The signaling nucleotide guanosine tetraphosphate, a potent inhibitor of chloroplast gene expression, mediates chloroplast and nucleocytoplasmic cooperation during plant growth and development.
Abstract
The chloroplast originated from the endosymbiosis of an ancient photosynthetic bacterium by a eukaryotic cell. Remarkably, the chloroplast has retained elements of a bacterial stress response pathway that is mediated by the signaling nucleotides guanosine penta- and tetraphosphate (ppGpp). However, an understanding of the mechanism and outcomes of ppGpp signaling in the photosynthetic eukaryotes has remained elusive. Using the model plant Arabidopsis thaliana, we show that ppGpp is a potent regulator of chloroplast gene expression in vivo that directly reduces the quantity of chloroplast transcripts and chloroplast-encoded proteins. We then go on to demonstrate that the antagonistic functions of different plant RelA SpoT homologs together modulate ppGpp levels to regulate chloroplast function and show that they are required for optimal plant growth, chloroplast volume, and chloroplast breakdown during dark-induced and developmental senescence. Therefore, our results show that ppGpp signaling is not only linked to stress responses in plants but is also an important mediator of cooperation between the chloroplast and the nucleocytoplasmic compartment during plant growth and development.
INTRODUCTION
More than one billion years ago, a eukaryotic cell engulfed and assimilated a cyanobacterium to give rise to a new organelle, the chloroplast, giving rise to all the photosynthetic eukaryotes, a vast complex of primary producing organisms (algae and plants) (Reyes-Prieto et al., 2007). Following endosymbiosis, many of the original cyanobacterial genes migrated to the nucleus, and the gene products were directed to the chloroplast. The ∼100 genes that remained in the chloroplast genome are involved in photosynthesis, metabolism, and organellar transcription and translation (Green, 2011; Jarvis and López-Juez, 2013). These processes involve proteins encoded by both the chloroplast and nuclear genomes. Tight coordination between chloroplastic and nuclear gene expression is therefore required for the biogenesis, operation, and differentiation of the chloroplast (Jarvis and López-Juez, 2013). Strikingly, chloroplasts have retained core elements of bacterial signaling pathways that are now thought to be involved in regulating chloroplast function (Puthiyaveetil et al., 2008; Masuda, 2012). One of these pathways is the stringent response, which is probably the most important stress signaling pathway in bacteria (Dalebroux and Swanson, 2012). In bacteria, the stringent response is characterized by the stress-induced accumulation of two nucleotides, guanosine penta- and tetraphosphate (hereafter referred to as ppGpp), that directly and indirectly modulate enzymes involved in proliferative processes such as transcription, translation, and replication to ensure the safe arrest of growth and the activation of adaptive responses (Dalebroux and Swanson, 2012). Over the last 10 years it has become clear that chloroplasts possess the factors necessary for a stringent-like response: ppGpp has been detected in plants and algae, and the nuclear genomes of photosynthetic eukaryotes have been discovered to encode chloroplast-targeted RelA and SpoT homologs (RSHs), named after the enzymes responsible for ppGpp homeostasis in Escherichia coli (van der Biezen et al., 2000; Takahashi et al., 2004; Atkinson et al., 2011; Tozawa and Nomura, 2011; Masuda, 2012).
In plants, ppGpp has been proposed to play a role during stress responses because it accumulates following environmental stress and the application of stress-related plant hormones such as abscisic acid (ABA) and jasmonic acid (Takahashi et al., 2004; Ihara et al., 2015). Studies using purified chloroplast enzymes and chloroplast extracts suggest that ppGpp may function in planta by inhibiting translation and/or transcription in a manner analogous to the bacterial stringent response (Takahashi et al., 2004; Sato et al., 2009; Masuda, 2012; Nomura et al., 2012; Nomura et al., 2014). However, there remains substantial uncertainty about both the principal targets and effects of ppGpp in the plant under physiological conditions.
In the photosynthetic eukaryotes, the RSH enzymes have diverged out into several broadly conserved families with distinct domain structures (Atkinson et al., 2011). Members of certain families are able to complement ppGpp deficient mutants of E. coli (Kasai et al., 2002; Tozawa et al., 2007; Mizusawa et al., 2008; Masuda et al., 2008). The four RSH genes found in Arabidopsis thaliana show diurnal expression rhythms in photosynthetic tissues, and their expression can be regulated by application of the jasmonate precursor 2-oxo-phytodienoic acid or ABA and during environmental stress (Mizusawa et al., 2008; Chen et al., 2014; Yamburenko et al., 2015). However, despite their potential importance, the contribution of the different RSH genes to plant growth and development and to plant stress responses has so far received surprisingly little attention. In Arabidopsis, CRSH, encoding a member of the calcium binding RSH family, has been proposed to be involved in flower development, although the mechanism is not yet clear (Masuda et al., 2008). RSH2 and RSH3 have also been implicated in the ABA-mediated downregulation of chloroplast transcription (Yamburenko et al., 2015).
In this study, we sought to gain a better understanding of the principal targets and effects of ppGpp in planta and, in turn, to shed more light on the roles of RSH enzymes during plant growth and development. Using plant lines that constitutively and conditionally accumulate ppGpp, we demonstrate that ppGpp is a potent regulator of chloroplast gene expression that directly reduces the quantity of chloroplast transcripts and chloroplast-encoded proteins. Then, using a collection of RSH mutants, we demonstrate that the antagonistic functions of different RSH enzymes together determine ppGpp levels and regulate chloroplast function during vegetative growth. We further show that RSH enzymes and ppGpp play important roles during developmental and dark-induced senescence where they are required for chlorophyll and Rubisco remobilization.
RESULTS
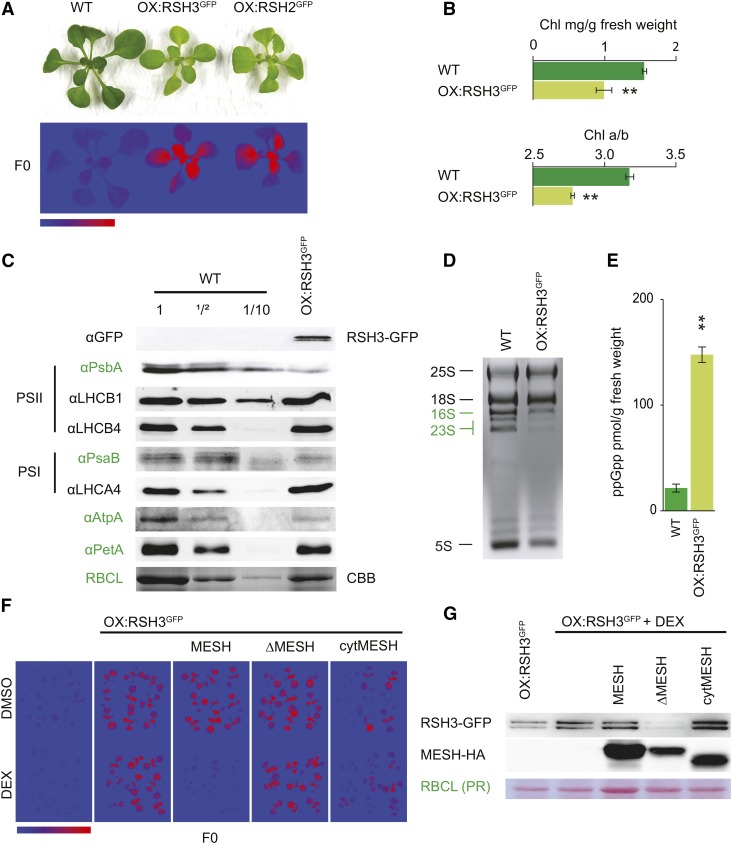
ppGpp Regulates Global Chloroplast Function
RSH2 and RSH3 are likely to function as the major ppGpp synthases in Arabidopsis because they possess conserved ppGpp synthase domains and are the most highly expressed of the RSH enzymes (Mizusawa et al., 2008). RSH2 and RSH3 also share 90% amino acid similarity and belong to the same RSH family (Atkinson et al., 2011) (Supplemental Figure 1). Therefore, as a first step toward understanding the role of ppGpp in Arabidopsis, we created plants overexpressing RSH2 and RSH3 with the addition of a C-terminal GFP tag. Because the activity and regulation of RSH enzymes can be sensitive to C-terminal tags, we verified that the GFP tag did not affect ppGpp biosynthesis activity by complementing ppGpp deficient E. coli strains with the native and fusion RSH proteins (Supplemental Figure 2). The selection of transgenic plants overexpressing RSH2 and RSH3 was challenging because of the low viability of transformants obtained and the frequent loss of transgene expression in later generations. At least one stable RSH2-GFP overexpressor line (OX:RSH2-GFP) and two stable RSH3-GFP overexpressor lines (OX:RSH3-GFP) that accumulated high levels of the fusion proteins were isolated (Supplemental Figure 3). These plants were pale and smaller than the wild-type control and produced small seeds that rapidly lost their ability to germinate (Figure 1A; Supplemental Figure 3). The photosynthetic parameters of the overexpressors were determined using chlorophyll fluorescence analysis. Overexpression lines have strong basal chlorophyll fluorescence, F0 (Figure 1A; Supplemental Figure 3), and a reduction in the maximal efficiency (or quantum yield, QY) of photosystem II (PSII): the average quantum yield was 0.86 ± 0.001 se in wild-type plants 12 d after stratification (DAS) versus 0.690 ± 0.002 se in OX:RSH2-GFP.1, 0.69 ± 0.006 se in OX:RSH3-GFP.1, and 0.73 ± 0.006 se in OX:RSH3-GFP.2 (n = 8). During preparation of this manuscript, similar phenotypes were reported for plant lines overexpressing RSH3 (Maekawa et al., 2015). Now focusing on plants overexpressing RSH3-GFP, we confirmed that chlorophyll levels are lower than in wild-type plants and found that this is accompanied by a reduction in the chlorophyll a/b ratio (Figure 1B). Previous work has shown that plants grown in the presence of lincomycin, an inhibitor of chloroplast translation, also have a high F0 and a low chlorophyll a/b ratio (Belgio et al., 2012). This distinctive phenotype is due to the lincomycin-mediated loss of the chloroplast-encoded reaction center subunits from PSII (RCII) and the retention of unattached nucleus-encoded PSII light-harvesting complexes (LHCII), which are rich in chlorophyll b and highly fluorescent (Belgio et al., 2012). Therefore, we suspected that RSH3-GFP overexpression leads to a reduction of chloroplast gene expression, in a similar manner to lincomycin treatment. In agreement with this notion, there were markedly reduced amounts of the majority of the signature chloroplast-encoded proteins that we tested (Figure 1C, in green).
Figure 1.
RSH3-GFP Overexpression Reduces Chloroplast Function.
(A) OX:RSH3-GFP.1 (OX:RSH3GFP) and OX:RSH2-GFP.1 (OX:RSH2GFP) plants are small and pale (above) and have a high basal chlorophyll fluorescence, F0 (below). Plants shown were grown on plates for 16 DAS. F0 false-color scale bar, 50 to 350 arbitrary units.
(B) OX:RSH3GFP.1 seedlings have significantly lower chlorophyll levels and chlorophyll a/b ratios than the wild type (n = 4 plants, 12 DAS).
(C) Immunoblots on equal quantities or dilutions of total protein from wild-type seedlings and seedlings overexpressing RSH3-GFP 12 DAS using the indicated antibodies against signature chloroplast proteins. Chloroplast-encoded proteins are indicated by green text. RBCL was revealed by Coomassie Brilliant Blue staining (CBB).
(D) Total RNA from wild-type plants and RSH3-GFP-overexpressing plants showing cytosolic (black) and chloroplastic rRNA (green).
(E) ppGpp was extracted from the leaves of soil-grown plants 32 DAS and quantified by ultraperformance liquid chromatography-mass spectrometry; P = 0.00013, n = 3 biological replicates.
(F) F0 images of wild-type plants, OX:RSH3-GFP.1 plants, and OX:RSH3-GFP.1 plants crossed with inducible MESH plants 12 DAS. Plants were grown on medium containing the carrier (DMSO) or 1 µM dexamethasone (DEX). MESH, catalytically active chloroplastic enzyme; ΔMESH, catalytically inactive chloroplastic enzyme; cytMESH, an active MESH targeted to the cytoplasm. cytMESH plants were segregating for OX:RSH3-GFP.1.
(G) Immunoblots showing the accumulation of RSH3-GFP and MESH proteins in total extracts from the same plants as analyzed for F0 in (F). For cytMESH, only plants overexpressing RSH3-GFP were selected for protein extraction. PR, Ponceau Red. Significance was tested using the two-way Student’s t test, ** P < 0.01. Error bars indicate se.
The greatest reduction was for PsbA, a subunit of RCII, which is reduced to less than one-tenth of wild-type levels. LHCB1, one of the major nucleus-encoded subunits of LHCII, remained at wild-type levels relative to total protein. The resulting >10-fold decrease in the RCII/LHCII ratio strongly suggests that a large fraction of LHCII is no longer attached to RCIIs, and it explains the high F0 and low QY of OX:RSH3-GFP.1 plants. In addition to reductions in the levels of chloroplast-encoded proteins, we also detected a marked reduction in the accumulation of chloroplast rRNA compared with cytosolic rRNA, indicating that there is a substantial drop in total chloroplast translation capacity (Figure 1D). Reduced translation and rRNA levels are hallmarks of the bacterial response to ppGpp accumulation (Dalebroux and Swanson, 2012). We verified that the overexpression of RSH3 increases ppGpp levels using a recently developed method (Figure 1E) (Ihara et al., 2015). To exclude the possibility that the observed phenotypes are due to other potential functions of RSH3 or indirect effects of overexpression, we introduced an inducible metazoan ppGpp-specific hydrolase, MESH (Sun et al., 2010), into OX:RSH3-GFP.1 plants. Expression of a chloroplast-targeted MESH was able to restore wild-type F0 and chlorophyll levels to OX:RSH3-GFP.1 plants, while RSH3-GFP overexpression was maintained (Figures 1F and 1G). Expression of a catalytically inactive chloroplastic MESH (ΔMESH) or an active cytoplasmic MESH was unable to restore a wild-type phenotype (Figures 1F and 1G). These findings indicate that the phenotype of OX:RSH3-GFP.1 plants is caused by the accumulation of ppGpp within the chloroplast.
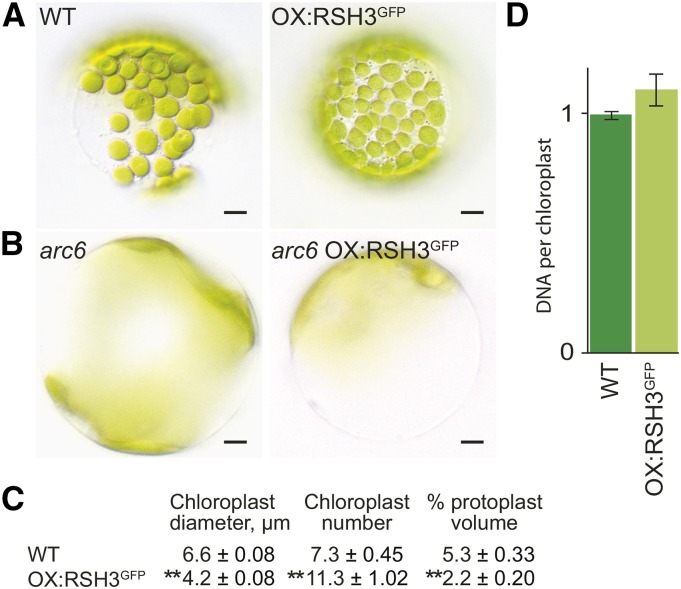
Chloroplasts in OX:RSH3-GFP Plants Are Smaller and More Numerous
Chloroplast size and number were analyzed in protoplasts from OX:RSH3-GFP.1 and wild-type plants (Figure 2). We found that OX:RSH3-GFP.1 chloroplasts were significantly smaller and more numerous than wild-type chloroplasts (Figures 2A and 2C). The change in chloroplast number could be suppressed by a mutation in the nuclear gene ACCUMULATION AND REPLICATION OF CHLOROPLASTS6 (ARC6), which encodes a component of the chloroplast division machinery (Figure 2B). DNA content per plastid was also constant, indicating that the increase in chloroplast number is due to increased chloroplast replication and division (Figure 2D) (Robertson et al., 1995). This contrasts with the situation in bacteria, where ppGpp inhibits proliferation by directly targeting DNA primase, the enzyme that initiates DNA replication (Dalebroux and Swanson, 2012). The inability of ppGpp to inhibit DNA replication in Arabidopsis chloroplasts could be explained by the recent observation that plants lack bacteria-like DNA primases and that chloroplast DNA replication is instead likely to be primed by a eukaryotic TWINKLE homolog (Diray-Arce et al., 2013). Importantly, we also found that, despite the increased chloroplast number, the percentage of total cell volume occupied by chloroplasts was significantly lower than that in wild-type plants (Figure 2C).
Figure 2.
ppGpp Accumulation in RSH3-GFP Plants Reduces Chloroplast Volume per Cell without Repressing Chloroplast Replication.
(A) and (C) Mesophyll protoplasts were isolated from the leaves of wild-type and mutant plants 35 DAS, and representative protoplasts are shown. Chloroplasts in OX:RSH3-GFP.1 (OX:RSH3GFP) plants were smaller than those in the wild type (A) and chloroplast number was consistently higher (C), even when adjusted for protoplast volume (expressed as chloroplasts per 10,000 µm3). Despite increased numbers of chloroplasts, the percentage chloroplast volume per protoplast in OX:RSH3-GFP.1 plants was significantly lower than that in wild-type plants. Significance was calculated using the Kruskal-Wallis test with the Dunn test post hoc, **P < 0.0001. Data are presented as means ± se; 255 to 323 chloroplasts were measured for chloroplast diameter, and 30 to 59 protoplasts were measured for chloroplast number and protoplast volume in OX:RSH3-GFP.1 and the wild type.
(B) Transfer of OX:RSH3-GFP.1 into the genetic background of the chloroplast division mutant arc6 suppressed the increased chloroplast number. arc6 OX:RSH3-GFP.1 plants also had a significantly lower chloroplast plan area per cell than arc6 plants alone (46% ± 2% se versus 62% ± 3% se, P < 0.0001, Kruskal-Wallis test, n = 13 to 16 protoplasts).
(D) Chloroplast DNA content was quantified by qRT-PCR on chloroplasts isolated from wild-type and OX:RSH3-GFP.1 plants at 24 DAS. Data are presented as means ± se for three independent biological replicates.
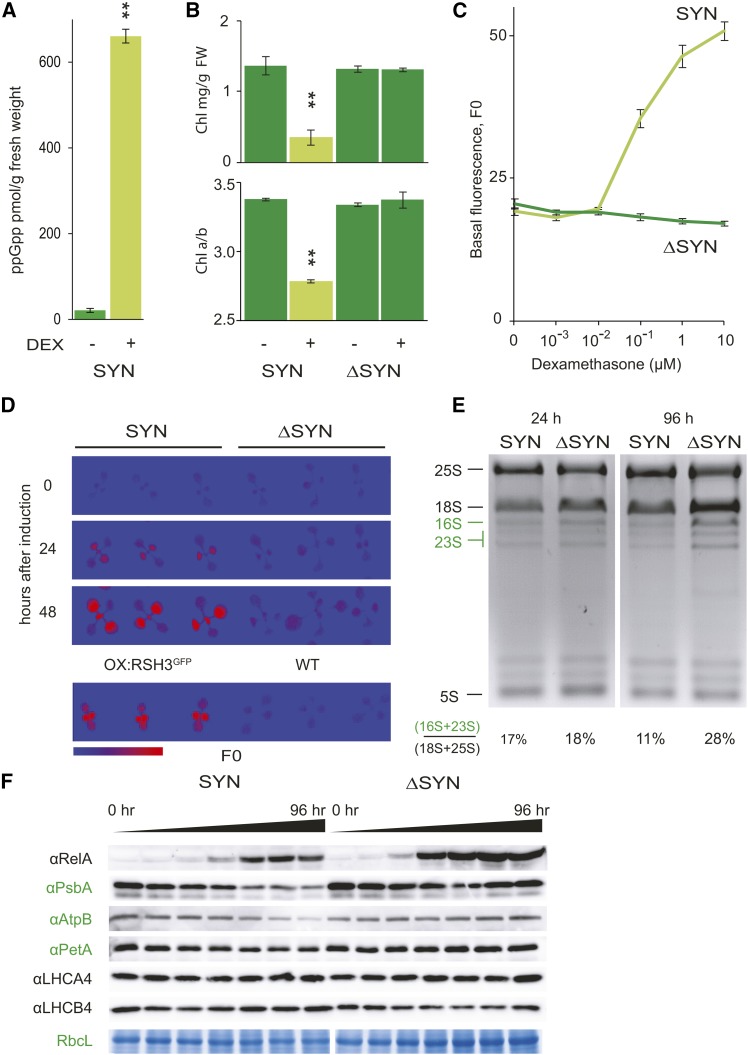
ppGpp Acts on Chloroplast Gene Expression
The pleiotropic phenotypes of RSH2- and RSH3-overexpressing plants makes it challenging to determine how ppGpp acts within the chloroplast. In particular, it is not clear to what extent the reduced chloroplast protein and RNA levels in these lines can be attributed to ppGpp rather than to the reduced total chloroplast volume per cell (Figure 2C). Therefore, we developed an inducible expression system that permits the conditional accumulation of ppGpp in phenotypically wild-type plants. For this approach, we created transgenic plants harboring a T-DNA insertion that encodes a dexamethasone-inducible ppGpp synthase domain from E. coli RelA (SYN), which is targeted to the chloroplast. The truncated RelA synthase domain used has constitutive ppGpp synthase activity in E. coli (Schreiber et al., 1991). Induction of SYN expression by dexamethasone application led to the accumulation of high levels of ppGpp (Figure 3A). This was accompanied by a reduction in chlorophyll levels, a reduction in the chlorophyll a/b ratio, and an increase in F0 in a dose- and time-dependent manner (Figures 3B to 3D). There was also a modest but significant repression of plant growth (Supplemental Figure 4). Inducible expression of catalytically inactivated ppGpp synthase (ΔSYN) had no effect on these parameters, indicating that the SYN phenotypes can be attributed directly to SYN-catalyzed ppGpp accumulation. The SYN phenotypes are very similar to those observed in OX:RSH3-GFP.1 plants or in plants treated with lincomycin, confirming that ppGpp causes a global suppression of chloroplast gene expression. In agreement, we found a reduction in the levels of signature chloroplast-encoded proteins and chloroplast rRNAs in plants expressing SYN (Figures 3E and 3F). Nucleus-encoded proteins in the chloroplast and cytosolic rRNAs were not affected. Taken together, these results indicate that both RSH3-GFP and SYN overexpression cause the accumulation of ppGpp in the chloroplast and that ppGpp accumulation can rapidly lead to reductions in the amounts of chloroplast-encoded rRNA and protein. In both cases, the marked depletion in chloroplast rRNA indicates that ppGpp accumulation severely constrains chloroplast translation capacity.
Figure 3.
Conditional Expression of a Bacterial ppGpp SYN Reduces Chloroplast Function.
SYN plants contain a transgene encoding a chloroplast-targeted ppGpp synthase from bacteria under the control of a dexamethasone-inducible promoter. ΔSYN plants contain a transgene encoding a catalytically inactive variant of SYN.
(A) SYN induction results in a large increase in ppGpp levels, P = 0.000003. ppGpp was extracted and quantified 72 h after induction of SYN seedlings grown on plates for 12 DAS by submersing with either the carrier (DMSO) or 30 µM dexamethasone (DEX) for 3 min. Data are presented as the means of three independent biological replicates.
(B) and (C) SYN and ΔSYN seedlings were analyzed for chlorophyll content and chlorophyll a/b ratios 4 days after induction with dexamethasone, **P < 0.001, versus DMSO control (n = 4 plants) (B) and F0 after 8 d growth on plates containing different concentrations of dexamethasone (n = 18 plants) (F0, arbitrary units) (C).
(D) to (F) After induction of SYN and ΔSYN plants 12 DAS, changes in F0 (F0 false-color scale bar, 50 to 350 arbitrary units) (D), rRNA (E), and chloroplast proteins (F) were followed. Chloroplast proteins were detected by immunoblots on equal quantities of protein using the indicated antibodies. Anti-RelA detects the SYN and ΔSYN proteins. Samples were taken at 0, 2, 4, 8, 24, 48, and 96 h after induction. RBCL was revealed by Coomassie Brilliant Blue staining. Chloroplast-encoded proteins and rRNAs are in green. Significance was calculated using the two-way Student’s t test. Error bars indicate se.
ppGpp Regulates Chloroplast Gene Expression by Reducing Steady State Transcript Levels in the Chloroplast
In bacteria, many of the principal physiological effects of ppGpp are caused by the inhibition of transcription, which can occur by at least two distinct mechanisms (Dalebroux and Swanson, 2012). In E. coli, ppGpp directly interacts with RNA polymerase in cooperation with the transcription factor DnaK suppressor (DksA) to alter promoter selection. Transcription from rRNA genes is subject to particularly strong inhibition in the presence of ppGpp. In contrast, in Bacillus subtilis, RNA polymerase is insensitive to ppGpp (Krásný and Gourse, 2004), and ppGpp instead causes a decrease in the GTP pool by direct inhibition of GTP biosynthesis enzymes such as guanylate kinase (GK) (Kriel et al., 2012). The decrease in GTP levels inhibits gene transcription, and again this effect is particularly strong for the rRNA and tRNA genes where GTP is the initiating nucleotide (Krásný and Gourse, 2004). In plants, ppGpp has also been linked to the regulation of chloroplast transcription, although so far this has not been directly demonstrated in vivo (Maekawa et al., 2015; Yamburenko et al., 2015). There is also evidence for both E. coli-like and B. subtilis-like mechanisms for the inhibition of transcription by ppGpp in plants. Despite the absence of a homolog of DksA, in vitro studies on chloroplast extracts have shown that ppGpp specifically binds to and inhibits the bacterial-like polymerase encoded by the chloroplast genome (Plastid-Encoded Polymerase [PEP]) (Takahashi et al., 2004; Sato et al., 2009). However, the 50% inhibitory concentrations (IC50s) are rather high (1 mM [Sato et al., 2009]; 2 mM [Takahashi et al., 2004]). Chloroplasts also contain an alternative Nucleus-Encoded Polymerase (NEP), which plays a minor role in green tissues and is not inhibited by ppGpp. A recent study also provides support for a B. subtilis-like mechanism by showing that recombinant chloroplastic GK enzymes from rice (Oryza sativa) and Arabidopsis are as sensitive to inhibition by ppGpp in vitro as the B. subtilis GK with IC50s of around 30 µM (Nomura et al., 2014).
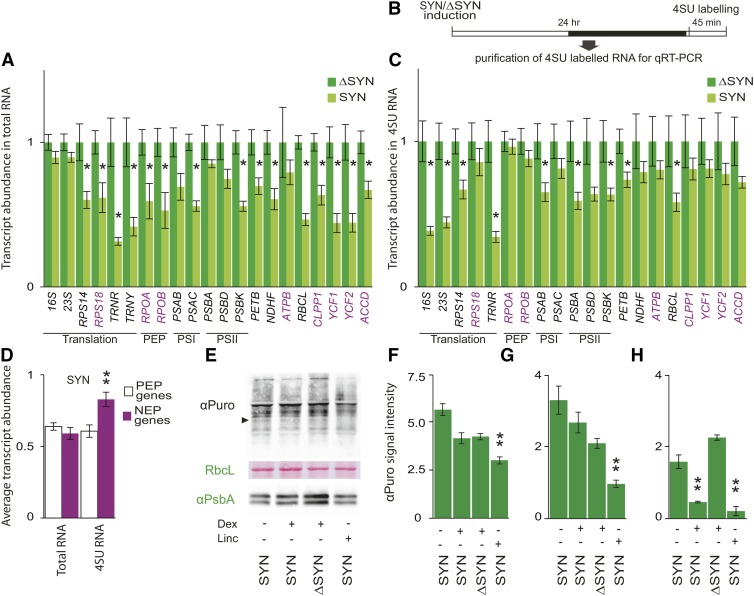
To assess the role of ppGpp in chloroplast transcription, we therefore quantified the steady state levels of a broad range of chloroplast transcripts at 24 h after induction of SYN (Figure 4A). This is an early time point when we detect only a small change in F0 and no change in rRNA levels (Figures 3D and 3E). Strongly supporting a role for ppGpp in directly regulating plastid transcript accumulation in vivo, we observed a significant reduction in the steady state levels of a broad range of chloroplast transcripts. Interestingly, transcript levels were also reduced for genes that are thought to be exclusively or significantly transcribed by NEP in green tissues such as RIBOSOMAL PROTEIN18; RNA POLYMERASE SUBUNIT ALPHA; RNA POLYMERASE SUBUNIT BETA; PHOTOSYNTHETIC ELECTRON TRANSFER B; YCF1, encoding a subunit of the translocon on the inner envelope of chloroplasts; YCF2, encoding an ATPase of unknown function; and ACCD, encoding acetyl-CoA carboxylase (Börner et al., 2015). In OX:RSH3-GFP.1 plants, where ppGpp levels are somewhat lower and at equilibrium, there was also a significant reduction in the accumulation of chloroplast transcripts (Supplemental Figure 5). However, in contrast to the situation in induced SYN plants, the reduction in transcript accumulation was limited to a subset of PEP-dependent genes, and NEP-dependent genes were not significantly affected. This may suggest that ppGpp levels in OX:RSH3-GFP.1 plants are not sufficient for the effects seen in SYN plants or that constitutive accumulation of ppGpp can lead to the activation of compensatory mechanisms.
Figure 4.
ppGpp Accumulation Reduces Chloroplast Transcript Levels but Does Not Have a Major Direct Effect on Chloroplast Translation.
(A) qRT-PCR for selected chloroplast-encoded transcripts 24 h after the induction of ΔSYN and SYN seedlings grown on plates for 12 DAS. Transcripts produced only or significantly by NEP in green tissue (NEP genes) are indicated in purple, *P < 0.05, SYN versus ΔSYN for a single transcript. Data are presented as means ± se for four independent biological replicates, and transcript abundance was normalized to the nucleus-encoded 18S, APT1, PP2A, and ULP7 reference transcripts.
(B) and (C) The transcription rates of chloroplast genes in induced SYN and ΔSYN plants were measured by labeling new transcripts with 4SU in vivo (B) and quantifying the abundance of purified 4SU transcripts by qRT-PCR (C). Data are presented as means ± se for four independent biological replicates, and transcript abundance was normalized to 18S, PP2A, and ULP7 reference genes.
(D) The induction of SYN had significantly less effect on the transcription of NEP genes than it did on PEP genes; P = 0.0011, ANOVA with post-hoc Dunnett test, n = 8 to 11. Twenty-four fours after induction of SYN and ΔSYN seedlings, translation rates were also analyzed by quantifying the incorporation of puromycin into nascent proteins during 1 h.
(E) Puromycin incorporation was assessed by immunoblot analysis on equal quantities of total chloroplast proteins (10 µg) using an antibody against puromycin. Plants treated with lincomycin for 24 h were used as a control. The black arrow indicates PsbA. RbcL is a loading and transfer control and is shown by Ponceau red staining on the same membrane used for puromycin detection.
(F) Incorporation was quantified across five biological replicates. Lincomycin-treated SYN plants showed a significant drop in puromycin incorporation compared with induced SYN plants, **P < 0.01. No significant difference could be detected in the incorporation of puromycin between induced SYN and ΔSYN plants.
(G) and (H) Puromycin incorporation into PsbA was also quantified at 24 h (G) and 72 h (H) after treatment, **P < 0.01, versus ΔSYN. Samples were normalized to total chloroplast protein. Unless stated otherwise, significance was calculated using the two-way Student’s t test. Error bars indicate se.
Steady state transcript levels are a function of the transcription and degradation rate. To test whether ppGpp specifically downregulates transcription under in vivo conditions, we used a metabolic labeling strategy with the base analog 4-thio uridine (4SU). Efficient and nontoxic labeling of total RNA, including plastid RNA, was recently demonstrated using this approach in Arabidopsis (Sidaway-Lee et al., 2014). We labeled newly synthesized RNA 24 h after SYN and ΔSYN induction. Labeled RNA was then isolated and the quantity of newly synthesized chloroplast transcripts from SYN and ΔSYN plants was analyzed by qRT-PCR using nucleus-encoded reference genes (Figure 4B). Consistent with ppGpp-mediated transcriptional downregulation, we found that the quantity of newly synthesized RNA was significantly lower in SYN plants for the majority of those genes that are principally transcribed by PEP (Figure 4C). In contrast, we found that SYN induction had significantly less effect on the transcription of genes that are principally transcribed by NEP (Figure 4D). This would suggest that the accumulation of certain transcripts may also be regulated posttranscriptionally. Similar differential regulation of the turnover for PEP- and NEP-derived transcripts has also been observed in maize (Zea mays; Cahoon et al., 2004). In a manner strikingly reminiscent to bacteria, the effect of ppGpp was strongest on the transcription of the chloroplast rRNAs (16S and 23S) and the arginine tRNA (TRNR). The very slight reduction in steady state levels of chloroplast rRNA 24 h after induction can be explained by its high stability, and we indeed did see a large drop in steady state levels after 96 h (Figure 3E) (Rapp et al., 1992). While we demonstrate that ppGpp is directly involved in the inhibition of plastidial transcription, our data do not allow us to clearly discern whether this is PEP dependent or not. However, using previous estimates of stroma volume in spinach chloroplasts, we calculate that stromal ppGpp concentrations in induced SYN plants can reach up to ∼30 µM (Gerhardt et al., 1987). This is more than an order of magnitude lower than inhibitory concentrations obtained for PEP in vitro. GK, on the other hand, is likely to be significantly inhibited at these concentrations, favoring the idea that a B. subtilis-like mechanism may inhibit transcription in chloroplasts. Notably, GTP is the initiating nucleotide for the principal P1 and minor PC promoters of the chloroplast rRNA operon containing the 16S and 23S rRNAs in Arabidopsis, and this appears to be conserved across a wide range of other plants including the monocots (Suzuki et al., 2003; Swiatecka-Hagenbruch et al., 2007).
ppGpp Accumulation Does Not Have a Rapid and Direct Effect on Chloroplast Translation
In bacteria, ppGpp directly inhibits translation through interaction with translation initiation and elongation factors (Dalebroux and Swanson, 2012). Chloroplasts contain a bacterial-like translation machinery, and ppGpp has also been shown to inhibit chloroplast translation in in vitro assays (Nomura et al., 2012). We therefore tested whether ppGpp directly represses chloroplast translation in vivo in SYN plants. Despite the inhibition of transcription by ppGpp, there was only a small reduction in rRNA levels 24 h after SYN induction; thus, a near wild-type translational capacity should be present (Figures 3E and 4A). Therefore, total chloroplast translational rates were quantified 24 h after induction using metabolic labeling with puromycin, an aminoacyl tRNA analog that can be incorporated into nascent polypeptide chains (Schmidt et al., 2009). We were first able to show that puromycin is taken up by plants in a time-dependent manner and is efficiently incorporated into nascent cytosolic and chloroplastic proteins (Supplemental Figure 6). Next, we analyzed total chloroplast translation rates in plants expressing SYN and ΔSYN (Figures 4E and 4F). No significant reduction in total chloroplast translation could be observed in plants expressing SYN at 24 h after induction compared with plants expressing ΔSYN (Figures 4E and 4F). However, chloroplast translation was significantly reduced by the application of the translation inhibitor lincomycin. The PSII RC subunit was an even more sensitive reporter of translation probably due to its high turnover rates (Figures 4G and 4H). PsbA translation was similar in induced SYN and ΔSYN plants at 24 h after induction and then dropped sharply only in induced SYN plants after 72 h. The effect of lincomycin on PsbA translation was strong at both 24 h and 72 h after treatment. These results show that ppGpp accumulation does not have a large direct effect on chloroplast translation under our conditions. This could be explained by a lower sensitivity of the translational machinery to ppGpp, a possibility that is supported by the existing in vitro data that suggest an IC50 of >400 µM (Nomura et al., 2012).
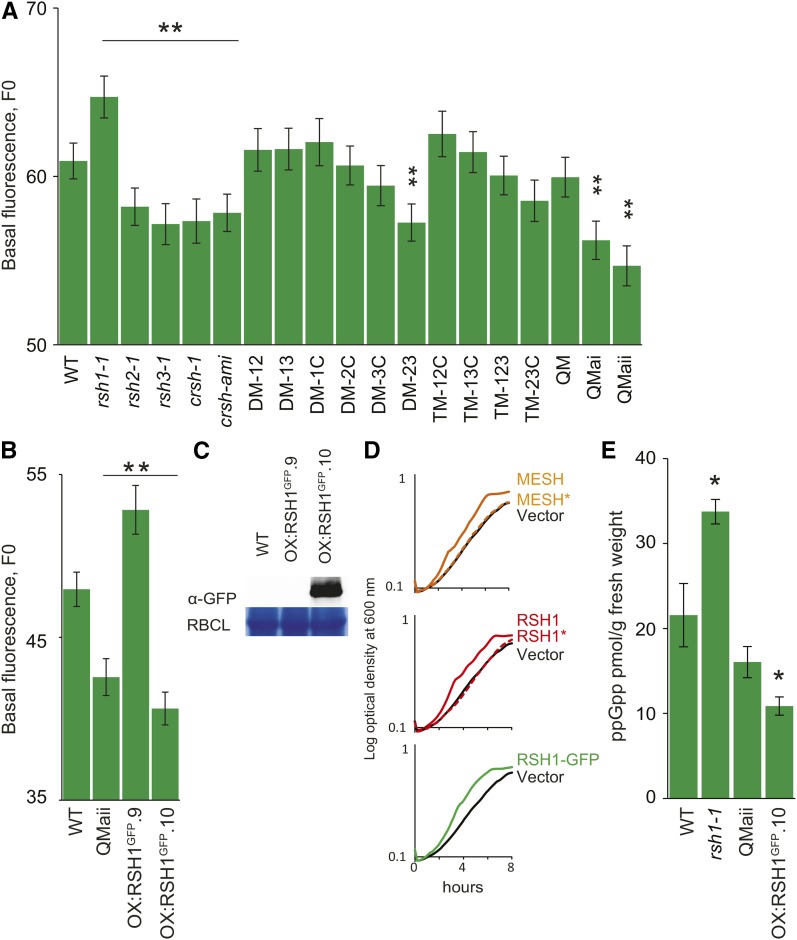
RSH Mutants Have Altered Chlorophyll Fluorescence and ppGpp Levels
We next sought to understand the role of RSH enzymes in regulating ppGpp levels in planta and their function during plant growth and development. The four Arabidopsis RSH proteins (RSH1, RSH2, RSH3, and CRSH) are likely to be the principal mediators of ppGpp homeostasis because they possess well known ppGpp synthase and hydrolase domains (Supplemental Figure 1) and because RSH2, RSH3, and CRSH show ppGpp biosynthetic activity in E. coli assays (Masuda et al., 2008; Mizusawa et al., 2008). Here, we also show that RSH2 and RSH3 overexpression results in ppGpp accumulation in planta (Figure 1E), and a recent study also confirms these findings for RSH3 (Maekawa et al., 2015).
We therefore isolated single insertion mutants for RSH1, RSH2, RSH3, and CRSH (referred to here as rsh1-1, rsh2-1, rsh3-1, and crsh-1, respectively) (Supplemental Figure 7A). The T-DNA insertions in rsh1-1, rsh2-1, and rsh3-1 are upstream of the regions encoding the ppGpp metabolizing domains and result in a complete loss of mature mRNA. We could also detect no or little ectopic transcription in the regions downstream of the T-DNA insertions (Supplemental Figure 7B). The T-DNA insertion in crsh-1 is downstream of the regions encoding the ppGpp metabolizing domains, and there is only a partial reduction in mature CRSH transcript levels. Therefore, we used an artificial micro RNA to knock down CRSH expression to very low levels (crsh-ami; Supplemental Figure 7C). No clear visible phenotypes could be observed in rsh1-1, rsh2-1, rsh3-1, crsh-1, and crsh-ami. A previous study showed that there was altered flower development and reduced fertility in a transgenic line where CRSH was silenced by cosuppression (Masuda et al., 2008). This phenotype was attributed to the loss of CRSH-mediated ppGpp accumulation. However, we did not observe altered flower development or fertility defects in the considerable number of independent crsh-ami lines generated during this study. Our different results could be explained by different levels of CRSH silencing or by the presence of a linked mutation or off-target silencing that reduced fertility in the original CRSH cosuppression line. Furthermore, overexpression of the RSH1 ppGpp hydrolase or induction of the MESH ppGpp hydrolase before and throughout flowering did not cause any detectable changes in flower development or fertilization. Thus, it is currently not clear whether CRSH regulates fertilization by modulating ppGpp levels. Next, due to likely redundancy for ppGpp biosynthesis and degradation, the RSH single mutants were crossed to make all the double mutant (DM) and triple mutant (TM) combinations as well as the quadruple mutants (QM for rsh1-1 rsh2-1 rsh3-1 crsh-1 and QMai and QMaii for rsh1-1 rsh2-1 rsh3-1 with independent crsh-ami insertions).
We reasoned that alterations in the ppGpp levels in the different RSH mutants could affect the stoichiometry of PSII in a manner that would be detectable as changes in chlorophyll fluorescence, F0, as we observed in OX:RSH2, OX:RSH3, and SYN plants (Figures 1 and 2). We therefore measured the F0 in each of the 18 RSH mutants (Figure 5A). Strikingly, we discovered that the single mutants for genes encoding the ppGpp biosynthetic enzymes RSH2, RSH3, and CRSH have a significantly lower F0 than the wild-type control and that the effect, which was very robust, increased when the mutations were combined in the quadruple mutants (QMai and QMaii). A low F0 is consistent with low ppGpp levels: A reduction in ppGpp would be expected to derepress plastid gene expression and thus increase the proportion of chloroplast-encoded PSII RC subunits relative to nucleus-encoded LHCII. Higher proportions of PSII RC are known to increase the efficiency of excitation transfer to photochemistry and therefore to directly diminish the proportion of excitation energy that is released as fluorescence (Engelmann et al., 2005). Interestingly, we found that rsh1-1 has a higher F0 than the wild type, a similar phenotype to SYN and OX:RSH3 plants that overaccumulate ppGpp. RSH1 lacks a functional ppGpp synthase domain and has a conserved ppGpp hydrolase domain, although ppGpp hydrolysis activity has not previously been demonstrated (Supplemental Figure 1) (Mizusawa et al., 2008). The fluorescence data therefore support the idea that RSH1 may act as a ppGpp hydrolase and that loss of RSH1 in rsh1-1 results in greater ppGpp accumulation and the consequent repression of PSII RC expression. Critically, and as would be expected for a mutation in a ppGpp hydrolase, the rsh1-1 mutant phenotype is epistatic to mutations in the ppGpp synthases. Mutations in RSH2, RSH3, and CRSH are sufficient to completely suppress rsh1-1 in QMai and QMaii (Figure 5A). These fluorescence experiments were repeated multiple times, and we found that F0 is a robust readout for the presumed action of ppGpp in the chloroplast under physiological conditions. F0 measurements on plants overexpressing RSH1-GFP provided additional evidence that RSH1 acts as a ppGpp hydrolase. OX:RSH1-GFP.10, an overexpression line that accumulates high levels of RSH1-GFP, has a significantly lower F0 than the wild-type control (Figure 5B). In contrast, OX:RSH1-GFP.9, a line where RSH1 appears to be silenced by cosuppression, has a higher F0 (Figures 5B and 5C). To provide evidence that is completely independent of chlorophyll fluorescence measurements, we also tested the hydrolase functions of RSH enzymes by expression in a slow growing E. coli mutant that overaccumulates ppGpp (Figure 5D) (My et al., 2013). The known ppGpp hydrolase MESH1 was capable of rescuing the slow growth phenotype. Expression of RSH1 was also able to rescue the mutant, demonstrating that RSH1 can indeed function as a ppGpp hydrolase. Furthermore, expression of the same RSH1-GFP fusion protein as that expressed in OX:RSH1-GFP.10 plants demonstrated that this protein was also capable of rescuing the slow growth phenotype of the E. coli mutant.
Figure 5.
RSH Enzymes Mediate ppGpp Equilibrium during Vegetative Growth.
(A) Basal chlorophyll fluorescence (F0, arbitrary units) was measured in the seedlings of a panel of 18 RSH mutants grown on plates for 12 DAS (n = 60 to 72 individual plants). crsh-ami, plants where CRSH is silenced by an artificial microRNA; DM-xy, double mutant for RSHx and RSHy; TM-xyz, triple mutant for RSHx, RSHy, and RSHz; QM, quadruple mutant with crsh-1 mutation; QMai and QMaii, quadruple mutants where CRSH is silenced by independent crsh-ami alleles.
(B) and (C) The 12-DAS seedlings from RSH1-GFP overexpression lines were analyzed for F0 (n = 60 to 72 individual plants) (B) and RSH1-GFP protein accumulation by immunoblotting (C). The ppGpp hydrolase activity of different RSH enzymes was tested by expression in a slow-growing E. coli strain that overaccumulates ppGpp.
(D) Bacterial growth curves were obtained by measuring optical density every 10 min over 8 h (average of four biological replicates). The expression of the ppGpp hydrolase MESH resulted in a significant acceleration of growth (doubling time, TD 1.84 h ± 0.003 h se for MESH versus 2.33 h ± 0.04 h se for the vector only control, P < 0.0001, two-way Student’s t test). RSH1 and RSH1-GFP also significantly accelerated growth of the mutant, indicating that they also act as ppGpp hydrolases (RSH1 TD 1.79 h ± 0.013 h se and RSH1-GFP TD 1.67 h ± 0.011 h se, P < 0.0001 versus vector only control, two-way Student’s t test). Mutation of the ppGpp hydrolase domains (MESH* and RSH1*) restored a slow growth phenotype indistinguishable from that of the vector only control. CRSH showed no activity in the same test, and RSH2 and RSH3 transformants could not be obtained to test, presumably due to overproduction of ppGpp.
(E) Quantification of ppGpp in different mutant lines. ppGpp was extracted from the leaves of soil-grown plants 35 DAS and quantified by ultraperformance liquid chromatography-mass spectrometry, *P < 0.05, two-way Student’s t test. Data are presented as the means of three biological replicates. Large-scale extractions confirmed that ppGpp levels were significantly lower in than the wild type in QMaii and OX:RSH1-GFP.10 (Supplemental Figure 8). Unless otherwise stated, data were analyzed by ANOVA with post-hoc Dunnett tests versus the wild-type controls, *P < 0.05 and **P < 0.01. Error bars indicate se.
We next sought to confirm our evidence for altered ppGpp levels by direct measurements of ppGpp. In agreement with our data, a significant increase in ppGpp levels could be detected for rsh1-1, and a significant decrease in ppGpp could be detected for OX:RSH1-GFP.10 (Figure 5E). Lower levels were also detected in QMaii, although the statistical significance was borderline. However, by scaling up the extraction procedure and analyzing more concentrated extracts, we could confirm that ppGpp levels were indeed significantly lower than the wild type in both QMaii and OX:RSH1.10 (Supplemental Figure 8). Together, these results indicate that RSH1 is antagonistic to RSH2 and RSH3 and that, together, the RSH enzymes appear to maintain ppGpp levels in equilibrium. Other enzymes may also be involved in maintaining this equilibrium; indeed, the absence of a runaway increase in ppGpp levels in rsh1-1 plants may be due to the presence of specific hydrolases such as the moiety X (Nudix) phosphohydrolase 26 (NUDX26) (Ito et al., 2012).
Chloroplast Function and Vegetative Growth Are Affected in RSH Mutants
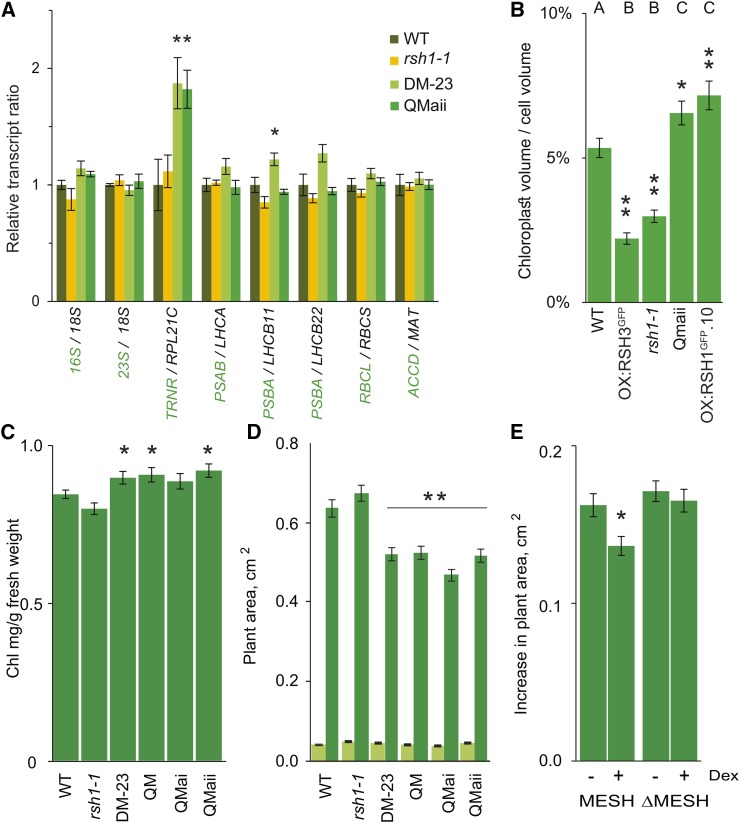
As we show above, in addition to perturbing the stoichiometry of PSII, the ectopic accumulation of ppGpp inhibits chloroplast gene expression by reducing steady state levels of chloroplast transcripts and reduces chloroplast size (Figures 1 to 3). If ppGpp acts on chloroplast transcription during vegetative growth, we might expect to see alterations in the ratio between chloroplast-encoded and nucleus-encoded transcripts for chloroplast complexes and pathways in plants with lower ppGpp levels. We therefore quantified the expression ratios for a range of such transcript pairs, including those for chloroplast and cytosolic rRNA (16S/18S and 23S/18S), the arginine tRNA TRNR and chloroplast 60S ribosomal protein L21 (RPL21C), transcripts encoding the RC and LHC subunits of PSI and PSII (PSAB/LHCA1, PSBA/LHCB11, and PSBA/LHCB22), transcripts encoding the small and large subunits of Rubisco (RBCL/RBCS), and transcripts encoding the acetyl-CoA carboxylase β subunit and malonyl/acetyltransferase of fatty acid biosynthesis (ACCD/MAT) (Figure 6A). Consistent with the idea that ppGpp regulates chloroplast transcription during vegetative growth, we found evidence for significant increases in the TRNR/RPL21C and PSBA/LHCB11 ratios for lines with lower ppGpp levels (DM-23 and QMaii). The increase in TRNR was robust, as it could also be observed when normalized against nucleus-encoded reference genes (Supplemental Figure 9A). Interestingly, TRNR was also the most affected transcript in SYN lines at the steady state and transcriptional levels (Figures 4A and 4B). The absence of detectable changes in levels for the remaining genes suggests that the altered ppGpp levels in the RSH mutants may cause effects that are too small for quantification by qRT-PCR (<25%) or alternatively that feedback mechanisms might regulate nuclear gene expression to maintain an expression ratio at close to wild-type levels.
Figure 6.
RSH Enzymes Are Required for Regulating Chloroplast Function, Volume, and Plant Growth.
(A) Ratios of chloroplast (green) to nucleus-encoded (black) transcripts in different RSH mutants. qRT-PCR was performed on cDNA extracted from seedlings grown on plates for 12 DAS. Data are presented as means ± se for five independent biological replicates. Significance was calculated using ANOVA with post-hoc Dunnett tests versus the wild-type controls.
(B) Total chloroplast volume per cell was calculated in protoplasts isolated from fully expanded leaves of plants grown on soil at 35 DAS. Representative protoplast images are shown in Supplemental Figure 9B. Statistical significance was calculated using Kruskal-Wallis with the Dunn test post hoc, and the resulting groups are indicated above each bar. Thirty protoplasts were analyzed for OX:RSH3-GFP.1 (OX:RSH3GFP) and 47 to 59 for the other lines. Similar results were also obtained using an independent approach on intact cells (Supplemental Figure 9C).
(C) Chlorophyll levels were measured in selected RSH mutants grown on soil at 24 DAS. DM-23 and the QMs have a higher chlorophyll content than the wild type; two-way Student’s t test, n = 8 plants.
(D) Plant surface area for wild-type and mutant seedlings grown on plates at 6 (light green) and 12 DAS (dark green) after stratification. Except for rsh1-1, which was larger (P < 0.0001), there were no significant differences between the mutants and the wild type at 6 DAS. Similar results were also obtained for plants grown in soil (Supplemental Figure 9D). Significance was calculated using ANOVA with post-hoc Dunnett tests versus the wild-type controls; n = 50 plants per line.
(E) At 8 DAS, MESH and ΔMESH plants were transferred onto medium containing DMSO (control) or 1 µM dexamethasone (induced) and the increase in plant area was measured 4 d after transfer. The two-way Student’s t test was used to compare noninduced and induced plants. n = 36 plants,*P < 0 .05 and **P < 0.01, error bars indicate se.
We next examined chloroplast size and number in protoplasts isolated from different RSH mutants and overexpression lines (Figure 6B; Supplemental Figure 9B). Strikingly, we found that chloroplast volume per cell was closely correlated to measured ppGpp levels. To exclude potential artifacts due to the protoplast isolation procedure, this result was also confirmed in intact cells using a different approach (Pyke and Leech, 1991) (Supplemental Figure 9C). Further in support of these results, we found that plants with mutations in the ppGpp biosynthetic enzymes (DM-23, QMai, and QMaii) have a significantly higher chlorophyll content than wild-type or rsh1-1 plants (Figure 6C).
Further analysis of selected mutants showed that plants lacking multiple RSH ppGpp synthase genes are significantly smaller than wild-type plants when grown in Phytagel or in soil (Figure 6C; Supplemental Figure 9D). Plants expressing the ppGpp hydrolase MESH were also smaller (Figure 6E). The reduced size was not due to altered developmental timing because leaf emergence and flowering time were not different in the mutants. These differences were even more marked in flowering plants that had been grown under short-day conditions for 95 d. Among these plants, rsh1-1 is visibly paler and DM-23 and QMaii plants are darker (Supplemental Figure 10).
Together, these results strongly suggest that the ppGpp hydrolase RSH1 acts antagonistically with the ppGpp synthase activities of RSH2, RSH3, and CRSH to regulate ppGpp levels during vegetative growth. The differences in F0 and steady state chloroplast transcript ratios in the different RSH mutants suggest that the small quantities of ppGpp found in growing plants are sufficient to regulate the expression of at least a subset of chloroplast genes and consequently to alter the stoichiometry of nucleus- and chloroplast-encoded proteins within the PSII supercomplex and other chloroplast complexes. The presence of functional ppGpp synthases and hydrolases is also important for regulating chloroplast volume per cell as well as vegetative growth.
ppGpp Regulates Senescence and Nutrient Remobilization
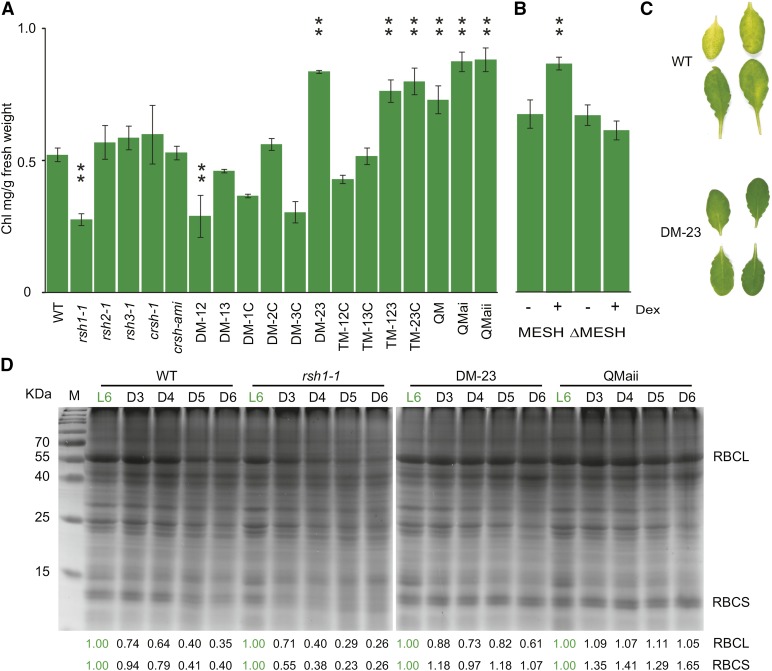
The expression of RSH2 and RSH3 has been shown to increase in ageing leaves in several studies (Schmid et al., 2005; Mizusawa et al., 2008; Breeze et al., 2011) (Supplemental Figure 11). This suggests that there may be specific roles for ppGpp during leaf senescence, when nutrients are recycled and redirected to the developing seeds (Lim et al., 2007). We therefore tested the 18 RSH mutants using a widely used dark-induced senescence assay on detached leaves that reproduces many of the phenotypes of developmental senescence and shows a large overlap in gene expression (Buchanan-Wollaston et al., 2005). We found a striking delayed senescence (or stay-green) phenotype in all the mutants containing insertions in both RSH2 and RSH3 (Figures 7A and 7C). CRSH may also contribute to some extent because in QMai and QMaii plants where CRSH is silenced, there was a significantly stronger stay-green phenotype than DM-23 when analyzed at later developmental time-points (Supplemental Figure 12A). These phenotypes are likely to be due to a reduction in ppGpp biosynthetic capacity by the mutations in the RSH genes. Induction of MESH expression 24 h before the senescence assay also caused a stay-green phenotype, indicating that removal of ppGpp alone is sufficient (Figure 7B). In agreement with our identification of RSH1 as a ppGpp hydrolase, we also observed an accelerated senescence phenotype in rsh1-1 (Figure 7A) and a delayed senescence phenotype in OX:RSH1-GFP.10 (Supplemental Figure 12B).
Figure 7.
The Antagonistic Activity of RSH Enzymes Is Critical for Senescence and Nutrient Remobilization.
(A) and (B) Senescence was induced by incubating detached leaves in the darkness, and chlorophyll levels were measured after 5 d in a panel of 18 RSH mutants (A) and induced and noninduced MESH plants (B). MESH plants were induced by spraying plants with 10 µM dexamethasone (Dex) or the vehicle (DMSO) 48 h before the dark-induced senescence assay. Plants were grown for 48 d under short-day conditions, and chlorophyll levels were not significantly different between untreated lines. Data are presented as means ± se for extractions from three biological replicates and were analyzed by ANOVA with post-hoc Dunnett tests versus the wild-type controls, **P < 0.01.
(C) A photograph of leaves from single plants 5 DAS treatment in (A).
(D) Equal quantities of total protein were separated by SDS-PAGE and visualized by Coomassie Brilliant Blue staining after extraction from the leaves of selected lines after 3 (D3), 4 (D4), 5 (D5), and 6 d (D6) of darkness. Untreated leaves on the plant were used as a control (L6). Relative pixel densities for RBCL and RBCS are shown below the gel image. For each plant line, pixel density is normalized to the L6 control.
Furthermore, the accelerated senescence phenotype of rsh1-1 is epistatic to mutations in both RSH2 and RSH3. Therefore, the ppGpp hydrolase RSH1 appears to be required to constrain an increase in ppGpp that may be driven by the transcriptional upregulation of RSH2 and RSH3 expression during senescence. However, ppGpp accumulation alone is not sufficient to trigger senescence because OX:RSH3-GFP plants and induced SYN plants do not show obvious senescence symptoms in vegetative tissues (Figure 1A), although they do show accelerated senescence during the seed filling stage. The induced senescence phenotypes of the RSH mutants are also relevant during natural plant growth. We saw differences in natural senescence in plants grown under long-day conditions that became very strong under short-day conditions (Supplemental Figure 10). QMaii plants were particularly striking because rather than becoming pale, old leaves crumpled and died while still green (Supplemental Figure 13). A similar phenotype was also observed in OX:RSH1-GFP.10 plants. Senescence is necessary for the remobilization of nutrients and their reallocation to developing fruit. We found that the seeds of DM-23 and QM mutant plants were significantly lighter in weight than the seeds of wild-type plants (Supplemental Figure 14), suggesting that nutrient reallocation is defective during senescence and/or that ppGpp additionally plays a role during seed development. Supporting the idea that nutrient allocation is defective, we also observed a striking retention of the Rubisco small and large subunits during senescence for mutants deficient in ppGpp biosynthetic enzymes such as DM-23 and QMaii (Figure 7D). As we observed for chlorophyll, Rubisco retention in these lines can be directly linked to ppGpp levels because the degradation of Rubisco was accelerated by overexpression of RSH3-GFP and greatly slowed by the overexpression of RSH1-GFP during the dark-induced senescence assay (Supplemental Figure 15A). The retention of Rubisco in DM-23 could also be reversed by complementation with RSH3, indicating that during senescence, RSH2 and RSH3 are redundant for Rubisco degradation as they are for chlorophyll degradation (Supplemental Figure 15B). Together, these data show that the antagonistic activity of RSH ppGpp synthases and hydrolases is required for chlorophyll degradation and nutrient remobilization during senescence.
DISCUSSION
Chloroplast gene expression changes dramatically during development and in response to environmental signals such as light or temperature (Liere et al., 2011; Pfannschmidt and Munné-Bosch, 2013; Rochaix, 2013; Tiller and Bock, 2014). Numerous mechanisms regulating the expression of specific chloroplast genes at the transcriptional and posttranscriptional levels have been identified (Liere et al., 2011; Kindgren et al., 2012; Pfannschmidt and Munné-Bosch, 2013; Rochaix, 2013; Tiller and Bock, 2014). However, few factors are known that regulate global chloroplast gene expression from within the chloroplast. Here, we used two independent approaches to show that ppGpp directly suppresses the accumulation of chloroplast transcripts and proteins in vivo and is thus a potent inhibitor of global chloroplast gene expression. ppGpp appears to act principally through the inhibition of chloroplast transcription to reduce the quantities of individual transcript available for translation and the total translational capacity of the chloroplast by reducing rRNA and tRNA transcript levels (Figures 4A and 4C). These results are broadly in agreement with previous in vitro data and a recent study with the phytohormone ABA that suggested a link between RSH gene function and chloroplast gene transcription (Sato et al., 2009; Nomura et al., 2012; Yamburenko et al., 2015). A less extensive study that leads to similar conclusions was also published during the final preparation of this manuscript (Maekawa et al., 2015). Notably, we did not detect a major direct effect on translation, suggesting that direct inhibition of the translation apparatus does not contribute significantly to the suppression of plastid gene expression that we observed in the presence of ppGpp. Our results also indicate that, although the effect of ppGpp appears to be global, the chloroplast does not respond monotonically and that the expression of rRNA and tRNA genes may be more affected than others (Figures 4A and 4B; Supplemental Figure 5). This shows that the main characteristics of the stringent response are conserved between plants and bacteria. We also propose on the basis of the data that we present here, the in vitro sensitivity of plant GKs to ppGpp (Nomura et al., 2014), and the GTP initiation of plant rRNA genes (Suzuki et al., 2003; Swiatecka-Hagenbruch et al., 2007) that ppGpp is most likely to inhibit transcription by a similar mechanism to that found in B. subtilis.
The conditional expression of SYN has also allowed us to uncouple the action of ppGpp from other signaling pathways and effects on chloroplast volume. This is relevant for the hormones ABA and methyl jasmonate, which induce the accumulation of ppGpp but also have large effects on nuclear gene expression that can extensively modify chloroplast function. This is apparent for PsbA, which we show to be downregulated at the transcriptional and steady state levels by ppGpp (Figures 4A and 4B) but which is little affected at the transcriptional level in response to ABA treatment (Yamburenko et al., 2015).
In addition to its effects on the chloroplast gene expression machinery, we also found that, although ppGpp overaccumulation strongly constrains chloroplast size and volume per cell, it does not inhibit DNA replication as in bacteria (Figure 2). This is likely to be because, in plants, the cyanobacterial DNA primase has been replaced by a eukaryotic TWINKLE homolog (Diray-Arce et al., 2013). This event has important implications for chloroplast evolution because it resulted in the transfer of the control of replication and division from the chloroplast to the nucleocytoplasmic compartment.
We also reveal important roles for ppGpp and RSH enzymes during plant growth and development. We first show an unexpected role for ppGpp in regulating chloroplast function during vegetative growth (Figures 5 and 6). Although small quantities of ppGpp have previously been detected in vegetative tissues (Takahashi et al., 2004; Ihara et al., 2015) ppGpp is usually thought to be involved in stress responses in plants, as it is in bacteria. However, we show that the antagonistic activity of RSH enzymes is required for maintaining ppGpp levels in vegetatively growing plants (Figures 5 and 6). The resulting ppGpp pool appears to be involved in fine-tuning chloroplast gene expression, and alteration of the pool by mutations in RSH genes or overexpression of the RSH1 hydrolase perturbs chloroplast transcription and can negatively affect plant growth (Figures 5 and 6). Growth may be affected by imbalances in chloroplast volume to cell volume (Figure 6B), as well as in the composition of protein complexes and metabolic pathways that involve proteins of nuclear and chloroplast origin. Indeed, we show that one of the major functions of ppGpp is in regulating the stoichiometry of the chloroplast-encoded RC subunits with the nucleus-encoded LHCII subunits of the PSII complex. This suggests that PSII assembly may be coupled to the regulation of RSH function in an autoregulatory feedback loop within the chloroplast.
Chloroplasts contain 70% of leaf nitrogen, mostly as photosynthetic proteins. During senescence, the size and activity of chloroplasts are reduced, and they are then broken down as part of a tightly regulated process that remobilizes nutrients to the developing seeds (Lim et al., 2007; Pfannschmidt and Munné-Bosch, 2013). Here, we show that ppGpp biosynthesis by RSH2, RSH3, and CRSH is constrained by the ppGpp hydrolase activity of RSH1 and is required for the timely initiation of senescence and for the breakdown of chlorophyll and Rubisco (Figure 7). Rubisco, which is the most abundant protein in the cell and alone accounts for 20 to 30% of total nitrogen (Feller et al., 2008), is subject to a complex degradation pathway during senescence that involves intraorganellar degradation as well as the intervention of extraplastidic pathways such as autophagy (Lim et al., 2007; Ishida et al., 2014). The retention of Rubisco by plants lacking the RSH2/RSH3 ppGpp synthases or overexpressing the ppGpp hydrolase RSH1 is therefore remarkable, and it suggests that ppGpp is specifically involved in the regulation of the progression of senescence and may therefore be a key player in nitrogen remobilization.
The expression level of RSH genes in the nucleus appears to govern the capacity of chloroplasts to synthesize ppGpp (Figures 1 and 5; Supplemental Figure 11). Intriguingly, regulation of RSH expression may also be involved in modulating ppGpp homeostasis during the circadian period and in response to abiotic stress and phytohormones (Takahashi et al., 2004; Mizusawa et al., 2008; Yamburenko et al., 2015). The C-terminal regions of bacterial RSH enzymes are involved in the regulation of enzyme activity (Potrykus and Cashel, 2008). Recent evidence suggests that the Arabidopsis RSH enzymes are regulated in a similar manner: The C-terminal domain of RSH1 has a conserved TGS domain that was shown to interact with the small GTPase ObgC in a yeast two-hybrid assay (Bang et al., 2012; Chen et al., 2014), and calcium binding at the C-terminal EF-hand domain of CRSH activates ppGpp synthase activity in vitro (Masuda et al., 2008). RSH2 and RSH3 also have extended C-terminal domains that are highly conserved in plants and so may also be involved in regulatory interactions. Although we show that RSH3-GFP and RSH1-GFP appear to function as constitutive ppGpp synthases/hydrolases in bacteria and in plants, it is likely that overexpression and/or the fusion with GFP modify the regulatory properties of these enzymes relative to their native counterparts. The role of small-molecule and protein interactions in modulating the function of RSH enzymes in plants therefore remains very much an open question. Notably, such mechanisms would permit the rapid modulation of ppGpp levels in response to changes in chloroplast status, for example, in response to redox conditions, hormone signaling, temperature, or changes in nutrient availability as occurs in bacteria.
ppGpp signaling is likely to operate in a similar manner in all photosynthetic eukaryotes due to the broad conservation of both ppGpp targets and RSH genes (Atkinson et al., 2011). Indeed, ppGpp signaling may have been critical for taming the bacterial ancestor of the chloroplast by preventing its growth rate from outstripping the capacity of the eukaryotic host to provide nutrients. Important questions now remain: How are ppGpp biosynthesis and degradation regulated within the chloroplast? What is the role of ppGpp during different stress responses? How is ppGpp signaling integrated with other chloroplast regulatory mechanisms? Answering these questions will help us to further understand the central role of the chloroplast in photosynthetic eukaryotes.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana T-DNA insertion mutants were provided by the Signal Insertion Mutant Library (http://signal.salk.edu) and were obtained via the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info) (Supplemental Figure 7). Homozygous insertion mutants were isolated by PCR genotyping (see Supplemental Data Set 1 for primers). Mutant lines were combined by crossing and confirmed by PCR genotyping. qRT-PCR was used to determine the accumulation of transcripts in the mutants (Supplemental Figure 7). The arc6 allele was the T-DNA insertion line SAIL_693_G04 (kindly provided by C. Laloi). For a given experiment, the seeds for each line to be analyzed were harvested and bulked from multiple individual plants that had grown alongside all the other lines analyzed. For in vitro growth, seeds were surface-sterilized with 75% ethanol, dried, plated onto Petri dishes containing growth medium (0.5× Murashige and Skoog salts [Sigma-Aldrich], 1% sucrose, 0.5 g/L MES, and 0.4% Phytagel [Sigma-Aldrich], adjusted to pH 5.7 with KOH), and placed at 4°C for 2 d in the darkness for stratification. Plates were then transferred to a 16-h-light/8-h-dark photoperiod at 22°C/19.5°C with 80 µmol m−2 s−1 PAR fluorescent lighting. For growth in soil, seeds were stratified as above, germinated in soil, and then transferred into pots at 4 d after germination. The plants were then grown under a 16-h-light/8-h-dark photoperiod at 18/22°C with 115 µmol m−2 s−1 PAR fluorescent lighting and a weekly application of Coïc-Lesaint fertilizer solution. Unless stated otherwise, for chemical treatments, seedlings grown on plates for 12 DAS were treated by submersion in water containing 30 µM dexamethasone (Sigma-Aldrich; from a 0.01 M stock in DMSO) or 1 mM lincomycin (Sigma-Aldrich) for 3 min and then returned to growing conditions.
Cloning and Plant Transformation
RSH Overexpression Lines
RSH1, RSH2, and RSH3 sequences were amplified from Arabidopsis genomic or cDNA using Phusion polymerase (New England Biolabs) (see Supplemental Data Set 1 for primers). The PCR products were then introduced by Invitrogen BP Gateway recombination (Life Technologies) into pDONR207. The entry clones were confirmed by sequencing and recombined by Invitrogen LR Gateway recombination (Life Technologies) into pEarleyGate103 under the control of the constitutive 35S promoter and with a C-terminal GFP tag (Earley et al., 2006). The resulting constructs were transferred into Agrobacterium tumefaciens (strain GV3101) and used to transform wild-type plants by floral dipping (Clough and Bent, 1998). Transgenic plants were then selected by screening the resulting seeds on soil for resistance to the herbicide BASTA. Lines stably expressing RSH genes across multiple generations were then identified by immunoblotting.
Genomic RSH3 Complementation Lines
The genomic RSH3 sequence including the 3′ untranslated region, 5′ untranslated region, and 3.4 kb of upstream sequence containing the promoter was amplified from Arabidopsis genomic DNA using Phusion polymerase (New England Biolabs). The PCR product was then introduced by Invitrogen BP Gateway recombination into pDONR207. The entry clone was confirmed by sequencing and recombined by Invitrogen LR Gateway recombination into pGGW6 (Field and Osbourn, 2008) (kindly provided by Alan Herr). The resulting constructs were transferred into Agrobacterium (strain GV3101) and used to transform DM-23 plants by floral dipping.
Inducible SYN and ΔSYN Plants
A fragment corresponding to amino acids 1 to 386 of RelA was amplified from E. coli K-12 MG1655 by PCR. Fragments of RelA that lack the C terminus have constitutive ppGpp synthase activity in E. coli (Schreiber et al., 1991). The RelA fragment was then fused by PCR to a genomic sequence coding for the 80-amino acid Rubisco small subunit 1A (RBCS1A) target peptide that is able to target chimeric proteins to the chloroplast (Lee et al., 2002). The fused PCR product (SYN) was then introduced into pENTR/D-Topo (Life Technologies). The entry clone was confirmed by sequencing. ΔSYN was then created using site-directed mutagenesis to convert the codon encoding aspartate 275 of RelA to glycine, thereby inactivating the ppGpp synthase domain (Hogg et al., 2004). SYN and ΔSYN were then recombined by Invitrogen LR Gateway recombination into the plant steroid inducible expression vector pOPOn2.1 (kindly provided by Ian Moore) (Craft et al., 2005). The resulting constructs were transferred into Agrobacterium (strain GV3101) and used to transform wild-type plants by floral dipping to give SYN and ΔSYN inducible plants. Independent lines with stable inducible expression across multiple generations were selected. All SYN lines showed similar phenotypes. One SYN (43A10) and one ΔSYN line (44B13) were used in this study. The T-DNA insertion sites were identified by HIT PCR (Liu and Chen, 2007): 43A10 after Chr3 23000651; 44B13 after Chr3 23185643.
Inducible MESH and ΔMESH Plants
The Drosophila melanogaster MESH1 was PCR amplified from cDNA clone IP06414 (provided by the Drosophila Genomics Resource Center). The MESH1 PCR fragment was fused by PCR to a genomic sequence coding for the RBCS1A target peptide and introduced into pENTR/D-Topo. The entry clone (MESH) was confirmed by sequencing. ΔMESH was created using site-directed mutagenesis to convert the codon encoding histidine 62 of MESH to phenylalanine, thereby inactivating the ppGpp hydrolase domain (Sun et al., 2010). cytMESH was constructed as for MESH but without the Rubisco small subunit target peptide. The resulting clones were then recombined by Invitrogen LR Gateway recombination into the plant expression vector pOPOn2, transferred into Agrobacterium (strain GV3101), and used to transform wild-type plants by floral dipping to give inducible MESH, ΔMESH, and cytMESH plants. Independent lines with stable inducible expression across multiple generations were selected.
Artificial MicroRNA Lines
An artificial microRNA targeting CRSH was constructed as previously described (Schwab et al., 2006) and introduced into pDONR207. The clones were sequenced, recombined into pEarleyGate 103 under the control of the constitutive 35S promoter and used to transform TM-123 and wild-type plants by floral dipping to give QMa and crsh-ami plants. Twenty independent lines were selected, and reduction of CRSH expression was confirmed by qRT-PCR in lines used for further experiments (Supplemental Figure 7).
Plasmids for Escherichia coli Hydrolase Tests
MESH and ΔMESH sequences were amplified from plasmids pENTR-MESH and pENTR-ΔMESH (see above). The DNA fragments were digested with restriction enzymes EcoRI and XhoI and introduced into pBAD24 (Guzman et al., 1995) opened with EcoRI and SalI. The mature RSH1, RSH2, RSH3, and CRSH coding sequences were amplified from Arabidopsis cDNA using Phusion polymerase (New England Biolabs), and the mature RSH1-GFP, RSH2-GFP, and RSH3-GFP coding sequences were amplified from the pEarleyGate103 constructs described above for plant transformation or assembled by fusion PCR. The PCR fragments were digested with PciI and PstI and introduced into pBAD24 opened with NcoI and PstI. Vectors encoding inactive forms of the enzymes were constructed by mutating essential residues in the synthase domains in RSH2 (D451G) and RSH3 (D452G) and the hydrolase domain in RSH1 (R166A) (Hogg et al., 2004). Introduced sequences were confirmed by sequencing.
RNA Isolation and qRT-PCR Analysis
RNA was extracted from plant tissue using TriReagent (Sigma-Aldrich), quality was confirmed by gel electrophoresis, and genomic DNA removed by treatment with DNase. cDNA was then synthesized from 500 ng of RNA using a Primescript RT Reagent Kit (Takara Bio) with random hexamer primers. qRT-PCR was performed on 1 μL of 1 in 40 diluted cDNA in 15-μL reactions using SYBR Premix Ex-Taq II reagent (Takara Bio) in a Bio-Rad CFX96 real-time system (see Supplemental Data Set 1 for primer pairs). Relative quantification of gene expression adjusted for efficiency was performed using PCR Miner (Zhao and Fernald, 2005). For each analysis, multiple validated reference genes were tested (Czechowski et al., 2005). Reference genes were excluded if their stability values were not within advised limits (M < 0.5 and Cv < 0.25) (Vandesompele et al., 2002). qRT-PCR was also used to measure plastid DNA in isolated chloroplasts as described previously (Rowan and Bendich, 2011). For RNA gels (Figures 1D and 3E), total RNA was denatured by heating at 70°C for 10 min in 47.5% formamide, 0.25 mM EDTA, and 0.0125% SDS before loading.
Extraction and Quantification of ppGpp by Ultraperformance Liquid Chromatography-Tandem Mass Spectrometry
ppGpp extraction was performed according to Ihara et al. (2015) with minor modifications. Approximately 100 mg of plant tissue was extracted in 3 mL 2 M formic acid on ice. After 30 min, 3 mL of 50 mM ammonium acetate, pH 4.5, was added and the sample was split into two portions, to one of which was added 25 μL 500 nM ppGpp (Trilink). The samples were then passed through prepared 1-mL Oasis WAX columns (Waters), washed with 1 mL 50 mM ammonium acetate, pH 4.5, and 1 mL methanol, and eluted with 1 mL methanol/water/NH4OH (20:70:10). The eluate was lyophilized and resuspended in 200 μL water, and then it was filtered through a NucleoSpin column (Machery and Nagel). The eluate was then adjusted to 6% acetonitrile and 10 μL was injected into an Acquity UPLC system (Waters) and separated on a Kinetex C18 (100 × 2.10 mm) with 2.6-µm particle size (Phenomenex). Mass spectrometric detection was performed with a SYNAP G2S mass spectrometer (Waters) with the ESI ion source set to negative ion mode. ppGpp was detected in tof MRM mode. The mass of the chosen parent ion (601.95 m/z) was selected by the quadrupole and fragmented in the collision cell to the target ion (158.95 m/z). The cone voltage was at 30 V, and the collision energy followed a power ramp from 15 to 40eV. ppGpp levels were then quantified against a standard curve and adjusted using the recovery rate calculated for individual samples. To avoid positive quantification bias in samples containing little ppGpp (such as the wild type), the calibration curve was modified to the form y = ax rather than y = ax+b, which was used previously (Ihara et al., 2015). This approach produced results that corresponded well with ppGpp measurements on more concentrated samples derived from large scale extractions and with previous measurements of ppGpp in plants (Takahashi et al., 2004). Large-scale extractions were performed on 500 mg of plant sample using 5-fold greater volumes and purification on 5-mL Oasis WAX columns. After lyophilization, the samples were suspended in 200 μL volume of water, as above, to give a 5-fold increase in analyte concentration.
Metabolic Labeling of Newly Synthesized RNA
Newly synthesized RNA was labeled with 4SU as described previously with some modifications (Sidaway-Lee et al., 2014). The 12-DAS seedlings were labeled 15 min after dawn by flooding with 1.5 mM 4SU (Carbosynth) in 0.5× Murashige and Skoog salts and 0.01% Silwet. Seedlings were frozen in liquid nitrogen after exactly 45 min. Total RNA was then extracted using TriReagent (Life Technologies). Seventy-five micrograms of total RNA was biotinylated in 10 mM Tris-Cl, pH 7.4, 1 mM EDTA, and 0.2 mg/mL in EZ-Link HPDP-Biotin (Life Technologies) for 1.5 h at room temperature. Unbound biotin was removed by chloroform extraction using phase lock gel (5 Prime), and the RNA was precipitated from the aqueous phase by adding one-tenth volume of 5 M NaCl and 1.1 volumes of isopropanol. Biotinylated RNA was then was separated from unlabeled RNA using streptavidin-coated magnetic beads (New England Biolabs). Biotinylated RNA (75 to 100 µg) was added to the beads, and the solution was incubated for 20 min at room temperature. The beads were washed three times with 1 mL of 65°C washing buffer (1 M NaCl, 100 mM Tris-Cl, pH 7.4, and 10 mM EDTA) and three times with 1 mL of room temperature washing buffer. Labeled RNA was then eluted by the addition of two portions of 5% β-mercaptoethanol. RNA was precipitated in the presence of glycogen by adding one-tenth volume of 5 M NaCl and 1.1 volumes of isopropanol, resuspended in 10 μL water, and quantified using QUBIT RNA HS (Thermo Fisher Scientific).
Metabolic Labeling of Newly Synthesized Proteins with Puromycin
The 12-DAS in vitro grown plants were treated by flooding the plates with 30 µM dexamethasone or 1 mM lincomycin for 3 min and then returned to growing conditions. After a fixed time, the plants were removed from the plates and vacuum infiltrated with the labeling mixture (1 mM KH2PO4, pH 6.3, 0.1% Tween 20, 50 µg/mL puromycin [Apollo Scientific], and 100 µg/mL cycloheximide). Plants were incubated in Petri dishes for exactly 1 h under growing conditions before being frozen in liquid nitrogen. A fraction highly enriched in whole chloroplasts was then extracted from the frozen tissue essentially as previously described by homogenization in homogenization buffer (10 mM Tricine KOH, pH 7.5, 0.4 M sucrose, 10 mM NaCl, 5 mM MgCl2, 100 mM ascorbate, 0.2 mM PMSF, 1 mM benzamidine, 5 mM aminocaproic acid, and 1 mM lincomycin), filtration through a 30-µm mesh, and centrifugation (Pesaresi, 2011). This chloroplast extract was then used directly for protein extraction and immunoblotting.
Chlorophyll Quantification
Frozen plant powder or leaf discs were extracted with ice-cold 90% acetone saturated with sodium carbonate. The extract was adjusted to 80% acetone and the absorbance measured between 350 and 750 nm in a Varian Cary 300 spectrophotometer (Agilent). Chlorophyll concentrations and chlorophyll a/b ratios were calculated using a fitting algorithm as described previously (Croce et al., 2002).
Chlorophyll Fluorescence
Plants were dark adapted for 20 min, and chlorophyll fluorescence was measured in a Fluorcam FC 800-O imaging fluorometer (Photon System Instruments). The standard protocol included in the supplied Fluorcam 7 software was used to image F0 and Fm. PSII maximum quantum yield was calculated as (Fm − F0)/Fm.
Protein Separation and Immunoblotting
Proteins were extracted in 2× SDS sample buffer (100 mM Tris-HCl, pH 6.8, 25 mM EDTA, 4% SDS, and 20% glycerol) by heating at 85°C for 5 min. Protein concentration was measured using the BCA assay (Sigma-Aldrich). Proteins were then reduced with 5% β-mercaptoethanol and equal quantities separated by SDS-PAGE and either stained with Coomassie Brilliant Blue or transferred onto nitrocellulose membranes according to the manufacturer’s instructions (Bio-Rad). Transfer homogeneity was confirmed by Ponceau Red staining. After incubation with 5% nonfat milk in TBST (10 mM Tris, pH 8.0, 150 mM NaCl, and 0.1% Tween 20) for 60 min, the membrane was incubated in the same buffer with antibodies at a 1/8000 dilution unless otherwise stated for 1 h at room temperature. Antibodies were used against AtpA (Agrisera; polyclonal, catalog number AS08 304, lot 1006), GFP (Roche; catalog number 11814460001, clones 7.1 and 13.1), HA (Sigma-Aldrich; catalog number H9658, clone HA-7), LHCA1 (Agrisera; polyclonal, catalog number AS01 005, lot 0512), LHCA4 (Agrisera; polyclonal, catalog number AS01 008, lot 0508), LHCB1 (Agrisera; polyclonal, catalog number AS01 004, lot 0806), LHCB4 (Agrisera; polyclonal, catalog number AS04 045, lot 0601), PetA (Agrisera; polyclonal, catalog number AS08 306, lot 0912), PsaC (Agrisera; polyclonal, catalog number AS04 042, lot 0601), PsbA (Agrisera; polyclonal, catalog number AS05 084, lot 1201), puromycin (1/1000 dilution of 10 mg/mL stock; kindly provided by P. Pierre and E. Gatti, clone 12D10; Schmidt et al., 2009), and RelA (1/2000 dilution, raised against E. coli RelA and kindly provided by M. Cashel). The membrane was then washed three times for 5 min in TBST and incubated in 5% nonfat milk in TBST with horseradish peroxidase-conjugated anti-mouse (Cell Signaling Technology; catalog number 7076, lot 25) or anti-rabbit (Cell Signaling Technology; catalog number 7074, lot 25) antibodies at a 1/10,000 dilution for 1 h at room temperature. The membrane was then washed a further three times in TBST, developed using Immobilon ECL substrate (Millipore), and imaged with a Fusion FX7 imager (Vilber Lourmat). For quantitative analysis, bands or lanes from the raw 16-bit TIFF images were integrated using ImageJ analysis software (National Institutes of Health).
Chloroplast Number and Volume Analysis
Protoplasts were made from leaves by digestion with cellulase and macerozyme (Yoo et al., 2007) and examined in resuspension solution within 16 h using a light microscope (Axio Imager M2; Zeiss). Chloroplast volume was approximated to a hemisphere (2/3 πr3), and the Feret diameter was used to calculate the radius. Average chloroplast volume was calculated for 300 chloroplasts for each sample within an experiment. This was then used to calculate total chloroplast volume in individual protoplasts. Chloroplast area was also analyzed in fixed plant cells as described previously (Pyke and Leech, 1991).
Synthase and Hydrolase Tests in E. coli
For testing ppGpp synthase activity, plasmids were transformed either into E. coli strain EB425 (MG1655∆relA∆spoT) (Wahl et al., 2011) and grown at 37°C on plates of M9 minimal medium without amino acids or into E. coli strain EB421 (MG1655∆relA) (Wahl et al., 2011) and grown at 37°C on SMG medium as described previously (Battesti and Bouveret, 2006).
For testing ppGpp hydrolase activity, plasmids were transformed into E. coli strain EB544 (MG1655∆relAspoT203) (My et al., 2013). Transformants could not be obtained for plasmids containing RSH2 or RSH3 presumably due to leaky expression and the accumulation of lethal levels of ppGpp. Precultures from independent colonies for each replicate were diluted in 150 μL Luria-Bertani medium containing ampicillin in a 96-well microplate and grown in a TECAN automated plate reader at 37°C, and optical density was measured at 600 nm every 10 min.
Senescence Induction
For senescence induction, all fully expanded leaves were detached from 3- to 4-week-old long-day-grown or 6- to 8-week-old short-day-grown plants and placed together in individual Petri dishes with moistened filter paper. The Petri dishes were then wrapped in foil and placed in the dark at 18 to 22°C. Leaves were analyzed after 3 to 6 d. For analysis, all the leaves from each plant were ground to a fine powder with liquid nitrogen before measurement of chlorophyll levels or extraction of total proteins. At least three plants were analyzed per line and per treatment.
Statistical Testing
Sample sizes were chosen to identify the smallest effect size that was practically obtainable. The two-way Student’s t test was used to compare control samples with treatment samples. ANOVA was used to compare multiple sample means, with the Dunnett test post hoc. For samples with non-normal distributions (Jarque-Bara test), the nonparametric Kruskal-Wallis test was used with the Dunn test post hoc.
Image Processing
Digitally acquired images were processed in Adobe Photoshop or Net.Paint and assembled into figures in Adobe Illustrator. The Adobe Photoshop white point function was used for the images in Figures 1A, 2A, and 7C. For visualization of immunoblotting results, the levels of raw nonsaturated 16-bit TIFF images were adjusted in a linear fashion to accurately reveal the bands, converted to 8-bit, black-to-white inverted, and cropped before placing into the figure panels.
Accession Numbers
Sequence data from this article can be found for Arabidopsis genes in The Arabidopsis Information Resource (http://www.arabidopsis.org/) under the following accession numbers: At4g02260 (RSH1), At3g14050 (RSH2), At1g54130 (RSH3), At3g17470 (CRSH), AtCg00020 (PsbA), At1g29910- At1g29920- At1g29930 (LHCB1), At2g40100-At3g08940-At5g01530 (LHCB4), AtCg00340 (PsaB), At3g47479 (LHCA4), AtCg00120 (AtpA), AtCg00540 (PetA), AtCg00490 (RBCL), At1g67090 (RBCS1A), and At5g42480 (ARC6); for E. coli genes in EcoCyc (http://ecocyc.org/) under the accession numbers EG10835 (RelA),EG10966 (SpoT), and EG10230 (DksA); and for Drosophila genes in Flybase (http://flybase.org/) under accession number FBgn0039650 (Mesh1). Accession numbers for genes used in qRT-PCR experiments can be found in Supplemental Data Set 1.
Supplemental Data
Supplemental Figure 1. Arabidopsis RSH domain structure.
Supplemental Figure 2. Complementation of ppGpp-deficient E. coli mutants by RSH2-GFP and RSH3-GFP.
Supplemental Figure 3. Phenotypes of RSH2-GFP and RSH3-GFP overexpression lines.
Supplemental Figure 4. Growth of SYN and ∆SYN following induction.
Supplemental Figure 5. qRT-PCR analysis of plants overexpressing RSH3-GFP.
Supplemental Figure 6. Proof of concept for puromycin labeling in plants.
Supplemental Figure 7. Insertion sites and gene expression in the RSH mutants.
Supplemental Figure 8. ppGpp levels in QMaii and RSH1-GFP-overexpressing plants.
Supplemental Figure 9. Phenotypes of RSH mutants during vegetative growth.
Supplemental Figure 10. Phenotypes of RSH mutants under short-day conditions.
Supplemental Figure 11. Developmental expression profiles of RSH genes.
Supplemental Figure 12. Additional dark-induced senescence phenotypes.
Supplemental Figure 13. Natural senescence is affected in RSH mutants.
Supplemental Figure 14. RSH mutants have altered seed weight.
Supplemental Figure 15. Rubisco degradation is regulated by ppGpp during dark-induced senescence.
Supplemental Data Set 1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank F.-A. Wollman, S. England, A. Matthes, B. Menand, and M. Montane for critical comments on the article, M. Cashel and H. Fakhfakh for support, and K. Alvarez, M. Reissolet, J. Ferrandi, M. Dussenne, and J. Garcia for assistance. We also thank the Plateau Technique of the Max Mousseron Institute of Biomolecules at Montpellier for ppGpp quantification. The project was supported by the Agence National de la Recherche (ANR-10-JCJC-1201), by an AMIDEX grant (ANR-11-IDEX-0001-02), and by base funding from Aix-Marseille University, the CNRS, and the CEA. B.F. was a recipient of a FEBS postdoctoral fellowship and now receives a salary from the CNRS, C.R. and S.C. receive a salary from Aix-Marseille University, M.S. was supported by ANR-10-JCJC-1201, H.A. received mobility grants from the Tunisian Ministry of Higher Education, and H.K. is supported by a PhD studentship from the China Scholarship Council.
AUTHOR CONTRIBUTIONS
B.F. and C.R. developed the initial concept. B.F. and M.S. designed research for all parts, S.C. for chlorophyll and photosynthesis analysis and E.B. for microbiology. B.F., M.S., H.A., H.K., and E.B. performed experiments and analyzed results. B.F. and M.S. wrote the article.
Glossary
- ppGpp
guanosine penta- and tetraphosphate
- RSH
RelA and SpoT homolog
- ABA
abscisic acid
- DAS
days after stratification
- GK
guanylate kinase
- IC50
50% inhibitory concentration
- 4SU
4-thio uridine
Footnotes
Articles can be viewed online without a subscription.
References
- Atkinson G.C., Tenson T., Hauryliuk V. (2011). The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6: e23479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang W.Y., Chen J., Jeong I.S., Kim S.W., Kim C.W., Jung H.S., Lee K.H., Kweon H.S., Yoko I., Shiina T., Bahk J.D. (2012). Functional characterization of ObgC in ribosome biogenesis during chloroplast development. Plant J. 71: 122–134. [DOI] [PubMed] [Google Scholar]
- Battesti A., Bouveret E. (2006). Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol. Microbiol. 62: 1048–1063. [DOI] [PubMed] [Google Scholar]
- Belgio E., Johnson M.P., Jurić S., Ruban A.V. (2012). Higher plant photosystem II light-harvesting antenna, not the reaction center, determines the excited-state lifetime-both the maximum and the nonphotochemically quenched. Biophys. J. 102: 2761–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner T., Aleynikova A.Y., Zubo Y.O., Kusnetsov V.V. (2015). Chloroplast RNA polymerases: Role in chloroplast biogenesis. Biochim. Biophys. Acta 1847: 761–769. [DOI] [PubMed] [Google Scholar]
- Breeze E., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V., Page T., Harrison E., Breeze E., Lim P.O., Nam H.G., Lin J.F., Wu S.H., Swidzinski J., Ishizaki K., Leaver C.J. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 42: 567–585. [DOI] [PubMed] [Google Scholar]
- Cahoon A.B., Harris F.M., Stern D.B. (2004). Analysis of developing maize plastids reveals two mRNA stability classes correlating with RNA polymerase type. EMBO Rep. 5: 801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Bang W.Y., Lee Y., Kim S., Lee K.W., Kim S.W., Son Y.S., Kim D.W., Akhter S., Bahk J.D. (2014). AtObgC-AtRSH1 interaction may play a vital role in stress response signal transduction in Arabidopsis. Plant Physiol. Biochem. 74: 176–184. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Craft J., Samalova M., Baroux C., Townley H., Martinez A., Jepson I., Tsiantis M., Moore I. (2005). New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. Plant J. 41: 899–918. [DOI] [PubMed] [Google Scholar]
- Croce R., Canino G., Ros F., Bassi R. (2002). Chromophore organization in the higher-plant photosystem II antenna protein CP26. Biochemistry 41: 7334–7343. [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux Z.D., Swanson M.S. (2012). ppGpp: magic beyond RNA polymerase. Nat. Rev. Microbiol. 10: 203–212. [DOI] [PubMed] [Google Scholar]
- Diray-Arce J., Liu B., Cupp J.D., Hunt T., Nielsen B.L. (2013). The Arabidopsis At1g30680 gene encodes a homologue to the phage T7 gp4 protein that has both DNA primase and DNA helicase activities. BMC Plant Biol. 13: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Engelmann E.C., Zucchelli G., Garlaschi F.M., Casazza A.P., Jennings R.C. (2005). The effect of outer antenna complexes on the photochemical trapping rate in barley thylakoid photosystem II. Biochim. Biophys. Acta 1706: 276–286. [DOI] [PubMed] [Google Scholar]
- Feller U., Anders I., Mae T. (2008). Rubiscolytics: fate of Rubisco after its enzymatic function in a cell is terminated. J. Exp. Bot. 59: 1615–1624. [DOI] [PubMed] [Google Scholar]
- Field B., Osbourn A.E. (2008). Metabolic diversification--independent assembly of operon-like gene clusters in different plants. Science 320: 543–547. [DOI] [PubMed] [Google Scholar]
- Gerhardt R., Stitt M., Heldt H.W. (1987). Subcellular metabolite levels in spinach leaves: Regulation of sucrose synthesis during diurnal alterations in photosynthetic partitioning. Plant Physiol. 83: 399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green B.R. (2011). Chloroplast genomes of photosynthetic eukaryotes. Plant J. 66: 34–44. [DOI] [PubMed] [Google Scholar]
- Guzman L.M., Belin D., Carson M.J., Beckwith J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177: 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg T., Mechold U., Malke H., Cashel M., Hilgenfeld R. (2004). Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response [corrected]. Cell 117: 57–68. [DOI] [PubMed] [Google Scholar]
- Ihara Y., Ohta H., Masuda S. (2015). A highly sensitive quantification method for the accumulation of alarmone ppGpp in Arabidopsis thaliana using UPLC-ESI-qMS/MS. J. Plant Res. 128: 511–518. [DOI] [PubMed] [Google Scholar]
- Ishida H., Izumi M., Wada S., Makino A. (2014). Roles of autophagy in chloroplast recycling. Biochim. Biophys. Acta 1837: 512–521. [DOI] [PubMed] [Google Scholar]
- Ito D., Kato T., Maruta T., Tamoi M., Yoshimura K., Shigeoka S. (2012). Enzymatic and molecular characterization of Arabidopsis ppGpp pyrophosphohydrolase, AtNUDX26. Biosci. Biotechnol. Biochem. 76: 2236–2241. [DOI] [PubMed] [Google Scholar]
- Jarvis P., López-Juez E. (2013). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14: 787–802. [DOI] [PubMed] [Google Scholar]
- Kasai K., Usami S., Yamada T., Endo Y., Ochi K., Tozawa Y. (2002). A RelA-SpoT homolog (Cr-RSH) identified in Chlamydomonas reinhardtii generates stringent factor in vivo and localizes to chloroplasts in vitro. Nucleic Acids Res. 30: 4985–4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindgren P., Kremnev D., Blanco N.E., de Dios Barajas López J., Fernández A.P., Tellgren-Roth C., Kleine T., Small I., Strand A. (2012). The plastid redox insensitive 2 mutant of Arabidopsis is impaired in PEP activity and high light-dependent plastid redox signalling to the nucleus. Plant J. 70: 279–291. [DOI] [PubMed] [Google Scholar]
- Krásný L., Gourse R.L. (2004). An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 23: 4473–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.H., Kim D.H., Lee S.W., Kim Z.H., Hwang I. (2002). In vivo import experiments in protoplasts reveal the importance of the overall context but not specific amino acid residues of the transit peptide during import into chloroplasts. Mol. Cells 14: 388–397. [PubMed] [Google Scholar]
- Liere K., Weihe A., Börner T. (2011). The transcription machineries of plant mitochondria and chloroplasts: Composition, function, and regulation. J. Plant Physiol. 168: 1345–1360. [DOI] [PubMed] [Google Scholar]
- Lim P.O., Kim H.J., Nam H.G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58: 115–136. [DOI] [PubMed] [Google Scholar]
- Liu Y.G., Chen Y. (2007). High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43: 649–650, 652, 654 passim. [DOI] [PubMed] [Google Scholar]
- Maekawa M., Honoki R., Ihara Y., Sato R., Oikawa A., Kanno Y., Ohta H., Seo M., Saito K., Masuda S. (2015). Impact of the plastidial stringent response in plant growth and stress responses. Nat. Plants 1: 15167. [DOI] [PubMed] [Google Scholar]
- Masuda S. (2012). The stringent response in phototrophs. In Advances in Photosynthesis: Fundamental Aspects, Najafpour M., ed (Croatia: InTech; ), pp. 488–500. [Google Scholar]
- Masuda S., Mizusawa K., Narisawa T., Tozawa Y., Ohta H., Takamiya K. (2008). The bacterial stringent response, conserved in chloroplasts, controls plant fertilization. Plant Cell Physiol. 49: 135–141. [DOI] [PubMed] [Google Scholar]
- Mizusawa K., Masuda S., Ohta H. (2008). Expression profiling of four RelA/SpoT-like proteins, homologues of bacterial stringent factors, in Arabidopsis thaliana. Planta 228: 553–562. [DOI] [PubMed] [Google Scholar]
- My L., Rekoske B., Lemke J.J., Viala J.P., Gourse R.L., Bouveret E. (2013). Transcription of the Escherichia coli fatty acid synthesis operon fabHDG is directly activated by FadR and inhibited by ppGpp. J. Bacteriol. 195: 3784–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura Y., Izumi A., Fukunaga Y., Kusumi K., Iba K., Watanabe S., Nakahira Y., Weber A.P., Nozawa A., and Tozawa Y. (2014). Diversity in guanosine 3′,5′-bisdiphosphate (ppGpp) sensitivity among guanylate kinases of bacteria and plants. J. Biol. Chem. 289: 15631–15641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura Y., Takabayashi T., Kuroda H., Yukawa Y., Sattasuk K., Akita M., Nozawa A., Tozawa Y. (2012). ppGpp inhibits peptide elongation cycle of chloroplast translation system in vitro. Plant Mol. Biol. 78: 185–196. [DOI] [PubMed] [Google Scholar]
- Pesaresi P. (2011). Studying translation in Arabidopsis chloroplasts. Methods Mol. Biol. 774: 209–224. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T., and Munné-Bosch S. (2013). Plastid signaling during the plant life cycle. In Plastid Development in Leaves during Growth and Senescence, Biswal B., Krupinska K., and Biswal U.C., eds (The Netherlands: Springer; ), pp. 503–528. [Google Scholar]
- Potrykus K., Cashel M. (2008). (p)ppGpp: still magical? Annu. Rev. Microbiol. 62: 35–51. [DOI] [PubMed] [Google Scholar]
- Puthiyaveetil S., Kavanagh T.A., Cain P., Sullivan J.A., Newell C.A., Gray J.C., Robinson C., van der Giezen M., Rogers M.B., Allen J.F. (2008). The ancestral symbiont sensor kinase CSK links photosynthesis with gene expression in chloroplasts. Proc. Natl. Acad. Sci. USA 105: 10061–10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K.A., Leech R.M. (1991). Rapid image analysis screening procedure for identifying chloroplast number mutants in mesophyll cells of Arabidopsis thaliana (L.) Heynh. Plant Physiol. 96: 1193–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp J.C., Baumgartner B.J., Mullet J. (1992). Quantitative analysis of transcription and RNA levels of 15 barley chloroplast genes. Transcription rates and mRNA levels vary over 300-fold; predicted mRNA stabilities vary 30-fold. J. Biol. Chem. 267: 21404–21411. [PubMed] [Google Scholar]
- Reyes-Prieto A., Weber A.P., Bhattacharya D. (2007). The origin and establishment of the plastid in algae and plants. Annu. Rev. Genet. 41: 147–168. [DOI] [PubMed] [Google Scholar]
- Robertson E.J., Pyke K.A., Leech R.M. (1995). arc6, an extreme chloroplast division mutant of Arabidopsis also alters proplastid proliferation and morphology in shoot and root apices. J. Cell Sci. 108: 2937–2944. [DOI] [PubMed] [Google Scholar]
- Rochaix J.D. (2013). Redox regulation of thylakoid protein kinases and photosynthetic gene expression. Antioxid. Redox Signal. 18: 2184–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan B.A., Bendich A.J. (2011). Isolation, quantification, and analysis of chloroplast DNA. Methods Mol. Biol. 774: 151–170. [DOI] [PubMed] [Google Scholar]
- Sato M., Takahashi K., Ochiai Y., Hosaka T., Ochi K., Nabeta K. (2009). Bacterial alarmone, guanosine 5′-diphosphate 3′-diphosphate (ppGpp), predominantly binds the beta’ subunit of plastid-encoded plastid RNA polymerase in chloroplasts. ChemBioChem 10: 1227–1233. [DOI] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506. [DOI] [PubMed] [Google Scholar]
- Schmidt E.K., Clavarino G., Ceppi M., Pierre P. (2009). SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6: 275–277. [DOI] [PubMed] [Google Scholar]
- Schreiber G., Metzger S., Aizenman E., Roza S., Cashel M., Glaser G. (1991). Overexpression of the relA gene in Escherichia coli. J. Biol. Chem. 266: 3760–3767. [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidaway-Lee K., Costa M.J., Rand D.A., Finkenstadt B., Penfield S. (2014). Direct measurement of transcription rates reveals multiple mechanisms for configuration of the Arabidopsis ambient temperature response. Genome Biol. 15: R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., et al. (2010). A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses. Nat. Struct. Mol. Biol. 17: 1188–1194. [DOI] [PubMed] [Google Scholar]
- Suzuki J.Y., Sriraman P., Svab Z., Maliga P. (2003). Unique architecture of the plastid ribosomal RNA operon promoter recognized by the multisubunit RNA polymerase in tobacco and other higher plants. Plant Cell 15: 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatecka-Hagenbruch M., Liere K., Börner T. (2007). High diversity of plastidial promoters in Arabidopsis thaliana. Mol. Genet. Genomics 277: 725–734. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Kasai K., Ochi K. (2004). Identification of the bacterial alarmone guanosine 5′-diphosphate 3′-diphosphate (ppGpp) in plants. Proc. Natl. Acad. Sci. USA 101: 4320–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller N., Bock R. (2014). The translational apparatus of plastids and its role in plant development. Mol. Plant 7: 1105–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozawa Y., Nomura Y. (2011). Signalling by the global regulatory molecule ppGpp in bacteria and chloroplasts of land plants. Plant Biol. (Stuttg.) 13: 699–709. [DOI] [PubMed] [Google Scholar]
- Tozawa Y., Nozawa A., Kanno T., Narisawa T., Masuda S., Kasai K., Nanamiya H. (2007). Calcium-activated (p)ppGpp synthetase in chloroplasts of land plants. J. Biol. Chem. 282: 35536–35545. [DOI] [PubMed] [Google Scholar]
- van der Biezen E.A., Sun J., Coleman M.J., Bibb M.J., Jones J.D. (2000). Arabidopsis RelA/SpoT homologs implicate (p)ppGpp in plant signaling. Proc. Natl. Acad. Sci. USA 97: 3747–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., and Speleman F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl A., My L., Dumoulin R., Sturgis J.N., Bouveret E. (2011). Antagonistic regulation of dgkA and plsB genes of phospholipid synthesis by multiple stress responses in Escherichia coli. Mol. Microbiol. 80: 1260–1275. [DOI] [PubMed] [Google Scholar]
- Yamburenko M.V., Zubo Y.O., Börner T. (2015). Abscisic acid affects transcription of chloroplast genes via protein phosphatase 2C-dependent activation of nuclear genes: repression by guanosine-3′-5′-bisdiphosphate and activation by sigma factor 5. Plant J. 82: 1030–1041. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhao S., Fernald R.D. (2005). Comprehensive algorithm for quantitative real-time polymerase chain reaction. J. Comput. Biol. 12: 1047–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.