolated and polyethylene glycol (PEG)-mediated transformation was performed by standard protocols (Yoo et al., 2007). Briefly, cell walls were digested using the fungal cellulase R10 and macerozyme R10 to release the protoplasts. Combinations of constructs were transformed into protoplasts by PEG-calcium transfection. After a 16-h incubation at room temperature, cells were harvested for further analysis. Signals of fluorescent proteins were detected with a LSM 710 inverted confocal microscope (Carl Zeiss).
Analysis of Protein Phosphorylation Status
FLAG-tagged eIF4AIII and UPF3 were constructed by introducing eIF4AIII and UPF3 into pEarleyGate 202 by LR reaction. The resulting constructs were transferred into Agrobacterium tumefaciens strain GV3101, and stable transgenic lines of C24 and fry2-1 were obtained by the flower-dipping method. The seedlings of T2 transgenic lines were harvested on selective media, and the whole proteins were extracted and subjected to Phos-tag (Alpha Laboratories) SDS-PAGE. Standard immunoblotting was performed using anti-FLAG M2 antibody (Sigma-Aldrich; F1804), and the signal was detected by the ImageQuant LAS4000 bimolecular imager. For CIP treatment, 20 units of CIP (New England Biolabs) was added to 200 μL protein extract supernatant of FLAG-tagged eIF4AIII transgenic plant tissues. Samples were cultured for different time at 37°C and then analyzed by Phos-tag SDS-PAGE and immunoblot using anti-FLAG antibody. For kinase inhibitor and phosphatase inhibitor treatment, 7-d-old seedlings of FLAG tagged eIF4AIII transgenic plants in the C24 or fry2-1 mutant background were divided into three aliquots for control and treatment with 1 μM kinase inhibitor staurosporine (Sigma-Aldrich) or 100 nM phosphatase inhibitor okadaic acid (Applichem; A2200). After 12-h incubation in MS solution, total proteins were extracted from the seedlings and subjected to Phos-tag SDS-PAGE and immunoblot analysis using the anti-FLAG antibody.
Analysis of Protein Localization
GFP was fused in frame with eIF4AIII at its C terminus and transiently expressed in C24 and fry2-1 mutant protoplasts by the PEG-mediated method (Yang and Shen, 2006). Transformed cells were divided into aliquots for control and treatment with 1 μM of the kinase inhibitor staurosporine (Sigma-Aldrich) or 100 nM of the phosphatase inhibitor OKD (Applichem; A2200). After a 16-h incubation in a growth chamber, the GFP signals were examined using a Zeiss LSM710 inverted confocal microscope.
Quantitative RT-PCR to Quantify 5′-Extended mRNAs and Other NMD Targets
Leaves of C24 and fry2-1 were excised from plants at the rosette stage. Protoplasts were isolated by treating the sample with cellulase and macerozyme as described (Yoo et al., 2007). C24 protoplasts were treated with the RNA polymerase inhibitor triptolide (Calbiochem) for 16 h, and protoplasts without treatment were used as the control. fry2-1 protoplasts were treated with triptolide or triptolide combined with staurosporine (Cayman Chemical) for 16 h, and protoplasts without treatment were used as the control. All samples were harvested by centrifuging at 100g for 2 min. Total RNAs were extracted using an RNeasy Mini Kit (Qiagen). Quantitative RT-PCR was performed on selected targets using the primers listed in Supplemental Data Set 3.
Analysis of mRNA Stability
Ten-day-old seedlings of C24 and fry2-1 were incubated in a half-strength MS liquid medium containing 200 μM TPL for 0, 4, and 8 h. Samples were harvested and total RNAs were extracted, followed by RT and real-time quantitative PCR as described above.
In Vitro Pull-Down Assays
In vitro pull-down assays were performed to test the interaction between FRY2 and eIF4AIII. The pTH24-FRY2 plasmid was constructed and transformed into the Escherichia coli Rosetta (DE3) strain. Soluble protein was purified by Ni2+ beads (Qiagen) according to the manufacturer’s protocol. Purified His-tagged proteins were eluted using an elution buffer and then dialyzed into IP buffer. Approximately 10 μg of purified His-tagged protein were bound to Ni-NTA beads (Qiagen) and incubated with 1 mL of the protein extract supernatant of FLAG-tagged eIF4AIII or AT3G62910 transgenic plant tissues at 4°C overnight on a rocker. The mixture was centrifuged at 2000 rpm for 30 s. After discarding the supernatant, beads were washed three times with IP buffer. The beads with the bound proteins were boiled in 2× SDS-PAGE sample buffer and analyzed by immunoblot using anti-FLAG and anti-HIS antibodies.
Accession Numbers
RNA-seq data from this article can be found in the SRA database (NCBI) under accession number SRP047445.
Supplemental Data
Supplemental Figure 1. Accumulation of PTC-containing FRY2 transcripts in the fry2-1 mutant.
Supplemental Figure 2. Mislocalization of eIF4AIII in fry2-1.
Supplemental Figure 3. Phosphorylation level of UPF3 could be regulated by FRY2 in vivo.
Supplemental Figure 4. Quality analyses of RNA-seq data from fry2-1 mutant and CHX-treated wild type (C24) and identification of AS events.
Supplemental Figure 5. Six representative AS events validated by RT-PCR and depicted by partial gene model.
Supplemental Figure 6. Quality analyses of the RNA-seq data for upf3-1 and identification and comparison of AS events.
Supplemental Figure 7. Accumulation of 5′-extended mRNAs in the fry2-1 mutant.
Supplemental Data Set 1. Alternative 5′ splice sites and 3′ splice sites that accumulated in fry2-1 mutant, upf3-1 mutant, and CHX-treated wild type (C24) plants
Supplemental Data Set 2. The 5′ extended mRNAs that accumulated in fry2-1 mutant and CHX-treated wild-type (C24) plants
Supplemental Data Set 3. Primers used for RT-PCR and qPCR.
Supplementary Material
Acknowledgments
This work was supported by King Abdullah University of Science and Technology Office of Sponsored Research (OSR) under Award URF/1/2283-01-01 and Faculty Baseline Funds BAS/1/1007-1-1. We thank the Bioscience Core Lab of KAUST for providing genome sequencing and imaging services.
AUTHOR CONTRIBUTIONS
L.X., P.C., and T.C. conceived the project idea. P.C. and F.D. performed data analysis. T.C. and T.Q. did the majority of the experiments. H.C. did the initial yeast two-hybrid screens and Z.W. did the initial BiFC assay. P.C. and L.X. wrote the manuscript. All authors read and approved the final manuscript.
Glossary
- uORF
upstream open reading frame
- NMD
nonsense-mediated decay
- PTC
premature translation-termination codon
- EJC
exon junction complex
- TPL
triptolide
- BiFC
bimolecular fluorescence complementation
- CIP
calf intestinal alkaline phosphatase
- OKD
okadaic acid
- STA
staurosporine
- AS
alternative splicing
- CHX
cycloheximide
- MS
Murashige and Skoog
- PEG
polyethylene glycol
Footnotes
Articles can be viewed online without a subscription.
References
- Arciga-Reyes L., Wootton L., Kieffer M., Davies B. (2006). UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 47: 480–489. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I., Izaurralde E. (2006). Quality control of gene expression: a stepwise assembly pathway for the surveillance complex that triggers nonsense-mediated mRNA decay. Genes Dev. 20: 391–398. [DOI] [PubMed] [Google Scholar]
- Blom N., Gammeltoft S., Brunak S. (1999). Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294: 1351–1362. [DOI] [PubMed] [Google Scholar]
- Brogna S., Wen J. (2009). Nonsense-mediated mRNA decay (NMD) mechanisms. Nat. Struct. Mol. Biol. 16: 107–113. [DOI] [PubMed] [Google Scholar]
- Brooks A.N., Yang L., Duff M.O., Hansen K.D., Park J.W., Dudoit S., Brenner S.E., Graveley B.R. (2011). Conservation of an RNA regulatory map between Drosophila and mammals. Genome Res. 21: 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.F., Imam J.S., Wilkinson M.F. (2007). The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 76: 51–74. [DOI] [PubMed] [Google Scholar]
- Chen T., Cui P., Chen H., Ali S., Zhang S., Xiong L. (2013). A KH-domain RNA-binding protein interacts with FIERY2/CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis. PLoS Genet. 9: e1003875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Cui P., Xiong L. (2015). The RNA-binding protein HOS5 and serine/arginine-rich proteins RS40 and RS41 participate in miRNA biogenesis in Arabidopsis. Nucleic Acids Res. 43: 8283–8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui P., Zhang S., Ding F., Ali S., Xiong L. (2014). Dynamic regulation of genome-wide pre-mRNA splicing and stress tolerance by the Sm-like protein LSm5 in Arabidopsis. Genome Biol. 15: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degtiar E., Fridman A., Gottlieb D., Vexler K., Berezin I., Farhi R., Golani L., Shaul O. (2015). The feedback control of UPF3 is crucial for RNA surveillance in plants. Nucleic Acids Res. 43: 4219–4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma M.K., Parker R. (2007). RNA quality control in eukaryotes. Cell 131: 660–668. [DOI] [PubMed] [Google Scholar]
- Drechsel G., Kahles A., Kesarwani A.K., Stauffer E., Behr J., Drewe P., Rätsch G., Wachter A. (2013). Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 25: 3726–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraiuolo M.A., Lee C.S., Ler L.W., Hsu J.L., Costa-Mattioli M., Luo M.J., Reed R., Sonenberg N. (2004). A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc. Natl. Acad. Sci. USA 101: 4118–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Li X., Spatrick P., Casillo R., Dong S., Jacobson A. (2003). Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell 12: 1439–1452. [DOI] [PubMed] [Google Scholar]
- Hori K., Watanabe Y. (2005). UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J. 43: 530–540. [DOI] [PubMed] [Google Scholar]
- Hsu IaW., Hsu M., Li C., Chuang T.W., Lin R.I., Tarn W.Y. (2005). Phosphorylation of Y14 modulates its interaction with proteins involved in mRNA metabolism and influences its methylation. J. Biol. Chem. 280: 34507–34512. [DOI] [PubMed] [Google Scholar]
- Ishigaki Y., Li X., Serin G., Maquat L.E. (2001). Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607–617. [DOI] [PubMed] [Google Scholar]
- Izaurralde E. (2013). A role for eIF4AII in microRNA-mediated mRNA silencing. Nat. Struct. Mol. Biol. 20: 543–545. [DOI] [PubMed] [Google Scholar]
- Jiang J., Wang B., Shen Y., Wang H., Feng Q., Shi H. (2013). The arabidopsis RNA binding protein with K homology motifs, SHINY1, interacts with the C-terminal domain phosphatase-like 1 (CPL1) to repress stress-inducible gene expression. PLoS Genet. 9: e1003625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M.J., He F., Spatrick P., Li C., Jacobson A. (2007). Association of yeast Upf1p with direct substrates of the NMD pathway. Proc. Natl. Acad. Sci. USA 104: 20872–20877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M., et al. (2012). Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 40: 2454–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerényi Z., Mérai Z., Hiripi L., Benkovics A., Gyula P., Lacomme C., Barta E., Nagy F., Silhavy D. (2008). Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J. 27: 1585–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerényi F., Wawer I., Sikorski P.J., Kufel J., Silhavy D. (2013). Phosphorylation of the N- and C-terminal UPF1 domains plays a critical role in plant nonsense-mediated mRNA decay. Plant J. 76: 836–848. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Koroleva O.A., Lewandowska D., Pendle A.F., Clark G.P., Simpson C.G., Shaw P.J., Brown J.W. (2009). Aberrant mRNA transcripts and the nonsense-mediated decay proteins UPF2 and UPF3 are enriched in the Arabidopsis nucleolus. Plant Cell 21: 2045–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwa H., Hausmann S., Bang W.Y., Ueda A., Kondo N., Hiraguri A., Fukuhara T., Bahk J.D., Yun D.J., Bressan R.A., Hasegawa P.M., Shuman S. (2004). Arabidopsis C-terminal domain phosphatase-like 1 and 2 are essential Ser-5-specific C-terminal domain phosphatases. Proc. Natl. Acad. Sci. USA 101: 14539–14544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwa H., et al. (2002). C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. Proc. Natl. Acad. Sci. USA 99: 10893–10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva O.A., Calder G., Pendle A.F., Kim S.H., Lewandowska D., Simpson C.G., Jones I.M., Brown J.W., Shaw P.J. (2009). Dynamic behavior of Arabidopsis eIF4A-III, putative core protein of exon junction complex: fast relocation to nucleolus and splicing speckles under hypoxia. Plant Cell 21: 1592–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y., et al. (2009). Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 2453–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A., Hansen K.D., Bullard J., Dudoit S., Sherlock G. (2008). Novel low abundance and transient RNAs in yeast revealed by tiling microarrays and ultra high-throughput sequencing are not conserved across closely related yeast species. PLoS Genet. 4: e1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Gatfield D., Izaurralde E., Moore M.J. (2001). The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20: 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd J.P.B., Davies B. (2013). SMG1 is an ancient nonsense-mediated mRNA decay effector. Plant J. 76: 800–810. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S., Jensen T.H. (2015). Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 16: 665–677. [DOI] [PubMed] [Google Scholar]
- Manavella P.A., Hagmann J., Ott F., Laubinger S., Franz M., Macek B., Weigel D. (2012). Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell 151: 859–870. [DOI] [PubMed] [Google Scholar]
- Maquat L.E. (2004). Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 5: 89–99. [DOI] [PubMed] [Google Scholar]
- Meijer H.A., Kong Y.W., Lu W.T., Wilczynska A., Spriggs R.V., Robinson S.W., Godfrey J.D., Willis A.E., Bushell M. (2013). Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science 340: 82–85. [DOI] [PubMed] [Google Scholar]
- Mendell J.T., Sharifi N.A., Meyers J.L., Martinez-Murillo F., Dietz H.C. (2004). Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 36: 1073–1078. [DOI] [PubMed] [Google Scholar]
- Mérai Z., Benkovics A.H., Nyikó T., Debreczeny M., Hiripi L., Kerényi Z., Kondorosi É., Silhavy D. (2013). The late steps of plant nonsense-mediated mRNA decay. Plant J. 73: 50–62. [DOI] [PubMed] [Google Scholar]
- Miura F., Kawaguchi N., Sese J., Toyoda A., Hattori M., Morishita S., Ito T. (2006). A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc. Natl. Acad. Sci. USA 103: 17846–17851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.J. (2005). From birth to death: the complex lives of eukaryotic mRNAs. Science 309: 1514–1518. [DOI] [PubMed] [Google Scholar]
- Nyikó T., Kerényi F., Szabadkai L., Benkovics A.H., Major P., Sonkoly B., Mérai Z., Barta E., Niemiec E., Kufel J., Silhavy D. (2013). Plant nonsense-mediated mRNA decay is controlled by different autoregulatory circuits and can be induced by an EJC-like complex. Nucleic Acids Res. 41: 6715–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios I.M., Gatfield D., St Johnston D., Izaurralde E. (2004). An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 427: 753–757. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J., Raes J., Izaurralde E. (2006). Nonsense-mediated mRNA decay: Target genes and functional diversification of effectors. Trends Biochem. Sci. 31: 639–646. [DOI] [PubMed] [Google Scholar]
- Shaul O. (2015). Unique aspects of plant nonsense-mediated mRNA decay. Trends Plant Sci. 20: 767–779. [DOI] [PubMed] [Google Scholar]
- Shi C., Baldwin I.T., Wu J. (2012). Arabidopsis plants having defects in nonsense-mediated mRNA decay factors UPF1, UPF2, and UPF3 show photoperiod-dependent phenotypes in development and stress responses. J. Integr. Plant Biol. 54: 99–114. [DOI] [PubMed] [Google Scholar]
- Shibuya T., Tange T.O., Sonenberg N., Moore M.J. (2004). eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat. Struct. Mol. Biol. 11: 346–351. [DOI] [PubMed] [Google Scholar]
- Singh G., Kucukural A., Cenik C., Leszyk J.D., Shaffer S.A., Weng Z., Moore M.J. (2012). The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 151: 750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thebault P., Boutin G., Bhat W., Rufiange A., Martens J., Nourani A. (2011). Transcription regulation by the noncoding RNA SRG1 requires Spt2-dependent chromatin deposition in the wake of RNA polymerase II. Mol. Cell. Biol. 31: 1288–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titov D.V., Gilman B., He Q.L., Bhat S., Low W.K., Dang Y., Smeaton M., Demain A.L., Miller P.S., Kugel J.F., Goodrich J.A., Liu J.O. (2011). XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat. Chem. Biol. 7: 182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toesca I., Nery C.R., Fernandez C.F., Sayani S., Chanfreau G.F. (2011). Cryptic transcription mediates repression of subtelomeric metal homeostasis genes. PLoS Genet. 7: e1002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Lee H., Ishitani M., Tanaka Y., Stevenson B., Koiwa H., Bressan R.A., Hasegawa P.M., Zhu J.K. (2002). Repression of stress-responsive genes by FIERY2, a novel transcriptional regulator in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 10899–10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Shen M.H. (2006). Polyethylene glycol-mediated cell fusion. Methods Mol. Biol. 325: 59–66. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.