Abstract
Only a small percentage of individuals living in endemic areas develop severe malaria suggesting that host genetic factors may play a key role. This study has determined the frequency of single nucleotide polymorphisms (SNPs) in some pro and anti-inflammatory cytokine gene sequences: IL6 (-174; rs1800795), IL12p40 (+1188; rs3212227), IL4 (+33; rs2070874), IL10 (-3575; rs1800890) and TGFb1 (+869; rs1800470), by means of PCR-RFLP. Blood samples were collected from 104 symptomatic and 37 asymptomatic subjects. Laboratory diagnosis was assessed by the thick blood smear test and nested-PCR. No association was found between IL6 (-174), IL12p40 (+1188), IL4 (+33), IL10 (- 3575), TGFb1 (+869) SNPs and malaria symptoms. However, regarding the IL10 -3575 T/A SNP, there were significantly more AA and AT subjects, carrying the polymorphic allele A, in the symptomatic group (c2 = 4.54, p = 0.01, OR = 0.40 [95% CI - 0.17- 0.94]). When the analysis was performed by allele, the frequency of the polymorphic allele A was also significantly higher in the symptomatic group (c2 = 4.50, p = 0.01, OR = 0.45 [95% CI - 0.21-0.95]). In conclusion, this study has suggested the possibility that the IL10 - 3575 T/A SNP might be associated with the presence and maintenance of malaria symptoms in individuals living in endemic areas. Taking into account that this polymorphism is related to decreased IL10 production, a possible role of this SNP in the pathophysiology of malaria is also suggested, but replication studies with a higher number of patients and evaluation of IL10 levels are needed for confirmation.
Keywords: Malaria, Genetic polymorphism, Polymerase Chain Reaction, Cytokines, Restriction enzymes
INTRODUCTION
Malaria is a disease that affects about 300-500 million people annually around the world, and in Brazil the 2013 year data reported 178,613 new cases1 , 2.
Clinical manifestations of malaria vary from asymptomatic to poly-symptomatic and only a small proportion of patients (1% to 2%) will have life-threatening disease3 , 4. The outcome of infection seems to depend on a balance between pro and anti-inflammatory cytokine production allowing the onset and expansion of the immune response and, at the same time, limiting the potential pathogenicity of the parasite2 , 5 , 6.
The identification of single nucleotide polymorphisms (SNPs) within cytokine gene promoters that are associated with the symptomatic forms of malaria could be useful for the patients' care and for vaccine development. In previous unpublished studies performed in our laboratory, several SNPs located at the IL6, IL12p40, IL4, IL10 and TGFb1 genes did not show any differences between symptomatic and asymptomatic malaria patients living in endemic areas for many years. Therefore, we decided to finish this investigation by means of the evaluation of a few remaining SNPs that had not yet been investigated. The choice of cytokines and SNPs was also based on the role of these cytokines in the pathophysiology of malaria and the lack of information for most of the SNPs in the context of malaria. Thus, the aim of this study was to determine and compare the genotype and allele frequencies of the following cytokine gene SNPs: IL6 (-174 rs1800795), IL12p40 (+1188; rs3212227), IL4 (+33; rs2070874), IL10 (- 3575; rs1800890), TGFb1 (+869; rs1800470), in people living in an endemic area of the Brazilian Amazon, with symptomatic or asymptomatic malaria.
PATIENTS, MATERIAL AND METHODS
Studied area: Blood samples were collected in the city of Peixoto de Azevedo, Mato Grosso, Brazil. At the time of blood collection (1995-1996) the transmission of malaria in the region was perennial due to the emergence of gold mines in the region. The index of bites/man/hour was 13.5 in the gold mining area and 3.1 in the peri-urban region7 , 8 , 9.
Symptomatic patients. The group of symptomatic malaria was composed of 104 unrelated individuals who reported fever, malaise, muscle pain and headache in the last seven days (one week), but not necessarily at the time of the medical appointment, but they did not have cerebral malaria, intense anemia, or other signs/symptoms of severe malaria. They should not be under treatment for malaria or any other infectious disease for at least seven days before the consultation. These patients presented a positive thick blood smear test or nested-PCR and a hematocrit > 20%10.
Asymptomatic patients. The group of asymptomatic malaria comprised 37 unrelated subjects with a positive thick blood smear test or PCR, hematocrit > 20% and without any symptoms that could be associated with malaria or other infectious disease for at least 60 consecutive days before the study. To recruit these asymptomatic patients the researchers conducted an active search, i.e., toured several neighborhoods and homes in the same town looking for asymptomatic subjects, but with the same level of exposure of the symptomatic ones. The complexity of performing this active search and the fact that the city chosen for this research was part of a known highly endemic area of malaria at the time the study was conducted has determined the lower number of cases in this group.
Ethical approvals: Blood samples were collected in a previous study, approved by the Institutional Research Ethics Committee (CAPPesq process number 968/06). The present research was approved by the Ethics Committee of the Hospital das Clínicas da Faculdade de Medicina da Universidade de São Paulo (CAPPesq process number 0633/10).
Laboratory diagnosis of malaria. Malaria infection was confirmed by the thick blood smear test and PCR11 , 12 , 13. Parasitemia was estimated by the thick blood smear test and expressed as low (< 1,000 parasites/mm3 of blood), moderate (1,000 to 10,000 parasites/mm3 of blood) or intense (> 10,000 parasites/mm3 of blood). Both tests were performed by trained technicians and were able to determine the genus and species of Plasmodium.
Genomic DNA extraction. EDTA-blood samples from symptomatic and asymptomatic malaria cases were submitted to DNA extraction following a phenol-chloroform protocol13, modified by Ferreira et al. (1998)14.
Polymerase Chain Reaction (PCR). To detect the cytokine gene SNPs, amplifications were carried out in 25 mL of final volume containing 50 ng of genomic DNA, 1U of Taq DNA polymerase (Fermentas, Thermo Fisher Scientific, USA), 100 mM of Tris-HCl (pH 8.3), 500 mM of KCL, 1.5 mM of MgCl2, 40 mM of dNTP (Fermentas, Thermo Fisher Scientific, USA) and 0.25 mM of the forward and reverse primers. A total of 35 to 40 cycles were performed with annealing temperatures varying from 51 to 62 0C (Table 1).
Table 1. Description of the studied cytokines, polymorphisms (SNP and rs) restriction enzymes, primers sequences, annealing temperatures and the expected RFLP patterns.
| Cytokine RFLP enzyme Reference | SNP rs | PRIMER SEQUENCE Annealing temperature | RFLP pattern wild type homozygous heterozygous |
|---|---|---|---|
| IL6 NlaIII 15 | -174 G/C (rs1800795) | F: TGACTTCAGCTTTACTCTTGT R: CTGATTGGACTTATTAAG 51 °C | GG 167 e 31bp CC 122, 45, 31bp GC 167, 122, 45, 31bp |
| IL12p40 TaqaI 16 | +1188 A/C (rs3212227) | F:CTGATCCAGGATGAAAATTTGG R:CCCATGGCAACTTGAGAGCTGG 58 °C | AA 226 bp CC 185 e 41bp AC 226, 185,41bp |
| IL4 MnlI 17 | +33 C/T, (rs2070874) | F:CTCATTTTCCCTCGGTTTCAGC R:GAAGCAGTTGGGAGGTGAGA 60 °C | TT 126bp CC 82 e 44bp TC 126, 82 e 44bp |
| IL10 ApoI 18 | -3575 T/A (rs1800890) | F: GGTTTTCCTTCATTTGCAGC R: ACACTGTGAGCTTCTTGAGG 62 °C | TT 121 e 107 bp AA 228 bp TA 228, 121, 107 bp |
| TGFb1 NotI 19 | +869 T/C (rs1800470) | F:GTGCCGGGGGGCGCCGCCTCCCCCATGCCTCCCTC R:CAGCACCAGTAGCCACAGCAGCGGTAGCCGCAGC 60 °C | TT 199 bp CC 179 e 20 bp TC 199,179, 20 bp |
SNP - Single Nucleotide Polymorphism; F=forward; R= Reverse; bp= base pair; rs (SNP accession number)
Restriction Fragment Length Polymorphism (RFLP). Ten microliters of the amplification products were cleaved with 2 to 3 units of the appropriate restriction enzyme in 3 hours of incubation at 37 ºC, excepting for the IL10 -3575 SNP (incubation at 50 ºC), generating DNA fragments that were able to discriminate the wild genotype, the homozygous polymorphic genotype and the heterozygous genotype (Table 1).
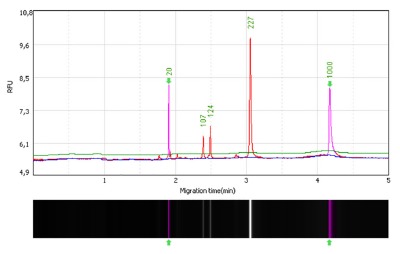
RFLP analysis. After the restriction enzyme cleavage, each of the cytokine SNPs amplification products were visualized by means of a capillary gel electrophoresis apparatus (GelBot, Loccus Biotecnologia, São Paulo, Brazil). The high resolution cartridge S1 kit was applied (Loccus Biotecnologia, São Paulo, Brazil), which is able to discriminate amplification products differing in only 2 to 4 base pairs. After the injection of the alignment marker MA-1 (20 to 1000 bp), 1 uL of the sample was injected into the cartridge, followed by 1 uL of the pBR322 DNA-MspI digest size marker (15-622 pb). The procedure was performed according to the M-4-2-06-300 method (2 seconds of sample injection at 4 kV followed by separation for 300 seconds at 6 kV) and data were analyzed by means of the software Q-Analyzer.Figure 1 illustrates the IL10 -3575 SNP capillary electrophoresis. Considering the higher accuracy of the capillary electrophoresis in comparison to the horizontal electrophoresis performed in agarose gels, the molecular weights reported for the amplification products corresponding to the wild, heterozygous and homozygous polymorphic genotypes may differ in 1 to 3 base pairs with respect to those mentioned in Table 1, that were based on detection in agarose gels.
Fig. 1. - IL10 -3575 SNP (rs1800890) after RFLP. The heterozygous genotype (TA) generates fragments of 227, 124, 107 bp. The first image on the left displays the capillary electrophoresis analysis of the IL10 -3575 (T/A) SNP after RFLP performed with the ApoI enzyme. The image shows in the X axis, a sample carrying a heterozygous genotype (TA) generating, after RFLP, three fragments of 227, 124, 107 bp (in red); the alignment markers (20 to 1000 bp, in pink) and their corresponding migration times (in minutes). The electropherogram also shows the relative fluorescent unit (RFU) of each fragment (semi-quantitation results) in the Y axis. The second image (below) shows the three DNA fragments located within the region delineated by the alignment markers, as they would appear in an agarose gel.
Statistical analysis. The required number of individuals in the control group would be two to three times the number of sick individuals, but this situation would apply to studies of patients versus healthy controls. In the present study we assessed only patients with malaria and stratified them into two groups according to the presence or absence of infection symptoms. It is also noteworthy that we were unable to accomplish a case-control study with the same number of cases in both groups due to the scarcity of asymptomatic individuals in the studied region that was, at that time, experiencing a malaria epidemic. Thus, we analyzed a convenience sample composed of the maximum number of cases (symptomatic patients) and controls (asymptomatic patients) that were available, provided that they shared the same environmental and genetic factors.
The statistical analysis was performed by means of the Sigma Stat software, version 3.5 (Systat Software Inc., London, UK) and the GraphPad Prism, version 6.0 (GraphPad Prism Software Inc., CA, USA). Genotype and allele frequencies were calculated by direct counting. Comparisons between the frequencies and proportions in the two groups (symptomatic or asymptomatic individuals) were made by the chi-square test (c2) or the Fisher exact test (for comparisons with n £ 5). The probabilities of tests considered a confidence interval of 95% and a p-value £ 0.05. Whenever a p-value £ 0.05 was found, the Yates correction (pc) was applied.
To determine if the genotype frequencies were in Hardy-Weinberg equilibrium (HW) we used the equation available at http://www.oege.org/software/hwe-mr-calc.shtml.
RESULTS
In this preliminary report a total of 141 subjects, 104 symptomatic and 37 asymptomatic were analyzed. In the symptomatic group there were 74 male and 30 female subjects with a median age of 30 years. In the asymptomatic group 30 subjects were male and seven were female, the median age was 33 years. This research was performed with a convenience sample consisting of a majority of young adults (18- 45 years old), either in the symptomatic group (80/104 - 76.92%), or in the asymptomatic one (30/37 - 81.08%). In both study groups (symptomatic and asymptomatic) the population was predominantly composed of people with mixed ethnicity, a mixture of Caucasians and Brazilian indigenous people (Amerindians) who were born and lived in this endemic geographical area. There was no significant difference in symptomatic and asymptomatic subjects regarding the age and gender. However, even though in symptomatic and asymptomatic malaria cases the number of years living in the endemic area was similar (nine years), when the number of malaria episodes was compared, the symptomatic group presented more episodes than the asymptomatic one as demonstrated by the comparison of the median values (Mann Whitney test, p = 0.0123) (Table 2).
Table 2. Characteristics of the Plasmodium-infected individuals in both groups (symptomatic and asymptomatic) .
| Symptomatic N=104 | Asymptomatic N=37 | Total N=141 | p value* | |
|---|---|---|---|---|
| Age years | 30 (2-71) | 33 (11-67) | 31 (2-71) | 0.0822 |
| Gender | ||||
| Male (%) | 74 (71.2) | 30 (81) | 104 | 0.2820 |
| Female (%) | 30 (28.8) | 07 (19) | 37 | |
| Number of previous malaria episodes** | 20 (0-20) | 15 (0-20) | 20 (0-20) | 0.0123 |
| Parasitemia (microscopy) | ||||
| Detectable | 60 | 9 | 69 | 0.0005 |
| Non-Detectable | 44 | 28 | 72 | |
| Plasmodium species | ||||
| P. falciparum | 38 | 9 | 47 | |
| P. vivax | 34 | 15 | 49 | |
| P. malariae | 0 | 2 | 2 | |
| P. falciparum and P. vivax | 32 | 11 | 43 |
Age and number of previous malaria episodes were expressed as median (minimum-maximum). *Statistical significance was determined by Mann Whitney and Chi square tests. **All subjects that reported more than 20 malaria episodes were arbitrarily designated as having 20, this is the reason the maximum value of the number of previous malaria episodes was 20 in either symptomatic and asymptomatic groups.
With respect to the Plasmodium species in the symptomatic group (n = 104), P. falciparum was found in 38 patients; P. vivax in 35 patients; and mixed infections by P. falciparum and P. vivax in 31 patients. In the asymptomatic group (n = 37), P. falciparum was found in nine patients; P. vivax in 15 patients; P. malariae in two patients; and mixed infections by P. falciparum and P. vivax in 11 patients.
In the symptomatic group, among the 104 subjects, six (5.76%) presented intense parasitemia in the thick blood smear test, 21 (20.20%) had moderate parasitemia, 33 (31.73%) had low parasitemia and 44 (42.31%) had non-detectable parasitemia (these individuals were diagnosed by PCR). In the asymptomatic group, among the 37 subjects, nine patients had detectable parasitemia in the thick blood smear test (24.3%) and 28 patients were diagnosed by PCR (75.7%). This difference was statistically significant (chi-square test, p = 0.0005) (Table 2).
Considering the five studied SNPs the genotypic distribution was in agreement with the Hardy-Weinberg equilibrium.
In the present study, no association between IL6 (-174), IL12p40 (+1188), IL4 (+33), TGFb1 (+869) SNPs and malaria symptoms was found (Table 3).
Table 3. Genotypic and allelic frequencies of the five studied SNPs: IL6 -174; IL12p40 +1188; IL4 +33; IL10 -3575; TGFb1+869 in the groups of symptomatic and asymptomatic patients .
| IL6 -174 G/C (rs1800795) | Symptomatic | Asymptomatic | Total | c2p | OR [95% CI] | c2pc | |
|---|---|---|---|---|---|---|---|
| Wild genotype | GG | 48 (46.15) | 21 (56.75) | 69 | 1.228 0.1339 | 0.6531 [0.30-1.39] | |
| Grouped genotypes | GC+CC | 56 (53.84) | 16 (43.24) | 72 | NC | ||
| Total | 104 | 37 | 141 | ||||
| Wild type alelle | G | 146 (70.19) | 57 (77.02) | 203 | 1.264 0.1304 | 0.7023 [0.37-1.30] | |
| Polymorphic alelle | C | 62 (29.80) | 17 (22.97) | 79 | NC | ||
| Total | 208 | 74 | 282 | ||||
| IL-12p40 A/C (rs3212227) | Symptomatic | Asymptomatic | Total | c 2 p | OR [95% CI] | c 2 pc | |
| Wild genotype | AA | 37 (35.57) | 17 (45.94) | 54 | 0.1242 0.1326 | 0.6497 [0.30-1.39] | |
| Grouped genotypes | AC+CC | 67 (64.42) | 20 (54.05) | 87 | NC | ||
| Total | 104 | 37 | 141 | ||||
| Wild type alelle | A | 129 (62.01) | 51 (68.91) | 180 | 1.125 0.1444 | 0.7364 [0.41-1.29] | |
| Polymorphic alelle | C | 79 (37.98) | 23 (31.08) | 102 | NC | ||
| Total | 208 | 74 | 282 | ||||
| IL-4 +33 C/T (rs2070874) | Symptomatic | Asymptomatic | Total | c 2 p | OR [95% CI] | c 2 pc | |
| Wild genotype | CC | 45 (43.26) | 14 (37.83) | 59 | 0.3308 0.2826 | 1.253 [0.58-2.70] | |
| Grouped genotypes | CT+TT | 59 (56.73) | 23 (62.16) | 82 | NC | ||
| Total | 104 | 37 | 141 | ||||
| Wild type alelle | C | 136 (65.38) | 41 (55.40) | 177 | 2.326 0.0636 | 1.520 [0.88-2.60] | |
| Polymorphic alelle | T | 72 (34.61) | 33 (44.59) | 105 | NC | ||
| Total | 208 | 74 | 282 | ||||
| IL10 -3575 T/A (rs1800890) | Symptomatic | Asymptomatic | Total | c 2 p | OR [95% CI] | c 2 pc | |
| Wild genotype | TT | 58 (55.76) | 28 (75.67) | 86 | 4.545 0.0165 | 0.4053 [0.17-0.94] | 0.3747 |
| Grouped genotypes | TA+AA | 46 (44.23) | 09 (24.32) | 55 | 0.0264 | ||
| Total | 104 | 37 | 141 | ||||
| Wild type alelle | T | 155 (74.51) | 64 (86.48) | 219 | 4.506 0.0169 | 0.4570 [0.21-0.95] | 3.842 |
| Polymorphic alelle | A | 53 (25.48) | 10 (13.51) | 63 | 0.0250 | ||
| Total | 208 | 74 | 282 | ||||
| TGFb1 +869 T/C (rs1800470) | Symptomatic | Asymptomatic | Total | c 2 p | OR [95% CI] | c 2 pc | |
| Wild genotype | TT | 44 (42.30) | 16 (43.24) | 60 | 0.00977 0.4606 | 0.9625 [0.45-2.05] | |
| Grouped genotypes | TC+CC | 60 (57.69) | 21 (56.75) | 81 | NC | ||
| Total | 104 | 37 | 141 | ||||
| Wild type alelle | T | 125 (60,09) | 45 (60,81) | 170 | 0.01164 0.4570 | 0.9705 [0.56-1.67] | |
| Polymorphic alelle | C | 83 (39,90) | 29 (39.18) | 112 | NC | ||
| Total | 208 | 74 | 282 | ||||
The c2 test with the Yates correction (pc) was used to determine differences between genotype and allele frequencies; OR - Odds Ratio, CI - confidence interval, pc= p value with Yates' correction. NC- not calculated.
Only the IL10 -3575 T/A SNP showed statistical differences as there were significantly more individuals with genotypes containing the polymorphic allele (AA and AT) in the symptomatic group (c2 = 4.54, p = 0.01, OR = 0.40 [95% CI- 0.17- 0.94]). When the analysis was performed by allele, the frequency of the polymorphic allele A was also higher in the symptomatic group (c2 = 4.50, p = 0.01, OR = 0.45 [95% CI - 0.21-0.95]). These results are displayed in Table 3.
The genotype and allele frequencies of the studied individuals with respect to the five SNPs (IL6 -174; IL12p40 +1188; IL4 +33; IL10 -3575; TGFb1 +869), were also analyzed according to the parasitemia level. As the diagnosis in endemic areas is performed by the thick blood smear test, the subjects were re-divided into two new groups: the first one composed of subjects with non-detectable parasitemia (n = 72) and the second one with detectable parasitemia (low, moderate and intense) with 69 subjects. As shown in Table 4, no association was observed between the IL6 (-174), IL12p40 (+1188), IL4 (+33), IL10 (- 3575), TGFb1 (+869), SNPs and the parasitemia level. We also analyzed if there were differences between subjects according to different IL10 genotypes and the density of parasitemia. No significant difference was observed (Kruskal-Wallis test,p = 0.4597; data not shown).
Table 4. Genotypic and allelic frequencies of the five SNPs in the 141 recruited subjects according to parasitemia determined by the thick blood smear test .
| Polymorphisms | Detectable Parasitemia N=69 | Non-Detectable Parasitemia N=72 | Total | c 2 p | OR [95% CI] | |
|---|---|---|---|---|---|---|
| IL6 -147 G/C | ||||||
| Genotypic frequency | GG | 33 | 36 | 69 | 0.224 | NC |
| GC | 33 | 32 | 65 | 0.893 | ||
| CC | 3 | 4 | 7 | |||
| Wild genotype | GG | 33 | 36 | 69 | 0.066 | 0.916 |
| Grouped genotype | GC+CC | 36 | 36 | 72 | 0.796 | [0.47-1.77] |
| Allelic frequency | G | 99 | 104 | 203 | 0.081 | 0.976 |
| C | 39 | 40 | 79 | 0.928 | [0.58-1.64] | |
| IL-12p40 A/C | ||||||
| Genotypic frequency | AA | 23 | 31 | 54 | 1.689 | NC |
| AC | 39 | 33 | 72 | 0.429 | ||
| CC | 7 | 8 | 15 | |||
| Wild genotype | AA | 23 | 31 | 54 | 1.409 | 0.661 |
| Grouped genotype | AC+CC | 46 | 41 | 87 | 0.235 | [0.33-1.31] |
| Allelic frequency | A | 85 | 95 | 180 | 0.585 | 0.827 |
| C | 53 | 49 | 102 | 0.444 | [0.50-1.47] | |
| IL-4 +33 C/T | ||||||
| Genotypic frequency | CC | 27 | 32 | 59 | 0.556 | NC |
| CT | 31 | 28 | 59 | 0.757 | ||
| TT | 11 | 12 | 23 | |||
| Wild genotype | CC | 27 | 32 | 59 | 0.408 | 0.803 |
| Grouped genotype | CT+TT | 42 | 40 | 82 | 0.522 | [0.41-1.57] |
| Allelic frequency | C | 85 | 92 | 177 | 0.158 | 0.906 |
| T | 53 | 52 | 105 | 0.690 | [0.55-1.47] | |
| IL10 -3575 T/A | ||||||
| Genotypic frequency | TT | 39 | 47 | 86 | 1.372 | NC |
| TA | 25 | 22 | 47 | 0.503 | ||
| AA | 5 | 3 | ||||
| Wild genotype | TT | 39 | 47 | 86 | 1.135 | 0.691 |
| Grouped genotype | TA+AA | 30 | 25 | 55 | 0.286 | [0.35-1.36] |
| Allelic frequency | T | 103 | 116 | 219 | 1.422 | 0.710 |
| A | 35 | 28 | 63 | 0.233 | [0.40-1.24] | |
| TGFβ1 +869 T/C | ||||||
| Genotypic frequency | TT | 27 | 33 | 60 | 0.826 | NC |
| TC | 25 | 25 | 50 | 0.661 | ||
| CC | 17 | 14 | 31 | |||
| Wild genotype | TT | 27 | 33 | 60 | 0.647 | 0.759 |
| Grouped genotype | CT+CC | 42 | 39 | 81 | 0.421 | [0.38-1.48] |
| Allelic frequency | T | 79 | 91 | 170 | 1.041 | 0.779 |
| C | 59 | 53 | 112 | 0.307 | [0.48-1.25] |
Note: The c2test was used to determine statistical significance differences between genotypic and allelic frequencies; OR - Odds Ratio, CI - confidence interval; NC - not calculated.
Figure 1 shows the IL10 -3575 (T/A) SNP amplification product after cleavage with the ApoI restriction enzyme of a sample corresponding to a heterozygous genotype (TA) generating, after RFLP, three fragments of 227, 124 and 107 bp. In the IL10 -3575 SNP, the wild genotype has two fragments and the homozygous polymorphic genotype does not suffer cleavage (one fragment of around 227-228 bp, depending on the detection system resolution).
DISCUSSION
The relevance of host genetic factors in malaria was proposed more than 50 years ago when it was observed that the presence of thalassemia acted as a protective factor against malaria20. It has also been suggested that anti-inflammatory Th2-type cytokines (IL4, IL10, and TGF-b) down regulate Th1-type cytokines and the pro-inflammatory response in malaria21.
At the time of blood collection in the highly endemic studied region, it was not possible to find healthy (non-infected) subjects in the studied area that shared the same ethnicity (Amerindians) and environmental factors. Due to the aim of the study, it would not be recommended to use another control group, for example, of healthy blood bank donors from a non-endemic area because they would not have the same genetic features of the studied symptomatic individuals, and also they would not have been exposed to the same environmental factors and number of mosquito bites. As a matter of fact, it was difficult to find asymptomatic individuals for a period of at least 60 days prior to the study, justifying the lower number of asymptomatic patients that were recruited to this group.
Several studies have demonstrated the importance of the Plasmodium interaction with the human host to produce more or less severe clinical conditions associated with greater or lesser control of parasitemia through the production of cytokines22. Most studies performed comparisons between patients with severe malaria (cerebral form, with or without severe anemia) and asymptomatic subjects, or people without malaria. However, considering that severe malaria is fortunately uncommon in most endemic countries, we decided to compare symptomatic and asymptomatic patients. To reinforce the differences between symptomatic and asymptomatic individuals in this study, it is noteworthy that half of the symptomatic patients required an initial hospitalization, although they did not present severe malaria. It was observed that in the symptomatic group isolates of P. falciparum were predominant in comparison with P. vivax, while in the group of asymptomatic subjects, P. vivax prevailed, as expected. According to Barbieri et al. (2005)8, in the city of Peixoto de Azevedo, from 1992 to 1995, a higher prevalence of P. falciparum was observed in the gold mining area. These data corroborate our findings in relation to the amount of P. falciparuminfections detected in the symptomatic group, and in fact, the majority of individuals with malaria evaluated in our study were gold mining workers.
Even if we did not find differences between symptomatic and asymptomatic malaria patients concerning the IL6 -174 C/G SNP, Sortica et al. (2014)23 have investigated the influence of IL6, IL12p40 and VDR SNPs in uncomplicated Plasmodium vivax infections according to the intensity of symptoms, parasitemia and gametocytemia levels in a Brazilian Amazonian population. A total of 167 malaria patients infected by P. vivax had 14 signs and symptoms recorded, aside from parasitemia and gametocytemia levels, all of the parameters evaluated before treatment. Patients were genotyped for IL6 -174C/G, IL12p40 735T/C, 458A/G, 159A/C, and VDR FokI, TaqI, BsmI SNPs by Taqman 5' nuclease Real Time PCR. Higher parasitemia levels were observed in IL6 -174 C carriers (C is the wild type allele) whereas IL12p40 CGT haplotype carriers had lower parasitemia levels. VDR TaqIC/BsmIA haplotype carriers showed higher gametocyte levels than non-carriers. Based on the clinical index values, the IL6 -174 C carriers presented a more severe clinical index when compared to GG homozygotes (G is the polymorphic allele). The authors concluded that IL6, IL12 and VDR do influence severity, parasitemia and gametocytemia clearance in P. vivax infections, and highlighted their potential role in the immune response against malaria.
We did not find differences between the two studied groups with respect to the IL12p40 +1188 A/C (rs3212227) SNP. However, Phawong et al. (2010)24 have studied the genetic role of IL12p40 SNPs in Thai adult patients and whether these SNPs might be associated with severe malaria. To this end, the functional association between IL12p40 SNPs [i.e. IL12p40 pro (rs17860508) and IL12p40 3' UTR T/G (rs3212227)], were examined in 355 patients with Plasmodium falciparum infections recruited in northwest Thailand. While both, circulating IL12p40 and IFN-g were increased in patients with severe malaria, only IL12p40 was significantly higher in severe malaria associated with hyperparasitemia. Carriage of the IL12p40 pro1.1 genotype was associated with enhanced severity of malaria and hyperparasitemia, relative to the IL12p40 pro2.2 genotype (wild type). Individuals with the IL12p40 pro1.1 genotype also had the lowest IL12p40 levels and the highest IFN-g levels. The construction of haplotypes revealed that carriage of the IL12p40 pro-2/3' UTR-T haplotype was associated with protection against severe malaria and reduced circulating IFN-g. The authors concluded that the variation of genotypes and haplotypes at IL12p40 pro and IL12p40 3' UTR in the Thai population has influenced the susceptibility to severe malaria and functional changes in circulating IL12p40 and IFN-g.
In addition, Ong'echa et al. (2011)25 had previously shown that suppression of IL12 in African children is associated with severe malaria and anemia. Afterwards, these authors have decided to investigate the association between the SNP (1188A/C, rs3212227), severe malaria and anemia (Hb < 6.0 g/dL), circulating IL12p40/p70 levels, and the clinical outcomes of 756 Kenyan children from an endemic Plasmodium falciparummalaria transmission area. In the 544 children with acute malaria, it was demonstrated that carriers of the polymorphic C allele had increased susceptibility to severe malaria in comparison to the ones carrying the wild genotype (AA). Although children with severe malaria and anemia had lower IL12p40/p70 levels than the others, the levels did not differ significantly according to genotype. Regarding the entire cohort of 756 children, the authors failed to show any significant relationships between the 1188A/C, rs3212227 genotypes and either susceptibility to severe malaria and anemia or other causes of mortality during a three years' follow-up. The authors concluded that the IL12p40 1188A/C (rs3212227) SNP is a marker of susceptibility to severe malaria and anemia in children with acute disease, but does not seem to mediate functional changes in IL12 production.
This study did not find differences between symptomatic and asymptomatic malaria patients with respect to the IL4 (+ 33; rs2070874) C/T SNP. Nonetheless, Lokossouet al. (2013)26 have studied IL4, IL10 and IL13 gene SNPs and antibody levels of 576 mothers and their newborns in the context of P. falciparum malaria infection determined in placentas. The study was conducted in a semi-rural area of Benin, where malaria is endemic. TaqMan(r) Real Time PCR was employed to genotype the following SNPs: IL4 (rs2243250, rs2070874 or +33 C/T), IL10 (rs1800896, rs1800871, rs1800872) and IL13 (rs1800925) genes. Antibody responses to several P. falciparum recombinant proteins (MSP-1, MSP-2, MSP-3, GLURP-R0, GLURP-R2 and AMA-1) were evaluated by ELISA. The results have evidenced that the maternal haplotype IL4 (-590)*T/ IL4 (+33)*T (one or two copies), i.e., carrying the IL4 +33 polymorphic allele T that we have also investigated in the present study, was associated with favorable maternal condition at delivery (high hemoglobin levels, absence of placental parasites) and one of its component, the IL4 (-590) TT genotype, was related to low IgG levels to MSP-1, MSP-2/3D7 and MSP-2/FC27. On the contrary, the maternal IL10 (-1082) AA was positively associated with P. falciparum placenta infection at delivery. As a consequence, the IL10 (-819)*T allele (in CT and TT genotypes), as well as the IL10 (-1082)*A/ IL10 (-819)*T/ IL10 (-592)*A haplotype (one or two copies) were related to an increased risk for anemia in newborns. The maternal IL10 (-1082) AA genotype was related to high IgG levels to MSP-2/3D7 and AMA-1 in mothers and newborns, respectively. The IL13 gene polymorphism was only involved in the newborn's antibody response to AMA-1.
Also in relation to IL4 SNPs, Cabantous et al. (2015)27 have associated the heterozygous form of the IL4 variable-number tandem repeat (VNTR) (rs8179190) or the IL4 - 33 SNP (rs2070874), i.e, not the same SNP that was investigated in this study, with a higher risk for severe malaria in children, whereas homozygous patients were protected suggesting that these SNPs may participate in the genetic control of the pathophysiology in severe malaria. In a following investigation, the best-fit model revealed three haplotype combination that were associated with different levels of risk to severe malaria. The highest risk was observed for subjects carrying at least one copy of both IL4 -33 allele T and IL4 VNTR allele 1, who exhibited higher IL4 plasma levels. Children homozygous for IL4 VNTR allele 2 had a lower risk of severe malaria, as well as lower IL4 plasma levels.
Concerning TGFb1, although we did not find differences in the present study when the frequency of the +869 T/C (rs1800470) SNP was investigated in the group of symptomatic and asymptomatic malaria patients, Frade et al. (2011)28 investigated TGFb1 gene SNPs at positions -509 C/T and +869 T/C in patients with visceral leishmaniasis compared to individuals with asymptomatic infection, but positive delayed hypersensitivity test (DTH +), and healthy control with DTH-. Regarding the SNP -509 C/T there was no association between the presence of the polymorphic allele T and the symptomatic forms of visceral leishmaniasis or a higher production of TGFb1 that would in turn modulate negatively the inflammatory response. In the same study, the TGFb1 +869 T/C SNP, the same we have included in our study, was analyzed in the same three groups and the polymorphic allele C was found in 50% of patients with visceral leishmaniasis, 52% of asymptomatic patients with DTH +, and 48% of healthy control subjects DTH-, therefore revealing that there were no statistically significant differences when the groups were compared in pairs, or in combination.
Continuing the discussion on the TGFb1 gene SNPs, Sepanjnia et al. (2015)29 have investigated three SNPs in the promoter region of the TGFb1 gene (-509 C/T [rs1800469], +869 T/C [rs1800470], and + 913 G/C [rs1800471]) because this cytokine plays a crucial role in the regulation of microbial replication and host responses to Brucellaspp. A total of 282 Iranian subjects including 153 patients with active brucellosis, and 128 age- and sex-matched healthy individuals were recruited. TGFb1 -509 C/T and + 869 T/C genotyping was performed using a multiplex-PCR and the + 913 G/C SNP was genotyped using an allele-specific PCR. The results demonstrated that the TGFb1 + 869 T/C polymorphic homozygote genotype (CC) was associated with the risk of developing brucellosis. Moreover, considering the TGFb1 + 869 T/C and + 913 G/C SNPs forming the haplotype (CC/GG), there was an association with an increased risk of brucellosis. Other TGFb1 variants did not increase the risk of brucellosis infection.
IL10 is an anti-inflammatory cytokine and its encoding gene is a good candidate to study different SNPs located at its promoter region in the context of malaria since IL10 is an immune regulatory cytokine, exerting a central role in the control of inflammation, apoptosis, and the maintenance of CD4 + helper cells30.
It has been demonstrated that 75% of the differences in IL10 production in human beings are due to inherited genetic factors31. Single nucleotide polymorphisms (SNPs) located at the IL10 promoter region have been considered as susceptibility or resistance markers in the context of diseases that impair the immune system such as systemic lupus erythematosus31 , 32, cancer33 , 34 , 35, and also some infectious diseases such as mycobacterium infections, especially leprosy36 , 37 , 38 , 39.
Mörmann et al. (2004)40 have hypothesized that some haplotypes in the IL10 promoter region may regulate the IL10 secretion by influencing the transcription level of this cytokine.
Regarding the leprosy model, Santos et al. (2002)39 have found that the frequency of the IL10 -819 T/T SNP was increased in Brazilian leprosy patients. Then, Moraes et al. (2003)18 analyzed the frequency of five SNPs in the IL10 promoter region (-3575/-2849/-2763/-1082/-819) in two healthy populations, one from the Netherlands and the other from Rio de Janeiro, Brazil. A total of 321 Dutch Caucasian individuals and 293 Brazilians, described as African-Brazilians and Euro-Brazilians were enrolled. Although the genotype frequencies in the Brazilian and the Dutch populations were different, the comparison of genotype frequencies between African-Brazilians and Euro-Brazilians did not show any differences. When the five SNPs were grouped in haplotypes -3575T/-2849G/-2763C/-1082A/-819C, the one formed by the three SNPs -3575T/-2849G/-2763C, showed a strong linkage disequilibrium. The TGC haplotype (-3575T/-2849G/-2763C) was the most frequent, while the AAA haplotype was much less represented in the Brazilian population.
In another study by Moraes et al. (2004)37 the authors analyzed the frequencies of the same five IL10 SNPs (-3575/-2849/2763/-1082/-819) and whether they could act as susceptibility or severity markers in leprosy. The studied parameters were the type (multibacillary or paucibacillary), and the severity of leprosy. Significant differences between controls and leprosy patients were found for three SNPs (-3575/-2849/-2763). The haplotype AGC (-3575A/-2849G/-2763C) was associated with resistance to leprosy and paradoxically with the development of severe forms of disease. The haplotype TGC (-3575T/-2849G/-2763C) was associated with susceptibility but not with severity of leprosy.
Malhotra et al. (2005)36 have investigated six SNPs in the IL10 promoter region of 282 Indian leprosy patients and 266 healthy controls. When the isolated allelic and genotypic SNPs frequencies have been considered, T-3575A, G-2849A, and C-2763A SNPs did not show any significant differences between cases and controls. However, the extended haplotype -3575T/-2849G/-2763C/-1082A/-819C/-592C, i.e, containing the -3575 wild type T allele has been associated with resistance to leprosy per se, but surprisingly also to the development of severe forms of leprosy, whereas the haplotype -3575T/-2849G/-2763C/-1082A/-819T/-592A, or in other words, with different alleles only regarding the last two SNPs, has been associated with the risk of developing severe forms of leprosy, in contrast to the minor risk haplotype -3575T/-2849A/-2763C that has been described in Brazilian patients37. The results of the IL10 promoter SNPs in Brazilians and Indians have strongly suggested the involvement of the IL10 locus in the outcome of leprosy.
In southwest China, Chen et al. (2013)41 have studied the frequency of IL10 haplotypes and have found that the haplotype -3575A/2849G/2763A/1082G /819C/592C, i.e., containing the polymorphic allele A at the -3575 position, was significantly more frequent among leprosy patients and also among paucibacillary patients in comparison to control subjects.
Gibson et al. (2001)31 have studied patients with systemic lupus erythematosus and have described eight novel polymorphisms that are located at the 4 kb distal region of the IL10 promoter, including the -3575 T/A SNP. These SNPs in an isolated way or forming haplotypes have consistently been associated with IL10 production. Regarding the -3575 T/A SNP, both alleles are frequently found in different populations, and phenotypic studies have associated high IL10 production with the presence of the wild type allele T and low IL10 production with the polymorphic allele A.
The association studies of individual promoter SNPs have suggested that the effect of proximal promoter SNPs (-1082A/G, -819C/T, -592C/A) in determining risk/ protection to leprosy is more pronounced than distal SNPs (-3575T/A, -2849G/A, -2763C/G). The proximal promoter polymorphisms have been reported to define 'high' (-1082G/-819C/-592C), 'medium' (-1082A/-819C/-592C) and 'low' (-1082A/-819T/-592A) expressing genotypes for IL1042. To corroborate these data, Pereira et al. (2009)38 have observed that the -819T allele is associated with leprosy susceptibility. Haplotypes combining promoter SNPs have also implicated a haplotype carrying the allele -819T in leprosy susceptibility. Finally, these authors have tested the IL10 production in peripheral blood mononuclear cells stimulated with Mycobacterium leprae antigens and found that -819T carriers (mutants) produced lower levels of IL10 when compared to non-carriers. These data have suggested that low levels of IL10 during the disease development can drive patients to a chronic and inadequate inflammatory and immune response that culminates with leprosy. These results corroborate with the fact that the outcome of mycobacterial infections such as leprosy involves complex interactions between several other host genes and also highlight the role of IL10 in the early and late phases of the leprosy infection. The involvement of IL10 SNPs in the disease outcome in two ethnically distinct populations (Brazilian and Indian), suggests that the IL10 promoter region needs to be further investigated both in genetic and functional studies by means of the evaluation of the IL10 production.
In the present study the IL10 -3575 T/A SNP (rs1800890) was investigated, and higher frequencies of the genotypes carrying the polymorphic allele A were found among symptomatic individuals suggesting a possible association between the presence of the polymorphic allele A at the IL10 promoter gene, -3575 position, with the presence of malaria symptoms in people living in an endemic malaria area for as long as nine years. It is possible that in the group of asymptomatic malaria patients in whom more individuals carried the wild type allele T, the modulation (containment) of the inflammatory process by means of a higher IL10 production could be beneficial to the patients.
It would have been interesting to confirm this finding by the evaluation of IL10 production in the patients. Unfortunately, the patients' serum samples had already been frozen and thawed several times, making the IL10 dosage unreliable.
In conclusion, this study has suggested the possibility that the IL10 - 3575 T/A SNP might be associated with the presence and maintenance of malaria symptoms in individuals living in endemic areas for many years. Taking into account that the presence of this polymorphism is related to decreased IL10 production, the results also suggest a possible role of IL10 SNPs in the pathophysiology of malaria, but replication studies with a higher number of patients and the simultaneous evaluation of IL10 levels are needed to confirm these findings.
Acknowledgements
This work was supported by the São Paulo Research Foundation - FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo) grant number 2010/15022-1.
The authors thank Dr. Sandra do Lago Moraes for providing us with the malaria repository and part of the molecular biology reagents, and Dr. Fabiana M. S. Leoratti for helpful scientific assistance. The parasitemia data were obtained in her PhD thesis under the supervision of Dr. Sandra do Lago Moraes.
REFERENCES
- 1.de Pina-Costa A, Brasil P, Di Santi SM, de Araujo MP, Suaréz-Mutis MC, Santelli AC. Malaria in Brazil: what happens outside the Amazonian endemic region. Mem Inst Oswaldo Cruz. 2014;109:618–633. doi: 10.1590/0074-0276140228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franklin BS, Parroche P, Ataide MA, Lauw F, Ropert C, de Oliveira RB. Malaria primes the innate immune response due to interferon-gamma induced enhancement of toll-like receptor expression and function. Proc Natl Acad Sci USA. 2009;106:5789–5794. doi: 10.1073/pnas.0809742106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leoratti FM, Farias L, Suárez-Mútis MC, Coura JR, Kalil J, Camargo EP. Variants in the toll-like receptor signaling pathway and clinical outcomes of malaria. J Infect Dis. 2008;198:772–780. doi: 10.1086/590440. [DOI] [PubMed] [Google Scholar]
- 4.Nkoghe D, Akue JP, Gonzalez JP, Leroy EM. Prevalence of Plasmodium falciparum infection in asymptomatic rural Gabonese populations. Malar J. 2011;10:33–33. doi: 10.1186/1475-2875-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leoratti FM, Trevelin SC, Cunha FQ, Rocha BC, Costa PA, Gravina HD. Neutrophil paralysis in Plasmodium vivax malaria. PLoS Negl Trop Dis. 2012;6:e1710. doi: 10.1371/journal.pntd.0001710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCall MB, Netea MG, Hermsen CC, Jansen T, Jacobs L, Golenbock D. Plasmodium falciparum infection causes proinflammatory priming of human TLR responses. J Immunol. 2007;179:162–171. doi: 10.4049/jimmunol.179.1.162. [DOI] [PubMed] [Google Scholar]
- 7.Alves FP, Durlacher RR, Menezes MJ, Krieger H, Pereira da Silva LH, Camargo EP. High prevalence of asymptomatic Plasmodium vivax and Plasmodium falciparum infections in native Amazonian populations. Am J Trop Med Hyg. 2002;66:641–648. doi: 10.4269/ajtmh.2002.66.641. [DOI] [PubMed] [Google Scholar]
- 8.Barbieri AF, Sawyer DO, Soares-Filho BS. Population and land use effects on malaria prevalence in the southern Brazilian Amazon. Hum Ecol. 2005;33:847–874. [Google Scholar]
- 9.Suárez-Mutis MC, Cuervo P, Leoratti FM, Moraes-Avila LS, Ferreira AW, Fernandes O. Cross sectional study reveals a high percentage of asymptomatic Plasmodium vivax infection in the Amazon Rio Negro area, Brazil. Rev Inst Med Trop Sao Paulo. 2007;49:159–164. doi: 10.1590/s0036-46652007000300005. [DOI] [PubMed] [Google Scholar]
- 10.Lee SJ, Stepniewska K, Anstey N, Ashley E, Barnes K, Binh TQ. The relationship between the haemoglobin concentration and the haematocrit in Plasmodium falciparum malaria. Malar J. 2008;7:149–149. doi: 10.1186/1475-2875-7-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura M, Kaneko O, Qing L, Mian Z, Kawamoto F, Wataya Y. Identification of the four species of human malaria parasites by nested PCR that targets variant sequences in the small subunit rRNA gene. Parasitol Int. 1997;46:91–95. [Google Scholar]
- 12.Brasil.Ministério da Saúde. Guia prático de tratamento da malária no Brasil, 2010/ Malaria treamtent in Brazil: practical guide; 2010. [Google Scholar]
- 13.Snounou G, Viriyakosol S, Jarra W, Thaithong S, Brown KN. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol Biochem Parasitol. 1993;58:283–292. doi: 10.1016/0166-6851(93)90050-8. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira MU, Liu Q, Zhou M, Kimura M, Kaneko O, Thien HV. Stable patterns of allelic diversity at the merozoite surface protein-1 locus of Plasmodium falciparum in clinical isolates from Southern Vietnam. J Eukaryot Microbiol. 1998;45:131–136. doi: 10.1111/j.1550-7408.1998.tb05080.x. [DOI] [PubMed] [Google Scholar]
- 15.Sainz J, Pérez E, Gómez-Lopera S, López-Fernández E, Moratalla L, Ovonarte S. Genetic variants of IL6 gene promoter influence on C-reactive protein levels but are not associated with susceptibility to invasive pulmonary aspergillosis in hematological patients. Cytokine. 2008;41:268–278. doi: 10.1016/j.cyto.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 16.Thurairajah PH, Hegazy D, Chokshi S, Shaw S, Demaine A, Kaminski ER. Hepatitis C virus (HCV)-specific T cell responses in injection drug users with apparent resistance to HCV infection. J Infect Dis. 2008;198:1749–1755. doi: 10.1086/593337. [DOI] [PubMed] [Google Scholar]
- 17.Shibata N, Ohnuma T, Takahashi T, Baba H, Ishizuka T, Ohtsuka M. The effect of IL4 +33C/T polymorphism on risk of Japanese sporadic Alzheimer's disease. Neurosci Lett. 2002;323:161–163. doi: 10.1016/s0304-3940(02)00008-3. [DOI] [PubMed] [Google Scholar]
- 18.Moraes MO, Santos AR, Schonkeren JJ, Vanderborght PR, Ottenhoff TH, Moraes ME. Interleukin-10 promoter haplotypes are differently distributed in the Brazilian versus the Dutch population. Immunogenetics. 2003;54:896–899. doi: 10.1007/s00251-003-0543-3. [DOI] [PubMed] [Google Scholar]
- 19.Wong TY, Poon P, Chow KM, Szeto CC, Cheung MK, Li PK. Association of transforming growth factor-beta (TGF-beta) T869C (Leu 10Pro) gene polymorphisms with type 2 diabetic nephropathy in Chinese. Kidney Int. 2003;63:1831–1835. doi: 10.1046/j.1523-1755.2003.00919.x. [DOI] [PubMed] [Google Scholar]
- 20.Haldane JBS. The rate of mutations of human genes. Proceedings of the Eighth International Congress of Genetics and Heredity. Hereditas. 1948;(35):267–273. [Google Scholar]
- 21.de Mendonça VR, Gonçalves MS, Barral-Neto M. The host genetic diversity in malaria infection. J Trop Med. 2012:940616–940616. doi: 10.1155/2012/940616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomes AP, Vitorino RR, Costa AP, Mendonça EG, Oliveira MGA, Siqueira-Batista R. Severe Plasmodium falciparum malaria. Rev Bras Ter Intensiva. 2011;23:358–369. doi: 10.1590/s0103-507x2011000300015. [DOI] [PubMed] [Google Scholar]
- 23.Sortica VA, Cunha MG, Ohnishi MD, Souza JM, Ribeiro-dos-Santos ÂK, Santos SE. Role of IL6, IL12B and VDR gene polymorphisms in Plasmodium vivax malaria severity, parasitemia and gametocytemia levels in an Amazonian Brazilian population. Cytokine. 2014;65:42–47. doi: 10.1016/j.cyto.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 24.Phawong C, Ouma C, Tangteerawatana P, Thongshoob J, Were T, Mahakunkijcharoen Y. Haplotypes of IL12B promoter polymorphisms condition susceptibility to severe malaria and functional changes in cytokine levels in Thai adults. Immunogenetics. 2010;62:345–356. doi: 10.1007/s00251-010-0439-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ong'echa JM, Raballah EO, Kempaiah PM, Anyona SB, Were T, Davenport GC. Polymorphic variability in the 3' untranslated region (UTR) of IL12B is associated with susceptibility to severe anaemia in Kenyan children with acute Plasmodium falciparum malaria. BMC Genet. 2011;12:69–69. doi: 10.1186/1471-2156-12-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lokossou AG, Dechavanne C, Bouraïma A, Courtin D, Le Port A, Ladékpo R. Association of IL-4 and IL-10 maternal haplotypes with immune responses to P.falciparum in mothers and newborns. BMC Infect Dis. 2013;13:215–215. doi: 10.1186/1471-2334-13-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cabantous S, Ranque S, Poudiougou B, Traore A, Berbache S, Vitte J. Genotype combinations of two IL4 polymorphisms influencing IL-4 plasma levels are associated with different risks of severe malaria in the Malian population. Immunogenetics. 2015;67:283–288. doi: 10.1007/s00251-015-0836-3. [DOI] [PubMed] [Google Scholar]
- 28.Frade AF, Oliveira LC, Costa DL, Costa CH, Aquino D, Van Weyenbergh J. TGFB1 and IL8 gene polymorphisms and susceptibility to visceral leishmaniasis. Infect Genet Evol. 2011;11:912–916. doi: 10.1016/j.meegid.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 29.Sepanjnia A, Eskandari-Nasab E, Moghadampour M, Tahmasebi A, Dahmardeh F. TGFb1 genetic variants are associated with an increased risk of acute brucellosis. Infect Dis (Lond) 2015;47:458–464. doi: 10.3109/23744235.2015.1016298. [DOI] [PubMed] [Google Scholar]
- 30.Duell BL, Tan CK, Carey AJ, Wu F, Cripps AW, Ulett GC. Recent insights into microbial triggers of interleukin-10 production in the host and the impact on infectious diseases pathogenesis. FEMS Immunol Med Microbiol. 2012;64:295–313. doi: 10.1111/j.1574-695X.2012.00931.x. [DOI] [PubMed] [Google Scholar]
- 31.Gibson AW, Edberg JC, Wu J, Westendorp RG, Huizinga TW, Kimberly RP. Novel single nucleotide polymorphisms in the distal IL-10 promoter affect IL-10 production and enhance the risk of systemic lupus erythematosus. J. Immunol. 2001;166:3915–3922. doi: 10.4049/jimmunol.166.6.3915. [DOI] [PubMed] [Google Scholar]
- 32.Chong WP, Ip WK, Wong WH, Lau CS, Chan TM, Lau YL. Association of interleukin-10 promoter polymorphisms with systemic lupus erythematosus. Genes Immun. 2004;5:484–492. doi: 10.1038/sj.gene.6364119. [DOI] [PubMed] [Google Scholar]
- 33.Shih CM, Lee YL, Chiou HL, Hsu WF, Chen WE, Chou MC. The involvement of genetic polymorphism of IL-10 promoter in non-small cell lung cancer. Lung Cancer. 2005;50:291–297. doi: 10.1016/j.lungcan.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Wang YC, Sung WW, Wu TC, Wang L, Chien WP, Cheng YW. Interleukin-10 haplotype may predict survival and relapse in resected non-small cell lung cancer. PLoS One. 2012;7:e39525. doi: 10.1371/journal.pone.0039525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei YS, Kuang XH, Zhu YH, Liang WB, Yang ZH, Tai SH. Interleukin-10 gene promoter polymorphisms and the risk of nasopharyngeal carcinoma. Tissue Antigens. 2007;70:12–17. doi: 10.1111/j.1399-0039.2007.00806.x. [DOI] [PubMed] [Google Scholar]
- 36.Malhotra D, Darvishi K, Sood S, Sharma S, Grover C, Relhan V. IL-10 promoter single nucleotide polymorphisms are significantly associated with resistance to leprosy. Hum Genet. 2005;118:295–300. doi: 10.1007/s00439-005-0042-8. [DOI] [PubMed] [Google Scholar]
- 37.Moraes MO, Pacheco AG, Schonkeren JJ, Vanderborght PR, Nery JA, Santos AR. Interleukin-10 promoter single-nucleotide polymorphisms as markers for disease susceptibility and disease severity in leprosy. Genes Immun. 2004;5:592–595. doi: 10.1038/sj.gene.6364122. [DOI] [PubMed] [Google Scholar]
- 38.Pereira AC, Brito-de-Souza VN, Cardoso CC, Dias-Baptista IM, Parelli FP, Venturini J. Genetic, epidemiological and biological analysis of interleukin-10 promoter single-nucleotide polymorphisms suggests a definitive role for -819C/T in leprosy susceptibility. Genes Immun. 2009;10:174–180. doi: 10.1038/gene.2008.97. [DOI] [PubMed] [Google Scholar]
- 39.Santos AR, Suffys PN, Vanderborght PR, Moraes MO, Vieira LM, Cabello PH. Role of tumor necrosis factor-alpha and interleukin-10 promoter gene polymorphisms in leprosy. J Infect Dis. 2002;186:1687–1691. doi: 10.1086/345366. [DOI] [PubMed] [Google Scholar]
- 40.Mörmann M, Rieth H, Hua TD, Assohou C, Roupelieva M, Hu SL. Mosaics of gene variations in the Interleukin-10 gene promoter affect interleukin-10 production depending on the stimulation used. Genes Immun. 2004;5:246–255. doi: 10.1038/sj.gene.6364073. [DOI] [PubMed] [Google Scholar]
- 41.Chen XH, Xiong JH, Ning Y, Wen Y, Liu J, Mao C. IL-10 promoter SNPs and susceptibility to leprosy in ethnic groups from southwest China. Genet Mol Res. 2013;12:2876–2885. doi: 10.4238/2013.August.12.3. [DOI] [PubMed] [Google Scholar]
- 42.Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24:1–8. doi: 10.1111/j.1365-2370.1997.tb00001.x. [DOI] [PubMed] [Google Scholar]


