The HD-ZIP transcription factor HOX12 positively regulates ELONGATED UPPERMOST INTERNODE1 and plays a vital role in panicle exsertion from the flag leaf sheath, thereby affecting hybrid seed production.
Abstract
Bioactive gibberellins (GAs) are key endogenous regulators of plant growth. Previous work identified ELONGATED UPPERMOST INTERNODE1 (EUI1) as a GA-deactivating enzyme that plays an important role in panicle exsertion from the flag leaf sheath in rice (Oryza sativa). However, the mechanism that regulates EUI1 activity during development is still largely unexplored. In this study, we identified the dominant panicle enclosure mutant regulator of eui1 (ree1-D), whose phenotype is caused by the activation of the homeodomain-leucine zipper transcription factor HOX12. Diminished HOX12 expression by RNA interference enhanced panicle exsertion, mimicking the eui1 phenotype. HOX12 knockdown plants contain higher levels of the major biologically active GAs (such as GA1 and GA4) than the wild type. The expression of EUI1 is elevated in the ree1-D mutant but reduced in HOX12 knockdown plants. Interestingly, both HOX12 and EUI1 are predominantly expressed in panicles, where GA4 is highly accumulated. Yeast one-hybrid, electrophoretic mobility shift assay, and chromatin immunoprecipitation analyses showed that HOX12 physically interacts with the EUI1 promoter both in vitro and in vivo. Furthermore, plants overexpressing HOX12 in the eui1 mutant background retained the elongated uppermost internode phenotype. These results indicate that HOX12 acts directly through EUI1 to regulate panicle exsertion in rice.
INTRODUCTION
Rice (Oryza sativa) is a staple food for nearly half of the world’s population. With the increasing challenges of food security caused by rapid global population growth and decreasing availability of arable land, meeting the demand for high-yielding rice is always an urgent task for breeders. Producing hybrid rice cultivars is considered to be one of the most effective strategies for improving rice yield (Li and Yuan, 2010; Luo et al., 2013). However, male-sterile lines of hybrid rice often have a defect in the elongation of the uppermost internode, leading to panicle enclosure, which greatly reduces seed production of hybrid rice due to its blocking of normal pollination (Shen et al., 1987; Yang et al., 2002). This syndrome is effectively overcome by the presence of extended upper internodes resulting from the ELONGATED UPPERMOST INTERNODE1 (EUI1) mutation (Zhang and Yang, 2003; Yang et al., 2005; Chen et al., 2013). The eui1 mutants are morphologically normal until drastic elongation of the uppermost internode occurs at the heading stage (Rutger and Carnahan, 1981). EUI1 encodes a cytochrome P450 monooxygenase that deactivates bioactive gibberellins (GAs). The inactivation of EUI1 causes the accumulation of GAs such as GA1 and GA4 in the uppermost internode, which consequently increases internode elongation and plant height (Luo et al., 2006; Zhu et al., 2006; Magome et al., 2013). Moreover, overexpression of EUI1 leads to the GA-deficient dwarf phenotypes due to inhibited internode elongation (Luo et al., 2006; Zhu et al., 2006). Accordingly, fine-tuning and flexible tuning of EUI1 levels could effectively reduce or increase internode elongation. EUI1, also annotated as CYP714D1, belongs to the CYP714 subfamily, which includes several members, such as CYP714B1, CYP714B2, CYP714C1, CYP714C2, and CYP714C3 (Luo et al., 2006; Zhu et al., 2006; Magome et al., 2013). It has been suggested that CYP714B1 and CYP714B2 play a predominant role in GA 13-hydroxylation in rice (Magome et al., 2013). Transgenic Arabidopsis thaliana plants overexpressing rice CYP714B1 or CYP714B2 show semidwarfism (Magome et al., 2013). Arabidopsis contains two closely related EUI1-like P450s, CYP714A1 and CYP714A2 (Zhang et al., 2011; Nomura et al., 2013). Overexpression of either CYP714A1 or CYP714A2 also results in dwarfism, which is similar to the phenotype of plants overexpressing the rice EUI1 gene (Zhang et al., 2011; Nomura et al., 2013). These results suggest that the regulation of internode elongation by EUI1 is evolutionarily conserved. However, the direct regulation of EUI1 at the transcriptional level is poorly understood.
Homeodomain-leucine zipper (HD-ZIP) transcription factors are unique to the plant kingdom (Ruberti et al., 1991; Schena and Davis, 1992; Ariel et al., 2007; Elhiti and Stasolla, 2009). HD-ZIP genes play diverse functions in plant development and plant adaptation to environmental stresses (Arce et al., 2011; Brandt et al., 2014). A common feature of HD-ZIP transcription factors is the presence of a leucine zipper (ZIP) domain adjacent to the homeodomain (HD); the HD domain is responsible for the specificity of protein-DNA interactions, and the ZIP domain is important for homo- and heterodimerization. Based on their molecular characteristics, the HD-ZIP family is classified into four classes: HD-ZIP I to IV (Ariel et al., 2007; Agalou et al., 2008). There are 17 HD-ZIP I proteins in Arabidopsis and 14 in rice; however, only a few have been characterized (Henriksson et al., 2005; Agalou et al., 2008; Harris et al., 2011). Members of HD-ZIP I increase plant plasticity by mediating external signals and regulating growth in response to environmental conditions (Wang et al., 2003; Lin et al., 2008; Manavella et al., 2008; Brandt et al., 2014; Chang et al., 2014; Zhao et al., 2014; Capella et al., 2015). For example, Arabidopsis HB7 and HB12 encode potential regulators of growth in response to water deficit (Olsson et al., 2004; Valdés et al., 2012). HB6 is a target of phosphatase ABI1 and regulates hormone responses (Himmelbach et al., 2002). Activation tagging of HB13 in Arabidopsis confers broad-spectrum disease resistance (Gao et al., 2014a). In rice, the HD-ZIP I gene HOX22 affects abscisic acid (ABA) biosynthesis and positively regulates drought and salt responses through ABA-mediated signal transduction pathways (Zhang et al., 2012).
HD-ZIP genes also play important roles in plant development and domestication in crops. Six-rowed spike1 (Vrs1), which regulates the change from two-rowed spikes in the wild progenitor of barley (Hordeum vulgare) to six-rowed spikes in domesticated barley, has been identified as a domestication gene (Komatsuda et al., 2007; Pourkheirandish et al., 2007). Maize (Zea mays) HD-ZIP I transcription factor grassy tillers1 (gt1) promotes lateral bud dormancy and suppresses elongation of lateral ear branches (Whipple et al., 2011). In rice, HOX4 is predominantly expressed in vascular tissues (Agalou et al., 2008; Dai et al., 2008). Overexpression of HOX4 in rice gives rise to a semidwarf phenotype with upregulated expression of the GA oxidase gene GA2ox3 (Dai et al., 2008; Zhou et al., 2015). However, our knowledge of the HD-ZIP I family is still fragmentary and far from comprehensive.
To analyze the regulatory role of EUI1, we conducted a large-scale screening of our rice activation-tagged mutant populations (Ma et al., 2009b) and identified a dominant mutant, regulator of eui1 (ree1-D), with a particularly striking defect in internode development. The ree1-D mutant exhibits incompletely differentiated internodes without panicle exsertion, which is confirmed to be caused by the activation of the HD-ZIP I transcription factor HOX12. In contrast, knockdown of HOX12 enhances the panicle exsertion, mimicking the phenotype of eui1. Based on dual-luciferase reporter assays, yeast one-hybrid assays, chromatin immunoprecipitation, and genetic analysis, we demonstrate that HOX12 is a positive regulator of EUI1 that plays a crucial role in panicle exsertion in rice.
RESULTS
The ree1-D Mutant Displays Defects in Panicle Exsertion
We previously demonstrated that mutation of EUI1 leads to an extremely elongated panicle exsertion resulting from an increase in bioactive GA levels, while EUI1 overexpression lines exhibit GA-deficient phenotypes, with shortened internodes and poor panicle exsertion (Luo et al., 2006). To further investigate the genetic components that govern panicle exsertion, we screened our T-DNA activation-tagged lines for mutants with defects in uppermost internode development. One such mutant, ree1-D, with a dwarf phenotype and deficiency in panicle exsertion, was identified.
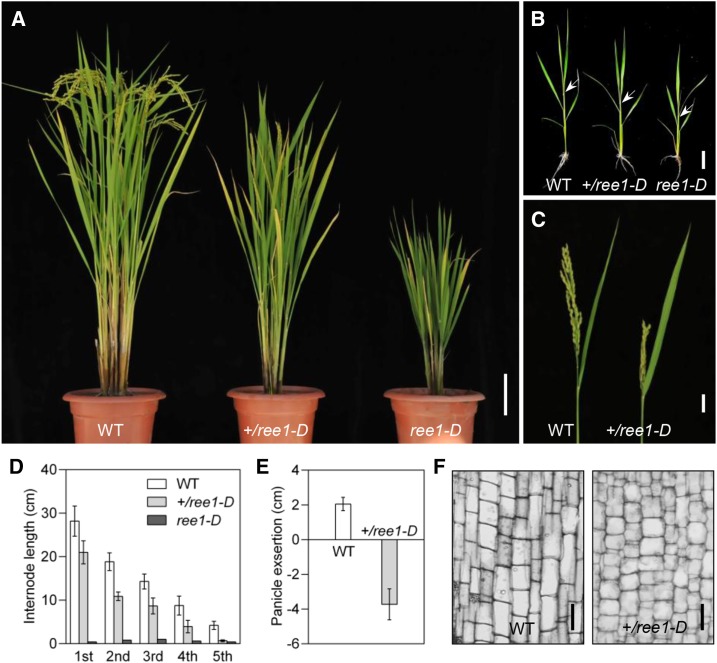
Genetic analysis demonstrated that there are two types of phenotypes in heterozygous segregating populations, which correspond to the heterozygous or homozygous states of the ree1-D allele, suggesting that the effect of this allele is dose dependent and that ree1-D is a gain-of-function mutant (Figure 1A). ree1-D mutants exhibited retarded growth beginning at the seedling stage. Shoot elongation was suppressed and a shorter plastochron for subsequently formed leaves was observed in the ree1-D mutants (Figure 1B). In rice, internode elongation is activated by the onset of reproductive growth. The shoot apical meristem of a wild-type plant normally generates five vegetative internodes before forming an inflorescence meristem. By contrast, the ree1-D mutants remained dwarf. Eventually, at the reproductive stage, the ree1-D mutants exhibited incompletely differentiated internodes in the stem, with no panicle exsertion from the leaf sheath, and they did not produce seeds. Since the homozygotes failed to complete their life cycle, the heterozygous ree1-D mutants (+/ree1-D) were used for further characterization. We found that each internode of the heterozygous mutants was shorter than that of the wild type, with panicles partly enclosed in the flag leaf sheath (Figures 1C to 1E). Detailed observations of longitudinal sections of the uppermost internodes revealed that the stem parenchyma cells of ree1-D were significantly shorter than those of the wild type (Figure 1F).
Figure 1.
Phenotypes of the ree1-D Mutant.
(A) The 120-d-old wild-type (WT; cv Nipponbare), heterozygous (+/ree1-D), and homozygous ree1-D plants. Bar = 10 cm.
(B) Seedling phenotype of 30-d-old wild-type, +/ree1-D, and ree1-D plants. Arrows indicate the fourth leaf sheath. Bar = 4 cm.
(C) Panicle exsertion of wild-type and +/ree1-D plants at the mature stage. Bar = 2 cm.
(D) Individual internode lengths of wild-type, +/ree1-D, and ree1-D plants. Error bars indicate the sd (n = 15).
(E) Quantification of panicle exsertion of wild-type and +/ree1-D plants. Error bars indicate the sd (n = 15).
(F) Longitudinal sections of the elongated zones of the uppermost internodes of wild-type and +/ree1-D plants at the mature stage. Bars = 50 μm.
GA Suppresses the ree1-D Mutation
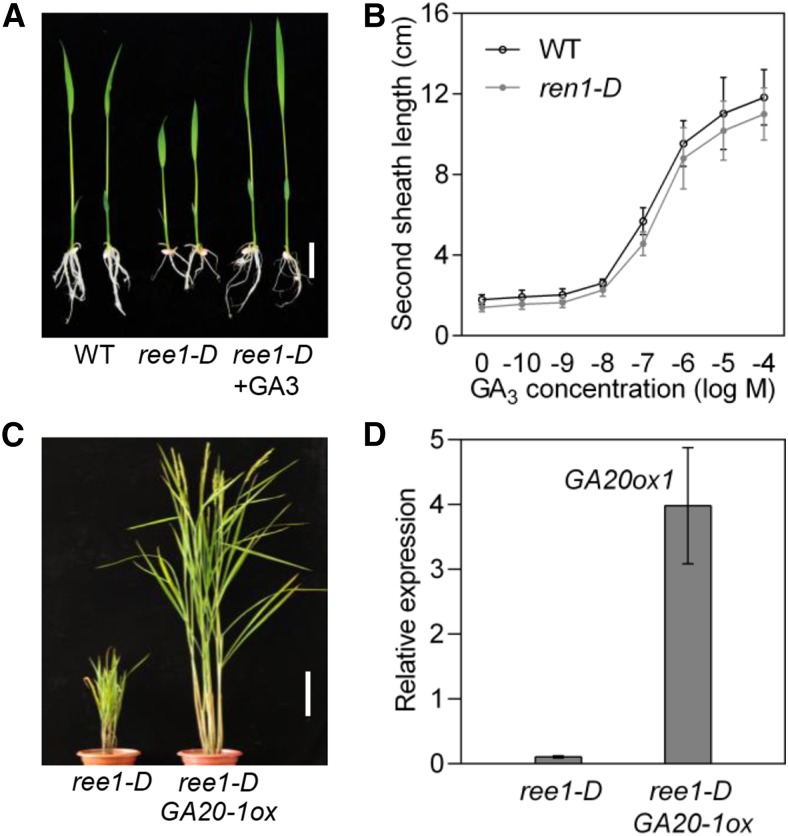
Dwarf phenotypes are often associated with malfunctions in the biosynthesis or perception of phytohormones such as GAs and brassinosteroids (BRs). As shown in Figure 1A, ree1-D showed a dwarf phenotype, indicating that the mutation in ree1-D might affect GA or BR-regulated processes. However, the typical features of BR mutants, including altered leaf angles, were not observed in ree1-D. To this end, we further tested the ree1-D mutants using exogenous GA3 treatment. The semidwarf phenotype of the ree1-D mutants could be restored to the wild type by exogenous GA application (Figure 2A). Treatment of ree1-D with different concentrations of GA revealed that the response of ree1-D to GA is quite similar to that of the wild type, although the leaf sheath of ree1-D is shorter than that of the wild type (Figure 2B). These results suggest that the ree1-D mutants can respond to exogenous GA3.
Figure 2.
Response of the ree1-D Mutant to GA.
(A) Semidwarf phenotype of ree1-D mutant can be rescued by GA3. The ree1-D mutant was treated with exogenous 10−8 M GA3 for 5 d. Bar = 2 cm.
(B) Elongation of the second leaf sheath in response to different concentrations of GA3. Error bars indicate sd (n = 12).
(C) Morphological phenotypes of ree1-D and ree1-D GA20-1-ox plants at the mature stage. Bar = 20 cm.
(D) qRT-PCR analysis of the expression level of GA20ox1 in ree1-D GA20-1-ox plants. Error bars indicate sd (n = 3).
To further confirm that the dwarf phenotype of the mutant plants is caused by GA deficiency, we crossed the ree1-D heterozygous mutant with a GA20ox1 overexpression line (GA20-1ox) in which the bioactive GA levels are strongly increased (Oikawa et al., 2004). The ree1-D GA20-1ox double mutant plants (Figures 2C and 2D) exhibited GA overproduction phenotypes, with a plant height similar to that of GA20-1ox plants. These results demonstrate that the ree1-D mutant lines also respond normally to endogenous GA, further supporting the notion that the ree1-D mutants are deficient in active GAs rather than signaling.
The Phenotypic Defects in ree1-D Are Caused by Activation of HOX12
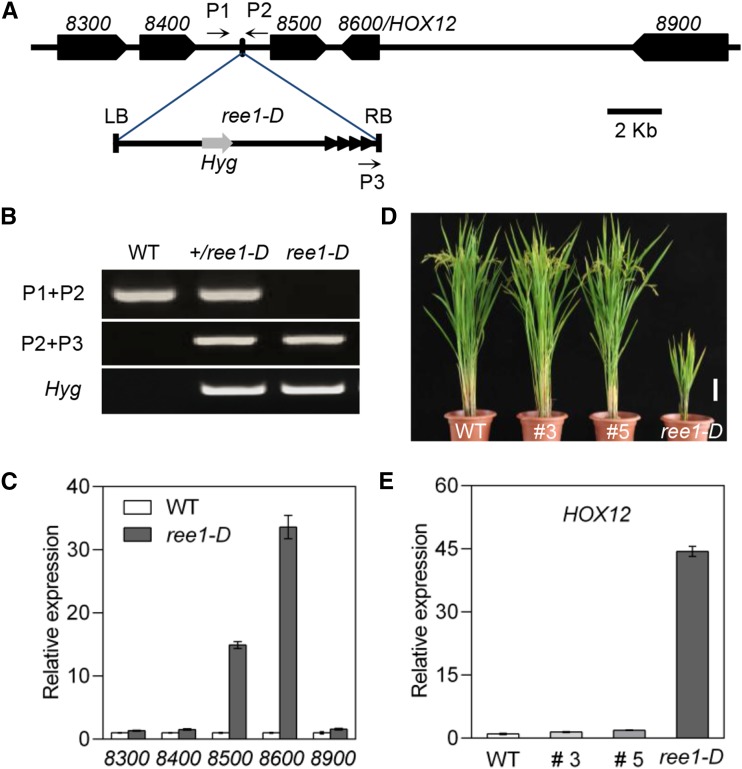
Given that ree1-D was isolated from the T-DNA insertion population, we performed genetic linkage analysis, finding that the dwarf phenotype indeed cosegregated with the T-DNA and that the mutation behaved as a dominant Mendelian trait. When a heterozygous plant was selfed, the phenotypes of the progeny segregated in an ∼1: 2: 1 ratio, suggesting that the dominance of the dwarf trait likely results from activated tagging of the target gene. To isolate the gene responsible for the ree1-D phenotype, the SiteFinding-PCR method (Wang et al., 2011) was employed to identify the T-DNA flanking sequence, and genotyping with a pair of gene-specific primers (P1 and P2) coupled with a T-DNA-specific primer (P3) revealed that the T-DNA insertion cosegregated with the dwarf phenotype (Figures 3A and 3B). Among the 418 T2 plants, 108 were wild type without the T-DNA insertion, 98 were homozygous for the T-DNA insertion, and 212 were heterozygous for the T-DNA insertion. Expression analysis of the five genes located within regions 50 kb upstream and downstream of the T-DNA insertion site revealed that the transcript level of Os03g0198500 and Os03g0198600 was increased, while the other three were not altered (Figure 3C). Hence, Os03g0198500 and Os03g0198600 around the T-DNA insertion site appeared to be candidates for the causal gene of the ree1-D mutation.
Figure 3.
Molecular Identification and Confirmation of HOX12.
(A) A diagram of the genomic region flanking the T-DNA insertion site in the ree1-D mutant. 8300, 8400, 8500, 8600, and 8900 represent Os03g0198300, Os03g0198400, Os03g0198500, Os03g0198600, and Os03g0198900, respectively. The small black arrows represent the P1, P2, and P3 primers used in cosegregation analysis. LB, T-DNA left border; RB, T-DNA right border; Black arrowheads are four tandem copies of the CaMV 35S enhancers near the T-DNA RB; Hyg, hygromycin resistance gene.
(B) Genotyping of T2 seedlings performed via PCR using the primers shown in (A).
(C) Expression analysis of genes surrounding the T-DNA insertion by qRT-PCR. The transcript level of the respective genes in the wild type was set as 1.0. Error bars indicate sd (n = 3).
(D) Suppression of ree1-D phenotypes by the HOX12-RNAi construct. The 120-d-old ree1-D homozygous mutant transformed with the HOX12-RNAi construct (#3 and #5) is shown. Bar = 10 cm.
(E) Expression levels of HOX12 in ree1-D-RNAi transgenic plants (#3 and #5) detected by qRT-PCR. The transcript level of HOX12 in the wild type was set as 1.0. Error bars indicate sd (n = 3).
Next, we generated transgenic plants carrying Os03g0198500 or Os03g0198600 under the control of the maize Ubiquitin promoter, finding that only transgenic plants carrying Os03g0198600 exhibited the growth retardation phenotype (Supplemental Figures 1 and 2). The severity of the phenotype in these transgenic plants was correlated with the expression level of the transgene (Supplemental Figure 2). This is consistent with previous observations of the heterozygous or homozygous states of the ree1-D allele. To further confirm that overexpression of Os03g0198600 caused the ree1-D phenotypes, we transformed ree1-D with an RNAi construct for Os03g0198600. Nine of these independent transgenic lines in the ree1-D background exhibited a wild-type phenotype (Figures 3D and 3E). Taken together, these results confirm that the mutant phenotype is caused by activation of Os03g0198600, which was annotated as HOX12 (Agalou et al., 2008).
HOX12 Acts as a Transcriptional Activator
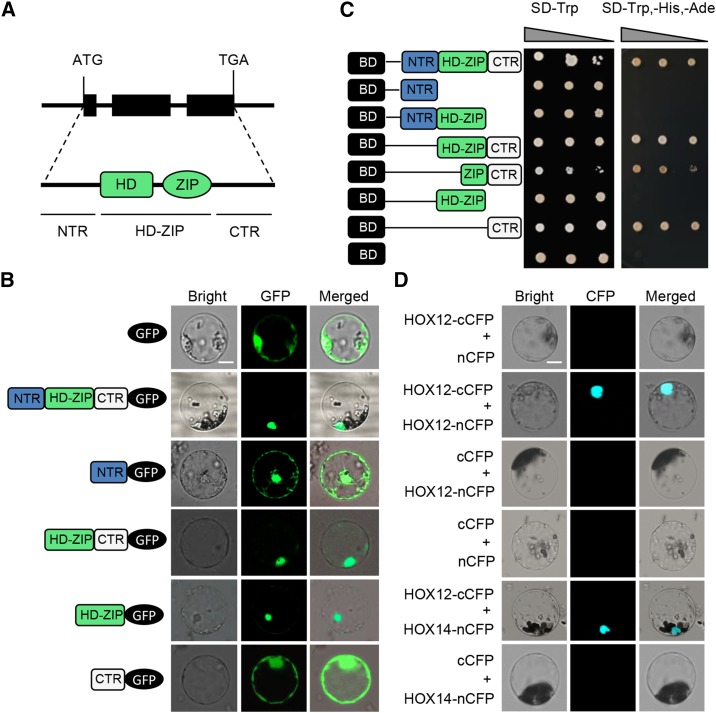
The full-length cDNA of HOX12 is 1167 nucleotides with an open reading frame of 720 nucleotides, encoding a protein of 239 amino acid residues with a predicted molecular mass of 26 kD. The coding region of HOX12 consists of three exons interrupted by two introns (Figure 4A). HOX12, together with HOX14, belongs to HD-ZIP I clade δ subgroup (Harris et al., 2011), with 46.1% identity at the protein level.
Figure 4.
Protein Structure, Subcellular Localization, Transactivation, and Dimerization of HOX12.
(A) Top: HOX12 genomic structure. Black boxes indicate exons. Bottom: conserved domains of HOX12.
(B) GFP fusion constructs of HOX12 and its truncated variants. The domains of HOX12 are indicated by NTR (N-terminal domain; 1 to 59 amino acids), HD-ZIP (HD-ZIP domain; 60 to 160 amino acids), and CTR (C-terminal domain; 161 to 239 amino acids). Bar = 10 μm.
(C) Transcription activity assay. The full-length cDNA of HOX12 and DNA fragments responsible for different truncated deletions were introduced into the pGBKT7 vector. BD represents the GAL4 DNA binding domain. The empty pGBKT7 was used as a negative control. The domains of HOX12 are indicated by NTR, HD-ZIP, and CTR. Yeast cultures were diluted (1:10 successive dilution series), spotted onto plates without Trp and without Trp, His, and Ade.
(D) BiFC assay to detect dimerization of HOX12. The C-terminal part of CFP was fused with HOX12 (HOX12-cCFP), while the N-terminal part of CFP was fused with HOX12 and HOX14 (HOX12-nCFP and HOX14-nCFP). Bar = 10 μm.
Phylogenetic analysis indicated that rice HOX12 shares high sequence similarity with related proteins identified from maize, millet (Setaria italica), and sorghum (Sorghum bicolor) (Supplemental Figure 3). HOX12 shares 76.52% identity with maize homolog gt1, which is highly expressed in immature tassels, meiotic tassels, and anthers (Whipple et al., 2011). HOX12 shares extensive sequence homology with Vrs1, a HD-ZIP protein encoded by barley Vrs1, which is responsible for the six-rowed spike phenotype in barley (Komatsuda et al., 2007).
We further investigated the subcellular localization of HOX12 by expressing HOX12-enhanced GFP (eGFP) fusion proteins under the control of the CaMV 35S promoter in rice protoplasts (Figure 4B). Empty eGFP vector was transfected into rice protoplasts as a control. As expected, eGFP itself was distributed evenly in the cytoplasm and the nucleus, whereas the HOX12-eGFP fusion protein was preferentially localized to the nucleus. To functionally analyze the nucleus-targeting domain of HOX12, we prepared a series of HOX12 truncated proteins fused with eGFP under the control of the CaMV 35S promoter, including the N terminus of HOX12 (amino acids 1 to 59), the HD-ZIP domain and C terminus of HOX12 (amino acids 60 to 239), the HD-ZIP domain of HOX12 (amino acids 60 to 160), and the C terminus of HOX12 (amino acids 161 to 239) (Figures 4A and 4B). Interestingly, HD-ZIP-eGFP localized to the nucleus, but the truncated proteins lacking the HD-ZIP domain localized to both the cytosol and nucleus, suggested that the nuclear localization signal is located within the HD-ZIP domain.
To investigate whether HOX12 can serve as a transcriptional activator, the full-length HOX12 coding sequence was fused in frame to the GAL4 DNA binding domain in yeast expression vector pGBKT7, and the construct containing only the GAL4 DNA binding domain was used as a negative control. Then, all constructs were transformed into yeast strain AH109, which carries the ADE2 and HIS3 reporter genes under the control of heterologous GAL4-responsive upstream activating sequences and promoter elements (Figure 4C). Yeast cells transformed with pGBKT7 or pGBKT7-HOX12 derivatives were selected on selective medium lacking Trp, His, and Ade and assayed visually using dilution growth tests. As shown in Figure 4C, little growth was observed when the strain (negative control) was grown on medium lacking Trp, His, and Ade. In contrast, strong growth was observed with the reconstructed pGBKT7-HOX12 strain at 10−2 and even 10−3 dilutions. These results indicate that HOX12 has transcriptional activation activity.
To identify the portions of the motif responsible for transcription activation, a series of HOX12 deletion constructs fused with the GAL4-DNA binding domain were constructed for transactivation analysis in yeast, including the N terminus of HOX12 (amino acids 1 to 59), the N terminus and HD-ZIP domain of HOX12 (amino acids 1 to 160), the entire HOX12 protein lacking only the 59 N-terminal amino acids (amino acids 60 to 239), the ZIP domain and C terminus of HOX12 (amino acids 124 to 239), the HD-ZIP domain of HOX12 (amino acids 60 to 160), and the C terminus of HOX12 (amino acids 161 to 239). Yeast cells harboring the constructs lacking the C terminus did not grow on medium lacking Trp, His, and Ade. However, cells harboring the constructs containing the C terminus all grew well (Figure 4C), indicating that the C-terminal region of HOX12 is responsible for its transcriptional activation activity.
HD-ZIP factors generally form dimeric proteins that function in the regulation of gene expression (Meijer et al., 2000). We therefore investigated whether HOX12 could form homo- or heterodimers with another member of the same δ clade, HOX14 (Harris et al., 2011). To test for homodimerization, the bimolecular fluorescence complementation (BiFC) system was used (Figure 4D). We observed that in the BiFC system, HOX12 could form homodimers and could also interact with HOX14 to form heterodimers.
Attenuated Expression of HOX12 Leads to Enhanced Panicle Exsertion
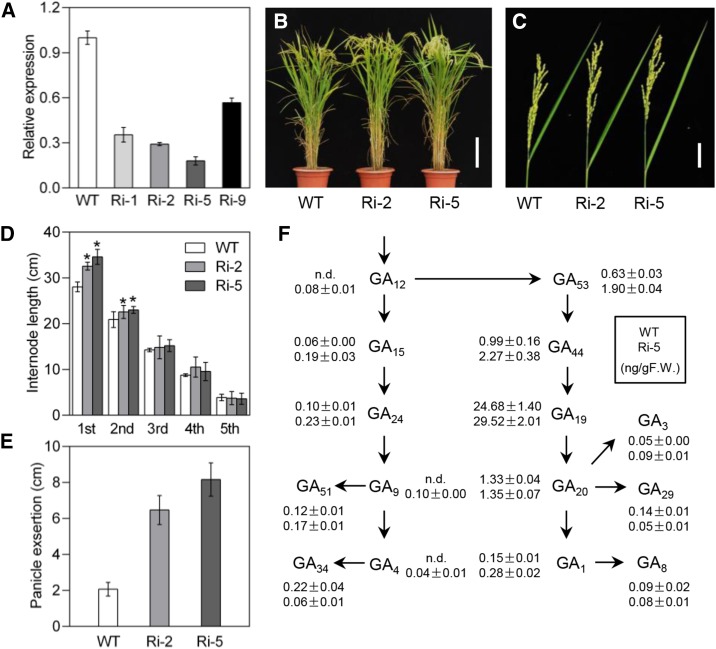
To further elucidate the biological function of HOX12 in planta, two independent HOX12-RNAi homozygous lines with the greatest reduction in HOX12 gene expression, #2 (Ri-2) and #5 (Ri-5), were selected for detailed phenotypic analysis (Figure 5A). The RNAi plants were morphologically similar to the wild type at the seedling and tillering stages, but taller than the wild type, with enhanced panicle exsertion, at the mature stage (Figure 5B). qRT-PCR analysis revealed a correlation between the observed internode phenotypes and HOX12 expression levels (Figure 5C). In the wild type, the internodal lengths were 3.9, 8.7, 14.3, 20.9, and 28.0 cm (Figure 5D). However, the internodal lengths were 3.7, 10.5, 14.8, 22.6, and 32.6 cm in Ri-2 plants and 3.6, 9.6, 15.2, 23.1, and 34.6 cm in Ri-5 plants. It is worth noting that the panicle exsertion of Ri-2 and Ri-5 was 6.5 cm and 8.2 cm, which is 3.1- and 3.9-fold longer than the 2.1 cm exsertion observed in the wild type, respectively (Figure 5E). These data suggest that the knockdown of HOX12 expression has a positive effect on panicle exsertion.
Figure 5.
Analysis of HOX12-RNAi Transgenic Plants.
(A) qRT-PCR analysis showing downregulation of HOX12 in four independent HOX12-RNAi lines. The transcript level of HOX12 in the wild type was set as 1.0. Error bars indicate sd (n = 3).
(B) Comparison of wild-type and HOX12-RNAi lines after bolting. Bar = 20 cm.
(C) Panicle exsertion of wild-type and HOX12-RNAi lines after bolting. Bar = 5 cm.
(D) The length of each internode of wild-type and HOX12-RNAi plants. Error bars indicate sd (n = 15). Asterisks indicate P < 0.05, as determined by Student’s t test analysis.
(E) Quantification of panicle exsertion in wild-type and HOX12-RNAi plants. Error bars indicate sd (n = 15).
(F) Levels of each of the indicated molecules in the GA biosynthesis pathway in wild-type and Ri-5 plants (top and bottom numbers, respectively). F.W., fresh weight. The numbers shown are the averages from three independent replicates. n.d., not detected (below the detection limit). The sd is indicated (n = 3).
GAs are essential regulators affecting internode length in rice. To test whether HOX12 affects GA homeostasis, we quantified the endogenous GAs levels in the elongating uppermost internodes of both wild-type and Ri-5 plants (Figure 5F). The levels of GA4 and its precursor GA9 were 0.04 ng/g and 0.10 ng/g, respectively, whereas both GA4 and GA9 were undetectable in the wild type. The levels of all other GAs tested in the non-13-hydroxylation pathway (GA12, GA15, GA24, and GA51) were also higher in Ri-5 than in the wild type. GA1 levels were ∼2-fold higher in Ri-5 (0.28 ng/g) than in the wild type (0.15 ng/g). The endogenous levels of GAs in the early 13-hydroxylation pathway (GA53, GA44, GA19, and GA3) were higher in ree1-D than in the wild type. The levels of GA20, a precursor of GA1, did not obviously differ between Ri-5 and the wild type. Taken together, these results indicate that HOX12 plays a major role in both the non-13-hydroxylation pathway and the early 13-hydroxylation pathway.
HOX12 Binds to and Activates the Promoter of EUI1
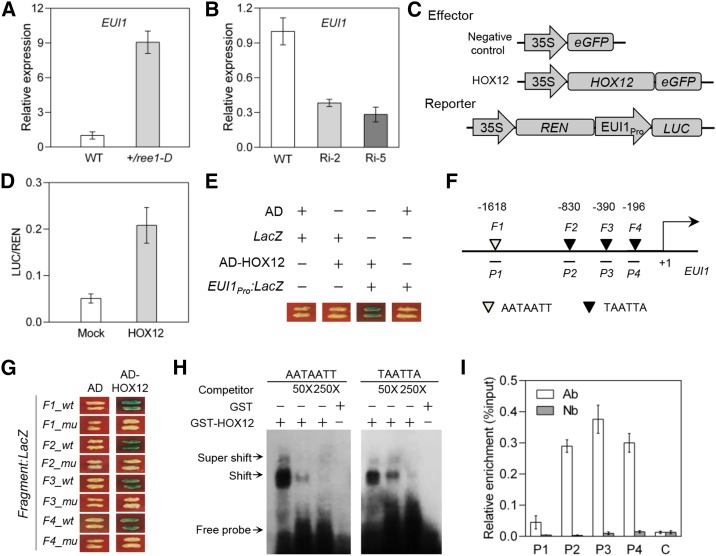
The eui1 mutants exhibit an increased panicle length in comparison with the wild type and accumulate much higher levels of GA1 and GA4 (Luo et al., 2006; Zhu et al., 2006). HOX12-RNAi plants show enhanced panicle exsertion and increased GA4 content, which is reminiscent of the eui1 mutants. Moreover, qRT-PCR analysis indicated that EUI1 mRNA was also more abundant in the ree1-D mutants than in wild-type plants and showed reduced levels in two HOX12-RNAi lines, Ri-2 and Ri-5 (Figures 6A and 6B). To investigate whether HOX12 directly affects EUI1 expression, the firefly luciferase reporter (EUI1Pro:LUC) driven by the EUI1 promoter and Renilla luciferase driven by the 35S promoter (35S Pro:REN; as an internal control) were constructed in the same plasmid and transiently expressed in rice protoplasts (Figure 6C). The LUC and REN activities were then measured and the LUC activity was normalized to REN activity. The LUC:REN ratio reflects in vivo EUI1 activity. As expected, coexpression of HOX12 with EUI1Pro:LUC increased the LUC:REN ratio. LUC activity in protoplasts transformed with LUC under the control of the EUI1 promoter was 4-fold that of the control (Figure 6D), suggesting that HOX12 is a positive regulator of EUI1.
Figure 6.
HOX12 Is a Transcriptional Activator of EUI1.
(A) qRT-PCR analysis of the expression levels of EUI1 in the panicles of +/ree1-D plants. The transcript level of HOX12 in the wild type was set as 1.0. Error bars indicate sd (n = 3).
(B) qRT-PCR analysis of the expression levels of EUI1 in the panicles of HOX12-RNAi plants. The transcript level of HOX12 in the wild type was set as 1.0. Error bars indicate sd (n = 3).
(C) Schematic diagrams of the effector and reporter plasmids used in the transient transactivation assay in rice protoplasts. REN, Renilla luciferase; LUC, firefly luciferase.
(D) The 35S:REN-EUI1Pro:LUC reporter construct (C) transiently expressed in rice protoplasts together with control vector (Mock) or HOX12 effector, respectively. Error bars indicate sd (n = 5).
(E) Yeast one-hybrid assay testing the binding of HOX12 to the EUI1 promoter. Yeast cells containing EUI1Pro:LacZ were transformed with HOX12 fused with the AD and grown on medium containing X-Gal. Coexpression of AD/LacZ, AD-HOX12/LacZ, and AD/EUI1Pro:LacZ was used as the negative controls.
(F) Schematic diagram of the EUI1 promoter showing the potential HOX12 binding sites (white and black triangles). The translational start sites (ATG) are shown as +1. Numbers above the diagram indicate the distance away from ATG. Probes (F1, F2, F3, and F4) indicate DNA fragments (F1, F2, F3, and F4) used for yeast one-hybrid experiments. DNA fragments (P1, P2, P3, and P4) were used for ChIP.
(G) Yeast one-hybrid assay showing that HOX12 binds to the promoter regions of EUI1. wt and mu indicate wild-type and mutant forms of the fragments, respectively. mu, mutated fragments in which the AATAATT and TAATTA motifs were replaced with GGTGGTT and TGGTTG, respectively.
(H) EMSA showing that HOX12 binds to the AATAATT and TAATTA motifs of the EUI1 promoter. Competition for binding was performed using 50× and 250× competitive probes; GST was used as a negative control.
(I) ChIP assays showing that HOX12 binds to the promoter of EUI1 in vivo. Immunoprecipitation was performed with anti-HOX12 antibody. Immunoprecipitated chromatin was analyzed by qRT-PCR using primers corresponding to the amplicons represented by the schematic diagram of the EUI1 promoter (F). qRT-PCR enrichment was calculated by normalizing to Ubq2 and to the total input of each sample. Segment C (located in the EUI1 coding region) was used as a negative control. Values are means ± sd of three technical replicates.
To test whether HOX12 has DNA binding specificity for the EUI1 promoter, the EUI1 promoter region was cloned in front of a LacZ reporter gene to form reporter construct, and the HOX12 coding sequence was fused with the yeast activation domain (AD) to form the effector AD-HOX12 construct (Figure 6E). Both the effector AD-HOX12 (AD alone as a negative control) and the reporter constructs were cotransformed into yeast. As shown in Figure 6E, HOX12 could bind to the EUI1 promoter, while AD alone did not. These results suggest that this EUI1 region contains HOX12 binding sequences (Figure 6F). Previous studies suggested HD-ZIP I proteins directly bind target genes with the pseudopalindromic binding site CAATNATTG or the consensus binding site (TAATTA) in their promoter sequences (Sessa et al., 1993; Ades and Sauer, 1994; Johannesson et al., 2001). Promoter analysis revealed that the EUI1 promoter contains similar cis-element binding sequences. Therefore, four EUI1 promoter regions (F1, F2, F3, and F4) containing predicted similar cis-elements were selected for the yeast one-hybrid assays. The F1 promoter (50 bp) fragment contained the 7-bp sequence AATAATT, whereas the others (F2, F3, and F4; 50 bp) contained the 6-bp sequence TAATTA. Yeast one-hybrid assays showed that AD-HOX12 fusion protein, but not AD alone, could bind to a 50-bp fragment containing the AATAATT or TAATTA motifs of EUI1 and strongly activated the expression of the LacZ reporter gene. Point mutations in the cis-elements (AATAATT changed to GGTGGTT and TAATTA changed to TGGTTG) of EUI gene indeed abolished LacZ activation (Figure 6G). To further investigate in vitro binding of HOX12 to the EUI1 promoter region, electrophoretic mobility shift assays (EMSAs) were conducted with GST-HOX12 protein. Shifted bands were clearly detected when probes containing AATAATT and TAATTA in the EUI1 promoter region were incubated with HOX12 protein. By contrast, shifted bands were not observed when the probes were incubated with GST protein (Figure 6H). Finally, we used chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) to demonstrate that HOX12 also binds in vivo to the EUI1 promoter (Figure 6I; Supplemental Figure 4). Thus, our data demonstrate that HOX12 is an upstream transcriptional regulator of EUI1.
HOX12 Is Preferentially Expressed in the Panicle, Overlapping with EUI1
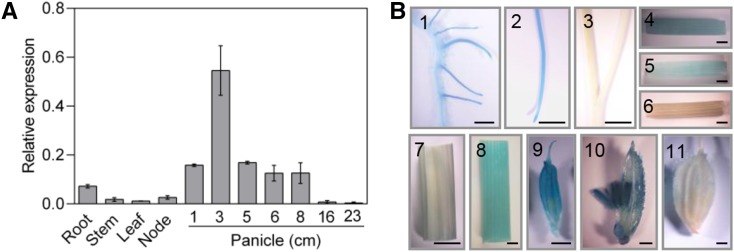
As reported previously, EUI1 is expressed preferentially in young panicles (Luo et al., 2006). To examine whether the spatiotemporal activity of HOX12 and EUI1 overlaps and meets the requirement for their interactions in vivo, we investigated the spatiotemporal expression pattern of HOX12 by qRT-PCR assays using total RNA samples from different organs. The results reveal that HOX12 was preferentially expressed in panicles, followed by roots, while low expression was detected in leaves (Figure 7A). In addition, HOX12 mRNA levels were higher in the node than in the stem. We then examined the developmental regulation of HOX12 expression in panicles using panicles 1 to 23 cm in length. The expression of HOX12 was lower at the early stage, peaked at 3 cm, and then gradually decreased as the panicle matured (Figure 7A).
Figure 7.
Expression Analysis of HOX12.
(A) qRT-PCR analysis of HOX12 expression in different tissues. Numbers indicate the panicle length in centimeters. Error bars indicate sd (n = 3).
(B) HOX12Pro:GUS expression patterns in transgenic rice plants. (1) Primary root and lateral roots. Bar = 1 mm. (2) Root tip. Bar = 5 mm. (3) Shoot of the seedling. Bar = 5 mm. (4) Divisional zone of the uppermost internode. Bar = 1 mm. (5) Elongating region of the uppermost internode. Bar = 1 mm. (6) Elongated region of the uppermost internode. Bar = 1 mm. (7) Mature leaf blade. Bar = 1 cm. (8) Mature leaf sheath. Bar = 1 mm. (9) Young floret. Bar = 1 mm. (10) Anthers. Bar = 1 mm. (11) Mature floret. Bar = 1 mm.
To obtain more detailed information about the spatial pattern of HOX12 expression, the promoter fragment of HOX12 was fused with the GUS reporter gene and transgenic plants were generated (Figure 7B). Eight HOX12Pro:GUS transgenic lines showed similar staining patterns. During the vegetative phase, GUS was expressed in roots (Figure 7B, parts 1 and 2). GUS signals were very weak in the leaves and the basal parts of shoots at the seedling stage (Figure 7B, part 3). At the reproductive phase, GUS signals were detected in the divisional and elongating regions of the uppermost internode (Figure 7B, parts 4 to 6; Supplemental Figure 5). Also, strong GUS activity was observed in the floral organs of young panicles, including anthers, but not in mature leaves or florets (Figure 7B, parts 7 to 11). These results demonstrate that the overlapping expression patterns of HOX12 and EUI1 meet the requirements for their interactions in vivo.
A Functional EUI1 Is Required for HOX12 to Regulate Panicle Exsertion
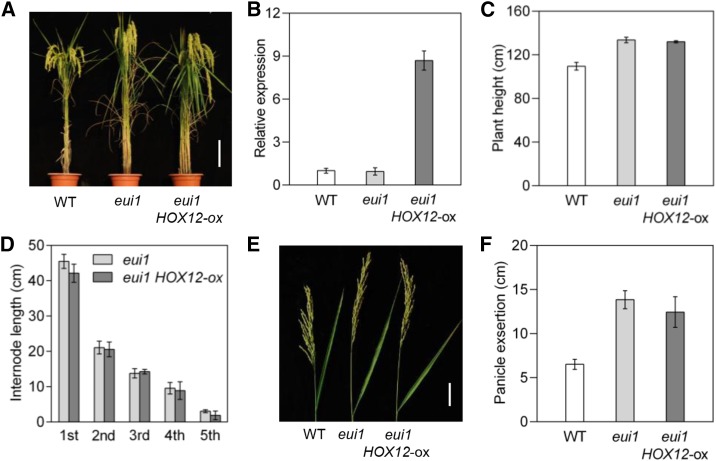
Previously, it was shown that mutation of EUI1 leads to increased bioactive GA levels and an extremely elongated uppermost internode (Luo et al., 2006; Zhu et al., 2006). To substantiate our premise that panicle exsertion is modulated via the HOX12-EUI1 regulatory cascade, HOX12 was overexpressed in the eui1 mutant background (Figure 8A). qRT-PCR analysis confirmed that HOX12 expression was markedly increased in these plants compared with the wild type and the eui1 mutant (Figure 8B). Intriguingly, the eui1 HOX12-ox plants consistently showed increased plant height compared with wild-type plants, a phenotype similar to that of the rice eui1 mutant (Figures 8C and 8D). In addition, the enhanced panicle exsertion phenotype of eui1 HOX12-ox was comparable to that of the eui1 mutant, although these plants were slightly shorter than the eui1 mutant (Figures 8E and 8F). These observations are consistent with the model that HOX12 acts upstream of EUI1 in a regulatory cascade for panicle exsertion.
Figure 8.
HOX12 Genetically Mediates EUI1 Activity.
(A) Morphological phenotypes of wild-type (cv Zhonghua11), eui1, and eui1 HOX12-ox plants. Bar = 20 cm.
(B) Expression levels of HOX12 in wild-type, eui1, and eui1 HOX12-ox plants by qRT-PCR analysis. The transcript level of HOX12 in the wild type was set as 1.0. Error bars indicate sd (n = 3).
(C) Quantification of the height of wild-type, eui1, and eui1 HOX12-ox plants. Error bars indicate sd (n = 15).
(D) Individual internode lengths of eui1 and eui1 HOX12-ox plants at the mature stage. Error bars indicate sd (n = 15).
(E) Panicle exsertion of wild-type, eui1, and eui1 HOX12-ox plants at the mature stage. Bar = 5 cm.
(F) Quantification of panicle exsertion of wild-type, eui1, and eui1 HOX12-ox plants. Error bars indicate sd (n = 15).
DISCUSSION
Our findings demonstrate that the HD-ZIP I transcription factor HOX12 is involved in regulating panicle exsertion. Gain-of-function ree1-D mutants show a dwarfism phenotype with no panicle exsertion. In contrast, diminished HOX12 expression by RNAi enhances panicle exsertion. Panicle exsertion principally depends on elongation of the uppermost internode. Internodal elongation is based on increased cell division and/or cell elongation. GAs are crucial phytohormones that play a critical role in this regulatory mechanism. GA1 and GA4, which are biosynthesized by plants, are thought to function as the bioactive forms of GA (Schomburg et al., 2003; Eriksson et al., 2006). The bioactivity of GA4 is higher than that of GA1 in both Arabidopsis and rice, which is presumably attributed to their binding affinity to the GA receptor GIBBERELLIN INSENSITIVE1 (Ueguchi-Tanaka et al., 2005). The presence of GAs with different activities (weak or strong) may be helpful for fulfilling different growth requirements (Hedden and Thomas, 2012). In rice, GA1 is the predominant bioactive form in vegetable tissues, while GA4 strongly accumulates in panicles (Hirano et al., 2008). EUI1 encodes a P450 monooxygenase that epoxidizes GAs (GA4, GA9, and GA12) in a deactivation reaction. The eui1 mutants exhibit enhanced internode elongation and panicle exsertion; accordingly, increased amounts of bioactive GA4 (a 13-H GA) and GA1 (a 13-OH GA) accumulate in the uppermost internodes of these mutants (Luo et al., 2006; Zhu et al., 2006). EUI1 is considered to function mainly in the uppermost internode. In fact, this gene is highly expressed in anthers and spikelets but is expressed at relatively low levels in the uppermost internodes (Magome et al., 2013). Deactivation of bioactive GA4 in anthers is catalyzed by EUI1, which epoxidizes GAs and regulates the influx of GA4 into the stem from panicles (Hedden and Thomas, 2012). Consequently, when EUI1 loses its function, increased amounts of GA4 flow into the uppermost internode, leading to the elongation of this internode. Recently, two homologous genes of EUI1, CYP714B1 and CYP714B2, were functionally characterized in rice. Compared with EUI1, CYP714B1 and CYP714B2 are highly expressed in spikelets and in the uppermost internodes of adult plants. However, cyp714b1 or cyp714b2 single mutant seedlings show no obvious change in 13-OH GA levels compared with the wild type, but the cyp714b1 cyp714b2 double mutant has a longer uppermost internode (Magome et al., 2013). These findings indicate that different CYP714 subfamilies play distinct roles in the growth and development of rice.
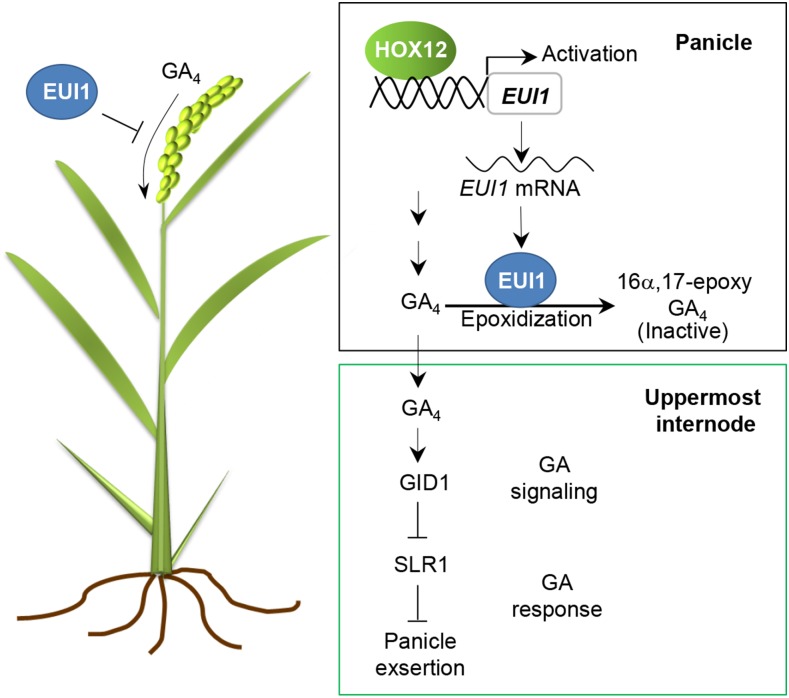
Our results demonstrate that HOX12 participates in the regulatory network orchestrating panicle exsertion by directly regulating EUI1 expression. Activation of HOX12 leads to GA deficiency, with a dwarf phenotype and defective panicle exsertion, which is also observed in EUI1 overexpressors. On the contrary, HOX12 knockdown plants exhibit elongated internodes with enhanced panicle exsertion, which is similar to the eui1 mutant. Both HOX12 knockdown and eui1 plants are morphologically normal until increased elongation of the uppermost internode occurs at the heading stage. Quantitative analysis of GA biosynthetic intermediates demonstrated that non-13-hydroxy GAs (GA12, GA15, GA24, GA9, and GA4) accumulate in the uppermost internodes of HOX12 knockdown plants, which was also observed in eui1 mutants (Luo et al., 2006; Zhu et al., 2006). qRT-PCR analysis indicated that EUI1 mRNA was more abundant in ree1-D mutants than in wild-type plants, and its levels were reduced in HOX12 knockdown lines. The expression level of EUI1 is positively regulated by HOX12, which was confirmed by transient transcriptional activity assays in rice protoplasts. Moreover, yeast one-hybrid analysis, EMSA, and ChIP-qPCR assays demonstrated that HOX12 can directly bind to the EUI1 promoter. Expression profiling revealed that EUI1 and HOX12 are both preferentially expressed in young panicles. These overlapping expression patterns meet the requirement for their interactions in vivo. Notably, when HOX12 was overexpressed in the absence of a functional EUI1 gene (in the eui1 mutant background), the elongated uppermost internode phenotype still existed (Figure 8A), revealing that EUI1 is a key target of HOX12. These results are consistent with the model that HOX12 acts upstream of EUI1 in a regulatory cascade for panicle exsertion. Based on these results, we propose a working model of the role of HOX12 in developmental process (Figure 9). EUI1 acts as a switch and regulates the influx of GA4 into the stem from panicle. HOX12 directly regulates the expression of EUI1 by binding to its promoter. Enhanced HOX12 levels promote EUI1 expression and inhibit GA4 accumulation. By contrast, diminished HOX12 expression downregulates EUI1 activity and consequently promotes the influx of GA4 into the stem, leading to enhanced panicle exsertion.
Figure 9.
A Proposed Working Model of the Molecular Actions of HOX12 during Development.
During the reproductive stage, GA4 accumulates in the panicle. EUI1 encodes a cytochrome P450 monooxygenase that deactivates bioactive GAs, including GA4. EUI1 acts as a switch that regulates the influx of GA4 into the stem from the panicle. HOX12 binds to the promoter and activates the expression of EUI1. Enhanced HOX12 levels promote EUI1 expression and inhibit GA4 accumulation. In contrast, diminished HOX12 expression downregulates EUI1 activity and consequently promotes the influx of GA4 into the stem, leading to enhanced panicle exsertion.
It should be pointed out that the GA4 levels in HOX12 knockdown plants are modest compared with those of the eui mutant (Zhu et al., 2006), which might be partly due to different genetic backgrounds, as well as the different tissues sampled and the different sampling times. In this study, the entire uppermost internode of the plant was harvested 1 d before flowering, and the GA4 levels changed greatly during the reproductive stage. Previously, we found that the increased lengths of panicle exsertion and uppermost internode elongation of the eui1 mutants significantly differ in the indica and japonica backgrounds (Luo et al., 2006).
Notably, the dwarf phenotype of homozygous ree1-D mutants can be rescued by crossing to the GA20ox1 overexpressor. GA20ox1 is involved in the late steps of the GA biosynthetic pathway to form bioactive GAs, including GA1 and GA4. As a result, the ree1-D GA20-1ox double mutant exhibited a GA overdose phenotype. However, the phenotypes of eui1 HOX12-ox were still not completely identical to those observed in eui1. Moreover, heterozygous rather than homozygous ree1-D mutants can be rescued by crossing to eui1 (Supplemental Figure 6). We speculate that severe dwarfing in homozygous ree1-D plants during the vegetable stage cannot be modified by the eui1 mutation, as EUI1 mainly acts during the reproductive stage. Undoubtedly, EUI1 is not the only target of HOX12. HOX12 likely has other targets during the regulation of plant development. Indeed, we identified other GA metabolism genes that were also affected in the HOX12 knockdown plants (Supplemental Figure 6E). Whether HOX12 interacts with these genes to regulate a specific developmental process requires further investigation.
HD-ZIP I transcription factors are involved in many developmental and stress-related processes, such as plant development, responses to stress and other environmental stimuli, and so on. Our results show that an increased level of HOX12 transcripts is induced by salt, drought, and cold stress (Supplemental Figure 7A). HOX12 expression is also induced within 15 min of ABA treatment in roots and reaches maximum levels after 2 h in both roots and shoots (Supplemental Figure 7B). In addition, indole-3-acetic acid and jasmonic acid (JA) trigger HOX12 expression in roots (Supplemental Figure 7C). Moreover, GA3 and JA slightly induce HOX12 expression after 2 h of treatment in shoots (Supplemental Figure 7C). These observations suggest that HOX12 might be involved in multiple hormone responses. In Arabidopsis, at least seven of the nine type 2C protein phosphatases (PP2Cs) from clade A act as negative regulators of the ABA pathway (Ma et al., 2009a). Os PYL/RCAR5, an ortholog of At PYL/RCAR, acts as a functional ABA receptor in rice, interacting with Os PP2Cs, which are orthologs of Arabidopsis subclass A PP2C. Notably, Arabidopsis HD-ZIP I protein HB6 is a target of the PP2C phosphatase ABI1 and regulates hormonal responses (Himmelbach et al., 2002). Ten genes encoding subfamily A PP2Cs are present in the rice genome. PP2C30 interacts with ABA receptor PYL/RCAR5 and SnRK2 subclass II gene SAPK2 to form a core ABA signaling pathway, which regulates ABA-dependent gene expression (Kim et al., 2012). We found that the HOX12 protein interacted with PP2C30 in yeast two-hybrid and BiFC experiments (Supplemental Figures 7D and 7E). PP2C30 is a possible candidate that acts as an upstream regulator of HOX12 expression. However, detailed functional analyses are required to unravel under which physiological conditions and in which cells PP2Cs interact and regulate HOX12 expression.
Plants adjust their final height in response to the prevailing environmental conditions by increasing or decreasing their growth rate in response to external and internal signals. Therefore, plants have evolved complex strategies to perceive stress signals and to further translate the perception into effective plant responses. Also, panicle exsertion is a dynamic process that is governed by two major counteracting phytohormones, ABA and GA, in response to various environmental factors (Muthurajan et al., 2011). As these two hormones act through complex crosstalk rather than through independent pathways, integration of their mutual interaction is critical for determining whether a plant should initiate development. An important aspect of our findings is that HOX12 expression itself is stimulated by ABA, thereby constituting a positively acting ABA-HOX12-EUI1 activator loop for regulating cell expansion upon detection of external and internal signals. EUI1 activity is induced by endogenous and exogenous ABA and GA (Zhang et al., 2008; Yaish et al., 2010; Magome et al., 2013). AP2-39, encoding a member of the AP2 family, regulates key interactions between ABA and GA in rice. EUI1 is directly regulated by Os AP2-39, which in turn regulates plant growth (Yaish et al., 2010). Our results demonstrate that HOX12 is not only a key player that regulates the GA pathway, but it also functions as a junction for the crosstalk between the ABA and GA signaling pathways. This mechanism may enable plants to regulate EUI1 activity with great flexibility and accuracy. However, this hypothesis requires additional support.
Previous studies have shown that HOX12 is highly expressed in panicles (Agalou et al., 2008). The HOX12 expression pattern is conserved in rice, barley, sorghum, and maize, suggesting that the ancestral function of HOX12 might be associated with shaping the initial steps of grass inflorescence architecture. In the rice ree1-D mutant, inflorescence formation is repressed. There are 14 putative HD-ZIP I genes in the rice genome. An interesting point raised by Valdés et al. (2012) is that HD-ZIP genes that share targets but have different expression patterns or different reactions to specific external conditions may play similar roles in modulating ABA signal perception. The data regarding the expression of HD-ZIP I HOXs in rice through different developmental stages obtained from the Rice Expression Profile Database (RiceXPro) show that different HD-ZIP I genes have different expression patterns (Sato et al., 2013; RiceXPro, http://ricexpro.dna.affrc.go.jp), indicating their different biological functions. HOX12 and its closest homolog HOX14 have identical binding domains, suggesting that they could bind to the same DNA sequences. Indeed, both proteins can interact with the EUI1 promoter in yeast (Supplemental Figure 8). However, they are not interchangeable, since HOX12 and HOX14 have distinct expression patterns. A difference in their expression patterns is observed during the reproductive phase, as HOX12 is strongly expressed in anther, while HOX14 transcripts are barely detected (RiceXPro).
In summary, our data allow us to incorporate the role of HOX12 into the regulatory network that orchestrates panicle exsertion. A further understanding of the mechanisms that sustain panicle development under stressful conditions will deepen our understanding of how plants adapt to environmental changes. Our findings not only provide important genetic evidence of the functions of an HD-ZIP I transcription factor, but they also help unravel the mechanism that functions in the regulation of panicle exsertion in rice. Identification of HOX12-interacting proteins and genes that are directly regulated by HOX12 will further elucidate its function in the GA pathway.
METHODS
Plant Materials and Growth Conditions
The activation-tagging rice (Oryza sativa) mutant ree1-D was isolated by screening our rice T-DNA activation population in the Nipponbare (japonica) background. The mutant and its origin cultivar, Nipponbare, were grown in the field under natural conditions or in Kimura nutrient solution in a growth chamber at 30°C/25°C (day/night) with a 14-h-light/10-h-dark cycle with ∼200 μmol m−2 s−1 photon density and 70% humidity. Field management adhered to normal agricultural practice. Seeds were immersed in water for 2 d and sown in a nursery bed. One-month-old seedlings were transplanted to a paddy field at a spacing of 20 × 35 cm. Mutant segregants were distinguished from normal segregants based on their dwarf phenotype. The eui1-4 mutant (japonica variety Zhonghua11 background), referred to herein as eui1, was described previously (Luo et al., 2006). To generate the double mutant, ree1-D heterozygotes were crossed with GA20-1ox or eui1, and F2 plants were phenotypically screened and genotyped.
PCR Genotyping of the ree1-D Allele
Genomic DNA was extracted from selected plants as described and used for PCR genotyping. The T-DNA flanking sequence of the ree1-D was determined by site-finding using the specific and arbitrary degenerate primers as described (Wang et al., 2011; Gao et al., 2014b). T-DNA flanking sequences were searched against the NCBI database using the BLASTN program (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Three primers, P1, P2, and P3, were designed for cosegregation analysis, with P1 and P2 corresponding to rice genomic sequences flanking the T-DNA insertion and P3 corresponding to the sequence of the T-DNA vector. All of the primers are listed in Supplemental Data Set 1.
Constructs for Rice Transformation
To generate the overexpression vector, the coding regions of Os03g0198500 and Os03g0198600 were amplified by PCR, cloned into the pEASY-Blunt vector (Transgen), and subcloned into the pCAMBIA2300 vector (Cambia) downstream of the maize (Zea mays) Ubiquitin promoter following KpnI and BamHI digestion.
To generate the HOX12Pro:GUS vector, the promoter region of HOX12 (2286 bp upstream of ATG) was amplified by PCR (see PCR primer sequences Pro-F and Pro-R in Supplemental Data Set 1) using Nipponbare genomic DNA as the template and inserted into the pCAMBIA2391Z (Cambia) binary vector containing the GUS reporter gene.
To generate the HOX12-RNAi vector, a 300-bp specific cDNA fragment of the HOX12 sequence was amplified using the first-strand cDNA derived from rice roots as the template and primers Ri-F and Ri-R. The PCR fragments were sequentially cloned into the BamHI-SalI and BglII-XhoI sites of the pUCC-RNAi vector (Luo et al., 2006) in the sense and antisense orientations. The fragment containing the inverted repeat segment of HOX12 was transferred into the PstI-SalI site of pCAMBIA2300 containing the rice ACTIN1 promoter and OCS terminator sequence, yielding the binary HOX12-RNAi vector.
All of the primers used to generate the above-mentioned constructs are listed in Supplemental Data Set 1, and all of the constructs were confirmed by sequencing. The constructs were introduced into Agrobacterium tumefaciens strain AGL1. Wild-type calli were used as the recipients for Agrobacterium-mediated transformation as described previously (Liu et al., 2007).
Total RNA Isolation and qRT-PCR Analysis
Total RNA from various tissues was extracted using TRIzol reagent (Invitrogen). One microgram of total RNA was used to synthesize cDNA using ReverTra Ace qPCR RT Master Mix (Toyobo). PCR amplification was performed with SYBR Green Real-Time PCR Master Mix reagent (Toyobo) on a real-time PCR detection system according to manufacturer's instructions (Bio-Rad CFX96). The rice housekeeping gene Ubiquitin2 (Ubq2) was used as an internal reference. Three biological repeats were performed for each analysis. The primers used for qRT-PCR are listed in Supplemental Data Set 1. The expression of each transcript was normalized against the amount of Ubq2 control transcript in each sample. Values are means ± sd of three biological repeats.
Hormone and Stress Treatments
For GA sensitivity analysis, the effect of GA3 on shoot elongation was quantified as previously described (Matsukura et al., 1998) with some modifications. Seeds of wild-type and mutant plants were dehusked and sterilized with 3% NaClO solution for 0.5 h, washed three times with sterile distilled water, and incubated in 6.9 μM uniconazole for 24 h to block endogenous GA biosynthesis, followed by sterile distilled water for an additional 24 h after washing out the uniconazole. These seeds were sown on 0.3% Phytagel-solidified half-strength Murashige and Skoog medium supplemented with GA3 at 10−10 to 10−4 M and grown for 5 d. The lengths of the second leaf sheaths were measured.
For abiotic and biotic stress treatments, the plants were grown hydroponically in normal Kimura solution for 7 d and transferred to nutrient solution containing 200 mM NaCl, 200 mM mannitol, 10 μM GA3, 50 μM ABA, 50 μM JA, 10 μM indole-3-acetic acid, or fresh nutrient solution (control plants). All treatments were performed with three independent biological replicates. For low-temperature treatment, plants were grown hydroponically in Kimura solution for 7 d under normal conditions and then transferred to a growth chamber at 4°C. After each treatment, tissues were immediately frozen in liquid nitrogen and stored at −80°C until further use.
Phylogenetic Analysis
The HOX12 protein sequence was used to search for the closest homologs from other plant species using the BLASTP program. Multiple sequence alignment of full-length protein sequences was performed using the ClustalW program, and the final alignment is available as Supplemental File 1. A neighbor-joining tree was constructed using MEGA 6 (Tamura et al., 2013).
Subcellular Localization
To construct GFP fusions with HOX12 and its variations, the coding sequence of HOX12 and truncated cDNA were fused in frame to the N terminus of the enhanced GFP coding sequence under the control of the CaMV 35S promoter and the OCS terminator in the pCAMBIA2300 vector. Primers used are listed in Supplemental Data Set 1. All of the constructs and the control were transfected into rice protoplasts using a polyethylene glycol-calcium-mediated method followed by an 18-h incubation to allow transient expression (Bart et al., 2006). Fluorescence imaging of the protoplasts was performed using a Leica TCS SP5 confocal laser scanning system. The excitation line for imaging the eGFP fusion was 488 nm.
Transactivation Activity Assay
To construct the serial vectors, the full-length coding sequence of HOX12 and sequences encoding N-terminal and C-terminal truncation products were amplified and cloned into pGBKT7 fused with the GAL4 DNA binding domain. The vectors were then transformed into Saccharomyces cerevisiae strain AH109. Transactivation activity assays were performed using the GAL4-based Matchmaker Two-Hybrid System 3 (Clontech). The transformed yeast was cultured in complete synthetic yeast medium without Trp and subsequently grown on selective medium lacking Trp, His, and Ade. Primers are listed in the Supplemental Data Set 1.
Dual-Luciferase Assay
For the transient transcriptional activity assay, a construct harboring LUC under the control of the EUI1 promoter (2 kb upstream of ATG) sequence in the pGreenII 0800-LUC vector was generated as a reporter (Hellens et al., 2005). The Renilla luciferase (REN) gene under the control of the 35S promoter in the pGreenII 0800-LUC vector was used as an internal control. The HOX12-eGFP construct described above was used as the effector. Empty eGFP vector was used as the control for the effector. Rice shoot protoplasts were prepared and transfected using a polyethylene glycol-calcium-mediated method followed by an 18-h incubation to allow transient expression (Bart et al., 2006). Firefly LUC and REN activities were measured with a Dual-Luciferase reporter assay kit using a GloMax 20/20 luminometer (Promega). The LUC activity was normalized to REN activity and LUC/REN ratios were calculated. For each plasmid combination, five independent transformations were performed. All primers used for these constructs are listed in Supplemental Data Set 1.
GUS Staining
Plant tissues were collected from transgenic plants containing the HOX12Pro:GUS transgenes. Histochemical GUS staining was performed as described previously (Jefferson, 1989). The tissues were incubated in 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid solution for 8 h at 37°C and dehydrated in an ethanol series (70, 85, 95, and 100%) to remove the chlorophyll. The stained tissues were viewed under a stereo microscope (Olympus SZX16) and photographed using a digital camera (Nikon D700).
Yeast Assays
To prepare constructs for the yeast one-hybrid assay, the promoter region of EUI1 (2 kb upstream of ATG) was amplified and cloned into the EcoRI-XhoI sites in the pLacZi2μ vector (Lin et al., 2007), resulting in the EUI1Pro:LacZ reporter constructs. To generate AD-HOX12, the full-length coding sequence of HOX12 was amplified by PCR with the respective primers and cloned into the EcoRI-XhoI sites of the pJG4-5 vector (Clontech). The yeast one-hybrid assay was performed according to the Yeast Protocols Handbook (Clontech). Briefly, the AD fusion constructs were cotransformed with various LacZ reporter plasmids into yeast strain EGY48. Transformants were grown on SD/-Trp-Ura dropout plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) for blue color development.
Yeast two-hybrid assays were conducted using the Matchmaker GAL4 two-hybrid system 3 (Clontech). The full-length HOX12 and truncated cDNA PCR products were cloned into pGADT7 using an In-Fusion HD cloning kit (Clontech) and sequenced. To confirm the interaction, the full-length encoding sequence of PP2C30 was cloned into the vector pGBKT7 to yield PP2C30-BD. Primers used for generating various clones are listed in Supplemental Data Set 1.
EMSA
To construct plasmids for the expression of recombinant HOX12 protein in Escherichia coli, the full-length coding sequence of HOX12 was cloned into pGEX-4T-3 (GE Healthcare) and transformed into E. coli strain BL21 (DE3). The recombinant proteins were affinity purified using Glutathione Sepharose 4B beads (GE Healthcare). Oligonucleotide probes containing AATAATT and TAATTA motifs were synthesized and labeled with biotin at the 3′ end (Invitrogen). For nonlabeled probe competition, nonlabeled probe was added to the reactions. EMSA was performed using a chemiluminescent EMSA kit (Beyotime). Probe sequences are shown in Supplemental Data Set 1.
BiFC Assays
The full-length coding sequence of HOX12 was PCR amplified and cloned into pSCYNE and pSCYCE (Waadt et al., 2008), resulting in HOX12-nCFP and HOX12-cCFP, respectively. Full-length HOX14 and PP2C30 coding sequences were inserted into pSCYCE to generate HOX14-nCFP and PPC30-nCFP, respectively. Primers used for generating fusion constructs are listed in Supplemental Data Set 1. Rice protoplast preparation and plasmid transformation were performed according to previous methods (Bart et al., 2006). Empty vectors of BiFC constructs were used as a negative control. Fluorescence imaging of the protoplasts was performed using a Leica TCS SP5 confocal laser scanning system.
Quantification of Endogenous GA
For GA measurements, the uppermost internodes of the HOX12-RNAi transgenic and wild-type plants were harvested 1 d before flowering and stored at −80°C until further use. Quantification of endogenous GAs was conducted as previously described (Chen et al., 2012). Each series of experiments was performed in biological triplicates.
ChIP-qPCR
The peptide epitope located between amino acid 78 and 87 of the HOX12 sequence (KKERKLETPR) was used to produce a specific mouse monoclonal antibody, which was performed by Abmart. Briefly, ∼2 g of rice seedlings was cross-linked in 1% formaldehyde under a vacuum, and cross-linking was stopped with 0.125 M glycine. The sample was ground to a powder in liquid nitrogen, and their nuclei were isolated. Anti-HOX12 (1:150 dilution) was used to immunoprecipitate the protein-DNA complex, and the precipitated DNA was recovered and analyzed by PCR. Chromatin precipitated without antibody was used as a negative control, while the isolated chromatin before precipitation was used as an input control. Primers used for ChIP-qPCR are listed in Supplemental Data Set 1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database under the following accession numbers: HOX12 (Os03g0198600), HOX14 (Os07g0581700), EUI1 (Os05g0482400), Ubq2 (Os02g0161900), PP2C30 (Os03g0268600), GA20ox1 (Os03g0856700), GA2ox3 (Os01g0757200), GA2ox4 (Os05g0514600), GA2ox5 (Os07g0103500), and GA2ox8 (Os05g0560900).
Supplemental Data
Supplemental Figure 1. Phenotypes of Os01g0198500 Overexpression Transgenic Plants.
Supplemental Figure 2. Phenotypes of Os01g0198600/HOX12 Overexpression Transgenic Plants.
Supplemental Figure 3. A Phylogenetic Tree of the HOX12-Like Proteins in Plants.
Supplemental Figure 4. Specificity of the Rice HOX12 Monoclonal Antibody.
Supplemental Figure 5. Representative Stem of a HOX12Pro:GUS Plant.
Supplemental Figure 6. EUI1 Is Required for HOX12 to Regulate Panicle Exsertion.
Supplemental Figure 7. HOX12 Expression Is Regulated by Environmental and Hormonal Stimuli.
Supplemental Figure 8. Yeast One-Hybrid Assay Testing the Binding of HOX14 to the EUI1 Promoter.
Supplemental Data Set 1. List of Primers Used in This Study.
Supplemental File 1. Alignment Used for Phylogenetic Analysis.
Supplementary Material
Acknowledgments
We thank Jiayang Li for the pSCYNE and pSCYCE plasmids, Xiaoya Chen for the pGreenII 0800-LUC plasmid, and Rongcheng Lin for the pJG4-5 and pLacZi2μ plasmids. We thank Shouyun Cao for all rice transformation and Gupo Li for plant management in the field. We thank Baodong Cai for the GA measurements. We also thank the Chu laboratory members for their helpful comments and discussions during the article preparation. This research was supported by grants from the National Natural Science Foundation of China (31430063 and 91335203), the Transgenic Research Program of the Ministry of Agriculture (2014ZX08001-004-001), and the State Key Laboratory of Plant Genomics.
AUTHOR CONTRIBUTIONS
C.C. conceived and supervised the whole project, analyzed data, and wrote the article. S.G. designed and performed experiments, analyzed data, and wrote the article. J.F. conceived the project and analyzed the data. F.X. and W.W. assisted in BiFC and subcellular localization assays. All authors discussed the results and contributed to the final article.
Glossary
- GA
gibberellin
- ABA
abscisic acid
- BR
brassinosteroid
- BiFC
bimolecular fluorescence complementation
- EMSA
electrophoretic mobility shift assay
- ChIP-qPCR
chromatin immunoprecipitation-quantitative PCR
- JA
jasmonic acid
Footnotes
Articles can be viewed online without a subscription.
References
- Ades S.E., Sauer R.T. (1994). Differential DNA-binding specificity of the engrailed homeodomain: the role of residue 50. Biochemistry 33: 9187–9194. [DOI] [PubMed] [Google Scholar]
- Agalou A., et al. (2008). A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol. Biol. 66: 87–103. [DOI] [PubMed] [Google Scholar]
- Arce A.L., Raineri J., Capella M., Cabello J.V., Chan R.L. (2011). Uncharacterized conserved motifs outside the HD-Zip domain in HD-Zip subfamily I transcription factors; a potential source of functional diversity. BMC Plant Biol. 11: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F.D., Manavella P.A., Dezar C.A., Chan R.L. (2007). The true story of the HD-Zip family. Trends Plant Sci. 12: 419–426. [DOI] [PubMed] [Google Scholar]
- Bart R., Chern M., Park C.J., Bartley L., Ronald P.C. (2006). A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt R., Cabedo M., Xie Y., Wenkel S. (2014). Homeodomain leucine-zipper proteins and their role in synchronizing growth and development with the environment. J. Integr. Plant Biol. 56: 518–526. [DOI] [PubMed] [Google Scholar]
- Capella M., Ribone P.A., Arce A.L., Chan R.L. (2015). Arabidopsis thaliana HomeoBox 1 (AtHB1), a Homedomain-Leucine Zipper I (HD-Zip I) transcription factor, is regulated by PHYTOCHROME-INTERACTING FACTOR 1 to promote hypocotyl elongation. New Phytol. 207: 669–682. [DOI] [PubMed] [Google Scholar]
- Chang X., Donnelly L., Sun D., Rao J., Reid M.S., Jiang C.Z. (2014). A Petunia homeodomain-leucine zipper protein, PhHD-Zip, plays an important role in flower senescence. PLoS One 9: e88320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Jiang S., Zheng J., Lin Y. (2013). Improving panicle exsertion of rice cytoplasmic male sterile line by combination of artificial microRNA and artificial target mimic. Plant Biotechnol. J. 11: 336–343. [DOI] [PubMed] [Google Scholar]
- Chen M.L., Fu X.M., Liu J.Q., Ye T.T., Hou S.Y., Huang Y.Q., Yuan B.F., Wu Y., Feng Y.Q. (2012). Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC-ESI-Q-TOF-MS analysis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 905: 67–74. [DOI] [PubMed] [Google Scholar]
- Dai M., Hu Y., Ma Q., Zhao Y., Zhou D.X. (2008). Functional analysis of rice HOMEOBOX4 (Oshox4) gene reveals a negative function in gibberellin responses. Plant Mol. Biol. 66: 289–301. [DOI] [PubMed] [Google Scholar]
- Elhiti M., Stasolla C. (2009). Structure and function of homodomain-leucine zipper (HD-Zip) proteins. Plant Signal. Behav. 4: 86–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S., Böhlenius H., Moritz T., Nilsson O. (2006). GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18: 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D., Appiano M., Huibers R.P., Chen X., Loonen A.E., Visser R.G., Wolters A.-M.A., Bai Y. (2014a). Activation tagging of ATHB13 in Arabidopsis thaliana confers broad-spectrum disease resistance. Plant Mol. Biol. 86: 641–653. [DOI] [PubMed] [Google Scholar]
- Gao S., Fang J., Xu F., Wang W., Sun X., Chu J., Cai B., Feng Y., Chu C. (2014b). CYTOKININ OXIDASE/DEHYDROGENASE4 integrates cytokinin and auxin signaling to control rice crown root formation. Plant Physiol. 165: 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J.C., Hrmova M., Lopato S., Langridge P. (2011). Modulation of plant growth by HD-Zip class I and II transcription factors in response to environmental stimuli. New Phytol. 190: 823–837. [DOI] [PubMed] [Google Scholar]
- Hedden P., Thomas S.G. (2012). Gibberellin biosynthesis and its regulation. Biochem. J. 444: 11–25. [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson E., Olsson A.S., Johannesson H., Johansson H., Hanson J., Engström P., Söderman E. (2005). Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiol. 139: 509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A., Hoffmann T., Leube M., Höhener B., Grill E. (2002). Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 21: 3029–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K., Aya K., Hobo T., Sakakibara H., Kojima M., Shim R.A., Hasegawa Y., Ueguchi-Tanaka M., Matsuoka M. (2008). Comprehensive transcriptome analysis of phytohormone biosynthesis and signaling genes in microspore/pollen and tapetum of rice. Plant Cell Physiol. 49: 1429–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A. (1989). The GUS reporter gene system. Nature 342: 837–838. [DOI] [PubMed] [Google Scholar]
- Johannesson H., Wang Y., Engström P. (2001). DNA-binding and dimerization preferences of Arabidopsis homeodomain-leucine zipper transcription factors in vitro. Plant Mol. Biol. 45: 63–73. [DOI] [PubMed] [Google Scholar]
- Kim H., Hwang H., Hong J.W., Lee Y.N., Ahn I.P., Yoon I.S., Yoo S.D., Lee S., Lee S.C., Kim B.G. (2012). A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 63: 1013–1024. [DOI] [PubMed] [Google Scholar]
- Komatsuda T., et al. (2007). Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc. Natl. Acad. Sci. USA 104: 1424–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yuan L. (2010). Hybrid rice: genetics, breeding, and seed production. Plant Breed. Rev. 17: 15–158. [Google Scholar]
- Lin R., Ding L., Casola C., Ripoll D.R., Feschotte C., Wang H. (2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Hong Y., Yin M., Li C., Zhang K., Grierson D. (2008). A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 55: 301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Bai X., Wang X., Chu C. (2007). OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 164: 969–979. [DOI] [PubMed] [Google Scholar]
- Luo A., et al. (2006). EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol. 47: 181–191. [DOI] [PubMed] [Google Scholar]
- Luo D., et al. (2013). A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat. Genet. 45: 573–577. [DOI] [PubMed] [Google Scholar]
- Ma Y., et al. (2009b). Molecular analysis of rice plants harboring a multi-functional T-DNA tagging system. J. Genet. Genomics 36: 267–276. [DOI] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009a). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068. [DOI] [PubMed] [Google Scholar]
- Magome H., Nomura T., Hanada A., Takeda-Kamiya N., Ohnishi T., Shinma Y., Katsumata T., Kawaide H., Kamiya Y., Yamaguchi S. (2013). CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc. Natl. Acad. Sci. USA 110: 1947–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella P.A., Dezar C.A., Bonaventure G., Baldwin I.T., Chan R.L. (2008). HAHB4, a sunflower HD-Zip protein, integrates signals from the jasmonic acid and ethylene pathways during wounding and biotic stress responses. Plant J. 56: 376–388. [DOI] [PubMed] [Google Scholar]
- Matsukura C., Itoh S., Nemoto K., Tanimoto E., Yamaguchi J. (1998). Promotion of leaf sheath growth by gibberellic acid in a dwarf mutant of rice. Planta 205: 145–152. [Google Scholar]
- Meijer A.H., de Kam R.J., d’Erfurth I., Shen W., Hoge J.H. (2000). HD-Zip proteins of families I and II from rice: interactions and functional properties. Mol. Gen. Genet. 263: 12–21. [DOI] [PubMed] [Google Scholar]
- Muthurajan R., Shobbar Z.-S., Jagadish S.V., Bruskiewich R., Ismail A., Leung H., Bennett J. (2011). Physiological and proteomic responses of rice peduncles to drought stress. Mol. Biotechnol. 48: 173–182. [DOI] [PubMed] [Google Scholar]
- Nomura T., Magome H., Hanada A., Takeda-Kamiya N., Mander L.N., Kamiya Y., Yamaguchi S. (2013). Functional analysis of Arabidopsis CYP714A1 and CYP714A2 reveals that they are distinct gibberellin modification enzymes. Plant Cell Physiol. 54: 1837–1851. [DOI] [PubMed] [Google Scholar]
- Oikawa T., Koshioka M., Kojima K., Yoshida H., Kawata M. (2004). A role of OsGA20ox1, encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice. Plant Mol. Biol. 55: 687–700. [DOI] [PubMed] [Google Scholar]
- Olsson A.S., Engström P., Söderman E. (2004). The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol. Biol. 55: 663–677. [DOI] [PubMed] [Google Scholar]
- Pourkheirandish M., Wicker T., Stein N., Fujimura T., Komatsuda T. (2007). Analysis of the barley chromosome 2 region containing the six-rowed spike gene vrs1 reveals a breakdown of rice-barley micro collinearity by a transposition. Theor. Appl. Genet. 114: 1357–1365. [DOI] [PubMed] [Google Scholar]
- Ruberti I., Sessa G., Lucchetti S., Morelli G. (1991). A novel class of plant proteins containing a homeodomain with a closely linked leucine zipper motif. EMBO J. 10: 1787–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutger J., Carnahan H. (1981). A fourth genetic element to facilitate hybrid cereal production-a recessive tall in rice. Crop Sci. 21: 373–376. [Google Scholar]
- Sato Y., Takehisa H., Kamatsuki K., Minami H., Namiki N., Ikawa H., Ohyanagi H., Sugimoto K., Antonio B.A., Nagamura Y. (2013). RiceXPro version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 41: D1206–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M., Davis R.W. (1992). HD-Zip proteins: members of an Arabidopsis homeodomain protein superfamily. Proc. Natl. Acad. Sci. USA 89: 3894–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg F.M., Bizzell C.M., Lee D.J., Zeevaart J.A., Amasino R.M. (2003). Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15: 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G., Morelli G., Ruberti I. (1993). The Athb-1 and -2 HD-Zip domains homodimerize forming complexes of different DNA binding specificities. EMBO J. 12: 3507–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Yang C., He Z. (1987). Studies on eliminating panicle enclosure in WA Type MS line of rice (Oryza sativa subsp. indica). Chin. J. Rice Sci. 1: 95–99. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., Chow T.Y., Hsing Y.I., Kitano H., Yamaguchi I., Matsuoka M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698. [DOI] [PubMed] [Google Scholar]
- Valdés A.E., Overnäs E., Johansson H., Rada-Iglesias A., Engström P. (2012). The homeodomain-leucine zipper (HD-Zip) class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities. Plant Mol. Biol. 80: 405–418. [DOI] [PubMed] [Google Scholar]
- Waadt R., Schmidt L.K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56: 505–516. [DOI] [PubMed] [Google Scholar]
- Wang H., Fang J., Liang C., He M., Li Q., Chu C. (2011). Computation-assisted SiteFinding- PCR for isolating flanking sequence tags in rice. Biotechniques 51: 421–423. [DOI] [PubMed] [Google Scholar]
- Wang Y., Henriksson E., Söderman E., Henriksson K.N., Sundberg E., Engström P. (2003). The Arabidopsis homeobox gene, ATHB16, regulates leaf development and the sensitivity to photoperiod in Arabidopsis. Dev. Biol. 264: 228–239. [DOI] [PubMed] [Google Scholar]
- Whipple C.J., Kebrom T.H., Weber A.L., Yang F., Hall D., Meeley R., Schmidt R., Doebley J., Brutnell T.P., Jackson D.P. (2011). grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc. Natl. Acad. Sci. USA 108: E506–E512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaish M.W., El-Kereamy A., Zhu T., Beatty P.H., Good A.G., Bi Y.M., Rothstein S.J. (2010). The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 6: e1001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Zhang S., Huang R., Zhang Q. (2005). The research and development of eui-hybrid rice. Hybrid Rice 20: 11–14. [Google Scholar]
- Yang R., Zhang S., Huang R., Yang S., Zhang Q. (2002). Breeding technology of eui hybrids of rice. Sci. Agric. Sin. 35: 233–237. [Google Scholar]
- Zhang Q., Yang R. (2003). The effect of different eui genes on biological characters of e-hybrid rice. Sci. Agric. Sin. 36: 735–739. [Google Scholar]
- Zhang S., Haider I., Kohlen W., Jiang L., Bouwmeester H., Meijer A.H., Schluepmann H., Liu C.M., Ouwerkerk P.B. (2012). Function of the HD-Zip I gene Oshox22 in ABA-mediated drought and salt tolerances in rice. Plant Mol. Biol. 80: 571–585. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang B., Yan D., Dong W., Yang W., Li Q., Zeng L., Wang J., Wang L., Hicks L.M., He Z. (2011). Two Arabidopsis cytochrome P450 monooxygenases, CYP714A1 and CYP714A2, function redundantly in plant development through gibberellin deactivation. Plant J. 67: 342–353. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhu Y., Peng Y., Yan D., Li Q., Wang J., Wang L., He Z. (2008). Gibberellin homeostasis and plant height control by EUI and a role for gibberellin in root gravity responses in rice. Cell Res. 18: 412–421. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Ma Q., Jin X., Peng X., Liu J., Deng L., Yan H., Sheng L., Jiang H., Cheng B. (2014). A novel maize homeodomain-leucine zipper (HD-Zip) I gene, Zmhdz10, positively regulates drought and salt tolerance in both rice and Arabidopsis. Plant Cell Physiol. 55: 1142–1156. [DOI] [PubMed] [Google Scholar]
- Zhou W., Malabanan P.B., Abrigo E. (2015). OsHox4 regulates GA signaling by interacting with DELLA-like genes and GA oxidase genes in rice. Euphytica 201: 97–107. [Google Scholar]
- Zhu Y., et al. (2006). ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18: 442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.