Abstract
Small non-coding RNAs have emerged as key modulators of viral infection. However, with the exception of hepatitis C virus, which requires the liver-specific microRNA (miRNA)-122, the interactions of RNA viruses with host miRNAs remain poorly characterized. Here, we used crosslinking immunoprecipitation (CLIP) of the Argonaute (AGO) proteins to characterize strengths and specificities of miRNA interactions in the context of 15 different RNA virus infections, including several clinically relevant pathogens. Notably, replication of pestiviruses, a major threat to milk and meat industries, critically depended on the interaction of cellular miR-17 and let-7 with the viral 3’UTR. Unlike canonical miRNA interactions, miR-17 and let-7 binding enhanced pestivirus translation and RNA stability. miR-17 sequestration by pestiviruses conferred reduced AGO binding and functional de-repression of cellular miR-17 targets, thereby altering the host transcriptome. These findings generalize the concept of RNA virus dependence on cellular miRNAs and connect virus-induced miRNA sequestration to host transcriptome regulation.
Keywords: hepatitis C virus, bovine viral diarrhea virus, pestivirus, sponge, Argonaute, miRNA, HiTS-CLIP, CLIP-Seq
Introduction
Direct and indirect interactions with host miRNAs play important roles for DNA viruses, many of which encode their own miRNAs (Kincaid and Sullivan, 2012). For RNA viruses, interactions with host miRNAs remain poorly characterized with the exception of HCV, which requires the liver-specific miR-122 (Wilson and Sagan, 2014). The discovery of interactions between miR-122 and the HCV 5’ UTR (Jopling et al., 2005) at two binding sites stimulating viral replication was unexpected, as miRNAs typically interact with the 3’ UTRs of mRNAs to destabilize transcripts or repress translation (Bartel, 2009). The HCV/miR-122 interaction has proven promising as therapeutic target (Janssen et al., 2013). In contrast, Eastern equine encephalitis virus (EEEV) is an example of canonical miRNA action, where conserved miR-142-3p sites limits viral replication and thereby activation of innate immunity specifically in hematopoietic cells; a mechanism that enhances neuropathogenesis (Trobaugh et al., 2014).
Studies of miRNA action were enhanced by AGO-CLIP methods (Chi et al., 2009; Hafner et al., 2010), which were used to characterize miRNA expression and regulation for several DNA viruses (Haecker et al., 2012; Riley et al., 2012; Skalsky et al., 2012). More recently, we generated small RNA binding landscapes for HCV RNA, confirming the 5’ UTR miR-122 interaction, and unexpectedly demonstrating that sequestration of miR-122 by HCV RNA leads to transcriptome-wide de-repression of cellular miR-122 targets (Luna et al., 2015). A major drawback of standard AGO-CLIP methods is the inability to link miRNAs directly to their targets. miRNA-target chimeras produced through direct ligation allow unambiguous identification of such interactions (Grosswendt et al., 2014; Helwak et al., 2013). Through Covalent Ligation of Endogenous Argonuate-bound RNAs (CLEAR)-CLIP on endogenous AGO complexes, we recently improved target identification efficiency (Moore et al., 2015).
In the current study, we used AGO-CLIP and analysis of miRNA-target chimeras to examine a broad panel of 15 different RNA viruses for miRNA interactions in mammalian cells. Of particular interest were uncovered interactions between miR-17 and let-7 and the 3’ UTR of bovine viral diarrhea virus (BVDV), which were critical for viral replication. BVDV and classical swine fever virus (CSFV) are members of the Pestivirus genus, an important group of animal pathogens distantly related to HCV within the Flaviviridae family. By examining host transcriptomes in vitro and ex vivo, we observed reduced AGO binding and functional mRNA de-repression of cellular miR-17 targets but not let-7 targets during pestivirus infection. This virus-host interaction paradigm provides a mechanism by which viruses can selectively regulate hundreds of cellular genes with minimal investment of genomic information.
Results
AGO-CLIP and miRNA-target chimeras unambiguously identify HCV-miRNA interactions
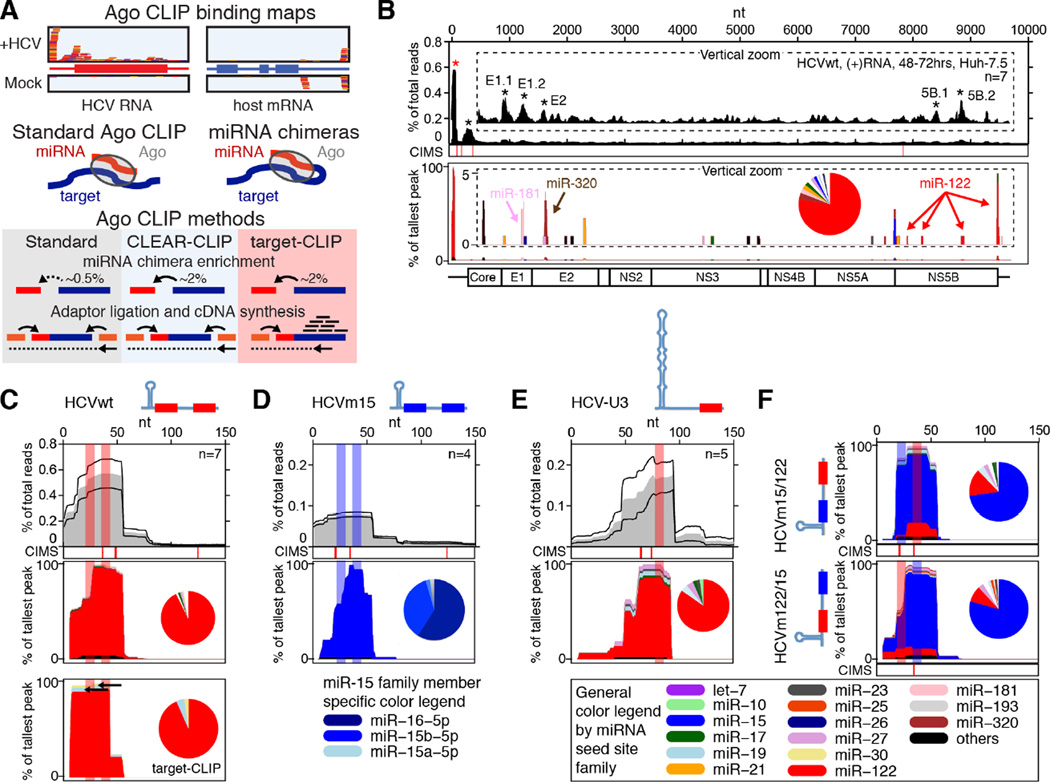
As proof-of-principle, we set out to experimentally verify HCV-miRNA interactions. To this end, we combined our previous approaches, generating viral miRNA binding landscapes using AGO-CLIP (Luna et al., 2015) and unambiguously identifying miRNA-target interactions (Moore et al., 2015) (Figure 1A). Analyzing miRNA chimeras from standard AGO-CLIP or CLEAR-CLIP experiments on HCV infected Huh-7.5 hepatoma cells, we confirmed that miR-122 constitutes the vast majority of miRNA binding on the HCV 5’ UTR and at sites in the NS5B region and in the 3’ UTR (Figure 1B–C, and Tables S1–S2). A minority of overall binding was attributed to other miRNAs, including miR-181-5p and miR-320a peaks in the envelope region (Figure S1A). We did not observe any of the non-miR-122 miRNAs previously suggested to interact with HCV. For HCVm15, where the miR-122 seed sites were substituted for miR-15 (Luna et al., 2015), the 5’ UTR was bound by miR-15b and related family member miR-16 (Figure 1D, and Tables S1–S2). For HCV-U3, where a single large cellular U3-snoRNA-derived stem-loop replaces stem-loop 1 and seed site 1 (Li et al., 2011), our analysis confirmed miR-122 binding at seed site 2 (Figure 1E, and Tables S1–S2), although viral growth is largely independent of miR-122 (Figure S1B and (Luna et al., 2015)). Specific miRNA binding at individual seed sites was discernable using viable recombinants harboring a site each for miR-15 and miR-122 in both configurations (Figure 1F and S1C).
Figure 1.
Method definition and validation on HCV. (A) Schematic of methods. Covalent ligation of miRNAs to their cognate targets creates chimeras. These are enriched in CLEAR-CLIP and target-CLIP. A further enrichment for targets of interest is achieved in target-CLIP, where RT-PCR reverse primers are designed inside AGO binding peaks identified by standard CLIP. (B) Standard AGO (top) and miRNA-specific chimera-derived (bottom) binding map on HCV. Vertical zoom (insert) highlights less abundant peaks. Pie chart shows miRNA abundance across all chimeras. See color legend below. CIMS: Crosslink-Induced Mutation Sites provide nucleotide resolution for AGO-binding. *: significant peak, p<0.05. (C) Horizontal zoom of (B) to the HCV 5’ end. Mean of n replicates (grey) +/− SEM (black lines) is shown. miRNA binding sites are highlighted in shaded bars. The miRNA-specific chimera-derived binding map and abundance pie chart is shown (middle). Target-CLIP using indicated reverse primers is shown (bottom). (D–F) miRNA binding maps as in (C) for HCVm15 (D), HCV-U3 (E), and HCVm15/122 and HCVm122/15 (F). Schematics of the HCV 5’ end with miRNA seed sites indicated are depicted in panels (C–F). See also Figure S1 and Table S1–2.
To specifically enrich for chimeras associated with known AGO peaks, we developed “target-CLIP”. Here, chimera enrichment is similar to CLEAR-CLIP but a set of nested primers ensures enrichment of a specific target (Figure 1A). Target-CLIP confirmed miR-122 binding to the HCV 5’ UTR (Figure 1C). These analyses validated our approach as a robust way to identify specific virus-miRNA interactions.
miRNA binding landscapes for 15 viruses
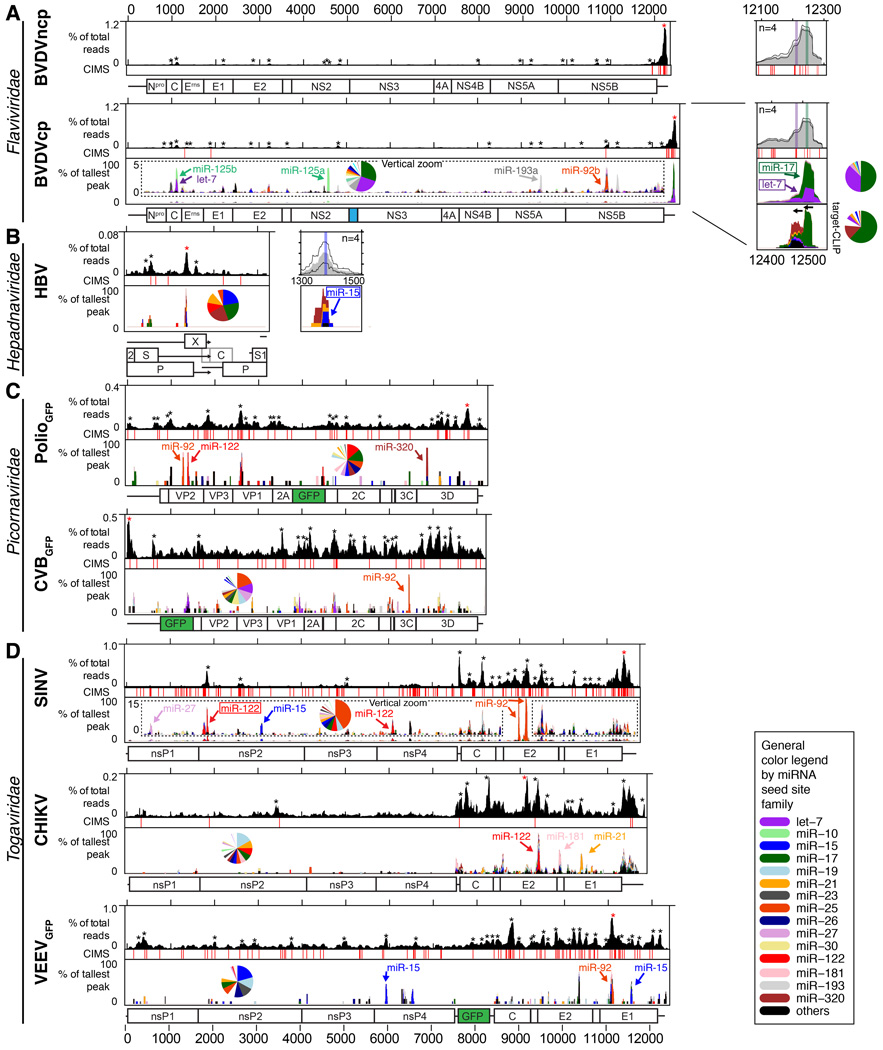
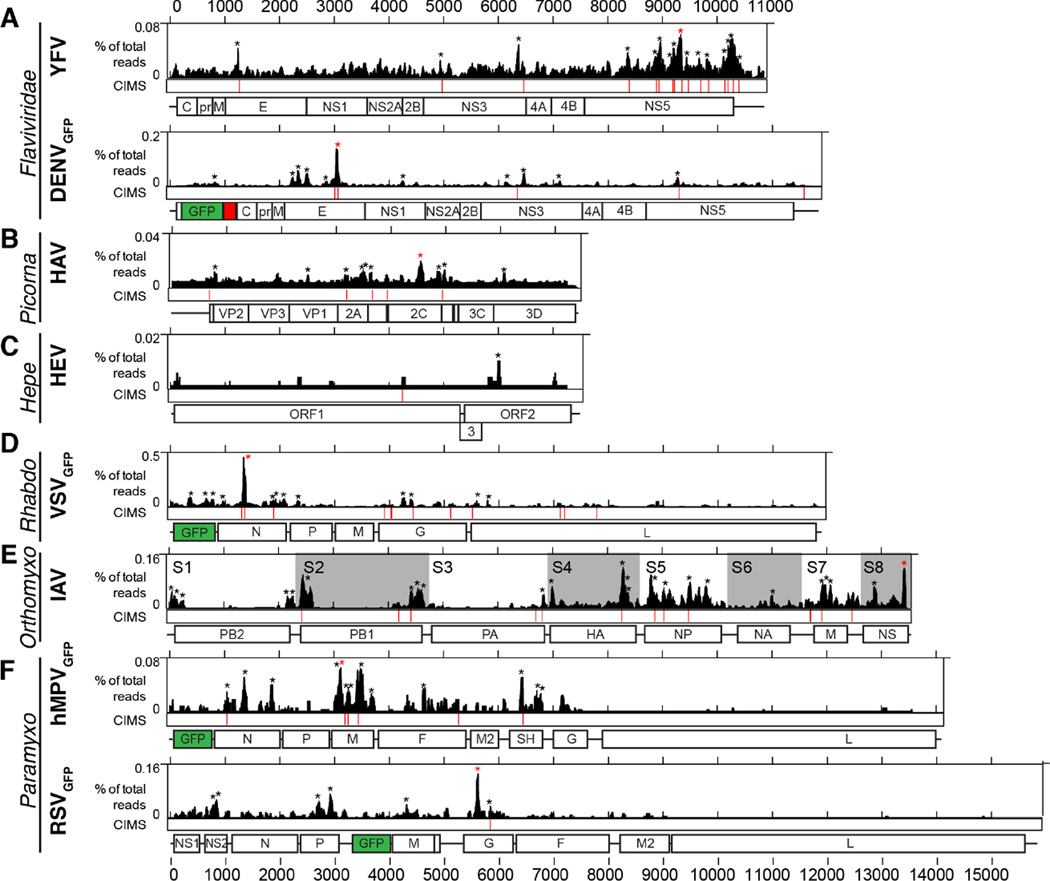
We next characterized AGO binding landscapes for a number of medically important RNA viruses and for hepatitis B virus (HBV). For both cytopathic (cp) and non-cytopathic (ncp) BVDV biotypes (differing in their natural acquisition of integrated host RNA by recombination), HBV, poliovirus (PV), coxsackie virus B3 (CVB), Sindbis virus (SINV), chikungunya virus (CHIKV), and Venezuelan equine encephalitis virus (VEEV), the read depth from AGO-viral RNA interactions was sufficient for characterization of miRNA chimeras (Figure 2, S2–S3, and Tables S1–S2). The lower AGO-viral RNA read depth for yellow fever virus (YFV), dengue virus (DENV), hepatitis A virus (HAV), hepatitis E virus (HEV), influenza A virus (IAV), vesicular stomatitis virus (VSV), human metapneumovirus (hMPV) and respiratory syncytial virus (RSV) only allowed mapping of AGO binding (Figure 3, Table S1).
Figure 2.
AGO binding maps for viruses with miRNA-specific information. Standard AGO (top) and miRNA-specific chimera-derived (bottom) binding maps are shown for BVDV in MDBK cells (A), HBV in Huh-7.5 cells (B), PolioGFP in Huh-7.5 cells and CVBGFP in HeLa cells (C), and SINV in Huh-7.5, CHIKV in Huh-7 cells and VEEVGFP in Huh-7.5 cells (D). Note the different y-scales for AGO binding maps. Vertical zoom (insert) highlights less abundant peaks for BVDV and SINV. miRNA chimera peaks overlapping conserved canonical seed sites are indicated with boxed names (Figure S2). Pie charts show miRNA abundance across all chimeras. CIMS: Crosslink-Induced Mutation Sites provide nucleotide resolution for AGO-binding. Horizontal zoom is shown for BVDV and HBV. Mean of n replicates (grey) +/− SEM is shown. miRNA binding sites are highlighted in colored bars. Genome schematics are shown below with GFP in green and cellular Jiv90 in blue. Stars indicate significant peaks (p<0.05); red stars indicate the tallest peak. See also Figure S2–3 and Table S1–2.
Figure 3.
AGO binding maps for viruses without miRNA-specific information. Standard AGO maps are shown for YFV and DENVGFP in Huh-7.5 cells (A), HAV in Huh-7.5 cells (B), HEV in Huh-7.S10.3 cells (C), VSVGFP in A549 cells (D), IAV in A549 cells (E), and hMPVGFP and RSVGFP in A549 cells (F). Note the different scales for AGO binding maps. CIMS: Crosslink-Induced Mutation Sites provide nucleotide resolution for AGO-binding. Genome schematics are shown below with GFP in green and FMDV-2A in red. See also Table S1.
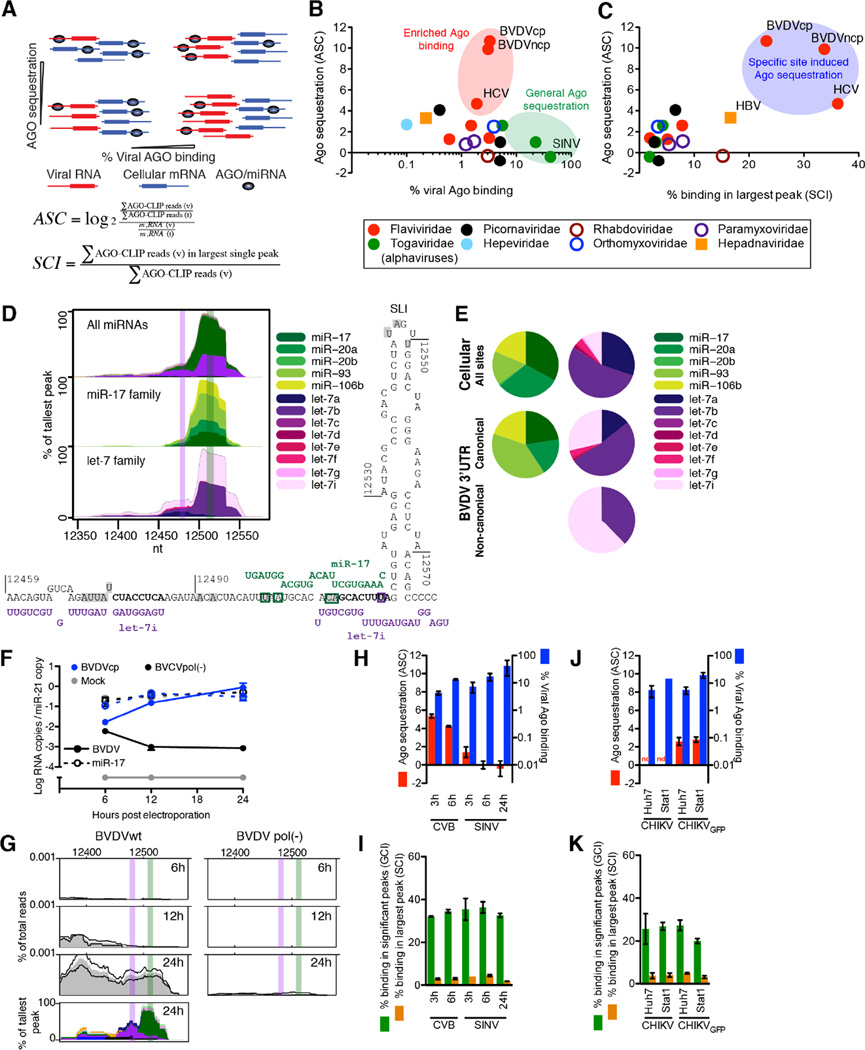
To quantitatively describe the extent of AGO binding to viral vs. host RNA, we calculated the viral CLIP read fraction normalized to total read depth. We further normalized by the fraction of viral RNA over the amount of total RNA determined by RT-qPCR (Figure S4A), and used this to define the AGO sequestration coefficient (ASC, Figure 4A). Whereas most viruses bound less than a few percent of the AGO pool and did not specifically sequester AGO, two outgroups were identified; those that bound significant proportions of the AGO pool due to extensive viral RNA amounts (low ASC, primarily alphaviruses, Togaviridae), and those that bound only a few percent of the AGO pool but were highly enriched for AGO binding relative to their replication level (high ASC, HCV and BVDV, Figure 4B). For HCV and BVDV, AGO/miRNA binding further was highly clustered with up to 40% of reads constituting the single largest binding peak (Figure 4C and Figure S4B). Although these parameters alone do not suffice for identifying interactions of biological significance, strong, specific and perhaps essential AGO interactions would be expected for viruses with high ASC and clustering, which was the case for HCV, for BVDV and, to a lesser degree, for HBV.
Figure 4.
Quantification of viral AGO/miRNA binding. (A) Schematics of AGO sequestration as a function of percentage viral AGO binding in scenarios of different viral RNA levels. Definitions of AGO sequestration coefficient (ASC) and specific clustering index (SCI) are given; v: viral, t: total. (B) ASC plotted as a function of viral AGO binding in percentage of total. (C) ASC plotted as a function of SCI. (D) miRNA-specific chimera maps for the BVDV 3’ UTR (top), broken down by family member for miR-17 (middle) or let-7 (bottom). Canonical miRNA binding sites are highlighted in colored bars. The predicted RNA structure of interactions is shown below. Significant CIMS sites are shaded in grey. Chimera-specific CIMS sites are boxed in green (miR-17) or purple (let-7). (E) Pie charts of miR-17 and let-7 family member abundance in cellular and BVDV chimeras. For let-7, data is divided between the two peaks of a putative canonical and non-canonical interaction. (F–G) BVDV electroporation time course showing BVDV (plain lines) and miR-17 (dashed lines) RNA quantification normalized to miR-21 levels (F) and AGO/miRNA binding to the BVDV 3’ UTR (G). (H) ASC and viral AGO binding in percentage of total binding for time course experiments of CVB and SINV. (I) SCI and general clustering index (GCI, binding in significant peaks) for time course experiments of CVB and SINV. (J) ASC and viral AGO binding as in (H) for cell line experiments with CHIKV. (K) AGO clustering as in (I) for cell line experiments with CHIKV. See also Figure S4.
miRNA chimera analyses establish specific virus-miRNA interactions
For BVDV, chimera analysis revealed that the strong AGO binding on the 3’ UTR was due to members of the let-7 and miR-17 families. The let-7 and miR-17 chimera peaks overlapped each other and canonical seed sites for both miRNAs (defined in (Bartel, 2009)), which were flanked by crosslink-induced mutation sites (CIMS, (Zhang and Darnell, 2011)) providing nucleotide resolution of AGO binding (Figure 2A and 4D). An additional let-7 peak suggested potential non-canonical let-7 binding overlapping the miR-17 site. Chimera-specific CIMS analysis supported the canonical miR-17 site and the non-canonical let-7 site (Figure 4D). Let-7 and miR-17 binding constituted >50% of the miRNA binding on BVDV and >80% of the binding on its 3’ UTR (Figure 2A). Relative to other miR-17 family members expressed in MDBK cells, miR-93 binding to the BVDV genome was enriched compared to binding to cellular RNA. Among expressed let-7 members, let-7i was enriched relative to 7a on the canonical BVDV site; on the putative non-canonical site, only 7b and 7i bound (Figure 4E). These findings corresponded to energetically favorable binding of miR-93 and let-7b and 7i (Figure S4C). The read depth of other peaks across the BVDV ORF constituted only 3–4% of the 3’UTR peak, and included let-7, miR-125b, miR-193a and miR-92b-3p (Figure 2A and S2A).
To assay BVDV-miRNA kinetics, we followed MDBK cells electroporated with BVDVcp or a replication deficient pol(-) mutant over time (Figure 4F). In contrast to observations for HCV and miR-122 (Luna et al., 2015), AGO binding to the BVDV 3’ UTR was observed only at late time points and never on pol(-) mutant RNA (Figure 4G).
Several direct miRNA interactions were previously suggested for HBV (Xie et al., 2014). The largest specific peak in our data set overlapped with a conserved miR-15 site (Figure 2B). The HBV miR-15 interaction was confirmed using target-CLIP and chimera data; a non-conserved, non-canonical miR-320a interaction may also contribute to AGO binding at this site (Figure S2B). These data confirmed previous observations that HBV can bind miR-15/16 at this site, possibly to downregulate this miRNA family and indirectly prevent apoptosis (Liu et al., 2013). A smaller peak overlapped a previously suggested miR-17 binding site (Xie et al., 2014), although no chimeras were present to support this. In Huh-7.5 cells we saw no evidence for binding of miR-122 or other miRNAs at previously suggested sites.
miRNA chimera data were also interpretable for PV, CVB, SINV, CHIKV and VEEV (Figure 2C–D and S2C–D). These viruses replicate to very high RNA copy numbers and had extensive AGO binding. To mitigate noise from non-specific binding, we analyzed AGO binding at early time points for CVB and SINV. Although the percentage of viral AGO binding increased over time, the ASC decreased due to the dramatic increase in viral RNA content (Figure 4H). AGO binding peaks generally remained at the same position and increased in read depth over time, although for SINV binding accumulated across the entire genome (Figure 4I and Figure S3A–B). Taken together, these data suggested that the extent of AGO binding on CVB and SINV RNA scales with RNA abundance, as does accumulation of non-specific binding.
To assess miRNA binding in different cellular contexts, we compared AGO binding and miRNA chimera profiles for CHIKV and CHIKV-GFP in Huh-7 hepatoma cells and STAT1−/− fibroblasts, and observed similar ASC and clustering values (Figure 4J–K). Overall binding landscapes were similar including chimera-supported interactions with miR-21 and miR-181. As expected, interactions with miR-122 were observed only in Huh-7 cells and with let-7 only in fibroblasts, reflecting the miRNA composition of these cell types (Figure S3C).
For the alphaviruses, we observed one particularly dominant interaction between the SINV E2 region and miR-92a-3p. Targeting this miRNA with a locked nucleic acid (LNA), however, had no effect on SINV replication or virus production (Figure S4D–E). These results are congruent with reporter gene assays showing that the predicted non-canonical binding of the miR-92a-3p 3’ region, as predicted for SINV (Figure S2D), did not functionally regulate reporter gene expression (Figure S4E). Thus, AGO-CLIP and miRNA chimeras identified important miRNA interactions for BVDV, confirmed the previously characterized HBV/miR-15 interaction and mapped miRNA binding landscapes for important picornaviruses and alphaviruses.
BVDV critically depends on miR-17 and let-7 for replication
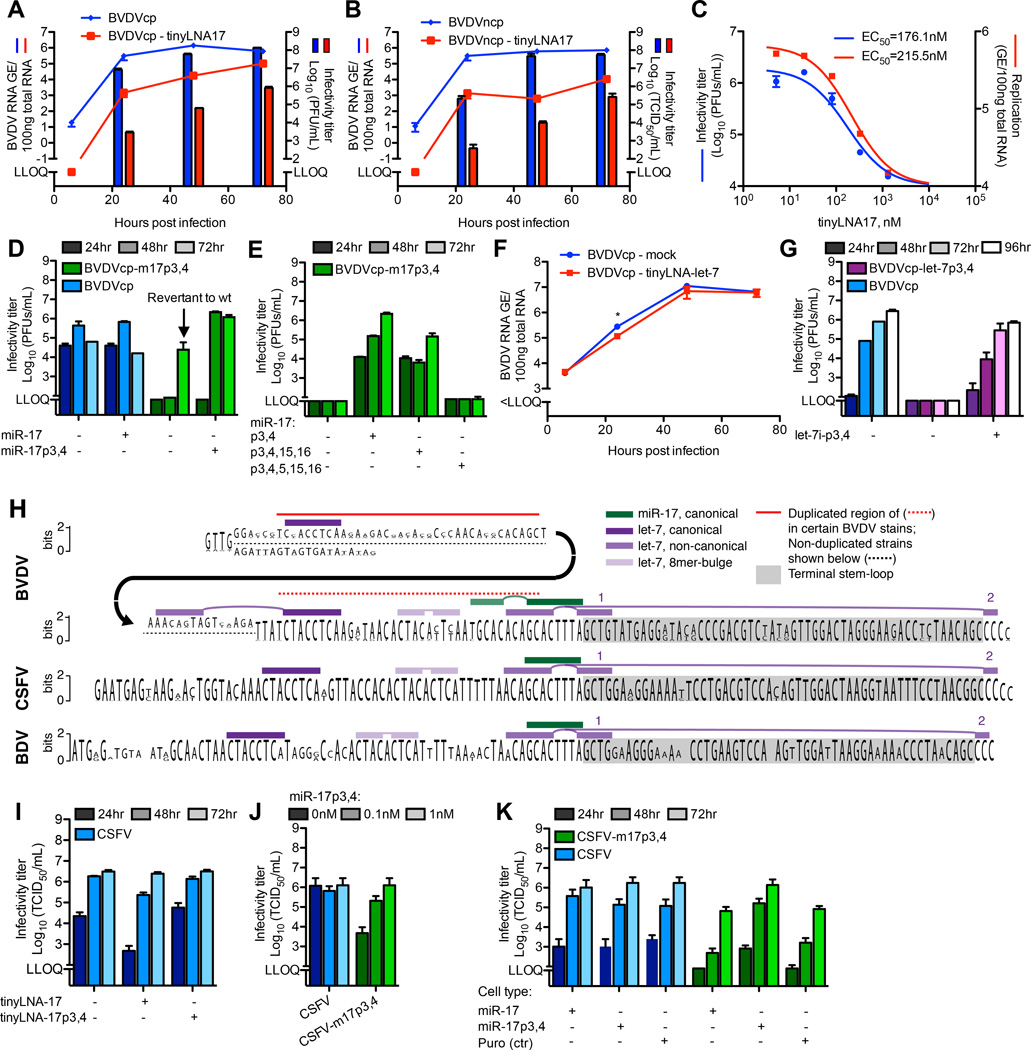
To determine the functional importance of the miR-17 interaction during BVDV infection, we inhibited the miR-17 family in MDBK cells using tinyLNA-17 (Obad et al., 2011)(Figure S5A). For both BVDV biotypes, tinyLNA-17 led to significant attenuation of replication and virus production (Figure 5A–B). This inhibition was reproducible in BT cells (Figure S5B) and occurred in a dose-dependent manner (Figure 5C), suggesting a therapeutic antiviral action similar to that of miR-122 inhibitors for HCV (Janssen et al., 2013).
Figure 5.
Pestivirus replication and virus production depend on miRNAs. (A–B) Replication and virus production after infection of MDBK cells with BVDVcp (A) or BVDVncp (B) (MOI=0.1) +/− tinyLNA-17 (0.3µM) (C) Dose-response of BVDVcp infection of MDBK cells (MOI=0.1) to tinyLNA-17. (D) Virus production after electroporation of BVDVcp and m17p3,4 mutant +/− trans-complementation with miR-17p3,4 (50nM). (E) Virus production as in (D) but with trans-complementing the m17p3,4 mutant with miR-17p3,4, miR-17p3,4,15,16 or miR-17p3,4,5,15,16. (F) BVDVcp replication after infection of MDBK cells (MOI=0.1) +/− tinyLNA-let-7 partially inhibiting let-7. (G) Virus production after electroporation of BVDVcp and let-7p3,4 mutant +/− trans-complementation with let-7i–p3,4 (0.1nM). (H) Conservation logo plot showing relative frequency of bases at every position in the 3’ end of BVDV (31 sequences), CSFV (68 sequences) and border disease virus (BDV, 7 sequences) downstream of the AT-rich region. Canonical and non-canonical miR-17 and let-7 sites are indicated. For BVDV, some strains (incl. NADL used in this study) carry a semi-conserved duplication (red line). (I) Virus production after CSFV-luc infection of SK-6 cells (MOI=0.1) +/− tinyLNA-17 or tinyLNA-17p3,4 (0.3µM). (J) Dose-response of CSFV wt or an m17p3,4 mutant 48hrs after transfection of SK-6 cells with miR-17p3,4. (K) Virus production after CSFV wt or m17p3,4 RNA transfection of SK-6 cells transduced with lentiviruses expressing miR-17 or miR-17p3,4 or with the lentivirus backbone (puro, ctr). See also Figure S5–6.
Following electroporation of MDBK cells with BVDV cp and ncp mutant RNAs that could no longer interact with miR-17 (m17p3,4 or p7,8 mutants), no virus production or cell death (cp variant) was observed (Figure 5D and S5C–E). In one experiment delayed virus production occurred; however, sequencing revealed reversion of the p3 site from day 5 post electroporation, and complete reversion to wt 5 days after passage to naïve cells. Trans-complementation with miR-17p3,4 or miR17p7,8 completely rescued virus production (Figure 5D and S5C–E), and with increasing concentrations BVDV replication reached a plateau (Figure S5D).
Using mutated miRNA mimics, we tested whether the BVDV/miR-17 interaction required auxiliary base pairing, as was observed for HCV/miR-122 (Machlin et al., 2011). This was not the case, since miR-17p3,4,15,16 mimic rescued BVDV-m17p3,4 virus production to a similar extent as miR-17p3,4. Virus production could not be rescued by the miR-17p3,4,5,15,16 mimic with an added mismatch in the seed (Figure 5E).
The let-7 family is highly abundant in MDBK cells and could be inhibited only to ~50% using tinyLNAs (Figure S5F). BVDV replication in cells partially devoid of let-7 activity was slightly decreased with significant difference only at 24hrs (Figure 5F). BVDV p3,4 mutants of the canonical let-7 site were highly attenuated in culture, and could be rescued by let-7p3,4 (Figure 5G). This suggested that both miR-17 and let-7 were critical for BVDV replication.
If the putative non-canonical binding of let-7 is important for replication, predictions suggest that a BVDV-m17p7,8 mutant would require miR-17p7,8 as well as trans-complementation with let-7-p17,18. However, BVDV-m17p7,8 virus production was unaffected by the presence of let-7i–p17,18 (Figure S5E). Further, a BVDV-m17p7,8 mutant with a swap of two base-pairs at the base of the terminal stem-loop was viable (Figure S5E), arguing against binding of let-7 to an open conformation of the stem-loop (Figure S4C). These data argue against a critical role for non-canonical let-7 interactions.
To evaluate the importance of weaker BVDV-miRNA interactions, we analyzed miR-92a-3p as an example, for which predictions allowed both canonical and auxiliary binding (Figure S2A). We did not observe any effect on BVDV replication or virus production from inhibition of this miRNA (Figure S5G–H), arguing against importance of minor miRNA interactions.
miRNA dependence is a general hallmark of pestivirus replication
The let-7 and miR-17 seed sites are highly conserved among pestiviruses (Figure 5H), suggesting that pestiviruses generally bind and depend on these host miRNAs. To test this hypothesis, we infected SK-6 cells with CSFV and observed decreased virus production (Figure 5I) after functional inhibition of miR-17 by tinyLNA-17 (Figure S6A). A CSFV-m17p3,4 mutant had reduced focus size and virus production after electroporation. This phenotype could be rescued by trans-complementation using miR-17p3,4 (Figure 5J and Figure S6B–C). We further established SK-6 cells stably expressing miR-17p3,4 by lentiviral transduction and fluorescent sorting (Figure S6D–E), which, in contrast to parental cells, efficiently propagated wt as well as m17p3,4 mutant CSFV (Figure 5K). Thus, dependence on host miRNAs appears to be a general phenomenon in pestivirus replication. We note that the near-universal tissue tropism of pestiviruses (Givens and Marley, 2013) is congruent with the ubiquitous expression of miR-17 and let-7 across a range of tissue types (Landgraf et al., 2007) and cell lines studied here (Figure S6F).
miR-17 enhances BVDV translation and RNA stability
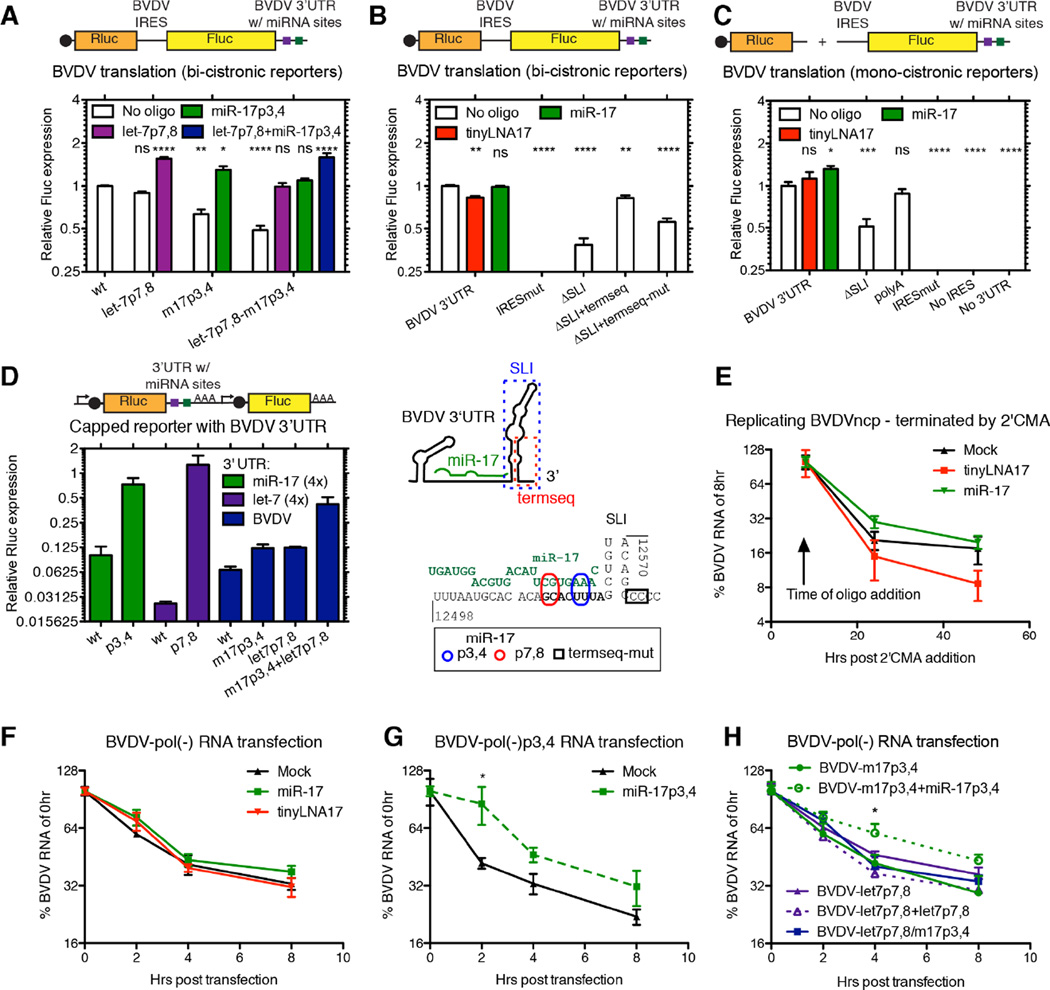
For HCV, miR-122 modestly stimulates viral translation (Henke et al., 2008) and protects the uncapped RNA genome from degradation (Sedano and Sarnow, 2014; Shimakami et al., 2012). Using bicistronic BVDV reporters, translation was non-significantly decreased after mutating the let-7 site and decreased to ~70% and ~50% after mutation of the miR-17 alone or in combination with let-7 (Figure 6A). Trans-complementation with high levels of the corresponding miRNAs rescued translation to ≥100%. Inhibition of miR-17 by tinyLNA-17 reduced translation to ~80% (Figure 6B). Translation was strongly inhibited by deleting the 3’ UTR terminal stem-loop. Retaining the terminal 19 nts of the 3’ UTR partially reversed this phenotype (Figure 6B). This suggested that both the terminal 3’ UTR sequence and the miRNA interactions positively regulate BVDV translation, with miR-17 playing a larger role than let-7.
Figure 6.
Functional characterization of BVDV miRNA interactions. (A) BVDV driven translation was measured from bicistronic reporters after introduction of let-7 or miR-17 seed site mutations and after trans-complementation with miRNA mimics (50nM). (B) Translation as in (A) measured after treatment with tinyLNA-17 (0.3µM) or miR-17 (50nM) or after modifications to the 3’ UTR as indicated in the schematic below. (C) Translation as in (B), but measured from monocistronic reporters with the exact BVDV termini. (D) Translation measured from RNAs harboring the BVDV 3’ UTR and/or miRNA seed sites in the capped, poly(A)-tailed context of the psiCHECK-2 system. In the schematics, caps are black circles, miRNA sites are squares, promoters are angled arrows, and poly(A)-tails are AAA’s. (E–G) Effect of miRNAs on BVDV RNA stability. Decay of BVDV RNA was measured +/− tinyLNA-17 (0.3µM) or miR-17 (50nM) after blocking active BVDV replication using 2’CMA (E), or after transfection of replication-deficient pol(-)BVDV RNA (F). (G–H) RNA decay assays as in (F), but for seed site mutants +/− corresponding miRNA complementation. See also Figure S6.
Because these results differ from canonical miRNA repression of cellular mRNAs, we confirmed the positive role for miR-17 on translation in context of monocistronic reporters containing the exact BVDV ends (Figure 6C). In contrast, in a capped, poly(A)-tailed mRNA-like context, seed site mutations led to increased translation. A combined let-7/miR-17 BVDV 3’ UTR mutant relieved translational repression to the greatest extent (Figure 6D). Thus, it appears that the positive role of let-7 and miR-17 on translation occurs specifically in context of a non-capped, non-poly(A)-tailed, IRES-containing RNA, like those of pestiviruses.
In a further attempt to analyze non-canonical binding of let-7, we measured translation from bicistronic BVDV-m17p7,8 reporters. Trans-complementation with miR-17p7,8, but not let-7p17,18, rescued translation (Figure S6G). Further, rescue of translation depended on an intact terminal stem-loop, and was not affected by swapping the stem base pairs (Figure S6G). Thus, putative non-canonical binding does not appear important for translation.
To assess the effect of miR-17 on intracellular BVDV RNA stability, we halted ongoing BVDV replication in MDBK cells in the presence or absence of tinyLNA-17 or excess miR-17. Measuring the relative level of BVDV RNA over time, we observed a small, non-significant stabilization by miR-17 and de-stabilization by tinyLNA-17 (Figure 6E). When BVDV-pol(-) RNA was transfected, excess miR-17 provided a small, non-significant protection from degradation (Figure 6F). A larger and significant effect was seen when BVDV-pol(-)m17p3,4 was stabilized by miR-17p3,4 (Figure 6G). A similar effect was not observed for BVDV-pol(-)let-7p7,8 (Figure 6H). These results indicate that the miR-17 but not let-7 interaction plays a small protective role in viral RNA degradation.
Pestiviruses functionally sequester the miR-17 but not let-7 family, indirectly regulating the cellular transcriptome
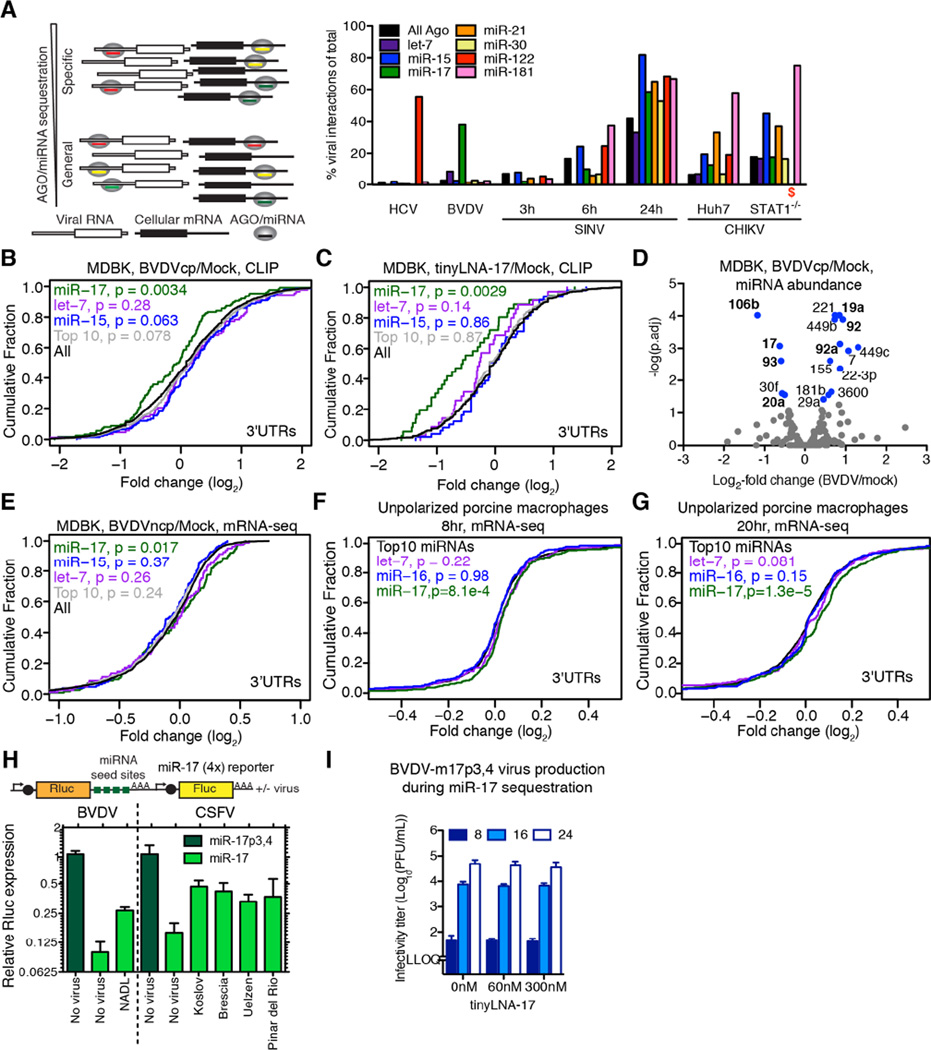
We reasoned that the number of miRNA chimeric reads mapping to the virus versus the host would be a direct way to evaluate specific miRNA sequestration during infection. For HCV, around 50% of miR-122 interactions mapped to the virus (Figure 7A), consistent with our modeling estimates of the available miR-122 pool during infection (Luna et al., 2015). For BVDV, around 40% of miR-17 interactions mapped to the virus, suggesting an analogous sponge effect. For let-7, this number was <10% due to the high abundance of this miRNA family (Figure S7A and Table S3). The sponge effects of HCV and BVDV were specific to miR-122 and miR-17, respectively; in contrast, SINV and CHIKV sequestered a broad range of miRNAs consistent with their general AGO sequestration (Figure 4B and Figure 7A).
Figure 7.
Impact of infection on cellular gene expression. (A) Percentage of virus-derived vs. total chimeras shown for selected miRNAs as a direct quantification of miRNA sponge effects. A schematic of specific vs. general miRNA/AGO sequestration is depicted to the left. (B) Cumulative density function (CDF) of the log2-fold change in CLIP binding between BVDV infected and uninfected cells for all 3’ UTR clusters containing indicated 8-mer seeds, from quadruplicate experiments. “Top 10” refers to the ten most abundant CLIP-derived AGO-bound miRNAs. Two-sided K-S test p-values of difference to “All” are shown. (C) CDF plot as in (B), but comparing tinyLNA-17 treated cells to mock. (D) Volcano plot for significant changes to AGO/miRNA profiles after infection with BVDV. miRNAs with a significant change in abundance are shown in blue and those of the miR-17 family or the miR-17-92 polycistron in boldface. (E) CDF plot for BVDV infection of MDBK cells as in (B) but measuring target expression via mRNA-seq, from duplicate experiments. (F–G) CDF plots as in (E) but for CSFV vEy-37-infected unpolarized porcine monocyte-derived macrophages 8hrs (F) and 20hrs (G) post infection, from triplicate experiments. (H) Reporter validation of target de-repression by BVDV in MDBK and CSFV in SK-6 cells. Schematics of reporters as in Figure 6. (I) Virus production after infection of miR-17p3,4 trans-complemented (10nM) MDBK cells with BVDVcp-m17p3,4 +/− tinyLNA-17. See also Figure S7 and Table S3–5.
Given specific interactions between BVDV and miR-17 and let-7, we analyzed AGO binding on the host transcriptome during infection (Figure S7B). We observed significantly reduced global AGO binding on 3’ UTR and coding sequence (CDS) targets of miR-17 but not let-7 when compared to targets of the miR-15 family, the top 10 miRNAs or all targets (Figure 7B, S7C–F and Table S4). A similar but stronger effect was observed when comparing tinyLNA-17 to mock treated cells (Figure 7C and S7G and Table S4). Furthermore, we observed that BVDV RNA genomes can approach miR-17 copy numbers (Figure 4F) and that the abundance of miR-17 family members available for regulation is decreased upon infection (Figure 7D). Taken together, these data suggest that miR-17 but not let-7 can be functionally inhibited by BVDV RNA.
Along these lines, we reasoned that the BVDV miR-17 sponge effect should result in specific de-repression of cellular miR-17 targets, measurable by mRNA-seq. Indeed, mRNAs harboring a CLIP-guided miR-17 site in their 3’ UTR were significantly de-repressed compared to those having a site for let-7, miR-15, or the top 10 miRNAs in MDBK cells (Figure 7E, S7H and Table S4). Among mRNAs with a CLIP-supported miR-17 8mer in their 3’ UTR, 19 were significantly upregulated during BVDV infection (Table S5). Using alternative parameters to identify miR-17 sponge regulated genes, 47 mRNAs had reduced AGO binding at 3’ UTR miR-17 6–8mer sites and were significantly upregulated during BVDV infection (Table S5). Excess amounts of miR-17p3,4 dramatically repressed an arbitrary set of mRNAs carrying the corresponding seed site in their 3’ UTRs (Figure S7I), and BVDV-m17p3,4 mutants were not able to induce a sponge effect under these conditions (Figure S7J). Thus, the BVDV miR-17 sponge effect is functional, but only within a certain range of miRNA abundance.
We investigated the miRNA sequestration for another pestivirus by performing mRNA-seq 8 and 20 hours after CSFV infection of porcine monocyte-derived macrophages ex vivo. In both unpolarized and interferon-γ-polarized macrophages, we observed specific, time-dependent de-repression of mRNAs harboring miR-17 sites in the 3’ UTR (Figure 7F–G and S7K–L). Among genes expressed in unpolarized macrophages with a miR-17 8mer in the 3’ UTR, 32 were significantly upregulated upon CSFV infection (Table S5). The de-repression of miR-17 targets induced by BVDV and CSFV (both high and low virulence strains) was confirmed at the protein level using psiCHECK-2 reporters harboring miR-17 seed sites in the 3’ UTR (Figure 7H).
Lastly, to evaluate how the aggregate gene regulation by sponging may influence pestiviral replication, we decoupled the direct miR-17 requirement from sponging by infecting miR-17p3,4 trans-complemented MDBK cells with BVDV-m17p3,4 and mimicking sponging using tinyLNA-17 (Figure 7I). Observing no change in virus production, we concluded that pestiviral miR-17 sponging does not appear to directly influence infection kinetics in this context.
Taken together, these results demonstrated that pestiviruses in vitro and ex vivo induce a miR-17-specific de-repression, thereby regulating hundreds of cellular targets.
Discussion
Despite broad interest in HCV dependence on miR-122, little work has been done to look for similar interactions among other viruses. Here we employed a powerful combination of AGO-CLIP and miRNA chimera analysis to map miRNA interactions across 15 different viruses and discovered critical miR-17 and let-7 interactions for pestiviruses. Similarly to the matched tissue tropism of HCV and the liver-specific miR-122 (Wilson and Sagan, 2014), the broad tropism of pestiviruses (Givens and Marley, 2013) correlates well with the distribution of miR-17 and let-7. For other viruses, significant challenges remain for assessing the functional relevance of nonspecific AGO coating and interactions lacking specific miRNA chimeras. Identification of miRNA-dependent interactions could be enhanced by improvements of chimera methods. In addition, AGO-CLIP in cells devoid of mature miRNAs (Bogerd et al., 2014; Luna et al., 2015) could potentially identify miRNA-independent binding. Such experiments may help clarify the functional relevance, if any, of broad AGO coating for highly abundant RNA viruses such as alphaviruses. Sequestration of AGO to reduce activity of the small RNA pathway could be a parallel to subgenomic flavivirus RNA (sfRNA)-mediated inhibition of Dicer (Schnettler et al., 2012). Further explorations of timing, cell type and miRNA chimeras are likely to provide additional insights. Whereas essential miRNA interactions will be observed in any infection context, regulatory or inhibitory interactions, such as that of EEEV (Trobaugh et al., 2014), could be missed in certain cell subsets and in cell lines devoid of mature miRNAs (Bogerd et al., 2014).
In our previous studies on HCV, we demonstrated that an RNA virus can regulate hundreds of cellular transcripts through sequestration of a specific miRNA (Luna et al., 2015), analogous to the concept of competing endogenous RNAs (ceRNAs) (Salmena et al., 2011). However, whereas most cellular miRNA targets are degraded or repressed upon miRNA binding, viral RNA replication is stimulated, acting as a positive feed-back loop for additional miRNA sequestration. Here, we expanded this finding to show that pestiviruses sequester ~40% of the miR-17 pool to de-repress cellular genes. Given the absence of pro-viral effects of the hepaci- and pestivirus-induced miRNA sponge effects, it is yet unclear whether such sponge effects are byproducts of the direct viral miRNAs dependence, or important mechanisms for manipulating host gene expression.
Previously, miR-17 has been implicated in both positive and negative regulation of viral infection. Exogenous miR-17 levels were found to have a pro-viral effect for herpes simplex virus (HSV) and IAV, possibly through repression of a set of interferon-stimulated genes (Seo et al., 2013). Likewise, Epstein-Barr virus (EBV) appears to co-repress cellular miR-17 targets (Riley et al., 2012). In contrast, other studies showed that human cytomegalovirus (hCMV) and human immunodeficiency virus (HIV) sequester or down-regulate the miR-17~92 cluster to counteract antiviral effects (Lee et al., 2013; Triboulet et al., 2007). In the immune system, over-expression of the miR-17~92 cluster leads to lymphoproliferative disease and autoimmunity, whereas inhibition can lead to apoptosis (Mendell, 2008). Pestiviruses primarily replicate in proliferating lymphocytes (Chase, 2013), which aligns well with elevated levels of miR-17 during proliferation. It is therefore conceivable that pestivirus-mediated sequestration of miR-17 plays a role in immunopathological events, e.g. by increasing lymphocyte apoptosis or decreasing proliferation to limit the immune response. In this context, it is noteworthy that pestiviruses induce mild to severe lymphopenia (Chase, 2013).
Mechanistic studies showed that let-7 and miR-17 have small positive effects on BVDV translation, and that miR-17 exerts a small stabilizing effect on viral RNA. Similar observations for HCV were explained by the proximity and putative engagement of AGO/miR-122 with the translation initiation complex (Henke et al., 2008). Concurrently, the proximity of AGO/miR-122 to the uncapped HCV RNA 5’ end may protect viral RNA from the exonucleases Xrn1 and Xrn2 (Sedano and Sarnow, 2014; Shimakami et al., 2012). A miR-17-dependent regulation of BVDV translation could be envisioned through genome circularization. Putative protection from exonucleases could most easily be envisioned for the 3’ exosome since miR-17 binds to the BVDV 3’ UTR. For classical flaviviruses, stable structures in the 3’ UTR do arrest Xrn1 activity to generate sfRNAs, and binding of miRNAs to the pestivirus 3’ UTR could serve a similar function, although sfRNAs have not been identified for BVDV (Pijlman et al., 2008). Perhaps more intriguing, miRNA binding could serve as a switch between translation and replication and/or packaging. A recent study suggested that miR-122 competes with binding of cellular factors such as PCBP2 to the HCV 5’ UTR to act as such a switch (Masaki et al., 2015). Future work should also expand on the role, if any, of non-canonical interaction with let-7. Such an interaction could potentially protect the pestiviral 3’ end in a manner similar to how miR-122 protects the HCV 5’ end (Figure 4D).
In conclusion, this study outlines an effective framework for discovering virus-miRNA interactions. We determined that the HCV/miR-122 interaction is not unique, as pestiviruses critically rely on the host miR-17 and let-7 families. Antiviral therapy using miRNA antagonists may therefore also find use beyond that of HCV. Interestingly, the concept of RNA virus-induced miRNA sponging could be extended to pestiviruses in primary cell settings where sponge effects may influence the course of disease. This emerging concept in virus-host interactions holds alluring promise for future exploration for these and other viruses.
Experimental Procedures
Cell lines, virus reagents and infection conditions for CLIP
Cells were cultured as described in Supplemental Information (SI). The genotype 2 HCVwt (J6/JFH1-clone2) (Catanese et al., 2013), HCVm15 (Luna et al., 2015) and HCV-U3 (Li et al., 2011) recombinants were described. BVDVncp and BVDVcp of the NADL stain (Mendez et al., 1998) had differences compared to published sequences (Table S6). The CSFV vA187-1 (CSFV wt) and vEy-37 were described (Mayer et al., 2003; Ruggli et al., 1996). The highly virulent CSFV strain Koslov was from V. Kaden, Brescia and Uelzen (highly and moderately virulent strains, respectively) were from the Community Reference Laboratory for CSFV (Hannover, Germany), and the low virulent Pinar del Rio (Perez et al., 2012) was from L. Ganges (Centre de Recerca en Sanitat Animal CReSA-IRTA, Barcelona, Spain). The bicistronic CSFV-luc virus, A187-Npro-Luc-IRES-C, was derived from a replicon (Suter et al., 2011). For HBV, the 1.3x genome of isolate adw was used (Doitsh and Shaul, 2004). Recombinant clones were obtained of the (+)RNA viruses polio-GFP strain Mahoney (Teterina et al., 2010) (V. Racaniello), coxsackie virus B -GFP strain woodruff (Feuer et al., 2002), SINV strain toto1101 (Rice et al., 1987), CHIKV and CHIKV-GFP strain La Reunion (Tsetsarkin et al., 2006), VEEV-GFP strain TC83 (I. Frolov), YFV strain Asibi (McElroy et al., 2005) with differences as noted in Table S6, DENV-GFP of serotype 2 strain 16681 (Schoggins et al., 2012), HAV strain HM175/18f (Lemon et al., 1991), and HEV strain Kernow-P6 (Shukla et al., 2012). Infectious virus stocks were obtained of the (-)RNA viruses VSV-GFP strain Indiana (Ramsburg et al., 2005) (J. Rose), IAV strain A/WSN/33 (P. Palese), hMPV-GFP strain CAN97-83 (Biacchesi et al., 2004), and RSV-GFP strain (Munir et al., 2008) (P. Collins). Infection conditions for the AGO-CLIP experiments are described in SI.
Construction of virus, reporter and lentivirus plasmids
Modifications to virus plasmids and BVDV translation reporters were done using standard PCR and cloning methods. Details are in SI, and the introduced modifications are listed in Table S6.
RNA transcription, electroporation, transfection, purification, and titration
Viral RNA for electroporation and transfection was produced as detailed in SI. miRNA mimics (Table S6) (GE Dharmacon) and tinyLNAs (Exiqon) were transfected using RNAi/MAX (Life Technologies) and used at non-cytotoxic doses. RT-qPCRs were done as detailed in SI and the primers and probes given in Table S6. The details of RNA transcription, electroporation, transfection, purification, and titration are described in SI.
Viral infectivity titration, monitoring of infection and sequencing of virus from supernatant
Viral infectivity titration was done using plaque-forming unit (PFU), focus-forming unit (FFU) or tissue culture infectious dose-50 (TCID50) methods. Details on these assays, infectious center assay, focus size assay, monitoring of infection and sequencing virus from supernatant are described in SI. Primers for sequencing the BVDV ORF are given in Table S6.
Luciferase reporter assays, lentivirus production and establishment of modified cell lines
Luciferase reporter assays were done as described (Luna et al., 2015) with modifications as in SI. Lentivirus production and establishment of modified cell lines were performed as described in SI.
RNA degradation assays
To evaluate the kinetics of BVDV RNA degradation, active replication was either stopped using 2’-C-methyladenosine (2’CMA) or non-replicating genomes were transfected. Intracellular RNA was harvested at indicated time points and viral RNA was quantified as described above. Details of these assays are given in SI.
CLIP and RNA-seq procedures
Preparation of cell cultures for CLIP assays (Luna et al., 2015), standard AGO-CLIP (Moore et al., 2014), and CLEAR-CLIP (Moore et al., 2015) methods were performed as published. Expansion on these methods, including target-CLIP, as used here is available in SI and Table S6.
The mRNA-seq libraries were prepared from Trizol or Direct-zol (Zymo Research) extracted RNA following Illumina TruSeq protocols for poly-A selection, fragmentation, and adaptor ligation. Libraries were sequenced as 100bp single- or paired-end runs (Illumina). Bioinformatic analysis was done as described in SI. AGO-CLIP alignment statistics are given in Table S7.
Supplementary Material
Highlights.
miRNA binding profiles for 15 different viruses elucidated by Argonaute (AGO)-CLIP
Groups of viruses sequester specific miRNAs or AGO in general
Pestiviruses critically depend on cellular miR-17 and let-7
Pestiviral RNA functionally reduces miR-17 binding on endogenous mRNA targets
Acknowledgments
A special thanks to P. Ambrose, K. Bell, H. Chung, M. Dittmann, H. Hoffmann, M. Li, P. Liu, J. Schoggins, W. Schneider, T. Sheahan and Y. Yu for sharing reagents and protocols, C. Coffran and C. Zhang for bioinformatics support; the Rockefeller Genomics Resource Center for technical assistance, the NGS Platform of the University of Bern for performing the porcine macrophage RNA-seq library preparation and sequencing, and A. Summerfield, N. Tautz and members of the Rice and Darnell labs for valuable discussion. This study was supported by grants from the NIH, NIAID (AI116943, AI099284, AI091707, AI090055), NINDS (NS034389, NS081706), Office of the Director through the NIH Roadmap for Medical Research (DK085713), NCI (CA057973), The Rockefeller University Center for Clinical and Translational Science (UL1RR024143), the Center for Basic and Translational Research on Disorders of the Digestive System (Leona M. and Harry B. Helmsley Charitable Trust), the Greenberg Medical Research Institute, and the Starr Foundation. T.K.H.S. was supported by a Postdoctoral Fellowship and a Sapere Aude Research Talent Award from The Danish Council for Independent Research. J.M.L. was supported by a David Rockefeller Graduate Student Fellowship. K.R.G. was supported by The Rockefeller University’s Women & Science Fellowship Program and an NRSA Postdoctoral Fellowship (F32AI120579). M.L., G.A., and in part I.K. and R.B. were supported by grant #310030-141045 from the Swiss National Science Foundation to N.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
AGO-CLIP and RNA-seq data have been deposited in GEO under accession number GSE76967.
Author contributions
TKHS, JML, RBD and CMR conceived the study. TKHS, JML, ML, GA, NR and CMR designed the experiments, TKHS, JML, ML, EN, KRG, GA, MG and JF performed the experiments, TKHS, JML, ML, KRG, GA, IK and RB analyzed the data, and TKHS, JML, ML, NR and CMR wrote the paper. JML and ML contributed equally to this study.
References
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biacchesi S, Skiadopoulos MH, Tran KC, Murphy BR, Collins PL, Buchholz UJ. Recovery of human metapneumovirus from cDNA: optimization of growth in vitro and expression of additional genes. Virology. 2004;321:247–259. doi: 10.1016/j.virol.2003.12.020. [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Skalsky RL, Kennedy EM, Furuse Y, Whisnant AW, Flores O, Schultz KL, Putnam N, Barrows NJ, Sherry B, et al. Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs. J Virol. 2014 doi: 10.1128/JVI.00985-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanese MT, Loureiro J, Jones CT, Dorner M, von Hahn T, Rice CM. Different requirements for scavenger receptor class B type I in hepatitis C virus cell-free versus cell-to-cell transmission. J Virol. 2013;87:8282–8293. doi: 10.1128/JVI.01102-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase CC. The impact of BVDV infection on adaptive immunity. Biologicals. 2013;41:52–60. doi: 10.1016/j.biologicals.2012.09.009. [DOI] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsh G, Shaul Y. Enhancer I predominance in hepatitis B virus gene expression. Mol Cell Biol. 2004;24:1799–1808. doi: 10.1128/MCB.24.4.1799-1808.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuer R, Mena I, Pagarigan R, Slifka MK, Whitton JL. Cell cycle status affects coxsackievirus replication, persistence, and reactivation in vitro. J Virol. 2002;76:4430–4440. doi: 10.1128/JVI.76.9.4430-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givens MD, Marley MS. Immunology of chronic BVDV infections. Biologicals. 2013;41:26–30. doi: 10.1016/j.biologicals.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Grosswendt S, Filipchyk A, Manzano M, Klironomos F, Schilling M, Herzog M, Gottwein E, Rajewsky N. Unambiguous identification of miRNA:target site interactions by different types of ligation reactions. Mol Cell. 2014;54:1042–1054. doi: 10.1016/j.molcel.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haecker I, Gay LA, Yang Y, Hu J, Morse AM, McIntyre LM, Renne R. Ago HITS-CLIP expands understanding of Kaposi’s sarcoma-associated herpesvirus miRNA function in primary effusion lymphomas. PLoS Pathog. 2012;8:e1002884. doi: 10.1371/journal.ppat.1002884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M. microRNA-122 stimulates translation of hepatitis C virus RNA. Embo J. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, et al. Treatment of HCV Infection by Targeting MicroRNA. N Engl J Med. 2013 doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Kincaid RP, Sullivan CS. Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog. 2012;8:e1003018. doi: 10.1371/journal.ppat.1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Song J, Kim S, Kim J, Hong Y, Kim Y, Kim D, Baek D, Ahn K. Selective degradation of host MicroRNAs by an intergenic HCMV noncoding RNA accelerates virus production. Cell Host Microbe. 2013;13:678–690. doi: 10.1016/j.chom.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Lemon SM, Murphy PC, Shields PA, Ping LH, Feinstone SM, Cromeans T, Jansen RW. Antigenic and genetic variation in cytopathic hepatitis A virus variants arising during persistent infection: evidence for genetic recombination. J Virol. 1991;65:2056–2065. doi: 10.1128/jvi.65.4.2056-2065.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YP, Gottwein JM, Scheel TK, Jensen TB, Bukh J. MicroRNA-122 antagonism against hepatitis C virus genotypes 1–6 and reduced efficacy by host RNA insertion or mutations in the HCV 5’ UTR. Proc Natl Acad Sci U S A. 2011;108:4991–4996. doi: 10.1073/pnas.1016606108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Zhang J, Jiao T, Li Z, Peng J, Cui Z, Ye X. Hepatitis B virus inhibits apoptosis of hepatoma cells by sponging the MicroRNA 15a/16 cluster. J Virol. 2013;87:13370–13378. doi: 10.1128/JVI.02130-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna JM, Scheel TK, Danino T, Shaw KS, Mele A, Fak JJ, Nishiuchi E, Takacs CN, Catanese MT, de Jong YP, et al. Hepatitis C Virus RNA Functionally Sequesters miR-122. Cell. 2015;160:1099–1110. doi: 10.1016/j.cell.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlin ES, Sarnow P, Sagan SM. Masking the 5’ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc Natl Acad Sci U S A. 2011;108:3193–3198. doi: 10.1073/pnas.1012464108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki T, Arend KC, Li Y, Yamane D, McGivern DR, Kato T, Wakita T, Moorman NJ, Lemon SM. miR-122 Stimulates Hepatitis C Virus RNA Synthesis by Altering the Balance of Viral RNAs Engaged in Replication versus Translation. Cell Host Microbe. 2015;17:217–228. doi: 10.1016/j.chom.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer D, Thayer TM, Hofmann MA, Tratschin JD. Establishment and characterisation of two cDNA-derived strains of classical swine fever virus, one highly virulent and one avirulent. Virus Res. 2003;98:105–116. doi: 10.1016/j.virusres.2003.08.020. [DOI] [PubMed] [Google Scholar]
- McElroy KL, Tsetsarkin KA, Vanlandingham DL, Higgs S. Characterization of an infectious clone of the wild-type yellow fever virus Asibi strain that is able to infect and disseminate in mosquitoes. J Gen Virol. 2005;86:1747–1751. doi: 10.1099/vir.0.80746-0. [DOI] [PubMed] [Google Scholar]
- Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez E, Ruggli N, Collett MS, Rice CM. Infectious bovine viral diarrhea virus (strain NADL) RNA from stable cDNA clones: a cellular insert determines NS3 production and viral cytopathogenicity. J Virol. 1998;72:4737–4745. doi: 10.1128/jvi.72.6.4737-4745.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Scheel TK, Luna JM, Park CY, Fak JJ, Nishiuchi E, Rice CM, Darnell RB. miRNA-target chimeras reveal miRNA 3’-end pairing as a major determinant of Argonaute target specificity. Nat Commun. 2015;6:8864. doi: 10.1038/ncomms9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Zhang C, Gantman EC, Mele A, Darnell JC, Darnell RB. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat Protoc. 2014;9:263–293. doi: 10.1038/nprot.2014.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir S, Le Nouen C, Luongo C, Buchholz UJ, Collins PL, Bukreyev A. Nonstructural proteins 1 and 2 of respiratory syncytial virus suppress maturation of human dendritic cells. J Virol. 2008;82:8780–8796. doi: 10.1128/JVI.00630-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez LJ, Diaz de Arce H, Perera CL, Rosell R, Frias MT, Percedo MI, Tarradas J, Dominguez P, Nunez JI, Ganges L. Positive selection pressure on the B/C domains of the E2-gene of classical swine fever virus in endemic areas under C-strain vaccination. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2012;12:1405–1412. doi: 10.1016/j.meegid.2012.04.030. [DOI] [PubMed] [Google Scholar]
- Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi PY, Hall RA, et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4:579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Ramsburg E, Publicover J, Buonocore L, Poholek A, Robek M, Palin A, Rose JK. A vesicular stomatitis virus recombinant expressing granulocyte-macrophage colony-stimulating factor induces enhanced T-cell responses and is highly attenuated for replication in animals. J Virol. 2005;79:15043–15053. doi: 10.1128/JVI.79.24.15043-15053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice CM, Levis R, Strauss JH, Huang HV. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987;61:3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley KJ, Rabinowitz GS, Yario TA, Luna JM, Darnell RB, Steitz JA. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. Embo J. 2012;31:2207–2221. doi: 10.1038/emboj.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggli N, Tratschin JD, Mittelholzer C, Hofmann MA. Nucleotide sequence of classical swine fever virus strain Alfort/187 and transcription of infectious RNA from stably cloned full-length cDNA. J Virol. 1996;70:3478–3487. doi: 10.1128/jvi.70.6.3478-3487.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnettler E, Sterken MG, Leung JY, Metz SW, Geertsema C, Goldbach RW, Vlak JM, Kohl A, Khromykh AA, Pijlman GP. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and Mammalian cells. J Virol. 2012;86:13486–13500. doi: 10.1128/JVI.01104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Dorner M, Feulner M, Imanaka N, Murphy MY, Ploss A, Rice CM. Dengue reporter viruses reveal viral dynamics in interferon receptor-deficient mice and sensitivity to interferon effectors in vitro. Proc Natl Acad Sci U S A. 2012;109:14610–14615. doi: 10.1073/pnas.1212379109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedano CD, Sarnow P. Hepatitis C virus subverts liver-specific miR-122 to protect the viral genome from exoribonuclease Xrn2. Cell Host Microbe. 2014;16:257–264. doi: 10.1016/j.chom.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo GJ, Kincaid RP, Phanaksri T, Burke JM, Pare JM, Cox JE, Hsiang TY, Krug RM, Sullivan CS. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe. 2013;14:435–445. doi: 10.1016/j.chom.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, Lemon SM. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc Natl Acad Sci U S A. 2012;109:941–946. doi: 10.1073/pnas.1112263109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla P, Nguyen HT, Faulk K, Mather K, Torian U, Engle RE, Emerson SU. Adaptation of a genotype 3 hepatitis E virus to efficient growth in cell culture depends on an inserted human gene segment acquired by recombination. J Virol. 2012;86:5697–5707. doi: 10.1128/JVI.00146-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M, Nusbaum JD, Feederle R, Delecluse HJ, Luftig MA, et al. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 2012;8:e1002484. doi: 10.1371/journal.ppat.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter R, Summerfield A, Thomann-Harwood LJ, McCullough KC, Tratschin JD, Ruggli N. Immunogenic and replicative properties of classical swine fever virus replicon particles modified to induce IFN-alpha/beta and carry foreign genes. Vaccine. 2011;29:1491–1503. doi: 10.1016/j.vaccine.2010.12.026. [DOI] [PubMed] [Google Scholar]
- Teterina NL, Levenson EA, Ehrenfeld E. Viable polioviruses that encode 2A proteins with fluorescent protein tags. J Virol. 2010;84:1477–1488. doi: 10.1128/JVI.01578-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- Trobaugh DW, Gardner CL, Sun C, Haddow AD, Wang E, Chapnik E, Mildner A, Weaver SC, Ryman KD, Klimstra WB. RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature. 2014;506:245–248. doi: 10.1038/nature12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin K, Higgs S, McGee CE, De Lamballerie X, Charrel RN, Vanlandingham DL. Infectious clones of Chikungunya virus (La Reunion isolate) for vector competence studies. Vector borne and zoonotic diseases. 2006;6:325–337. doi: 10.1089/vbz.2006.6.325. [DOI] [PubMed] [Google Scholar]
- Wilson JA, Sagan SM. Hepatitis C virus and human miR-122: insights from the bench to the clinic. Curr Opin Virol. 2014;7:11–18. doi: 10.1016/j.coviro.2014.03.005. [DOI] [PubMed] [Google Scholar]
- Xie KL, Zhang YG, Liu J, Zeng Y, Wu H. MicroRNAs associated with HBV infection and HBV-related HCC. Theranostics. 2014;4:1176–1192. doi: 10.7150/thno.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol. 2011;29:607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.