Abstract
We propose a new tool named Autotract to automate fiber tracking in diffusion tensor imaging (DTI). Autotract uses prior knowledge from a source DTI and a set of corresponding fiber bundles to extract new fibers for a target DTI. Autotract starts by aligning both DTIs and uses the source fibers as seed points to initialize a tractography algorithm. We enforce similarity between the propagated source fibers and automatically traced fibers by computing metrics such as fiber length and fiber distance between the bundles. By analyzing these metrics, individual fiber tracts can be pruned. As a result, we show that both bundles have similar characteristics. Additionally, we compare the automatically traced fibers against bundles previously generated and validated in the target DTI by an expert. This work is motivated by medical applications in which known bundles of fiber tracts in the human brain need to be analyzed for multiple datasets.
Keywords: Tractography, automatic, diffusion tensor imaging, prior, atlas, registration
1. INTRODUCTION
Diffusion tensor imaging (DTI) is a magnetic resonance imaging method which allows measuring the anisotropic diffusion of water molecules in in-vivo biological tissue. Water molecules diffusion reveal the underlying structure of a tissue. In the brain, DTI data can be used to model the white matter (WM) and visually represent neural tracts. The WM tracts are reconstructed with a technique known as tractography, which estimates a tract trajectory by following likely directions of the tract at each voxel in the data.
There are two main tractography approaches, probabilistic1 (describe all possible trajectories) and streamline2 (describe the most probable trajectory). Tractography in this paper is focused on the latter approach. Tractography has found many applications over the past decade. Some applications include neurosurgical planning;3 assessment and study of neurological diseases such as multiple sclerosis4 or schizophrenia;5 and post-surgical validation. Tractography relays on the fiber-tracking algorithm and the estimation of accurate orientations. Complex fiber architecture is difficult to characterized and the quality of the results depend on the methods’ ability to resolve crossing of fibers, kissing, branching, and so on. Tractography is error-prone since entire fiber trajectories are calculated by stepping along the major eigenvector direction (principal diffusion direction) for a predefined step size (usually 0.5 mm) and using the Runge-Kutta numerical integration method of order 2 or higher.2
To reduce tracking variability, clustering and atlas-based methods are used. Clustering generates fiber bundles and validates fiber orientations. It is supported by the assumption that fiber trajectories that begin near each other, follow similar paths and terminate near each other belong to the same anatomical structure.6 On the other hand, atlas-based methods7, 8 use manually delineated fibers that are projected or transfered to the new subject being analyzed.
In this paper, we present Autotract, an automatic tractography tool that uses prior knowledge from manually corrected tracts in an atlas. Autotract produces fibers that are similar to the atlas fibers used as input. With Autotract, we seek to enable further clinical and research applications of tractography.
In summary, Autotract performs a pair-wise registration between a source and a target DTI. Using the source DTI fibers as seed points, new fibers are traced in the target DTI. To ensure the similarity of the fibers, Autotract computes similarity metrics such as the length of the fibers and the distance between individual fibers in the source and target bundles. Additionally, it enforces anatomical constraints from the target DTI to ensure the validity of the tracts. In other words, Autotract prunes fibers if they do not have similar length to the source, are far from it or pass through implausible regions such as the cerebral spinal fluid (CSF).
In order to validate this framework, we will compare the automatically traced fibers in the target DTI with fiber bundles previously traced and validated by an expert. We want to demonstrate that an automated tractography tool is feasible and could reduce the time spent by an expert doing this time consuming operation. We do not claim that the tracts produced by Autotract are correct but we wish to facilitate the task of propagating and cleaning fibers in a target DTI. The fibers bundles produced by this framework could be further refined or corrected by an expert.
The following section explains in detail the methods used in Autotract.
2. METHODS
This section explains in detail the tools used in Autotract. The first step is registration, followed by tractography and finally pruning of the fibers based on similarity metrics between the propagated target and source fiber bundles.
2.1 Registration
Autotract uses DTI-reg* to perform the registration between the source and target DTI. It is a two step approach were a deformation field is computed and the DTI data is resampled accordingly.
The registration method uses the fractional anisotropy (FA) map to calculate the deformation field. The FA is calculated as the normalized variance of the three eigenvalues at each voxel in the DTI. The FA value varies between 0 (isotropic diffusion) and 1 (infinite anisotropy). The deformation field is used as input for the DTI resampling. Optionally, the deformation field can also be provided as input.
Resampling DTI data is not a straight forward operation. Due to the directional nature of DTIs, using standard interpolation methods would result in losing the structural information of the image. To preserve it, the interpolation is done in euclidean space as shown by.9
After this registration step, random seed points selected from the input fibers are used to initialize the tractography algorithm explain the following section.
2.2 Tractography
The tractography algorithm is based on the second-order Runge-Kutta method. Points from the input fibers were seeded and the tractography was performed by following small steps along the principal diffusion direction. Tracking stops when the FA drops below a threshold value (default is 0.1) or when fiber direction changes rapidly, this could be due to noise or abnormal curvatures†.
The following section explains the fiber pruning step performed on the traced fibers.
2.3 Fiber pruning
The fiber pruning starts by cropping the fibers that go outside of the WM region. The WM region can be provided as input to Autotract or it is calculated using the FA map and thresholding with Otsu’s method.10 The CSF mask can also be provided as input or it is generated with a multi Otsu threshold segmentation. Specifically, the FA is divided into three clusters and the CSF is considered to lie in the first cluster. The fibers are pruned according to following criteria:
Using morphological operations, the dilated WM and eroded CSF masks are used to check weather a fiber remains inside the WM and does not go through the CSF.
Voxelizing the source fiber bundles and using them as a mask to check weather the newly generated tract remains inside the bundle. We check if there is more than 60% overlap between them.
Using the propagated source fiber bundle length to detect outliers in the automatically traced fibers.
Finally, we check if localized regions in the fiber do not deviate more than 10 mm from the source bundle.
The following section shows the results of this framework.
3. MATERIALS
We apply our method to DTI data from 15 healthy infants (scanned at 1 month, 1 year and 2 years approximately). A DTI atlas was generated for the 2 year old DTI scans with in house tool DTIAtlasBuilder‡. This atlas has a set of fiber bundles previously traced and validated by an expert and they will serve as ground truth for comparison.
A second atlas is generated with DTI data from 30 neonate infants. This atlas has fiber bundles previously traced, pruned and validated by an expert. These fiber bundles will be used as input to the framework.
The following fiber bundles were traced in both source and target atlas: cingulate cortex (CgC); cingulum of the hippocampal region (CgH); corpus callosum (CC); cortico fugal (CoF); cortico thalamic (CoTh); Fornix (Fo); inferior fronto-occipital fasciculus (IFO); inferior longitudinal fasciculus (ILF); superior longitudinal fasciculus (SLF); and optic radiation (OpR);
The following section shows the results of this framework and a comparison between the ground truth and automatically traced bundles.
4. RESULTS
Using the source atlas DTI and fiber bundles plus a target DTI, new fiber bundles are traced automatically using the framework described above. With Autotract we want to determine if the automated cleaning of a bundle produces results comparable to bundles validated by an expert user. We do not claim that the bundles produced by Autotract are correct, but we propose an automated tool to reduce the time consuming operation of tracing and cleaning bundles with available tools today.
We propose to compare the generated fibers by Autotract against the bundles validated by an expert in the target DTI.
Figure 1.a shows the source fiber bundles used as input to Autotract. The fiber bundles shown in Figure 1.b are are the fibers previously traced and Figure 1.c shows the resulting fibers using Autotract. The coloring of the bundles is done using tensor orientation.
Figure 1.
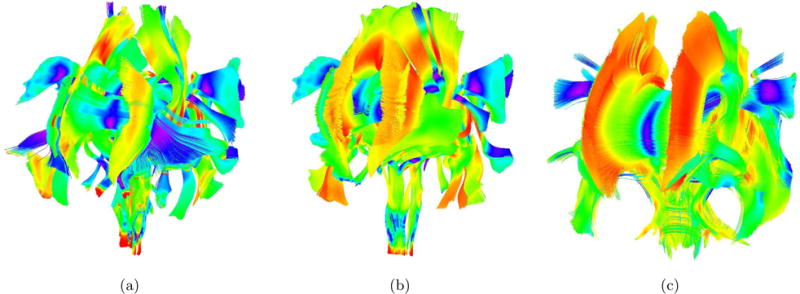
a) Propagated source fiber bundles validated by an expert. b) Propagated target fiber bundles validated by an expert. c) Propagated target fiber bundles generated by Autotract using (a) as input.
4.1 Average fiber length
We compute the average length for all fiber bundles.
Let α be a fiber bundle, then α = {A1, A2, …, AN} where AI is a single fiber in the bundle and N is the number of fibers in the bundle. Each fiber is defined as AI = {a1, a2, …, an} where n is the number of points in each fiber.
With this definition we can now define the average bundle length as shown in equation 2 where dist is the euclidean distance between two points.
| (1) |
| (2) |
Figures 2, 3 and 4 show the boxplots for every fiber bundle in the source, target (ground truth) and automatically traced bundles. The boxplot displays the distribution of fiber length per bundle based on the five number summary: minimum, first quartile, median, third quartile, and maximum.
Figure 2.
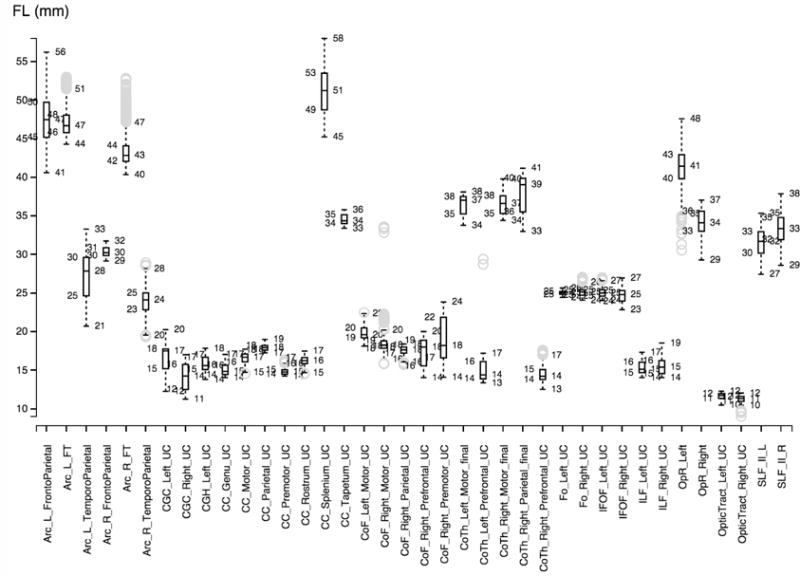
Boxplot visualization for the propagated source bundles.
Figure 3.
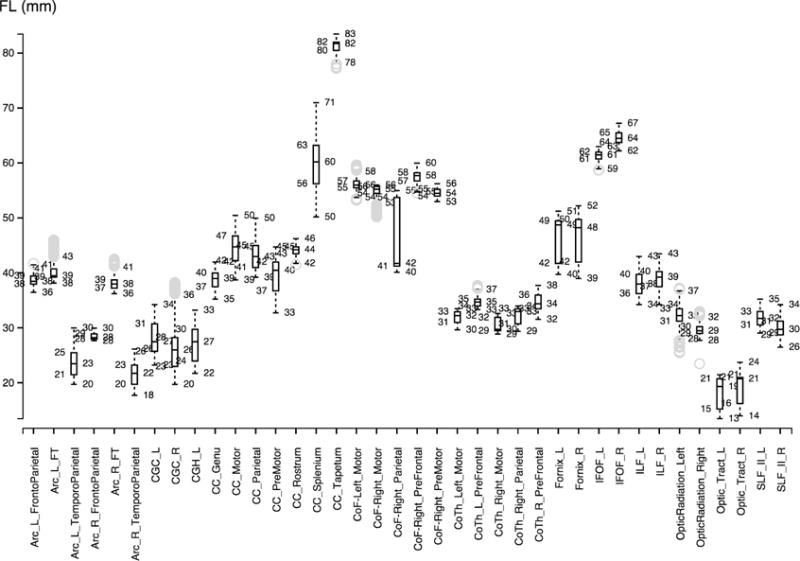
Boxplot visualization for the propagated target bundles.
Figure 4.
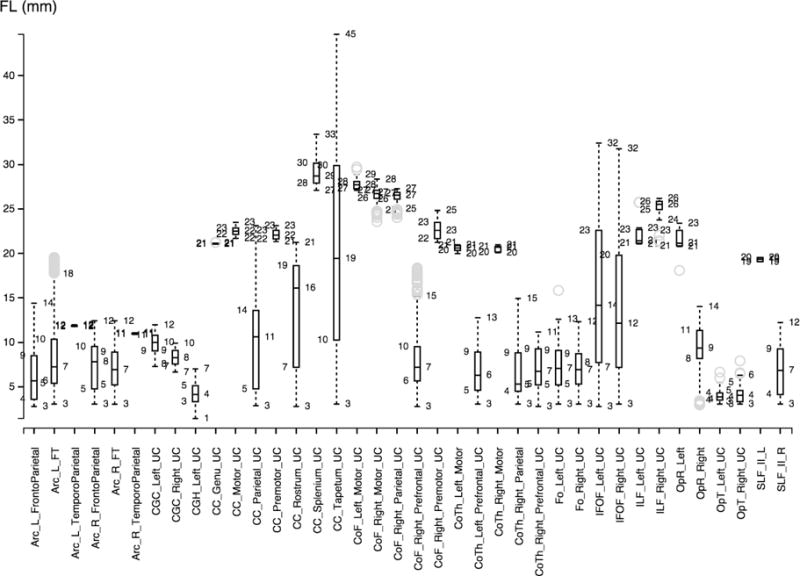
Boxplot visualization for the bundles produced by Autotract.
Figure 5 shows the average bundle length for the fibers traced by Autotract and those validated by the expert. Autotract produced fiber bundles smaller than expected. This suggest that new stopping criteria should be considered in the future.
Figure 5.
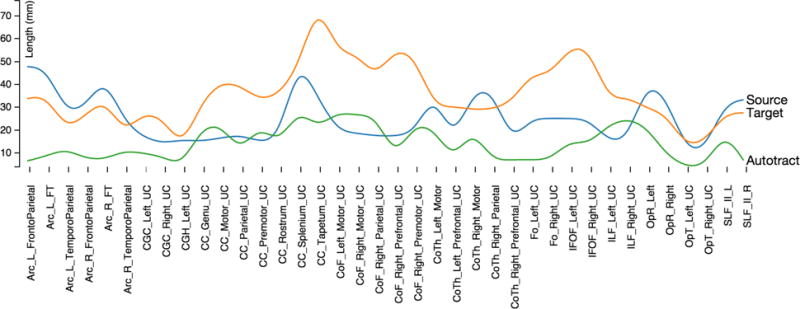
Average bundle length for the fibers traced automatically and the bundles validated by an expert.
4.2 Bundle similarity to ground truth
To assess the quality of the fibers produced by AutoTract, we will measure the distance from each fiber to the closest fiber in the ground truth. Equation 3 defines the minimum distance from a point a ∈ A to the fiber bundle b. Equation 5 defines the average minimum distance from ∀a ∈ A to bundle B.
| (3) |
| (4) |
| (5) |
Figure 6 shows a color map for the minimum distance of each point ai where the color is given by M(j) mapped to a rainbow color bar.
| (6) |
Equation 6 defines the Median Absolute Deviation (MAD) for a an array V with values v. Figure 7 shows the median minimum distance for every fiber bundle compared to the ground truth.
Figure 6.
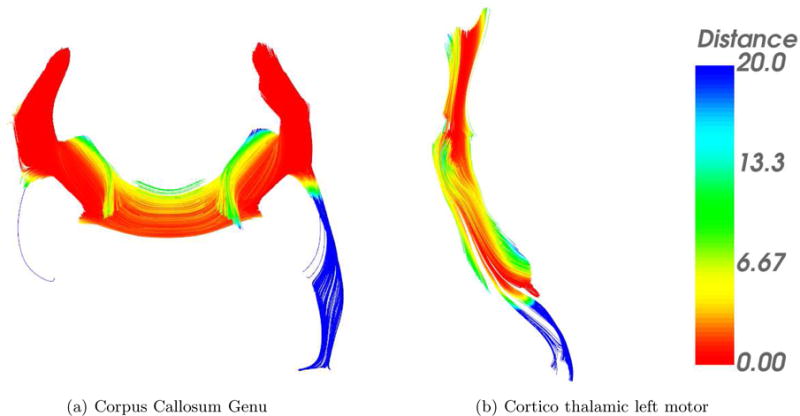
Distance in (mm) mapped to a unique color described by the color bar to the right of each figure. Each fiber traced by Autotract is compared against the corresponding ground truth bundles. The bundles shown are the output of Autotract.
Figure 7.
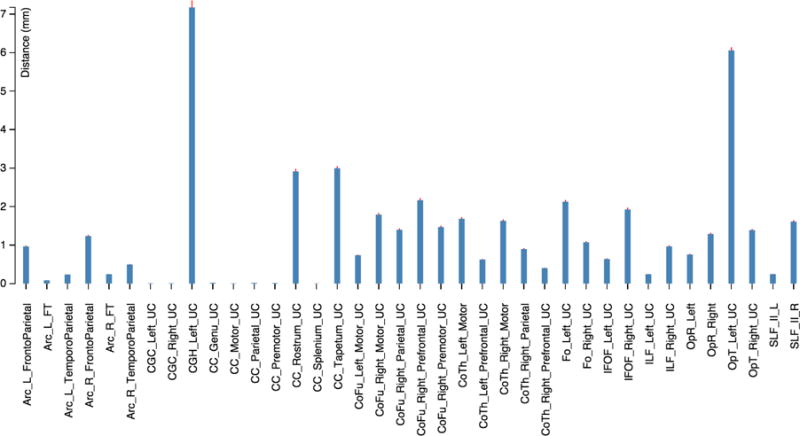
Median distance from the bundles traced automatically vs. the bundles validated by an expert. The median absolute deviation is depicted by the red line on top of each bar in the histogram.
5. CONCLUSIONS
We presented Autotract, a fully automated tool for fiber tracking and cleaning. The method uses fiber bundles previously traced as templates and produces new bundles in a target DTI.
We have demonstrated that the method is able to trace a wide variety of brain structures including cingulate cortex, cortico thalamic, corpus callosum among others.
Using an automatic procedure has several advantages such as saving time from the tedious procedure of cleaning and validating fiber tracts for different brain regions. Autotract speeds up the tracking and cleaning of fibers and enables new possibilities for research applications such as statistical white matter analysis using tractography. Segmenting the same white matter region across subjects allows neuroscientific hypotheses to be tested regarding group differences.
As shown by the results in this study, future work will focus on optimizing the approach to achieve similar fiber lengths. We need to investigate a new strategy to crop fibers based on shape descriptors, different similarity metrics, or improving stopping criteria for tractography. Further improvements to this framework could lead to a fully automated approach for tractography.
Acknowledgments
Thanks to Claudia Buss, UCL, for use of atlas data. Grants: RO1MH091645, RO1MH091351, RO1MHD055741, RO1MHD059854, RO1MH053000, RO1MH070890, RO1MH079124, RO1MH03110.
Footnotes
Contributor Information
Jean Y. Yang, Email: jyves@email.unc.edu.
François Budin, Email: fbudin@email.unc.edu.
Martin Styner, Email: styner@cs.unc.edu.
References
- 1.Friman O, Farnebäck G, Westin C-F. A bayesian approach for stochastic white matter tractography. Medical Imaging, IEEE Transactions on. 2006;25(8):965–978. doi: 10.1109/tmi.2006.877093. [DOI] [PubMed] [Google Scholar]
- 2.Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A. In vivo fiber tractography using dt-mri data. Magnetic resonance in medicine. 2000;44(4):625–632. doi: 10.1002/1522-2594(200010)44:4<625::aid-mrm17>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 3.Stippich C. Clinical Functional MRI. Springer; 2015. Presurgical functional mri and diffusion tensor imaging; pp. 1–12. [Google Scholar]
- 4.Hannoun S, Roch J-A, Durand-Dubief F, Vukusic S, Sappey-Marinier D, Guttmann CR, Cotton F. Weekly multimodal mri follow-up of two multiple sclerosis active lesions presenting a transient decrease in adc. Brain and Behavior. 2015;5(2) doi: 10.1002/brb3.307. n/a–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roalf DR, Gur RE, Verma R, Parker WA, Quarmley M, Ruparel K, Gur RC. White matter microstructure in schizophrenia: Associations to neurocognition and clinical symptomatology. Schizophrenia Research. 2015;161(1):42–49. doi: 10.1016/j.schres.2014.09.026. White Matter Pathology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moberts B, Vilanova A, Van Wijk JJ. Visualization 2005. VIS 05. IEEE. IEEE; 2005. Evaluation of fiber clustering methods for diffusion tensor imaging; pp. 65–72. [Google Scholar]
- 7.Maddah M, Mewes AU, Haker S, Grimson WEL, Warfield SK. Medical image computing and computer-assisted intervention–MICCAI 2005. Springer; 2005. Automated atlas-based clustering of white matter fiber tracts from dtmri; pp. 188–195. [DOI] [PubMed] [Google Scholar]
- 8.O’Donnell LJ, Westin C-F. Automatic tractography segmentation using a high-dimensional white matter atlas. Medical Imaging, IEEE Transactions on. 2007;26(11):1562–1575. doi: 10.1109/TMI.2007.906785. [DOI] [PubMed] [Google Scholar]
- 9.Budin F, Bouix S, Shenton M, Styner M, Oguz I. An itk implementation of a diffusion tensor images resampling filter. 2012;12 [Google Scholar]
- 10.Otsu N. A threshold selection method from gray-level histograms. Automatica. 1975;11(285–296):23–27. [Google Scholar]


