Abstract
Thiopeptides are posttranslationally processed macrocyclic peptide metabolites, characterized by extensive backbone and side chain modifications that include a six-membered nitrogeneous ring, thiazol(in)e/oxazol(in)e rings, and dehydrated amino acid residues. Thiostrepton A, one of the more structurally complex and well-studied thiopeptides, contains a second macrocycle bearing a quinaldic acid moiety. Antibacterial, antimalarial, and anticancer properties have been described for thiostrepton A and other thiopeptides, although the molecular details for binding the cellular target in each case are not fully elaborated. We previously demonstrated that a mutation of the TsrA core peptide, Ala4Gly, supported the successful production of the corresponding thiostrepton variant. To more thoroughly probe the thiostrepton biosynthetic machinery’s tolerance toward structural variation at the fourth position of the TsrA core peptide, we report here the saturation mutagenesis of this residue using a fosmid-dependent biosynthetic engineering method and the isolation of 16 thiostrepton analogs. Several types of side chain substitutions at the fourth position of TsrA, including those that introduce polar or branched, hydrophobic residues, are accepted, albeit with varied preferences. In contrast, proline and amino acid residues inherently charged at physiological pH are not well-tolerated at the queried site by the thiostrepton biosynthetic system. These newly generated thiostrepton analogs were assessed for their antibacterial activities and abilities to inhibit the proteolytic functions of the eukaryotic 20S proteasome. We demonstrate that the identity of the fourth amino acid residue in the thiostrepton scaffold is not critical for either ribosome or proteasome inhibition.
The discovery of the first thiopeptide in 1948, a micrococcin, launched numerous investigations into thiopeptide structures, chemistry, modes of biological action, and biosyntheses.(1–3) Thiopeptides are highly modified, macrocyclic peptide metabolites (Figure 1) and are produced by diverse genera of Gram-positive bacteria.(2) The core macrocycle of a thiopeptide is characterized by the presence of a central nitrogen-containing six-membered ring, thiazol(in)e/oxazol(in)e moieties, and can harbor additional modifications, including dehydrated amino acid residues.(2, 3) Thiopeptides display potent activity against Gram-positive bacterial pathogens, including methicillin-resistant Staphylococcus aureus (MRSA) and penicillin-resistant Streptococcus pneumoniae (PRSP), and also demonstrate antimalarial and anticancer properties.(4–6) Despite thiopeptides’ promise as lead compounds for drug development, their clinical application is currently limited due, at least in part, to poor water solubility and bioavailability. Until recently, access to thiopeptide derivatives relied predominantly on semi-synthetic strategies.(7–12) The approaches taken for thiopeptide-based antibacterial development include C-terminal tail modifications to introduce functional groups that enhance aqueous solubility and have led to a GE2270A derivative being investigated for the treatment of gastrointestinal Clostridium difficile infections.(7, 13) The limitations imposed by de novo synthesis and the naturally available chemical handles for the semi-synthetic modification of thiopeptides have prevented the structure-activity relationships of these complicated molecules from being fully explored. In 2009, it was revealed that thiopeptides are ribosomally synthesized and posttranslationally modified peptides (RiPPs), suggesting that thiopeptide analogs could be obtained through the site-directed mutagenesis of their precursor peptides.(14–17) Biosynthetic engineering of thiopeptides has rapidly emerged as an effective strategy to provide analogs that could ultimately support improved pharmacological and pharmacokinetic parameters.(18–24)
Figure 1.
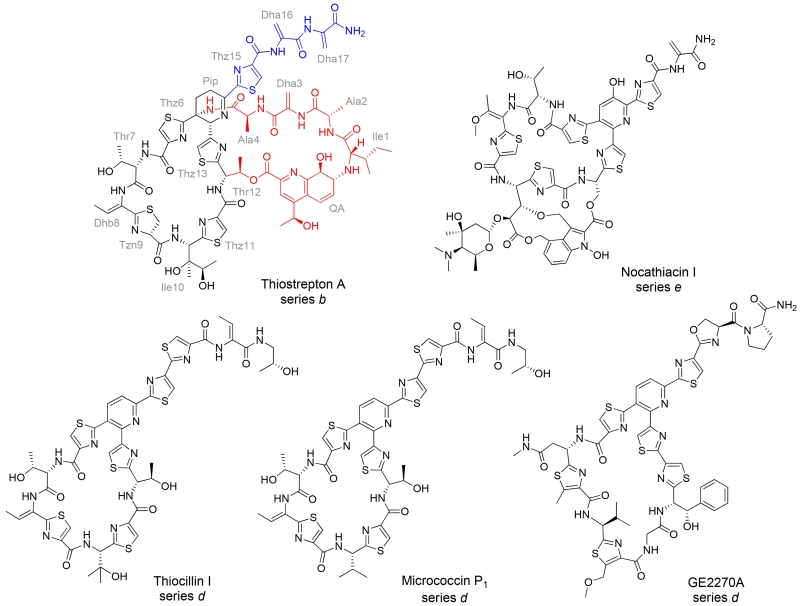
Examples of thiopeptides. The thiostrepton A residues are abbreviated using a three letter code and labeled in grey. Dha and Dhb refer to dehydroalanine and dehydrobutyrine, respectively. The core macrocycle of thiostrepton A is shown in black, while the quinaldic acid (QA)-containing loop and the C-terminal tail are highlighted in red and blue, respectively.
Thiostrepton A (Figure 1) is one of the more extensively studied metabolites of this family and is among the thiopeptides that have exhibited antibacterial, antimalarial, and anticancer properties.(4, 25–27) Structurally, thiostrepton A is a series b thiopeptide, distinguished by a central dehydropiperidine ring and a second, quinaldic acid (QA)-containing macrocycle. The peptidic loop encompassed by the QA linkage appears in a limited number of thiopeptides from three structural subfamilies, series a-c, and the loop contains four amino acid residues N-terminal to the piperidine/dehydropiperidine ring.(2) The vast majority of thiopeptides characterized to date contain a pyridine ring, and lack the residues corresponding to those found in thiostrepton A’s quinaldic acid loop, as exemplified by nocathiacin I, thiocillin I, micrococcin P1, and GE2270A (Figure 1).(2) To inhibit bacterial growth, thiostrepton A binds to the 50S ribosomal subunit between the ribosomal protein L11 and the 23S rRNA near the elongation factor G (EF-G) binding site and impedes ribosomal translocation along the mRNA transcript.(26) The other ribosome-binding thiopeptides, including micrococcin P1 (Figure 1) also bind to this site of the 50S subunit.(26) The antimalarial properties of thiostrepton A draw upon two distinct mechanisms of action: inhibition of the prokaryotic-like ribosome housed within the apicoplast of Plasmodium falciparum and inhibition of the cytosolic proteasome, a complex essential to protein degradation and recycling in eukaryotes.(11, 25, 27, 28) For its recently discovered anticancer activity, thiostrepton A appears to interfere directly with both proteasome function and forkhead box M1 (FOXM1) transcription factor binding to its affiliated promoter regions.(4, 29, 30) In contrast to the general structural knowledge of how thiopeptides affect protein translation, rather limited information is available concerning how thiostrepton A is also able to engage the recently recognized 20S proteasome and FOXM1 targets. It is therefore unknown whether overlapping or differing structural regions of thiostrepton A are critical for each of its three major biological activities.
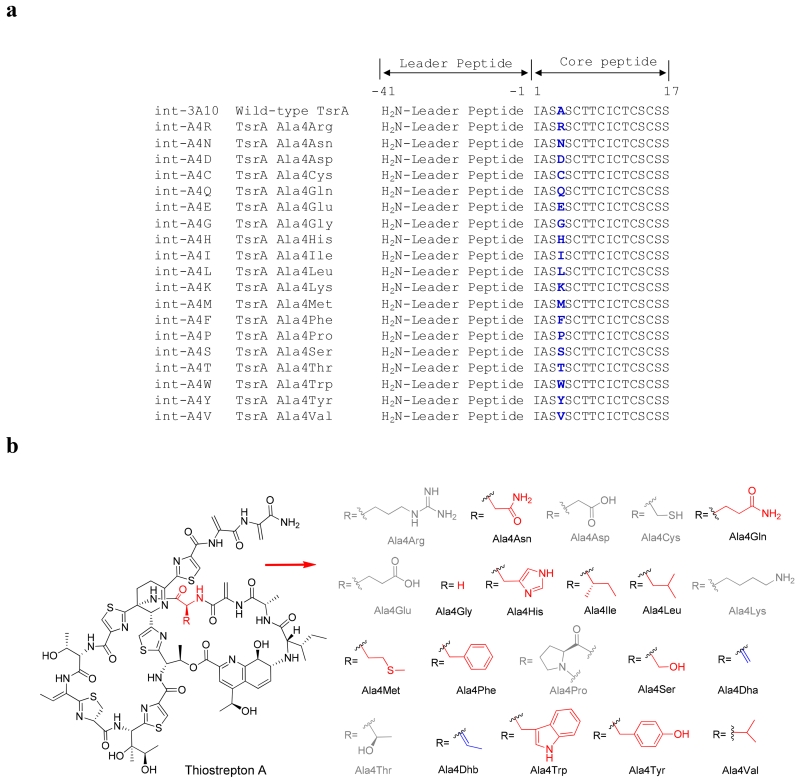
The thiostrepton A precursor peptide TsrA is composed of two regions: a 41 amino acid N-terminal leader peptide that is removed during the maturation process and a C-terminal 17 amino acid core peptide that is incorporated into the mature metabolite (Figure 2).(14) We recently developed a biosynthetic engineering platform to produce thiostrepton analogs in Streptomyces laurentii ATCC 31255 (S. laurentii) by the site-directed mutagenesis of TsrA.(20, 21) In this initial study, mutation of the fourth residue of the TsrA core peptide from alanine to glycine supported the production of a thiostrepton Ala4Gly variant that retained antibacterial activity, suggesting that this residue is amenable to substitution.(20) Herein, we thoroughly probe the range of proteinogenic amino acid residues tolerated at the fourth position of the TsrA core peptide by the saturation mutagenesis of Ala4. The newly generated thiostrepton variants are then interrogated for their antibacterial and proteasome inhibition properties to determine whether or not the identity of the fourth residue is critical for either activity.
Figure 2.
Thiostrepton A and the analogs to be generated by site-directed mutagenesis of TsrA. a) Comparison of wild-type TsrA with the TsrA Ala4 variants encoded in the mutant fosmids. b) Structures of thiostrepton Ala4 analogs. The amino acid residue is abbreviated using a three-letter code. The analogs in grey are not produced. The observed analogs possessing no modification of the variant residue are highlighted in red, while the ones resulting from a modification of the variant residue are highlighted in blue.
RESULTS AND DISCUSSION
Engineering tsrA variants in S. laurentii
The thiostrepton biosynthetic engineering platform utilizes a tsrA deletion mutant of S. laurentii (S. laurentii NDS1) and an E. coli-Streptomyces shuttle fosmid, int-3A100, allowing for tsrA mutagenesis to be conducted in E. coli.(20) Fosmid int-3A100 is derived from the single-copy fosmid pCC1FOS (Epicentre Biotechnologies) and was engineered to include an integrase and and attP sequence for integration into the S. laurentii chromosome.(20, 31) The entire tsr biosynthetic gene cluster is harbored within fosmid int-3A100, except that the TsrA core peptide-encoding region is replaced by a dual-marker selection cassette consisting of chlR (a chloramphenicol resistance gene) and a levansucrase-encoding sacB gene.(20) The tsrA genes varying at the position encoding the core peptide’s fourth residue, and codon-optimized for expression in Streptomyces, were incorporated into the fosmid by PCR-targeted gene replacement and the resulting fosmids were introduced into S. laurentii NDS1 by intergeneric conjugation.(32–34) By this method, 18 different S. laurentii NDS1/int-A4X (where “X” is the one-letter amino acid code for the introduced mutation) variants were successfully generated, in addition to the previously constructed S. laurentii NDS1/int-A4G (Figure 2A).(20)
Production and structural characterization of thiostrepton analogs
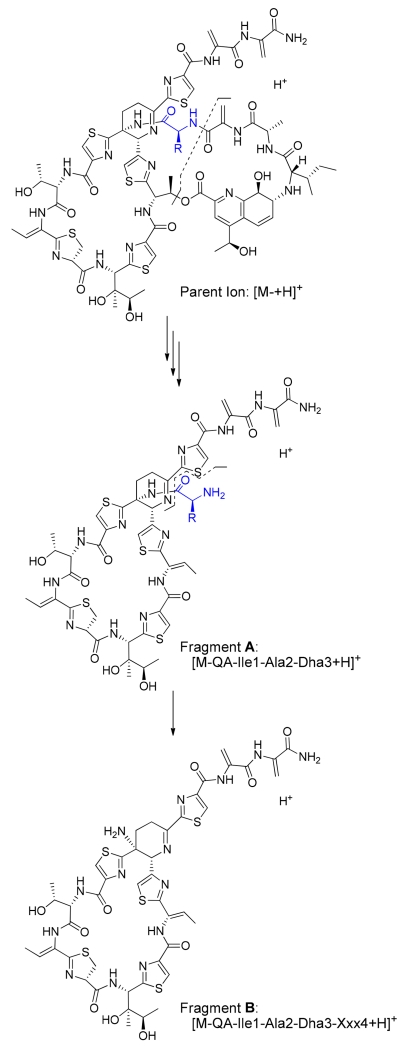
Culture extracts of the S. laurentii strains encoding TsrA Ala4 variants were evaluated for thiopeptide production by HPLC and HPLC-MS. Structures of the anticipated thiostrepton analogs are shown in Figure 2B. Mature thiostrepton analogs were not detected in the crude extracts of S. laurentii NDS1/int-A4D, A4E, A4K, and A4R, either by HPLC or HPLC-MS, suggesting that amino acid residues inherently charged at physiological pH are not well-tolerated by the thiostrepton biosynthetic system at the fourth position of the TsrA core peptide. The Ala4Pro substitution, which introduces conformational constraints on the precursor peptide backbone, is also not processed into a mature thiostrepton derivative, as thiostrepton Ala4Pro was not observed in the S. laurentii NDS1/int-A4P extract. A majority of the Ala4 substitutions in TsrA, however, did yield the expected mature thiostrepton analogs, including Ala4Asn, Ala4Gln, Ala4Gly, Ala4His, Ala4Ile, Ala4Leu, Ala4Met, Ala4Phe, Aal4Trp, Ala4Tyr, and Ala4Val (Figure 2B). These analogs were purified by semi-preparative HPLC and their identities confirmed by HR-MS and MALDI-MS/MS (Figures S1, S35–S40, S42–S44, and reference 20 for the Ala4Gly variant). Loss of the quinaldic acid (QA) moiety and sequential fragmentation of the newly exposed N-terminal residues are consistently observed in most thiostrepton analogs during MALDI-MS/MS, often allowing for unambiguous identification of the residues in the QA-containing loop.(21) The mass difference between two key fragments is particularly diagnostic for the identity of the fourth residue: fragment A corresponds to the loss of the QA moiety and the three N-terminal residues (Ile1, Ala2, and Dha3 (Dha: dehydroalanine)) from the parent ion, and fragment B accounts for the subsequent loss of the fourth residue (Xxx4) (Figures 3, S1, S35–40, and S42–44).
Figure 3.
Two characteristic fragments from MALDI-MS/MS used to confirm the identity of the fourth residue in a thiostrepton analog. Fragment A corresponds to the loss of the quinaldic acid (QA) moiety and the three N-terminal residues Ile1, Ala2, and Dha3 (Dha: dehydroalanine) from the parent ion and fragment B additionally lacks the fourth residue (Xxx4).
The variant peptides TsrA Ala4Thr and Ala4Ser both led to thiostrepton analogs bearing a modification at the newly introduced residue. For S. laurentii NDS1/int-A4T, a major metabolite displaying a similar UV-visible absorption spectrum to that of thiostrepton A was detected by HPLC analysis of the culture extract. Further investigation by HPLC-MS revealed a species 18 Da less than that expected for thiostrepton Ala4Thr, suggesting thiostrepton Ala4Dhb (Dhb: dehydrobutyrine) as the dehydration product of the newly introduced threonine residue (Figure 2B). Meanwhile, two thiostrepton analogs were readily observed by HPLC analysis of the S. laurentii NDS1/int-A4S culture extract. Initial HPLC-MS analyses of the extract revealed masses consistent with the anticipated thiostrepton Ala4Ser in addition to thiostrepton Ala4Dha, resulting from dehydration of the introduced serine residue (Figure 2B). These three analogs were purified and then analyzed by HR-MS and MALDI-MS/MS (Figures S19, S27, and S41). The proposed structures of thiostreptons Ala4Dha and Ala4Dhb were further verified by one- and two-dimensional NMR analyses (Figures S20–26, S28–34, and Tables S3 and S4). Relative to thiostrepton A, the Ala4Dha analog revealed an additional quaternary carbon (δC: 138.2) and an unsaturated CH2 (δC: 102.9 and δH: 5.82, 5.25), each characteristic of a Dha residue (Table S3). Additional resonances corresponding to a quaternary carbon (δC: 131.8) and an unsaturated CH (δC: 121.8 and δH: 5.87), suggestive of a Dhb residue, were observed in the NMR spectra of thiostrepton Ala4Dhb (Table S4).
Dehydration of a Ser or Thr residue at the fourth position of thiostrepton could be catalyzed by the dehydratase(s) encoded within the thiostrepton biosynthetic gene (tsr) cluster: TsrC, TsrD, and TsrL.(14) TsrC and TsrD, homologous to NisB and the LanB family of lanthipeptide dehydratases, have homologs in all reported thiopeptide biosynthetic systems, and are proposed to dehydrate serine and threonine residues of the core peptide during thiopeptide maturation.(31, 35) The additional LanB-type dehydratase, TsrL, is not conserved among all thiopeptide biosynthetic gene clusters, and its role is currently unclear.(14) In contrast to other families of lanthipeptide dehydratases, which employ phosphorylation to activate the β-hydroxyl as a leaving group, NisB dehydrates the serine and threonine residues of the nisin precursor peptide NisA via a glutamylated intermediate, and the LanB-like dehydratases involved in thiopeptide maturation likely adopt a similar catalytic strategy.(35–37) The LanB dehydratases demonstrate a broad tolerance toward non-native peptide substrates and are flexible in their abilities to dehydrate serine and threonine residues introduced by precursor peptide engineering.(38-40) For example, the Lys12Ser and Lys12Thr mutants of NisA both led to the production of a mixture of nisin analogs either unmodified or dehydrated at the twelfth position, with dehydration of the threonine residue predominating.(41) Furthermore, an in silico analysis of 37 lanthipeptides suggested that their affiliated dehydratases may be more likely to incompletely process the side chain of serine than that of threonine.(42) Consistent with these lanthipeptide observations, S. laurentii expressing TsrA Ala4Thr exclusively yielded the Ala4Dhb derivative, but the strain expressing TsrA Ala4Ser supported the production of both thiostreptons Ala4Ser and Ala4Dha at a 1:14 ratio (Table S6). All thiopeptides isolated thus far from the structural series a and b contain an unmodified Ala4 residue, but the lone member reported for series c, Sch 40832, possesses a Dha4.(2) Precursor peptides identified thus far for series d and e thiopeptides also contain a conserved Ala residue at the position N-terminal to the pyridine ring, however, this residue is lost, along with the remainder of the leader peptide, during the maturation process. The naturally-occurring and engineered thiopeptides from series a–c suggest that the dehydratases for at least the QA loop-containing thiopeptides may be biased toward dehydration if a suitable residue presents at the fourth position of the core peptide. In addition, insertion of a serine into the thiocillin precursor peptide led not only to ring-expanded analogs, but also to dehydration of the newly introduced residue in the series d thiopeptide.(18) It therefore appears that, like their lanthipeptide dehydratase counterparts, thiopeptide dehydratases are not highly restrictive in their substrate specificities and this flexibility can be further explored for directing the introduction of dehydrated amino acids into engineered thiopeptides.
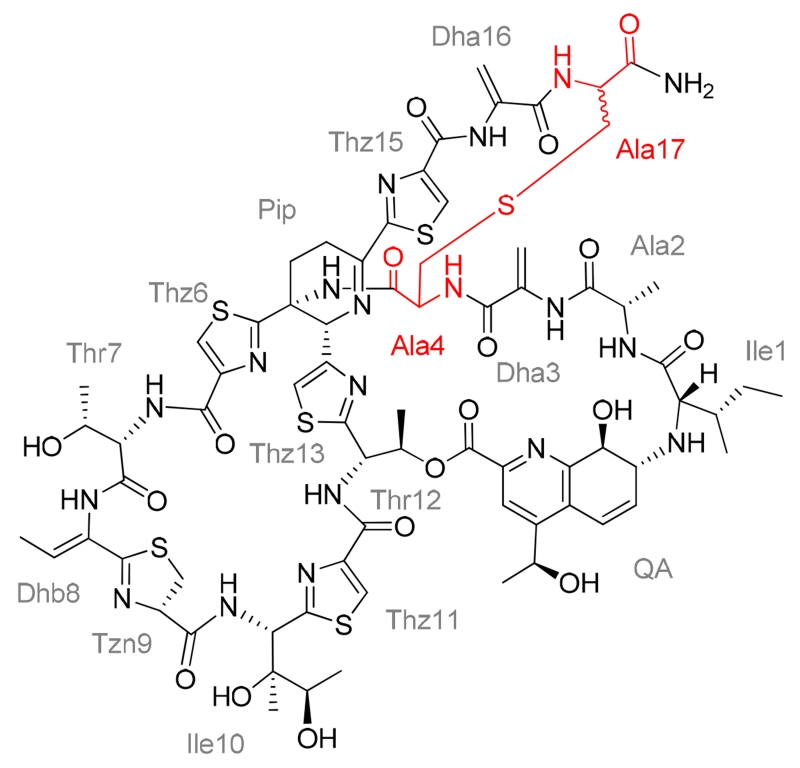
Several metabolites with UV-visible absorption spectra comparable to that of thiostrepton A were observed during the HPLC analyses of the S. laurentii NDS1/int-A4C culture extract. We focused on two of the more abundant metabolites whose masses were consistent with that expected for thiostrepton Ala4Cys and those two compounds were assigned as thiostreptons Ala4Cys F1 and F2. These thiopeptide products could result either from the retention of an unmodified Cys4 thiol or, via a scenario wherein the thiol attacked one of the thiostrepton dehydro residues by a Michael addition, forming a lanthionine or methyllanthioine (Lan or MeLan, respectively) as observed in lanthipeptides.(43) To distinguish between these two possibilities, both Ala4Cys analogs were analyzed by MALDI-MS/MS. If the newly introduced Cys4 residue remains unmodified, the two distinguishing A and B fragments would be observed (Figure 3). The MS/MS fragments of thiostreptons Ala4Cys F1 and F2, however, both only revealed the presence of fragment A (Figures S3 and S11). Thus, the Cys4 residue in both analogs was probably contained within either a Lan or MeLan residue. The thioether ring most likely does not involve the neighboring Dha3 side chain, which would preclude fragment A formation and only permit the observation of fragment B by MS/MS analysis. There are three additional dehydrated amino acid residues to consider in the thiostrepton scaffold: Dhb8, Dha16, and Dha17. Cys4 is not expected to form a thioether linkage with Dhb8 due to the relative positioning of the core macrocycle and the QA loop, which places the two residues on opposing faces of the molecule.(44) The C-terminal tail, on the other hand, does sample a variety of conformations in solution and could access a position poised for nucleophilic attack of the Dha16 or Dha17 side chain by the Cys4 thiol.(45) It is also worth noting that Lan or MeLan formation could precede the installation of one or both macrocycles of the native thiostrepton architecture. To examine the possibilities for the location of the thioether bridge, one- and two-dimensional NMR studies were performed with thiostreptons Ala4Cys F1 and F2 (Figures S4–S10, S12–S18, and Tables S1 and S2). The expected 1H and 13C resonances for Dha3 and Dhb8 in both F1 and F2 analogs were indeed present, confirming that these two residues remain intact.(46) In contrast, the resonances corresponding to one of the C-terminal Dha residues of thiostrepton A changed dramatically. The β proton resonances of a Dha residue typically appear between 6.7 and 5.2 ppm, whereas the resonances of an aliphatic Lan residue’s β protons occur upfield, around 3 ppm.(20, 21, 46–48) The NMR analyses revealed only two unsaturated CH2’s in both thiostreptons Ala4Cys F1 and F2, as opposed to three in thiostrepton A (Dha3, Dha16, and Dha17). The absence of proton and carbon resonances corresponding to one of the C-terminal Dha residues and the appearance of new resonances near 3 ppm in the 1H spectra of thiostreptons Ala4Cys F1 and F2 suggested that the thioether bridges are formed between Cys4 and either Dha16 or Dha17. The lack of HMBC signals diagnostic for the location of the Lan residue within the thiopeptide backbone, however, prevented the identification of the participating Dha side chain.
To differentiate whether the 16th or 17th dehydro residue of thiostreptons Ala4Cys F1 and F2 is incorporated into the thioether ring of each metabolite, the analogs were subjected to a diethylamine-mediated truncation of their C-termini and the reactions monitored by HPLC-MS (Figure S2).(49, 50) Using this method, the two C-terminal Dha residues can be sequentially removed from the thiostrepton to provide two truncated products lacking Dha17 or both Dha17 and Dha16.(49, 50) If the Lan residue was formed with Dha17, no truncated thiostrepton would be observed; however, if instead, Dha16 was attacked by the Cys4 thiol, a product shortened only by the loss of Dha17 would be generated. The expected Dha17 and Dha17 plus Dha16 truncation products were readily identified by HPLC-MS in the control reactions containing thiostrepton A and thiostrepton Ala4Asn (Figure S2). Following exposure of thiostreptons Ala4Cys F1 and F2 to the truncation conditions, ions suggestive of the corresponding diethylamine adducts of the intact metabolites were observed, but no products consistent with a C-terminal cleavage event were detected (Figure S2). Based on the MS, NMR, and chemical modification studies, we propose that the Lan residue in both thiostreptons Ala4Cys F1 and F2 arise from Cys4 and Dha17 (Figures 4, S3–10, S11–18, and Tables S1 and S2).
Figure 4.
Structures of thiostreptons Ala4Cys F1 and F2. The two analogs are proposed to be diastereomers differing in configuration at the α carbon of the 17th residue. The residues comprising the lanthionine are each annotated in the structure as “Ala” and are highlighted in red.
The thiazoline residue (Tzn9) of thiostrepton A occurs in a d-configuration (Figure 1), and nonenzymatic epimerization at this site has been reported.(46, 51) For a d-configured Tzn9, the chemical shift of the α proton appears at approximately 4.8 ppm and the β proton resonances appear at about 3.5 ppm and 3.0 ppm.(46, 51) In contrast, the corresponding resonances for an l-configured Tzn9 appear at about 4.6, 3.4, and 3.1 ppm, respectively.(51) The 1H resonances of Tzn9 in the two Ala4Cys analogs are closely aligned to those of thiostrepton A and previously reported thiostrepton analogs, all of which presumably adopt the d-configuration (Tables S1 and S2).(20, 21, 46) Collectively, these data suggest that thiostreptons Ala4Cys F1 and F2 are diastereomers differing in the configuration at the α-carbon of the 17th residue, however, their absolute configurations were not determined (Figure 4).
During class I lanthipeptide biosynthesis, Lan and MeLan rings are introduced by a two-step process. First, a LanB-type dehydratase acts upon a serine or threonine side chain to generate Dha and Dhb residues.(35, 43) Next, a LanC-type cyclase directs the attack of a cysteine thiol upon the dehydrated amino acid to close the thioether ring.(43) There is no lanthionine cyclase homolog encoded in the tsr cluster, leaving enzyme-mediated Lan installation for the thiostrepton Ala4Cys variants unlikely. It is worth noting that Lan formation between a free cysteine thiol and a dehydrated residue of a synthetic peptide is readily achieved near physiological pH.(52, 53) An adventitious 1,4-conjugate addition of the Cys4 β-thiol to the α,β-unsaturated alkene of Dha17 therefore seems to be the most reasonable explanation for the Lan residue of thiostreptons Ala4Cys F1 and F2, and this addition could occur either before or after biosynthesis of the core and QA-containing macrocycles. In addition to these two TsrA Ala4Cys metabolites, several minor species were detected in the crude culture extract possessing masses suggestive of an oxidized cysteine side chain or adducts consistent with the spontaneous capture of a the free thiol rather than an enzymatic modification (Data not shown). Among these species, there were no indications that the cysteine side chain was efficiently subjected to direct enzymatic posttranslational modification, such as thiazoline ring formation. Although all five Cys residues in the wild-type TsrA core peptide are processed by the cyclodehydratase TsrH, it appears that either a Cys introduced at the fourth position is not a preferred substrate for the modification or that the conformational restraints imposed by a thiazoline ring at this site preempt QA loop formation.
In total, sixteen thiostrepton analogs were successfully isolated following the saturation mutagenesis of TsrA Ala4, however, the titers of these analogs varied significantly (Table S6). TsrA Ala4 variant peptides with hydrophobic alkyl or linear thioether chains (Ile, Leu, Met, and Val) are well-tolerated by the thiostrepton biosynthetic system, as suggested by their robust production titers (> 40 mg L−1) compared to that of wild-type thiostrepton A (115 ± 35 mg L−1). Exchanging Ala4 of TsrA with a β-heteroatom-containing residue also enables maturation of the precursor peptide to a thiostrepton at a reasonable overall quantity. TsrA Ala4Ser and Ala4Thr are efficiently channeled to thiopeptide derivatives dehydrated at the engineered site whereas the Ala4Cys metabolites detected suggest that this residue largely escaped posttranslational processing by the thiopeptide cyclodehydratase. When the polar residues Asn, Gln, and His are used to replace Ala4, the anticipated thiostrepton analogs are generated, but these species were produced at levels ranging from abundant (near 40 mg L−1 for Asn) to relatively impaired (< 5 mg L−1 for Gln and His). Likewise, substitution of Ala4 with Phe, Trp, and Tyr only supported analog production reduced nearly 40-fold from wild-type thiostrepton A, suggesting that, while permitted, aromatic side chains at the fourth residue are not favored by the collective thiostrepton biosynthetic machinery. Finally, TsrA core peptides bearing a positively or negatively charged residue (Arg, Lys, Asp, and Glu) or Pro at the fourth position were not transformed into a mature thiopeptide. At this time, it is not known which step(s) contribute(s) to low or failed thiostrepton analog production, and it is certainly possible that multiple enzymes are not able to efficiently accommodate the alternate substrates.
Antibacterial activities of the thiostrepton analogs
After the aqueous solubilities for the thiostrepton analogs were determined (Table S7), the antibacterial activities of the purified thiostrepton Ala4 variants were evaluated using a previously described liquid microdilution method.(20, 21) When Ala4 is substituted by nonpolar aliphatic or aromatic amino acids, the resulting thiostrepton analogs showed minimum inhibitory concentrations (MICs) comparable to the values for thiostrepton A against three indicator strains of Staphylococcus aureus, Bacillus sp. and Enterococcus faecium (Table 1). The antibacterial activities of thiostreptons Ala4His, Ala4Ser, Ala4Dha, and Ala4Dhb decreased moderately, by approximately 10 to 50-fold, whereas the amide side-chain containing Ala4Asn and Ala4Gln variants were reduced in activity between 100 to 300-fold. Among all the Ala4 analogs tested, thiostreptons Ala4Cys F1 and F2 suffered the greatest impairments in their MIC values, by as much as 300-fold. Neither thiostrepton A nor its Ala4 variants displayed any antibacterial activity against the E. coli indicator strain ATCC 27856 (Data not shown).
Table 1.
Antibacterial activities of thiostrepton analogs.
| Compound | MICa (μg mL−1) |
||
|---|---|---|---|
| MRSAb | VREc | Bacillus d | |
| Thiostrepton A | 0.012 | 0.012 | 0.025 |
| Ala4Asn | 1.6 | 3.3 | 3.3 |
| Ala4Cys F1 | >3.4 | >3.4 | >3.4 |
| Ala4Cys F2 | >3.4 | >3.4 | >3.4 |
| Ala4Dha | 0.10 | 0.20 | 0.40 |
| Ala4Dhb | 0.25 | 0.25 | 0.50 |
| Ala4Gln | 1.3 | 2.7 | 2.7 |
| Ala4Gly | 0.12 | 0.12 | 0.46 |
| Ala4His | 0.16 | 0.43 | 1.30 |
| Ala4Ile | 0.026 | 0.026 | 0.013 |
| Ala4Leu | 0.015 | 0.015 | 0.029 |
| Ala4Met | 0.014 | 0.014 | 0.029 |
| Ala4Phe | 0.013 | 0.013 | 0.013 |
| Ala4Ser | 0.42 | 0.21 | 1.04 |
| Ala4Trp | 0.014 | 0.007 | 0.014 |
| Ala4Tyr | 0.028 | 0.007 | 0.055 |
| Ala4Val | 0.015 | 0.015 | 0.015 |
| Thr7Ala | >3.4 | >3.4 | >3.4 |
| SL105-1 | >3.4 | >3.4 | >3.4 |
| Thr7Val | >3.4 | >3.4 | >3.4 |
| SL106-1 | >3.4 | >3.4 | >3.4 |
| Vancomycin | 0.39 | NDe | ND |
| Chloramphenicol | ND | 3.9 | 0.98 |
Minimum inhibitory concentration.
Staphylococcus aureus ATCC 10537.
Enterococcus faecium ATCC 12952.
Bacillus sp. ATCC 27859.
Not determined.
The observed loss of antibacterial activity could either be due to a diminished capacity of the newly generated analogs to bind the target ribosomal site, or be instead caused by off-target effects. To distinguish between these possibilities and to more directly assess the inhibition of protein synthesis by the thiopeptide derivatives, a coupled in vitro transcription-translation assay was employed using a luciferase reporter system. Under these conditions, the half-maximal inhibitory concentration (IC50) obtained for thiostrepton A is in agreement with a previously reported value (0.63 ± 0.01 μM, Table 2).(45) Surprisingly, all thiostrepton Ala4 derivatives revealed IC50 parameters against in vitro protein synthesis comparable to that of the parent compound, thiostrepton A (Table 2). The IC50 for the Ala4Trp variant could not be measured due to solubility limitations, however its MIC values against the indicator strains imply that binding of this analog to the ribosome is not dramatically impaired (Tables 1 and 2). To rule out any non-specific inhibition of the in vitro transcription-translation system by the thiostrepton scaffold, four Thr7 variants were examined, and the aqueous solubilities of these analogs are included in Table S7. The unmodified threonine residue is highly conserved among ribosome-binding thiopeptides and it is expected to contribute key interactions for inhibition of translation.(26, 45, 54) An earlier mutagenesis effort confirmed that thiostreptons Thr7Ala, Thr7Val, and the corresponding shunt metabolites lacking an intact QA loop, SL105-1 and SL106-1 (for shunt metabolite structures, see Figure S45), were substantially diminished in their abilities to inhibit bacterial growth (Table 1).(21) Consistent with their poor MIC values, the abilities of the Thr7 analogs to inhibit in vitro protein synthesis were also abrogated (Table 2). Collectively, the bacterial growth and protein synthesis inhibition assays indicate that the identity of the fourth residue within the architecture of thiostrepton is not critical for ribosome binding, but may impact other factors key to potent in vivo activity, such as cell membrane permeability or metabolic stability. The X-ray crystal structure of thiostrepton A bound to the large ribosomal subunit reveals a solvent-exposed Ala4 residue that does not engage in any contacts with the ribosome.(26) This positioning of the fourth residue likely accounts for antibacterial activity of thiostreptons presenting an assortment of modifications at this site. It is noted that the pyridine-containing thiopeptides (Figure 1) altogether lack a residue corresponding to Ala4 of thiostrepton A, yet still effectively bind to the same region of the ribosome to inhibit function.(26)
Table 2.
In vitro translation inhibition by thiostrepton analogs.
| Compound | IC50 (μM) |
|---|---|
| Thiostrepton A | 0.63 ± 0.01 |
| Ala4Asn | 0.64 ± 0.01 |
| Ala4Cys F1 | 0.66 ± 0.03 |
| Ala4Cys F2 | 0.64 ± 0.01 |
| Ala4Dha | 0.44 ± 0.02 |
| Ala4Dhb | 0.69 ± 0.02 |
| Ala4Gln | 0.41 ± 0.01 |
| Ala4Gly | 0.44 ± 0.02 |
| Ala4His | 0.35 ± 0.02 |
| Ala4Ile | 0.49 ± 0.02 |
| Ala4Leu | 0.50 ± 0.04 |
| Ala4Met | 0.33 ± 0.01 |
| Ala4Phe | 0.67 ± 0.01 |
| Ala4Ser | 0.71 ± 0.05 |
| Ala4Trp | > 0.19a |
| Ala4Tyr | 0.56 ± 0.02 |
| Ala4Val | 0.32 ± 0.01 |
| Thr7Ala | > 19a |
| SL105-1 | > 6a |
| Thr7Val | > 17a |
| SL106-1 | > 11a |
IC50 not determined due to solubility limitations.
Structural modeling of thiostrepton Ala4Cys analogs
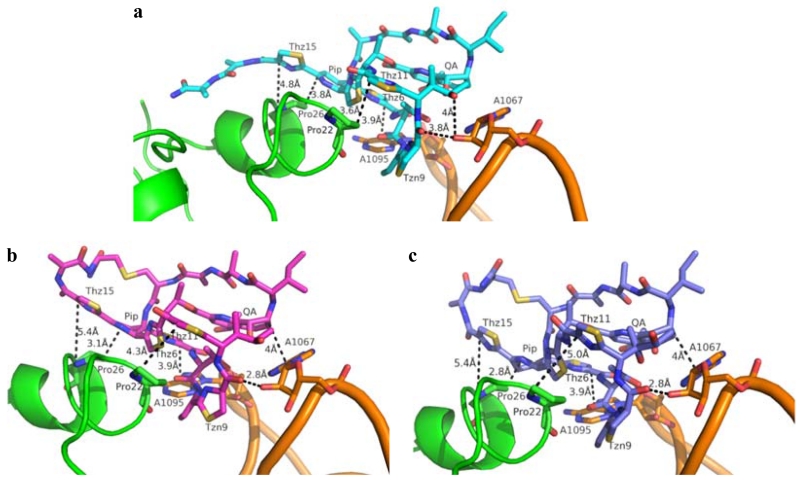
Although the in vitro activity of most thiostrepton Ala4 analogs could be rationalized from a structure-based knowledge of thiopeptide-ribosome interactions, it is less obvious how thiostreptons Ala4Cys F1 and F2, containing a third macrocycle, preserve their abilities to inhibit protein synthesis. To explain the apparent inconsistency between the in vitro experiments and the poor MIC values, we modeled the R and S stereoisomers for the Ala17 α-carbon of the Ala4Cys analogs into the 50S ribosome, adapting the reported crystal structure in which thiostrepton A is bound.(26) Figure 5 shows a local view of thiostrepton A complexed to the ribosome and the proposed interactions between the thiostrepton Ala4Cys analogs and the ribosome. Based on these models, the core macrocycle and QA-containing loops of thiostrepton A and its analogs are expected to adopt similar overall conformations (Figures 5 and S46). In thiostrepton A, the Thr7 hydroxyl group contributes to the overall fold of the peptide by participating in a hydrogen bond with the quinaldic acid 9-OH.(44) This hydrogen bond appears to be maintained in the models for the Ala4Cys analogs, as the distances between the Thr7 and quinaldic acid hydroxyl groups remain within hydrogen bonding distances, at 2.8 and 2.9 Å in the R-form and S-form variants, respectively (Figure S46). In addition, Thr7 of the three residues approach N1 of A1095 of the 23S rRNA similarly, within 5 Å, in both the reported X-ray crystal structure and the models (Figure 5).(26) Thz11 and Thz15 sit above Pro22 and Pro26 of the ribosomal protein L11, respectively, and are postulated to contribute hydrophobic interactions when thiostrepton A binds the ribosome (Figure 5A).(26) In the structures modeled for the Ala4Cys variants, Thz11 and Thz15 move away from the two L11 prolines, while the dehydropiperidine ring (Pip) approaches Pro26 and may retain a hydrophobic interaction for the complex (Figures 5B and 5C). More importantly, the carbonyl of Tzn9 in the Ala4Cys variants are now within hydrogen bonding distance (2.8 Å) to the A1067 2’-OH of 23S rRNA, an interaction not observed in the crystal structure of thiostrepton A bound to the ribosome due to the greater distance between the two groups (3.8 Å) (Figure 5).(26) In addition, the stacking interaction between Thz6 and A1095 as seen in the original complex is retained in the modeled structures.(26) The quinaldic acid moiety of the thiostrepton Ala4Cys analogs also appears to remain in proximity with A1067 and may provide a hydrophobic interaction as reported previously for the parent thiopeptide.(26) Overall, the major binding interactions between the core macrocycle and the QA moiety of thiostrepton A and the ribosome are expected to be conserved for the two Ala4Cys variants. As a result, these interactions, coupled with an additional hydrogen bond, may account for the retained in vitro translation inhibition efficacy of the thiostrepton Ala4Cys analogs.
Figure 5.
a) Thiostrepton A (Cyan) bound to the ribosome adapted from PDB 3CF5.(26) b) Thiostrepton Ala4Cys with the R configuration at the α-carbon of the 17th residue (magenta) modeled into the ribosome. c) Thiostrepton Ala4Cys with the S configuration at the α-carbon of the 17th residue (blue) modeled into the ribosome. Helices 43 and 44 of the 23S rRNA are colored in orange and the ribosomal protein L11 is shown in green.
20S proteasome inhibitory activities of the thiostrepton analogs
The 20S proteasome is a multi-subunit complex that plays a key role in eukaryotic protein degradation.(55) Different proteolytic activities (trypsin-, chymotrypsin-, and caspase-like) are conferred by three distinct types of β-subunits, and most reported nonpeptidic molecules inhibit the chymotrypsin-like activity.(55) Prior investigations by Arndt and coworkers revealed that thiostrepton A displays 20S proteasome inhibitory properties, and that this activity contributes to its antimalarial activity.(11, 28) Using fluorogenic substrates, the thiostrepton analogs described here were tested for their abilities to inhibit each of the 20S proteasome’s three proteolytic functions. In this analysis, thiostrepton A demonstrates a similar trend against the various activities to that previously reported, with inhibition of the caspase-like activity being the strongest.(11) Overall, the identity of the fourth residue of the thiostrepton scaffold does not appear to be absolutely critical for proteasome inhibition, since the IC50 values for most of the thiostrepton Ala4 analogs are comparable to that of thiostrepton A (Table 3). The inhibitory properties of thiostreptons Ala4Met and Ala4Trp could not be fully addressed due to solubility limitations, however, the Ala4Met variant did retain anti-caspase activity (Table 3). Aside from these two variants, thiostrepton Ala4 analogs bearing an unmodified side chain, with the exception of thiostrepton Ala4Gln, are associated with similar or slightly enhanced abilities to inhibit the three proteolytic activities of the proteasome. The potencies of thiostrepton Ala4Gln against the chymotrypsin and trypsin-like functions were somewhat impaired relative to the parent compound, reduced by 4-and at least 11-fold, respectively, but this analog did reveal about a 2-fold improvement in inhibition of the caspase-like activity of the proteasome (Table 3). Modifications introduced at the fourth residue side chain, present in the Ala4Dha, Ala4Dhb, and the Ala4Cys variants, also do not contribute to any dramatic change in proteasome inhibition, as the IC50 values for this subset of analogs remained within a 5-fold range relative to that of thiostrepton A (Table 3). Semi-synthetic thiostrepton derivatives, including those with modifications to the C-terminus and an aromatic thiazole in the core macrocycle in lieu of the more reduced thiazoline (position 9), retain their proteasome inhibitory properties, and, similar to the parent compound, several exhibit a bias against the caspase-like activity.(11) Likewise, our current study revealed thiostrepton Ala4 analogs also tend to demonstrate a more pronounced effect on caspase-like function. These results suggest that thiostrepton A and its analogs either adopt differing binding modes to the individual β-subunits or may instead allosterically modulate 20S proteasome activity.
Table 3.
Inhibition of the 20S proteasome trypsin-, chymotrypsin-, and caspase-like activities by thiostrepton analogs
| Compound | IC50 (μM) |
||
|---|---|---|---|
| Trypsin | Chymotrypsin | Caspase | |
| Thiostrepton A | 0.95 ± 0.15 | 1.02 ± 0.12 | 0.59 ± 0.06 |
| Ala4Asn | 0.38 ± 0.06 | 0.89 ± 0.17 | 0.86 ± 0.14 |
| Ala4Cys F1 | 1.75 ± 0.23 | 1.54 ± 0.18 | 0.32 ± 0.03 |
| Ala4Cys F2 | 0.83 ± 0.05 | 1.38 ± 0.23 | 0.70 ± 0.06 |
| Ala4Dha | 2.46 ± 0.43 | 2.73 ± 0.20 | 2.43 ± 0.30 |
| Ala4Dhb | 0.29 ± 0.03 | 0.25 ± 0.02 | 0.28 ± 0.02 |
| Ala4Gln | > 10.7a | 4.19 ± 0.48 | 0.31 ± 0.02 |
| Ala4Gly | 0.41 ± 0.05 | 0.77 ± 0.04 | 0.48 ± 0.04 |
| Ala4His | 0.51 ± 0.09 | 0.76 ± 0.15 | 0.53 ± 0.03 |
| Ala4Ile | 0.24 ± 0.05 | 0.27 ± 0.03 | 0.20 ± 0.04 |
| Ala4Leu | 0.34 ± 0.04 | 0.53 ± 0.02 | 0.31 ± 0.03 |
| Ala4Met | > 0.59a | > 0.59a | 0.46 ± 0.05 |
| Ala4Phe | 0.24 ± 0.03 | 0.37 ± 0.09 | 0.27 ± 0.05 |
| Ala4Ser | 0.39 ± 0.03 | 1.11 ± 0.06 | 0.40 ± 0.06 |
| Ala4Trp | > 0.19a | > 0.19a | > 0.19a |
| Ala4Tyr | 0.25 ± 0.02 | 0.63 ± 0.09 | 0.36 ± 0.03 |
| Ala4Val | 0.59 ± 0.04 | 0.49 ± 0.03 | 0.18 ± 0.03 |
| Thr7Ala | 0.71 ± 0.07 | 0.57 ± 0.05 | 0.28 ± 0.04 |
| SL105-1 | 0.77 ± 0.12 | 0.77 ± 0.12 | 0.56 ± 0.09 |
| Thr7Val | 1.33 ± 0.27 | 5.38 ± 0.97 | 1.68 ± 0.19 |
| SL106-1 | > 11a | 6.30 ± 0.95 | 6.72 ± 0.40 |
| Bortezomib | 1.62 ± 0.20 | 0.005 ± 0.001 | 0.049 ± 0.003 |
IC50 not determined due to solubility limitations.
The thiostrepton Thr7 variants and their two shunt metabolites were also evaluated as 20S proteasome inhibitors. Unlike its key role for ribosome binding, a residue with hydrogen bonding capability at the seventh position of thiostrepton A does not appear to be obligatory in order to disrupt proteasome function.(21) Both thiostrepton Thr7A variants retained activity against the proteasome, with the inhibitory properties of thiostrepton Thr7Ala slightly enhanced and those of thiostrepton Thr7Val reduced relative to the wild-type metabolite (Table 3). Previously, it was demonstrated that a thiostrepton A derivative in which the ester bond connecting Thr12 to the QA moiety was opened via methanolysis resulted in a less effective proteasome inhibitor, as the IC50 against caspase-like activity increased over 10-fold.(11) Our current study reinforces this observation: the analogs SL105-1 and SL106-1 lacking the intact QA loop, yet conserving thiostrepton’s ester linkage between Thr12 and QA (Figure S45), still act as proteasome inhibitors, albeit with decreased efficacy for the Thr7Val derivative SL106-1 (Table 3). Thiostrepton A and siomycin, both series b thiopeptides containing a QA loop, are active proteasome inhibitors, whereas series d thiopeptides lacking this additional macrocycle, such as micrococcin P1 and thiocillin I, have not yet revealed any proteasome inhibitory properties.(4, 29) It appears that the QA macrocycle is indeed important for a thiopeptide’s inhibitory activity and that the proper orientation of the QA moiety contributes to binding interactions with the proteasome, as the analogs lacking an intact QA loop do vary in their IC50 values. Before a comprehensive understanding of thiopeptide-based proteasome inhibition can be developed, structural studies are required to provide the molecular details into how thiostrepton A binds to the 20S proteasome and to rationalize the observed activities for the varied thiostrepton analogs reported thus far.
In conclusion, we isolated sixteen thiostrepton analogs following saturation mutagenesis at the fourth position of the TsrA core peptide. To our knowledge, this is the first report that thoroughly probes the permissiveness of the collective biosynthetic machinery toward modifications to the fourth residue in the quinaldic acid loop of a series b thiopeptide by substituting the parent alanine residue with the remaining 19 proteinogenic amino acid residues. The in vitro inhibition assays of the Ala4 variants reveal that the fourth residue of this thiopeptide is not essential for either ribosome or proteasome binding. The wide range of thiostrepton analogs that retain the ability to complex with these two macromolecular targets, including Ala4Dha, Ala4Dhb, and Ala4Ser, provide promise for the incorporation of unnatural amino acids and for coupling an engineered thiopeptide with semi-synthetic modification to acquire a biologically active analog with enhanced water solubility. Such a strategy could also facilitate the inclusion of a cross-linking functional group or fluorescent label to probe binding interactions, which are currently unknown, when thiostrepton A is complexed to the proteasome. Modifications to the C-terminal tail region of thiostrepton A and other thiopeptides, often by conjugate addition to a dehydroalanine residue, have been utilized to increase water solubility through the introduction of polar functional groups.(9–11, 28, 50) These and other types of semi-synthetic modifications of a thiopeptide scaffold, when introduced at an amenable position, can be broadly accepted with retained antibacterial or antimalarial potencies. The results described here support further manipulation of the quinaldic acid-containing macrocycle of thiostrepton A and related thiopeptides to generate novel analogs for structure-function studies and potential clinical applications.
METHODS
Engineering TsrA Ala4 Variants in S. laurentii
A synthetic ultramer, containing a degenerate codon (NNS) at the fourth position of the tsrA core peptide-encoding region and regions homologous to fosmid int-3A100 (Table S9), was used as the template for the initial amplification of mutant tsrA genes by polymerase chain reaction (PCR), using the primers Amp-TsrA-SP-F and -R (Table S9). The amplicons were cloned into pSC-B-amp/kan and the resulting plasmids were analyzed by DNA sequencing. Inserts containing the highest percentage of synonymous codon usage for Streptomyces were selected for further studies (Table S8).(34) The mutant plasmids served as the templates for amplification for each mutant tsrA gene, using the primers Amp-TsrA-SP-F and -R. The resulting amplicons were used to replace the dual-marker disruption cassette in fosmid int-3A100 by PCR-targeted gene replacement.(32, 33) Fosmids from the chloramphenicol-sensitive and sucrose-tolerant colonies were transformed into chemically competent E. coli EPI300 cells to induce high copy number before isolation and further analysis by restriction digestion with EcoRI. Samples that displayed the same restriction digestion pattern as fosmid int-3A10 were selected for DNA sequence analysis to confirm the allelic replacement of wild-type tsrA by each mutant tsrA gene. Mutant int-A4X fosmids (Table S8) were transformed into E. coli ET12567/pUZ8002, and then introduced into S. laurentii NDS1 through intergeneric conjugation.(32, 56) Colonies resistant to apramycin were confirmed by PCR using the primers SD3-F and -R (Table S9), and the strains were annotated as S. laurentii NDS1/int-A4X, according to the introduced mutation.
Evaluation of Thiostrepton Ala4 Analog Production in S. laurentii
Growth of S. laurentii was carried out in a three-step process as described previously.(14) After fermentation for 4 days, the whole culture was extracted twice with an equal volume of chloroform. The chloroform layers were pooled together and the solvent was removed in vacuo. The solid residue was dissolved in 4 mL chloroform. Samples were analyzed by high-performance liquid chromatography (HPLC) on a Beckman Coulter System Gold instrument with a Phenomenex Luna C18(2) column (250 × 4.6 mm, 5 μm) which was developed at 1 mL min−1 using a gradient of 0–100% (v/v) acetonitrile in water over 30 min. Absorbance was monitored at 254 nm. Purification of the thiostrepton Ala4 analogs is described in the Supporting Information as Supplemental Methods.
Antibacterial and In vitro Trancsription-Translation Coupled Assays
Minimum inhibitory concentrations (MICs) of thiostrepton analogs against indicator strains (MRSA, VRE, Bacillus, and E. coli ATCC 27854) were determined following a liquid microdilution method described previously.(20) The in vitro transcription-translation coupled assays were conducted using the E. coli S30 Extract System for Circular DNA and the Luciferase Assay System from Promega. Reactions were conducted according to the manufacturer’s protocol, except that the experiment was performed using a 10 μL reaction volume containing 1 μL amino acid mixture, 4 μL S30 premix, 3 μL S30 E. coli extract, 1.25 μL nuclease-free water, 0.25 μL thiostrepton analog, and 0.5 μL pBESTLuc™ template. The luciferase substrate was reconstituted as recommended by the manufacturer. Thiostrepton A and its analogs were prepared in a range of concentrations in DMSO and quantified by absorbance at 280 nm or HPLC as described in the Supplemental Methods. The reaction was incubated at 37 °C for 1 h, at which time 5 μL of the reaction mixture was transferred to a well in a 96-well black with clear, flat bottom plate. Luciferase substrate (50 μL) was added to each well immediately before luminescence was measured. Relative activity, obtained by normalizing luminescence to that of a DMSO control, was plotted against compound concentration. Each assay was performed in triplicate and the half maximal inhibitory concentration (IC50) was calculated for each compound by fitting the data to the Hill equation using GraphPad Prism 5.
Structural Modeling
The crystal structure of thiostrepton A complexed with the ribosome from Deinococcus radiodurans (D. radiodurans) (PDB entry: 3CF5) was used as the source to build the in silico model of the thiostrepton Ala4Cys analogs bound to the ribosome.(26) Thiostrepton A interacts with helices 43 and 44 (H43/44) of D. radiodurans 23S rRNA and ribosomal protein L11. By visual inspection, the local complex structure, including thiostrepton A, H43/44, and L11, was extracted from the crystal structure and exported to ChemDraw 3D Ultra 12.0 (CambridgeSoft). The thiostrepton analogs, Ala4Cys F1 and F2, were manually built in ChemDraw 3D using the crystal structure of thiostrepton A complexed to the ribosome (PDB entry: 3CF5) as the template.(57) The analog structures were energy-minimized using the MM2 force field in ChemDraw 3D, where H43/44 and L11 were immobilized during minimization.(58) The interactions between the energy minimized structures of the thiostrepton Ala4Cys analogs and the ribosome were visualized using PyMOL.(59)
20S Proteasome Inhibition Assay
Components used to assay the inhibition of the 20S proteasome were acquired from Enzo Life Sciences, including the three 7-amino-4-methylcoumarin (AMC)-based fluorogenic substrates Boc-Leu-Arg-Arg-AMC, Suc-Leu-Leu-Val-Tyr-AMC, and Ac-Nle-Pro-Nle-Asp-AMC, which were used to measure the trypsin-, chymotrypsin-, and caspase-like activities of the 20S proteasome, respectively. Thiostrepton A and its analogs were prepared in DMSO and quantified by absorption at 280 nm or HPLC as described above. DMSO and bortezomib were included as controls. The assay was executed in a final volume of 50 μL in a 384-well black plate consisting of: 5 μL test compound, 10 μL 1 μg mL−1 20S proteasome, 10 μL 50 μM fluorogenic substrate and 25 μL assay buffer (20 mM Tris-HCl, 1 mM EDTA, pH 7.5) and each assay was performed in triplicate. The 20S proteasome and the test compound were incubated together for 15 min at 37 °C, followed by the addition of the appropriate fluorogenic substrate. Fluorescence was measured using an excitation wavelength of 360 nm and an emission wavelength of 460 nm. Emissions were documented every 50 s for 1 h and the arbitrary fluorescence units (AFU) were plotted against time for each compound to acquire the slope of the linear fit. Relative activity, obtained by normalizing the compound slope to the DMSO control slope, was plotted against compound concentration and fit to the Hill equation using GraphPad Prism 5 to calculate IC50.
Supplementary Material
ACKNOWLEDGEMENT
This work was funded by the Defense Advanced Research Projects Agency (N6601-09-2086) and National Institutes of Health (R01GM090327). We thank K. Rommel and C. Li, for insightful discussions and S. Nguyen for assistance with bacterial cultures. We also thank C. Hsiao (currently at National Taiwan University) for his assistance with the modeling of the thiostrepton analogs. We are grateful to D. Bostwick for performing mass spectrometry and L. Gelbaum for helpful suggestions with NMR experiments.
Footnotes
ASSOCIATED CONTENT
Supporting methods, tables of strains, plasmids, primers, NMR spectra, mass spectra, and supplemental tables. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Su TL. Micrococcin, an antibacterial substance formed by a strain of Micrococcus. Br. J. Exper. Pathol. 1948;29:473–481. [PMC free article] [PubMed] [Google Scholar]
- 2.Bagley MC, Dale JW, Merritt EA, Xiong X. Thiopeptide antibiotics. Chem. Rev. 2005;105:685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- 3.Li C, Kelly WL. Recent advances in thiopeptide antibiotic biosynthesis. Nat. Prod. Rep. 2010;27:153–164. doi: 10.1039/b922434c. [DOI] [PubMed] [Google Scholar]
- 4.Bhat UG, Halasi M, Gartel AL. Thiazole antibiotics target FoxM1 and induce apoptosis in human cancer cells. PLoS One. 2009;4:e5592. doi: 10.1371/journal.pone.0005592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li W, Leet JE, Ax HA, Gustavson DR, Brown DM, Turner L, Brown K, Clark J, Yang H, Fung-Tomc J, Lam KS. Nocathiacins, new thiazolyl peptide antibiotics from Nocardia sp. I. Taxonomy, fermentation and biological activities. J. Antibiot. 2003;56:226–231. doi: 10.7164/antibiotics.56.226. [DOI] [PubMed] [Google Scholar]
- 6.Rogers MJ, Cundliffe E, McCutchan TF. The antibiotic micrococcin is a potent inhibitor of growth and protein synthesis in the malaria parasite. Antimicrob. Agents Chemother. 1998;42:715–716. doi: 10.1128/aac.42.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LaMarche MJ, Leeds JA, Amaral A, Brewer JT, Bushell SM, Deng G, Dewhurst JM, Ding J, Dzink-Fox J, Gamber G, Jain A, Lee K, Lee L, Lister T, McKenney D, Mullin S, Osborne C, Palestrant D, Patane MA, Rann EM, Sachdeva M, Shao J, Tiamfook S, Trzasko A, Whitehead L, Yifru A, Yu D, Yan W, Zhu Q. Discovery of LFF571: an investigational agent for Clostridium difficile infection. J. Med. Chem. 2012;55:2376–2387. doi: 10.1021/jm201685h. [DOI] [PubMed] [Google Scholar]
- 8.LaMarche MJ, Leeds JA, Dzink-Fox J, Mullin S, Patane MA, Rann EM, Tiamfook S. 4-Aminothiazolyl analogs of GE2270 A: Design, synthesis and evaluation of imidazole analogs. Bioorg. Med. Chem. Lett. 2011;21:3210–3215. doi: 10.1016/j.bmcl.2011.04.048. [DOI] [PubMed] [Google Scholar]
- 9.Myers CL, Hang PC, Ng G, Yuen J, Honek JF. Semi-synthetic analogues of thiostrepton delimit the critical nature of tail region modifications in the control of protein biosynthesis and antibacterial activity. Bioorg. Med. Chem. 2010;18:4231–4237. doi: 10.1016/j.bmc.2010.04.098. [DOI] [PubMed] [Google Scholar]
- 10.Naidu BN, Sorenson ME, Matiskella JD, Li W, Sausker JB, Zhang Y, Connolly TP, Lam KS, Bronson JJ, Pucci MJ, Yang H, Ueda Y. Synthesis and antibacterial activity of nocathiacin I analogues. Bioorg. Med. Chem. Lett. 2006;16:3545–3549. doi: 10.1016/j.bmcl.2006.03.079. [DOI] [PubMed] [Google Scholar]
- 11.Schoof S, Pradel G, Aminake MN, Ellinger B, Baumann S, Potowski M, Najajreh Y, Kirschner M, Arndt HD. Antiplasmodial thiostrepton derivatives: proteasome inhibitors with a dual mode of action. Angew. Chem. Int. Ed. Engl. 2010;49:3317–3321. doi: 10.1002/anie.200906988. [DOI] [PubMed] [Google Scholar]
- 12.LaMarche MJ, Leeds JA, Dzink-Fox J, Gunderson K, Krastel P, Memmert K, Patane MA, Rann EM, Schmitt E, Tiamfook S, Wang B. 4-Aminothiazolyl analogues of GE2270 A: antibacterial lead finding. J. Med. Chem. 2011;54:2517–2521. doi: 10.1021/jm101602q. [DOI] [PubMed] [Google Scholar]
- 13.Xu L, Farthing AK, Dropinski JF, Meinke PT, McCallum C, Hickey E, Liu K. Synthesis and antibacterial activity of novel water-soluble nocathiacin analogs. Bioorg. Med. Chem. Lett. 2013;23:366–369. doi: 10.1016/j.bmcl.2012.10.065. [DOI] [PubMed] [Google Scholar]
- 14.Kelly WL, Pan L, Li C. Thiostrepton biosynthesis: prototype for a new family of bacteriocins. J. Am. Chem. Soc. 2009;131:4327–4334. doi: 10.1021/ja807890a. [DOI] [PubMed] [Google Scholar]
- 15.Liao R, Duan L, Lei C, Pan H, Ding Y, Zhang Q, Chen D, Shen B, Yu Y, Liu W. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem. Biol. 2009;16:141–147. doi: 10.1016/j.chembiol.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wieland Brown LC, Acker MG, Clardy J, Walsh CT, Fischbach MA. Thirteen posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin. Proc. Natl. Acad. Sci. U.S.A. 2009;106:2549–2553. doi: 10.1073/pnas.0900008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris RP, Leeds JA, Naegeli HU, Oberer L, Memmert K, Weber E, LaMarche MJ, Parker CN, Burrer N, Esterow S, Hein AE, Schmitt EK, Krastel P. Ribosomally synthesized thiopeptide antibiotics targeting elongation factor Tu. J. Am. Chem. Soc. 2009;131:5946–5955. doi: 10.1021/ja900488a. [DOI] [PubMed] [Google Scholar]
- 18.Bowers AA, Acker MG, Young TS, Walsh CT. Generation of thiocillin ring size variants by prepeptide gene replacement and in vivo processing by Bacillus cereus. J. Am. Chem. Soc. 2012;134:10313–10316. doi: 10.1021/ja302820x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowers AA, Acker MG, Koglin A, Walsh CT. Manipulation of thiocillin variants by prepeptide gene replacement: structure, conformation, and activity of heterocycle substitution mutants. J. Am. Chem. Soc. 2010;132:7519–7527. doi: 10.1021/ja102339q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li C, Zhang F, Kelly WL. Heterologous production of thiostrepton A and biosynthetic engineering of thiostrepton analogs. Mol. BioSyst. 2011;7:82–90. doi: 10.1039/c0mb00129e. [DOI] [PubMed] [Google Scholar]
- 21.Li C, Zhang F, Kelly WL. Mutagenesis of the thiostrepton precursor peptide at Thr7 impacts both biosynthesis and function. Chem. Commun. 2012;48:558–560. doi: 10.1039/c1cc14281j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Acker MG, Bowers AA, Walsh CT. Generation of thiocillin variants by prepeptide gene replacement and in vivo processing by Bacillus cereus. J. Am. Chem. Soc. 2009;131:17563–17565. doi: 10.1021/ja908777t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malcolmson SJ, Young TS, Ruby JG, Skewes-Cox P, Walsh CT. The posttranslational modification cascade to the thiopeptide berninamycin generates linear forms and altered macrocyclic scaffolds. Proc. Natl. Acad. Sci. U.S.A. 2013;110:8483–8488. doi: 10.1073/pnas.1307111110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young TS, Dorrestein PC, Walsh CT. Codon randomization for rapid exploration of chemical space in thiopeptide antibiotic variants. Chem. Biol. 2012;19:1600–1610. doi: 10.1016/j.chembiol.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clough B, Strath M, Preiser P, Denny P, Wilson IR. Thiostrepton binds to malarial plastid rRNA. FEBS Lett. 1997;406:123–125. doi: 10.1016/s0014-5793(97)00241-x. [DOI] [PubMed] [Google Scholar]
- 26.Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, Spahn CMT, Fucini P. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol. Cell. 2008;30:26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 27.McConkey GA, Rogers MJ, McCutchan TF. Inhibition of Plasmodium falciparum protein synthesis. Targeting the plastid-like organelle with thiostrepton. J. Biol. Chem. 1997;272:2046–2049. doi: 10.1074/jbc.272.4.2046. [DOI] [PubMed] [Google Scholar]
- 28.Aminake MN, Schoof S, Sologub L, Leubner M, Kirschner M, Arndt HD, Pradel G. Thiostrepton and derivatives exhibit antimalarial and gametocytocidal activity by dually targeting parasite proteasome and apicoplast. Antimicrob. Agents Chemother. 2011;55:1338–1348. doi: 10.1128/AAC.01096-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pandit B, Bhat U, Gartel AL. Proteasome inhibitory activity of thiazole antibiotics. Cancer Biol. Ther. 2011;11:43–47. doi: 10.4161/cbt.11.1.13854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hegde NS, Sanders DA, Rodriguez R, Balasubramanian S. The transcription factor FOXM1 is a cellular target of the natural product thiostrepton. Nat. Chem. 2011;3:725–731. doi: 10.1038/nchem.1114. [DOI] [PubMed] [Google Scholar]
- 31.Zhang F, Kelly WL. In vivo production of thiopeptide variants. Methods Enzymol. 2012;516:3–24. doi: 10.1016/B978-0-12-394291-3.00022-8. [DOI] [PubMed] [Google Scholar]
- 32.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright F, Bibb MJ. Codon usage in the G+C-rich Streptomyces genome. Gene. 1992;113:55–65. doi: 10.1016/0378-1119(92)90669-g. [DOI] [PubMed] [Google Scholar]
- 35.Garg N, Salazar-Ocampo LMA, van der Donk WA. In vitro activity of the nisin dehydratase NisB. Proc. Natl. Acad. Sci. U.S.A. 2013;110:7258–7263. doi: 10.1073/pnas.1222488110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goto Y, Li B, Claesen J, Shi Y, Bibb MJ, van der Donk WA. Discovery of unique lanthionine synthetases reveals new mechanistic and evolutionary insights. PLoS Biol. 2010;8:e1000339. doi: 10.1371/journal.pbio.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goto Y, Ökesli A, van der Donk WA. Mechanistic studies of Ser/Thr dehydration catalyzed by a member of the LanL lanthionine synthetase family. Biochemistry. 2011;50:891–898. doi: 10.1021/bi101750r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bierbaum G, Szekat C, Josten M, Heidrich C, Kempter C, Jung G, Sahl HG. Engineering of a novel thioether bridge and role of modified residues in the lantibiotic Pep5. Appl. Environ. Microbiol. 1996;62:385–392. doi: 10.1128/aem.62.2.385-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuipers OP, Rollema HS, Yap WM, Boot HJ, Siezen RJ, de Vos WM. Engineering dehydrated amino acid residues in the antimicrobial peptide nisin. J. Biol. Chem. 1992;267:24340–24346. [PubMed] [Google Scholar]
- 40.Rink R, Wierenga J, Kuipers A, Kluskens LD, Driessen AJM, Kuipers OP, Moll GN. Production of dehydroamino acid-containing peptides by Lactococcus lactis. Appl. Environ. Microbiol. 2007;73:1792–1796. doi: 10.1128/AEM.02350-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molloy EM, Field D, O’Connor PM, Cotter PD, Hill C, Ross RP. Saturation mutagenesis of lysine 12 leads to the identification of derivatives of nisin A with enhanced antimicrobial activity. PLoS One. 2013;8:e58530. doi: 10.1371/journal.pone.0058530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rink R, Kuipers A, de Boef E, Leenhouts KJ, Driessen AJM, Moll GN, Kuipers OP. Lantibiotic structures as guidelines for the design of peptides that can be modified by lantibiotic enzymes. Biochemistry. 2005;44:8873–8882. doi: 10.1021/bi050081h. [DOI] [PubMed] [Google Scholar]
- 43.Chatterjee C, Paul M, Xie L, van der Donk WA. Biosynthesis and mode of action of lantibiotics. Chem. Rev. 2005;105:633–683. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- 44.Bond CS, Shaw MP, Alphey MS, Hunter WN. Structure of the macrocycle thiostrepton solved using the anomalous dispersion contribution of sulfur. Acta Crystallogr. D Biol. Crystallogr. 2001;57:755–758. doi: 10.1107/s0907444901003134. [DOI] [PubMed] [Google Scholar]
- 45.Jonker HRA, Baumann S, Wolf A, Schoof S, Hiller F, Schulte KW, Kirschner KN, Schwalbe H, Arndt HD. NMR structures of thiostrepton derivatives for characterization of the ribosomal binding site. Angew. Chem. Int. Ed. Engl. 2011;50:3308–3312. doi: 10.1002/anie.201003582. [DOI] [PubMed] [Google Scholar]
- 46.Mocek U, Beale JM, Floss HG. Reexamination of the 1H and 13C NMR spectral assignments of thiostrepton. J. Antibiot. 1989;42:1649–1652. doi: 10.7164/antibiotics.42.1649. [DOI] [PubMed] [Google Scholar]
- 47.van de Kamp M, van den Hooven HW, Konings RNH, Bierbaum G, Sahl HG, Kuipers OP, Siezen RJ, de Vos WM, Hilbers CW, van de Ven FJM. Elucidation of the primary structure of the lantibiotic epilancin K7 from Staphylococcus epidermidis K7. Cloning and characterisation of the epilancin-K7-encoding gene and NMR analysis of mature epilancin K7. Eur. J. Biochem. 1995;230:587–600. doi: 10.1111/j.1432-1033.1995.tb20600.x. [DOI] [PubMed] [Google Scholar]
- 48.Chan WC, Bycroft BW, Leyland ML, Lian LY, Yang JC, Roberts GCK. Sequence-specific resonance assignment and conformational analysis of subtilin by 2D NMR. FEBS Lett. 1992;300:56–62. doi: 10.1016/0014-5793(92)80163-b. [DOI] [PubMed] [Google Scholar]
- 49.Nicolaou KC, Zak M, Safina BS, Estrada AA, Lee SH, Nevalainen M. Total synthesis of thiostrepton. Assembly of key building blocks and completion of the synthesis. J. Am. Chem. Soc. 2005;127:11176–11183. doi: 10.1021/ja052934z. [DOI] [PubMed] [Google Scholar]
- 50.Schoof S, Baumann S, Ellinger B, Arndt HD. A fluorescent probe for the 70 S-ribosomal GTPase-associated center. ChemBioChem. 2009;10:242–245. doi: 10.1002/cbic.200800642. [DOI] [PubMed] [Google Scholar]
- 51.Schoof S, Arndt HD. D-Cysteine occurrence in thiostrepton may not necessitate an epimerase. Chem. Commun. 2009:7113–7115. doi: 10.1039/b912733j. [DOI] [PubMed] [Google Scholar]
- 52.Okeley NM, Zhu Y, van der Donk WA. Facile chemoselective synthesis of dehydroalanine-containing peptides. Org. Lett. 2000;2:3603–3606. doi: 10.1021/ol006485d. [DOI] [PubMed] [Google Scholar]
- 53.Burrage S, Raynham T, Williams G, Essex JW, Allen C, Cardno M, Swali V, Bradley M. Biomimetic synthesis of lantibiotics. Chem. Eur. J. 2000;6:1455–1466. doi: 10.1002/(sici)1521-3765(20000417)6:8<1455::aid-chem1455>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 54.Lentzen G, Klinck R, Matassova N, Aboul-ela F, Murchie AIH. Structural basis for contrasting activities of ribosome binding thiazole antibiotics. Chem. Biol. 2003;10:769–778. doi: 10.1016/s1074-5521(03)00173-x. [DOI] [PubMed] [Google Scholar]
- 55.Borissenko L, Groll M. 20S proteasome and its inhibitors: crystallographic knowledge for drug development. Chem. Rev. 2007;107:687–717. doi: 10.1021/cr0502504. [DOI] [PubMed] [Google Scholar]
- 56.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. John Innes Foundation; Norwich, UK: 2000. [Google Scholar]
- 57.Cousins KR. Computer review of ChemDraw Ultra 12.0. J. Am. Chem. Soc. 2011;133:8388. doi: 10.1021/ja204075s. [DOI] [PubMed] [Google Scholar]
- 58.Allinger NL. Conformational analysis. 130. MM2. A hydrocarbon force field utilizing V1 and V2 torsional terms. J. Am. Chem. Soc. 1977;99:8127–8134. [Google Scholar]
- 59.The PyMOL Molecular Graphics System, Version 1.5.0.4. Schrodinger, LLC; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.